Published online Jun 15, 2003. doi: 10.3748/wjg.v9.i6.1256
Revised: February 4, 2003
Accepted: February 16, 2003
Published online: June 15, 2003
AIM: To analyze the amino acid sequences of hypervariable region 1 (HVR1) of HCV isolates in China and to construct a combinatorial chimeric HVR1 protein having a very broad high cross-reactivity.
METHODS: All of the published HVR1 sequences from China were collected and processed with a computer program. Several representative HVR1's sequences were formulated based on a consensus profile and homology within certain subdivision. A few reported HVR1 mimotope sequences were also included for a broader representation. All of them were cloned and expressed in E.coli. The cross-reactivity of the purified recombinant HVR1 antigens was tested by ELISA with a panel of sera from HCV infected patients in China. Some of them were further ligated together to form a combinatorial HVR1 chimera.
RESULTS: Altogether 12 HVR1s were selected and expressed in E.coli and purified to homogeneity. All of these purified antigens showed some cross-reactivity with sera in a 27 HCV positive panel. Recombinant HVR1s of No. 1, 2, 4, and 8# showing broad cross-reactivities and complementarity with each other, were selected for the ligation elements. The chimera containing these 4 HVR1s was highly expressed in E.coli. The purified chimeric antigen could react not only with all the HCV antibody positive sera in the panel but also with 90/91 sera of HCV -infected patients.
CONCLUSION: The chimeric antigen was shown to have a broad cross-reactivity. It may be helpful for solving the problem caused by high variability of HCV, and in the efforts for a novel vaccine against the virus.
- Citation: Xiu BS, Ling SG, Song XG, Zhang HQ, Chen K, Zhu CX. Cross-reactivity of hypervariable region 1 chimera of hepatitis C virus. World J Gastroenterol 2003; 9(6): 1256-1260
- URL: https://www.wjgnet.com/1007-9327/full/v9/i6/1256.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i6.1256
Hepatitis C virus (HCV) is a major etiological agent of non-A, non-B hepatitis worldwide[1-3], and HVR1, the N-terminal 27 amino acid residues of the putative HCV envelope protein E2, is known as the principal neutralization epitopes up to date[4-7]. Antibodies to HVR1 in human sera have been shown to block viral attachment to human cell lines in vitro and to protect chimpanzees from HCV infection in vivo[8-10]. The HVR1 sequence is highly variable, and is the greatest obstacle for the vaccine development and immune therapy[11,12]. However, the highly variable HVR1s have been shown to have some cross-reactivities with each other, indicating that a broadly cross-reactive HVR1 peptide or their cocktails are helpful to solving the problem[13]. Data were accumulated in this study all over the world[14-17].
In China, HVR1 sequences of different HCV isolates have been reported by many authors, but few studies were on HVR1 cross-reactivity. Integrating the HVR1 sequences reported in China together with some published mimotopes, 12 representative HVR1 sequences were selected using bioinformatics technology. All of the representative HVR1 sequences were cloned and expressed, and their cross-reactivity was studied with a panel of 27 HCV positive sera. Finally we obtained an HVR1 fusion antigen broadly cross-reactive with the HCV-infected sera.
Samples of HCV-infected sera were obtained from blood donor applicants in Beijing Red Cross Blood Center and from chronic HCV-infected patients from 302 Hospital of PLA. All were positive for HCV antibodies using the 2nd-generation ELISA kit. (Ortho Diagnostics, Raritan N.J).
All of the HVR1 sequences published in China were loaded into database and their consensus sequence was obtained by BASIC program according to the frequency of amino acid residues. All of these HVR1 sequences were divided into several groups according to their alignment, and one sequence was chosen as the representative from each group. All of the work above was operated on Goldkey (a molecular biology software developed by our institute). Some HVR1 sequences or mimotopes published were chosen as representative ones for their high cross-reactivity with sera of HCV infected patients from other countries.
The representative HVR1 sequences were modified considering the Escherischia coli's favorable codon usage. The coding genes were synthesized chemically and to facilitate further ligation, two linkers with a specially designed restriction endonuclease site were incorporated into their N- and C- terminals respectively. The N terminal arm is F1 (5'-gcctcgagggtggtggatct-3'), The C terminal arm is R1 (5'-gctctagaacctccaccact-3'). The fragments were digested with XhoI and XbaI enzymes and inserted into the expressing plasmid pBVIL1 digested by the same restrictive enzymes. In the same way, 12 different pBVIL1-HVR1 constructs were prepared and the HVR1 genes were expressed as fusion protein with IL1β in E. coli.
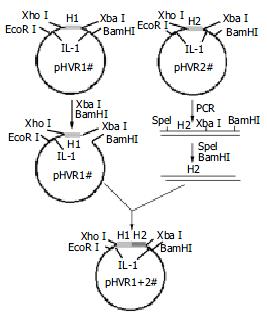
According to the cross-reactivity with the HCV antibodies positive sera panel, several HVR1s were chosen to ligate together one by one as illustrated in Figure 1. The plasmid pBVIL1-HVR1-1# (pHVR1#) was chosen as a vector digested by Xba I and BamH I, while the plasmid pBVIL1-HVR1-2#, was chosen as the donor of pattern, amplified using constant primer F2 (5'-gcactagtggtggtggatct-3') and R2 (5'-cgggatccttaggaagacacaaa-3') which annealed to C-terminal of IL1β. The PCR product was digested with Spe I and BamH I, and inserted into the digested vector, pBVIL1-HVR1-1#. Owe to the same cohesive end of the endonuclease Xba I and Spe I, the digested PCR fragment could accurately linked to the digested plasmid and the new ligated site could be digested by neither of them.
The pBVIL1-HVR1-1+2# had the same enzyme sites with pHVR1# and so it could be used as a new vector and connected with other HVR1 gene fragments. In this way, the pBVIL1-chimeric-HVR1 was constructed to contain four HVR1 genes, HVR1-1#, HVR1-4#, HVR1-6# and HVR1-8#.
The plasmids carrying HVR1 fragments were transformed into HB101 as routine, and were examined for their orientation and nucleic acid sequences. The transformed HB101 was grown overnight, diluted 1:20 with fresh LB-medium and further incubated at 37 °C to an OD600 of 0.6. After induction for 4 h at 42 °C, the bacteria were harvested by centrifugation, and lysed by sonication. All of the recombinant proteins existed in inclusion bodies, and could be dissolved in a solution containing 8 M urea. The recombinant proteins were isolated and purified consecutively by Q-Sepharose-FF and Sephadex G50 chromatography.
Microplates were coated with 0.3 μg recombinant HVR1 peptide in 100 mM phosphate buffer (pH7.4) by incubation overnight at 4 °C. The plates were then blocked with the phosphate buffer containing 0.2% BSA at 4 °C for 3 h, and then incubated with 100 μL of the serum sample 1:10 diluted with a sample buffer (100 mM sodium phosphate buffer, pH7.5 containing 10% goat serum and 0.05% Tween) at 37 °C for 1 h. After being washed for five times with 100 mM phosphate buffer (pH7.5) containing 0.05% Tween, the plates were then incubated for 30 min at 37 °C with 1:25000 diluted HRP-conjugated monoclonal antibody against human IgG. After washing, the reaction was visualized in the substrate buffer (50 mM sodium phosphate-citric acid buffer, pH5.0 containing 0.4 mg/mL TMB and 0.4 μL/mL of 30% hydrogen peroxide). The reaction was stopped by adding 50 μL of 2 M sulfuric acid, and the absorbance was measured in a microplates ELISA reader at 450 nm.
A total number of 123 sequences on HVR1 were reported in China, and the derived consensus profile of them is shown in Figure 2A. Some amino acid residues of HVR1 were shown to be hypervariable, while those at position 385, 389, 406, 409 were conserved. The sequence on first line was defined as CCS (Chinese consensus sequence), whose amino acid residues emerged most frequently. CCS was chosen as the first representative sequence, named HVR1-1#, being different at some positions from Puntoriero's consensus sequence[13] (Figure 2B). The homology of the 123 sequences was analyzed using the Goldkey software, and divided into 6 groups, HVR1-2 to 7# according to their alignment to CCS. In this way 6 sequences named were obtained. HVR1-8# 9# were from GenBank (L19383, S24080), both being broadly cross-reactive with mice sera induced by mimotopes. HVR1-10# and 11# were sequences for the mimotope R9 and M122 respectively (Puntoriero et al[14], 1998), and HVR1-12# reported by Watanabe.
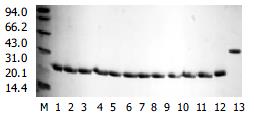
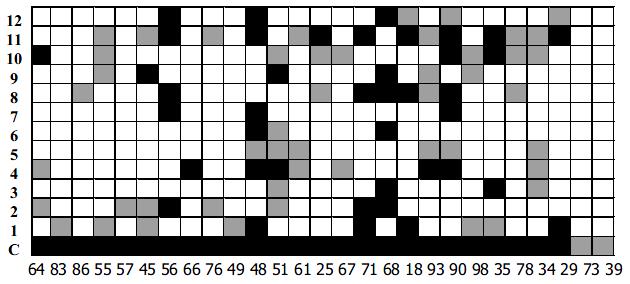
Twelve representative HVR1 gene as shown in Figure 3, were expressed in E.coil fused with human IL-1β. The HVR1/IL-1β fusion protein migrated at the expected position of about 21 kD (Figure 4). Twenty-seven HCV patients sera were used as panel I to show the cross-reactivities of 12 representative HVR1 by ELISA. As shown in Figure 5, all of the HVR1 peptide reacted with more than one serum. No reactivity was detected to IL-1β in sera of panel I, and none of the anti-HCV negative sera reacted with the 12 recombinant peptides. The most broadly cross-reactive HVR1 was HVR1-11#, and the Chinese consensus sequence (CCS) which showed a higher cross-reactivity too.
We took HVR1-1, 2, 4, 8# as components for the best cocktail, because these 4 HVR1 peptides showed complementary reactivities to the sera in panel I, as showed in Figure 5. There overall cross-reactivity was found to be 25/27.
HVR1-1#, 2#, 4# and 8# were ligated one by one in tandem within plasmid pBVIL1. The chimeric protein was expressed in HB101 and further purified (Figure 4).
As expected, a broader reactive spectrum was observed for the chimeric HVR1 antigen. It was shown to be reactive with all of the sera of panel I, including sera 73# and 39# which were not reactive with any single HVR1 (Figure 5). For more data 91 sera from HCV-infected patients were also used for the assay as panel II, with 90 reactive with chimera antigen (Table 1). The data indicated that application of the chimera protein helped to acquire a higher cross-reactivity.
| OD difference between sera and co | No. of reactive sera | The adding up percent of total sera |
| > 2.0 | 56 | 61.5 |
| 1.5~2.0 | 14 | 76.9 |
| 1.0~1.5 | 13 | 91.2 |
| 0.5~1.0 | 6 | 97.8 |
| 0.371 | 1 | 98 |
| 0.065 | 1 |
HVR1, which contains a principal neutralization epitope in HCV, is important for the development of HCV vaccine[4-7]. Due to the high mutation rate of HVR1, there are now hundreds of HVR1 isolates reported, presenting a great obstacle for HCV vaccine development[18-21]. It was suggested that to select a highly cross-reactive HVR1 antigen could solve the variability problem[22-27], thus highlighted the importance to study the cross-reactivity of HVR1.
Most of the work about the cross-reactivity of HVR1 focused on single HVR1 antigen. However we think the cross-reactivity of single HVR1 is limited. Recently, multi-epitope chimeric antigen was used to improve the sensitivity of HCV immunoassay reagents[28,29]. Here we provided evidence for enhancing the cross-reactivity by constructing a chimeric antigen that incorporates several representative HVR1 peptides.
Considering geographical variation of HVR1[30-33], we gave priority to Chinese sequence when we selected representative HVR1 sequences. The differences between CCS and Puntoriero's suggest the HVR1 variant found in China differs to a certain extent from what occurred elsewhere[14]. The chimeric antigen contains 3 representative HVR1 sequences coming from China, and showed broad cross-reactivity with sera of the HCV-infected patients.
The reported HVR1 antigen or mimotope could cross-react with no more than 80% of sera containing HCV antibodies[22-27]. Chimeric HVR1 antigen could cross-react with 90/91 (98%) of tested sera. The results also suggested that most of HCV infected patients could generate some antibodies against HVR1. The possible association between HVR1 antibody and the self-limiting course of HCV infection and a more favorable response to interferon[34-40], remains to be evaluated in the following study.
Evidently, the reaction spectrum of the chimera HVR1 antigen include the total cross-reactivity of the representative HVR1 antigen contained. Interestingly, the chimera HVR1 antigen could react with sera 73# and 39#, which are not definitely reactive with any of the four representative HVR1. In our consideration, those samples might react with some of the representative HVR1 used for ligation, but reactions are too weak to be detected. The OD value would be elevated when 4 HVR1 is added up together.
In this study, we used a prepared chimeric antigen instead of synthetic peptides[41-43]. The antigen may also be used in the study for the HCV vaccine. In addition, the chimeric antigen is fused with IL-1β. The latter part contains a nano-peptide sequence. It may act as an immune adjuvant[44-46], promoting a strong immune response when injected.
In summary, the chimeric HVR1 antigen, containing several representative HVR1 fragments, can show very high cross-reactivity, which may be helpful to overcome the variability of HCV. The chimeric HVR1 antigen is of potential application for HCV vaccination and immune therapy.
Edited by Su Q and Wang XL
| 2. | Farci P. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244: 359-362]. J Hepatol. 2002;36:582-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4655] [Article Influence: 129.3] [Reference Citation Analysis (0)] |
| 3. | Sela B. [New approaches to immune against hepatitis C virus]. Harefuah. 2002;141:1076-180, 1089. [PubMed] |
| 4. | Esumi M, Rikihisa T, Nishimura S, Goto J, Mizuno K, Zhou YH, Shikata T. Experimental vaccine activities of recombinant E1 and E2 glycoproteins and hypervariable region 1 peptides of hepatitis C virus in chimpanzees. Arch Virol. 1999;144:973-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 44] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A. 1994;91:7792-7796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 347] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 6. | Cerino A, Meola A, Segagni L, Furione M, Marciano S, Triyatni M, Liang TJ, Nicosia A, Mondelli MU. Monoclonal antibodies with broad specificity for hepatitis C virus hypervariable region 1 variants can recognize viral particles. J Immunol. 2001;167:3878-3886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter HJ, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci U S A. 1996;93:15394-15399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 454] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 8. | Zhou YH, Takekoshi M, Maeda F, Ihara S, Esumi M. Recombinant antibody Fab against the hypervariable region 1 of hepatitis C virus blocks the virus adsorption to susceptible cells in vitro. Antiviral Res. 2002;56:51-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 9. | Goto J, Nishimura S, Esumi M, Makizumi K, Rikihisa T, Nishihara T, Mizuno K, Zhou Y, Shikata T, Fujiyama S. Prevention of hepatitis C virus infection in a chimpanzee by vaccination and epitope mapping of antiserum directed against hypervariable region 1. Hepatol Res. 2001;19:270-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 168] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 11. | Korenaga M, Hino K, Katoh Y, Yamaguchi Y, Okuda M, Yoshioka K, Okita K. A possible role of hypervariable region 1 quasispecies in escape of hepatitis C virus particles from neutralization. J Viral Hepat. 2001;8:331-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Gao G, Buskell Z, Seeff L, Tabor E. Drift in the hypervariable region of the hepatitis C virus during 27 years in two patients. J Med Virol. 2002;68:60-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 13. | Scarselli E, Cerino A, Esposito G, Silini E, Mondelli MU, Traboni C. Occurrence of antibodies reactive with more than one variant of the putative envelope glycoprotein (gp70) hypervariable region 1 in viremic hepatitis C virus-infected patients. J Virol. 1995;69:4407-4412. [PubMed] |
| 14. | Puntoriero G, Meola A, Lahm A, Zucchelli S, Ercole BB, Tafi R, Pezzanera M, Mondelli MU, Cortese R, Tramontano A. Towards a solution for hepatitis C virus hypervariability: mimotopes of the hypervariable region 1 can induce antibodies cross-reacting with a large number of viral variants. EMBO J. 1998;17:3521-3533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 113] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Watanabe K, Yoshioka K, Ito H, Ishigami M, Takagi K, Utsunomiya S, Kobayashi M, Kishimoto H, Yano M, Kakumu S. The hypervariable region 1 protein of hepatitis C virus broadly reactive with sera of patients with chronic hepatitis C has a similar amino acid sequence with the consensus sequence. Virology. 1999;264:153-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Mondelli MU, Cerino A, Segagni L, Meola A, Cividini A, Silini E, Nicosia A. Hypervariable region 1 of hepatitis C virus: immunological decoy or biologically relevant domain. Antiviral Res. 2001;52:153-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Rispeter K, Lu M, Behrens SE, Fumiko C, Yoshida T, Roggendorf M. Hepatitis C virus variability: sequence analysis of an isolate after 10 years of chronic infection. Virus Genes. 2000;21:179-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Fan X, Di Bisceglie AM. Genetic characterization of hypervariable region 1 in patients chronically infected with hepatitis C virus genotype 2. J Med Virol. 2001;64:325-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 19. | Lin HJ, Seeff LB, Barbosa L, Hollinger FB. Occurrence of identical hypervariable region 1 sequences of hepatitis C virus in transfusion recipients and their respective blood donors: divergence over time. Hepatology. 2001;34:424-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Lu L, Nakano T, Orito E, Mizokami M, Robertson BH. Evaluation of accumulation of hepatitis C virus mutations in a chronically infected chimpanzee: comparison of the core, E1, HVR1, and NS5b regions. J Virol. 2001;75:3004-3009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Ray SC, Wang YM, Laeyendecker O, Ticehurst JR, Villano SA, Thomas DL. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999;73:2938-2946. [PubMed] |
| 22. | Zucchelli S, Roccasecca R, Meola A, Ercole BB, Tafi R, Dubuisson J, Galfré G, Cortese R, Nicosia A. Mimotopes of the hepatitis C virus hypervariable region 1, but not the natural sequences, induce cross-reactive antibody response by genetic immunization. Hepatology. 2001;33:692-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Roccasecca R, Folgori A, Ercole BB, Puntoriero G, Lahm A, Zucchelli S, Tafi R, Pezzanera M, Galfre G, Tramontano A. Induction of cross-reactive humoral immune response by immunization with mimotopes of the hypervariable region 1 of the hepatitis C virus. Int Rev Immunol. 2001;20:289-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Roccasecca R, Folgori A, Ercole BB, Puntoriero G, Lahm A, Zucchelli S, Tafi R, Pezzanera M, Galfre G, Tramontano A. Mimotopes of the hyper variable region 1 of the hepatitis C virus induce cross-reactive antibodies directed against discontinuous epitopes. Mol Immunol. 2001;38:485-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Li C, Candotti D, Allain JP. Production and characterization of monoclonal antibodies specific for a conserved epitope within hepatitis C virus hypervariable region 1. J Virol. 2001;75:12412-12420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Shang D, Zhai W, Allain JP. Broadly cross-reactive, high-affinity antibody to hypervariable region 1 of the hepatitis C virus in rabbits. Virology. 1999;258:396-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Zhou YH, Moriyama M, Esumi M. Multiple sequence-reactive antibodies induced by a single peptide immunization with hypervariable region 1 of hepatitis C virus. Virology. 1999;256:360-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Yagi S, Kashiwakuma T, Yamaguchi K, Chiba Y, Ohtsuka E, Hasegawa A. An epitope chimeric antigen for the hepatitis C virus serological screening test. Biol Pharm Bull. 1996;19:1254-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 29. | Paolini R, Marson P, Vicarioto M, Ongaro G, Viero M, Girolami A. Anti-hepatitis C virus serology in patients affected with congenital coagulation defects: a comparative study using three second generation ELISA tests. Transfus Sci. 1994;15:303-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 30. | Bosch FX, Ribes J. Epidemiology of liver cancer in Europe. Can J Gastroenterol. 2000;14:621-630. [PubMed] |
| 31. | Simmonds P. Viral heterogeneity of the hepatitis C virus. J Hepatol. 1999;31 Suppl 1:54-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 155] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 32. | Cai Q, Zhang X, Tian L, Yuan M, Jin G, Lu Z. Variant analysis and immunogenicity prediction of envelope gene of HCV strains from China. J Med Virol. 2002;67:490-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 33. | Oliveira ML, Bastos FI, Sabino RR, Paetzold U, Schreier E, Pauli G, Yoshida CF. Distribution of HCV genotypes among different exposure categories in Brazil. Braz J Med Biol Res. 1999;32:279-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Zibert A, Meisel H, Kraas W, Schulz A, Jung G, Roggendorf M. Early antibody response against hypervariable region 1 is associated with acute self-limiting infections of hepatitis C virus. Hepatology. 1997;25:1245-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 78] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 35. | Lechner S, Rispeter K, Meisel H, Kraas W, Jung G, Roggendorf M, Zibert A. Antibodies directed to envelope proteins of hepatitis C virus outside of hypervariable region 1. Virology. 1998;243:313-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 36. | Isaguliants MG, Widell A, Zhang SM, Sidorchuk A, Levi M, Smirnov VD, Santantonio T, Diepolder HM, Pape GR, Nordenfelt E. Antibody responses against B-cell epitopes of the hypervariable region 1 of hepatitis C virus in self-limiting and chronic human hepatitis C followed-up using consensus peptides. J Med Virol. 2002;66:204-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 37. | Del Porto P, Puntoriero G, Scottà C, Nicosia A, Piccolella E. High prevalence of hypervariable region 1-specific and -cross-reactive CD4(+) T cells in HCV-infected individuals responsive to IFN-alpha treatment. Virology. 2000;269:313-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 38. | Hjalmarsson S, Blomberg J, Grillner L, Pipkorn R, Allander T. Sequence evolution and cross-reactive antibody responses to hypervariable region 1 in acute hepatitis C virus infection. J Med Virol. 2001;64:117-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 39. | Hattori M, Yoshioka K, Aiyama T, Iwata K, Terazawa Y, Ishigami M, Yano M, Kakumu S. Broadly reactive antibodies to hypervariable region 1 in hepatitis C virus-infected patient sera: relation to viral loads and response to interferon. Hepatology. 1998;27:1703-1710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 40. | Pawlotsky JM, Germanidis G, Frainais PO, Bouvier M, Soulier A, Pellerin M, Dhumeaux D. Evolution of the hepatitis C virus second envelope protein hypervariable region in chronically infected patients receiving alpha interferon therapy. J Virol. 1999;73:6490-6499. [PubMed] |
| 41. | Zhou YH, Sugitani M, Esumi M. Sequences in the hypervariable region 1 of hepatitis C virus show only minimal variability in the presence of antibodies against hypervariable region 1 during acute infection in chimpanzees. Arch Virol. 2002;147:1955-1962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 42. | Wang Y, Hao F, Huang Y. [Molecular design and immunoreactivity studies of multiple antigen peptide corresponding to hypervariable region 1 sequence of hepatitis C virus]. Zhonghua Ganzangbing Zazhi. 2000;8:48-50. [PubMed] |
| 43. | Dueñas-Carrera S, Viña A, Garay HE, Reyes O, Alvarez-Lajonchere L, Guerra I, González LJ, Morales J. Immunological evaluation of Escherichia coli-derived hepatitis C virus second envelope protein (E2) variants. J Pept Res. 2001;58:221-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 44. | Böckmann S, Mohrdieck K, Paegelow I. Influence of interleukin-1 beta on bradykinin-induced responses in guinea pig peritoneal macrophages. Inflamm Res. 1999;48:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 45. | Akasu T, Tsurusaki M. Effects of interleukin-1 beta on neurons in mammalian pelvic ganglia. Kurume Med J. 1999;46:143-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 46. | Boraschi D, Tagliabue A. Interleukin-1 and interleukin-1 fragments as vaccine adjuvants. Methods. 1999;19:108-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |