Published online Nov 15, 2003. doi: 10.3748/wjg.v9.i11.2474
Revised: June 1, 2003
Accepted: June 12, 2003
Published online: November 15, 2003
AIM: To establish an efficient, sensitive, cell-based assay system for NS3 serine protease in an effort to study further the property of hepatitis C virus (HCV) and develop new antiviral agents.
METHODS: We constructed pCI-neo-NS3/4A-SEAP chimeric plasmid, in which the secreted alkaline phosphatase (SEAP) was fused in-frame to the downstream of NS4A/4B cleavage site. The protease activity of NS3 was reflected by the activity of SEAP in the culture media of transient or stable expression cells. Stably expressing cell lines were obtained by G418 selection. Pefabloc SC, a potent irreversible serine protease inhibitor, was used to treat the stably expressing cell lines to assess the system for screening NS3 inhibitors. To compare the activity of serine proteases from 1b and 1a, two chimeric clones were constructed and introduced into both transient and stable expression systems.
RESULTS: The SEAP activity in the culture media could be detected in both transient and stable expression systems, and was apparently decreased after Pefabloc SC treatment. In both transient and stable systems, NS3/4A-SEAP chimeric gene from HCV genotype 1b produced higher SEAP activity in the culture media than that from 1a.
CONCLUSION: The cell-based system is efficient and sensitive enough for detection and comparison of NS3 protease activity, and screening of anti-NS3 inhibitors. The functional difference between NS3/4A from 1a and 1b subtypes revealed by this system provides a clue for further investigations.
- Citation: Mao HX, Lan SY, Hu YW, Xiang L, Yuan ZH. Establishment of a cell-based assay system for hepatitis C virus serine protease and its primary applications. World J Gastroenterol 2003; 9(11): 2474-2479
- URL: https://www.wjgnet.com/1007-9327/full/v9/i11/2474.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i11.2474
The infection of hepatitis C virus (HCV) is associated with a high frequency of chronic hepatitis (up to 80%), which often progresses to liver cirrhosis and hepatocellular carcinoma (up to 20%)[1]. Until now, there has been no vaccine against HCV and the most efficacious pharmacological treatment, a combined therapy of interferon alpha and ribavirin, which could lead to sustained remission only in a minority of cases[2,3]. Considerable efforts have been therefore undertaken to develop specific antiviral agents.
HCV is a positive-sense, single-stranded RNA virus and belongs to the flaviviridae family. Its genome is about 9.6 kb in length and encodes the structural proteins C, E1, E2, and the non-structural proteins NS2, NS3, NS4A, NS4B, NS5A and NS5B, which are released by action of both host cell and virally encoded proteases[4,5]. The N-terminal domain of the NS3 protein contains a serine protease, belonging to the chymotrypsin family[6], which is responsible for the proteolytic cleavage at the NS3/4A, NS4A/NS4B, NS4B/NS5A and NS5A/5B junctions of the viral polyprotein[4]. The NS3 thus plays a pivotal role in the viral maturation and replication. It is also known to affect normal cellular functions, such as cell proliferation and cell death, suggesting its involvement either direct or indirect in HCV hepatocarcinogenesis[7-9]. Therefore, the NS3 protease has become one of the most attractive targets for the development of HCV specific antiviral agent.
To establish an efficient, sensitive, cell-based assay system for NS3 serine protease in an effort to study further the property of HCV and develop new antiviral agents, we first constructed the pCI-neo-NS3/4A-SEAP chimeric plasmid, in which the secreted alkaline phosphatase (SEAP) gene was fused in-frame to the downstream of NS4A/4B cleavage site. The protease activity of NS3 was reflected by the activity of SEAP in the culture media of transient or stably expressing cells. Stably expressing cell lines were obtained by G418 selection. To explore the possibility of applying this system to the screening of NS3 inhibitors, Pefabloc SC, a potent irreversible serine protease inhibitor, was then used to treat the stably expressing cell lines. After that, we investigated the difference of serine protease activity between HCV genotype 1b and 1a by constructing two pCI-neo-NS3/4A-SEAP chimeric clones and studying them in both transient and stable expression systems.
To amplify NS3/4A region (HCV 1b) using PCR from a near-full length HCV cDNA (from a patient with 1b genotype), two pairs of primers were used: NS3/4A5’, 5’-CCGGAATTCATGGCCCCCATCACAGCCTAT-3’ (nucleotides [nt] 3420-3437), and NS3/4A3’, 5’-CAGTCTCGAGGAGGTGTGAGGCGCA-3’ (nucleotides [nt] 5472-5486); NS3/4A (TGA)3’, 5’-CCGCTCGAGTCAGCACTCCTCCATCTCATC-3’ (nucleotides [nt] 5457-5474, with stop codon, as negative control). RNS3/4A (HCV 1a) was amplified with primer RNS3/4A5’, 5’-ATGAATTCATGGCGCCCATCACGGCGTAC-3’ (nucleotides [nt] 3420-3437) and primer RNS3/4A3’, 5’-GCCAAGCTTTAAGTGCTGAGAGCA-3’ (nucleotides [nt] 5472-5486) from p90/HCV FL-long pU (AF009606) (a generous gift from Professor Charle M Rice from the Center for the Study of Hepatitis C, Rockefellor University). SEAP fragment was obtained by PCR with two pairs of primers: SEAP (Xho I)5’, 5’-ATCTCGAGATGCTGCTGCTGCTGCTGCTG-3’ (nucleotides [nt] 54-74), SEAP (Hind III)5’, 5’-ATAAGCTTATGCTGCTGCTGCTGCTGCTGCTG-3’ (nucleotides [nt] 54-77), and SEAP3’, 5’-ATGCGGCCGCTCAGGGAGCAGTGGCCGTC-3’ (nucleotides [nt] 1628-1647) from pCMV-SEAP (generously provided by Professor Byan R. Cullen of the Medical Center, Duke University).
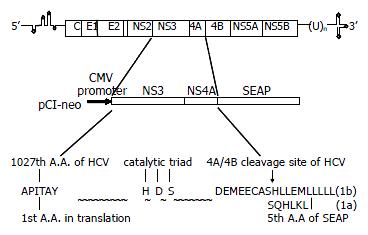
The NS3/4A fragment from HCV 1b genotype and digested with EcoR I and Xho I, was ligated into EcoR I/Not I site of pCI-neo vector (Stratagene) with the SEAP fragment digested with Xho I and Not I to generate pCI-neo-NS3/4A-SEAP. The HCV 1a RNS3/4A fragment digested with EcoR I and Hind III and the SEAP fragment digested with Hind III and Not I were ligated into EcoR I/Not I site of pCI-neo vector to generate pCI-neo-RNS3/4A-SEAP. The schematic diagram is shown in Figure 1. The EcoR I/Xho I fragment from the pET28a-NS3/4A (TGA) was ligated into EcoR I/Sal I site of pCI-neo vector and the clone was named as pCI-neo-NS3/4A (TGA).
HepG2 cells or Cos7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with penicillin (100 U/mL) and streptomycin (100 μg/mL), supplemented with 10% fetal calf serum. Recombinant plasmid DNA used for transfection was extracted and purified with the Qiagen Midiprep kits. DNA (5 μg) was used to transfect HepG2 or Cos7 cells in 6-well NUNC multidish by the calcium phosphate precipitation method as reported previously[10]. Duplicate dishes were used for all samples, and 3 μg of reporter plasmid pcDNA3.1-CAT expressing chloramphenicol acetyltransferase (CAT) was cotransfected as an internal control to normalize the transfection efficiency among dishes. Forty-eight hours after transfection, culture media and cells were collected separately to monitor the secretion of SEAP in the supernatant and NS3 protease expression in transfected cells. pCI-neo-NS3/4A (TGA) was used as a mock transfection control. Transfection experiments were repeated twice on separate days.
The SEAP activity of the culture media was measured at 48 h after transfection by performing a colorimetric assay according to the author’s recommendation[11]. Briefly, 20 μL heat-treated (at 65 °C for 5 min) medium was adjusted to 1 × SEAP assay buffer (1.0 M diethanolamine pH9.8, 0.5 mM MgCl2, 10 mM L-homoarginine) in a final volume of 200 μL and prewarmed to 37 °C for 10 min in a 96-well flat-bottom culture dish. Twenty μL of prewarmed 120 mM p-nitrophenylphosphate dissolved in SEAP assay buffer was then added with mixing. The A405 of the reaction mixture was read in a BIO-RAD (Benchmark) microplate reader at 5-min intervals. The change in absorbance was plotted and the maximum linear reaction rate determined. The SEAP activity was expressed in milliunits (mU) per mL. One mu equals an increase of 0.04 A405 units per min. Each SEAP assay was performed in triplicate.
Two different cell lines of Cos7 and HepG2 cells were transfected with the plasmid pCI-neo-NS3/4A-SEAP, pCI-neo-RNS3/4A-SEAP or pCI-neo-NS3/4A (TGA), respectively by calcium phosphate precipitation method mentioned above. At 48 h after transfection, the cells were subcultured and selected with the 600 mg/L of G418. After 3 wk of selection, the colonies were picked up and amplified in the presence of 200 mg/L G418.
Cos7 cell lines (1b and 1a type) were seeded into a 6-well NUNC multidish at a density of 105 cells/mL in 3 mL medium and cultured overnight at 37 °C in a 50 mL·L-1 CO2 incubator. The overnight culture media were changed with fresh one containing Pefabloc SC at the concentration of 0.1 mM, 0.2 mM, 0.4 mM, 0.5 mM, or 0.6 mM. The cells without Pefabloc SC treatment were set as control. After 24 h, the media were collected for the colorimetric assay for SEAP activity. Effect of Pefabloc SC on the viability of Cos7 stable cells was evaluated by the alamarBlueTM assay (Biosource International Corp.)[12]. Alamar Blue was added in an amount equal to 10% of the culture volume and after 2 h of incubation at 37 °C, the panels were read spectrofluorometrically (excitation, 544 nm; emission, 590 nm) by Ascent FL. The experiment was performed twice and each sample was detected in duplicate.
Total RNA was extracted using GIBCO-BRL TRIZOL Reagent (Cat No. 15596 Gibco Life Tech) according to the instructions of manufacturers. It was digested with RNase free DNase (Promega) to wipe off the transfected plasmid DNA. After electrophoresis in a 1% agarose gel, 10 μg total RNA was blotted onto a positively charged Nylon membrane (Roche). HCV NS3/4A (100 ng of PCR products, 1a and 1b respectively) and random primers were used to prepare the [32P] dCTP labled probes for hybridization. The signals were detected by autoradiography and scanned with densitometry for semiquantification of the intensity of signals. The corresponding intensity of β-actin signals was used as controls.
To assess the expression levels of the HCV NS3/4A of two HCV genotypes in the transfected HepG2 or Cos7, Western blot analysis was conducted as described[13] with some modifications. Briefly, cells were lysed in 200 μL TSA buffer (50 mM Tris-HCl, 140 mM NaCl, 5 mL•L-1 NP40, 1 mM PMSF, 1 mM DTT). Ten μL aliquot was subjected to 7.5% SDS-PAGE eletrophoresis. The separated proteins were transferred to nitrocellulose membrane in transfer buffer (20 mM Tris-HCl, 190 mM Glycine, 200 mL•L-1 methanol, pH8.3) using Mini Trans-blot transfer system (Bio-Rad) at 100V for 1 h. The membrane was blocked with 50 mg•L-1 nonfat dried milk in PBST (Tris-buffered saline containing 0.5 mL•L-1 Tween-20) and then incubated with mouse anti-NS3 multiclonal antibody. After three washes in PBST, the membrane was incubated with peroxidase-conjugated goat anti-mouse second antibody (A90-131P, BETHYL Co.). The blot was incubated with TMB substrate to reveal the antigen bands. Cellular protein La (a phosphoprotein, 47 KD, present at proximately 2 × 107 molecules per cell) was used as a quantificational control. The expression levels of the NS3 of two HCV isolates were compared by the semi-quantification of scanned intensity of antigen bands.
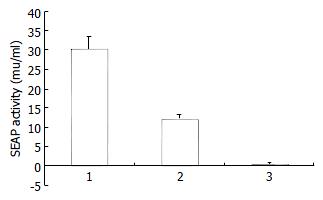
The plasmids pCI-neo-NS3/4A-SEAP, pCI-neo-NS3/4A (TGA) and pCMV-SEAP were transfected into HepG2 or Cos7 cells by calcium phosphate method and the SEAP activity of the culture media was measured at 48 h after transfection. As expected, in HepG2 cells, the pCMV-SEAP produced SEAP activity about 30.16 ± 3.46 mu/mL, while pCI-neo-NS3/4A (TGA) produced 0.34 ± 0.48 mu/mL, which is almost the background level (Figure 2). The culture media from cells transfected with pCI-neo-NS3/4A-SEAP showed a SEAP activity of about 12.05 ± 1.25 mu/mL. Similar results were obtained in Cos7 cells (data not shown). The processing of fusion proteins expressed in transfected cells was examined by immunoblotting with anti-HCV human sera and mouse derived multiple-clonal sera. Processed NS3 protein of 70 KD was detected (Figure 3). The results indicate that, depending on the cleavage activity of the NS3 protease, the SEAP could be cleaved and secreted into the extracellular media.
To establish the NS3/4A-SEAP assay system in stable expression cells, HepG2 and Cos7 cells transfected with the pCI-neo-NS3/4A-SEAP were selected for stable expression ones. During 600 mg/L G418 selection, most transfected HepG2 or Cos7 cells died out after 4 changes of media and the selected colonies were picked up and amplified in the presence of 200 mg/L G418. Results of SEAP assay showed that all selected stably expressing cell lines produced SEAP in their supernatant. However, the media from Cos7-derived stably expressing cell lines showed higher SEAP activity than those from HepG2-derived cell lines. Initially, the two kinds of stably expressing cell lines did not show any significant differences. Then, the HepG2-derived cell lines appeared to lose supernatant SEAP activity, depending on the passages and showed finally, very low SEAP activity (data not shown) while selected cell lines derived from Cos7 appeared to express high levels of SEAP during continuous passages over 4 mo.
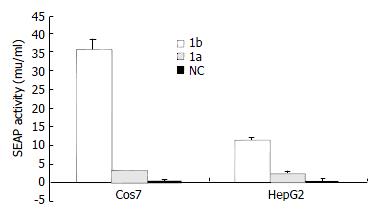
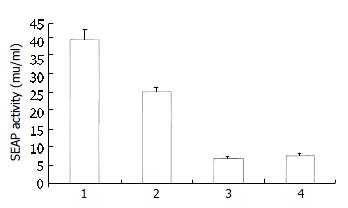
To compare the activity of serine proteases from HCV subtype 1b and 1a, two chimeric clones were constructed and studied in both transient and stable expression systems. Results of SEAP assay showed that, in transient expression system, the Cos7 cells transfected with the pCI-neo-NS3/4A (1b)-SEAP produced higher supernatant SEAP activity (37.49 ± 3.06 mu/mL) than those transfected with pCI-neo-RNS3/4A (1a)-SEAP (3.48 ± 0.15 mu/mL). The overall assay results were similar in HepG2 cells with pCI-neo-NS3/4A (1b)-SEAP at 11.82 ± 0.92 mu/mL and pCI-neo-RNS3/4A (1a)-SEAP at 2.49 ± 0.67 mu/mL. The above results indicated that cells transfected with pCI-neo-NS3/4A-SEAP from HCV 1b isolate produced 5-10 folds higher supernatant SEAP activity than those transfected with pCI-neo-RNS3/4A-SEAP from HCV 1a isolate (Figure 4). Consistently, the 1b derived stable cell lines also produced higher SEAP activity in media (39.11 ± 2.97 mu/mL, 25.36 ± 1.41 mu/mL) than those derived from 1a (6.92 ± 0.56 mu/mL, 7.85 ± 0.57 mu/mL, Figure 5).
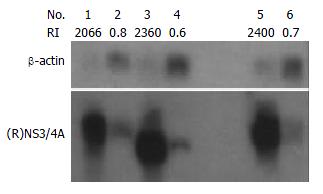
To compare the amounts of NS3/4A mRNAs in transfected HepG2 and Cos7 cells, Northern blotting was carried out using the isotope-labeled DNA probes corresponding to NS3/4A regions of 1b and 1a. The NS3/4A specific signals could be detected in both HepG2 and Cos7 cells transfected with pCI-neo-NS3/4A-SEAP (1b), pCI-neo-NS3/4A (TGA), or pCI-neo-RNS3/4A-SEAP (1a) respectively. The hybridization signals of NS3/4A mRNA were similar between conspecific cells transfected with 1b or 1a constructs as the ratio of signal densities of NS3/4A to β-actin in pCI-neo-NS3/4A-SEAP (1b), pCI-neo-NS3/4A (TGA) and pCI-neo-RNS3/4A-SEAP (1a) transfected cells were similar (2066/2360/2400 in transfected Cos7 cells and 0.8/0.6/0.7 in transfected HepG2 cells) (Figure 6).
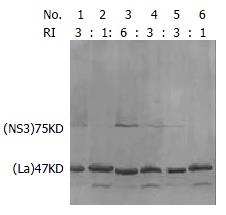
To compare the levels of NS3 proteins in transfected cells, Western blotting was carried out to detect the HCV protease expression with mouse anti-NS3 antibody. The expression levels of the NS3 were compared by the semiquantification of bands detected by immunoblotting. The relative intensities (RI) of NS3 to cellular protein La are indicated in Figure 3. The results showed that NS3 level expressed by pCI-neo-NS3/4A-SEAP (1b) was similar to the one expressed by pCI-neo-RNS3/4A-SEAP (1a) (3/3 in transfected Cos7 cells and 1/1 in transfected HepG2 cells) in transfected cells (Figure 6).
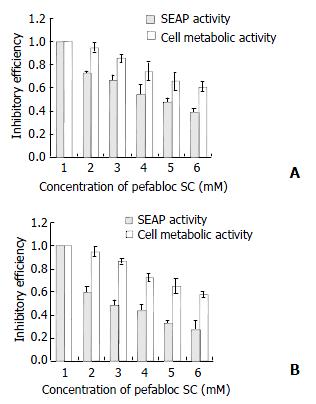
Pefabloc SC, a serine protease inhibitor was added into the media of Cos7-derived stable expression cells at the concentration of 0.1 mM, 0.2 mM, 0.4 mM, 0.5 mM, or 0.6 mM. After 24 h, the media were collected for the colorimetric assay of SEAP activity. The effect of Pefabloc SC on the viability of stable expression cells was evaluated by the alamarBlueTM assay. Results showed that, although the Pefabloc SC was relatively toxic to cells, it had some specific inhibitory effect on the protease activity of NS3/4A as the secretion of SEAP, which was dependent on the active HCV protease, was decreased more than the metabolic activity of the cells after treatment, and it was 60%-70% when the treated cells still kept almost 100% metabolic activity (Figure 7).
It has been reported that the cleavage activity of NS3 could be detected by in vitro transcription/translation and transient expression experiments[14-16], or using purified active NS3 protease[17,18]. But they are known to be difficult to apply for screening specific NS3 inhibitors at large scale. ANother problem has to be considered referring to the rather featureless substrate binding pocket of NS3[19], which may make the development of specific inhibitors rather difficult. Alternative in vivo assay systems for NS3 protease, which can differentiate the inhibitors’ effects on NS3 protease and cellular metabolic activity (including cellular serine-type protease), will accelerate the screening of specific inhibitors. Several chimeric viral replication systems had been developed to screen HCV inhibitors in tissue culture[20,21]. These systems have some disadvantages regarding instability of chimeric viruses and difficulty in evaluating the inhibition quantitatively. Comparably, our cell-based NS3/4A-SEAP expression system is safe, easy to handle and the report gene of SEAP can be sensitively and quantitatively measured continuously without killing cells[11].
In the scheme of pCI-neo-NS3/4A-SEAP constructs, the NS4A/SEAP junction has the sequence DEMEEC↓ASHL (HCV 1b subtype)[22] or DEMEEC↓SQHL (HCV 1a subtype) which contains NS4A/4B cleavage site. The rationale for this system was based on the assumption that the secretion of SEAP protein into the culture media depends on the cleavage between NS4A protein and SEAP protein by HCV NS3 protease. The unusual substrate specificity, which is quite distinct from cellular serine-type proteases, makes it possible for the system to generate inhibitors with a high degree of selectivity[23,24]. To make the system more suitable for the screening of NS3 inhibitors, we also established the stably expressing cell lines. During the screening, the inhibitor could be introduced into the stable cells by transfection or simply adding in culture media[25] according to their own characteristics.
In the process of establishing both transient and stable expression systems, we found that Cos7-derived cells always showed higher SEAP activity in media than HepG2-derived ones. It is consistent with the previous report that Cos7 cells and other cell lines expressing the SV40 large T-antigen, have been of great benefit to the transfection studies using transient expression vectors containing the SV40 origin of replication, as these cells yield plasmid replication to a high copy number[26]. In addition, we also observed that the two kinds of cell lines initially did not show any significant differences in cell growth and morphology, but depending on the passages, the HepG2-derived cell lines appeared to lose SEAP activity and finally showed very low SEAP activity (data not shown). However, the Cos7-derived cell lines appeared to express high levels of SEAP activity during continuous passages even after 4 mo. It is likely that the expression of active protease is relatively toxic in some cells, leading to selection of cells with low protease activity in the process of G418 selection and passage[22].
When this assay system was used to compare the activity of HCV serine proteases from different genotypes (1b and 1a), the cells transfected with 1b type plasmid DNA showed 5-10 folds higher SEAP activity in culture media than those with 1a type one in transient system. And there were no significant differences in protein and mRNA levels of NS3/4A between them in either transiently transfected Cos7 or HepG2 cells. The results indicated that the cleavage efficiency of 1b type NS3 protease was 5-10 folds higher than that of 1a type. Consistently, we observed similar results in the stable expression system. Since the NS3 serine protease activity is required for cleavages at the downstream 3/4A, 4A/4B, 4B/5A, 5A/5B sites, its cleavage efficiency will affect the assembly of functional HCV RNA replication complex, viral particle maturation and host-virus interaction. It has been reported that the success in establishing efficient HCV replication depended on the particular genotype 1b consensus cDNA clone studied[27]. Whether the higher protease activity of 1b subtype NS3 contributes to the above phenomenon is worthy of further investigations. The availability of replication system[27-29] will promote such studies.
In this study, the cell-based system was also successfully used to evaluate the inhibition effect of Pefabloc SC on the NS3 protease in vivo. Pefabloc SC[30] is one of the most potent inhibitors of the class of sulfonyl fluorides like phenylmethylsulfonylfluoride (PMSF). It has been widely used to inhibit all kinds of serine proteases (including HCV NS3)[31,32]. It was reported that 8 mM of Pefabloc SC showed strong effects on NS3-4A junction in a trans-cleavage assay system[33]. Due to its toxicity to cells, the concentrations of Pefabloc SC used in our system were only from 0.1 mM to 0.6 mM. However, results here still showed that it had some specific inhibitory effect on the protease activity of the HCVNS3/4A as the secretion of SEAP, which depended on the active HCV protease, was decreased more than the metabolic activity of the cells after treatment (Figure 6). As shown in Figure 6, when the cells treated with 0.1 mM Pefabloc SC still kept near 100% metabolic activity, the SEAP activity in culture media had decreased to 60%-70%. These data reflected the sensitivity of our system and its practicability for the detection and comparison of NS3 protease activity and the screening anti-NS3 inhibitors.
In summary, we reported the establishment of the cell-based NS3/4A-SEAP expression system in both transiently transfected and stable cell lines. It was further applied to the evaluation of Pefabolc SC, a known protease inhibitor and the comparison of activities of NS3 protease from 1a, 1b genotypes. It could be concluded that this cell-based system is efficient and sensitive enough for the detection and comparison of NS3 protease activity and the screening of anti-NS3 inhibitors. The functional difference between NS3/4A from 1a and 1b subtypes revealed by this system provides a clue for further investigations.
Edited by Ma JY
| 1. | Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547-6549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 840] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 2. | Saracco G, Ciancio A, Olivero A, Smedile A, Roffi L, Croce G, Colletta C, Cariti G, Andreoni M, Biglino A. A randomized 4-arm multicenter study of interferon alfa-2b plus ribavirin in the treatment of patients with chronic hepatitis C not responding to interferon alone. Hepatology. 2001;34:133-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Kronenberger B, Rüster B, Elez R, Weber S, Piiper A, Lee JH, Roth WK, Zeuzem S. Interferon alfa down-regulates CD81 in patients with chronic hepatitis C. Hepatology. 2001;33:1518-1526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 4. | Neddermann P, Tomei L, Steinkühler C, Gallinari P, Tramontano A, De Francesco R. The nonstructural proteins of the hepatitis C virus: structure and functions. Biol Chem. 1997;378:469-476. [PubMed] |
| 5. | Bartenschlager R. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J Viral Hepat. 1999;6:165-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 106] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 6. | Lesk AM, Fordham WD. Conservation and variability in the structures of serine proteinases of the chymotrypsin family. J Mol Biol. 1996;258:501-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 104] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 7. | Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893-3896. [PubMed] |
| 8. | Zemel R, Gerechet S, Greif H, Bachmatove L, Birk Y, Golan-Goldhirsh A, Kunin M, Berdichevsky Y, Benhar I, Tur-Kaspa R. Cell transformation induced by hepatitis C virus NS3 serine protease. J Viral Hepat. 2001;8:96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 9. | Fujita T, Ishido S, Muramatsu S, Itoh M, Hotta H. Suppression of actinomycin D-induced apoptosis by the NS3 protein of hepatitis C virus. Biochem Biophys Res Commun. 1996;229:825-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 65] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Lin X, Yuan ZH, Wu L, Ding JP, Wen YM. A single amino acid in the reverse transcriptase domain of hepatitis B virus affects virus replication efficiency. J Virol. 2001;75:11827-11833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Berger J, Hauber J, Hauber R, Geiger R, Cullen BR. Secreted placental alkaline phosphatase: A powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 520] [Cited by in RCA: 554] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 12. | Nakayama GR, Caton MC, Nova MP, Parandoosh Z. Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J immunol Methods. 1997;204:205-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 461] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 13. | Kong WH, Zheng G, LU JN, Tso JK. Temperature dependent expression of cdc2 and cyclin B1 in spermatogenic cells during spermatogenesis. Cell Res. 2000;10:289-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Du GX, Hou LH, Guan RB, Tong YG, Wang HT. Establishment of a simple assay in vitro for hepatitis C virus NS3 serine protease based on recombinant substrate and single-chain protease. World J Gastroenterol. 2002;8:1088-1093. [PubMed] |
| 15. | Kolykhalov AA, Agapov EV, Rice CM. Specificity of the hepatitis C virus NS3 serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J Virol. 1994;68:7525-7533. [PubMed] |
| 16. | Bartenschlager R, Ahlborn-Laake L, Yasargil K, Mous J, Jacobsen H. Substrate determinants for cleavage in cis and in trans by the hepatitis C virus NS3 proteinase. J Virol. 1995;69:198-205. [PubMed] |
| 17. | Bianchi E, Steinkühler C, Taliani M, Urbani A, De Francesco R, Pessi A. Synthetic depsipeptide substrates for the assay of human hepatitis C virus protease. Anal Biochem. 1996;237:239-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Steinkühler C, Urbani A, Tomei L, Biasiol G, Sardana M, Bianchi E, Pessi A, De Francesco R. Activity of purified hepatitis C virus protease NS3 on peptide substrates. J Virol. 1996;70:6694-6700. [PubMed] |
| 19. | Pizzi E, Tramontano A, Tomei L, La Monica N, Failla C, Sardana M, Wood T, De Francesco R. Molecular model of the specificity pocket of the hepatitis C virus protease: implications for substrate recognition. Proc Natl Acad Sci USA. 1994;91:888-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 75] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Hahm B, Back SH, Lee TG, Wimmer E, Jang SK. Generation of a novel poliovirus with a requirement of hepatitis C virus protease NS3 activity. Virology. 1996;226:318-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Cho YG, Moon HS, Sung YC. Construction of hepatitis C-SIN virus recombinants with replicative dependency on hepatitis C virus serine protease activity. J Virol Methods. 1997;65:201-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Cho YG, Yang SH, Sung YC. In vivo assay for hepatitis C viral serine protease activity using a secreted protein. J Virol Methods. 1998;72:109-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832-2843. [PubMed] |
| 24. | Komoda Y, Hijikata M, Sato S, Asabe S, Kimura K, Shimotohno K. Substrate requirements of hepatitis C virus serine proteinase for intermolecular polypeptide cleavage in Escherichia coli. J Virol. 1994;68:7351-7357. [PubMed] |
| 25. | Heintges T, Encke J, zu Putlitz J, Wands JR. Inhibition of hepatitis C virus NS3 function by antisense oligodeoxynucleotides and protease inhibitor. J Med Virol. 2001;65:671-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | Harvey TJ, Macnaughton TB, Gowans EJ. The development and characterisation of a SV40 T-antigen positive cell line of human hepatic origin. J Virol Methods. 1997;65:67-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 27. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2294] [Cited by in RCA: 2251] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 28. | Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972-1974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1154] [Cited by in RCA: 1150] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 29. | Guo JT, Bichko VV, Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J Virol. 2001;75:8516-8523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 383] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 30. | Dentan C, Tselepis AD, Chapman MJ, Ninio E. Pefabloc, 4-[2-aminoethyl] benzenesulfonyl fluoride, is a new, potent nontoxic and irreversible inhibitor of PAF-degrading acetylhydrolase. Biochim Biophys Acta. 1996;1299:353-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Angelloz-Nicoud P, Harel L, Binoux M. Recombinant human insulin-like growth factor (IGF) binding protein-3 stimulates prostate carcinoma cell proliferation via an IGF-dependent mechanism. Role of serine proteases. Growth Regul. 1996;6:130-136. [PubMed] |
| 32. | Hiramatsu N, Ichikawa N, Fukada H, Fujita T, Sullivan CV, Hara A. Identification and characterization of proteases involved in specific proteolysis of vitellogenin and yolk proteins in salmonids. J Exp Zool. 2002;292:11-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 56] [Article Influence: 2.4] [Reference Citation Analysis (0)] |