Published online Apr 15, 2001. doi: 10.3748/wjg.v7.i2.289
Revised: September 27, 2000
Accepted: September 29, 2000
Published online: April 15, 2001
- Citation: Zhang WZ, Han TQ, Tang YQ, Zhang SD. Rapid detection of sepsis complicating acute necrotizing pancreatitis using polymerase chain reaction. World J Gastroenterol 2001; 7(2): 289-292
- URL: https://www.wjgnet.com/1007-9327/full/v7/i2/289.htm
- DOI: https://dx.doi.org/10.3748/wjg.v7.i2.289
Acute necrotizing pancreatitis usually takes a severe clinical course and is associated with multiple organ dysfunction. With the further understanding of pathophysiological events of acute pancreatitis and the therapeutic measuses taken by the clinicians, the patients can pass through the critical early stages, and then the septic complication caused by translocated bacteria, mostly gram-negative microbes from the intestines ensues[1]. During this stage, the clinical manifestation is not specific and is characterized by systemic inflammatory response, but bacterial cultures are often negative.
Identification of minute quantities of microbial-specific DNA has been made possible by using polymerase chain reaction techniques[2-19] and this method has been used to detect and identify specific pathogen in clinical specimens. It has been shown that PCR method is more sensitive than conventional blood cultures for detecting microbial products in blood[20-22].
The current study was performed to evaluate the technique of PCR with the universal primers targeting bacterial 16S rRNA genes in diagnosing the systemic infection secondary to acute necrotizing pancreatitis.
Between May 1998 and May 1999, 22 blood samples were obtained from 13 patients with CT or surgically confirmed acute necrotizing pancreatitis who were admitted consecutively to surgical ICU in Ruijin Hospital, Shanghai. There were 8 men and 5 women, the average age was 56.6 ± 8.9 years, and the average APACHE II scores were 10.5 ± 2.2 points.
The blood samples were drawn if the patients presented two or more of the following conditions: ① temperature more than 38 °C or less than 36 °C, ② elevated heart rate more than 90 beats per minute, ③ respiratory rate more than 20 breaths per minute or PaCO2 less than 32 mmHg, and ④ white blood cell count more than 12000/cu mm, less than 4000/cu mm, or more than 10% immature band forms . And the foci of infection were documented[23-26].
Twelve mL of blood was drawn from each patient, of which 2 milliliter was collected in sterile Na2 EDTA anticoagulant Eppendorf tubes and stored at 4 °C until DNA extraction was performed, 10 milliliter was sent fo rconventional blood cultures.
At the same time, 10 blood samples were obtained from 10 healthy volunteers for controlled study.
The bacterial strains used were clinical isolates collected from Ruijin Hospital and identified by automated Vitek system. The strains were cultured at 37 °C on blood agar plates until DNA extraction was performed.
Blood was transferred from Na2EDTA tubes to sterile 1.5 mL Eppendorf tubes, red cells were lysed in 0.32M sugar-5 mmol MgCl2-0.01M Tris-HCl -1% Triton-x for 10 min at room temperature. After centrifugation for 5 min at 5000 rpm, the supernatant was discarded and sediment was preserved for DNA extraction.
The sediment was lysed in 10% Chelex-100 (Sigma) -0.03% Sodium dodecyl sulfate -1% Tween 20-1% Nonidetp-40 for 5 min at 95 °C. After centrifugation (5000 rpm) for 10 s, 5 μL of the supernatant was directly used for PCR amplification[2].
One set of oligonucleotide primer pair was synthesized by the Promega Company, Shanghai Office. The target DNA sequence was the 16S rRNA gene. This set of primers was 5’-GGCGGACGGGTGAGTAA-3’ and 5’-ACTGCTGCCTCCCGTAG-3’ to amplify a 255 bp region.
DNA from clinical isolates of E.coli was extracted in the same manner as outlined previously. This DNA was used in PCR reactions to determine if the PCR reaction was successful. In addition to a positive control, each PCR experiment contained a reagent negative control that consisted of all PCR reagents but without DNA to determine whether the potential contamination was present.
PCR assay was established according to the protocols described by Widjojoatmodjo et al[2]. The PCR mixture (50 μL) contained 50 mM Tris-HCl, 200 mM each deoxynucleoside triphosphate (dNTP), 0.4 μM each primer and 1.0 u of Super-Taq Polymerase (Promega Company, Shanghai Office ) and 7 mM MgCl2.
The PCR was performed in a DNA Thermal Controller (MJ, Research, INC, USA) as follows. The first step of 5 min at 94 °C was followed by 30 cycles of 30 s at 94 °C, 10 s at 72 °C and 1 min at 55 °C; and extension period of 2 min at 55 °C completed the cycling squence.
After amplification, 5 μL of PCR products was run on a 1% agarose gel in 0.5 × TBE. DNA bands were detected by ethidium bromide staining and visualized by UV light photography.
Blood obtained for culture was collected from patients in a sterile manner and inoculated directly into aerobic and anaerobic bottles. The procedure was performed in the department of clinical diagnosis, Ruijin Hospital.
Statistical analysis was done by using the Chi-square test. The difference was considered significant at P < 0.05.
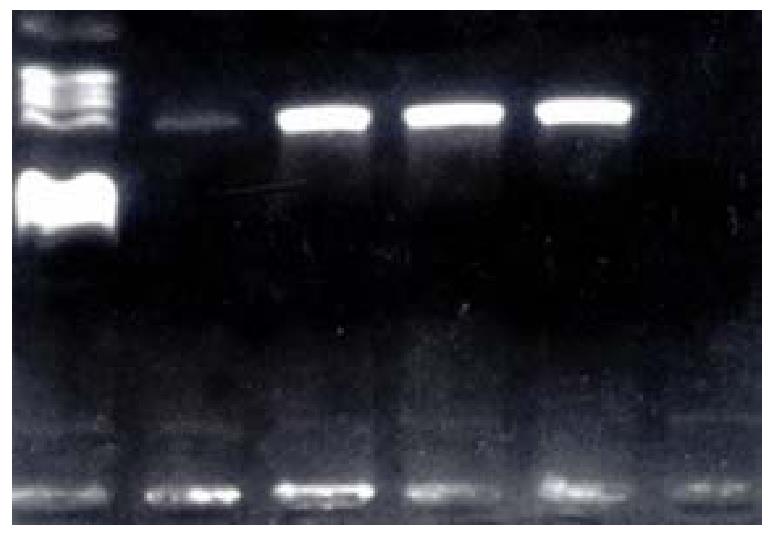
There was only 1 positive blood culture in the 22 blood samples of 13 patients (4.55%). The organism was Escherichia coli(Table 1).But PCR amplification was positive for 8 samples (36.36% P < 0.05 vs culture) from patients and all clinical isolates, yielding the 255 bp band (Figure 1). No DNA amplification occurred in the blood samples from volunteers.
| Sample No. | Age (yr) | Gender | Blood culture | PCR | T (°C) | HR (beats /min) | R (breaths /min) | WBC (109/L) |
| 1 | 53 | M | - | + | 39 | 120 | 20 | 13.5 |
| 2* | 36 | M | + | + | 39.1 | 116 | 22 | 14.3 |
| - | + | 38.6 | 130 | 25 | 17.9 | |||
| - | - | 38.1 | 114 | 22 | 9.8 | |||
| 3* | 51 | M | - | + | 39.1 | 130 | 24 | 17.1 |
| - | - | 39.1 | 140 | 24 | 18.3 | |||
| 4 | 55 | M | - | + | 38.3 | 100 | 22 | 17.5 |
| 5 | 67 | F | - | - | 38.2 | 110 | 24 | 12.8 |
| 6* | 54 | M | - | - | 38.5 | 116 | 34 | 14.4 |
| - | - | 37.9 | 116 | 22 | 12.2 | |||
| - | - | 37.2 | 95 | 18 | 18.7 | |||
| - | - | 37.9 | 118 | 23 | 18.3 | |||
| 7 | 57 | M | - | - | 39.5 | 120 | 26 | 18.6 |
| 8* | 51 | M | - | + | 39.7 | 180 | Mechanic | 19.2 |
| - | - | 39.2 | 130 | Mechanic | 13.4 | |||
| 9* | 54 | F | - | + | 39.6 | 128 | Mechanic | 37.7 |
| - | - | 41.3 | 130 | Mechanic | 18.5 | |||
| 10 | 60 | F | - | - | 39.3 | 170 | Mechanic | 9.3 |
| 11* | 61 | F | - | - | 39.1 | 126 | 28 | 20.4 |
| - | + | 38.5 | 116 | 22 | 8.9 | |||
| 12 | 60 | M | - | - | 38.5 | 108 | 22 | 16 |
| 13 | 74 | F | - | - | 38 | 116 | Mechanic | 18.6 |
During the late course of acute necrotizing pancreatitis, starting from the second week, local and systemic complication caused by translocated bacteria from intestines are dominant. The infection occurs in 30% to 40% of patients with acute necrotizing pancreatitis. Around 80% of deaths in patients with acute necrotizing pancreatitis are caused by septic complication[1]. But during this stage, the clinical manifestation is not specific and characterized by systemic inflammatory reaction and the blood culture is usually negative; this will levy a heavy toll on the clinician for the prompt management of the patients.
Recent studies showed that blood culture techniques, such as volume of inoculated blood, culture media could significantly influence the recovery of bacteria in clinically suspected septic patients and culture is more time-consuming[27-30].
Molecular biology techniques, such as PCR have been used in making a specific and sensitive diagnosis of bacterial infection[2,8,9,12,13,15,16]. The 16S rRNA sequence is highly conserved through the phylogenetic tree. The conserved sequences of the 16S rRNA have led to the development of conserved primers for PCR for the detection of eubacteria. Recently the PCR with universal primers targeting 16S rRNA genes has been used widely to define bacteria[2,15,16,31-41].
With the protocol described by Widjojoatmodjo et al[2,31], we developed PCR assay by using the 16S rRNA genes as the amplification targets. In this assay, we found no DNA amplification in healthy blood cells, suggestive of high specificity of these primer pairs. The disadvantage of PCR technique is the contamination of DNA templates, and therefore we employed negative controls at each PCR experiment to safeguard against the potential contamination of stock PCR reagants with microbial DNA products in the environment, and this study showed no false-positive results (Figure 1).
The gold standard of identifying sepsis is blood culture; however, the clinical sepsis is observed in the absence of documented infection in more than 50% of patients with MOF[21] and the prevalence of positive blood culture is around 12%. The positive rate of blood cultures in our study was 4.55% (propably due to small sample), whereas the PCR-positive rate was 36.36% (P < 0.05), which signifies that this detection method has higher sensitivity than blood culture.
Another advantage of this PCR assay is its ability to perform serial measurements in the same patient for detection of bacterial DNA in the blood, as shown in patients 2, 3, 6, 8, 9 and 11 (Table 1), because PCR is time-saving (less than 8 h) and blood cultures usually take much longer time (at least 2 days).
However, this detection method cannot identify whether it represents living invading organisms or dead ones engulfed by phagocytes, so this approach cannot differentiate between controlled and invasive infections . Until methods that quantitate bacterial DNA are developed[36], we should combined the results of PCR assays with relevant clinical information to determine whether the sepsis is present. Furthermore, if we apply multiple oligonucleotide primers in the PCR assay[21], there would be a higher PCR-positive rate.
In conclusion, the PCR assay with universal primers targeting 16S rRNA genes is more sensitive in detecting the sepsis secondary to acute necrotizing pancreatitis and this may prompt us to take more aggressive approach to the disease.
Edited by Lu HM proofread by Ma JY
| 1. | Beger HG, Rau B, Mayer J, Pralle U. Natural course of acute pancreatitis. World J Surg. 1997;21:130-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 324] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 2. | Widjojoatmodjo MN, Fluit AC, Verhoef J. Molecular identification of bacteria by fluorescence-based PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1995;33:2601-2606. [PubMed] |
| 3. | An P, Li SY, Han LX. The value PCR direct detection in HBsAg negative liver diseases. Xin Xiaohuabingxue Zazhi. 1996;4:385-386. |
| 4. | Shi FJ, Li XZ, Wang CS. The detection of HBV DNA in digestive carcinomatous tissue by PCR. Xin Xiaohuabingxue Zazhi. 1996;4:408-409. |
| 5. | Liang YR, Cheng JD. Nested PCR demonstrated: hepatocellular carcinoma had HBV-DNA replication. Xin Xiaohuabingxue Zazhi. 1996;4:424-426. |
| 6. | Cheng JD, Liang YR. A study on HBVDNA in hepatocellular car-cinoma and para-carcinomatous tissue by location PCR. Xin Xiaohuabingxue Zazhi. 1996;4:427-429. |
| 7. | Ou YH, Yao QX, Kang AN, Jiang ZH, Niu Y. Investigation of HBV infection in blood donors with PCR. Xin Xiaohuabingxue Zazhi. 1997;5:108-109. |
| 8. | Li RP, Zhang ZK, Yao XX, Ren XL, Li LG. Evaluation of PCR method in detection of Helicobacter pylori infection. Xin Xiaohuabingxue Zazhi. 1997;5:301-302. |
| 9. | Ji XH, Xu GM, Li ZS, Man XH, Zhang HF, Xu AF. Clinical diag-nostic value of PCR in gastric H . pylori infection. Xin Xiaohuabingxue Zazhi. 1997;5:364-366. |
| 10. | Tang W, Du SC, Tao QM, Zhu L. A study on anti contamination of RT-PCR in detection of HCV-RNA. Xin Xiaohuabingxue Zazhi. 1997;5:638-639. |
| 11. | Zheng N, Yu ZY, Zhu SN. Detection of hepatitis B virus DNA in liver cancer tissue by in situ polymerase chain reaction. Huaren Xiaohua Zazhi. 1998;6:371-373. |
| 12. | Hua JS, Zheng PY, Bow H, Megraud F. Detection of Helicobacter pylori in gastric biopsy by polymerase chain reaction. Huaren Xiaohua Zazhi. 1998;6:377-379. |
| 13. | Hua JS, Zheng PY, Bow H. Species differentiation and identification in the genus of Helicobacter. World J Gastroenterol. 1999;5:7-9. [PubMed] |
| 14. | Yamashita Y, Kohno S, Koga H, Tomono K, Kaku M. Detection of Bacteroides fragilis in clinical specimens by PCR. J Clin Microbiol. 1994;32:679-683. [PubMed] |
| 15. | Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335-351. [PubMed] |
| 16. | Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Dymock D, Wade WG. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795-799. [PubMed] |
| 17. | Walsh TJ, Francesconi A, Kasai M, Chanock SJ. PCR and single-strand conformational polymorphism for recognition of medically important opportunistic fungi. J Clin Microbiol. 1995;33:3216-3220. [PubMed] |
| 18. | Mannarelli BM, Kurtzman CP. Rapid identification of Candida albicans and other human pathogenic yeasts by using short oligonucleotides in a PCR. J Clin Microbiol. 1998;36:1634-1641. [PubMed] |
| 19. | Zhang WZ, Han TQ, Tang YQ, Wan ZM, Zhang SD. Study on rapid diagnosis of fungal infection in the patients with acute ne-crotizing pancreatitis by polymerase chain reaction. Zhongguo Shiyan Zhenduanxue. 2000;4:109-111. |
| 20. | Laforgia N, Coppola B, Carbone R, Grassi A, Mautone A, Iolascon A. Rapid detection of neonatal sepsis using polymerase chain reaction. Acta Paediatr. 1997;86:1097-1099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 56] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Kane TD, Alexander JW, Johannigman JA. The detection of microbial DNA in the blood: a sensitive method for diagnosing bacteremia and/or bacterial translocation in surgical patients. Ann Surg. 1998;227:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 84] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Kane TD, Johnson SR, Alexander JW, Babcock GF, Ogle CK. Detection of intestinal bacterial translocation using PCR. J Surg Res. 1996;63:59-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6211] [Cited by in RCA: 6522] [Article Influence: 197.6] [Reference Citation Analysis (0)] |
| 24. | Bone RC, Sibbald WJ, Sprung CL. The ACCP-SCCM consensus conference on sepsis and organ failure. Chest. 1992;101:1481-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 498] [Cited by in RCA: 509] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 25. | Salvo I, de Cian W, Musicco M, Langer M, Piadena R, Wolfler A, Montani C, Magni E. The Italian SEPSIS study: preliminary results on the incidence and evolution of SIRS, sepsis, severe sepsis and septic shock. Intensive Care Med. 1995;21 Suppl 2:S244-S249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 139] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Rangel-Frausto MS, Pittet D, Costigan M, Hwang T, Davis CS, Wenzel RP. The natural history of the systemic inflammatory response syndrome (SIRS). A prospective study. JAMA. 1995;273:117-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1196] [Cited by in RCA: 1047] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 27. | Murray PR, Traynor P, Hopson D. Critical assessment of blood culture techniques: analysis of recovery of obligate and facultative anaerobes, strict aerobic bacteria, and fungi in aerobic and anaerobic blood culture bottles. J Clin Microbiol. 1992;30:1462-1468. [PubMed] |
| 28. | Weinstein MP, Mirrett S, Wilson ML, Reimer LG, Reller LB. Controlled evaluation of 5 versus 10 milliliters of blood cultured in aerobic BacT/Alert blood culture bottles. J Clin Microbiol. 1994;32:2103-2106. [PubMed] |
| 29. | Wilson ML, Weinstein MP, Mirrett S, Reimer LG, Feldman RJ, Chuard CR, Reller LB. Controlled evaluation of BacT/alert standard anaerobic and FAN anaerobic blood culture bottles for the detection of bacteremia and fungemia. J Clin Microbiol. 1995;33:2265-2270. [PubMed] |
| 30. | McDonald LC, Fune J, Gaido LB, Weinstein MP, Reimer LG, Flynn TM, Wilson ML, Mirrett S, Reller LB. Clinical importance of increased sensitivity of BacT/Alert FAN aerobic and anaerobic blood culture bottles. J Clin Microbiol. 1996;34:2180-2184. [PubMed] |
| 31. | Widjojoatmodjo MN, Fluit AC, Verhoef J. Rapid identification of bacteria by PCR-single-strand conformation polymorphism. J Clin Microbiol. 1994;32:3002-3007. [PubMed] |
| 32. | Weiss J, Mecca J, da Silva E, Gassner D. Comparison of PCR and other diagnostic techniques for detection of Helicobacter pylori infection in dyspeptic patients. J Clin Microbiol. 1994;32:1663-1668. [PubMed] |
| 33. | Rådström P, Bäckman A, Qian N, Kragsbjerg P, Påhlson C, Olcén P. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and streptococci using a seminested PCR strategy. J Clin Microbiol. 1994;32:2738-2744. [PubMed] |
| 34. | West B, Wilson SM, Changalucha J, Patel S, Mayaud P, Ballard RC, Mabey D. Simplified PCR for detection of Haemophilus ducreyi and diagnosis of chancroid. J Clin Microbiol. 1995;33:787-790. [PubMed] |
| 35. | Battles JK, Williamson JC, Pike KM, Gorelick PL, Ward JM, Gonda MA. Diagnostic assay for Helicobacter hepaticus based on nucleotide sequence of its 16S rRNA gene. J Clin Microbiol. 1995;33:1344-1347. [PubMed] |
| 36. | Wang RF, Cao WW, Cerniglia CE. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242-1247. [PubMed] |
| 37. | Karttunen TJ, Genta RM, Yoffe B, Hachem CY, Graham DY, el-Zaatari FA. Detection of Helicobacter pylori in paraffin-embedded gastric biopsy specimens by in situ hybridization. Am J Clin Pathol. 1996;106:305-311. [PubMed] |
| 38. | Goldenberger D, Künzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35:2733-2739. [PubMed] |
| 39. | Ley BE, Linton CJ, Longhurst S, Jalal H, Millar MR. Eubacterial approach to the diagnosis of bacterial infection. Arch Dis Child. 1997;77:148-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 40. | Messick JB, Berent LM, Cooper SK. Development and evaluation of a PCR-based assay for detection of Haemobartonella felis in cats and differentiation of H. felis from related bacteria by restriction fragment length polymorphism analysis. J Clin Microbiol. 1998;36:462-466. [PubMed] |
| 41. | Matar GM, Sidani N, Fayad M, Hadi U. Two-step PCR-based assay for identification of bacterial etiology of otitis media with effusion in infected Lebanese children. J Clin Microbiol. 1998;36:1185-1188. [PubMed] |