Published online Apr 15, 2000. doi: 10.3748/wjg.v6.i2.228
Revised: August 26, 1999
Accepted: September 5, 1999
Published online: April 15, 2000
AIM: In order to study the association between the null genotypes of GSTM1 and GSTT1 and the genetic susceptibility to hepatocellular carcinoma (HCC).
METHODS: The genotypes of GSTM1 and GSTT1 of 63 cases of HCC and 88 controls were detected with the multiple PCR technique.
RESULTS: The frequency of GSTM1 null genotype was 57.1% among the cases, and 42.0% among the controls, the difference being statistically significant (χ² = 3.35, P = 0.067), but χ² value approaching the significance level. The odds ratio was 1.84 (95%CI = 0.91-3.37). The frequency of GSTT1 non-null genotype was 87.3% among the cases and 62.5% among the controls, the difference being statistically significant (χ² = 11.42, P = 0.0007274). The oddsn ratio was 4.13 (95%CI = 1.64-10.70). According to the cross analysis, the GSTT1 non-null genotype was more closely associated with HCC than GSTM1 null genotype, and these two factors play an approximate additive interaction in the occurrence of HCC.
CONCLUSION: The persons with GSTM1 null genotype and GSTT1 non-null genotype have the increased risk to HCC.
- Citation: Bian JC, Shen FM, Shen L, Wang TR, Wang XH, Chen GC, Wang JB. Susceptibility to hepatocellular carcinoma associated with null genotypes of GSTM1 and GSTT1. World J Gastroenterol 2000; 6(2): 228-230
- URL: https://www.wjgnet.com/1007-9327/full/v6/i2/228.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i2.228
Some previous studies have shown that glutathione s-transferase M1 (GSTM1) null genotype is a susceptible genotype to hepatocellular carcinoma (HCC)[1]. A recent research has found that glutathiones transferase T1 (GSTT1) also has the null genotype[2], which is similar to GSTM1. GSTM1 and GSTT1 are the members of GST family, which can detoxify some extraneous chemicals[3]. Persons with two null genotypes have no ability to do so[2-4]. Few reports have been found on the association between GSTT1 null genotype and HCC as well as the interaction between GSTT1 and GSTM1 null genotype to HCC. We used the multiple PCR technique to detect GSTM1 and GSTT1 genotypes of 63 HCC cases and 88 healthy controls in an attempt to provide scientific ground on which the screening among high risk population is based.
The specimens of non-cancerous liver tissue and peripheral blood of 63 HCC cases were provided by Qidong Institute for Liver Cancer, Zhongshan Hospital attached to Shanghai Medical University and the teaching hospital Nantong Medical College. The peripheral blood specimens of 88 healthy controls were provided by Qidong Institute for Liver Cancer, Haimen Municipal Anti-Epidemic and Health Station and Clinic of Shanghai Medical University. The cases were pathologically diagnosed as HCC. No consanguineous relationship existed among controls. The subjects mainly came from east China. There were 47 males and 16 females among the 63 cases, and 67 males and 21 females among the 88 controls.
The extraction of genomic DNA from liver tissues and peripheral blood was carried out according to the methods by Hoelzel[5] and Tas[6].
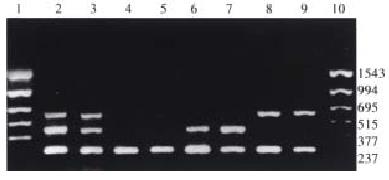
The corresponding fragments of GSTM1, GSTT1 and β-globulin genes were amplified with 3 pairs of primers synthesized according to the method recommended in the literature (Table 1)[7-9]. The amplificate of β-globulin gene was designed as the internal control with the purpose to remove false negative outcome. Whether GSTM1 null genotype existed or not was judged by the fragment of 650 bp, while GSTT1 null genotype by the fragment of 480 bp. The total reaction volume was 50 μL containing 100 ng-600 ng templets, 0.5 μmol/L primers (3 pairs), 200 μmol/L dNTP, 2.0 mmol/L MgCl2, 2.5 U Taq DNA polymerase (Promega) and corresponding buffers. The reaction condition was: predenaturation at 95 °C for 5 min; denaturation at 95 °C for 1 min; annealing at 58 °C for 1 min; extention at 72 °C for 1.5 min; and extention at 72 °C for 5 min after 30 cycles. The amplificates were separated with agarose gel electrophoresis for 50 min at 120 V, and inspected by ultraviolet reflector and pictured by ImageMaster VDS (Pharmacia) (Figure 1).
| Gene | Location | Sequence | Length of amplificates (bp) |
| GSTM1 | 5′ | 5′-CTCCTGATTATGACAGAAGCC-3′ | 650 |
| 3′ | 5′-CTGGATTGTAGCAGATCATGC-3′ | ||
| GSTT1 | 5′ | 5′-TTCCTTACTGGTCCTCACATCTC-3′ | 480 |
| 3′ | 5′-TCACCGGATCATGGCCAGCA-3′ | ||
| β-globulin | 5′ | 5′-CAACTTCATCCACGTTCACC-3′ | 268 |
| 3′ | 5′-GAAGAGCCAAGGACAGGTAC-3′ |
The DNA templates used by the multiple PCR was extracted from the noncancerous liver tissues and the peripheral blood of HCC cases and the controls. The frequency of GSTM1 null genotype was 57.1% among cases and 42.0% among controls. The difference was not statistically significant, but χ² value approached the significance level (χ² = 3.35, P = 0.067). When GSTM1 null genotype was regarded as the exposure factor, odds ratio (OR) was 1.84 (95%CI = 0.91-3.73). It was suggested that persons with GSTM1 null genotype had a 84% of increased risk to HCC as compared with persons with GSTM1 non-null genotype (Table 2).
| Genotypes | Cases | Controls | Total | ||
| n | % | n | % | ||
| Null | 36 | 57.1 | 37 | 42.0 | 73 |
| Non-null | 27 | 42.9 | 51 | 58.0 | 78 |
| Total | 63 | 100.0 | 88 | 100.0 | 151 |
Table 3 shows that the frequency of GSTT1 null genotype was 12.7% among cases and 37.5% among controls. The difference was statistically significant (χ² = 11.42, P = 0.0007274). The odds ratio was 4.13 (95%CI = 1.64-10.70) when GSTT1 non-null genotype was regarded as the exposure factor, suggesting that in persons with GSTT1 non-null genotype the risk to HCC increased by 3.13 times as compared with persons with GSTT1 null genotype.
| Genotype | Cases | Controls | Total | ||
| n | % | n | % | ||
| Null genotype | 55 | 87.3 | 55 | 62.5 | 110 |
| Non-null genotype | 8 | 12.7 | 33 | 37.5 | 41 |
| Total | 63 | 100.0 | 88 | 100.0 | 151 |
To study the interaction of GSTM1 and GSTT1 in the occurrence of HCC, the cross analysis was carried out. The results indicated that when GSTM1 null genotype and GSTT1 non-null genotype were regarded as exposure factors, and that GSTM1 non-null genotype and GSTT1 null genotype as non-exposure factors, for those only exposed to GSTM1 null genotype, only exposed to GSTT1 non-null genotype and exposed to both, their OR was 6.95, 11.8 and 23.00, respectively. It was suggested that GSTT1 non-null genotype was more closely associated with HCC than GSTM 1 null genotype and these two factors exerted an additive interaction in the occurrence of HCC. According to trend χ² test, the rank association existed among these exposure factors (Table 4).
| Exposure level | Cases | Control | OR | |||
| GSTM1 | GSTT1 | n | % | n | % | |
| - | - | 1 | 1.6 | 16 | 18.2 | 1.00 |
| + | - | 7 | 11.1 | 17 | 19.3 | 6.59 |
| - | + | 26 | 41.3 | 35 | 39.8 | 11.89 |
| + | + | 29 | 46.0 | 20 | 22.7 | 23.20 |
| Total | 63 | 100.0 | 88 | 100.0 | ||
Numerous epidemiological studies have shown that HBV, aflatoxin and pollutants in drinking water are the main environmental hazard factors to HCC in China. The association between HBV and HCC has been universally accepted. But only 20%-25% of chronic HBV infected persons developed HCC. Therefore attention should be paid to the effect of chemical carcinogen in the occurrence of hepatocarcinoma.
GST is a supergene family composed of 4 kinds of isoenzymes (α, μ, π, theta;), which plays an important role in the second stage of biotransformation by conjugating extraneous chemicals with glutathione. Moreover, GST can com bine itself with chemicals directly. After being absorbed, aflatoxin B1 is transfered to the ultimate carcinogen, i.e. aflatoxin B1-8, 9-epoxide, by the catalyzation of cytochrome P450. GSTM1 can transfer aflatoxin B1-8, 9-epoxide to be untoxic metabolite which is highly-soluble and can be excreted out of body[10]. Three alleles exist on GSTM1 gene locus, including a, band null. In persons with null genotype, GSTM1 can not be expressed in the liver and therefore has no ability to detoxify AFB1 and other chemicals[10].
With regard to the research into the association of GSTM1 null genotype and HCC, 4 of the 5 case-control studies showed significant difference[1,11]. The result from one study is almost identical to ours. It is likely that the sample size plays a role in it. Summarizing these results, we believe that GSTM1 null genotype is a susceptible genotype to HCC.
GSTT1 is also a member of GST family. GSTT1 and GSTM1 are mutual isoenzymes and both have the null genotypes. There has been no report on whether GSTT1 participates in the metabolism of aflatoxin or not. Our research shows that GSTT1 null genotype frequency is 12.7% among cases, and 37.5% among controls, the difference being significant (P = 0.0007274). The association between GSTT1 null genotype and HCC was reverse to that of GSTM1 null genotype and HCC. GSTT1 null genotype is a protective factor to HCC, thus lowering the risk to HCC. In other words, GSTT1 non-null genotype is a hazard factor to HCC. Its OR is 4.13 indicating that persons with GSTT1 non-null genotype increased the risk to HCC by 3.13 times as against persons with GSTT1 null genotype. The results imply that certain procarcinogens such as AFB1 or other chemicals can be activated by GSTT1 and the metabolites are carcinogeous. It was reported that GSTT1 could activate dicholoromethane into mutagen, inducing lung and liver cancer in mice[2]. This report has provided evidence for our study.
We used cross analysis to study the interaction of GSTM1 and GSTT1 in the occurrence of HCC. When GSTM1 non-null genotype and GSTT1 null genotype were regarded as the non-exposure factors, for those only exposed to GSTM1 null genotype, only exposed to GSTT1 non-null genotype and exposed to both, their OR are 6.59, 11.89 and 23.20, respectively. GSTT1 non-null genotype is more closely associated with HCC than GSTM1 null genotype and those two factors exert an additive interaction in the occurrence of HCC. According to the trend χ² test, the rank association existed among the different exposures.
In summary, we think that both GSTM1 and GSTT1 are the susceptible loci to HCC. The degree of accordance between their genotype and phenotype was as high as 98.5%-100%[2,7]. Persons with GSTM1 null genotype have an increased susceptibility to HCC on account of lacking for GSTM1 activity in liver. Likewise, persons with GSTT1 non-null genotype, who possess GSTT1 activity in liver, have an increased susceptibility to HCC in which procarcinogen is activated by GSTT1.
We sincerely thank JIANG Feng, LU Meng and LIU Cha-Zhen for their help in this work.
Edited by Ma JY
| 1. | Bian JC, Wang JB, Wu Y, Zhang BC, Shen FM. Relationship between GSTM1 null genotype and the genetic susceptibility to primary hepatocellular carcinoma. Zhonghua Yixue Yichuanxue Zazhi. 1996;13:353-356. |
| 2. | Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300:271-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 888] [Cited by in RCA: 885] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 3. | Hollstein MC, Wild CP, Bleicher F, Chutimataewin S, Harris CC, Srivatanakul P, Montesano R. p53 mutations and aflatoxin B1 exposure in hepatocellular carcinoma patients from Thailand. Int J Cancer. 1993;53:51-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Seidegård J, Vorachek WR, Pero RW, Pearson WR. Hereditary differences in the expression of the human glutathione transferase active on trans-stilbene oxide are due to a gene deletion. Proc Natl Acad Sci USA. 1988;85:7293-7297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 463] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 5. | Hoelzel AR (editors). Molecular genetic analysis of population: a practical approach. 1st editor. New York: Oxford University Press 1992; 166-167. |
| 7. | Brockmöller J, Kerb R, Drakoulis N, Nitz M, Roots I. Genotype and phenotype of glutathione S-transferase class mu isoenzymes mu and psi in lung cancer patients and controls. Cancer Res. 1993;53:1004-1011. [PubMed] |
| 8. | Chen H, Sandler DP, Taylor JA, Shore DL, Liu E, Bloomfield CD, Bell DA. Increased risk for myelodysplastic syndromes in individuals with glutathione transferase theta 1 (GSTT1) gene defect. Lancet. 1996;347:295-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 187] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 9. | Bell DA, Taylor JA, Paulson DF, Robertson CN, Mohler JL, Lucier GW. Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J Natl Cancer Inst. 1993;85:1159-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 423] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 10. | Pearson WR, Vorachek WR, Xu SJ, Berger R, Hart I, Vannais D, Patterson D. Identification of class-mu glutathione transferase genes GSTM1-GSTM5 on human chromosome 1p13. Am J Hum Genet. 1993;53:220-233. [PubMed] |
| 11. | McGlynn KA, Rosvold EA, Lustbader ED, Hu Y, Clapper ML, Zhou T, Wild CP, Xia XL, Baffoe-Bonnie A, Ofori-Adjei D. Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin B1. Proc Natl Acad Sci USA. 1995;92:2384-2387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 155] [Article Influence: 5.2] [Reference Citation Analysis (0)] |