Published online Jun 7, 2025. doi: 10.3748/wjg.v31.i21.106808
Revised: April 15, 2025
Accepted: May 12, 2025
Published online: June 7, 2025
Processing time: 91 Days and 2.1 Hours
We read with great interest the study by Huang et al. Cholangiocarcinoma (CC) is the second most common type of primary liver tumor worldwide. Although surgical resection remains the primary treatment for this disease, almost 50% of patients experience relapse within 2 years after surgery, which negatively affects their prognosis. Key predictors can be used to identify several factors (e.g., tumor size, tumor location, tumor stage, nerve invasion, the presence of intravascular emboli) and their correlations with long-term survival and the risk of posto
Core Tip: Machine learning-driven preoperative risk stratification enhances surgical planning in intrahepatic cholangiocarcinoma. Huang et al demonstrated that the concept of the textbook outcome can be predicted preoperatively using artificial intelligence models, which outperform traditional prognostic methods. Their study underscored the importance of dynamic, data-driven approaches for improving disease-free survival and optimizing patient selection for curative resection.
- Citation: Morales-Galicia AE, Rincón-Sánchez MN, Ramírez-Mejía MM, Méndez-Sánchez N. Outcome prediction for cholangiocarcinoma prognosis: Embracing the machine learning era. World J Gastroenterol 2025; 31(21): 106808
- URL: https://www.wjgnet.com/1007-9327/full/v31/i21/106808.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i21.106808
Cholangiocarcinoma is a highly malignant and heterogeneous biliary tract cancer classified into intrahepatic, perihilar, and extrahepatic subtypes[1]. Among these, intrahepatic cholangiocarcinoma (ICC) is the second most common primary liver malignancy, accounting for 10%-15% of all primary liver cancers. Its incidence has been rising globally, particularly in regions with high HBV and hepatitis C virus prevalence, reflecting an increasing public health burden[2].
Despite advances in surgical and oncological care, ICC remains a challenging disease due to late-stage diagnosis and high recurrence rates. Surgical resection is the only curative option, yet long-term survival outcomes remain poor, with 5-year survival rates ranging between 20% and 40%. Even after curative-intent surgery, recurrence rates exceed 50% within 2 years, underscoring the need for improved preoperative risk stratification and outcome prediction[3]. Risk factors, which are conditions or exposures that increase the likelihood of developing ICC, include chronic liver disease, cirrhosis, primary sclerosing cholangitis, and liver fluke infections. These factors contribute to chronic inflammation, fibrosis, and malignant transformation of biliary epithelial cells and further complicate disease management[4,5].
One of the most critical challenges in ICC is the unpredictability of surgical outcomes, even when patients meet the criteria for resection[6]. Surgery remains the only curative therapeutic option for ICC, offering the best opportunity of prolonged survival by removing the main tumor and reducing the overall tumor burden[7]. Nevertheless, only a minority of patients are eligible for surgery because of the high frequency of advanced-stage disease at diagnosis and tumor unresectability at presentation. Achieving complete resection (R0) is crucial as it significantly improves disease-free survival (DFS) and overall survival, and thus surgical intervention is a cornerstone of ICC management[6].
Traditionally, oncological success has been evaluated according to metrics such as margin status and lymph node involvement[8]. Nonetheless, the concept of textbook outcome (TO), which incorporates multiple perioperative success indicators, has gained traction as a more holistic measure of surgical quality[9]. In their study, Huang et al[10] proposed that TO can be predicted preoperatively using machine learning (ML). This represents a paradigm shift in surgical planning.
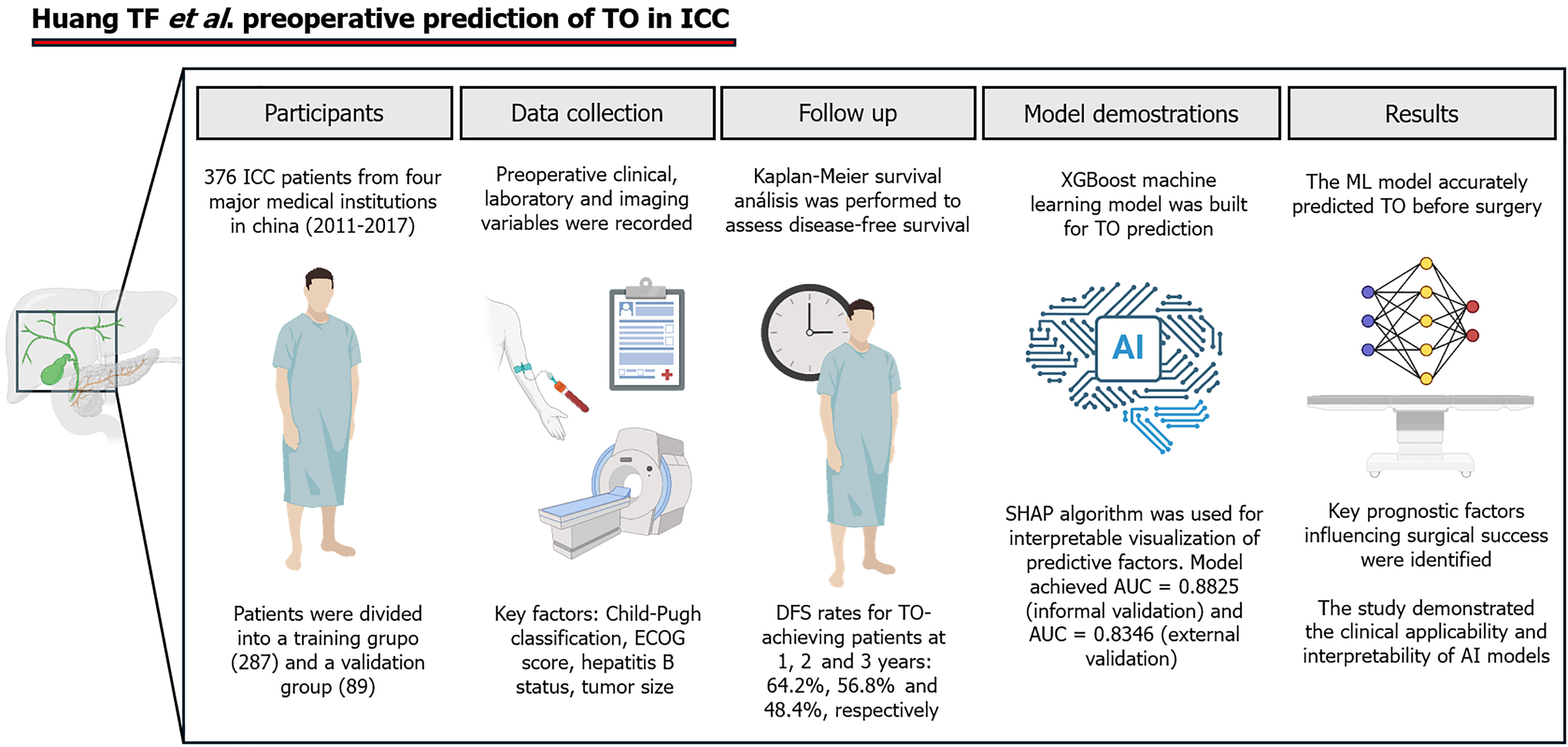
To develop their model, they retrospectively analyzed 376 patients with ICC from four major medical institutions in China (2011–2017). Logistic regression identified key preoperative variables associated with TO, which were then used to construct an Extreme Gradient Boosting (XGBoost) model. To enhance interpretability, they applied the SHapley Additive exPlanations (SHAP) algorithm to visualize the contribution of each variable to the predictions of the model. The XGBoost package in R was used to build the prediction model, with 70% of the data randomly assigned to the training set and 30% to the validation set. The model was constructed using variables identified through logistic regression, such as Child-Pugh grade, Eastern Cooperative Oncology Group (ECOG) score, HBV status, and tumor size. The XGBoost model demonstrated strong predictive performance, yielding an area under the curve of 0.882 in the training group and 0.834 in the validation group, indicating high accuracy and robustness. Additionally, a Kaplan-Meier survival analysis was performed to compare the DFS between the TO-achieving and non-TO-achieving groups, highlighting the clinical relevance of the model in stratifying long-term outcomes (Figure 1).
The integration of artificial intelligence (AI) and ML in predictive oncology has transformed the landscape of clinical decision-making, surpassing traditional statistical methods by leveraging multidimensional datasets (Table 1). Unlike conventional prognostic models that rely on fixed clinical and pathological variables, ML algorithms process vast amounts of data, recognize intricate patterns, and dynamically adapt to patient-specific factors. These capabilities enhance risk stratification, surgical planning, and treatment optimization, making AI-driven models particularly valuable in refining preoperative assessments for ICC.
| ML-driven approach | Description |
| Radiomics-based ML models | Extract imaging features from magnetic resonance or CT scans to predict tumor aggressiveness and microvascular invasion |
| Multiparametric clinical models | Integrate laboratory values, liver function scores, and tumor markers to assess perioperative risk |
| Hybrid AI models | Combine genomic, histopathological, and radiomic data to refine survival predictions and guide personalized treatment strategies |
Several ML-driven approaches utilize diverse data sources, such as imaging, histopathological features, molecular markers, and electronic health records, to improve prognostic accuracy[9]. In this context, the model created by Huang et al[10] represents a significant advancement in AI-driven risk stratification for patients with ICC, demonstrating a high degree of predictive accuracy and clinical utility. Incorporating AI-driven decision support systems into surgical planning frameworks could further enhance predictive accuracy and clinical decision-making[11].
As AI and ML continue to evolve, their role in oncology will likely expand, shaping the future of precision medicine and personalized patient care. ML-driven approaches have already demonstrated potential in various domains, including early cancer detection, recurrence prediction, and treatment response monitoring[9]. The study by Huang et al[10] exemplified how ML can improve preoperative risk stratification for patients with ICC, setting the stage for further exploration and refinement of AI-assisted surgical planning. Future research should prioritize prospective validation, integration with clinical decision-support systems, and continuous learning through real-time data incorporation to maximize the potential of AI in ICC management. This letter to the editor aimed to critically analyze the implications of AI-driven prognostic models in ICC, highlighting both their potential and the challenges that must be overcome for successful clinical integration.
One of the greatest challenges in the management of ICC is its high recurrence rate, which significantly affects long-term survival. Although surgical resection is the sole curative approach, more than 50% of patients develop recurrence within 2 years, even after R0 resection[2]. Factors that contribute to recurrence include tumor aggressiveness, microscopic residual disease, and the lack of effective adjuvant therapies[12]. Huang et al[10] emphasized that achieving a TO is strongly linked to better DFS as their study revealed that patients who met the TO criteria had significantly greater DFS rates at 1 year, 2 years, and 3 years. These findings underscore the need for early identification of high-risk patients and personalized perioperative strategies to reduce the risk of recurrence. Nevertheless, traditional preoperative risk stratification models remain limited as they rely on tumor size, nodal involvement, and margin status and fail to capture the complex biological behavior of ICC[9]. In response to these challenges Huang et al[10] proposed ML-based models as a more accurate, data-driven alternative as they integrate multiple dynamic factors to improve perioperative predictions.
Tumor characteristics, histopathological features, and surgical parameters are key prognostic factors in ICC. Larger tumors (> 5 cm) are associated with higher recurrence rates and worse survival as they are more likely to invade vascular structures and lymph nodes[9]. Additionally, centrally located tumors pose greater surgical challenges and increase the risk of positive margins and postoperative liver failure[9]. Huang et al[10] confirmed that tumor size is one of the strongest predictors of TO, and larger tumors are significantly less likely to meet TO criteria. This observation reinforces the importance of early detection and aggressive management, including neoadjuvant therapies and meticulous surgical planning.
Histopathological factors such as perineural invasion and intravascular emboli further contribute to early recurrence and poor survival, but traditional prognostic models fail to integrate these microscopic tumor characteristics preoperatively[12]. The ML model developed by Huang et al[10] addressed this limitation and incorporated preoperative variables that are indirectly correlated with tumor aggressiveness.
Still, concerns remain regarding the interpretability of ML predictions as complex models often function as "black boxes", making it difficult for clinicians to understand how specific features contribute to outcomes. Techniques such as SHAP have been implemented to enhance model transparency, fostering clinical trust and supporting decision-making. SHAP is based on game theory and explains model predictions by quantifying the contribution of each feature. Other interpretability methods, such as LIME, counterfactual explanations, and saliency maps, also help make complex models more understandable, which is essential in clinical contexts where transparency is key to adoption.
Surgical and perioperative factors also play crucial roles in ICC prognosis. Achieving R0 resection is associated with significant improvements in survival, yet approximately 20% of patients with ICC have positive surgical margins due to the proximity of the tumor to vascular structure[9,10]. Postoperative complications, such as bile leakage and liver failure, further impact overall survival and recurrence rates[12]. Huang et al[10] proposed that TO should be used as a benchmark for surgical quality as their study revealed that patients with prolonged hospitalization and perioperative complications were less likely to achieve a TO. This reinforced the need for optimized perioperative care and enhanced recovery protocols. Their ML model accurately predicted TO in high-risk patients and offered opportunities for preoperative optimization to improve surgical outcomes.
Traditional ICC prognostic models have several limitations. First, they lack real-time predictive power as most are retrospective and cannot dynamically adjust to new patient-specific variables[9]. Second, they fail to capture multidimensional interactions since traditional tools do not account for the relationships among clinical, radiological, and molecular factors and limits their predictive accuracy[9,10]. Third, their generalizability is restricted as many scoring systems are developed from single-institution datasets, which reduces their applicability to broader patient populations. Additionally, AI models must address ethical concerns and the possibility of algorithmic errors that could influence high-impact clinical decisions. Regulatory approvals and physician training also represent critical barriers to widespread AI adoption in oncology[11].
Huang et al[10] challenged the validity of these conventional models by demonstrating the superiority of ML-based predictions. Their study revealed that ML models that integrate multiple preoperative factors (Child-Pugh classification, ECOG score, HBV status, and tumor size) were significantly more accurate than traditional statistical models in predicting TO. These findings highlighted the need for a paradigm shift in ICC prognostication, with a transition toward AI-enhanced risk assessment frameworks that offer greater predictive accuracy and individualized patient care. Moreover, comparisons with other hepatobiliary malignancies, such as hepatocellular carcinoma and pancreatic cancer, suggest that AI-driven prognostic applications may have broader implications in oncological risk assessment.
ML has emerged as a transformative tool in predictive oncology, offering a data-driven approach to risk stratification and outcome prediction. Unlike traditional statistical models, ML algorithms can process vast amounts of multidimensional data, recognize complex patterns, and provide more precise risk estimations[9]. In ICC ML has the potential to revolutionize preoperative risk assessment by integrating clinical, radiological, and molecular parameters to predict surgical success, recurrence risk, and long-term survival[13]. Additionally, its ability to detect nonlinear relationships between prognostic factors allows for a more nuanced analysis compared with conventional regression models, uncovering subtle interactions that might otherwise be overlooked[14].
A comparable application of ML in oncology risk prediction was demonstrated in the study by Ke et al[15], who developed an ML-based model to predict early gastric cancer risk using a large-scale, population-based retrospective dataset. Their methodology integrated demographic, clinical, and biochemical variables to enhance early detection and stratification of high-risk individuals, which improved screening efficiency. Similar to the study by Huang et al[10] on ICC, Ke et al[15] emphasized the importance of ML-driven risk assessment, yet their focus was on early disease identification rather than surgical outcome prediction. Both studies highlighted the predictive power of ML algorithms. Huang et al[10] applied their model to perioperative decision-making, whereas Ke et al[15] used ML for preventive screening. These findings reinforce the versatility of AI in oncology and demonstrate its role not only in preoperative optimization but also in early cancer detection, which could be further refined with external validation and multicenter studies.
A similar approach has been explored for breast cancer prognosis and treatment outcome prediction as demonstrated by Zhang et al[16]. Their study applied ML models to multiomics data to enhance breast cancer survival predictions and integrated genomic, transcriptomic, and clinical variables for a more comprehensive prognostic assessment. While their findings highlight the potential of AI-driven models in oncology, challenges such as model generalizability, data bias, and the need for external validation remain comparable with those discussed in ICC prognosis. Unlike the study by Huang et al[10], which focused on preoperative risk stratification using clinical and radiological data, Zhang et al[16] incorporated multiomics analysis and underscored how AI can adapt to different cancer types and datasets. This comparison emphasizes the growing applicability of AI in personalized oncology but reinforces the need for continued validation and refinement to ensure clinical reliability across various malignancies.
The integration of AI in medicine presents several challenges that must be addressed to ensure its effective and ethical implementation. One major concern is the complexity of AI algorithms, particularly deep learning models, which can obscure the decision-making process, raising transparency and reliability issues.
Trusting and effectively using AI in patient care depends on two key elements: Knowledge and experience. The more data and clinical exposure available, the better equipped health care professionals are to make informed decisions. Data are derived from evidence-based sources such as textbooks and peer-reviewed studies, whereas experience is gained through real-world patient outcomes, including medical records, lab results, and imaging[17-19]. The potential for bias in training data further complicates equitable treatment outcomes. To mitigate these risks, legal frameworks must define liability when AI-driven recommendations result in adverse outcomes[20].
Despite its potential, AI should complement rather than replace clinical expertise. Clinicians must oversee and interpret AI-generated recommendations, ensuring that they align with individual patient needs. AI integration should support, rather than hinder, clinical workflows and should enhance decision-making rather than dictate it[21]. The “black box” nature of many AI algorithms has raised concerns about transparency and clinical trust, prompting the use of explainable AI techniques, such as SHAP, that enhances model transparency by quantifying the contribution of each input feature to a given prediction, facilitating clinician understanding and supporting informed decision-making. Other interpretability methods like LIME, counterfactual explanations, and saliency maps also contribute to making AI outputs more accessible and trustworthy in clinical contexts.
A major limitation of current ML models lies in their generalizability. While retrospective studies offer valuable insights, external validation in independent cohorts, especially those representing different geographic and ethnic backgrounds, is essential to confirm the robustness and real-world applicability of these models. Without rigorous validation AI models risk producing biased or inaccurate outcomes, particularly for underrepresented populations[22]. Moreover, biases introduced by region-specific datasets can compromise the performance and fairness of AI-driven predictions in broader clinical applications.
Ethical and legal considerations are central to the safe implementation of AI in oncology. Issues such as algorithmic fairness, accountability, and the potential for AI-induced diagnostic errors must be rigorously evaluated to ensure responsible use in high-stakes clinical decision-making. Addressing these concerns is essential to promote trust, minimize harm, and uphold patient rights within an evolving technological landscape[23].
AI technologies, particularly ML, are continuously advancing surgical practices by reducing errors, improving outcomes, and optimizing minimally invasive techniques that shorten hospital stays and lower complication rates[24]. Postoperative care also benefits from AI-driven predictive analytics, which anticipate potential complications or readmissions, enabling proactive interventions and personalized treatment adjustments[25].
A key advantage of AI is its role in risk stratification, leveraging large datasets from electronic health records, genetic information, and imaging to precisely identify high-risk patients. This allows multidisciplinary teams to tailor interventions and allocate resources efficiently, particularly in complex diseases such as cancer where personalized care is essential[26]. ML-based models have demonstrated their ability to predict lymph node metastasis in early-stage colorectal cancer, refining patient selection for more aggressive treatment strategies[27]. Similarly, ML algorithms using liver stiffness measurements show promise in predicting hepatocellular carcinoma risk, offering etiology-independent assessments that could be adapted for ICC surveillance[28]. AI also enhances disease progression prediction and treatment outcome forecasting, ensuring that high-risk patients receive prioritized attention[20].
To fully realize these benefits, ongoing efforts must focus on expanding and diversifying datasets to improve model generalizability and reduce bias. Large-scale, multi-institutional collaborations are necessary to develop more representative training datasets that capture variations in demographics, tumor biology, and treatment responses[18]. Additionally, real-world validation through prospective clinical trials is critical to confirm AI model performance outside of controlled settings. Establishing standardized evaluation frameworks will facilitate objective comparisons between AI tools, ensuring consistency across institutions[27]. Pilot testing and continuous monitoring must be integrated into clinical workflows, allowing AI systems to adapt to new data and evolving medical practices while maintaining safety and efficacy.
Huang et al[10] exemplified how ML-driven decision-making influences ICC management, particularly in preoperative TO prediction. Their model integrated tumor size, Child-Pugh classification, ECOG score, and HBV status to enhance risk assessment and surgical planning, demonstrating higher accuracy than conventional prognostic tools. By incorporating SHAP analysis, their AI-driven approach provided transparent and interpretable predictions, addressing a major barrier to AI adoption. This study underscored the potential of AI in refining surgical decision-making and optimizing patient selection for resection, highlighting its clinical relevance in hepatobiliary oncology.
Successful AI implementation also requires structured clinician involvement throughout model development and validation. Engaging medical professionals in AI training processes and decision support system design ensures that models align with real-world clinical workflows. Interdisciplinary collaboration between data scientists, bioinformaticians, and clinicians is essential to refine algorithms based on expert input. The integration of dual safety mechanisms where AI provides insights but final decisions remain physician-led can help bridge the trust gap and facilitate seamless AI adoption in daily medical practice. Finally, regulatory frameworks should be updated to establish guidelines for AI integration, ensuring both patient safety and ethical accountability in AI-driven clinical decision-making[29].
The integration of AI and ML in ICC prognosis and management represents a significant advancement in oncological decision-making. Huang et al[10] demonstrated that ML-based models outperformed traditional prognostic tools, offering more precise and individualized risk stratification. Their findings confirmed the feasibility and clinical relevance of preoperative TO prediction, with direct implications for optimizing surgical candidacy and improving long-term outcomes. While their model exhibited strong predictive performance, its generalizability remains a key concern. Future efforts should prioritize diverse, multi-institutional data and real-world validation to enhance AI model reliability and clinical relevance. Interdisciplinary collaboration and updated regulatory frameworks are essential to ensure ethical deployment and meaningful integration into ICC care.
| 1. | Nathan H, Pawlik TM, Wolfgang CL, Choti MA, Cameron JL, Schulick RD. Trends in survival after surgery for cholangiocarcinoma: a 30-year population-based SEER database analysis. J Gastrointest Surg. 2007;11:1488-96; discussion 1496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 191] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 2. | da Fonseca LG, Izquierdo-Sanchez L, Hashizume PH, Carlino Y, Baca EL, Zambrano C, Sepúlveda SA, Bolomo A, Rodrigues PM, Riaño I, Boonstra A, Debes JD, Bujanda L, Carrilho FJ, Arrese M, Roa JC, Carrera E, Ferrer JD, Balderramo D, Oliveira CP, Banales JM. Cholangiocarcinoma in Latin America: a multicentre observational study alerts on ethnic disparities in tumour presentation and outcomes. Lancet Reg Health Am. 2024;40:100952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 3. | Saeed A, Park R, Al-Jumayli M, Al-Rajabi R, Sun W. Biologics, Immunotherapy, and Future Directions in the Treatment of Advanced Cholangiocarcinoma. Clin Colorectal Cancer. 2019;18:81-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 4. | Kelley RK, Bridgewater J, Gores GJ, Zhu AX. Systemic therapies for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72:353-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 277] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 5. | Chinchilla-López P, Aguilar-Olivos NE, García-Gómez J, Hernández-Alejandro KK, Chablé-Montero F, Motola-Kuba D, Patel T, Méndez-Sánchez N. Prevalence, Risk Factors, and Survival of Patients with Intrahepatic Cholangiocarcinoma. Ann Hepatol. 2017;16:565-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 6. | Alaimo L, Endo Y, Pawlik TM. ASO Author Reflections: Benchmarks in Liver Resection for Intrahepatic Cholangiocarcinoma. Ann Surg Oncol. 2024;31:4464-4465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 7. | Ruzzenente A, Conci S, Valdegamberi A, Pedrazzani C, Guglielmi A. Role of surgery in the treatment of intrahepatic cholangiocarcinoma. Eur Rev Med Pharmacol Sci. 2015;19:2892-2900. [PubMed] |
| 8. | de Jong MC, Nathan H, Sotiropoulos GC, Paul A, Alexandrescu S, Marques H, Pulitano C, Barroso E, Clary BM, Aldrighetti L, Ferrone CR, Zhu AX, Bauer TW, Walters DM, Gamblin TC, Nguyen KT, Turley R, Popescu I, Hubert C, Meyer S, Schulick RD, Choti MA, Gigot JF, Mentha G, Pawlik TM. Intrahepatic cholangiocarcinoma: an international multi-institutional analysis of prognostic factors and lymph node assessment. J Clin Oncol. 2011;29:3140-3145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 557] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 9. | Miao G, Qian X, Zhang Y, Hou K, Wang F, Xuan H, Wu F, Zheng B, Yang C, Zeng M. An MRI-Based Radiomics Model for Preoperative Prediction of Microvascular Invasion and Outcome in Intrahepatic Cholangiocarcinoma. Eur J Radiol. 2025;183:111896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | Huang TF, Luo C, Guo LB, Liu HZ, Li JT, Lin QZ, Fan RL, Zhou WP, Li JD, Lin KC, Tang SC, Zeng YY. Preoperative prediction of textbook outcome in intrahepatic cholangiocarcinoma by interpretable machine learning: A multicenter cohort study. World J Gastroenterol. 2025;31:100911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 11. | Zhou SN, Jv DW, Meng XF, Zhang JJ, Liu C, Wu ZY, Hong N, Lu YY, Zhang N. Feasibility of machine learning-based modeling and prediction using multiple centers data to assess intrahepatic cholangiocarcinoma outcomes. Ann Med. 2023;55:215-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 12. | Xie H, Hong T, Liu W, Jia X, Wang L, Zhang H, Xu C, Zhang X, Li WL, Wang Q, Yin C, Lv X. Interpretable machine learning-based clinical prediction model for predicting lymph node metastasis in patients with intrahepatic cholangiocarcinoma. BMC Gastroenterol. 2024;24:137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 13. | Meng A, Zhuang Y, Huang Q, Tang L, Yang J, Gong P. Development and validation of a cross-modality tensor fusion model using multi-modality MRI radiomics features and clinical radiological characteristics for the prediction of microvascular invasion in hepatocellular carcinoma. Eur J Surg Oncol. 2025;51:109364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 14. | Mi S, Qiu G, Zhang Z, Jin Z, Xie Q, Hou Z, Ji J, Huang J. Development and validation of a machine-learning model to predict lymph node metastasis of intrahepatic cholangiocarcinoma: A retrospective cohort study. Biosci Trends. 2025;18:535-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Ke X, Cai X, Bian B, Shen Y, Zhou Y, Liu W, Wang X, Shen L, Yang J. Predicting early gastric cancer risk using machine learning: A population-based retrospective study. Digit Health. 2024;10:20552076241240905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 16. | Zhang ZY, Wang QL, Zhang JY, Duan YY, Liu JX, Liu ZS, Li CY. Machine learning applications in breast cancer survival and therapeutic outcome prediction based on multi-omic analysis. Yi Chuan. 2024;46:820-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 17. | Mintz Y, Brodie R. Introduction to artificial intelligence in medicine. Minim Invasive Ther Allied Technol. 2019;28:73-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 279] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 18. | Sinz FH, Pitkow X, Reimer J, Bethge M, Tolias AS. Engineering a Less Artificial Intelligence. Neuron. 2019;103:967-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 19. | Choi RY, Coyner AS, Kalpathy-Cramer J, Chiang MF, Campbell JP. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl Vis Sci Technol. 2020;9:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 252] [Reference Citation Analysis (2)] |
| 20. | Keskinbora KH. Medical ethics considerations on artificial intelligence. J Clin Neurosci. 2019;64:277-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 21. | Liyanage H, Liaw ST, Jonnagaddala J, Schreiber R, Kuziemsky C, Terry AL, de Lusignan S. Artificial Intelligence in Primary Health Care: Perceptions, Issues, and Challenges. Yearb Med Inform. 2019;28:41-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 22. | Howard J. Artificial intelligence: Implications for the future of work. Am J Ind Med. 2019;62:917-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 23. | Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. 2017;69S:S36-S40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 827] [Article Influence: 103.4] [Reference Citation Analysis (0)] |
| 24. | Chen M, Decary M. Artificial intelligence in healthcare: An essential guide for health leaders. Healthc Manage Forum. 2020;33:10-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 129] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 25. | Gumbs AA, Frigerio I, Spolverato G, Croner R, Illanes A, Chouillard E, Elyan E. Artificial Intelligence Surgery: How Do We Get to Autonomous Actions in Surgery? Sensors (Basel). 2021;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 26. | Zhou LQ, Wang JY, Yu SY, Wu GG, Wei Q, Deng YB, Wu XL, Cui XW, Dietrich CF. Artificial intelligence in medical imaging of the liver. World J Gastroenterol. 2019;25:672-682. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 192] [Cited by in RCA: 135] [Article Influence: 22.5] [Reference Citation Analysis (8)] |
| 27. | Cheong C, Kim NW, Lee HS, Kang J. Application of machine learning for predicting lymph node metastasis in T1 colorectal cancer: a systematic review and meta-analysis. Langenbecks Arch Surg. 2024;409:287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 28. | Lin H, Li G, Delamarre A, Ahn SH, Zhang X, Kim BK, Liang LY, Lee HW, Wong GL, Yuen PC, Chan HL, Chan SL, Wong VW, de Lédinghen V, Kim SU, Yip TC. A Liver Stiffness-Based Etiology-Independent Machine Learning Algorithm to Predict Hepatocellular Carcinoma. Clin Gastroenterol Hepatol. 2024;22:602-610.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Reference Citation Analysis (0)] |
| 29. | Whicher D, Rapp T. The Value of Artificial Intelligence for Healthcare Decision Making-Lessons Learned. Value Health. 2022;25:328-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |