Published online Feb 28, 2024. doi: 10.3748/wjg.v30.i8.919
Peer-review started: November 2, 2023
First decision: December 6, 2023
Revised: December 24, 2023
Accepted: January 27, 2024
Article in press: January 27, 2024
Published online: February 28, 2024
Processing time: 116 Days and 0.5 Hours
Hepatocellular carcinoma (HCC) is one of the most common types of tumors. The influence of lipid metabolism disruption on the development of HCC has been demonstrated in published studies.
To establish an HCC prognostic model for lipid metabolism-related long non-coding RNAs (LMR-lncRNAs) and conduct in-depth research on the specific role of novel LMR-lncRNAs in HCC.
Correlation and differential expression analyses of The Cancer Genome Atlas data were used to identify differentially expressed LMR-lncRNAs. Quantitative real-time polymerase chain reaction analysis was used to evaluate the expression of LMR-lncRNAs. Nile red staining was employed to observe intracellular lipid levels. The interaction between RP11-817I4.1, miR-3120-3p, and ATP citrate lyase (ACLY) was validated through the performance of dual-luciferase reporter gene and RIP assays.
Three LMR-lncRNAs (negative regulator of antiviral response, RNA transmembrane and coiled-coil domain family 1 antisense RNA 1, and RP11-817I4.1) were identified as predictive markers for HCC patients and were utilized in the construction of risk models. Additionally, proliferation, migration, and invasion were reduced by RP11-817I4.1 knockdown. An increase in lipid levels in HCC cells was significantly induced by RP11-817I4.1 through the miR-3120-3p/ACLY axis.
LMR-lncRNAs have the capacity to predict the clinical characteristics and prognoses of HCC patients, and the discovery of a novel LMR-lncRNAs, RP11-817I4.1, revealed its role in promoting lipid accumulation, thereby accelerating the onset and progression of HCC.
Core Tip: In the current study, we investigated the functions of lipid metabolism-related long noncoding RNAs (LMR-lncRNAs) in assessing and forecasting the prognosis of patients with hepatocellular carcinoma (HCC), and established an accurate and reliable lipid metabolism-related risk score model for prediction. In particular, we focused on a novel LMR-lncRNA, RP11-817I4.1, which proved its important regulatory role in HCC lipid metabolism and tumor progression and showed therapeutic potential. Our findings will be extremely beneficial in understanding the probable molecular biological processes of HCC and discovering novel prognostic indicators and molecular targets.
- Citation: Wang RY, Yang JL, Xu N, Xu J, Yang SH, Liang DM, Li JZ, Zhu H. Lipid metabolism-related long noncoding RNA RP11-817I4.1 promotes fatty acid synthesis and tumor progression in hepatocellular carcinoma. World J Gastroenterol 2024; 30(8): 919-942
- URL: https://www.wjgnet.com/1007-9327/full/v30/i8/919.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i8.919
Hepatocellular carcinoma (HCC) is among the most common malignant tumors and ranks as the fourth leading cause of cancer-related fatalities globally[1]. Although surgical resection is an effective therapy for HCC, even patients in the early stages experience higher recurrence rates[2]. HCC is characterized by its rarity, poor prognosis, and frequent recurrence. Despite significant advancements in improving the prognosis of HCC patients, the 5-year survival rate remains low[3]. Therefore, further exploration of the pathogenesis and development of HCC, along with the discovery of novel diagnostic and prognostic indicators, is essential to identify potential therapeutic targets for enhancing patient survival and achieving precision treatment.
Mounting evidence underscores the significance of metabolic reprogramming in both the initiation and progression of carcinomas[4,5]. Due to their rapid growth and adaptability to changes in the tumor microenvironment (TME), cancer cells necessitate metabolic reprogramming to meet their high-energy demands[6]. One of the most prominent metabolic dysregulations observed in tumor cells is lipid metabolic reprogramming, a phenomenon that has garnered increasing attention[7]. HCC serves as a significant site for lipid metabolism and is associated with various lipid metabolic ab-normalities[8]. Previous studies have established a connection between HCC and abnormal metabolic alterations, including increased de novo fatty acid synthesis, reduced oxidation, and modified phosphatidylcholine metabolic activity[9]. These metabolic pathways generate intermediate energy sources that facilitate HCC cell growth, proliferation, and metastasis[10]. An in-depth examination of the alterations in the TME is imperative for assessing the prognostic and therapeutic implications for HCC patients, as the proliferation of HCC cells in the body is intricately linked to the support provided by the tumor milieu[11]. Emerging evidence suggests that the TME of HCC may include numerous metabolic irregularities, with altered lipid metabolism being the most prominent among them, which has been gaining significant attention in recent years.
Noncoding RNAs longer than 200 nucleotides are long noncoding RNAs (lncRNAs)[12]. Recent studies suggest that lncRNAs may modulate fatty acid metabolism and affect tumor development[13]. Moreover, the sterol regulatory element binding protein (SREBP) transcription factor, apolipoprotein, triglyceride (TG) metabolism, and macrophage cholesterol absorption and efflux are other pathways through which lncRNAs impact lipid metabolism[14,15]. For example, it has been revealed that metastasis associated lung adenocarcinoma transcript 1 binds to SREBP-1c in hepatoma carcinoma and can maintain the stability of nuclear SREBP-1c protein through the ubiquitin-protease pathway, leading to lipid metabolism abnormalities in HepG2 hepatocytes[16]. In general, lncRNAs participate in the regulation of numerous genes associated with lipid metabolism in tumor cells[17,18].
The objective of this research was to assess the potential predictive value of lipid metabolism-related lncRNAs (LMR-lncRNAs) in HCC patients by integrating clinical data with the expression levels of relevant lncRNAs. Subsequently, three lncRNAs [negative regulator of antiviral response (NRAV), RNA transmembrane and coiled-coil domain family 1 antisense RNA 1 (TMCC1-AS1), and RP11-817I4.1] that were differentially expressed in the The Cancer Genome Atlas (TCGA) cohort and among lipid metabolism-related genes were used to establish the lipid metabolism-related risk score model (LMRRSM) for predicting the prognosis of HCC using univariate and multivariate Cox regression and Least absolute shrinkage and selection operator (LASSO) regression analyses. Receiver Operating Characteristic Curve (ROC) were generated to assess the specificity and sensitivity of the models. We developed a personalized prognostic characteristic model for patients with HCC by comprehensively analyzing the expression status and prognostic landscape of LMR lncRNAs. Quantitative real-time polymerase chain reaction (qRT–PCR) confirmed the differential expression of RP11-817I4.1 between HCC cells and adjacent normal tissues. Loss-of-function assays revealed the biological characteristics of the lncRNA RP11-817I4.1 in hepatoma cell lines. According to these results, a prognostic model and crucial LMR-lncRNAs might be developed as potential biomarkers and serve as indices for HCC prognosis.
Normalized RNA-seq data from 50 normal and 374 HCC samples were obtained from the TCGA[19]. Furthermore, the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Set Enrichment Analysis (GSEA) databases yielded a list of eight lipid metabolism-related pathways and 323 lipid metabolism-related genes. One study of clinical relevance omitted 135 patients from the TCGA cohort because of inadequate clinical information.
With the conditions of a |coefficient of correlation|> 0.6 and P < 0.001, Pearson’s correlation analyses were conducted to investigate the coexpression of lipid metabolism-related genes and lncRNAs. The “limma” package of R was used to search for lncRNAs that were differentially expressed between tumor and normal tissues. We defined the lncRNA difference analysis data as follows: “false discovery rate (FDR)< 0.05, log2| fold change (FC)|> 1, and P < 0.05.”
To identify LMR-lncRNAs associated with survival, we employed the R package “survival” to conduct univariate Cox regression analysis of the differentially expressed lncRNAs and overall survival (OS) in HCC patients. Subsequently, to mitigate overfitting and select the optimal LMR-lncRNAs, we applied a LASSO regression model. Following collinearity assessment, stepwise multivariate Cox regression analysis was employed to establish the LMR-lncRNA-derived risk signature in HCC patients. The following algorithm, which was based on the combination of Cox coefficients and lncRNA expression data, was used to calculate risk scores for the TCGA cohort. The risk score was calculated as follows: risk score = expression of lncRNA 1 α1 + expression of lncRNA 2 α2 + expression of the lncRNA n αn. The regression coefficient of the LMR-lncRNAs is represented by the symbol α in the risk signature. Using the median values of the risk scores, HCC patients in the TCGA cohort were categorized into either high-risk or low-risk groups. Survival analysis was subsequently performed to evaluate the survival rates of the high- and low-risk groups. Further investigation was conducted to analyze the differences in prognosis between the two risk groups. A reasonable cutoff threshold was defined as a P value < 0.05. The prediction accuracy and reliability were also assessed using ROC curves.
To validate the association between LMR-lncRNAs and clinical characteristics, we evaluated the associations between the risk scores and clinical features. In the univariate Cox regression analysis, factors, including age, sex, grade, stage, T stage, N stage, M stage, and the risk scores derived from the lipid metabolism-related signature were all taken into account to assess the prognostic relationship of LMRRSM within the TCGA cohort. Subsequently, through multivariate analysis, we assessed the independent prognostic potential of the risk scores. Next, we investigated the association between the risk score and clinicopathological features to determine the significance of the predictive models for the progression of HCC.
The GeneExpression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/) can be used for cancer or normal gene expression profiling and interaction analyses. We used the GEPIA database to evaluate LMR-lncRNA expression in HCC and normal tissue samples. Furthermore, GEPIA was used to evaluate the survival of patients with HCC stratified by the lipid metabolism-related. A P value less than 0.05 was used.
Functional enrichment analyses were performed using the clusterProfiler package. A P-value < 0.05 indicated significant enrichment. We employed the R packages “GOplot” and “ggplot2” to visualize the most prominently enriched GO terms and KEGG pathways, respectively. GSEA was used to investigate the connection between the expression of RP11-817I4.1 and signaling pathway activity.
To further illustrate the potential impact of the predictive risk score on the tumor immune microenvironment, immune-related scores for different risk groups were calculated using the R software tool ESTIMATE. Single sample gene set enrichment analysis (ssGSEA), which measures the enrichment levels of 29 immune characteristics for every HCC patient through the form of ssGSEA scores, was carried out using the R package “GSVA.” Previous studies have furnished evidence regarding immune cell signatures[20,21]. Subsequently, in the high- and low-risk groups, we explored the relationship between immune cell infiltration and immune-related pathways.
The TIMER online database (https://cistrome.shinyapps.io/timer/) was used to reanalyze gene expression data from the TCGA to assess the abundance of six subtypes of tumor-infiltrating immune cells[22]. As a result, we determined the extent of immune infiltration in HCC patients and examined the relationship between the risk score and immune cell infiltration.
Between June 2021 and July 2022, we obtained 80 pairs of HCC samples and their corresponding nontumor tissues with explicit consent from HCC patients at the Second Affiliated Hospital in Kunming, China. None of the participants in this study received preoperative chemotherapy or radiation. The diagnosis of HCC was independently confirmed by two pathologists. The ethical committee of the Second Affiliated Hospital of Kunming Medical University approved this study, which was conducted in accordance with the Helsinki Declaration.
HCC cell lines (Hep-G2, Hep-3B, Li-7, Huh-7, and HCC-LM3) and the normal human hepatic cell line L02 were sourced from the Chinese Academy of Sciences Cell Bank in Shanghai, China. Li-7, Huh-7, HCC-LM3, Hep-G2, Hep-3B, and L02 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). All cell culture media were supplemented with penicillin (100 U/mL) and streptomycin (100 U/mL) and maintained at 37 °C in a 5% CO2 incubator.
TRIzol reagent was used to collect and lyse the cells (Invitrogen, CA, United States). A PrimeScript RT Kit (TaKaRa, Osaka, Japan) and RNA (1 µg) were used to generate reverse-transcribed complementary DNA (cDNA). For real-time-quantitative polymerase chain reaction, Fast SYBR Green Master Mix was used (Bio-Rad, CA, United States). The reaction cycle conditions were as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. GAPDH served as the internal loading control, and the relative quantification 2-ΔΔCT technique was employed to calculate the relative levels. The primer sequences used in this study were as follows: RP11-817I4.1: Forward: 5'-GTAGTGGCTGCTGCTGTTAGG-3’ Reverse: 5'-TTCAACGGTGGCAAACTCAAAG-3’ Actin sequence: Forward: 5’-ATCATGTTTGAGACCTTCAACA-3’ Reverse: 5'-CATCTCTTGCTCGAAGTCCA-3'-Actin was utilized as the internal control.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis is commonly used to separate extracted proteins. The samples were loaded into the wells of the gel, and an electric current was applied to separate the proteins based on their size and charge, resulting in the formation of distinct bands. Subsequently, the separated proteins were transferred onto a PVDF membrane by applying an electric current to the membrane. The transferred membrane was treated with a protein-blocking agent (bovine serum albumin) to prevent nonspecific binding. ATP citrate lyase (ACLY) recombinant rabbit monoclonal antibody (702731; Thermo Scientific) was added to a mixture containing the blocking agent, after which the proteins were allowed to bind to the transferred membrane. The antibody specifically binds to the target protein, leading to the formation of an immune complex. To remove unbound antibodies and other non-specific binding substances, the transferred membrane underwent multiple TBST washes. Images were captured using a chemiluminescence imaging system, and quantitative analysis was conducted. The expression levels of the target proteins in the samples were quantified based on optical density or fluorescence intensity.
Nuclear and cytoplasmic RNA was separated from each fraction using a PARIS Kit (Life Technologies, United States). Next, the nuclear and cytoplasmic RNA molecules isolated from each sample were identified via qRT-PCR. The nuclear and cytoplasmic markers GAPDH and Neat1 were identified.
To downregulate RP11-817I4.1, three human small interfering RNAs (RP11-817I4.1-sh1, UAAAUCAUUACCACUUUGGUU; RP11-817I41-sh2, AUUAUCACUACCAAUUUCCUU; RP11-817I41-sh3, UUGUUUGCCACUUCUGCCCUU; and control shRNA (si-NC, TTCTCCGAACGTGTCACGT) were manufactured by Qingke (SL100568, SignaGen, United States). Lipofectamine® 3000 (Sigma, United States) was employed for all cell transfections, which were carried out for 48 h at 37 °C. The cells were then utilized for subsequent experiments 48 h after transfection.
After 24 h of transfection, 500 tumor cells/well were plated into 6-well plates and cultivated for colony formation at 37 °C with 5% CO2. The cells were fixed for 5 min with 10% formaldehyde and stained for 1 h with 0.1% crystal violet. The count of colonies was conducted after a 2-wk period. Each experiment was performed in triplicate, and the means of the results were computed.
Cell migration was assessed via wound healing assays. Transfected cells were placed in 6-well plates [5 × 105 cells/well containing 5% (v/v) FBS]. Subsequently, a 200 µL pipette tip was employed to gently scrape the cell monolayer that had reached 95%-100% confluence. Using a light inverted microscope, the wound was inspected at 0 and 24 h, and the results were recorded as W0 and 24, respectively (magnification, 40 ×). Cell migration (%) was estimated using the following formula: (W024)/W0 × 100.
The transfected cells were centrifuged and suspended before being introduced into the upper layer of the Transwell system for use in the Transwell migration test. Following that, 600 µL of complete medium containing 20% FBS was added to the lower compartment. After 24-48 h of incubation, the transplanted membranes were stained with 0.1% crystal violet and preserved in methanol. Subsequently, the stained cells were counted and photographed under a microscope, and the average value was determined. Similarly, in the Transwell invasion test, the Matrigel was removed, and the top chamber was precoated for the migration experiment. The migration assay was carried out in the following stages.
After the tissues were homogenized, the cells were placed in 6-well plates with 4% paraformaldehyde solution, washed with phosphate-buffered saline (PBS), and stained with Nile red solution (7385-67-3; Solarbio, Beijing, China) in the dark. Images were acquired using IF microscopy after the samples had been subjected to two PBS washes and stained with 4’,6-diamidino-2-phenylindole.
We designed and synthesized a dual-luciferase reporter vector containing these gene sequences. This vector typically includes genes for two luciferases (firefly luciferase and green fluorescent protein), along with the promoter and terminator sequences of RP11-817I4.1 and ACLY. The constructed dual-luciferase reporter vector was transfected into liver cancer cells. After the addition of miRNA mimics or miRNA inhibitors, luciferase activity was assessed using a luciferase assay kit (16186, Thermo Scientific) following the manufacturer’s instructions. This involved separately measuring the activities of firefly luciferase and green fluorescent protein. The competitive relationship between miRNAs and mRNAs was evaluated by comparing the differences in luciferase activity among the various groups.
Cell lysates were harvested and prepared from liver cancer cells. Then, the specific anti-AGO2 antibody (MA5–23515; Thermo Scientific) was added to the protein A/G agarose beads to form antigen-antibody complexes. The cell lysate was coprecipitated with antigen-antibody complexes to enrich the complex containing AGO2. The coprecipitated complexes were washed and dissolved, followed by colloidal sol/colloidal removal. The proteins were removed, and the remaining RNA was extracted by proteinase treatment. The isolated RNA was reverse-transcribed into cDNA using reverse transcriptase (RT), and quantitative polymerase chain reaction was employed to detect the presence and quantification of the target mRNA and miRNA within the AGO2 complex. Specific primers and probes were used to quantitatively measure the expression levels of the target mRNAs, lncRNAs, and miRNAs.
The samples (cells or tissues) were prepared, and RNA was extracted. The isolated RNA was transcribed to synthesize cDNA using RT and random primers. This conversion process allowed the mRNA to be transformed into cDNA. An RNA library suitable for sequencing was subsequently constructed by adding adapter sequences and performing PCR amplification. The prepared RNA library was subjected to high-throughput sequencing (via the Illumina HiSeq platform). Afterward, we carried out preprocessing of the sequencing data, which involved the elimination of low-quality reads and adapter sequences. Bioinformatic tools were employed to align and quantify the preprocessed sequencing data against a reference genome or transcriptome. Significantly differentially expressed genes were determined through differential expression analysis (log-fold change > 1, FDR value < 0.05).
We chose 6-wk-old male BALB/c mice from the Animal Experimental Center of Kunming Medical University, ensuring that the animals were purebred, healthy, and met the experimental criteria. Tumor cell lines were transfected with the RP11-817I4.1-sh1 plasmid for in vitro cultivation, ensuring the health and purity of the cells. The cultured tumor cells were harvested, and an appropriate cell suspension was obtained through washing and centrifugation. The sterile operating room was filled, and the operating area and tools were prepared in advance. Isoflurane was used to anesthetize the mice, ensuring that they were painless. The tumor cell suspension was subcutaneously injected into the left axilla of each animal. After a period of 3 wk, the mice were humanely euthanized, and specimens were collected. All procedures and steps related to animal experiments were subject to rigorous review and approval by the Ethics Review Committee for Animal Experiments at Kunming Medical University (Approval Number: kmmu20211188).
For all the statistical studies, GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, United States) and SPSS 24.0 (SPSS, Inc., Chicago, IL, United States) were used. At least three separate experiments were conducted for each group. Continuous data are reported as the mean ± SD. For statistical analysis, the unpaired Student’s t-test was employed to compare continuous variables. Variations across experimental groups were assessed using either Student’s t-test or one-way ANOVA. Survival times across groups were evaluated using Kaplan-Meier (K-M) survival analysis or univariate Cox regression analysis. P-values were calculated for both sides, and a confidence threshold of 0.05 was considered statistically significant. aP < 0.05, bP < 0.01, cP < 0.001.
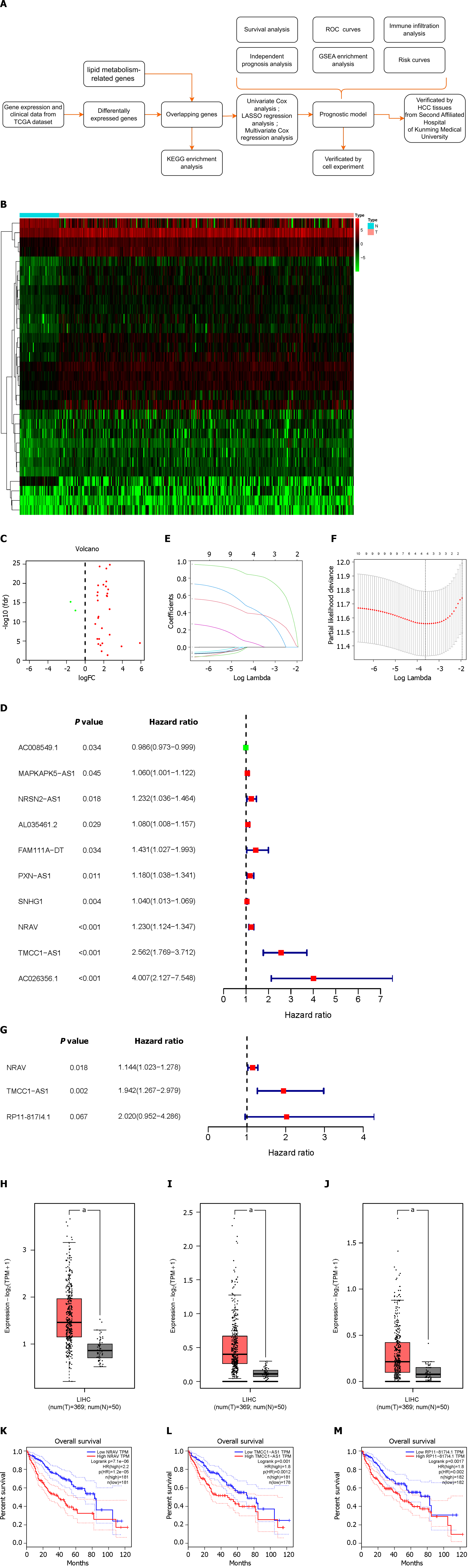
The study’s workflow is depicted in Figure 1A, with each step elaborated upon in the subsequent sections.
Using RNA sequencing data from HCC patients and 323 lipid metabolism-related genes, we identified 31 differentially expressed LMR-lncRNAs via the R “limma” package; 29 of these lncRNAs were upregulated, and 2 were downregulated. The results are shown as heatmaps and volcano plots (Figure 1B and C).
To further understand the connection between LMR-lncRNAs and patient prognosis, we constructed a three-lncRNA model based on LMR-lncRNAs to predict disease prognosis and survival in patients with HCC. The expression of ten LMR-lncRNAs was found to be strongly correlated with OS via univariate Cox regression analysis. A forest plot of the hazard ratio (HR) revealed that, whereas one lncRNA was protective, nine lncRNAs were risk factors (Figure 1D). Subsequently, LASSO regression analysis of the LMR lncRNAs associated with OS was performed to evaluate the predictive ability of the model (Figure 1E and F). Thus, after LASSO regression, only four lncRNAs were found among the 10 significant LMR-lncRNAs in the univariate Cox regression model. Ultimately, a predictive signature consisting of three LMR-lncRNAs, NRAV, TMCC1-AS1, and RP11-817I4.1 was chosen to construct a prognostic model via multivariate Cox regression analysis (Figure 1G). Similarly, the coefficients of the three lncRNAs were 0.134336124, 0.663916403, and 0.703331257. The three lncRNAs had HRs of 1.143777206, 1.942384618, and 2.020472221. The total risk score was calculated as follows: (0.134336124 × expression level of NRAV) + (0.663916403 × expression level of TMCC1-AS1) + (0.703331257 × expression level of RP11-817I4.1). Figure 1H-J shows that the expression of the three lncRNAs was upregulated in HCC tissues according to the GEPIA database. Additionally, GEPIA revealed that HCC patients with higher NRAV, TMCC1-AS1, or RP11-817I4.1 expression had worse prognoses (Figure 1K-M).
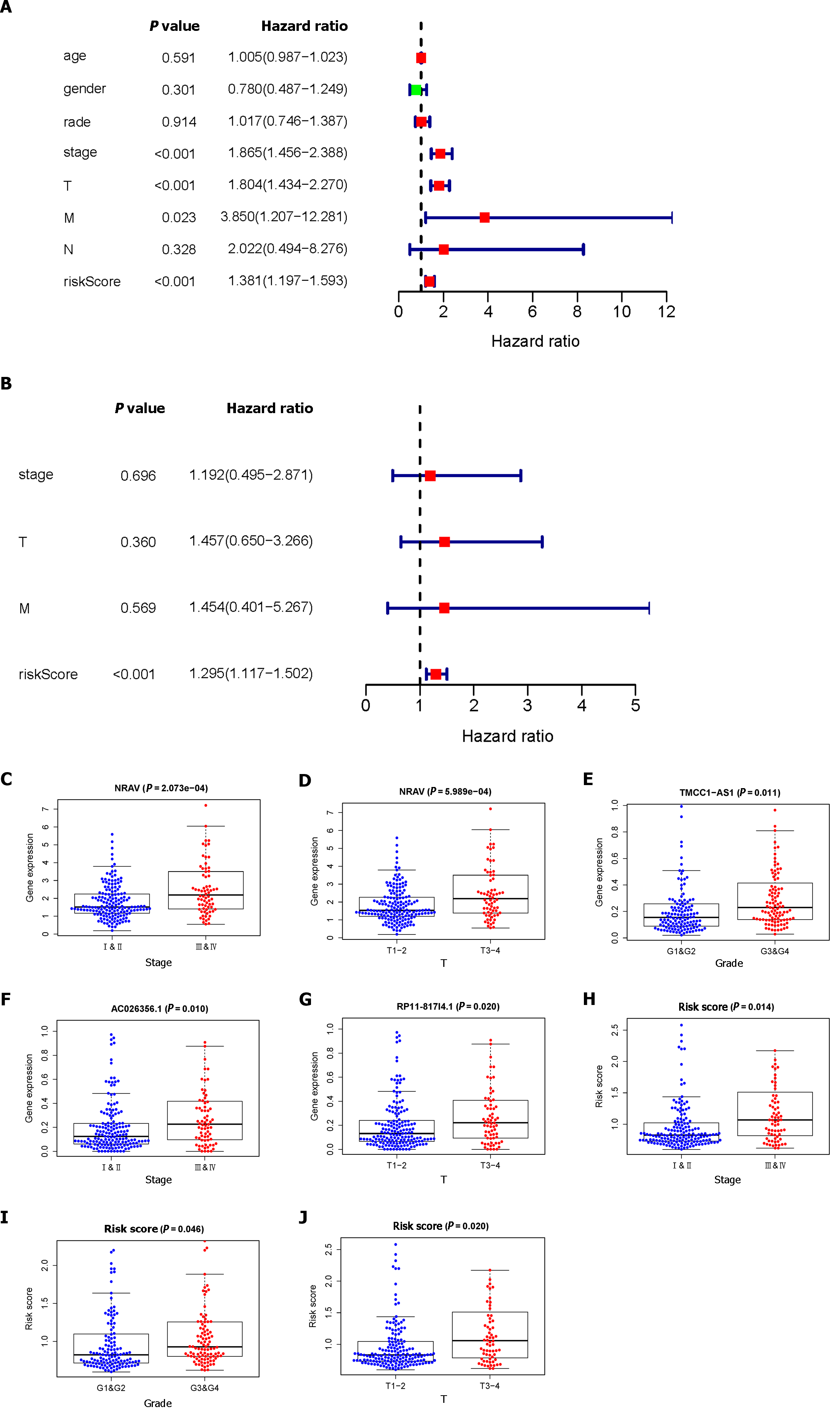
Through univariate and multivariate Cox regression analyses, we established that risk scores could serve as independent prognostic factors. As illustrated in Figure 2A, the univariate Cox regression analysis demonstrated significant associations between OS and stage, T stage, M stage, and risk score. However, according to our multivariate analysis, only the risk score was significantly associated with OS (Figure 2B). We observed that the advanced-stage (Figure 2C) and T-stage (Figure 2D) tumors had higher expression levels of NRAV; the expression levels of TMCC1-AS1 increased in the advanced-stage tumors (Figure 2E); and the advanced-stage (Figure 2F) and T-stage (Figure 2G) tumors had higher expression levels of RP11-817I4.1. In addition, we determined that the risk score could be used as an independent prognostic factor. Additionally, correlation analysis revealed strong associations between risk score and disease stage (Figure 2H), grade (Figure 2I), and T stage (Figure 2J) (P < 0.05).
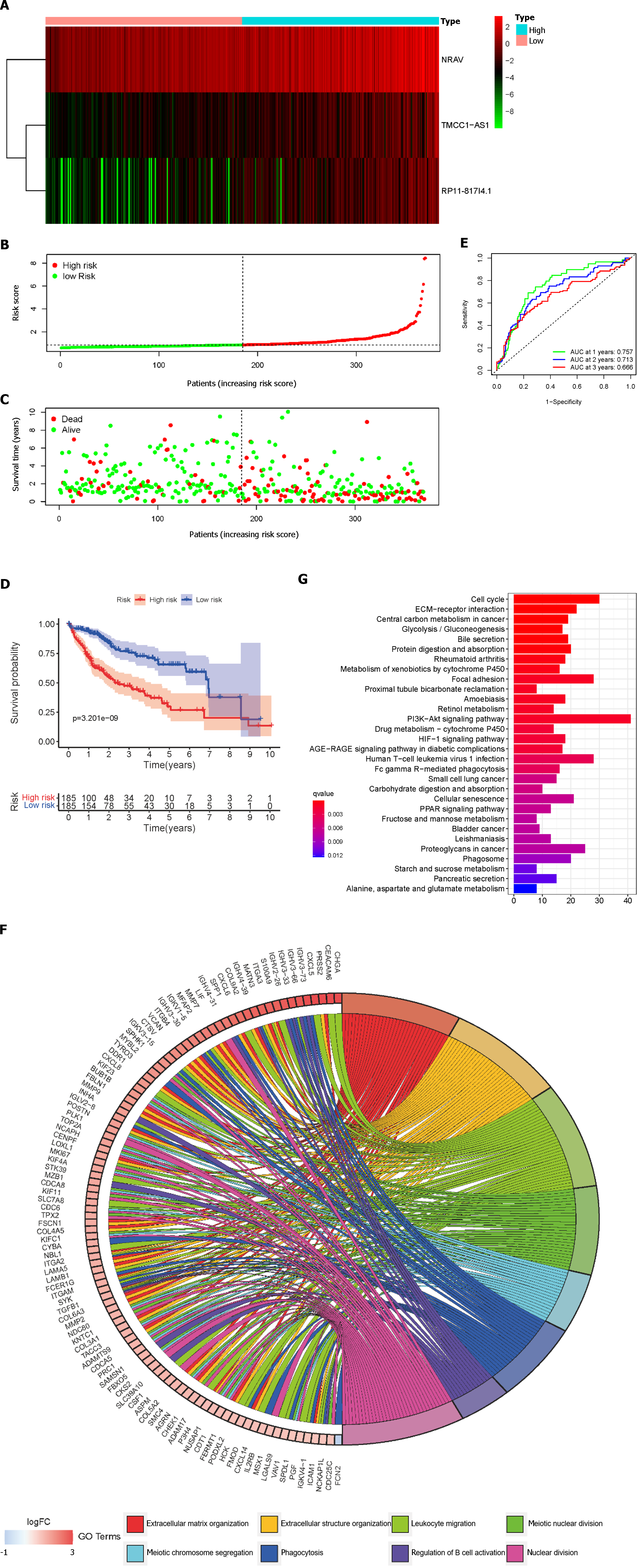
Based on the median risk scores, HCC patients were divided into low- and high-risk groups. The prognostic risk scores for each HCC sample were computed using the formula mentioned above. Figure 3A-C show the dispersion of the survival status, risk score, and matching heatmap of the expression of the three lncRNAs in the TCGA cohort. As the risk score increased, the expression levels of NRAV, TMCC1-AS1, and RP11-817I4.1 increased (Figure 3A). Figure 3C demonstrates that patients with a higher risk score had a considerably greater mortality rate than those with a lower risk score. In addition, K-M survival curve analysis revealed that patients with HCC in the high-risk subgroup had poorer outcomes than those in the low-risk subgroup (Figure 3D). Subsequently, we evaluated the time-dependent ROC curves to confirm the accuracy of the LMRRSM. At 1, 2, and 3 years, the AUC values of the prognostic model in the TCGA cohort were 0.757, 0.713, and 0.666, respectively (Figure 3E). Overall, our findings indicate that the LMRRSM can accurately predict the occurrence and progression of HCC.
To further explore the biological processes and pathways associated with the risk score, GO and KEGG analyses were performed on the differentially expressed genes (DEGs) between the high- and low-risk groups. According to the GO-BP analysis, the genes with differential expression were mostly enriched in extracellular matrix organization, extracellular structure organization, and nuclear division (Figure 3F). The DEGs identified via KEGG analysis were mostly related to the PI3K/Akt signaling pathway, the cell cycle, and focal adhesion (Figure 3G).
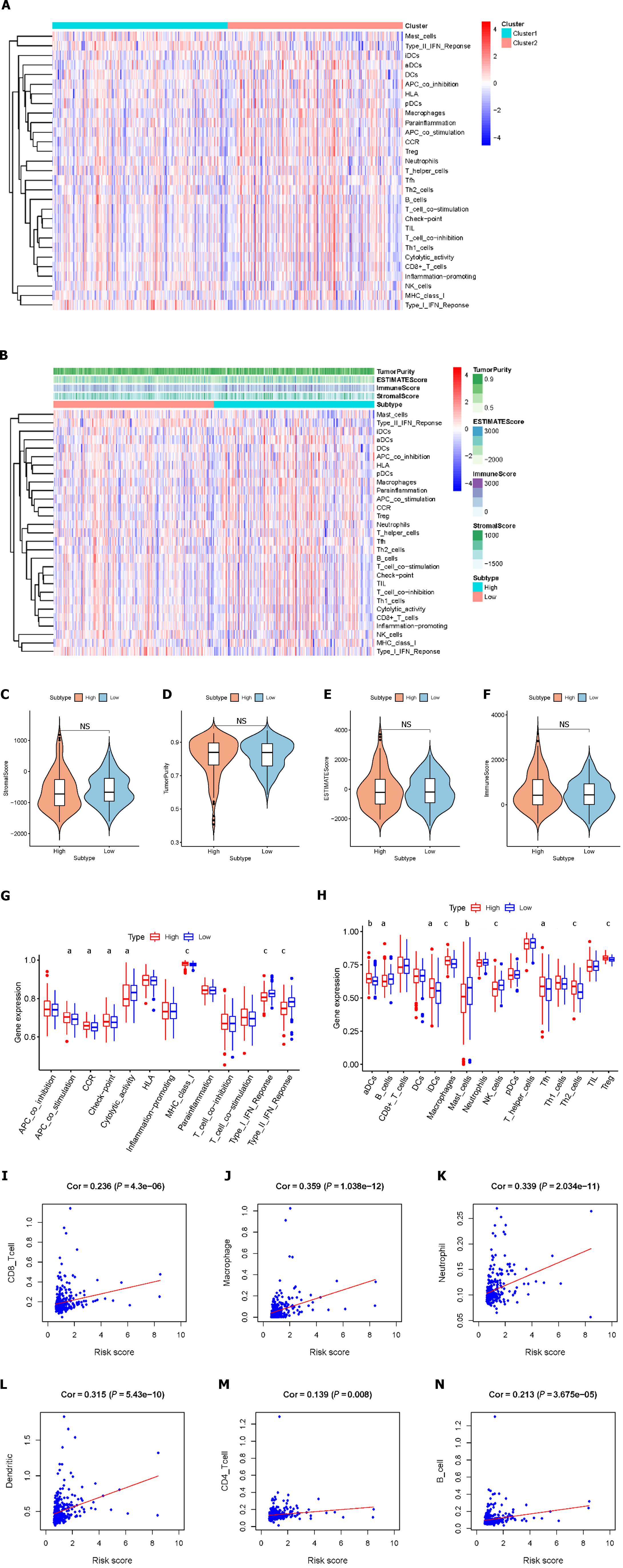
We assessed the activity or enrichment levels of immune cells, functions, or pathways in the different risk score groups using ssGSEA scores, which were derived from the analysis of 29 immune-associated gene sets (Figure 4A). We effectively calculated scores for immune cell infiltration, stromal content, and tumor purity using the ESTIMATE methodology (Figure 4B). By comparing stromal content, tumor purity, and immune cell infiltration scores, we elucidated the relationship between distinct risk groups and immune cell infiltration. The results showed that there were no significant differences in stromal scores, estimated scores, ESTIMATE scores, tumor purity, or immune scores between the high-risk and low-risk patients (Figure 4C-F). According to the ssGSEA results, the percentages of aDCs, iDCs, macrophages, Tfhs, Th2 cells, and Tregs were significantly greater in the high-risk group than in the control group, whereas the percentages of infiltrating B cells, mast cells, and NK cells were significantly lower. Additionally, we found that when risk scores improved, some immunological traits (such as antigen presenting cell costimulatory, C-C chemokine receptor, checkpoint, and major histocompatibility complex class I classes) were significantly more active, whereas others [such as cytolytic activity and interferon (IFN) classes I and II] were repressed (Figure 4G and H). We conducted a further analysis of the relationship between the risk score and immune cell infiltration. Correlation analysis revealed that the risk score exhibited strong associations with CD8+ T cells (r = 0.236; P = 4.3e-6), macrophages (r = 0.359; P = 1.038e-12), neutrophils (r = 0.339; P = 2.034e-11), dendritic cells (r = 0.315; P = 5.43e-10), CD4+ T cells (r = 0.139; P = 0.008), and B cells (r = 0.213; P = 3.675e-05) (Figure 4I-N). These findings suggest that risk signatures based on the LMRRSM may provide new insights into the TME, immune responses, and immune infiltration in HCC patients.
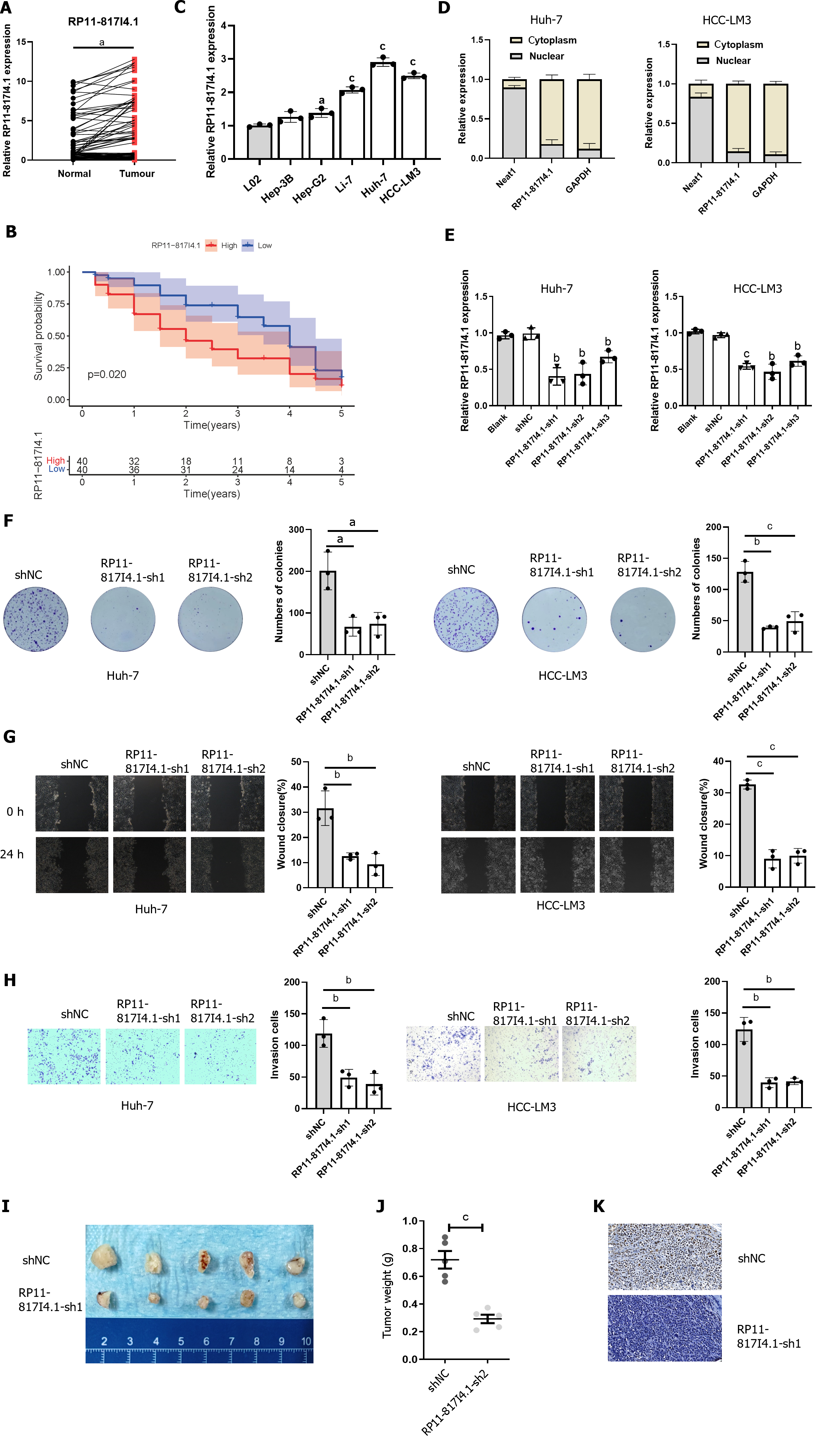
qRT-PCR was employed to assess the expression of RP11-817I4.1 in human HCC tissues and HCC cell lines. The qRT-PCR analysis revealed that RP11-817I4.1 exhibited upregulation in HCC tissues compared to adjacent normal tissues (Figure 5A). Additionally, we separated the HCC patients into two groups based on the median RP11-817I4.1 expression level: High- and low-expression groups. K-M survival analysis revealed that the high-expression group had a considerably worse OS than the low-expression group (Figure 5B). And RP11-817I4.1 was overexpressed in HCC cell lines (Hep-3B, Li-7, HCC-LM3, and Huh-7) but not in the normal hepatic cell line L02 (Figure 5C). Among the six cell lines, RP11-817I4.1 displayed significantly higher expression in HCC-LM3 and Huh-7 cells; consequently, these cell lines were selected for subsequent experiments. To further investigate the potential function of RP11-817I4.1 in HCC development, we evaluated the subcellular localization of RP11-817I4.1 in HCC cells. Figure 5D demonstrates that RP11-817I4.1 was primarily localized in the cytoplasm of Huh-7 and HCC-LM3 cells.
To further explore the role of RP11-817I4.1 in the occurrence and development of HCC, we used three shRNAs to specifically inhibit the expression of RP11-817I4.1 in Huh-7 and HCC-LM3 cells. The efficiency of RP11-817I4.1 transfection in Huh-7 and HCC-LM3 cells was confirmed by qRT-PCR, and RP11-817I4.1-sh1 and RP11-817I4.1-sh2 were confirmed to be the most efficient (Figure 5E). Colony formation assays demonstrated that the silencing of RP11-817I4.1 inhibited HCC cell growth and colony formation (Figure 5F). Subsequently, RP11-817I4.1 silencing significantly reduced the migratory and invasive capabilities of Huh-7 and HCC-LM3 cells (Figure 5G and H). The subcutaneous tumor experiment further provided evidence that RP11-817I4.1 knockdown led to a suppression of tumor size and weight in vivo (Figure 5I and J). Immunostaining with Ki67 revealed that RP11-817I4.1 knockdown significantly restrained the in vivo proliferative capacity of HCC cells (Figure 5K). Taken together, these findings confirmed the involvement of RP11-817I4.1 in the progression of HCC both in vitro and in vivo.
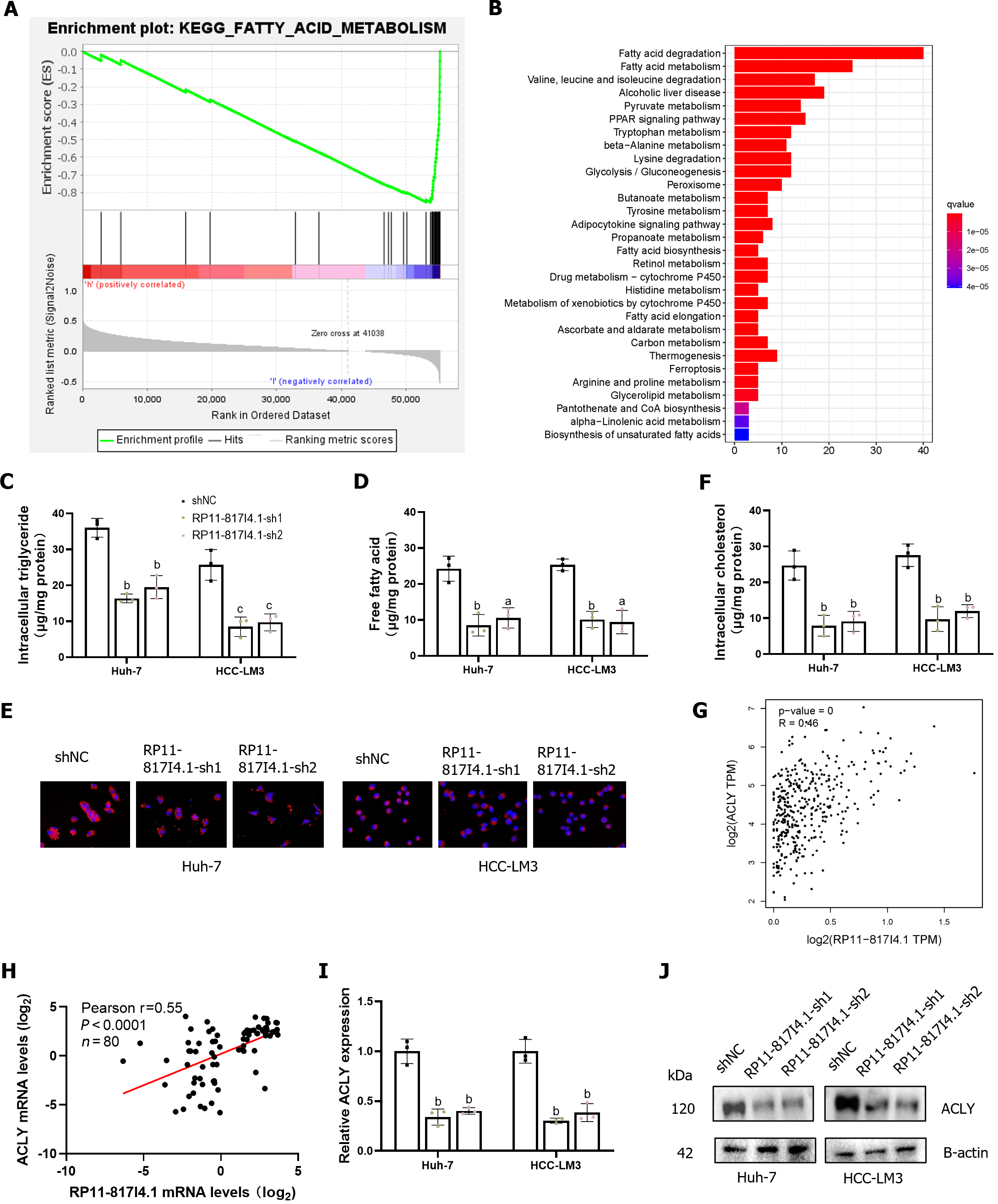
Patients with HCC were divided into high- and low-expression groups according to the expression of RP11-817I4.1. Using GSEA, we detected enrichment pathways for genes that were coexpressed with high- or low-expression groups in HCC. The results indicated a negative correlation between the activity of fatty acid metabolism signaling pathways and the expression of RP11-817I4.1. A heatmap showed that the genes related to the signaling pathway of fatty acid metabolism were considerably active in the low-expression groups (Figure 6A). To gain further insight into the function of RP11-817I4.1, we conducted transcriptomic analysis on HCC-LM3 cells transfected with shNC or RP11-817I4.1-sh1 plasmids and subsequently conducted KEGG enrichment analysis of the DEG. As shown in Figure 6B, these genes were enriched mainly for fatty acid degradation and fatty acid metabolism. These pathways indicate a crucial role for RP11-817I4.1 in the regulation of lipid metabolism. Consequently, we assessed the intracellular TG content in HCC cell lines transfected with siRP11-817I4.1. The results demonstrated that the levels of intracellular TGs and free fatty acids (FFA) were significantly lower in the RP11-817I4.1-silenced cells compared to the control cells (Figure 6C and D). Nile red staining also revealed that the suppression of RP11-817I4.1 Led to reduced cellular lipid accumulation in Huh7 and HCC-LM3 cells (Figure 6E), the same changes were found in intracellular cholesterol (Figure 6F). According to the correlation analysis of mRNA expression profiles based on TCGA data, RP11-817I4.1 and ACLY, the key genes involved in lipid synthesis, exhibited the highest correlation (r = 0.46) (Figure 6G). These findings were further substantiated by the analysis of 80 patient samples from the Second Affiliated Hospital of Kunming Medical University, where the correlation between RP11-817I4.1 and ACLY was even more pronounced (r = 0.55) (Figure 6H). We then observed that both the mRNA and protein expression of ACLY were inhibited by RP11-817I4.1 knockdown (Figure 6I and J). In summary, our findings indicate that RP11-817I4.1 fosters lipid accumulation in HCC cells.
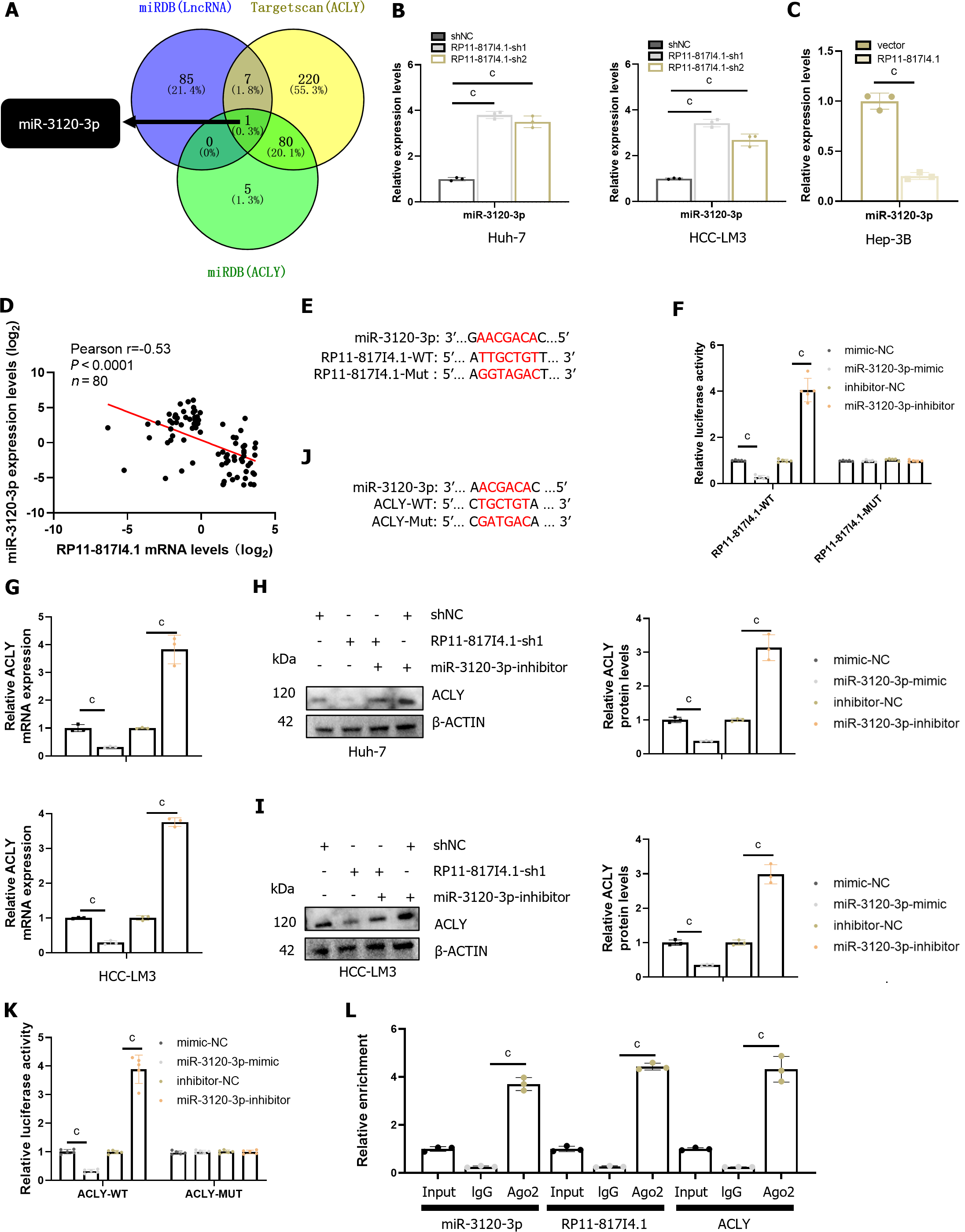
Through nucleocytoplasmic fractionation, we previously established that RP11-817I4.1 is primarily located in the cytoplasm (Figure 5D), suggesting its potential role in regulating ACLY expression through a ceRNA mechanism[23]. Subsequently, utilizing the miRDB and TargetScan databases, we identified miR-3120-3p as a potential interacting miRNA that binds to both RP11-817I4.1 and ACLY mRNAs (Figure 7A). qRT-PCR demonstrated that knockdown of RP11-817I4.1 Led to an upregulation in the expression of miR-3120-3p, while overexpression of RP11-817I4.1 had the opposite effect (Figure 7B and C). Moreover, analysis of clinical specimens unveiled a strong negative correlation between the expression levels of RP11-817I4.1 and miR-3120-3p (Figure 7D). Subsequently, we designed luciferase reporter gene plasmids based on the binding sites of RP11-817I4.1 and miR-3120-3p to validate their interactions (Figure 7E). The results indicated that the miR-3120-3p mimic significantly reduced the luciferase activity of RP11-817I4.1-WT cells, whereas the miR-3120-3p inhibitor had the opposite effect (Figure 7F). Neither the miR-3120-3p mimic nor the miR-3120-3p inhibitor had an impact on the luciferase activity of RP11-817I4.1-MUT cells (Figure 7F). These findings collectively demonstrated that RP11-817I4.1 binds to miR-3120-3p and inhibits its expression.
Subsequently, we assessed the regulatory role of miR-3120-3p in ACLY. We observed that the miR-3120-3p mimic significantly reduced the protein level of ACLY mRNA, while the miR-3120-3p inhibitor had the opposite effect (Figure 7G-I). Following this, we designed luciferase reporter gene plasmids based on the binding sites of ACLY and miR-3120-3p (Figure 7J). Luciferase reporter gene assays revealed that the miR-3120-3p mimic significantly diminished the luciferase activity of ACLY-WT cells, whereas the miR-3120-3p inhibitor had the opposite effect (Figure 7K). Neither the miR-3120-3p mimic nor the miR-3120-3p inhibitor impacted the luciferase activity of the ACLY-MUT cells (Figure 7K). Additionally, we conducted RNA immunoprecipitation (RIP) experiments with the Argonaute 2 protein, the binding partner of miRNAs, which confirmed the direct interaction between RP11-817I4.1, miR-3120-3p, and ACLY (Figure 7L). These findings collectively demonstrated that RP11-817I4.1 regulates ACLY expression by acting as a sponge for miR-3120-3p.
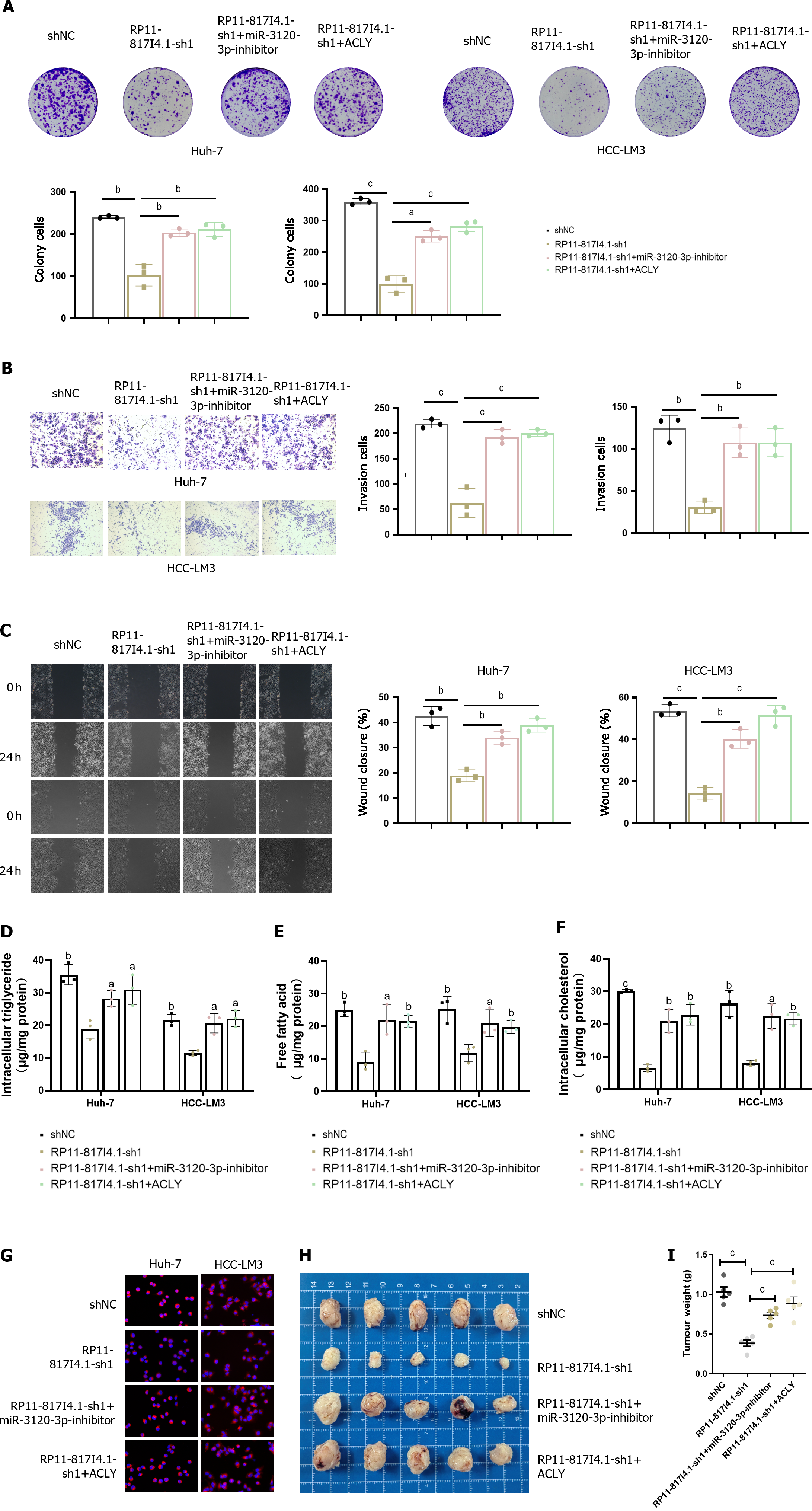
To confirm the functional role of the RP11-817I4.1/miR-3120-3p/ACLY axis in HCC, functional rescue experiments were conducted. Clonogenic assays demonstrated that the decreased proliferation capacity of HCC cells due to RP11-817I4.1 knockdown could be rescued by a miR-3120-3p inhibitor or overexpression of ACLY (Figure 8A). Furthermore, the reduced invasion and migration abilities of HCC cells resulting from RP11-817I4.1 knockdown could be rescued by the miR-3120-3p inhibitor or ACLY overexpression (Figure 8B and C). In terms of lipid metabolism, the decreases in the intracellular TG, FFA, and total cholesterol levels caused by RP11-817I4.1 knockdown were reversed by the miR-3120-3p inhibitor or ACLY overexpression (Figure 8D-F). Moreover, Nile red staining illustrated that the decrease in neutral lipids resulting from RP11-817I4.1 knockdown could be reversed by the miR-3120-3p inhibitor or ACLY overexpression (Figure 8G). Subcutaneous tumor assays provided evidence that the reduced in vivo proliferation capability of HCC cells upon RP11-817I4.1 knockdown could be rescued by a miR-3120-3p inhibitor or ACLY overexpression (Figure 8H and I).
HCC is a type of primary liver tumor that is common worldwide and has a poor prognosis. Research on the molecular etiology and biological features of this disease is critical[8]. A growing body of research suggests that metabolic repro-gramming linked to lipid metabolism is a characteristic feature of malignant tumors[24]. Cancer cells employ lipid metabolism pathways to proliferate, survive, invade, and metastasize, as well as to obtain nutrients, biofilm constituents, and signaling molecules essential for the TME[25]. lncRNAs regulate various biological processes through intricate interactions among multiple mechanisms. Previous studies have identified several lncRNAs as crucial regulatory factors in lipid metabolism and signaling processes[16]. Therefore, lipid metabolism-related markers hold significant potential as prognostic indicators. Research has shown that lipid metabolism-related lncRNA signatures can serve as independent prognostic markers in various solid tumors[26-29]. However, the prognostic significance and relationship between lipid metabolism status and risk signatures in HCC patients have not been determined. Hence, it is imperative to conduct additional fundamental and clinical investigations to establish the clinical applications of LMR-lncRNAs in diagnosis and treatment. This research aims to elucidate the link between lipid metabolism and HCC development and lay the foundation for utilizing LMR-lncRNAs as novel targets for HCC treatment and prognosis in the future.
In this study, LMR-lncRNAs were identified via bioinformatic analysis of HCC data from TCGA, and a lipid metabolism-related prognostic model was established. Following the analysis, 31 LMR-lncRNAs were identified. Among them, three LMR-lncRNAs (NRAV, TMCC1-AS1, and RP11-817I4.1) were validated as components of the risk signature through Cox regression and LASSO regression analysis. This risk signature effectively stratified HCC patients into low- and high-risk groups. Furthermore, the risk score displayed significant predictive value for OS in both univariate and multivariate Cox regression analyses. Furthermore, the risk signature based on three lncRNAs performed well in classifying the risk groups of HCC patients in the TCGA cohort, according to KM, ROC curve, and risk diagram analyses.
Recent research has shown that risk signatures based on lncRNAs linked to lipid metabolism can serve as a dependable indicator for categorizing risk groups in various types of tumors. Similar to our study, Wu et al[30] revealed that lncRNA signatures connected to lipid metabolism may stratify patients with colon cancer into various risk categories. Similarly, another study demonstrated that LMR-lncRNA signatures could effectively categorize patients with human lung adenocarcinoma into different response groups, where the risk scores from these signatures emerged as the most substantial predictors of pathological remission[31]. In our investigation, a prognostic model built with LMR-lncRNAs, referred to as LMRRSM, displayed the capability to accurately forecast the prognosis of HCC patients. According to these findings, individuals with HCC who fell into the high-risk score category had a worse prognosis than those with HCC in the low-risk score category (P < 0.001). Additionally, according to univariate and multivariate Cox regression analyses, the risk score maintained its independent predictive significance in comparison to well-established clinicopathological prognostic markers. In the TCGA cohort, the prognostic model had a high level of accuracy for 1-, 2-, and 3-year OS. According to our biological function analysis, the genes associated with the DEGs among the risk score groups were enriched mainly in the PI3K-Akt signaling pathway. This indirectly indicated that the risk score was closely related to the PI3K-Akt signaling pathway. It is worth noting that the PI3K/Akt signaling pathway plays a crucial role in the proliferation and survival of cancer cells. According to previous reports, the PI3K/Akt signaling pathway is abnormally activated in 30%-50% of patients with HCC[32].
Our focus was directed towards the TME, a vital component in the context of cancer. The TME has been recognized as a significant factor in tumor biology[33]. Variations in the lipid metabolism of tumors, which comprises an abundance of metabolites and lipid metabolic products, may be responsible for both local immunosuppression in the TME and tumor progression[34]. Zhang et al[35] conducted an in vitro assay to mimic the interaction between tumor-associated macrophages and HCC within the TME. Their research revealed that M2 monocyte-derived macrophages promoted HCC cell migration in a manner dependent on fatty acid oxidation, which was linked to increased interleukin-1β secretion. Notably, immune checkpoint protein PD-L1, along with other immunosuppressive signals such as Toll-like receptor 4 and CD48/2B4, are expressed by macrophages and impact local immunity by deactivating CD8+ T cells, facilitating the recruitment of Tregs, and suppressing NK cell activity[36,37]. These findings align with our own results. The patients were typically divided into two groups according to LMRRSM. We then examined the connection between immune infiltration and LMRRSM. There were more aDCs, iDCs, macrophages, Tfh cells, Th2 cells, and Tregs in the high-risk group. In the low-risk group, there were significantly higher numbers of B cells, mast cells, and NK cells. Consequently, LMRRSM is closely connected to the immunological condition of patients with HCC.
In this study, three lncRNAs were identified as risk signatures: NRAV, TMCC1-AS1, and RP11-817I4.1. Previous reports have linked certain molecular components of this signature to the development and regulation of tumor growth. NRAV, for instance, is primarily found in the cytoplasm and plays a role in regulating vesicle-transporting protein activity[38]. NRAV reduction is part of the human antiviral innate immune response to viral infections and is achieved through the control of IFN-stimulated gene transcription[39]. Additionally, the lncRNA NRAV is elevated in HCC and may promote cell growth and migration by suppressing miR-199a-3p, thereby enhancing the expression of CISD2 and activating the Wnt/β-catenin signaling pathway[40]. In patients with HCC, the lncRNA TMCC1-AS1 has been repeatedly associated with prognosis. Elevated expression of TMCC1-AS1 is strongly correlated with poor OS and disease-free survival in HCC patients. At the molecular level, epithelial-mesenchymal transition is activated by upregulating the expression of TMCC1AS1 in HCC cell lines, which decreases E-cadherin expression and elevates Ki67, proliferating cell nuclear antigen, Ncadherin, and vimentin expression[41].
Currently, there are no studies on the lncRNA RP11-817I4.1, and its biological function and potential mechanism in HCC remain unclear. However, this study has provided insights into RP11-817I4.1. It revealed that the expression of RP11-817I4.1 was significantly increased in HCC tissues and cell lines, suggesting that RP11-817I4.1 may play a role in promoting the progression of HCC. Notably, Kaplan-Meier survival curves were generated for RP11-817I4. The expression of RP11-817I4.1 was found to be strongly correlated with OS and has the potential to serve as a standalone predictive biomarker for patients with HCC. Additionally, this study showed that RP11-817I4.1 enhanced the proliferation, migration, and invasion of HCC-LM3 and Huh-7 cells, indicating that RP11-817I4.1 plays a critical role in the regulation of hepatocellular tumorigenesis. The present study focused on the signaling pathway involved in fatty acid metabolism to investigate the potential mechanism by which RP11-817I4.1 promotes abnormalities in lipid metabolism in HCC cells. The present study showed that inhibiting RP11-817I4.1 expression resulted in a considerable increase in intracellular TG levels in HCC lines. Specifically, we demonstrated that RP11-817I4.1 positively regulates the expression of ACLY, a key gene involved in lipid synthesis, by sponging miR-3120-3p, thereby promoting lipid synthesis and HCC progression. These findings suggest that RP11-817I4.1 promotes HCC progression by promoting lipid synthesis.
In the present study, we investigated the functions of LMR-lncRNAs in assessing and predicting the prognosis of patients with HCC and established an accurate and reliable LMRRSM for prediction. In particular, we focused on a novel LMR-lncRNA, RP11-817I4.1, which has been proven to play an important regulatory role in HCC lipid metabolism and tumor progression and has therapeutic potential. Our findings will be extremely beneficial for understanding the probable molecular biological processes involved in HCC and discovering novel prognostic indicators and molecular targets.
Published studies have demonstrated the impact of disturbances in lipid metabolism on the progression of hepatocellular carcinoma (HCC), which is a prevalent form of cancer.
Long noncoding RNAs (lncRNAs) modulate fatty acid metabolism and influence HCC development through pathways involving transcription factors and lipid-related processes. Understanding the role of metabolic reprogramming and lncRNAs in HCC may lead to novel diagnostic markers and therapeutic targets.
This study aims to investigate a novel lncRNA, RP11-817I4.1, revealed its role in promoting lipid accumulation, thereby accelerating the onset and progression of HCC.
The study identified three lipid metabolism-related lncRNAs (LMR-lncRNAs), negative regulator of antiviral response (NRAV), RNA transmembrane and coiled-coil domain family 1 antisense RNA 1 (TMCC1-AS1), and RP11-817I4.1, as predictive markers for HCC and used them to construct risk models. Knockdown of RP11-817I4.1 decreased proliferation, migration, and invasion in HCC cells. RP11-817I4.1 was found to increase lipid levels in HCC cells through the miR-3120-3p/ATP citrate lyase (ACLY) axis, highlighting its role in lipid metabolism and HCC progression.
The study identified three LMR-lncRNAs, NRAV, TMCC1-AS1, and RP11-817I4.1, as predictive markers for HCC patients and utilized them in constructing risk models. Knockdown of RP11-817I4.1 resulted in reduced proliferation, migration, and invasion of HCC cells. Moreover, RP11-817I4.1 was found to significantly increase lipid levels in HCC cells through the miR-3120-3p/ACLY axis. These findings provide valuable insights into the molecular processes of HCC, uncover novel prognostic indicators and molecular targets, and highlight the therapeutic potential of RP11-817I4.1 in the context of HCC treatment.
In this study, the functions of LMR-lncRNAs were investigated for their role in assessing and predicting the prognosis of HCC patients, leading to the establishment of an accurate and reliable lipid metabolism-related risk score model for prediction. Of particular interest was the novel LMR-lncRNA, RP11-817I4.1, which was found to regulate HCC lipid metabolism and tumor progression, showing potential as a therapeutic target. The findings hold significant promise for understanding the molecular processes in HCC, identifying novel prognostic indicators, and uncovering molecular targets for potential therapeutic interventions.
Lipid metabolism-related lncRNA RP11-817I4.1 may be a potential therapeutic target, and the development of small molecule drugs targeting RP11-817I4.1 may help improve the prognosis of HCC patients.
We appreciate and acknowledge our colleagues for their feedback, opinions, and technical support for this project.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bredt LC, Brazil S-Editor: Qu XL L-Editor: A P-Editor: Chen YX
| 1. | Dai Y, Qiang W, Lin K, Gui Y, Lan X, Wang D. An immune-related gene signature for predicting survival and immunotherapy efficacy in hepatocellular carcinoma. Cancer Immunol Immunother. 2021;70:967-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 105] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 2. | Famularo S, Di Sandro S, Giani A, Lauterio A, Sandini M, De Carlis R, Buscemi V, Uggeri F, Romano F, Gianotti L, De Carlis L. Recurrence Patterns After Anatomic or Parenchyma-Sparing Liver Resection for Hepatocarcinoma in a Western Population of Cirrhotic Patients. Ann Surg Oncol. 2018;25:3974-3981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 3. | Ingle PV, Samsudin SZ, Chan PQ, Ng MK, Heng LX, Yap SC, Chai AS, Wong AS. Development and novel therapeutics in hepatocellular carcinoma: a review. Ther Clin Risk Manag. 2016;12:445-455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 4. | Crunkhorn S. Targeting cancer cell metabolism in glioblastoma. Nat Rev Cancer. 2019;19:250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 5. | Rosario SR, Long MD, Affronti HC, Rowsam AM, Eng KH, Smiraglia DJ. Pan-cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas. Nat Commun. 2018;9:5330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 176] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 6. | Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 819] [Cited by in RCA: 1104] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 7. | Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1209] [Cited by in RCA: 1584] [Article Influence: 132.0] [Reference Citation Analysis (0)] |
| 8. | Hu B, Lin JZ, Yang XB, Sang XT. Aberrant lipid metabolism in hepatocellular carcinoma cells as well as immune microenvironment: A review. Cell Prolif. 2020;53:e12772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 9. | Nakagawa H, Hayata Y, Kawamura S, Yamada T, Fujiwara N, Koike K. Lipid Metabolic Reprogramming in Hepatocellular Carcinoma. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 10. | Wu JM, Skill NJ, Maluccio MA. Evidence of aberrant lipid metabolism in hepatitis C and hepatocellular carcinoma. HPB (Oxford). 2010;12:625-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Fu Y, Liu S, Zeng S, Shen H. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 297] [Article Influence: 49.5] [Reference Citation Analysis (0)] |
| 12. | Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2202] [Cited by in RCA: 2422] [Article Influence: 269.1] [Reference Citation Analysis (0)] |
| 13. | Shang C, Wang W, Liao Y, Chen Y, Liu T, Du Q, Huang J, Liang Y, Liu J, Zhao Y, Guo L, Hu Z, Yao S. LNMICC Promotes Nodal Metastasis of Cervical Cancer by Reprogramming Fatty Acid Metabolism. Cancer Res. 2018;78:877-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 107] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 14. | van Solingen C, Scacalossi KR, Moore KJ. Long noncoding RNAs in lipid metabolism. Curr Opin Lipidol. 2018;29:224-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 15. | Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 1122] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 16. | Duan J, Huang Z, Nice EC, Xie N, Chen M, Huang C. Current advancements and future perspectives of long noncoding RNAs in lipid metabolism and signaling. J Adv Res. 2023;48:105-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Chen Y, Wu X, Chen X, Guo D, Ma W, Guo Y, Xu K, Ma S, Yuan Y, Zhu Q. LncRNA TGFB2-OT1 Promotes Progression and Angiogenesis in Hepatocellular Carcinoma by Dephosphorylating β-Catenin. J Hepatocell Carcinoma. 2023;10:429-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 18. | Xu K, Xia P, Liu P, Zhang X. A six lipid metabolism related gene signature for predicting the prognosis of hepatocellular carcinoma. Sci Rep. 2022;12:20781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 19. | Song K, Li L, Zhang G. Bias and Correction in RNA-seq Data for Marine Species. Mar Biotechnol (NY). 2017;19:541-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 2956] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 21. | He Y, Jiang Z, Chen C, Wang X. Classification of triple-negative breast cancers based on Immunogenomic profiling. J Exp Clin Cancer Res. 2018;37:327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 379] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 22. | Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108-e110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2728] [Cited by in RCA: 4092] [Article Influence: 511.5] [Reference Citation Analysis (0)] |
| 23. | Xia P, Zhang H, Lu H, Xu K, Jiang X, Jiang Y, Gongye X, Chen Z, Liu J, Chen X, Ma W, Zhang Z, Yuan Y. METTL5 stabilizes c-Myc by facilitating USP5 translation to reprogram glucose metabolism and promote hepatocellular carcinoma progression. Cancer Commun (Lond). 2023;43:338-364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 81] [Reference Citation Analysis (0)] |
| 24. | Shang C, Huang J, Guo H. Identification of an Metabolic Related Risk Signature Predicts Prognosis in Cervical Cancer and Correlates With Immune Infiltration. Front Cell Dev Biol. 2021;9:677831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 25. | Bian X, Liu R, Meng Y, Xing D, Xu D, Lu Z. Lipid metabolism and cancer. J Exp Med. 2021;218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 580] [Article Influence: 145.0] [Reference Citation Analysis (0)] |
| 26. | Lin Y, Xiao Y, Liu S, Hong L, Shao L, Wu J. Role of a lipid metabolism-related lncRNA signature in risk stratification and immune microenvironment for colon cancer. BMC Med Genomics. 2022;15:221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 27. | Xu K, Dai C, Yang J, Xu J, Xia C, Li J, Zhang C, Xu N, Wu T. Disulfidptosis-related lncRNA signatures assess immune microenvironment and drug sensitivity in hepatocellular carcinoma. Comput Biol Med 2024; 169:107930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 28. | Wang H, Zhang Y, Guan X, Li X, Zhao Z, Gao Y, Zhang X, Chen R. An Integrated Transcriptomics and Proteomics Analysis Implicates lncRNA MALAT1 in the Regulation of Lipid Metabolism. Mol Cell Proteomics. 2021;20:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 29. | Xu K, Xia P, Gongye X, Zhang X, Ma S, Chen Z, Zhang H, Liu J, Liu Y, Guo Y, Yao Y, Gao M, Chen Y, Zhang Z, Yuan Y. A novel lncRNA RP11-386G11.10 reprograms lipid metabolism to promote hepatocellular carcinoma progression. Mol Metab. 2022;63:101540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 39] [Reference Citation Analysis (0)] |
| 30. | Wu S, Gong Y, Chen J, Zhao X, Qing H, Dong Y, Li S, Li J, Wang Z. Identification of fatty acid metabolism-related lncRNAs in the prognosis and immune microenvironment of colon adenocarcinoma. Biol Direct. 2022;17:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Zhao J, Li G, Zhao G, Wang W, Shen Z, Yang Y, Huang Y, Ye L. Prognostic signature of lipid metabolism associated LncRNAs predict prognosis and treatment of lung adenocarcinoma. Front Oncol. 2022;12:986367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 32. | Mikhail S, Cosgrove D, Zeidan A. Hepatocellular carcinoma: systemic therapies and future perspectives. Expert Rev Anticancer Ther. 2014;14:1205-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 33. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47138] [Article Influence: 3367.0] [Reference Citation Analysis (5)] |
| 34. | Hao Y, Li D, Xu Y, Ouyang J, Wang Y, Zhang Y, Li B, Xie L, Qin G. Investigation of lipid metabolism dysregulation and the effects on immune microenvironments in pan-cancer using multiple omics data. BMC Bioinformatics. 2019;20:195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 35. | Zhang Q, Wang H, Mao C, Sun M, Dominah G, Chen L, Zhuang Z. Fatty acid oxidation contributes to IL-1β secretion in M2 macrophages and promotes macrophage-mediated tumor cell migration. Mol Immunol. 2018;94:27-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 36. | Wu Y, Kuang DM, Pan WD, Wan YL, Lao XM, Wang D, Li XF, Zheng L. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology. 2013;57:1107-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 215] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 37. | Wu K, Kryczek I, Chen L, Zou W, Welling TH. Kupffer cell suppression of CD8+ T cells in human hepatocellular carcinoma is mediated by B7-H1/programmed death-1 interactions. Cancer Res. 2009;69:8067-8075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 304] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 38. | Li J, Li M, Wang X, Sun M, Ma C, Liang W, Gao X, Wei L. Long Noncoding RNA NRAV Promotes Respiratory Syncytial Virus Replication by Targeting the MicroRNA miR-509-3p/Rab5c Axis To Regulate Vesicle Transportation. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 39. | Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16:616-626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 286] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 40. | Wang Q, Tang Y, Ge Y, Zhang S, Zheng M. Long non-coding RNA NRAV enhances proliferation and invasion of hepatocellular carcinoma cells by modulating the Wnt/β-catenin signaling pathway. Bioengineered. 2022;13:10026-10037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 41. | Chen C, Su N, Li G, Shen Y, Duan X. Long non-coding RNA TMCC1-AS1 predicts poor prognosis and accelerates epithelial-mesenchymal transition in liver cancer. Oncol Lett. 2021;22:773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |