Published online Feb 14, 2024. doi: 10.3748/wjg.v30.i6.565
Peer-review started: December 4, 2023
First decision: December 8, 2023
Revised: December 20, 2023
Accepted: January 16, 2024
Article in press: January 16, 2024
Published online: February 14, 2024
Processing time: 62 Days and 22.2 Hours
Esophageal squamous cell carcinoma (ESCC) is a deadly malignancy with limited treatment options. Deubiquitinases (DUBs) have been confirmed to play a crucial role in the development of malignant tumors. JOSD2 is a DUB involved in con
To investigate the impact of JOSD2 on the progression of ESCC.
Bioinformatic analyses were employed to explore the expression, prognosis, and enriched pathways associated with JOSD2 in ESCC. Lentiviral transduction was utilized to manipulate JOSD2 expression in ESCC cell lines (KYSE30 and KYSE150). Functional assays, including cell proliferation, colony formation, drug sensitivity, migration, and invasion, were performed, revealing the impact of JOSD2 on ESCC cell lines. JOSD2's role in xenograft tumor growth and drug sensitivity in vivo was also assessed. The proteins that interacted with JOSD2 were identified using mass spectrometry.
Preliminary research indicated that JOSD2 was highly expressed in ESCC tissues, which was associated with poor prognosis. Further analysis demonstrated that JOSD2 was upregulated in ESCC cell lines compared to normal esophageal cells. JOSD2 knockdown inhibited ESCC cell activity, including proliferation and colony-forming ability. Moreover, JOSD2 knockdown decreased the drug resistance and migration of ESCC cells, while JOSD2 overexpression enhanced these phenotypes. In vivo xenograft assays further confirmed that JOSD2 promoted tumor proliferation and drug resistance in ESCC. Mechanistically, JOSD2 appears to activate the MAPK/ERK and PI3K/AKT signaling pathways. Mass spectrometry was used to identify crucial substrate proteins that interact with JOSD2, which identified the four primary proteins that bind to JOSD2, namely USP47, IGKV2D-29, HSP90AB1, and PRMT5.
JOSD2 plays a crucial role in enhancing the proliferation, migration, and drug resistance of ESCC, suggesting that JOSD2 is a potential therapeutic target in ESCC.
Core Tip: JOSD2, a deubiquitinating enzyme, is a key player in the aggressive pathogenesis of esophageal squamous cell carcinoma (ESCC). Elevated JOSD2 expression in ESCC tissues is associated with poor prognosis. Functional analyses, including in vivo xenograft assays, highlight JOSD2's role in promoting tumor proliferation and drug resistance. Mechanistically, JOSD2 activates the MAPK/ERK and PI3K/AKT signaling pathways. Mass spectrometry identified key interacting proteins, including USP47, IGKV2D-29, HSP90AB1, and PRMT5. This study underscores the potential role of JOSD2 as a therapeutic target in ESCC.
- Citation: Wang WP, Shi D, Yun D, Hu J, Wang JF, Liu J, Yang YP, Li MR, Wang JF, Kong DL. Role of deubiquitinase JOSD2 in the pathogenesis of esophageal squamous cell carcinoma. World J Gastroenterol 2024; 30(6): 565-578
- URL: https://www.wjgnet.com/1007-9327/full/v30/i6/565.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i6.565
Esophageal squamous cell carcinoma (ESCC) ranks among the deadliest malignancies worldwide, posing a significant public health concern[1]. Despite advances in early detection and therapeutic interventions, ESCC remains a formidable challenge due to its mostly late-stage diagnosis and limited treatment options. To explore the potential therapeutic targets of ESCC, there has been an increasing focus on the genetic underpinnings of this aggressive cancer. Deubiquitinases (DUBs), responsible for cleaving ubiquitin chains from their protein targets, are crucial for controlling protein ubiquitination and preserving protein homeostasis. DUBs influence important cellular processes such as tumor cell proliferation, drug resistance, distant metastasis, and immune evasion by stabilizing the expression of key cancer proteins[2-6]. The research on DUBs provides new avenues for developing treatment options for cancers lacking effective therapeutic strategies. However, the role of DUBs in malignant tumors is far from fully elucidated. Currently, an increasing number of small molecule inhibitors targeting DUBs are being developed and reported, with preclinical and clinical trials underway, demonstrating significant potential in this research field[7].
JOSD2, also known as Josephin domain-containing 2, is a member of the Machado-Joseph disease protein family. It consists of 188 amino acids and contains only one highly conserved catalytic Josephin domain, possessing enzymatic activity. Several recent studies have shed light on the involvement of JOSD2 in some malignant tumors[8-12]. JOSD2 has been found to interact with key signaling pathways, such as the Hippo pathway, Wnt/β-catenin pathway, and DNA repair mechanisms[8,9,11]. Dysregulation of JOSD2 expression has been implicated in cancer initiation, tumor growth, and resistance to chemotherapy[8,11]. There is a lack of relevant research on the association between JOSD2 and ESCC. The elucidation of the function of JOSD2 in ESCC will be helpful to identify individuals at higher risk and devise personalized treatment strategies. Therefore, we aimed to explore the function of JOSD2 in ESCC, shedding light on its potential as a promising avenue for further investigation and clinical applications.
The University of Alabama at Birmingham CANcer database (https://ualcan.path.uab.edu/) was used to analyze the differential expression of JOSD2 mRNA between ESCC and normal esophageal tissues[13,14]. The online survival database Kaplan–Meier Plotter (https://kmplot.com/analysis/) was used to assess the impact of high vs low JOSD2 mRNA expression on the survival prognosis of patients with ESCC[15]. Clinical and RNA-seq data from ESCC patients were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), and a nomogram predicting the 1-, 3- and 5-year survival probability of ESCC patients was constructed and visualized using the “survival” and “rms” packages in R (4.2.1).
The top 500 genes that have a similar expression pattern to JOSD2 in ESCC were downloaded from the Gene Expression Profiling Interactive Analysis 2 database (http://gepia2.cancer-pku.cn/#index) and listed in Supple
Gene Ontology (GO) (Supplementary Table 2) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Supplemen
A normal esophageal epithelial cell line, Het-1A, was obtained from the American Type Culture Collection and cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, United States) supplemented with 10% fetal bovine serum (FBS) (Sigma, United States), penicillin (100 U/mL) and streptomycin (0.1 mg/mL) (Sigma, United States). Four ESCC cell lines (KYSE30, KYSE140, KYSE150, and KYSE410) were obtained from the Chinese Academy of Sciences Cell Bank and cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Thermo Fisher Scientific, United States) supplemented with 10% FBS (Sigma, United States), penicillin (100 U/mL), and streptomycin (0.1 mg/mL) (Sigma, United States). These cells were cultured at 37°C in a 5% CO2 incubator.
Two independent small hairpin RNA (shRNA) sequences targeting JOSD2 (sh1: CGATGAGATCTG-CAAGAGGTT; sh2: GTGTCTACTACAACCTGGACT) were designed, cloned into a PSIH1 vector (GenePharma, China), and used for lentivirus packaging in 293T cells. The lentiviral supernatant was collected. KYSE150 cells (which had the highest JOSD2 expression among the ESCC cell lines) were transduced with the lentivirus and selected with puromycin 72 h later.
The JOSD2 gene was cloned into the lentiviral expression vector pLVX-IRES-Neo to create the overexpression plasmid pLVX-G418 JOSD2-Flag (GenePharma, China). The lentivirus was packaged and used to infect KYSE30 cells (which had the lowest JOSD2 expression among the ESCC cell lines), which were then selected with G418 72 h later.
The total RNA was extracted using an RNApure Tissue/Cell Kit (Cwbiotech, China). The isolated RNA was used as a template for reverse transcription reaction using a HiFiScript cDNA Synthesis Kit (Cwbiotech, China). Real-time fluorescent quantitative polymerase chain reaction (RT-qPCR) was performed using SYBR Fast qPCR Mix (TaKaRa, Japan) and a CFX96 Real-Time System (Bio-Rad, United States). The primer sequences for JOSD2 were as follows: Forward: 5’-CCCACCGTGTACCACGAAC-3’; reverse: 5’-CTCCTGGCTAAAGAGCTGCTG-3’. The primer sequences for GAPDH were as follows: Forward: 5’-GATTCCACCCATGGCAAATTC-3’; reverse: 5’-CTGGAAGATGGTGATGGGATT-3’.
Approximately 1 × 106 cells were placed in each well of a 6-well culture plate. The cells were lysed by adding radioimmunoprecipitation assay buffer (10 μL) and a phosphatase inhibitor (1 μL) for every 100000 cells. The protein lysate was centrifuged at 17000g for 30 min to obtain the supernatant. The protein concentration was measured using a bicinchoninic acid assay. A mixture containing 10 μg of protein was then boiled at 95 °C for 10 min to denature the proteins.
Gel electrophoresis and transfer were carried out using a Mini Gel Tank chamber system (Thermo Fisher Scientific, United States) following the detailed procedures and reagents provided in the manufacturer's instructions. Gel electrophoresis was performed with constant voltage, starting at 70 V for 20 min, followed by an adjustment to 100 V for 50 min. The proteins were then transferred to a polyvinylidene fluoride membrane using a constant voltage of 10 V for 50 min.
The membrane was blocked with 5% skim milk for 1 h. The membrane was then incubated with one of the following primary antibodies overnight at 4 °C: JOSD2 antibody (sab2103354, 1:500, Sigma-Aldrich, United States), phosphorylated p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (9101, 1:1000, Cell Signaling Technology, United States), p44/42 MAPK (Erk1/2) (L34F12) antibody (4696, 1:1000, Cell Signaling Technology, United States), phosphorylated Akt (Ser473) (D9E) antibody (4060, 1:2000, Cell Signaling Technology, United States), phosphorylated Akt (Thr308) antibody (13038, 1:1000, Cell Signaling Technology, United States), or Akt (pan) (40D4) antibody (2920, 1:2000, Cell Signaling Technology, United States). The membrane was then incubated with one of the following secondary antibodies at room temperature for 1 h: Anti-rabbit (7074, 1:1000, Cell Signaling Technology, United States) or anti-mouse (7076, 1:1000, Cell Signaling Technology, United States) horseradish peroxidase-linked antibody. The membrane was subjected to enhanced chemiluminescence (ECL) detection using SignalFire ECL reagent (Cell Signaling Technology, United States), and images were captured and saved using an automated imaging system. Anti-β-actin antibody (4967, 1:1000, Cell Signaling Technology, United States) was then added and incubated at room temperature for 1 h, followed by detection and image capture.
The Cell Counting Kit-8 (CCK-8) assay (Solarbio, China) was performed according to the manufacturer's instructions. Cells were seeded in a 96-well culture plate with approximately 1000 cells per well, and incubated at 37 °C in a 5% CO2 incubator for 24, 48, 72, or 96 h. Subsequently, 10 μL of CCK-8 reagent was added to the cells, and the absorbance at 450 nm was measured using a microplate reader.
After exposure to shRNA1, shRNA2, or negative control, 1000 cells in the logarithmic growth phase were suspended in RPMI-1640 medium with 10% FBS and seeded in each well of 6-well plates. Following a 12-d incubation, the cells were fixed with methanol for 15 min and then stained with 0.5% crystal violet for 3 min at room temperature. After three washes with distilled water, the plates were air-dried and the cell colonies were manually counted. A positive colony was defined as a cluster containing at least 50 cells.
Cells in the logarithmic growth phase, with knocked-down or overexpressed JOSD2, and their respective control cells, were divided into different groups and seeded into 96-well plates with each well containing 1 × 104 cells. After cell adhesion, cisplatin (Med-ChemExpress, United States) was added to each well at concentration gradients of 0, 10, 20, 40, 60, and 80 μg/mL in JOSD2 knock-down cell group. For the JOSD2 overexpression cell group, cisplatin was added at concentration gradients of 0, 20, 40, 60, 80, and 100 μg/mL. After 48 h, the sensitivity of the tumor cells to cisplatin was assessed using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay kit (Sangon Biotech, China).
Transwell chambers (Corning, United States) were preloaded with 5 × 104 cells suspended in 100 μL of RPMI-1640 medium without FBS. The chambers were then placed in a 24-well plate, with each well containing 500 μL of RPMI-1640 medium supplemented with 10% FBS, and incubated in a 37 °C, 5% CO2 incubator. After 24 h, the medium in the chambers was removed, and the cells in the chambers were gently wiped away with a cotton swab. The cells on the chambers were fixed on a new 24-well plate with 4% paraformaldehyde for 20 min. Next, the chambers were removed, followed by crystal violet staining for 20 min. Excess crystal violet solution was washed off with phosphate-buffered saline (PBS), and cell images were captured using a microscope.
Approximately 60 μL of diluted Matrigel (1:8, Becton, Dickinson and Company, United States) was added to Transwell chambers. The chambers were incubated in a 37 °C, 5% CO2 incubator for 3 h, excess liquid was removed from the chambers, and 100 μL of RPMI-1640 medium without FBS was added. The chambers were then placed in the 37 °C, 5% CO2 incubator for 30 min to hydrate the basement membrane. Subsequently, 100 μL of cell suspension comprising 5 × 104 cells in FBS-free RPMI-1640 medium was added to the chambers. The chambers were then placed in a 24-well plate, with each well containing 500 μL of RPMI-1640 medium with 10% FBS, and incubated in the 37 °C, 5% CO2 incubator. After 24 h, the medium in the chambers was removed, and the Matrigel and the cells in the chambers were gently wiped away with a cotton swab. As in the Transwell migration assays, a new 24-well plate with 4% paraformaldehyde was used to fix the cells on the chambers for 20 min, the chambers were removed, crystal violet staining was performed for 20 min, excess crystal violet solution was rinsed off with PBS, and cell images were captured using a microscope.
Xenograft assays were conducted by subcutaneously injecting JOSD2-knockdown KYSE150 cells and JOSD2-overexpressing KYSE30 cells under the armpits of BALB/c nude mice. Tumor dimensions, including length and width, were assessed using a vernier caliper every 3 d. Tumor volume was determined as 0.52 × length × width², and growth curves were plotted. From subcutaneous injection until tumor growth on the 19th day, the tumor tissues were harvested and their weights were recorded.
Additionally, the role of JOSD2 in cisplatin sensitivity in vivo was studied using xenograft and drug sensitivity assays. JOSD2-knockdown KYSE150 cells or JOSD2-overexpressing KYSE30 cells were injected under the armpits of BALB/c nude mice. When the tumor volume reached 10 mm³, cisplatin was intraperitoneally injected (6 mg/kg, every 3 d for 15 d) and the tumor volume was measured at the same time. At approximately 2 wk after the first administration of cisplatin, the tumor tissues were excised and weighed.
To explore the proteins that interact with JOSD2, KYSE30 cells with Flag-tagged JOSD2 (Flag-JOSD2 sequence: GATTACAAGGATGACGACGATAAG) were lysed with protein lysis buffer to obtain the total proteins. Flag-JOSD2 was then enriched by immunoprecipitation. After obtaining the protein precipitate interacting with Flag-JOSD2, the protein complex was subjected to SDS-PAGE, followed by silver staining for band visualization. Specific bands were then subjected to mass spectrometry analysis (Beijing Protein Innovation Co., Ltd., China).
ImageJ software was used to quantify the protein expression levels in Western blot analysis. Graphs were constructed and statistical analyses were performed using GraphPad Prism 10 software (GraphPad Software, Inc., United States). A P value less than 0.05 was considered statistically significant. Student's t-test was used to determine the significance of differences between two groups, while analysis of variance was employed to compare differences among more than two groups.
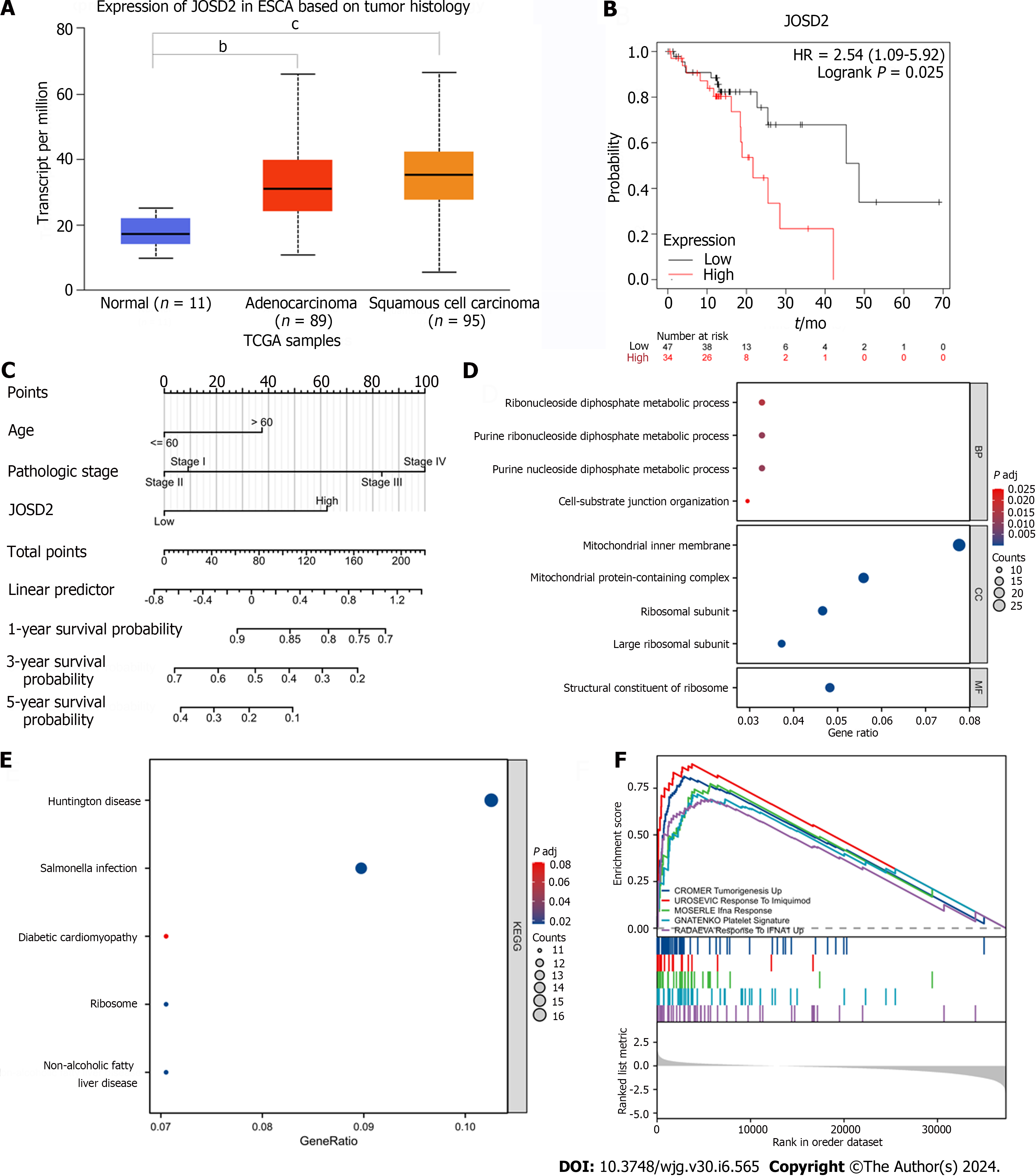
JOSD2 expression was significantly higher in ESCC tissues than normal esophageal tissues (P < 0.0001) (Figure 1A). Additionally, ESCC patients with high JOSD2 expression had a worse prognosis than those with low expression (P = 0.025), providing a basis for predicting the prognosis of ESCC (Figure 1B and C).
To determine the biological functions of JOSD2, GO and KEGG enrichment analyses were performed. The top five most enriched biological process, cellular component, and molecular function (MF) terms are shown in Figure 1D (only one MF term was enriched), and the top five most enriched KEGG pathways are shown in Figure 1E. The top five gene sets in the GSEA, comprising CROMER Tumorigenesis Up, UROSEVIC Response to Imiquimod, MOSERLE IFNA Response, GNATENKO Platelet Signature, and RADAEVA Response to IFNA1 Up, are shown in Figure 1F. A PPI diagram based on the top 100 genes associated with JOSD2 is depicted in Supplementary Figure 1.
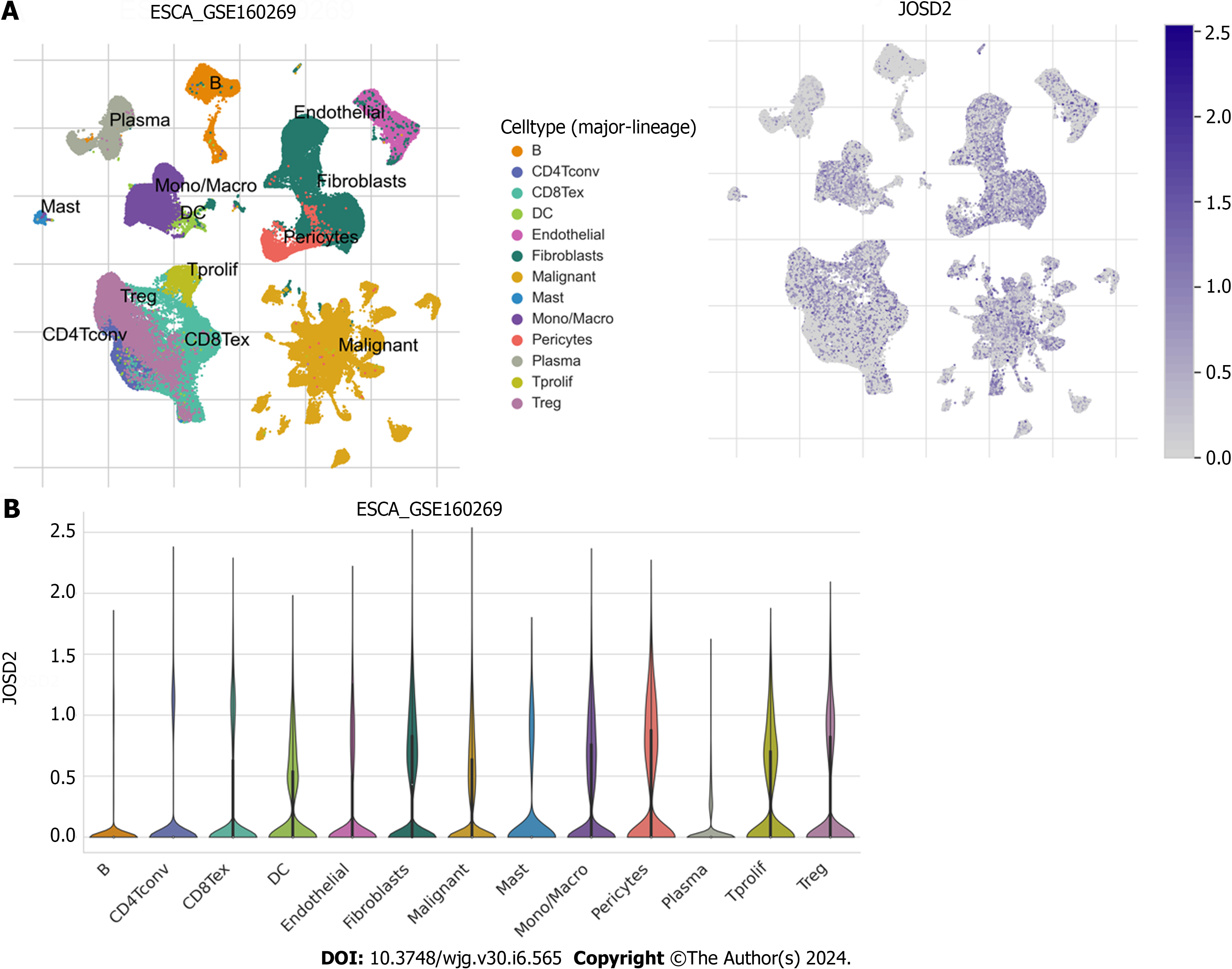
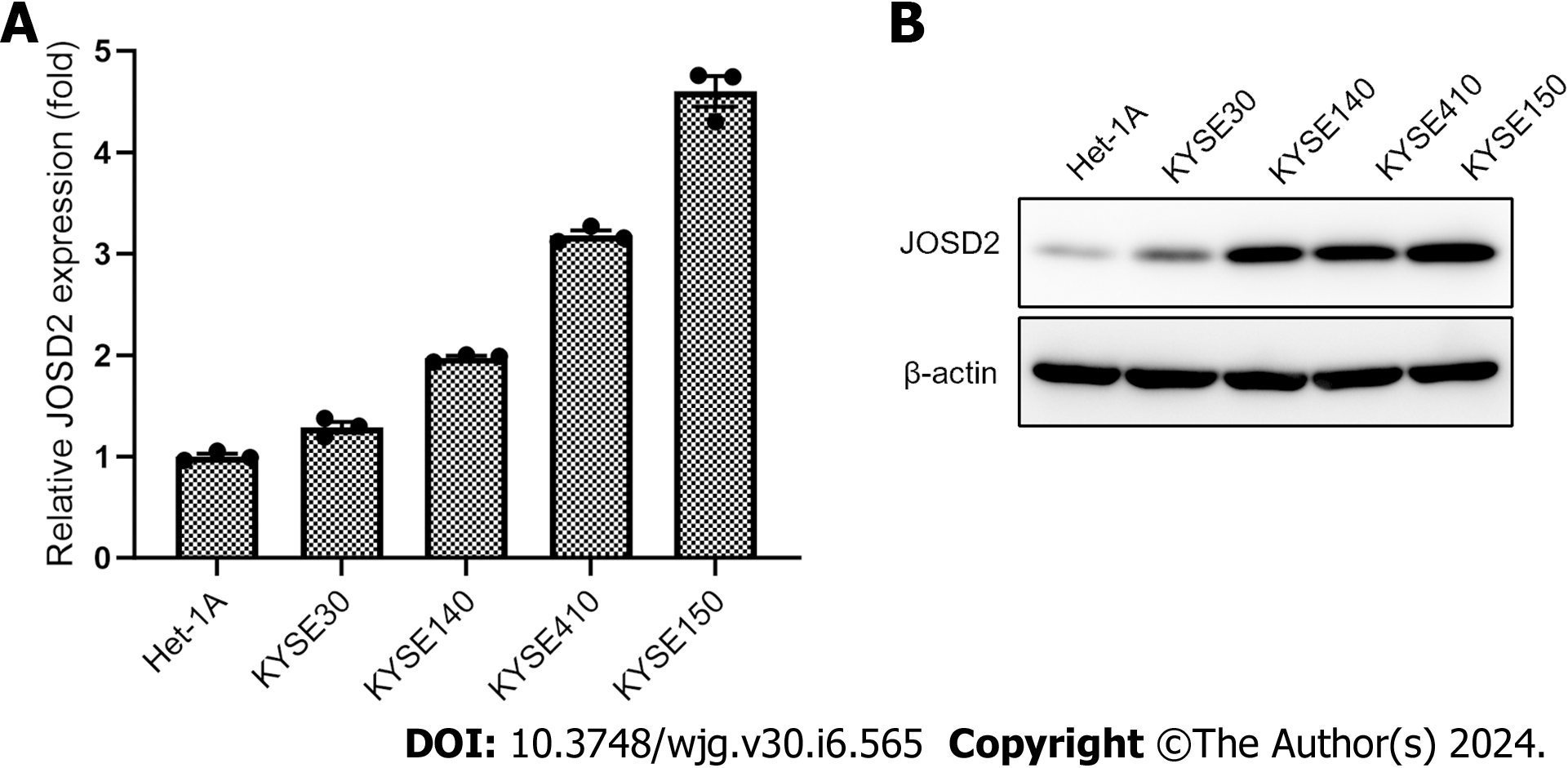
To determine whether JOSD2 is expressed in ESCC cells, single-cell sequencing data in GSE160269 was utilized and JOSD2 was highly expressed in ESCC cells (Figure 2). Additionally, JOSD2 mRNA and protein expression in a normal esophageal epithelial cell line (Het-1A) and ESCC cell lines (KYSE30, KYSE140, KYSE150, and KYSE410) was assessed using RT-qPCR and western blotting, respectively. The results showed that both JOSD2 mRNA (Figure 3A) and protein (Figure 3B) expression were consistently upregulated in ESCC cell lines compared to the normal esophageal cell line.
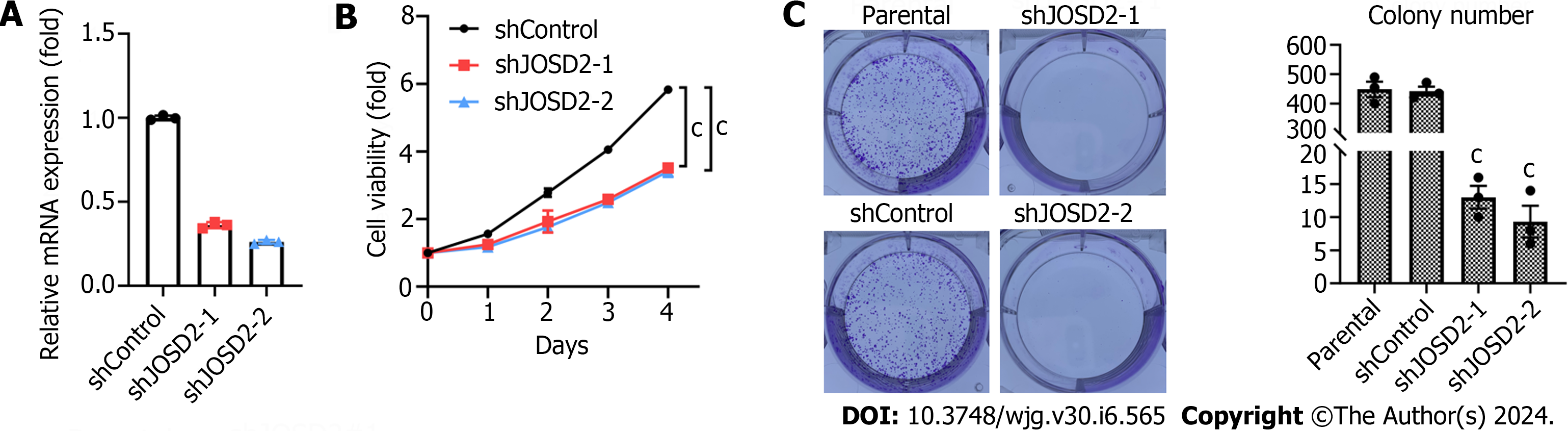
To confirm the function of JOSD2 in ESCC cells, two shRNAs targeting JOSD2 were designed and knockdown assays in the KYSE150 cell line (which had the highest JOSD2 expression among the ESCC cell lines) were conducted. RT-qPCR results showed that both shRNAs achieved effective knockdown (Figure 4A).
Subsequently, CCK-8 cell viability assays were performed to study the influence of JOSD2 knockdown on cell proliferation. The results indicated that both shRNAs significantly inhibited the proliferation of KYSE150 cells (P < 0.0001 for both) (Figure 4B). Colony formation assays were conducted to investigate the effect of JOSD2 knockdown on tumor-forming ability. JOSD2 knockdown significantly suppressed the colony-forming ability of KYSE150 cells (Figure 4C).
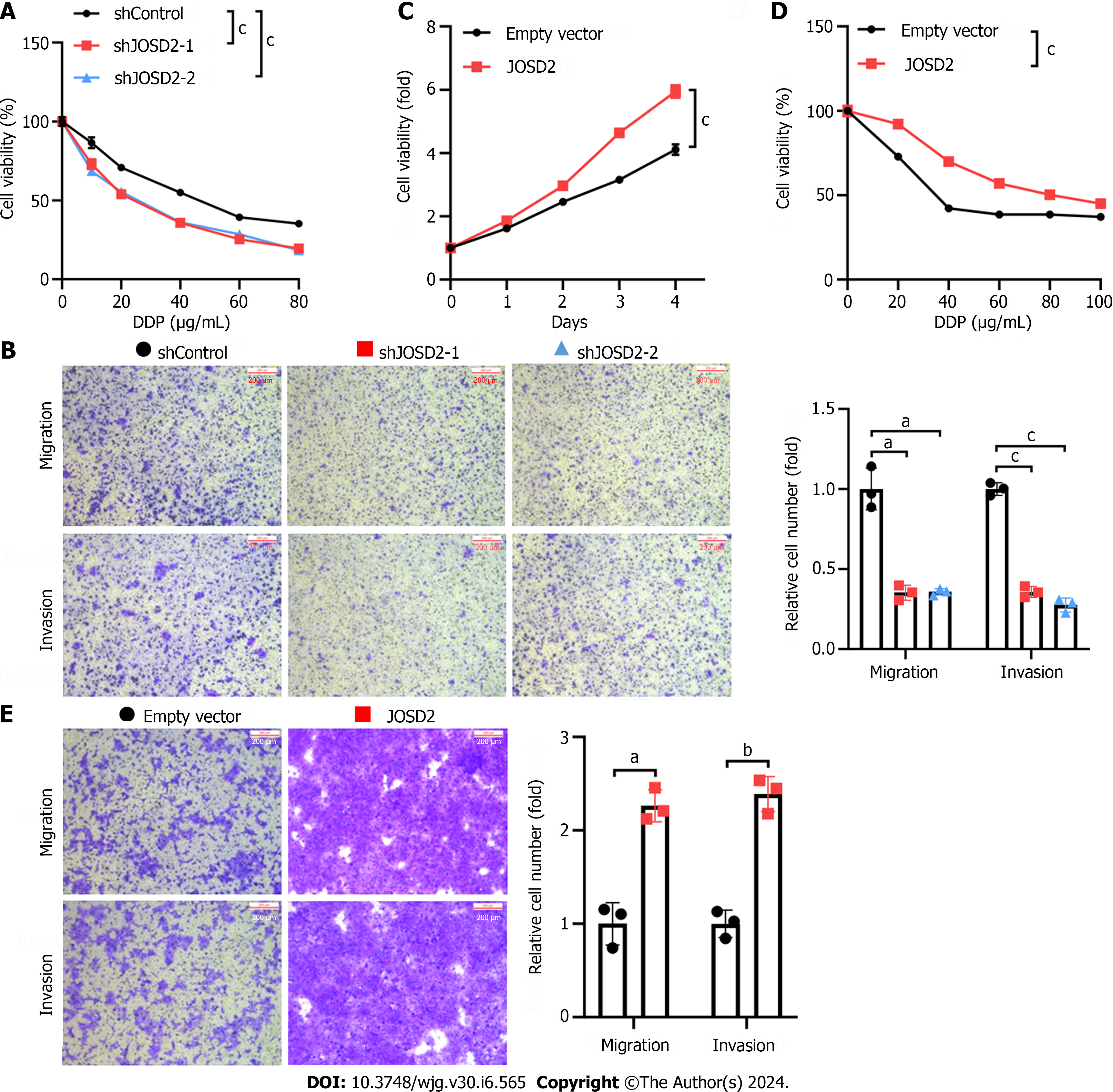
Chemoresistance and distant metastasis are major contributors to the poor prognosis of ESCC patients. Therefore, the next step was to investigate whether JOSD2 also affects the drug resistance and migratory ability of ESCC cells. JOSD2 knockdown increased the sensitivity of KYSE150 cells to 48-h treatment with various concentrations of cisplatin (P < 0.0001 for both) (Figure 5A). Transwell migration and invasion assays also demonstrated that JOSD2 knockdown significantly inhibited the migratory (P < 0.01) and invasion (P < 0.0001) ability of KYSE150 cells (Figure 5B). To validate the promoting role of JOSD2 in the development of ESCC cells, exogenous JOSD2 was overexpressed in KYSE30 cells (which had the lowest JOSD2 expression among the ESCC cell lines). The results showed that JOSD2 overexpression significantly promoted the proliferation (P < 0.0001), drug resistance (P < 0.0001), migration (P < 0.01), and invasion (P < 0.001) capability of KYSE30 cells (Figure 5C-E).
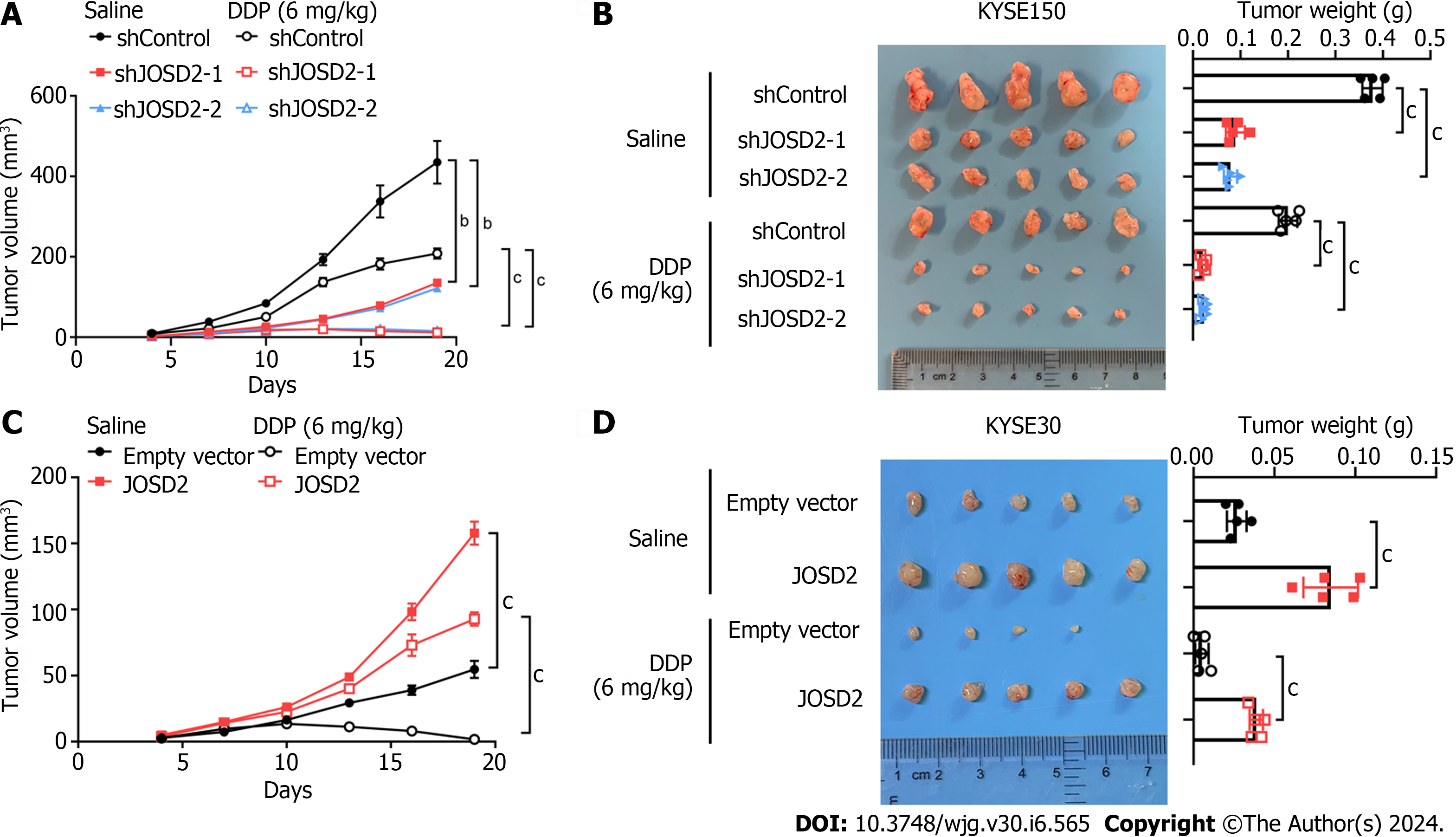
Xenograft assays were conducted in nude mice using JOSD2-knockdown KYSE150 cells, JOSD2-overexpressing KYSE30 cells, and their respective control cells. The mice in each group were divided into subgroups with and without cisplatin treatment to study the effects of JOSD2 on ESCC cell proliferation and drug sensitivity in vivo. JOSD2-knockdown KYSE150 cells exhibited significantly slower tumor growth and a more pronounced reduction in tumor volume under cisplatin treatment compared to control cells (P < 0.001 for tumor volume, P < 0.0001 for tumor weight) (Figure 6A and B). On the other hand, JOSD2-overexpressing KYSE30 cells not only had faster tumor growth but also exhibited significantly increased resistance to cisplatin (P < 0.0001 for tumor volume, P < 0.0001 for tumor weight) (Figure 6C and D). These findings suggested that JOSD2 has a vital role in promoting the development of ESCC.
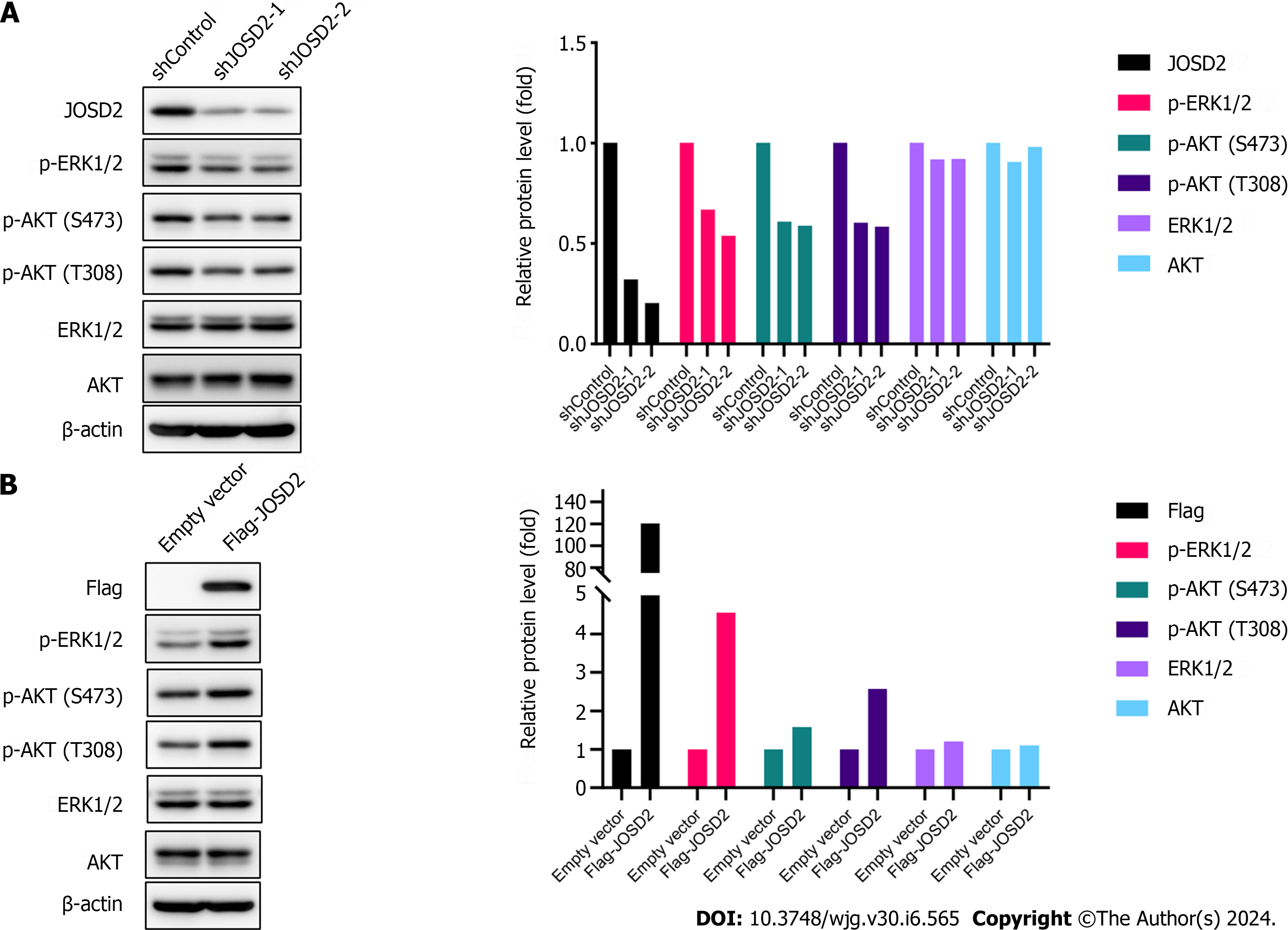
To explore the molecular mechanisms by which JOSD2 promotes ESCC, the effects of JOSD2 on the phosphorylation pathways MAPK/ERK and PI3K/AKT, which play important roles in carcinogenesis, were assessed. Western blotting results showed that JOSD2 knockdown in ESCC cells inhibited the phosphorylation levels of ERK1/2 and AKT (Figure 7A), while JOSD2 overexpression in ESCC cells led to the activation of MAPK/ERK and PI3K/AKT signaling pathways (Figure 7B). These results indicated that the activation of the MAPK/ERK and PI3K/AKT signaling pathways serves as a pivotal downstream mechanism in facilitating the oncogenic function of JOSD2.
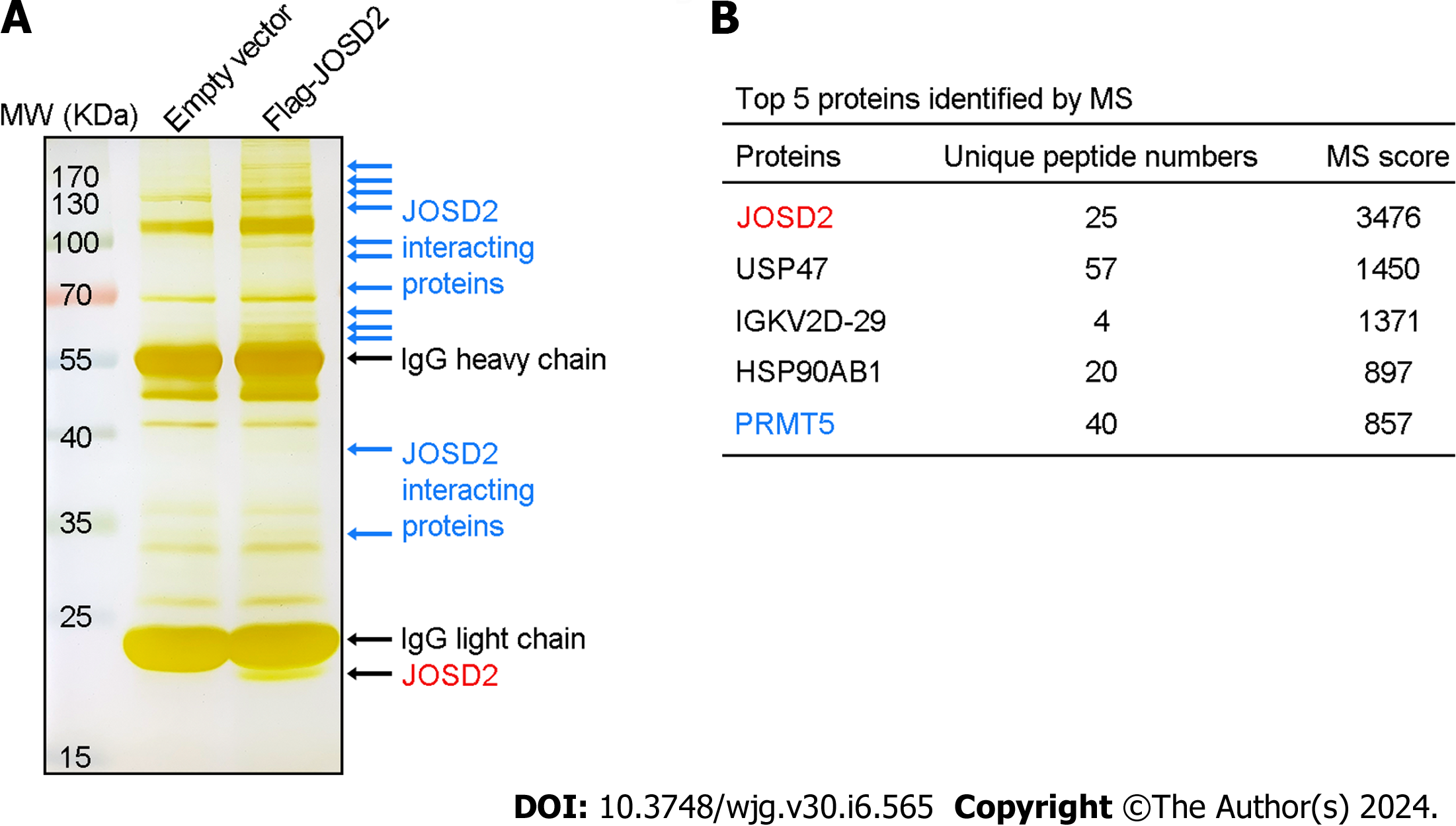
To identify key substrate proteins interacting with JOSD2, anti-Flag magnetic beads were used to enrich Flag-JOSD2 protein and its interacting proteins in KYSE30 cells with exogenous Flag-JOSD2 overexpression. As indicated by the blue arrows in Figure 8A, silver staining of the SDS-PAGE gel revealed numerous protein bands in the overexpression group but not in the control group, indicating that these proteins were specifically immunoprecipitated along with Flag-JOSD2. Mass spectrometry showed that JOSD2 had the highest score (validating the reliability of its enrichment) and the other top-ranked proteins were USP47, IGKV2D-29, HSP90AB1, and PRMT5 (Figure 8B), which indicated that these proteins may be the significant substrates that bind to JOSD2.
In recent years, an increasing number of studies have shown that DUBs play a crucial role in the development of malignant tumors[11,16]. There has been a substantial amount of research on DUBs in five common malignant tumors, namely non-small cell lung cancer, hepatocellular cancer, gastric cancer, colorectal cancer, and breast cancer[8,17-20]. For instance, USP9X, DUB3, and USP7 have been identified in these malignant tumors[21-23]. There has been less research on DUBs in ESCC, and this research has mostly focused on the impact of the known DUBs on the metastatic process of ESCC[24-27]. PSMD14, OTUB1, USP26, and EIF3H, by stabilizing Snail, promote the occurrence of metastasis[24-27]. This redundancy in the regulatory mechanism makes using individual DUBs as effective therapeutic targets challenging.
This study is the first to report on the role of JOSD2 in ESCC. The preliminary findings indicated that JOSD2 is not only highly expressed in ESCC tissues, but its high expression is significantly associated with a poor prognosis. Subsequent analyses revealed that JOSD2 significantly enhanced the proliferation, migration, and drug resistance of ESCC cells. The in vivo results confirmed that altering JOSD2 expression, either by overexpression or knockdown, modulates the resistance of ESCC to the chemotherapy drug cisplatin (one of the primary chemotherapy drugs used to treat ESCC). This highlights the potential significance of using JOSD2 as a therapeutic target in order to overcome cisplatin resistance in ESCC.
Norberg et al[28] reported the role of JOSD2 in lung adenocarcinoma. They analyzed the metabolic profile of lung adenocarcinoma and found that PHGDH, a critical rate-limiting enzyme in serine synthesis, was highly expressed in a subgroup with poor prognosis[28]. Tumors with high PHGDH expression exhibited rapid proliferation and migration. Subsequently, the authors discovered that the protein expression of PHGDH is regulated by the ubiquitin proteasome system pathway. The authors screened for DUBs that stabilize PHGDH by using a siRNA library targeting 99 DUBs for transient knockdown. Targeting the DUB JOSD2, among these 99 DUBs, led to the largest significant reduction (> 80%) in PHGDH protein expression. Thus, JOSD2 affected the metabolism of lung adenocarcinoma by stabilizing PHGDH and promoting tumor growth. The same research team further explored and revealed the relationship between JOSD2 and metabolism[29]. They found that both in vitro and in vivo, JOSD2 directly regulated the metabolic enzyme complex comprising aldolase A, phosphofructokinase 1, and PHGDH. Wild-type JOSD2, but not its enzymatic mutant, stabilized this complex via deubiquitination, enhancing its activity, and thereby increasing the glycolytic rate of cancer cells. The absence of JOSD2 inhibited various cancer cells (including non-small cell lung cancer, breast cancer, and ovarian cancer cells) and reduced glycolysis. In summary, JOSD2 effectively integrated glycometabolism and serine metabolism by stabilizing the metabolic enzyme complex. For cancer types that are highly reliant on glycolysis for their energy supply, increased JOSD2 expression significantly promoted cell proliferation and growth. This finding suggests that JOSD2 is a potential therapeutic target in cancer cells that are dependent on glycolytic metabolism.
Qian et al[11] reported the role of JOSD2 in the progression of cholangiocarcinoma (CCA)[11]. They identified JOSD2 as a crucial regulator that stabilizes Yes-associated protein/Transcriptional co-activator with PDZ-binding motif (YAP/TAZ), which are involved in the malignant progression of CCA. Depleting JOSD2 led to the degradation of YAP/TAZ and thereby significantly inhibited CCA proliferation both in vitro and in vivo. Additionally, there was a positive association between JOSD2 and YAP abundance in CCA patient samples, suggesting that JOSD2 is a potential target for treatment in patients with YAP/TAZ-related CCA. Moreover, Huang et al[9] identified JOSD2 as a novel prognostic indicator for individuals with hepatocellular cancer and identified CTNNB1 as a significant collaborator and downstream protein targeted by JOSD2[9]. However, Lei et al[10] reported that, in acute myeloid leukemia, JOSD2 is a tumor suppressor and PKM2 is a newfound JOSD2-interacting partner, which suggests that JOSD2 has different functions and mechanisms in different malignant tumors[10].
The MAPK/ERK and PI3K/AKT signaling pathways are crucial for key cancer characteristics, including cell proliferation, differentiation, migration, and genomic stability[30,31]. Therefore, we assessed the effects of JOSD2 on these pathways. The results revealed that the phosphorylation activation of MAPK/ERK and PI3K/AKT was a critical downstream event mediating the oncogenic function of JOSD2.
Regarding the key substrate proteins interacting with JOSD2, we found that USP47, IGKV2D-29, HSP90AB1, and PRMT5 were the top four binding proteins of JOSD2 and may also be substrates for JOSD2's activity. USP47, a DUB, can counteract the functions of E3 ubiquitin ligases, playing a role in cell growth and survival processes[32]. Several studies have provided evidence that USP47 is involved in the advancement of diverse cancer types[33-35]. There is limited research on the IGKV2D-29 gene, but polymorphism in this gene was shown to lower the recombination frequency in B cells and to be especially important for immune responses to Haemophilus influenzae type b polysaccharide[36]. HSP90AB1 is a crucial participant in oncogene activity and the preservation of cancer cell viability[37]. This is due to its chaperone mechanism in cancer cells, safeguarding significant amounts of mutated and excessively expressed oncogenic proteins from undergoing misfolding and degradation[37]. Lastly, PRMT5 plays a crucial oncogenic role in various malignancies and has been a key target in recent cancer therapies[38,39]. However, there have been no studies reporting its deubiquitination modification. PRMT5’s role in various malignancies implies that it is likely a key substrate protein for JOSD2's oncogenic function, and JOSD2's deubiquitination of PRMT5 may have significant implications for the treatment of ESCC.
In conclusion, this study reveals the tumorigenic role of JOSD2 in the advancement of ESCC. In terms of the mechanism, JOSD2 influences the phosphorylation activation of MAPK/ERK and PI3K/AKT. USP47, IGKV2D-29, HSP90AB1, and PRMT5 are the four primary proteins that interact with JOSD2 and may serve as substrates for JOSD2's functional activity, especially PRMT5. In 2019, Grasty et al[40] elucidated the molecular structure of the JOSD2 protein, which will facilitate the development of molecular targeted inhibitors of JOSD2. However, there are currently no report on JOSD2 inhibitors. Consequently, there is a need for further exploration of the effects of specific and potent JOSD2 inhibitors on the clinical outlook for ESCC patients.
Esophageal squamous cell carcinoma (ESCC) is a highly lethal malignancy with limited treatment options. Deubiquitinases (DUBs), crucial for maintaining protein homeostasis, are emerging as key players influencing vital cellular processes in ESCC, offering new treatment avenues. In addition, the ongoing development of small molecule inhibitors targeting DUBs shows significant promise, with several preclinical and clinical trials underway.
Recognizing the crucial involvement of DUBs in malignant tumor development, JOSD2, a specific DUB, has been identified as playing a pivotal role in controlling protein deubiquitination and impacting essential cellular processes in cancer. Nevertheless, the function of JOSD2 in ESCC remains uncertain.
The objective of this study was to explore the impact of JOSD2 on the progression of ESCC.
Bioinformatics analyses were used to investigate the expression patterns, prognosis, and enriched pathways of JOSD2 in ESCC tissues. Manipulation of JOSD2 expression in ESCC cell lines (KYSE30 and KYSE150) was achieved through lentiviral transduction. Comprehensive functional assays, encompassing cell proliferation, colony formation, drug sensitivity, migration, and invasion assays, were conducted to unveil the influence of JOSD2 on ESCC cell lines. Additionally, the effects of JOSD2 on xenograft tumor growth and drug sensitivity in vivo were assessed. Proteins interacting with JOSD2 were determined by mass spectrometry.
The initial results suggested that JOSD2 was highly expressed in ESCC tissues and was associated with a poor prognosis. Subsequent investigations revealed upregulation of JOSD2 in ESCC cell lines compared to normal esophageal cells. JOSD2 knockdown inhibited various ESCC cell activities, including proliferation, colony formation, and migration, as well as reducing drug resistance. Conversely, JOSD2 overexpression enhanced these phenotypes. In vivo xenograft assays confirmed that JOSD2 promoted tumor proliferation and drug resistance in ESCC. Mechanistically, JOSD2 appears to activate the MAPK/ERK and PI3K/AKT signaling pathways. Mass spectrometry identified four primary proteins interacting with JOSD2: USP47, IGKV2D-29, HSP90AB1, and PRMT5.
JOSD2 promotes cell proliferation, migration, and drug resistance in ESCC.
JOSD2 is a promising therapeutic target for the treatment of ESCC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Alcántara-Hernández R, Mexico S-Editor: Fan JR L-Editor: A P-Editor: Cai YX
| 1. | Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W, He J. Cancer incidence and mortality in China, 2016. J Nati Cancer Cent. 2022;2:1-9. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 852] [Cited by in RCA: 939] [Article Influence: 313.0] [Reference Citation Analysis (1)] |
| 2. | Ge Z, Leighton JS, Wang Y, Peng X, Chen Z, Chen H, Sun Y, Yao F, Li J, Zhang H, Liu J, Shriver CD, Hu H; Cancer Genome Atlas Research Network, Piwnica-Worms H, Ma L, Liang H. Integrated Genomic Analysis of the Ubiquitin Pathway across Cancer Types. Cell Rep. 2018;23:213-226.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 3. | Leznicki P, Kulathu Y. Mechanisms of regulation and diversification of deubiquitylating enzyme function. J Cell Sci. 2017;130:1997-2006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 4. | Zhang X, Smits AH, van Tilburg GB, Jansen PW, Makowski MM, Ovaa H, Vermeulen M. An Interaction Landscape of Ubiquitin Signaling. Mol Cell. 2017;65:941-955.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 5. | Zhang S, Zhang M, Jing Y, Yin X, Ma P, Zhang Z, Wang X, Di W, Zhuang G. Deubiquitinase USP13 dictates MCL1 stability and sensitivity to BH3 mimetic inhibitors. Nat Commun. 2018;9:215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 6. | Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang SS, Lin WC, Hsu JM, Hsu YH, Kim T, Chang WC, Hsu JL, Yamaguchi H, Ding Q, Wang Y, Yang Y, Chen CH, Sahin AA, Yu D, Hortobagyi GN, Hung MC. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 601] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 7. | Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57-78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 648] [Article Influence: 92.6] [Reference Citation Analysis (0)] |
| 8. | Ge F, Liu X, Zhang H, Yuan T, Zhu H, Yang B, He Q. Deubiquitinating enzyme JOSD2 affects susceptibility of non-small cell lung carcinoma cells to anti-cancer drugs through DNA damage repair. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2023;52:533-543. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (1)] |
| 9. | Huang Y, Zeng J, Liu T, Xu Q, Song X. Deubiquitinating enzyme JOSD2 promotes hepatocellular carcinoma progression through interacting with and inhibiting CTNNB1 degradation. Cell Biol Int. 2022;46:1089-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Lei H, Yang L, Wang Y, Zou Z, Liu M, Xu H, Wu Y. JOSD2 regulates PKM2 nuclear translocation and reduces acute myeloid leukemia progression. Exp Hematol Oncol. 2022;11:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 11. | Qian M, Yan F, Wang W, Du J, Yuan T, Wu R, Zhao C, Wang J, Lu J, Zhang B, Lin N, Dong X, Dai X, Yang B, Zhu H, He Q. Deubiquitinase JOSD2 stabilizes YAP/TAZ to promote cholangiocarcinoma progression. Acta Pharm Sin B. 2021;11:4008-4019. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 12. | Wang Y, Li ZX, Wang JG, Li LH, Shen WL, Dang XW. Deubiquitinating enzyme Josephin-2 stabilizes PHGDH to promote a cancer stem cell phenotype in hepatocellular carcinoma. Genes Genomics. 2023;45:215-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 13. | Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 4194] [Article Influence: 524.3] [Reference Citation Analysis (0)] |
| 14. | Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 1289] [Article Influence: 429.7] [Reference Citation Analysis (0)] |
| 15. | Győrffy B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience. 2023;45:1889-1898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 226] [Reference Citation Analysis (0)] |
| 16. | Jin X, Yan Y, Wang D, Ding D, Ma T, Ye Z, Jimenez R, Wang L, Wu H, Huang H. DUB3 Promotes BET Inhibitor Resistance and Cancer Progression by Deubiquitinating BRD4. Mol Cell. 2018;71:592-605.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 17. | Li L, Liu T, Li Y, Wu C, Luo K, Yin Y, Chen Y, Nowsheen S, Wu J, Lou Z, Yuan J. The deubiquitinase USP9X promotes tumor cell survival and confers chemoresistance through YAP1 stabilization. Oncogene. 2018;37:2422-2431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 18. | Wang J, Liu R, Mo H, Xiao X, Xu Q, Zhao W. Deubiquitinase PSMD7 promotes the proliferation, invasion, and cisplatin resistance of gastric cancer cells by stabilizing RAD23B. Int J Biol Sci. 2021;17:3331-3342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 19. | Wu J, Liu C, Wang T, Liu H, Wei B. Deubiquitinase inhibitor PR-619 potentiates colon cancer immunotherapy by inducing ferroptosis. Immunology. 2023;170:439-451. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Ning Z, Guo X, Liu X, Lu C, Wang A, Wang X, Wang W, Chen H, Qin W, Zhou L, Ma C, Du J, Lin Z, Luo H, Otkur W, Qi H, Chen D, Xia T, Liu J, Tan G, Xu G, Piao HL. USP22 regulates lipidome accumulation by stabilizing PPARγ in hepatocellular carcinoma. Nat Commun. 2022;13:2187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 111] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 21. | Zhang Q, Zhang ZY, Du H, Li SZ, Tu R, Jia YF, Zheng Z, Song XM, Du RL, Zhang XD. DUB3 deubiquitinates and stabilizes NRF2 in chemotherapy resistance of colorectal cancer. Cell Death Differ. 2019;26:2300-2313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 22. | Zhang FK, Ni QZ, Wang K, Cao HJ, Guan DX, Zhang EB, Ma N, Wang YK, Zheng QW, Xu S, Zhu B, Chen TW, Xia J, Qiu XS, Ding XF, Jiang H, Qiu L, Wang X, Chen W, Cheng SQ, Xie D, Li JJ. Targeting USP9X-AMPK Axis in ARID1A-Deficient Hepatocellular Carcinoma. Cell Mol Gastroenterol Hepatol. 2022;14:101-127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 23. | Dai X, Lu L, Deng S, Meng J, Wan C, Huang J, Sun Y, Hu Y, Wu B, Wu G, Lovell JF, Jin H, Yang K. USP7 targeting modulates anti-tumor immune response by reprogramming Tumor-associated Macrophages in Lung Cancer. Theranostics. 2020;10:9332-9347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 178] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 24. | Zhu R, Liu Y, Zhou H, Li L, Li Y, Ding F, Cao X, Liu Z. Deubiquitinating enzyme PSMD14 promotes tumor metastasis through stabilizing SNAIL in human esophageal squamous cell carcinoma. Cancer Lett. 2018;418:125-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 25. | Zhou H, Liu Y, Zhu R, Ding F, Cao X, Lin D, Liu Z. OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene. 2018;37:3356-3368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 26. | Li L, Zhou H, Zhu R, Liu Z. USP26 promotes esophageal squamous cell carcinoma metastasis through stabilizing Snail. Cancer Lett. 2019;448:52-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 27. | Guo X, Zhu R, Luo A, Zhou H, Ding F, Yang H, Liu Z. EIF3H promotes aggressiveness of esophageal squamous cell carcinoma by modulating Snail stability. J Exp Clin Cancer Res. 2020;39:175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 28. | Zhang B, Zheng A, Hydbring P, Ambroise G, Ouchida AT, Goiny M, Vakifahmetoglu-Norberg H, Norberg E. PHGDH Defines a Metabolic Subtype in Lung Adenocarcinomas with Poor Prognosis. Cell Rep. 2017;19:2289-2303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 29. | Krassikova L, Zhang B, Nagarajan D, Queiroz AL, Kacal M, Samakidis E, Vakifahmetoglu-Norberg H, Norberg E. The deubiquitinase JOSD2 is a positive regulator of glucose metabolism. Cell Death Differ. 2021;28:1091-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 30. | Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 836] [Cited by in RCA: 1291] [Article Influence: 129.1] [Reference Citation Analysis (0)] |
| 31. | Akbarzadeh M, Mihanfar A, Akbarzadeh S, Yousefi B, Majidinia M. Crosstalk between miRNA and PI3K/AKT/mTOR signaling pathway in cancer. Life Sci. 2021;285:119984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 32. | Shi J, Liu Y, Xu X, Zhang W, Yu T, Jia J, Liu C. Deubiquitinase USP47/UBP64E Regulates β-Catenin Ubiquitination and Degradation and Plays a Positive Role in Wnt Signaling. Mol Cell Biol. 2015;35:3301-3311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 33. | Peng J, Li W, Tan N, Lai X, Jiang W, Chen G. USP47 stabilizes BACH1 to promote the Warburg effect and non-small cell lung cancer development via stimulating Hk2 and Gapdh transcription. Am J Cancer Res. 2022;12:91-107. [PubMed] |
| 34. | Zhang S, Ju X, Yang Q, Zhu Y, Fan D, Su G, Kong L, Li Y. USP47 maintains the stemness of colorectal cancer cells and is inhibited by parthenolide. Biochem Biophys Res Commun. 2021;562:21-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 35. | Lei H, Xu HZ, Shan HZ, Liu M, Lu Y, Fang ZX, Jin J, Jing B, Xiao XH, Gao SM, Gao FH, Xia L, Yang L, Liu LG, Wang WW, Liu CX, Tong Y, Wu YZ, Zheng JK, Chen GQ, Zhou L, Wu YL. Targeting USP47 overcomes tyrosine kinase inhibitor resistance and eradicates leukemia stem/progenitor cells in chronic myelogenous leukemia. Nat Commun. 2021;12:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 36. | Padyukov L, Hahn-Zoric M, Blomqvist SR, Ulanova M, Welch SG, Feeney AJ, Lau YL, Hanson LA. Distribution of human kappa locus IGKV2-29 and IGKV2D-29 alleles in Swedish Caucasians and Hong Kong Chinese. Immunogenetics. 2001;53:22-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 37. | Haase M, Fitze G. HSP90AB1: Helping the good and the bad. Gene. 2016;575:171-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 38. | Jarrold J, Davies CC. PRMTs and Arginine Methylation: Cancer's Best-Kept Secret? Trends Mol Med. 2019;25:993-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 242] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 39. | Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 726] [Cited by in RCA: 846] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 40. | Grasty KC, Weeks SD, Loll PJ. Structural insights into the activity and regulation of human Josephin-2. J Struct Biol X. 2019;3:100011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |