Published online Sep 21, 2024. doi: 10.3748/wjg.v30.i35.3972
Revised: August 9, 2024
Accepted: August 27, 2024
Published online: September 21, 2024
Processing time: 122 Days and 5.2 Hours
Fusobacterium nucleatum (F. nucleatum) is a Gram-negative anaerobic bacterium that plays a key role in the development of oral inflammation, such as periodontitis and gingivitis. In the last 10 years, F. nucleatum has been identified as a prevalent bacterium associated with colorectal adenocarcinoma and has also been linked to cancer progression, metastasis and poor disease outcome. While the role of F. nucleatum in colon carcinogenesis has been intensively studied, its role in gastric carcinogenesis is still poorly understood. Although Helicobacter pylori infection has histo
Core Tip: Fusobacterium nucleatum (F. nucleatum) plays a key role in the development of oral inflammatory diseases and is a member of the oral microbiota. Recently, however, F. nucleatum has been associated with colorectal cancer and several other malignancies, including gastric cancer (GC). In particular, F. nucleatum has been associated with patient outcomes, suggesting its role in clinical practice. In this review, we summarize the current knowledge on the potential role of F. nucleatum in the development and progression of GC from a basic and translational perspective and highlight its potential therapeutic relevance.
- Citation: Petkevicius V, Lehr K, Kupcinskas J, Link A. Fusobacterium nucleatum: Unraveling its potential role in gastric carcinogenesis. World J Gastroenterol 2024; 30(35): 3972-3984
- URL: https://www.wjgnet.com/1007-9327/full/v30/i35/3972.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i35.3972
Gastric cancer (GC) is not only one of the most common and deadliest tumor types worldwide, but is probably also one of the most representative cancers related to the microbiome and microenvironment[1]. Historically, GC has been recognized as a multifactorial disease caused by the interaction of host, microbial and environmental factors[2,3]. The strongest known risk factor for the disease is chronic infection with Helicobacter pylori (H. pylori), which favors the development of premalignant lesions such as atrophic gastritis (AG) and intestinal metaplasia (IM), which in turn can lead to dysplasia and eventually GC[4-6]. H. pylori infection of the stomach is a complex disease that depends on bacterial virulence factors, host factors and probably other nutritional and environmental factors that can influence not only acid production but all physiological processes in the stomach. For example, H. pylori may contribute to increased acidity with a higher risk of peptic ulcer complications due to antrum-dominant gastritis, or lead to a decrease in pH due to corpus-dominant inflammation and subsequent progression of chronic gastritis (CG) to AG/IM[7]. H. pylori-dependent or -independent changes in the acidic gastric environment can therefore directly reduce the local defense of the highly proliferative gastric epithelial cells, but also the induction of local apoptosis and inflammation with accompanying changes can further contribute to changes in the gastric microbiota that favor the development of GC[8].
The gastric microbiota also has other diverse members besides H. pylori, but the acidic gastric environment and thick protective mucin layer strongly contribute to the limited growth of bacteria in the niches of the stomach. The microbiota of the stomach is therefore less rich than the microbiota of the intestine and oral cavity, but its potential effect on the mucosa is probably at least as great, if not more pronounced[9]. The latest research findings suggest that not only H. pylori but also the entire microbiota of the human stomach can have important effects on the development of GC[10-12].
The proximity to the oral microenvironment suggests that the oral microbiome may play a functionally relevant role in controlling local inflammation in the upper gastrointestinal (GI) tract and especially in the stomach. However, the alteration of the upper GI acid defense mechanism is also likely to affect the distal lower GI tract. Therefore, it is not surprising that the first reports analyzing the lower GI tumor microenvironment and stool samples from patients with colorectal carcinoma demonstrated an enrichment of Fusobacterium nucleatum (F. nucleatum)[13,14]. F. nucleatum is a widespread Gram-negative anaerobic bacterium that is common in the oral cavity and plays a key role in the develop
Before reviewing the existing data on F. nucleatum in GC, it is useful to summarize the evidence from other cancers and specifically from CRC. Initial studies, supported by subsequent reports, have convincingly shown that the level of F. nucleatum is significantly higher in CRC tissue than in adjacent normal tissue. A higher abundance of F. nucleatum has been found in stool samples from CRC patients compared to normal controls. The prevalence of F. nucleatum has been reported to be associated with CRC progression, metastasis and poor disease outcome[24,27-29].
From a molecular point of view, several mechanisms of the role of F. nucleatum in the pathogenesis of CRC have been explored[30]. F. nucleatum has been shown to accelerate cancer development by increasing cell proliferation via the FadA/E-cadherin/β-catenin pathway[31]. FadA binds to E-cadherin, activating β-catenin signaling, which increases the expression of inflammatory and oncogenic genes. This pathway also leads to the overexpression of chk2, a multifunctional enzyme involved in DNA damage[32]. F. nucleatum may promote CRC by regulating gene expression through noncoding RNAs. Yang et al[33] showed that F. nucleatum induces high expression of miR-21 in CRC, which promotes cell proliferation. F. nucleatum promotes the expression of several inflammatory genes and the release of inflammatory cytokines, particularly interleukin (IL)-8, IL-10 and tumor necrosis factor-α, which are associated with activation of the NF-κB pathway, thereby creating a tumor-promoting proinflammatory environment that stimulates colorectal tumor progression[34,35]. Fusobacterial Fap2 protein can inhibit immune cell activity and protect tumors from killing by natural killer cells and tumor-infiltrating T cells through activation of the TIGIT inhibitory receptor[36].
Bullman et al[37] successfully detected and cultured F. nucleatum not only from the primary CRC tumor, but also from the liver metastasis of the same patient. The average nucleotide similarity of F. nucleatum in tumor and metastasis was mostly > 90%. The discovery of nearly identical viable Fusobacterium strains in matched primary and metastatic CRCs confirms the persistence of viable Fusobacterium species during the metastatic process and suggests that Fusobacterium could potentially migrate with CRC cells to the metastatic site.
Abed et al[38] found that CRC-specific recognition by Fusobacteria is mediated by the fusobacterial Fap2 lectin, which specifically recognizes and binds tumor-derived D-galactose-b(1-3)-N-acetyl-D-galactosamine (Gal-GalNAc). Gal-GalNAc is overexpressed in colorectal tumors. High levels of Gal-GalNAc have also been found in adenocarcinomas of the prostate, ovary, colon, uterus, pancreas, breast, lung, esophagus and stomach[39].
It is now well established that the microbiota plays an important role in human gastric health, and changes in the composition of the microbiota may be associated with various diseases[12,40]. The microbiota of the human digestive tract varies in richness and diversity, with the highest numbers of bacteria in the oral cavity and gut, and lower numbers in the esophagus and stomach. Because of its acidic environment, the stomach[41-43] was thought to be a sterile organ until the discovery of H. pylori; however, recent advances in molecular techniques have shown that other microorganisms besides H. pylori can colonize the gastric epithelium[44]. The composition of the gastric microbiota varies considerably between individuals with different conditions, particularly in relation to H. pylori infection, as has been repeatedly shown[45-47]. Although the gastric microbiome can be strongly modified by medication use, dietary habits, age, gender and other factors, the key factor responsible for the gastric microbiome pattern is H. pylori infection[48-51]. Overall, the five most abundant phyla (Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Fusobacteria) were identified using culture dependent, mass spectrometry and sequencing techniques[8,9,52]. H. pylori colonization significantly reduces the overall diversity of the gastric microbiota[46,53]. For example, Maldonado-Contreras et al showed that four phyla (Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes) dominated in the stomach; however, H. pylori infection increased abundance of non-H. pylori-Proteobacteria, Spirochaete, and Acidobacteria, while decreasing the abundance of Actinobacteria, Bacteroidetes and Firmicutes[54]. Klymiuk et al[55] found that the genera Actinomyces, Granulicatella, Veillonella, Fusobacterium, Neisseria, Helicobacter, Streptococcus and Prevotella were significantly different between the H. pylori-positive and H. pylori-negative individuals[55].
Although H. pylori infection is the strongest risk factor for developing GC, other members of the gastric microbiota may influence disease progression. H. pylori infection plays a crucial role in the initial steps of carcinogenesis by causing inflammation of the gastric mucosa, leading to hypochlorhydria and mucosal atrophy[5,6]. A decrease in gastric acidity favors bacterial growth, which contributes to malignant transformation by maintaining inflammation and converting nitrates into N-nitrosamines[56]. Some studies have shown significant differences in the gastric microbiota of patients with gastritis, IM and GC, suggesting that changes in the microbiota are associated with the progression of gastric carcinogenesis[57-59]. Studies of the composition, richness and biodiversity of the microbiota in patients with GC have shown mixed results. Due to the large variation between study cohorts and associated factors, data on the predominant bacteria in the cancer-associated gastric microbiota vary widely.
Wang et al[60] found that five types of bacteria (Lactobacillus, Escherichia–Shigella, Nitrospirae, Burkholderia fungorum and Lachnospiraceae) were enriched in GC[60]. Ferreira et al[61] found that the GC microbiota had an over-representation of Actinobacteria and Firmicutes, and lower abundance of Bacteroidetes and Fusobacteria[61]. Some studies have shown the enrichment of Fusobacteria in GC compared to other disease stages[22,23,62]. Among the various bacteria reported above, Fusobacteria are among the most common associated with the gastric microbiome and are gaining further attention, particularly due to the concurrent association data from CRC and increasing understanding of molecular interactions.
As previously reported, Fusobacteria are one of the five major phyla that dominate a diverse gastric bacterial community, while having the lowest relative abundance of 5%–10%[9,52,53,55,63]. Bik et al[9] found 56 clones and 10 phylotypes of Fusobacteria in the stomach. However, the presence of Fusobacteria is influenced by the highly dominant presence of H. pylori in the stomach, which shows a lower abundance of Fusobacteria[53,55,64,65]. Klymiuk et al[55] showed that the relative abundance of Fusobacteria was significantly lower in H. pylori positive individuals infected with CagA-positive strains. The frequency of Fusobacteria in all samples examined was 2.53%, while it was 3.79% in the H. pylori-negative group. Schulz et al[53] also found a lower relative abundance of all four major phyla, including Fusobacteria, in gastric biopsy samples from H. pylori positive individuals. It has also been shown that operational taxonomic units with the annotation Fusobacterium are significantly reduced in patients treated with proton pump inhibitors[66]. Overall, based on the summarized evidence, Fusobacteria are among the most common phyla reproducibly identified in the gastric environment.
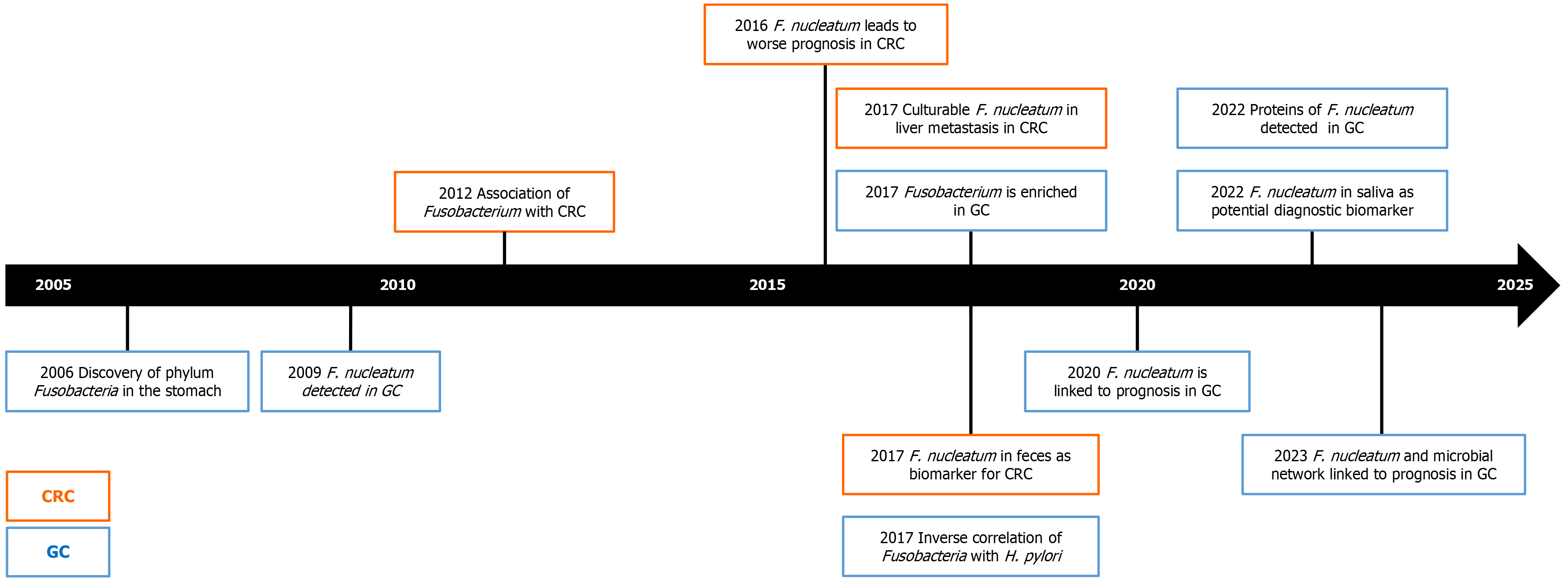
Despite the growing evidence, there are still few studies that have investigated the microbial composition in relation to gastric carcinogenesis. In Table 1, we have summarized the most relevant studies and their characteristics related to the Fusobacteria and F. nucleatum in the stomach and GC. While during the earlier years the focus of GC was rather on other entities, it is obvious that the strong and consistent data on the potential role of F. nucleatum in gastric carcinogenesis is gaining substantial attention. As shown in Figure 1 using a time trend, significant progress has been made in under
| Ref. | Year | Taxonomic rank | Methods | Sample type | Sample origin | No. of subjects | No. of controls | No. of preneoplastic conditions | No. of GC | GC | Major findings | Clinical value | Enriched in GC tissue |
| Bik et al[9] | 2006 | Fusobacteria | 16S rRNA seq | Gastric biopsy | United States | 23 | 11 H. pylori-negative, 12 H. pylori- positive | No | Frequent phyla: Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, and Fusobacteria | ||||
| Dicksved et al[63] | 2009 | Fusobacteria | 16S rRNA seq | Gastric biopsy | Sweden | 15 | 5 controls | 10 GC | Yes | GC microbiota not significantly different from controls, with the phyla Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, and Fusobacteria | |||
| Maldonado-Contreras et al[54] | 2011 | Fusobacteria | 16S rRNA gene microarray | Gastric biopsy | Venezuela | 12 | 12 healthy | No | Inverse correlation of Fusobacteria with Helicobacter | ||||
| Aviles-Jimenez et al[59] | 2014 | Fusobacteria | 16S rRNA gene microarray | Gastric biopsy | Mexico | 15 | 5 gastritis | 5 IM | 5 GC | Yes | Fusobacteria among the most prevalent phyla | ||
| Parsons et al[66] | 2017 | Fusobacterium | 16S rRNA seq | Gastric biopsy | UK | 95 | 20 healthy, 19 PPI treated, 22 gastritis | 34 AG | No | Fusobacterium lower in PPI treated patients | |||
| Yamamura et al[19] | 2017 | F. nucleatum | qPCR | Gastric tissue in paraffin | Japan | 40 | 20 controls | 20 GC | Yes | F. nucleatum detected in GC | |||
| Klymiuk et al[55] | 2017 | Fusobacterium | 16S rRNA seq | Gastric biopsy | Austria | 30 | 30 gastritis | No | Inverse correlation of Fusobacterium with H. pylori | ||||
| Castaño-Rodríguez et al[22] | 2017 | Fusobacterium | 16S rRNA seq | Gastric biopsy | Malaysia, Singapore | 32 | 20 functional dyspepsia | 12 GC | Yes | Fusobacterium enriched in GC | ↑ | ||
| Li et al[58] | 2017 | Fusobacterium | 16S rRNA seq | Gastric biopsy | Hong Kong (China) | 33 | 8 healthy, 9 gastritis | 9 IM | 7 GC | Yes | Fusobacterium is enriched after H. pylori eradication | ↑ | |
| Coker et al[62] | 2018 | Fusobacterium | 16S rRNA seq | Gastric biopsy | China | 207 | 77 gastritis | 74 AG; 17 IM | 39 GC | Yes | Fusobacterium enriched in GC | ↑ | |
| Schulz et al[53] | 2018 | Fusobacterium | 16S rRNA seq | Saliva, gastric juice, duodenal juice, gastric biopsy, duodenal biopsy | Germany | 24 | 8 H. pylori- positive, 16 H. pylori-negative | No | Inverse correlation of Fusobacterium with H. pylori | ||||
| Hsieh et al[23] | 2018 | F. nucleatum | 16S rRNA seq | Gastric biopsy | Taiwan | 27 | 9 gastritis | 7 IM | 11 GC | Yes | Fusobacterium enriched in GC | ↑ | |
| Ferreira et al[61] | 2018 | Fusobacteria | 16S rRNA seq | Gastric biopsy | Portugal | 135 | 81 gastritis | 54 GC | Yes | Fusobacteria lower in GC; correlation with of Helicobacter | ↓ | ||
| Chen et al[69] | 2019 | Fusobacterium | 16S rRNA seq | Gastric tissue | China | 62 | 62 GC + Adj | Yes | Fusobacterium enriched in GC | ↑ | |||
| Shao et al[70] | 2019 | Fusobacteria | 16S rRNA seq | Gastric tissue | China | 103 | 36 GC + Adj | Yes | Fusobacteria enriched in GC | ↑ | |||
| Gantuya et al[64] | 2019 | Fusobacterium | 16S rRNA seq | Gastric biopsy | Mongolia | 75 | 24 controls; 11 H. pylori- positive, 40 H. pylori-negative | No | Inverse correlation of Fusobacteria with H. pylori | ||||
| Guo et al[65] | 2020 | Fusobacterium | 16S rRNA seq | Gastric biopsy | China | 164 | 49 healthy; 115 H. pylori gastritis | No | Inverse correlation of Fusobacterium with H. pylori only in advanced gastric lesion patients | ||||
| Rodriguez et al[91] | 2020 | F. nucleatum | Exome sequencing | Gastric tissue | United States | 85 | 85 GC | Yes | F. nucleatum detected in GC | ||||
| Gantuya et al[74] | 2020 | Fusobacterium | 16S rRNA seq | Gastric biopsy | Mongolia | 168 | 20 gastritis | 40 IM, 40 AG | 48 GC | Yes | Fusobacterium enriched in GC regardless of H. pylori infection | ↑ | |
| Boehm et al[84] | 2020 | F. nucleatum | qPCR | Gastric tissue | Lithuania Germany | 125 | 18 controls; 17 gastritis | 9 AG/IM | 81 GC + Adj | Yes | F. nucleatum-positivity associated with prognosis; lower than in CRC; no difference in AG/IM | Prognosis | |
| Ravegnini et al[78] | 2020 | Fusobacteria | 16S rRNA seq | Gastric tissue in paraffin | Italy | 20 | 20 GC | Yes | signet-ring cell GCs were significantly enriched in the phyla Fusobacteria | ||||
| Zhang et al[72] | 2021 | Fusobacteria | 16S rRNA seq | Stool samples | China | 146 | 61 healthy | 85 GC | Yes | Fusobacteria enriched in GC; Fusobacteria reduced following chemotherapy | ↑ | ||
| Hsieh et al[86] | 2021 | F. nucleatum | Nested PCR | Gastric tissues | Taiwan | 60 | 60 GC | Yes | F. nucleatum+ and H. pylori+ associated with worse prognosis | Prognosis | |||
| Nascimento et al[82] | 2021 | F. nucleatum | qPCR | Gastric biopsies in paraffin | Brazil | 120 | 40 controls | 80 GC | Yes | No difference to controls; F. nucleatum associated with aging | |||
| Xu et al[80] | 2021 | Fusobacterium | 16S rRNA seq | Tongues coating | China | 293 | 112 controls | 181 GC | Yes | Fusobacterium shows diagnostic value | Diagnosis | ↑ | |
| Nie et al[88] | 2021 | Fusobacterium | 16S rRNA seq | Gastric tissue | China | 61 | 61 GC | Yes | Fusobacteria enriched in GC and common in elderly. Fusobacterium positive GC patients more likely to have tumour-infiltrating lymphocytes | ||||
| Gunathilake et al[47] | 2021 | F. nucleatum | 16S rRNA seq | Gastric biopsy | South Korea | 556 | 288 controls | 268 GC | Yes | Fusobacteria enriched in controls | ↓ | ||
| Conti et al[92] | 2021 | Fusobacteria | 16S rRNA seq | Gastric biopsy | Italy | 55 | 32 controls | 23 AG | No | Fusobacteria enriched in controls | |||
| Hsieh et al[87] | 2022 | F. nucleatum | TruSight Oncology 500 panel | Gastric tissue | Taiwan | 36 | 36 GC | Yes | F. nucleatum and high tumour mutation burden are associated with poor prognosis | Prognosis | |||
| Chen et al[93] | 2022 | F. nucleatum | qPCR | Saliva | China | 232 | 20 normal controls; 35 gastritis; 26 gastric polyps | 31 AG | 120 GC | Yes | F. nucleatum in saliva as biomarker for GC | Diagnosis | ↑ |
| Liu et al[94] | 2022 | Fusobacterium | 16S rRNA seq | Gastric biopsy | Columbia, China, Italy, Mexico, South Korea | 1270 | 109 healthy controls, 183 gastritis | 135 AG; 124 IM, 94 IN | 344 GC; 281 GC adj | Yes | Fusobacterium enriched in GC | ↑ | |
| Aziz et al[85] | 2022 | F. nucleatum | Mass spectrometry | Gastric biopsy | Pakistan | 75 | 12 normal controls, 38 gastritis, 12 ulcers | 13 GC | Yes | Protein of F. nucleatum detectable in GC and gastritis | |||
| Zhang et al[81] | 2022 | Fusobacterium | 16S rRNA seq | Saliva, stool, GC tissues | China | 146 | 70 healthy controls | 76 GC | Yes | Fusobacterium enriched in GC stool and saliva sample | ↑ | ||
| Li et al[95] | 2022 | Fusobacteria | 16S rRNA seq | Saliva, esophageal swab, gastric juice, gastric biopsy, stool sample | China | 40 | 22 healthy controls, 18 esophagitis | No | Fusobacteria in ~3% in all regions except faeces | ||||
| Lehr et al[83] | 2023 | F. nucleatum | 16S rRNA seq | Gastric tissue | Lithuania | 64 | 64 GC + Adj | Yes | Higher abundance of F. nucleatum in GC tumour tissue associated with poorer survival; microbial network | Prognosis | |||
| Liu et al[89] | 2023 | F. nucleatum | qPCR | Gastric tissue | China | 91 | 91 GC | Yes | Fusobacterium nucleatum conditions megakaryocytes by induction of extracellular vesicles | ||||
| Wei et al[73] | 2023 | Fusobacterium | 16S rRNA seq | Gastric juice | China | 139 | 61 healthy controls | 78 GC | Yes | Fusobacterium enriched in advanced GC | ↑ | ||
| Nikitina et al[46] | 2023 | Fusobacterium | 16S rRNA seq | Gastric biopsy | Lithuania | 105 | 29 healthy controls | 76 GC | Yes | Fusobacterium abundance increased in tumour DNA versus RNA | |||
| Jeong et al[96] | 2024 | Fusobacteria | Whole metagenome shotgun sequencing | Stool sample | Taiwan | 65 | 33 gastritis | 32 GC | Yes | Fusobacteria enriched in GC | ↑ | ||
| Meng et al[90] | 2024 | F. nucleatum | 16S rRNA seq | Gastric tissue | China | 30 | 30 GC + Adj | Yes | F. nucleatum is more abundant in tumor than in adjacent samples and extracellular vesicles of F. nucleatum promote the growth of GC tumors and metastasis in the liver |
As it has already been mentioned, there is a considerable variation in the results, which can be explained, at least in part, by the multiple factors, including the sample size, the origin of the patients, the H. pylori status, as well as the methodological factors. Therefore, taking into account the heterogeneity, it remains to be determined what role does the microbiome play in gastric carcinogenesis and whether the interaction may include H. pylori or may even go beyond directing carcinogenesis[67,68].
Several studies have been conducted to evaluate the alteration of the microbiome in GC. One of the earliest studies found that the microbiota did not differ significantly between controls and GC patients[63]. However, several studies later found a variety of differences, for example a group from Taiwan analyzed the gastric microbiota in patients with gastritis, IM and GC[23]. The results showed that the prevalence of H. pylori was significantly lower in the gastric microbiota of patients with GC compared to patients with noncancerous gastric diseases, while the abundance of Fusobacterium, Clostridium and Lactobacillus species was higher in patients with GC. Similar results have been reported by other groups. For instance, Chen et al[69] and Shao et al[70] reported that Fusobacterium was more abundant in tumor compared to nontumor tissue.
Wang et al[71] analyzed changes in the gastric microbiome at different stages of neoplastic progression. Bacterial diversity decreased progressively from nonatrophic CG to GC[71-80]. Actinobacteria, Bacteriodes, Firmicutes, Fusobacteria, SR1 and TM7 were more abundant in intraepithelial neoplasia and GC compared to other histological types. Zhang et al[81] reported that Fusobacteria are also enriched in GC, but also in patients who have undergone chemotherapy. In addition, Wei et al[73] found an increased abundance of F. nucleatum in the gastric juice of patients with advanced GC.
The study by Castaño-Rodríguez et al[76] conducted in ethnic Chinese subjects, compared the gastric microbiomes of GC patients and controls with functional dyspepsia. They found that the gastric microbiome was significantly different in GC, and these differences were independent of the contribution of H. pylori to relative abundance, which was also reported by Gantuya et al[74]. Bacterial genera commonly found in the oral cavity including Fusobacterium, Veillonella, Leptotrichia, Haemophilus and Campylobacter were enriched in GC patients. The authors discussed that Fusobacteria may induce inflammation, with Toll-like receptor (TLR)4 and autophagy playing an important role, while polymorphisms in TLR4 and autophagy have been shown to increase the risk of developing GC in Chinese individuals[75,76]. In addition, Fusobacteria have been shown to coaggregate with other bacteria to form biofilms, which play an important role in colorectal adenocarcinoma, and Veillonella species could be a plausible pro-oncogenic partner, as enrichment of these bacteria has been found in other types of cancer[77].
Ravegnini et al[78] identified potential microbial biomarkers associated with GC subtypes. In signet-ring cell carcinoma, the phyla Fusobacteria, Bacteroidetes and Patescibacteria were significantly enriched, whereas, in the adenocarcinoma type, Proteobacteria and Acidobacteria were found.
Based on the observation of an increased abundance of Fusobacteria or F. nucleatum in GC, efforts have been made to evaluate the potential value of the microbiome as a diagnostic biomarker. For example, the combination of F. nucleatum and Clostridium colicanis in tissue has been proposed as a diagnostic biomarker for GC with a sensitivity of 73% and specificity of 100%[23]. While the tumor microbiome may be of limited translational and clinical value for diagnostic purposes, the potential differences in the microbiome are still helpful in identifying potential species in saliva, blood or faces. In particular, the vicinity of the oral and gastric niches may be most exciting in terms of identifying non-invasive microbiome-based biomarkers for GC.
The over-representation of the members of the oral microbiome in GC was reported by Coker et al[62] in a Chinese population. They observed that 14 of the 21 bacterial taxa enriched in GC were bacteria commonly found in the oral cavity, such as Parvimonas micra, Peptostreptococcus stomatis, Gemella and F. nucleatum. After adjusting for potential confounders, including age, sex, tissue location and H. pylori status, the differences between GC and all precancerous stages remained statistically significant. The diversity and richness of the microbiota was increased in tumor tissue. The enriched bacterial taxa were represented by oral bacteria such as Peptostreptococcus, Streptococcus and Fusobacterium, while lactic acid producing bacteria such as Lactococcus lactis and Lactobacillus brevis were more abundant in adjacent nontumor tissue. The association between periodontal disease caused by oral microbial dysbiosis and GC was confirmed in a meta-analysis of observational studies[79].
Going one further Xu et al[80] and also Zhang et al[81] investigated the differences of the microbiome and specifically F. nucleatum in oral specimens from GC patients. They showed that both tongue coating and saliva samples can be used as potential biomarkers for GC patients.
Although several studies have reported the higher abundance of Fusobacterium in GC patients, there are also studies that have not been able to confirm the data. Specifically, Nascimento et al[82] found no differences in the abundance of F. nucleatum between GC patients and noncancer controls, but an association of F. nucleatum with age has been reported. Similarly, Ferreira et al[61] even reported a decrease in the relative abundance of Fusobacteria in patients with GC compared to patients with gastritis, 0.5% and 1.8%, respectively. The enrichment of other bacterial genera, mostly represented by intestinal commensals, was found. Two other studies reported a higher abundance of Fusobacterium in GC tissues compared to adjacent tissues, but the data did not reach significance using either quantitative qPCR analysis or 16S rRNA sequencing[83,84]. In addition, the authors also included the subcohort of subjects with normal mucosa, CG including H. pylori and preneoplastic conditions such as AG and IM, but although the variation in GC samples was much higher than in the non-neoplastic cohort, the differences did not reach quantitatively significant differences[84]. It is worth mentioning that all these studies did not include Asian population but were from Brazil, Portugal, Lithuania and Germany, suggesting that many factors may be attributed to the variation of microbiome and Fusobacteria in gastric mucosa and GC.
Most detections of F. nucleatum in the stomach are based on sequencing techniques, but these results do not guarantee that the bacteria detected are transcriptionally active. Recently, Aziz et al[85] found F. nucleatum proteins in gastric biopsies from patients with GC, demonstrating that these bacteria are metabolically active in this biological niche. This supports the hypothesis that F. nucleatum plays a mechanistic role in GC. Furthermore, Nikitina et al[46] have also reported differences between 16S rRNA data depending on the samples used, either DNA or RNA, suggesting that this variable may also contribute to the differences between studies.
Analysis of the microbiota in gastric tumors not only provides evidence of its potential role in cancer initiation, but may also lead to the identification of its potential impact on prognosis and prediction of treatment response. In particular, the first evidence has been reproducibly reported for CRC showing an association between F. nucleatum and worse prognosis of CRC patients, including poor overall survival[29].
Boehm et al[84] performed the first analysis to investigate the prognostic role of Fusobacterium and specifically F. nucleatum in GC patients. Using a qPCR-based quantitative targeted approach, the authors found that F. nucleatum positivity was associated with significantly worse overall survival in patients with GC, specifically Lauren's diffuse type GC. These findings were further supported by Hsieh et al[86] in a GC patient cohort from Taiwan. The authors also showed that the combination of F. nucleatum infection and high tumor mutation burden is a highly effective biomarker for poor prognosis[87]. In these studies, F. nucleatum was detected by qPCR, but last year Lehr et al[83] showed in a 16S rRNA-based study that F. nucleatum in tumor tissue, but not in adjacent tissue, was associated with worse overall survival, independent of GC classification. In particular, phylogenetic tree analysis linked F. nucleatum and in some cases Fusobacterium necrophorum to GC, while Fusobacterium peridonticum was also frequently observed in healthy controls[83].
The mechanistic reason for this effect on survival has not been elucidated and mechanistic data are generally limited. Abed et al[39] found high levels of Gal-GalNAc in normal and cancerous gastric cells, suggesting that Fusobacteria can enter and accumulate in gastric cells and may be involved in carcinogenesis by inducing inflammation and inhibiting immune cell activity. It has also recently been reported that GC patients infected with Fusobacterium sp. are more likely to have tumor-infiltrating lymphocytes and p53 expression[88].
Liu et al[89] recently demonstrated that F. nucleatum colonization is associated with the occurrence of portal vein thrombosis in patients with GC. However, the underlying mechanism by which F. nucleatum promotes thrombosis remained unclear. They also found that F. nucleatum-positive patients had increased neutrophil extracellular traps and thrombocyte counts. Extracellular vesicles from F. nucleatum-positive patients were able to promote the differentiation and maturation of megakaryocytes and had upregulated 14-3-3 proteins, particularly 14-3-3ε. Upregulation of 14-3-3ε promotes megakaryocyte differentiation and maturation in vitro[89].
The extracellular vesicles of F. nucleatum appear to play a major role in other molecular mechanisms in GC. In vivo analyses have shown that they not only promote the growth of GC tumors and metastasis in the liver, but also the resistance of GC tumors to oxaliplatin[90]. Together with the results of Liu et al[89], this makes the extracellular vesicles of F. nucleatum a highly exciting field of research for the future.
Although limited evidence is available to elucidate the translational role of microbial deregulation and F. nucleatum positivity, the evidence from other cancers may help to explore the pathogenic mechanisms of F. nucleatum in GC are unknown.
GC remains a common malignancy worldwide, characterized by high incidence and poor prognosis. This review provides a comprehensive overview of the current knowledge on microbiome deregulation, with a special focus on Fusobacteria and F. nucleatum in GC. Despite accumulating evidence implicating Fusobacterium in GC pathogenesis, several research gaps remain. First, there is considerable heterogeneity in F. nucleatum positivity among reported GC data, which may be due to differences in the demographics of the cohorts studied (Asian vs non-Asian populations). Therefore, future studies should aim to provide more robust evidence on this issue. Second, although sample sizes in studies, particularly those investigating the diagnostic role of F. nucleatum, are generally adequate, further improvements are warranted. In particular, analysis of microbiome data at higher taxonomic levels may not provide superior value given the predominant association of F. nucleatum with GC. Thirdly, the mere identification of F. nucleatum in tumor tissue is not sufficient to establish its carcinogenic role, especially as samples with preneoplastic conditions do not show significant differences from controls. One of the most intriguing aspects of F. nucleatum in GC is its potential prognostic significance. Although not yet translated into clinical implications for GC patients, it suggests the emergence of novel therapeutic strategies. These may include specific targeted therapeutics or even bacteriophages designed to selectively target F. nucleatum. In summary, our understanding of the contribution of the microbiome to GC has advanced significantly. F. nucleatum is emerging as a reproducible and clinically relevant species, providing insights beyond H. pylori infection. Going forward, high-quality data are essential to comprehensively characterize microbial alterations and explore translational implications, including novel therapeutic strategies and mechanistic insights.
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55853] [Article Influence: 7979.0] [Reference Citation Analysis (132)] |
| 2. | Malfertheiner P, Camargo MC, El-Omar E, Liou JM, Peek R, Schulz C, Smith SI, Suerbaum S. Helicobacter pylori infection. Nat Rev Dis Primers. 2023;9:19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 401] [Article Influence: 200.5] [Reference Citation Analysis (1)] |
| 3. | Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 726] [Article Influence: 103.7] [Reference Citation Analysis (1)] |
| 4. | Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136:487-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 697] [Article Influence: 69.7] [Reference Citation Analysis (0)] |
| 5. | Wroblewski LE, Peek RM Jr. Helicobacter pylori in gastric carcinogenesis: mechanisms. Gastroenterol Clin North Am. 2013;42:285-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 6. | Servetas SL, Bridge DR, Merrell DS. Molecular mechanisms of gastric cancer initiation and progression by Helicobacter pylori. Curr Opin Infect Dis. 2016;29:304-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | El-Omar EM. Mechanisms of increased acid secretion after eradication of Helicobacter pylori infection. Gut. 2006;55:144-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Dias-Jácome E, Libânio D, Borges-Canha M, Galaghar A, Pimentel-Nunes P. Gastric microbiota and carcinogenesis: the role of non-Helicobacter pylori bacteria - A systematic review. Rev Esp Enferm Dig. 2016;108:530-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 9. | Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 730] [Cited by in RCA: 780] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 10. | Wang LL, Yu XJ, Zhan SH, Jia SJ, Tian ZB, Dong QJ. Participation of microbiota in the development of gastric cancer. World J Gastroenterol. 2014;20:4948-4952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 57] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | Noto JM, Peek RM Jr. The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS Pathog. 2017;13:e1006573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 135] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 12. | Rajilic-Stojanovic M, Figueiredo C, Smet A, Hansen R, Kupcinskas J, Rokkas T, Andersen L, Machado JC, Ianiro G, Gasbarrini A, Leja M, Gisbert JP, Hold GL. Systematic review: gastric microbiota in health and disease. Aliment Pharmacol Ther. 2020;51:582-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 13. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1498] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 14. | Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol. 2015;46:1135-1140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 15. | Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13:25-36. [PubMed] |
| 16. | Shang FM, Liu HL. Fusobacterium nucleatum and colorectal cancer: A review. World J Gastrointest Oncol. 2018;10:71-81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 192] [Cited by in RCA: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 17. | Hashemi Goradel N, Heidarzadeh S, Jahangiri S, Farhood B, Mortezaee K, Khanlarkhani N, Negahdari B. Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J Cell Physiol. 2019;234:2337-2344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 18. | Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, Sawayama H, Kinoshita K, Ishimoto T, Iwatsuki M, Sakamoto Y, Yamashita Y, Yoshida N, Watanabe M, Baba H. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin Cancer Res. 2016;22:5574-5581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 324] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 19. | Yamamura K, Baba Y, Miyake K, Nakamura K, Shigaki H, Mima K, Kurashige J, Ishimoto T, Iwatsuki M, Sakamoto Y, Yamashita Y, Yoshida N, Watanabe M, Baba H. Fusobacterium nucleatum in gastroenterological cancer: Evaluation of measurement methods using quantitative polymerase chain reaction and a literature review. Oncol Lett. 2017;14:6373-6378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, Kimura Y, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209-7220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 288] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 21. | Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, Yajuk O, Isaacson B, Abed J, Maalouf N, Nissan A, Sandbank J, Yehuda-Shnaidman E, Ponath F, Vogel J, Mandelboim O, Granot Z, Straussman R, Bachrach G. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. 2020;11:3259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 403] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 22. | Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7:15957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 23. | Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, Deng YF, Hsu WT, Wu CS, Li C. Increased Abundance of Clostridium and Fusobacterium in Gastric Microbiota of Patients with Gastric Cancer in Taiwan. Sci Rep. 2018;8:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 24. | Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, Wan YJ, Nie YQ. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227-3233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 141] [Cited by in RCA: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 25. | Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S, Shindo Y, Hazama S, Oka M, Nagano H, Sakaida I, Yamasaki T. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. 2017;54:86-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 26. | Amitay EL, Werner S, Vital M, Pieper DH, Höfler D, Gierse IJ, Butt J, Balavarca Y, Cuk K, Brenner H. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis. 2017;38:781-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 27. | Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 385] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 28. | Chen S, Zhang L, Li M, Zhang Y, Sun M, Wang L, Lin J, Cui Y, Chen Q, Jin C, Li X, Wang B, Chen H, Zhou T, Wang L, Hsu CH, Zhuo W. Fusobacterium nucleatum reduces METTL3-mediated m(6)A modification and contributes to colorectal cancer metastasis. Nat Commun. 2022;13:1248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 145] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 29. | Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973-1980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 756] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 30. | Wang S, Liu Y, Li J, Zhao L, Yan W, Lin B, Guo X, Wei Y. Fusobacterium nucleatum Acts as a Pro-carcinogenic Bacterium in Colorectal Cancer: From Association to Causality. Front Cell Dev Biol. 2021;9:710165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 31. | Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1511] [Cited by in RCA: 1669] [Article Influence: 139.1] [Reference Citation Analysis (1)] |
| 32. | Guo P, Tian Z, Kong X, Yang L, Shan X, Dong B, Ding X, Jing X, Jiang C, Jiang N, Yu Y. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J Exp Clin Cancer Res. 2020;39:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 108] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 33. | Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer MP, Wang P, Cai S, Goel A, Qin H, Ma Y. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851-866.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 716] [Article Influence: 89.5] [Reference Citation Analysis (1)] |
| 34. | Quah SY, Bergenholtz G, Tan KS. Fusobacterium nucleatum induces cytokine production through Toll-like-receptor-independent mechanism. Int Endod J. 2014;47:550-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1659] [Cited by in RCA: 1919] [Article Influence: 159.9] [Reference Citation Analysis (0)] |
| 36. | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 978] [Article Influence: 97.8] [Reference Citation Analysis (0)] |
| 37. | Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 1064] [Article Influence: 133.0] [Reference Citation Analysis (0)] |
| 38. | Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 581] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 39. | Abed J, Maalouf N, Parhi L, Chaushu S, Mandelboim O, Bachrach G. Tumor Targeting by Fusobacterium nucleatum: A Pilot Study and Future Perspectives. Front Cell Infect Microbiol. 2017;7:295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 40. | Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2636] [Cited by in RCA: 2256] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 41. | Vilchez-Vargas R, Salm F, Znalesniak EB, Haupenthal K, Schanze D, Zenker M, Link A, Hoffmann W. Profiling of the Bacterial Microbiota along the Murine Alimentary Tract. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Vasapolli R, Schütte K, Schulz C, Vital M, Schomburg D, Pieper DH, Vilchez-Vargas R, Malfertheiner P. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology. 2019;157:1081-1092.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 43. | Alarcón T, Llorca L, Perez-Perez G. Impact of the Microbiota and Gastric Disease Development by Helicobacter pylori. Curr Top Microbiol Immunol. 2017;400:253-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 44. | Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. 2013;4:505-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 45. | Smet A, Kupcinskas J, Link A, Hold GL, Bornschein J. The Role of Microbiota in Gastrointestinal Cancer and Cancer Treatment: Chance or Curse? Cell Mol Gastroenterol Hepatol. 2022;13:857-874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 46. | Nikitina D, Lehr K, Vilchez-Vargas R, Jonaitis LV, Urba M, Kupcinskas J, Skieceviciene J, Link A. Comparison of genomic and transcriptional microbiome analysis in gastric cancer patients and healthy individuals. World J Gastroenterol. 2023;29:1202-1218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 15] [Article Influence: 7.5] [Reference Citation Analysis (2)] |
| 47. | Gunathilake M, Lee J, Choi IJ, Kim YI, Kim J. Association between bacteria other than Helicobacter pylori and the risk of gastric cancer. Helicobacter. 2021;26:e12836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 48. | Schütte K, Schulz C, Vilchez-Vargas R, Vasapolli R, Palm F, Simon B, Schomburg D, Lux A, Geffers R, Pieper DH, Link A, Malfertheiner P. Impact of healthy aging on active bacterial assemblages throughout the gastrointestinal tract. Gut Microbes. 2021;13:1966261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 49. | Vilchez-Vargas R, Skieceviciene J, Lehr K, Varkalaite G, Thon C, Urba M, Morkūnas E, Kucinskas L, Bauraite K, Schanze D, Zenker M, Malfertheiner P, Kupcinskas J, Link A. Gut microbial similarity in twins is driven by shared environment and aging. EBioMedicine. 2022;79:104011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 50. | Vesper BJ, Jawdi A, Altman KW, Haines GK 3rd, Tao L, Radosevich JA. The effect of proton pump inhibitors on the human microbiota. Curr Drug Metab. 2009;10:84-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 51. | Yang I, Nell S, Suerbaum S. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev. 2013;37:736-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 52. | Péré-Védrenne C, Flahou B, Loke MF, Ménard A, Vadivelu J. Other Helicobacters, gastric and gut microbiota. Helicobacter. 2017;22 Suppl 1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 53. | Schulz C, Schütte K, Koch N, Vilchez-Vargas R, Wos-Oxley ML, Oxley APA, Vital M, Malfertheiner P, Pieper DH. The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut. 2018;67:216-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 146] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 54. | Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 55. | Klymiuk I, Bilgilier C, Stadlmann A, Thannesberger J, Kastner MT, Högenauer C, Püspök A, Biowski-Frotz S, Schrutka-Kölbl C, Thallinger GG, Steininger C. The Human Gastric Microbiome Is Predicated upon Infection with Helicobacter pylori. Front Microbiol. 2017;8:2508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 56. | Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 484] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 57. | Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, Song KS, Kim YS, Kim JF. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19:407-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 220] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 58. | Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK. Alterations in Gastric Microbiota After H. Pylori Eradication and in Different Histological Stages of Gastric Carcinogenesis. Sci Rep. 2017;7:44935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 138] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 59. | Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. 2014;4:4202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 264] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 60. | Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, Dong Q. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. 2016;28:261-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 158] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 61. | Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 341] [Cited by in RCA: 476] [Article Influence: 68.0] [Reference Citation Analysis (1)] |
| 62. | Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, Yu J. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67:1024-1032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 476] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 63. | Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009;58:509-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 242] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 64. | Gantuya B, El-Serag HB, Matsumoto T, Ajami NJ, Oyuntsetseg K, Azzaya D, Uchida T, Yamaoka Y. Gastric Microbiota in Helicobacter pylori-Negative and -Positive Gastritis Among High Incidence of Gastric Cancer Area. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 65. | Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L, Vieth M, Ma JL, Bajbouj M, Suchanek S, Liu WD, Ulm K, Quante M, Li ZX, Zhou T, Schmid R, Classen M, Li WQ, You WC, Pan KF. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut. 2020;69:1598-1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 215] [Article Influence: 43.0] [Reference Citation Analysis (1)] |
| 66. | Parsons BN, Ijaz UZ, D'Amore R, Burkitt MD, Eccles R, Lenzi L, Duckworth CA, Moore AR, Tiszlavicz L, Varro A, Hall N, Pritchard DM. Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of Helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PLoS Pathog. 2017;13:e1006653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 67. | Stewart OA, Wu F, Chen Y. The role of gastric microbiota in gastric cancer. Gut Microbes. 2020;11:1220-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 68. | Yang J, Zhou X, Liu X, Ling Z, Ji F. Role of the Gastric Microbiome in Gastric Cancer: From Carcinogenesis to Treatment. Front Microbiol. 2021;12:641322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 69. | Chen XH, Wang A, Chu AN, Gong YH, Yuan Y. Mucosa-Associated Microbiota in Gastric Cancer Tissues Compared With Non-cancer Tissues. Front Microbiol. 2019;10:1261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 70. | Shao D, Vogtmann E, Liu A, Qin J, Chen W, Abnet CC, Wei W. Microbial characterization of esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high-risk region of China. Cancer. 2019;125:3993-4002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 71. | Wang Z, Gao X, Zeng R, Wu Q, Sun H, Wu W, Zhang X, Sun G, Yan B, Wu L, Ren R, Guo M, Peng L, Yang Y. Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis. Front Microbiol. 2020;11:997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 72. | Zhang Y, Shen J, Shi X, Du Y, Niu Y, Jin G, Wang Z, Lyu J. Gut microbiome analysis as a predictive marker for the gastric cancer patients. Appl Microbiol Biotechnol. 2021;105:803-814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 73. | Wei Q, Zhang Q, Wu Y, Han S, Yin L, Zhang J, Gao Y, Shen H, Zhuang J, Chu J, Liu J, Wei Y. Analysis of bacterial diversity and community structure in gastric juice of patients with advanced gastric cancer. Discov Oncol. 2023;14:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 74. | Gantuya B, El Serag HB, Matsumoto T, Ajami NJ, Uchida T, Oyuntsetseg K, Bolor D, Yamaoka Y. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Aliment Pharmacol Ther. 2020;51:770-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 75. | Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, Li Q, Wu J, Fu X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017;8:31802-31814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 76. | Castaño-Rodríguez N, Kaakoush NO, Pardo AL, Goh KL, Fock KM, Mitchell HM. Genetic polymorphisms in the Toll-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer. Hum Immunol. 2014;75:808-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 77. | Geng J, Song Q, Tang X, Liang X, Fan H, Peng H, Guo Q, Zhang Z. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 2014;6:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 78. | Ravegnini G, Fosso B, Saverio VD, Sammarini G, Zanotti F, Rossi G, Ricci M, D'Amico F, Valori G, Ioli A, Turroni S, Brigidi P, Hrelia P, Angelini S. Gastric Adenocarcinomas and Signet-Ring Cell Carcinoma: Unraveling Gastric Cancer Complexity through Microbiome Analysis-Deepening Heterogeneity for a Personalized Therapy. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 79. | Yin XH, Wang YD, Luo H, Zhao K, Huang GL, Luo SY, Peng JX, Song JK. Association between Tooth Loss and Gastric Cancer: A Meta-Analysis of Observational Studies. PLoS One. 2016;11:e0149653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 80. | Xu S, Xiang C, Wu J, Teng Y, Wu Z, Wang R, Lu B, Zhan Z, Wu H, Zhang J. Tongue Coating Bacteria as a Potential Stable Biomarker for Gastric Cancer Independent of Lifestyle. Dig Dis Sci. 2021;66:2964-2980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 81. | Zhang C, Hu A, Li J, Zhang F, Zhong P, Li Y, Li Y. Combined Non-Invasive Prediction and New Biomarkers of Oral and Fecal Microbiota in Patients With Gastric and Colorectal Cancer. Front Cell Infect Microbiol. 2022;12:830684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 82. | Nascimento Araujo CD, Amorim AT, Barbosa MS, Alexandre JCPL, Campos GB, Macedo CL, Marques LM, Timenetsky J. Evaluating the presence of Mycoplasma hyorhinis, Fusobacterium nucleatum, and Helicobacter pylori in biopsies of patients with gastric cancer. Infect Agent Cancer. 2021;16:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 83. | Lehr K, Nikitina D, Vilchez-Vargas R, Steponaitiene R, Thon C, Skieceviciene J, Schanze D, Zenker M, Malfertheiner P, Kupcinskas J, Link A. Microbial composition of tumorous and adjacent gastric tissue is associated with prognosis of gastric cancer. Sci Rep. 2023;13:4640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 84. | Boehm ET, Thon C, Kupcinskas J, Steponaitiene R, Skieceviciene J, Canbay A, Malfertheiner P, Link A. Fusobacterium nucleatum is associated with worse prognosis in Lauren's diffuse type gastric cancer patients. Sci Rep. 2020;10:16240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 85. | Aziz S, Rasheed F, Akhter TS, Zahra R, König S. Microbial Proteins in Stomach Biopsies Associated with Gastritis, Ulcer, and Gastric Cancer. Molecules. 2022;27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 86. | Hsieh YY, Tung SY, Pan HY, Chang TS, Wei KL, Chen WM, Deng YF, Lu CK, Lai YH, Wu CS, Li C. Fusobacterium nucleatum colonization is associated with decreased survival of helicobacter pylori-positive gastric cancer patients. World J Gastroenterol. 2021;27:7311-7323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 87. | Hsieh YY, Kuo WL, Hsu WT, Tung SY, Li C. Fusobacterium Nucleatum-Induced Tumor Mutation Burden Predicts Poor Survival of Gastric Cancer Patients. Cancers (Basel). 2022;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 88. | Nie S, Wang A, Yuan Y. Comparison of clinicopathological parameters, prognosis, micro-ecological environment and metabolic function of Gastric Cancer with or without Fusobacterium sp. Infection. J Cancer. 2021;12:1023-1032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 89. | Liu C, Zhang H, Li T, Jiang Z, Yuan Y, Chen X. Fusobacterium nucleatum Promotes Megakaryocyte Maturation in Patients with Gastric Cancer via Inducing the Production of Extracellular Vesicles Containing 14-3-3ε. Infect Immun. 2023;91:e0010223. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 90. | Meng X, Ma G, Zhang X, Yin H, Miao Y, He F. Extracellular vesicles from Fusobacterium nucleatum: roles in the malignant phenotypes of gastric cancer. Cell Cycle. 2024;23:294-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 91. | Rodriguez RM, Hernandez BY, Menor M, Deng Y, Khadka VS. The landscape of bacterial presence in tumor and adjacent normal tissue across 9 major cancer types using TCGA exome sequencing. Comput Struct Biotechnol J. 2020;18:631-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 92. | Conti L, Borro M, Milani C, Simmaco M, Esposito G, Canali G, Pilozzi E, Ventura M, Annibale B, Lahner E. Gastric microbiota composition in patients with corpus atrophic gastritis. Dig Liver Dis. 2021;53:1580-1587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 93. | Chen WD, Zhang X, Zhang MJ, Zhang YP, Shang ZQ, Xin YW, Zhang Y. Salivary Fusobacterium nucleatum serves as a potential diagnostic biomarker for gastric cancer. World J Gastroenterol. 2022;28:4120-4132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 94. | Liu D, Zhang R, Chen S, Sun B, Zhang K. Analysis of gastric microbiome reveals three distinctive microbial communities associated with the occurrence of gastric cancer. BMC Microbiol. 2022;22:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (1)] |
| 95. | Li M, Shao D, Zhou J, Gu J, Qin J, Li X, Hao C, Wei W. Microbial Diversity and Composition in Six Different Gastrointestinal Sites among Participants Undergoing Upper Gastrointestinal Endoscopy in Henan, China. Microbiol Spectr. 2022;10:e0064521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 96. | Jeong S, Liao YT, Tsai MH, Wang YK, Wu IC, Liu CJ, Wu MS, Chan TS, Chen MY, Hu PJ, Kao WY, Liu HC, Tsai MJ, Liu CY, Chang CC, Wu DC, Hsu YH. Microbiome signatures associated with clinical stages of gastric Cancer: whole metagenome shotgun sequencing study. BMC Microbiol. 2024;24:139. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |