Published online Sep 15, 1997. doi: 10.3748/wjg.v3.i3.139
Revised: April 10, 1997
Accepted: May 15, 1997
Published online: September 15, 1997
AIM: To establish the hepatoma cell–specific expression of human interferon (IFN) gene mediated by retroviral vectors
METHODS: Human interferon α and interferon β complementary DNA (IFN cDNA) were cloned into the polylinker site of pMNSM retroviral vector to construct recombinant retroviral vectors pMNSIFNA and pMNSIFNB, with the transcription of IFN gene being driven by Simian virus 40 early region promoter (SV40) early region promoter. IFN cDNAs were also cloned into pMNAIFNA, pAMNSIFNA, and pMNAIFNB, with the transcription of IFN gene being driven by SV40 early region promoter regulated by α-fetoprotein enhancer. Next, the retroviral constructs were introduced into retroviral amphotropic packaging cells using the lipofectamine-mediated gene transfer procedure. The rate of plasmid transfection was (4-40) × 103 colonies/μg DNA/106 PA317 cells. The rate of retrovirus infection was (5-500) × 104 colony forming units (CFU)/mL. Further, the recombinant retroviruses were used to infect human hepatoma cells, renal carcinoma cells, and melanoma cell lines in the presence of 4 μmg/L polybrene.
RESULTS: Northern and Dot hybridization of total RNA from the neomycin-resistant colonies and IFN expression assay indicated that human α fetoprotein enhancer induced efficient and specific transcription and expression of IFN genes driven by the promoter of different origins in human hepatoma cells, leading to high production of α fetoprotein.
CONCLUSION: Cis active element of α-fetoprotein gene can drive specific expression of IFN genes in human hepatoma cells, which provides some valuable data for the hepatoma-specific immune gene therapy.
- Citation: Cao GW, Jun-Gao, Du P, Qi ZT, Kong XT. Construction of retroviral vectors to induce a strong expression of human class interferon gene in human hepatocellular carcinoma cells in vitro. World J Gastroenterol 1997; 3(3): 139-142
- URL: https://www.wjgnet.com/1007-9327/full/v3/i3/139.htm
- DOI: https://dx.doi.org/10.3748/wjg.v3.i3.139
Class I interferon (IFN) is a potent immune-modulating factor in vivo that plays an important role in activating anti-tumor cytotoxic T lymphocytes and non-specific effector cells such as natural killer (NK) cells[1]. IFN genes and many other cytokine genes have been introduced into tumor cells, tumor interstitial cells, and tumor-infiltrating lymphocytes[2-4] in order to sustain an effective anti-tumor concentration of cytokine located at tumor sites. It has been shown that some cytokine-gene-modified tumor cells lost their tumorigenicity and inhibited the growth of parental tumor in animal models. Practical antitumor effects were obtained in the protocol. The morbidity of hepatoma is relatively high in China, and no effective procedure is currently available for the treatment of advanced hepatoma. In this study, hepatoma-specific IFN retroviral vectors were prepared and in vitro expression assays were conducted to establish an effective gene therapy against hepatoma and prevent the possible adverse effects of class I INF expression in non-tumoral tissues in addition to verifying the possibility of the enhancer of α-fetoprotein (AFP) “house-keeping gene” regulating viral promoter in retroviral vector.
Plasmids pCGS261 containing human leukocyte interferon (IFN-α) cDNA were obtained from the American Type Culture Collection (ATCC), (Rockville, United States). pSPGNHIFNB10 carrying the HuIFN-β-cDNA was purchased from LMBP (Gent University, Belgium). Plasmid pAF5.1-CAT containing the AFP enhancer and plasmid pGEM7z-AFPe containing human AFP enhancer core sequence were kindly provided by Professor Tamaoki (Calgary University, Canada). Retroviral vector pMNSM containing the neomycin phosphotransferase and the SV40 early region promoter was a gift from Dr. Tsuchiya (Tokyo Medical and Dental University, Japan).
Amphotropic retrovirus packaging cell line PA317, retrovirus titering cell line NIH3T3TK-, human hepatoma cell lines HepG2 and Hep3B, and IFN bioassay cell line WISH were obtained from ATCC. Retroviral packaging cell Psi2 was procured from Dr. Shigeki Kuriyama. Human hepatoma cell line SMMC7721 and human melanoma cell line M21 were obtained from Chinese Academy of Sciences. Human renal carcinoma cell line RZ94602 was established at our laboratory.
TdR, dNTP, polybrene, PEG8000, guanidine isothiocyanate, PVP2500, and MTT were from Sigma. New generation liposome transfection reagent LIPOFECTAMINE[TM], RPMI-1640, DMEM, FCS, random primers DNA labeling system, and G418 were from Gibco. Herring sperm DNA, restriction enzymes, T4 ligase, DNA polymerase Klenow fragment, and CIP were from Promega. α32P-dCTP was from Beijing Yahui Biomedical Co. Nitrocellulose membrane was from Amersham.
Plasmids extraction with alkali lysis, enzymes digestion, ligation, isolation and harvest of DNA fragments and transformation of Escherichia coli was carried out according to the standard methods[5].
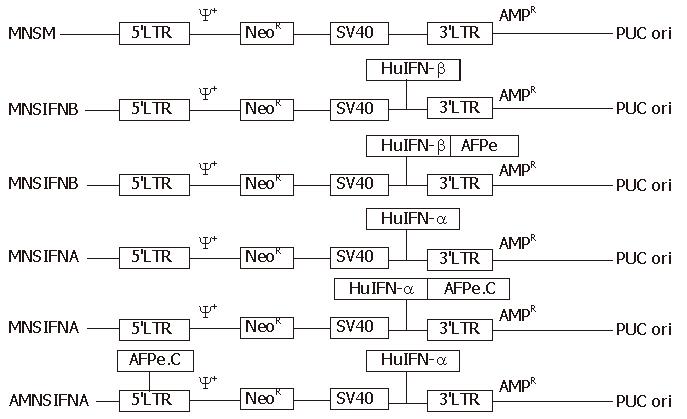
The procedures of construction are as follows. The plasmids pSPGNHIFNB10, pCGS261, pAF5.1-CAT, pGEM7z AFPe, and pMNSM were identified with enzyme digestion. pMNSM was linearized with Hind III/BamH I digestion. pSPGNHIFNB10 was digested with Hind III/BamHI to release 1.06-kb IFN-β cDNA. IFN-β cDNA was linked to the linearized pMNSM to construct pMNSIFNB. pAF5-1-CAT was fully digested with Aat II/SpeI/BamHI to release 5.5-kb human AFP enhancer sequence and blunted with Klenow. pMNSIFNB was linearized with EcoR I digestion and blunted with Klenow. The enhancer was linked with the linearized pMNSIFNB to construct pMNAIFNB. pMNSIFNA and pMNAIFNA were constructed using the above construction procedure with some modifications. The human IFN-β gene was replaced with human IFN-α gene, and 5.5-kb AFP enhancer sequence was replaced with 0.7-kb AFP enhancer core sequence. As for the construction of pAMNSIFNA, an 0.7-kb AFP enhancer core sequence was inserted into the long terminal repeat (LTR) region of the retroviral vector. Then, 1.1-kb human IFN-α gene was cloned into the polylinker site of pMNSM and under transcriptional control of SV40 early region promoter. The identity of all the retroviral constructs were confirmed by restricted digestion.
Packaging cells were pretreated with 2 μmg/L TdR to induce simultaneous cell division. Psi2 cells were transfected with the retroviral constructs by using the lipofectin-mediated gene transfer procedure, which was performed as per the manufacturer′s instructions. After full selection with G418, the supernatant of the modified Psi2 cells was harvested, tittered, and used to infect PA317 cells. Measurement of plasmid transfection and retrovirus infection rates was carried out according as described previously[6].
Northern blot and dot hybridization were performed using a previously described method[5]. The IFN bioactivities in the supernatant of 106 modified cells were calculated per 24 h. IFN bioassay was performed according to the routine procedure of our laboratory.
All the retroviral constructs are outlined in Figure 1. IFN genes in pMNSIFN B and pMNSIFNA were controlled by the SV40 early region promoter. IFN genes derived by SV40 early region promoter in pMNAIFNB and pMNAIFNA were controlled by [WTBZ] AFP enhancer. Then, 0.7-kb AFP enhancer core sequence was inserted into the LTR region in order to enactivate the intrinsic enhancer element located in the region and diminish the intervention of the intrinsic enhancer on AFP enhancer.
PA317 cells with division synchronized with TdR pretreatment were infected with the supernatants of Psi2 cells containing MNSM, MNSIFNB, MNAIFNB, MNSIFNA, MNAIFNA, and AMNSIFNA recombinant retroviruses, and the infection rate of the retroviruses in the PA317 supernatants harvested on the third day after the infection were 5.0 × 106, 6.0 × 105, 4.5 × 104, 8.0 × 106, 1.0 × 106, and 5.0 × 106 CFU/mL, respectively. Psi2 cells were transfected with the retroviral constructs by using a lipofectamine-mediating procedure. The modified Psi2 cells were split 1/20 onto medium containing 400 μmg/L active G418. After 10 d selection, the corresponding transfection rates were as follows: 2.5 × 104, 8.5 × 103, 4.0 × 103, 2.3 × 104, 3.6 × 104, and 3.4 × 104 CFU/μg DNA/106 PA317 cells. The greater the genome of the vectors, the lower the infection and transfection rates. However, all the highest retroviruses producing PA317 colonies of 6 different groups could produce 107 CFU recombinant retroviruses per 106 PA317 cells per 24 h. After being frozen in liquid nitrogen for one month, the modified PA317 did not show any significant decrease in the retrovirus production (P > 0.05)
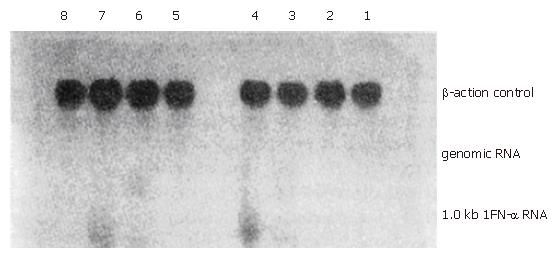
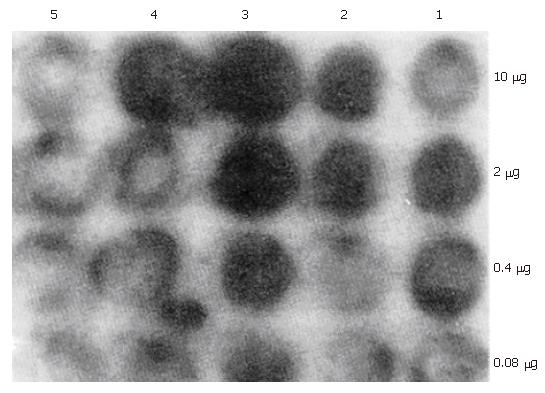
MNSM, MNSIFNB, MNAIFNB, MNSIFNA, MNAIFNA, and AMNSIFNA retroviruses were used to infect the tumor cells in vitro in the presence of 4 μg/L polybrene. The total RNA from the infected HepG2 and Hep3B hepatoma cells and M21 melanoma cells, after being fully selected with G418, were extracted by the guanidine isothiocyanate single-step methods. Total RNAs from MNSM, MNSIFNA, MNAIFNA, and AMNSIFNA modified tumor cells were electrophoresed on a 1% agarose/2.2 M formaldehyde gel and transferred onto nitrocellulose. Blots were hybridized with 32P-labeled HuIFN-α-cDNA probe. Total RNAs from MNSM, MNSIFNB, and MNAIFNB modified tumor cells were subjected to 5-fold serial dilution from 10 μg, transferred onto the nitrocellulose membrane, and probed with IFN-β-cDNA. The result is shown in Figures 2 and 3. In the hepatoma cell line HepG2, which produces high levels of AFP, the transcription level of IFNs cDNA, after infection with MNAIFNB, AMNSIFNA, and MNAIFNA retroviruses, was significantly higher than that of the cells infected with MNS-IFNB and MNSIFNA retroviruses. In the Hep3B cells, which produce intermediate levels of AFP, the difference in the IFN-β-cDNA transcription level between the two infection groups was not so significant as that in the HepG2 cells, indicating that the enhancing effect of the AFP enhancer on the SV40 early region promoter in the genome of the hepatoma cells correlated with the level of AFP produced in the hepatoma cells.
HepG2, Hep3B, SMMC7721, RZ94602, and M21 tumor cells were infected with MNSM, MNSIFNB, MNAIFNB, MNSIFNA, MNAIFNA, and AMNSIFNA retroviruses, respectively, and fully selected with G418. The bioactivity of IFNs secreted by 106 gene-modified tumor cells per 24 h was precisely determined with MTT measurement and CpEI50 method. The results are shown in Table 1. In the human hepatoma cell line HepG2, which produces high levels of AFP, an AFP enhancer significantly augmented the expression of IFNs gene induced by SV40 early region promoter. Specific expression of IFNs gene in the Hep3B cell line, which produces intermediate AFP levels, was not as significant as that in HepG2 cells. IFN gene expression in the M21 and RZ94602 cell lines, which did not produce AFP, was inhibited after infection with MNAIFNB, MNAIFNA and AMNSIFNA retroviruses.
| Cell lines | Origin | AFP1 | Genetic alteration | G418 resistant | Interferon bioactivity (unit/106 cells·24 h) |
| HepG2 | Human | 2044 | Parental2 | - | 0 |
| (n = 5) | hepatocyte | MNSM | + | 0 | |
| MNSIFNB | + | 218 ± 102 | |||
| MNAIFNB | + | 818 ± 35 | |||
| MNSIFNA | + | 128 ± 36 | |||
| MNAIFNA | + | 808 ± 180 | |||
| AMNSIFNA | + | 1200 ± 36 | |||
| Hep3B | Human | 311 | Parental | - | 0 |
| (n = 4) | hepatocyte | MNSM | + | 0 | |
| MNSIFNB | + | 156 ± 48 | |||
| MNAIFNB | + | 462 ± 104 | |||
| MNSIFNA | + | 136 ± 42 | |||
| MNAIFNA | + | 548 ± 120 | |||
| AMNSIFNA | + | 465 ± 204 | |||
| SMMC7721 | Human | ND | Parental | - | 0 |
| (n = 5) | hepatocyte | MNSM | + | 0 | |
| MNSIFNB | + | 280 ± 80 | |||
| MNAIFNB | + | 580 ± 56 | |||
| MNSIFNA | + | 242 ± 120 | |||
| MNAIFNA | + | 468 ± 84 | |||
| AMNSIFNA | + | 524 ± 132 | |||
| M21 | Human | Parental | - | 0 | |
| (n = 5) | melanocyte | MNSM | + | 0 | |
| MNSIFNB | + | 324 ± 66 | |||
| MNAIFNB | + | 21 ± 10 | |||
| MNSIFNA | + | 226 ± 128 | |||
| MNAIFNA | + | 42 ± 36 | |||
| AMNSIFNA | + | 0 ± 2.4 | |||
| RZ94602 | Human renal | Parental | - | 0 | |
| (n = 4) | cell | MNSM | + | 0 | |
| MNSIFNB | + | 280 ± 64 | |||
| MNAIFNB | + | 18 ± 8.4 | |||
| MNSIFNA | + | 320 ± 120 | |||
| MNAIFNA | + | 24 ± 48 | |||
| AMNSIFNA | + | 0 ± 2.4 |
In vivo gene therapy for cancer is more practicable than the ex vivo protocol in clinical trials. As for the in vivo protocol, the major problem is to induce the guaranteed specific expression of the gene of interest in tumor cells. In vivo gene therapy can be guaranteed if the transcription regulatory sequences (TRS) of “house-keeping gene” of the tumor characteristic proteins are employed to regulate gene expression. First, high expression levels of the gene of interest can be achieved if the gene is controlled by the TRS. The concentration of interferon or expression of IFN gene at the tumor location is correlated with the antitumor effect[1]. Second, possible microenvironmental disorders caused by the expression of Class I IFN gene in non-tumoral tissues can be prevented. This is of great significance in cancer gene therapy.
Human IFN α and IFN β bind to the same receptors and are called class I interferon. Class I IFN plays an important role in vivo in tumor immunosurveillance and tumor biotherapy. In the present study, human IFN α and IFN β genes were individually introduced into the established hepatoma, renal cell carcinoma, and melanoma cell lines by means of the retrovirus. Expression of IFN genes, under the transcription control of AFP enhancer, was in the hepatoma-cell-specific manner, and correlated with the AFP production levels in the hepatoma cells. Some transcription regulatory proteins existing in the nuclei of the hepatoma cells are believed to be related to the hepatoma-specific gene expression. In the AFP-producing hepatoma cells, the transcription regulatory sequence of AFP “house-keeping gene” is controlled by the tissue-specific transcription regulatory proteins. After the transcription regulatory sequence carried by the retroviruses is integrated into the genome of the hepatoma cells, the regulatory proteins could also activate AFP enhancer in the same manner. Since enhancer is not dependent on origin and orientation, AFP enhancer may transactivate the promoter of viral origin and increase the transcription rate of the promoter in certain cell types.
Promoter interference between heterologous promoter and retroviral promoter has been documented. Retroviral LTR promoter and enhancer may silence some internal heterologous promoters[7]. In this study, construction of pAMNS-IFNA was carried out by the insertion of AFP enhancer core sequence into retroviral LTR region in order to silence LTR enhancer activity and increase the hepatoma cell–specific expression of human IFN-α gene. However, the hepatoma cells–specific expression was not significantly increased after the treatment. It was demonstrated that enhancer elements in retroviral LTR region did not significantly interfere with the function of AFP “house-keeping” gene enhancer.
Tumor-tissue-specific gene therapy is carried out in the thymidine kinase prodrug transformation gene therapy, but not in cytokine gene therapy and other immune gene therapies. It has been recently demonstrated that the transcription regulatory sequences of AFP, carcinoembryonic antigen, and tyrosinase “house-keeping gene” regulated the thymidine kinase gene specifically expressed in the hepatocellular carcinoma, gastric cancer and melanoma cells[8-10].Tumor tissue specific gene therapy is considered to be an important direction of gene therapy against cancer in the near future. Probability of helper virus production, which is generated by homologous recombination, is expected to be decreased on the condition of the transcription regulatory sequences existing in the recombinant retrovirus. The future of gene therapy is subsequently guaranteed.
Guang Wen Cao, Associate Professor, having 36 papers and 2 books published, Director of the Division of Tumor & Virus Gene Therapy, Department of Microbiology, Second Military Medical University, Shanghai 200433, China
Original title:
S- Editor: Filipodia L- Editor: Jennifer E- Editor: Hu S
| 1. | Cao GW, Du P. Modern cancer biotherapology. Beijing: People Military Medical Publisher 1995; 179-288. |
| 2. | Colombo MP, Forni G. Cytokine gene transfer in tumor inhibition and tumor therapy: where are we now? Immunol Today. 1994;15:48-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 174] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 3. | Mizuno M, Yoshida J, Sugita K, Hayashi Y, Yagi K. [Basic research for interferon gene therapy against malignant glioma]. No Shinkei Geka. 1992;20:547-551. [PubMed] |
| 4. | Hwu P, Yannelli J, Kriegler M, Anderson WF, Perez C, Chiang Y, Schwarz S, Cowherd R, Delgado C, Mulé J. Functional and molecular characterization of tumor-infiltrating lymphocytes transduced with tumor necrosis factor-alpha cDNA for the gene therapy of cancer in humans. J Immunol. 1993;150:4104-4115. [PubMed] |
| 5. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. 2nd ed. New York: Cold Spring Harbar Laboratory Press 1989; 1-500. |
| 6. | Miller AD, Miller DG, Garcia JV, Lynch CM. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 1993;217:581-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 300] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 7. | Gunzburg WH, Salmons B. Retroviral vectors. Gene therapy. London: Bios Scientific Publisher 1996; 33-53. |
| 8. | Huber BE, Richards CA, Krenitsky TA. Retroviral-mediated gene therapy for the treatment of hepatocellular carcinoma: an innovative approach for cancer therapy. Proc Natl Acad Sci USA. 1991;88:8039-8043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 186] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Tanaka T, Kanai F, Okabe S, Yoshida Y, Wakimoto H, Hamada H, Shiratori Y, Lan K, Ishitobi M, Omata M. Adenovirus-mediated prodrug gene therapy for carcinoembryonic antigen-producing human gastric carcinoma cells in vitro. Cancer Res. 1996;56:1341-1345. [PubMed] |
| 10. | Vile RG, Hart IR. Use of tissue-specific expression of the herpes simplex virus thymidine kinase gene to inhibit growth of established murine melanomas following direct intratumoral injection of DNA. Cancer Res. 1993;53:3860-3864. [PubMed] |