Published online Jan 21, 2023. doi: 10.3748/wjg.v29.i3.508
Peer-review started: September 27, 2022
First decision: October 30, 2022
Revised: December 5, 2022
Accepted: December 27, 2022
Article in press: December 27, 2022
Published online: January 21, 2023
Processing time: 106 Days and 12.8 Hours
Inflammatory bowel diseases, namely ulcerative colitis and Crohn’s disease, are chronic and relapsing conditions that pose a growing burden on healthcare sys
Core Tip: Management of patients with inflammatory bowel disease is complex and costly. Therefore, in this field being able to improve clinical efficiency and optimize healthcare resources is of paramount importance. In this regard, artificial intelligence appears to be an extremely promising tool with a significantly wide range of potential applications that encompass diagnosis, treatment, and follow-up. This review summarized the most recent significant scientific findings regarding the application of artificial intelligence in inflammatory bowel diseases, providing a picture of the current state of the field and future perspectives.
- Citation: Da Rio L, Spadaccini M, Parigi TL, Gabbiadini R, Dal Buono A, Busacca A, Maselli R, Fugazza A, Colombo M, Carrara S, Franchellucci G, Alfarone L, Facciorusso A, Hassan C, Repici A, Armuzzi A. Artificial intelligence and inflammatory bowel disease: Where are we going? World J Gastroenterol 2023; 29(3): 508-520
- URL: https://www.wjgnet.com/1007-9327/full/v29/i3/508.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i3.508
Ulcerative colitis (UC) and Crohn’s disease (CD) are chronic and relapsing conditions that affect the gastrointestinal tract and are labelled as inflammatory bowel diseases (IBD)[1,2]. In the last few decades, IBD has emerged as a global disease with a conspicuous burden on public health and healthcare costs[3].
The diagnosis and management of IBD is complex and implies the interplay and synergy of various specialists, including clinical gastroenterologists, gastrointestinal endoscopists, radiologists, pathologists, surgeons, and clinical nutritionists[4].
Along with histological assessment, endoscopy has an important role in the diagnosis and the follow-up of IBD while also being the mainstay for colorectal cancer surveillance[5]. Due to the essential role of endoscopy and imaging in gastroenterology, and particularly in IBD, artificial intelligence (AI)-based image analysis can be utilized in numerous applications such as evaluation of endoscopic lesions, cancer detection, and assessment of disease activity (e.g., prognosis and response to treatment)[6].
AI is an umbrella term that encompasses any cognitive function developed by machines for learning or problem solving. A particular subset of AI is represented by machine learning (ML), a discipline that uses large datasets as an input in order to identify patterns of interaction among variables, allowing the possibility to apply these findings to new data[7].
A further subset and evolution of ML is deep learning (DL), which mimics the neuronal interaction in the human brain to develop artificial neural networks and subsequently convolution neural networks (CNN) that are then able to use the input data in an autonomous fashion with the aim of assessing predictive factors of a specific outcome through the development of multiple levels of abstractions[8].
Among the numerous implementations of AI in medicine, a promising field is that of automatic collection of complex and nuanced clinical data from electronic medical records through natural language processing, the subset of AI that studies the interpretation of the human language made by the computer[9,10].
Another fertile ground of application of AI is the interpretation of radiologic imaging. A computer model based on computed tomography enterography allows for accurate characterization of intestinal fibrosis in CD, in some instances outperforming human radiologists[11].
In the context of gastrointestinal endoscopy, AI has found application in two main fields, namely in the detection of mucosal lesions, with computer-aided detection (CADe) and in the characterization of mucosal lesions, with computer-aided diagnosis, along with the evaluation of the quality of the endoscopic procedure itself, with computer aided monitoring[12-20].
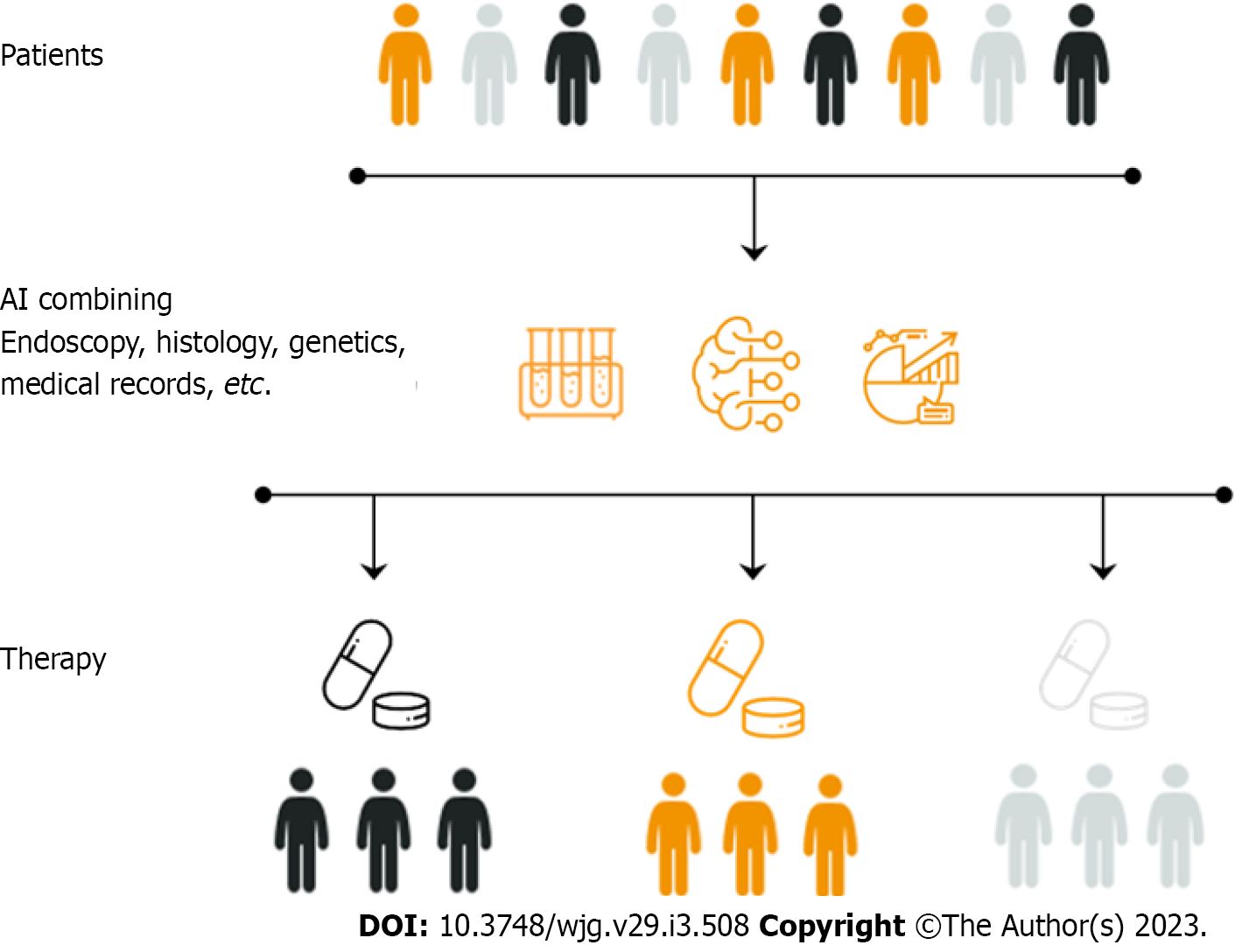
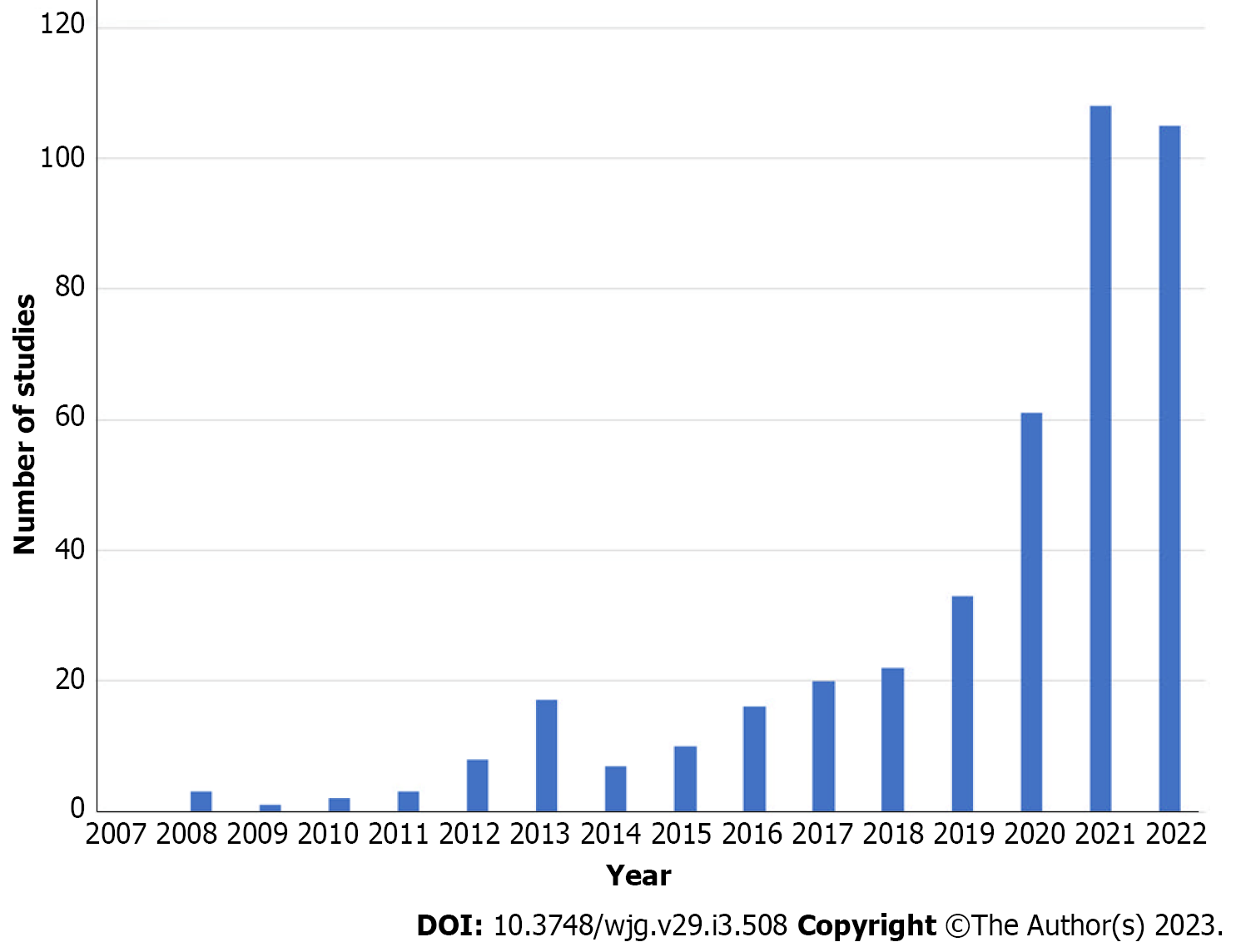
Therefore, implementation of AI in IBD is a promising tool for improving the assessment of disease activity and reducing the interobserver variability in grading such activity[21]. In addition, similar to what is already happening in the general population, CADe systems could eventually improve detection of IBD-associated dysplasia[22]. Finally, AI may also allow for application of precision medicine through the analysis of large databases, correlating differences in the biology of the patient with differences in the susceptibility to develop IBD, the activity of the disease, and the response to specific therapies[23] (Figure 1). Such considerations are further supported by the growing number of clinical studies on the application of AI for IBD in recent years (Figure 2).
Table 1 summarized the most impactful studies on AI in several fields of IBD that were identified after a literature review using PubMed (MEDLINE) from inception to November 6, 2022. The impact measure of the articles cited was assessed via the Reference Citation Analysis (RCA; Baishideng Publishing Group Inc.) tool.
| Ref. | Field | Study features | Main finding |
| Isakov et al[26], 2017 | IBD genetics | ML model to assess 16390 genes in IBD and healthy patients | Identified 347 IBD-risk genes (67 newly identified) |
| Cheng et al[27], 2019 | IBD genetics | Software analysis to assess the genetics of 32713 IBD patients | Identified several genes potentially involved in UC; identification of 11 common Gene Ontology terms for UC |
| Yuan et al[28], 2017 | IBD genetics | Software analysis to assess 12754 genes in IBD and healthy patients | Identified 41 genes closely associated with IBD |
| Mihajlović et al[30], 2021 | IBD and microbiota | ML classification algorithm to identify IBD from 1638 fecal samples | Confirmed strong connection between IBD and specific fecal microbial species |
| Manandhar et al[31], 2021 | IBD and microbiota | ML model analysis of fecal microbiota from 729 IBD patients and 700 healthy controls | Identified of 117 bacterial taxa with a potential role in diagnostic screening of IBD |
| Mossotto et al[32], 2017 | IBD diagnosis | ML model to assess 287 pediatric patients with IBD | Accuracy of 83.3% of the combined endoscopy-histology ML model in the classification of pediatric IBD patients |
| Quénéhervé et al[33], 2019 | IBD diagnosis | AI analysis of CLE images from 50 IBD patients and 9 healthy controls | AI analysis had 100% sensitivity and specificity for IBD diagnosis, 92% sensitivity and 91% specificity of IBD differential diagnosis |
| Ananthakrishnan et al[9], 2013 | IBD diagnosis and data collection | NLP model trained and validated on 700 UC patients and 700 CD patients to improve case definition and identification from EMRs | NLP model provided better accuracy (AUC 0.94-0.95) than models using only the International Classification of Diseases 9th revision for IBD case definition and identification |
| Stidham et al[39], 2019 | IBD endoscopy | DL model for UC severity trained on 16514 endoscopic images | Similar performance of the DL model and experienced human reviewers in grading UC endoscopic severity |
| Ozawa et al[40], 2019 | IBD endoscopy | CNN-based CADe system for UC severity trained on 26304 endoscopic images | CADe system had AUCs of 0.86 and 0.98 in the identification of Mayo score 0 and 0-1, respectively |
| Maeda et al[41], 2021 | IBD endoscopy | Endoscopic AI model used in real time on 135 UC patients in clinical remission | Endoscopic applications of real time AI predicted clinical relapse of UC with statistical significance |
| Gottlieb et al[42], 2021 | IBD endoscopy | DL algorithm to assess UC severity on 795 full-length endoscopy videos | DL algorithm showed significant inter-rater agreement to human central readers for prediction of UC severity |
| Yao et al[43], 2021 | IBD endoscopy | Endoscopic AI model (CNN) to assess UC grading used on 169 endoscopy videos and compared to dual central reader review | AI model approximated the scoring of experienced reviewers for grading of UC endoscopic activity |
| Byrne et al[44], 2021 | IBD endoscopy | DL model (CNN) to detect and assess UC activity leveraged on > 375000 frames | DL model resulted in well aligned scoring guidelines and experts’ performances |
| Takenaka et al[45], 2020 | IBD endoscopy | DL algorithm trained on endoscopic images and biopsy results and tested on 875 UC patients | DL model identified with an accuracy > 90% patients in endoscopic and histologic remission |
| Maeda et al[46], 2019 | IBD endoscopy (endocytoscopy) | CADe system to predict persistent histologic phlogosis from endocytoscopy validated on 100 UC patient | CADe system provided a diagnostic accuracy of 91% with perfect reproducibility for identification of persistent histologic inflammation |
| Bossuyt et al[47], 2020 | IBD endoscopy | AI algorithm based on pixel color data and pattern recognition from endoscopic images tested on 55 patients | AI algorithm (“red density”) provided an objective computer-based assessment of UC disease activity with good correlation with endoscopic and histological scoring systems |
| Aoki et al[51], 2019 | IBD endoscopy (VCE) | AI system (CNN) tested on 10440 small bowel images for detection of erosions and ulcers in CD | AI system showed an accuracy of 90.8% for detection of erosions and ulcers |
| Klang et al[52], 2020 | IBD endoscopy (VCE) | DL algorithm applied on 17640 VCE images for ulcer detection in CD | DL algorithm provided an accuracy ranging from 95.4% to 96.7% with an AUC of 0.99 for ulcer detection |
| Klang et al[53], 2021 | IBD endoscopy (VCE) | DL model applied on 27892 VCE images for identification of intestinal strictures in CD | DL model showed an accuracy of 93.5% in stricture identification and excellent differentiation between strictures and other lesions |
| Ferreira et al[54], 2022 | IBD endoscopy (VCE) | DL model trained and validated on 8085 VCE images for detection of erosions and ulcers in CD | DL model provided an accuracy of 92.4% and a precision of 97.1% for lesion detection |
| Aoki et al[55], 2020 | VCE | Comparison between standard endoscopist reading and reading after AI model screening of 20 full-length VCE videos | The mean VCE video reading time was significantly shorter after AI model (CNN) screening compared to standard reading |
| Maeda et al[56], 2021 | IBD endoscopy (surveillance) | Case report of dysplasia detection by AI system in a patient with long standing UC | AI system (EndoBRAIN) identified 2 colonic lesions that harbored low-grade dysplasia upon histological examination |
| Fukunaga et al[57], 2021 | IBD endoscopy (surveillance) | Case report of dysplasia detection by AI system in a patient with long standing UC | AI system (EndoBRAIN) identified rectal lesions that harbored high-grade dysplasia upon histological examination |
| Reddy et al[72], 2019 | IBD prognosis | ML model employed on 82 CD patients’ EMRs to predict disease course | ML model predicted inflammation severity with high accuracy (AUC 92.8%) from EMR data |
| Takenaka et al[73], 2022 | IBD prognosis | ML model validated on endoscopic images and biopsy results from 875 UC patients to predict disease course | Histologic remission detected by the ML model correlated with a significant reduction in clinical relapse, steroid use, hospitalization, and colectomy |
| Li et al[75], 2021 | Response to treatment | ML model employed on 174 CD patients to predict response to infliximab | ML model based on clinical and serological parameters showed an accuracy of 0.85 for prediction of response to infliximab |
| Waljee et al[76], 2018 | Response to treatment | AI model employed on 472 CD patients to predict response to vedolizumab | AI model based on clinical and serological parameters was able to identify patients that achieved a corticosteroid-free biologic remission at week 52 of vedolizumab |
| Waljee et al[77], 2018 | Response to treatment | ML algorithm employed on 491 UC patients to predict response to vedolizumab | ML algorithm based on clinical and serological parameters was able to identify patients that achieved a corticosteroid-free biologic remission at week 52 of vedolizumab |
| Doherty et al[78], 2018 | Response to treatment | AI model to assess response to treatment with ustekinumab in 306 patients with CD | AI model detected patients in remission based on clinical data and fecal microbiota at week 6 and 22 of ustekinumab |
Although it is not yet thoroughly understood, the etiology of IBD is known to depend upon the complex interplay of genetic, microbial, and environmental factors and the immune system[24]. In this context, AI allows for more effective data analysis to evaluate the role of specific genes in the predisposition and development of IBD.
Genome-wide association studies have been employed in order to identify sequence variations related with specific conditions, with over 200 genes found to be potentially implicated in IBD etiology[25]. Such complex and large genomic data, however, are difficult to assess with standard analytical tools. In this context, studies have shown that ML and DL can effectively analyze genome-wide association study data and overcome part of the inherent limitation of such methodology through algorithms of minimum Redundancy-Maximum Relevance and incremental feature selection[26-28].
Gut microbiota is thought to play a relevant role in the complex etiology and pathogenesis of IBD, partly concurring in providing a substrate that may range from protective to proinflammatory[29]. Moreover, in the context of dysbiosis, the taxonomic composition appears to vary between patients affected by IBD and unaffected subjects and between IBD subtypes, UC, and CD. Promising studies in this field show that the application of ML algorithms on the analysis of gut microbiome data may assist the clinician in diagnosing IBD[30,31].
AI has also been employed in the attempt of supporting the conventional diagnosis of IBD. In this regard, isolated and combined endoscopic and histological parameters were used to develop ML models, which was accurate in the classification of pediatric patients affected by IBD[32].
Additional models based on confocal laser endomicroscopy were developed and by quantitative analysis through cryptometry were able to provide a diagnosis of IBD with high sensitivity and specificity. Moreover, these models managed to successfully differentiate between UC and CD[33].
In order to rigorously define endoscopic disease activity through specific parameters and limit interobserver variability, a plethora of endoscopic scores have been developed in the field of IBD. The most relevant scores are the CD endoscopic index of severity and the simple endoscopic score for CD, the Mayo endoscopic score, the UC endoscopic index of severity, and the UC colonoscopic index[34-38].
Implementation of AI in this field might be a further step towards reproducibility and homogeneity of endoscopic findings. The first successful attempts in identifying the presence of mucosal remission or activity via AI were made using a dedicated CADe system based on CNN trained on large datasets of endoscopic still images in patients affected by UC[39,40].
A subsequent development was represented by the implementation of neural networks in order to assess disease activity not only on still images but on the entirety of colonoscopy videos in real time. Also in this field, AI was efficient in recognizing active disease, calculating scores, predicting the risk of clinical relapse, and supporting clinicians in real time decision making for treatment[41-44].
Moreover, deep neural networks trained on endoscopic images and histological reports of UC managed to identify patients in endoscopic remission and histological remission with such an accuracy that it may potentially obviate the need for biopsy collection and analysis to identify patients in remission[45].
With regards to prediction of in vivo histological activity, the first dedicated CADe system was recently developed for application in endocytoscopy, yielding a high accuracy when compared with the gold standard of the pathologist’s assessment[46].
Finally, in order to overcome the fact that conventional DL systems train CNN based on the subjective scoring of images done by clinicians, novel approaches have been developed based on algorithms that only analyze parameters such as pixel color data and vascular pattern recognition. Objective computer-based and operator-independent tools provided a dedicated score that significantly correlated with both endoscopic and histological scoring systems[47].
Video capsule endoscopy (VCE) represents one of the main modalities of investigating the small bowel in suspect or established CD[48]. The main scoring systems used for disease activity quantification are the Lewis score and the capsule endoscopy CD activity index, which consider parameters such as extension, grading of inflammation, and presence of strictures[49,50].
VCE video review, however, is time-consuming and requires a high level of attention during the observation of thousands of frames. In order to simplify this task, AI has been implemented with the objective of reducing the time needed for image assessment by selecting the most relevant frames or portions of the video. Several CNN models have been developed in order to recognize pathologic findings such as erosions, ulcers, and strictures with very high accuracy[51-54]. Moreover, a study that assessed the review time for VCE showed how the employment of AI systems allowed clinicians to complete the task in a fraction of the time with no differences in overall accuracy[55].
In addition to recognition of disease activity, AI may find a relevant role in the surveillance of colorectal neoplasia in patients with IBD since a history of long-standing disease is a significant risk factor in developing colorectal cancer[5]. While AI is proving increasingly useful in the detection of colonic neoplasia in the general population with dedicated CADe systems, first attempts at developing specific applications for surveillance in IBD are being made[13]. As of today, successful detection of dysplasia has been described in case reports regarding the application of EndoBRAIN, a CADe system, in endoscopy and endocytoscopy[56,57].
Along with CADe systems, AI may prove to be an effective and safe tool for real-time quality improvement of the examination as well (i.e. withdrawal time, checking for blind spots), thereby potentially increasing the adenoma detection rate also in IBD patients[58].
Histological remission is now considered an adjunct target of treatment in UC and arguably the most stringent way to assess remission[59]. Growing evidence shows that persistence of histologic disease activity, even in the absence of macroscopic endoscopic inflammation, is associated with worse clinical outcome and risk of relapse[60]. More than 30 histological scores have been proposed to grade UC histological activity, but their application in clinical practice remains minimal, mainly due to the impracticality of the scores[61,62]. Even when the scores are applied, for example in clinical trials, the interobserver variability is very high, limiting comparison and reproducibility. Indeed, clinical trials increasingly resort to expensive central reading systems so that all biopsies are evaluated by few highly-qualified pathologists to reduce variability. Therefore, AI-based systems to automatically read UC biopsies would be of great help standardizing the assessment and reducing interobserver variability[63].
Trials in this field are ongoing, and initial results are promising. The first attempt to develop a CADe model to assess UC biopsies focused on eosinophils. The system had a good agreement compared to the manual count performed by human pathologists (interclass correlation coefficient = 0.81-0.92) but did not demonstrate an association between eosinophils counted and overall inflammatory activity[64]. More recently, Gui et al[65] proposed to simply assess UC activity by taking into consideration the sole presence or absence of neutrophils, the hallmark of active inflammation. They proposed a simplified score, the PICaSSO histologic remission index that was then embedded into a CADe system that was able to distinguish histological activity from remission in biopsies of UC with good accuracy. Further improvements to the same CADe have been recently presented showing that the neutrophil-only assessment by the CADe is largely consistent with mainstay scores such as Robarts and Nancy histological indexes[66].
Other studies on CADe systems for assessment of UC are ongoing and preliminary results are promising[67,68] (Table 2). Further applications of CNN in computational pathology have been studied in order to empower the pathologist’s accuracy and efficiency, with models trained on whole slide images that were able to effectively support the pathologists by excluding non-diagnostic slides, while retaining optimal sensitivity[69].
| Ref. | Study design | Population | Outcome | Results |
| Vande Casteele et al[64], 2022 | Cohort study | Colonic biopsies from 88 UC patients with histologically active disease | To assess a DL machine in quantifying eosinophils in colonic biopsies and validate against a pathologist’s count | The AI system highly agreed with manual eosinophil count by pathologists (ICC 0.81-0.92) |
| Peyrin-Biroulet et al[67], 2022 | Cohort study | 200 histological images of UC biopsies | To evaluate an AI algorithm in assessing histological disease activity according to the Nancy index | The CNN model had an excellent agreement with pathologists in the assessment of the Nancy index (ICC 0.84) |
| Villanacci et al[66], 2022 | Cohort study | 614 biopsies from 307 UC patients | To test a CNN-based CADe system for evaluating HR based on PHRI, Robarts, and Nancy indexes | The CADe system accurately assessed HR (sensitivity 89%, specificity 85% for PHRI) and similar performance for Nancy and Robarts |
Being able to predict the course of disease with regards to severity and progression is of the utmost importance in IBD patients in order to implement specific management strategies accordingly[70].
Studies have showed that by employing natural language processing and ML algorithms trained on several clinical data from electronic medical records (i.e. demographics, laboratory tests, endoscopy reports) it was possible to predict disease severity and surrogate markers of disease flare such as outpatient steroid use or hospitalization[9,71,72].
Furthermore, initial studies on the use of ML to predict the prognosis of patients affected by UC have shown that the endoscopic mucosal healing predicted by a deep neural network model was associated with prognostic features (reduced risks of hospitalization and colectomy) with statistical significance[73].
Lack of response to biological therapies (around 1 out of 3 patients with anti-TNF-alpha therapy) or progressive decline in response over time is an issue of paramount importance in IBD, with a significant economic toll on healthcare costs[74]. For this reason, the possibility to anticipate the likelihood of a patient responding to a specific biological drug prior to its start represents a cost-effective approach to treatment individualization.
ML has been implemented to deal with the complexity of such topics with encouraging results. Through the use of ML on clinical data from trials, random forests were developed to predict the response to biologics such as infliximab, vedolizumab, and ustekinumab, both in CD and in UC with encouraging results[75-78]. The integration and analysis of patients’ molecular and clinical features via ML algorithms therefore appears to be a promising field with great potential in terms of patient management and optimization of healthcare costs[79].
Design and discovery of new drugs are complex processes hampered by high costs and time con
AI, however, is proving to be a crucial tool in the development of novel drug candidates modernizing this field as well. Among the various implementations of ML and DL in the drug discovery process we can list peptide synthesis, virtual screening (structure-based and ligand-based), prediction of toxicity, drug monitoring and release, quantitative structure-activity relationship, and drug repositioning[81].
In the field of IBD AI has already been employed in drug discovery with encouraging initial results. Computational approaches have been used to identify metabolite-target interactions using the dataset of the IBD cohort from the Human Microbiome Project 2, followed by ML analyses aimed at ranking metabolites according to their importance in IBD and identifying possible human targets through virtual ligand-based screening. Overall, 983 high quality connections between metabolites from the gut microbiota and human proteins possibly relevant to IBD were identified, thus providing multiple novel drug targets for potential immune therapies[82].
Another relevant application of DL was the construction of a scaffold-based molecular design workflow aimed at developing drug candidates targeted towards the discoidin domain receptor 1. Through a deep generative model, molecular docking and virtual profiling, a high-quality scaffold-based molecular library was established, subsequently leading to the synthesis of a molecular compound that effectively inhibited the expression of proinflammatory cytokines, showing extraordinary kinase selectivity and significant therapeutic protection in ex vivo and in vivo animal models, respectively[83].
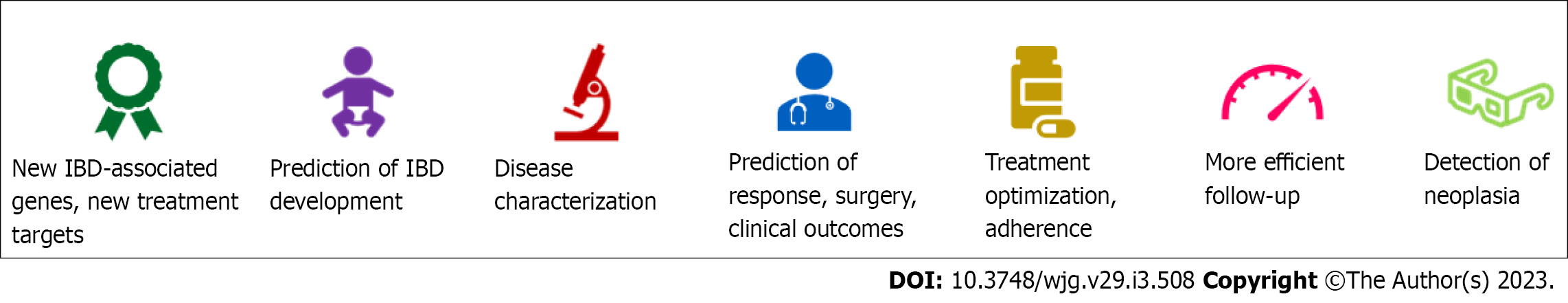
Based on the numerous advances in the field of IBD in recent years, it is foreseeable that AI will gain an ever-greater role in the standard patient care, ranging from evaluation of the risk of developing IBD to assistance in the assessment of mucosal activity or detection of dysplasia to support in histopathological reporting, prediction of disease course, and treatment efficacy (Table 1). At the same time, the integration of AI in daily practice will lead to changes in clinical practice itself, getting us closer to the concept of precision medicine and its subsequent improvement in the quality of care and optimization of healthcare costs (Figure 3).
As for research, AI is proving to be an irreplaceable aid in the collection and analysis of large data, while also limiting subjectiveness and interobserver variability, simplifying standardization and providing a feasible alternative to the need of independent central reading in clinical trials. The implementation of AI in everyday practice is expected to improve diagnostic accuracy and reproducibility by allowing for a better standardization of lesion features and classification and by increasing the detection rate of small and subtle mucosal abnormalities or lesions. Moreover, evidence shows how AI may have a role in advanced endoscopic techniques, such as confocal laser endomicroscopy or molecular imaging. Through these advanced application AI will support the clinician by detecting microscopic and even molecular details otherwise invisible, thereby paving the way to the potential incorporation of ultrastructural and molecular endpoints in IBD endoscopy.
Nonetheless, with regard to future applications of AI, there are several issues that will need solving, such as how to provide more transparency of the AI algorithms, how to assess and choose among different models with similar purposes, and how to effectively allocate resources in the plethora of models that will be available, to only list a few[84]. To the best of our knowledge, this is the first review on the roles and the future perspectives of AI in IBD that considers the first clinical applications in a real world setting that are now available from the most recent clinical studies. Nevertheless, the chief limitation is represented by the limited amount of evidence that support our review, which is inevitably due to the scarcity of data currently available for the topic itself from the literature. Undoubtedly further studies are direly needed to build a more robust and comprehensive foundation for future analyses. In brief, while the human component is unlikely to be substituted altogether in future clinical practice and while a few questions still await an answer, AI is an extremely promising means of improvement in patient care and resource optimization.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bao CH, China; Dai YC, China; Liu XQ, China S-Editor: Chen YL L-Editor: Filipodia A P-Editor: Chen YL
| 1. | Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389:1756-1770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2199] [Cited by in RCA: 2461] [Article Influence: 307.6] [Reference Citation Analysis (2)] |
| 2. | Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn’s disease. Lancet. 2017;389:1741-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1121] [Cited by in RCA: 1787] [Article Influence: 223.4] [Reference Citation Analysis (111)] |
| 3. | Mak WY, Zhao M, Ng SC, Burisch J. The epidemiology of inflammatory bowel disease: East meets west. J Gastroenterol Hepatol. 2020;35:380-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 406] [Article Influence: 81.2] [Reference Citation Analysis (0)] |
| 4. | Park J, Park S, Lee SA, Park SJ, Cheon JH. Improving the care of inflammatory bowel disease (IBD) patients: perspectives and strategies for IBD center management. Korean J Intern Med. 2021;36:1040-1048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 5. | Maaser C, Sturm A, Vavricka SR, Kucharzik T, Fiorino G, Annese V, Calabrese E, Baumgart DC, Bettenworth D, Borralho Nunes P, Burisch J, Castiglione F, Eliakim R, Ellul P, González-Lama Y, Gordon H, Halligan S, Katsanos K, Kopylov U, Kotze PG, Krustinš E, Laghi A, Limdi JK, Rieder F, Rimola J, Taylor SA, Tolan D, van Rheenen P, Verstockt B, Stoker J; European Crohn’s and Colitis Organisation [ECCO] and the European Society of Gastrointestinal and Abdominal Radiology [ESGAR]. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis. 2019;13:144-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1242] [Cited by in RCA: 1153] [Article Influence: 192.2] [Reference Citation Analysis (0)] |
| 6. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 323] [Article Influence: 64.6] [Reference Citation Analysis (1)] |
| 7. | Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1155] [Cited by in RCA: 1941] [Article Influence: 215.7] [Reference Citation Analysis (6)] |
| 8. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36149] [Cited by in RCA: 19943] [Article Influence: 1994.3] [Reference Citation Analysis (0)] |
| 9. | Ananthakrishnan AN, Cai T, Savova G, Cheng SC, Chen P, Perez RG, Gainer VS, Murphy SN, Szolovits P, Xia Z, Shaw S, Churchill S, Karlson EW, Kohane I, Plenge RM, Liao KP. Improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis. 2013;19:1411-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 10. | Nehme F, Feldman K. Evolving Role and Future Directions of Natural Language Processing in Gastroenterology. Dig Dis Sci. 2021;66:29-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Li X, Liang D, Meng J, Zhou J, Chen Z, Huang S, Lu B, Qiu Y, Baker ME, Ye Z, Cao Q, Wang M, Yuan C, Feng S, Zhang Y, Iacucci M, Ghosh S, Rieder F, Sun C, Chen M, Li Z, Mao R, Huang B, Feng ST. Development and Validation of a Novel Computed-Tomography Enterography Radiomic Approach for Characterization of Intestinal Fibrosis in Crohn’s Disease. Gastroenterology. 2021;160:2303-2316.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 12. | Sumiyama K, Futakuchi T, Kamba S, Matsui H, Tamai N. Artificial intelligence in endoscopy: Present and future perspectives. Dig Endosc. 2021;33:218-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 13. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. 2021;93:77-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 307] [Article Influence: 76.8] [Reference Citation Analysis (1)] |
| 14. | Mori Y, Wang P, Løberg M, Misawa M, Repici A, Spadaccini M, Correale L, Antonelli G, Yu H, Gong D, Ishiyama M, Kudo SE, Kamba S, Sumiyama K, Saito Y, Nishino H, Liu P, Glissen Brown JR, Mansour NM, Gross SA, Kalager M, Bretthauer M, Rex DK, Sharma P, Berzin TM, Hassan C. Impact of Artificial Intelligence on Colonoscopy Surveillance After Polyp Removal: A Pooled Analysis of Randomized Trials. Clin Gastroenterol Hepatol. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 15. | Rondonotti E, Hassan C, Tamanini G, Antonelli G, Andrisani G, Leonetti G, Paggi S, Amato A, Scardino G, Di Paolo D, Mandelli G, Lenoci N, Terreni N, Andrealli A, Maselli R, Spadaccini M, Galtieri PA, Correale L, Repici A, Di Matteo FM, Ambrosiani L, Filippi E, Sharma P, Radaelli F. Artificial intelligence-assisted optical diagnosis for the resect-and-discard strategy in clinical practice: the Artificial intelligence BLI Characterization (ABC) study. Endoscopy. 2023;55:14-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 69] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 16. | Areia M, Mori Y, Correale L, Repici A, Bretthauer M, Sharma P, Taveira F, Spadaccini M, Antonelli G, Ebigbo A, Kudo SE, Arribas J, Barua I, Kaminski MF, Messmann H, Rex DK, Dinis-Ribeiro M, Hassan C. Cost-effectiveness of artificial intelligence for screening colonoscopy: a modelling study. Lancet Digit Health. 2022;4:e436-e444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 115] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 17. | Wallace MB, Sharma P, Bhandari P, East J, Antonelli G, Lorenzetti R, Vieth M, Speranza I, Spadaccini M, Desai M, Lukens FJ, Babameto G, Batista D, Singh D, Palmer W, Ramirez F, Palmer R, Lunsford T, Ruff K, Bird-Liebermann E, Ciofoaia V, Arndtz S, Cangemi D, Puddick K, Derfus G, Johal AS, Barawi M, Longo L, Moro L, Repici A, Hassan C. Impact of Artificial Intelligence on Miss Rate of Colorectal Neoplasia. Gastroenterology. 2022;163:295-304.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 140] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 18. | Spadaccini M, Iannone A, Maselli R, Badalamenti M, Desai M, Chandrasekar VT, Patel HK, Fugazza A, Pellegatta G, Galtieri PA, Lollo G, Carrara S, Anderloni A, Rex DK, Savevski V, Wallace MB, Bhandari P, Roesch T, Gralnek IM, Sharma P, Hassan C, Repici A. Computer-aided detection versus advanced imaging for detection of colorectal neoplasia: a systematic review and network meta-analysis. Lancet Gastroenterol Hepatol. 2021;6:793-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 19. | Repici A, Spadaccini M, Antonelli G, Correale L, Maselli R, Galtieri PA, Pellegatta G, Capogreco A, Milluzzo SM, Lollo G, Di Paolo D, Badalamenti M, Ferrara E, Fugazza A, Carrara S, Anderloni A, Rondonotti E, Amato A, De Gottardi A, Spada C, Radaelli F, Savevski V, Wallace MB, Sharma P, Rösch T, Hassan C. Artificial intelligence and colonoscopy experience: lessons from two randomised trials. Gut. 2022;71:757-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 132] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 20. | Sinagra E, Badalamenti M, Maida M, Spadaccini M, Maselli R, Rossi F, Conoscenti G, Raimondo D, Pallio S, Repici A, Anderloni A. Use of artificial intelligence in improving adenoma detection rate during colonoscopy: Might both endoscopists and pathologists be further helped. World J Gastroenterol. 2020;26:5911-5918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 21. | Iacucci M, Furfaro F, Matsumoto T, Uraoka T, Smith S, Ghosh S, Kiesslich R. Advanced endoscopic techniques in the assessment of inflammatory bowel disease: new technology, new era. Gut. 2019;68:562-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 22. | Vinsard DG, Fetzer J, Raffals LH, Agrawal U, Singh J, Patel MH, Avvaru HK, Kaur MP, Lahori S, Sampath S, Arunachalam SP, Leggett CL, Sundaram DSB, Coelho-Prabhu N. Development of an artificial intelligence tool for detection of polypoid lesions in inflammatory bowel disease (IBD-CADE). Gastrointest Endosc. 2022;95:AB220-1. [DOI] [Full Text] |
| 23. | Borg-Bartolo SP, Boyapati RK, Satsangi J, Kalla R. Precision medicine in inflammatory bowel disease: concept, progress and challenges. F1000Res. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 24. | Zhang YZ, Li YY. Inflammatory bowel disease: pathogenesis. World J Gastroenterol. 2014;20:91-99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 751] [Cited by in RCA: 1031] [Article Influence: 93.7] [Reference Citation Analysis (18)] |
| 25. | Annese V. Genetics and epigenetics of IBD. Pharmacol Res. 2020;159:104892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 26. | Isakov O, Dotan I, Ben-Shachar S. Machine Learning-Based Gene Prioritization Identifies Novel Candidate Risk Genes for Inflammatory Bowel Disease. Inflamm Bowel Dis. 2017;23:1516-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 27. | Cheng B, Liang X, Wen Y, Li P, Zhang L, Ma M, Cheng S, Du Y, Liu L, Ding M, Zhao Y, Zhang F. Integrative analysis of transcriptome-wide association study data and messenger RNA expression profiles identified candidate genes and pathways for inflammatory bowel disease. J Cell Biochem. 2019;120:14831-14837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Yuan F, Zhang YH, Kong XY, Cai YD. Identification of Candidate Genes Related to Inflammatory Bowel Disease Using Minimum Redundancy Maximum Relevance, Incremental Feature Selection, and the Shortest-Path Approach. Biomed Res Int. 2017;2017:5741948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 29. | Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1021] [Cited by in RCA: 1173] [Article Influence: 146.6] [Reference Citation Analysis (0)] |
| 30. | Mihajlović A, Mladenović K, Lončar-Turukalo T, Brdar S. Machine Learning Based Metagenomic Prediction of Inflammatory Bowel Disease. Stud Health Technol Inform. 2021;285:165-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 31. | Manandhar I, Alimadadi A, Aryal S, Munroe PB, Joe B, Cheng X. Gut microbiome-based supervised machine learning for clinical diagnosis of inflammatory bowel diseases. Am J Physiol Gastrointest Liver Physiol. 2021;320:G328-G337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 32. | Mossotto E, Ashton JJ, Coelho T, Beattie RM, MacArthur BD, Ennis S. Classification of Paediatric Inflammatory Bowel Disease using Machine Learning. Sci Rep. 2017;7:2427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 33. | Quénéhervé L, David G, Bourreille A, Hardouin JB, Rahmi G, Neunlist M, Brégeon J, Coron E. Quantitative assessment of mucosal architecture using computer-based analysis of confocal laser endomicroscopy in inflammatory bowel diseases. Gastrointest Endosc. 2019;89:626-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 34. | Mary JY, Modigliani R. Development and validation of an endoscopic index of the severity for Crohn’s disease: a prospective multicentre study. Groupe d’Etudes Thérapeutiques des Affections Inflammatoires du Tube Digestif (GETAID). Gut. 1989;30:983-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 665] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 35. | Daperno M, D’Haens G, Van Assche G, Baert F, Bulois P, Maunoury V, Sostegni R, Rocca R, Pera A, Gevers A, Mary JY, Colombel JF, Rutgeerts P. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc. 2004;60:505-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 999] [Cited by in RCA: 1314] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 36. | Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1958] [Cited by in RCA: 2246] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 37. | Travis SP, Schnell D, Krzeski P, Abreu MT, Altman DG, Colombel JF, Feagan BG, Hanauer SB, Lémann M, Lichtenstein GR, Marteau PR, Reinisch W, Sands BE, Yacyshyn BR, Bernhardt CA, Mary JY, Sandborn WJ. Developing an instrument to assess the endoscopic severity of ulcerative colitis: the Ulcerative Colitis Endoscopic Index of Severity (UCEIS). Gut. 2012;61:535-542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 450] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 38. | Samuel S, Bruining DH, Loftus EV Jr, Thia KT, Schroeder KW, Tremaine WJ, Faubion WA, Kane SV, Pardi DS, de Groen PC, Harmsen WS, Zinsmeister AR, Sandborn WJ. Validation of the ulcerative colitis colonoscopic index of severity and its correlation with disease activity measures. Clin Gastroenterol Hepatol. 2013;11:49-54.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 39. | Stidham RW, Liu W, Bishu S, Rice MD, Higgins PDR, Zhu J, Nallamothu BK, Waljee AK. Performance of a Deep Learning Model vs Human Reviewers in Grading Endoscopic Disease Severity of Patients With Ulcerative Colitis. JAMA Netw Open. 2019;2:e193963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 40. | Ozawa T, Ishihara S, Fujishiro M, Saito H, Kumagai Y, Shichijo S, Aoyama K, Tada T. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest Endosc. 2019;89:416-421.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (1)] |
| 41. | Maeda Y, Kudo SE, Ogata N, Misawa M, Iacucci M, Homma M, Nemoto T, Takishima K, Mochida K, Miyachi H, Baba T, Mori K, Ohtsuka K, Mori Y. Evaluation in real-time use of artificial intelligence during colonoscopy to predict relapse of ulcerative colitis: a prospective study. Gastrointest Endosc. 2022;95:747-756.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 43] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 42. | Gottlieb K, Requa J, Karnes W, Chandra Gudivada R, Shen J, Rael E, Arora V, Dao T, Ninh A, McGill J. Central Reading of Ulcerative Colitis Clinical Trial Videos Using Neural Networks. Gastroenterology. 2021;160:710-719.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 95] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 43. | Yao H, Najarian K, Gryak J, Bishu S, Rice MD, Waljee AK, Wilkins HJ, Stidham RW. Fully automated endoscopic disease activity assessment in ulcerative colitis. Gastrointest Endosc. 2021;93:728-736.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 44. | Byrne M, East J, Iacucci M, Panaccione R, Kalapala R, Duvvur N, Rughwani H, Singh A, Henkel M, Berry S, Canaran L, Laage G, St-Denis L, Nikfal S, Asselin J, Monsurate R, Cremonese E, Soudan F, Travis S. DOP13 Artificial Intelligence (AI) in endoscopy-Deep learning for detection and scoring of Ulcerative Colitis (UC) disease activity under multiple scoring systems. J Crohns Colitis. 2021;15:S051-2. [DOI] [Full Text] |
| 45. | Takenaka K, Ohtsuka K, Fujii T, Negi M, Suzuki K, Shimizu H, Oshima S, Akiyama S, Motobayashi M, Nagahori M, Saito E, Matsuoka K, Watanabe M. Development and Validation of a Deep Neural Network for Accurate Evaluation of Endoscopic Images From Patients With Ulcerative Colitis. Gastroenterology. 2020;158:2150-2157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 174] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 46. | Maeda Y, Kudo SE, Mori Y, Misawa M, Ogata N, Sasanuma S, Wakamura K, Oda M, Mori K, Ohtsuka K. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. 2019;89:408-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 163] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 47. | Bossuyt P, Nakase H, Vermeire S, de Hertogh G, Eelbode T, Ferrante M, Hasegawa T, Willekens H, Ikemoto Y, Makino T, Bisschops R. Automatic, computer-aided determination of endoscopic and histological inflammation in patients with mild to moderate ulcerative colitis based on red density. Gut. 2020;69:1778-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 48. | Pennazio M, Spada C, Eliakim R, Keuchel M, May A, Mulder CJ, Rondonotti E, Adler SN, Albert J, Baltes P, Barbaro F, Cellier C, Charton JP, Delvaux M, Despott EJ, Domagk D, Klein A, McAlindon M, Rosa B, Rowse G, Sanders DS, Saurin JC, Sidhu R, Dumonceau JM, Hassan C, Gralnek IM. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Clinical Guideline. Endoscopy. 2015;47:352-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 558] [Article Influence: 55.8] [Reference Citation Analysis (1)] |
| 49. | Gralnek IM, Defranchis R, Seidman E, Leighton JA, Legnani P, Lewis BS. Development of a capsule endoscopy scoring index for small bowel mucosal inflammatory change. Aliment Pharmacol Ther. 2008;27:146-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 325] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 50. | Niv Y, Ilani S, Levi Z, Hershkowitz M, Niv E, Fireman Z, O’Donnel S, O’Morain C, Eliakim R, Scapa E, Kalantzis N, Kalantzis C, Apostolopoulos P, Gal E. Validation of the Capsule Endoscopy Crohn’s Disease Activity Index (CECDAI or Niv score): a multicenter prospective study. Endoscopy. 2012;44:21-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 135] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 51. | Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Tanaka S, Koike K, Tada T. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2019;89:357-363.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 173] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 52. | Klang E, Barash Y, Margalit RY, Soffer S, Shimon O, Albshesh A, Ben-Horin S, Amitai MM, Eliakim R, Kopylov U. Deep learning algorithms for automated detection of Crohn’s disease ulcers by video capsule endoscopy. Gastrointest Endosc. 2020;91:606-613.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 53. | Klang E, Grinman A, Soffer S, Margalit Yehuda R, Barzilay O, Amitai MM, Konen E, Ben-Horin S, Eliakim R, Barash Y, Kopylov U. Automated Detection of Crohn’s Disease Intestinal Strictures on Capsule Endoscopy Images Using Deep Neural Networks. J Crohns Colitis. 2021;15:749-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 54. | Ferreira JPS, de Mascarenhas Saraiva MJDQEC, Afonso JPL, Ribeiro TFC, Cardoso HMC, Ribeiro Andrade AP, de Mascarenhas Saraiva MNG, Parente MPL, Natal Jorge R, Lopes SIO, de Macedo GMG. Identification of Ulcers and Erosions by the Novel Pillcam™ Crohn’s Capsule Using a Convolutional Neural Network: A Multicentre Pilot Study. J Crohns Colitis. 2022;16:169-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (1)] |
| 55. | Aoki T, Yamada A, Aoyama K, Saito H, Fujisawa G, Odawara N, Kondo R, Tsuboi A, Ishibashi R, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Clinical usefulness of a deep learning-based system as the first screening on small-bowel capsule endoscopy reading. Dig Endosc. 2020;32:585-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 56. | Maeda Y, Kudo SE, Ogata N, Misawa M, Mori Y, Mori K, Ohtsuka K. Can artificial intelligence help to detect dysplasia in patients with ulcerative colitis? Endoscopy. 2021;53:E273-E274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 57. | Fukunaga S, Kusaba Y, Ohuchi A, Nagata T, Mitsuyama K, Tsuruta O, Torimura T. Is artificial intelligence a superior diagnostician in ulcerative colitis? Endoscopy. 2021;53:E75-E76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 58. | Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, Wang Z, Zhou W, An P, Huang X, Jiang X, Li Y, Wan X, Hu S, Chen Y, Hu X, Xu Y, Zhu X, Li S, Yao L, He X, Chen D, Huang L, Wei X, Wang X, Yu H. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 262] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 59. | Turner D, Ricciuto A, Lewis A, D’Amico F, Dhaliwal J, Griffiths AM, Bettenworth D, Sandborn WJ, Sands BE, Reinisch W, Schölmerich J, Bemelman W, Danese S, Mary JY, Rubin D, Colombel JF, Peyrin-Biroulet L, Dotan I, Abreu MT, Dignass A; International Organization for the Study of IBD. STRIDE-II: An Update on the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): Determining Therapeutic Goals for Treat-to-Target strategies in IBD. Gastroenterology. 2021;160:1570-1583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 1612] [Article Influence: 403.0] [Reference Citation Analysis (1)] |
| 60. | Bryant RV, Burger DC, Delo J, Walsh AJ, Thomas S, von Herbay A, Buchel OC, White L, Brain O, Keshav S, Warren BF, Travis SP. Beyond endoscopic mucosal healing in UC: histological remission better predicts corticosteroid use and hospitalisation over 6 years of follow-up. Gut. 2016;65:408-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 316] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 61. | Mosli MH, Feagan BG, Sandborn WJ, Dʼhaens G, Behling C, Kaplan K, Driman DK, Shackelton LM, Baker KA, Macdonald JK, Vandervoort MK, Geboes K, Levesque BG. Histologic evaluation of ulcerative colitis: a systematic review of disease activity indices. Inflamm Bowel Dis. 2014;20:564-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 62. | Mojtahed A, Khanna R, Sandborn WJ, D’Haens GR, Feagan BG, Shackelton LM, Baker KA, Dubcenco E, Valasek MA, Geboes K, Levesque BG. Assessment of histologic disease activity in Crohn’s disease: a systematic review. Inflamm Bowel Dis. 2014;20:2092-2103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 63. | Chateau T, Feakins R, Marchal-Bressenot A, Magro F, Danese S, Peyrin-Biroulet L. Histological Remission in Ulcerative Colitis: Under the Microscope Is the Cure. Am J Gastroenterol. 2020;115:179-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 64. | Vande Casteele N, Leighton JA, Pasha SF, Cusimano F, Mookhoek A, Hagen CE, Rosty C, Pai RK. Utilizing Deep Learning to Analyze Whole Slide Images of Colonic Biopsies for Associations Between Eosinophil Density and Clinicopathologic Features in Active Ulcerative Colitis. Inflamm Bowel Dis. 2022;28:539-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 65. | Gui X, Bazarova A, Del Amor R, Vieth M, de Hertogh G, Villanacci V, Zardo D, Parigi TL, Røyset ES, Shivaji UN, Monica MAT, Mandelli G, Bhandari P, Danese S, Ferraz JG, Hayee B, Lazarev M, Parra-Blanco A, Pastorelli L, Panaccione R, Rath T, Tontini GE, Kiesslich R, Bisschops R, Grisan E, Naranjo V, Ghosh S, Iacucci M. PICaSSO Histologic Remission Index (PHRI) in ulcerative colitis: development of a novel simplified histological score for monitoring mucosal healing and predicting clinical outcomes and its applicability in an artificial intelligence system. Gut. 2022;71:889-898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 89] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 66. | Villanacci V, Parigi TL, Del Amor R, Mesguer Esbrì P, Gui X, Bazarova A, Bhandari P, Bisschops R, Danese S, De Hertogh G, Ferraz JG, Götz M, Grisan E, Hayee B, Kiesslich R, Lazarev M, Mandelli G, Monica MAT, Panaccione R, Parra-Blanco A, Pastorelli L, Rath T, Røyset ES, Shivaji U, Tontini GE, Vieth M, Zardo D, Ghosh S, Naranjo V, Iacucci M. OP15 A new simplified histology artificial intelligence system for accurate assessment of remission in Ulcerative Colitis. J Crohns Colitis. 2022;16:i015-7. [DOI] [Full Text] |
| 67. | Peyrin-Biroulet L, Adsul S, Dehmeshki J, Kubassova O. DOP58 An artificial intelligence-driven scoring system to measure histological disease activity in Ulcerative Colitis. J Crohns Colitis. 2022;16:i105. [DOI] [Full Text] |
| 68. | Iacucci M, Cannatelli R, Parigi TL, Nardone OM, Tontini GE, Labarile N, Buda A, Rimondi A, Bazarova A, Bisschops R, Del Amor R, Meseguer P, Naranjo V, Ghosh S, Grisan E; PICaSSO group. A virtual chromoendoscopy artificial intelligence system to detect endoscopic and histologic activity/remission and predict clinical outcomes in ulcerative colitis. Endoscopy. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 69. | Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, Brogi E, Reuter VE, Klimstra DS, Fuchs TJ. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 1194] [Article Influence: 199.0] [Reference Citation Analysis (0)] |
| 70. | Marlicz W, Skonieczna-Żydecka K, Dabos KJ, Łoniewski I, Koulaouzidis A. Emerging concepts in non-invasive monitoring of Crohn’s disease. Therap Adv Gastroenterol. 2018;11:1756284818769076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 71. | Stidham RW, Yu D, Zhao X, Bishu S, Rice M, Bourque C, Vydiswaran VVG. Identifying the Presence, Activity, and Status of Extraintestinal Manifestations of Inflammatory Bowel Disease Using Natural Language Processing of Clinical Notes. Inflamm Bowel Dis. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 72. | Reddy BK, Delen D, Agrawal RK. Predicting and explaining inflammation in Crohn’s disease patients using predictive analytics methods and electronic medical record data. Health Informatics J. 2019;25:1201-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 73. | Takenaka K, Fujii T, Kawamoto A, Suzuki K, Shimizu H, Maeyashiki C, Yamaji O, Motobayashi M, Igarashi A, Hanazawa R, Hibiya S, Nagahori M, Saito E, Okamoto R, Ohtsuka K, Watanabe M. Deep neural network for video colonoscopy of ulcerative colitis: a cross-sectional study. Lancet Gastroenterol Hepatol. 2022;7:230-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 74. | Argollo M, Kotze PG, Kakkadasam P, D’Haens G. Optimizing biologic therapy in IBD: how essential is therapeutic drug monitoring? Nat Rev Gastroenterol Hepatol. 2020;17:702-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 75. | Li Y, Pan J, Zhou N, Fu D, Lian G, Yi J, Peng Y, Liu X. A random forest model predicts responses to infliximab in Crohn’s disease based on clinical and serological parameters. Scand J Gastroenterol. 2021;56:1030-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 76. | Waljee AK, Liu B, Sauder K, Zhu J, Govani SM, Stidham RW, Higgins PDR. Predicting Corticosteroid-Free Biologic Remission with Vedolizumab in Crohn’s Disease. Inflamm Bowel Dis. 2018;24:1185-1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (1)] |
| 77. | Waljee AK, Liu B, Sauder K, Zhu J, Govani SM, Stidham RW, Higgins PDR. Predicting corticosteroid-free endoscopic remission with vedolizumab in ulcerative colitis. Aliment Pharmacol Ther. 2018;47:763-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 78. | Doherty MK, Ding T, Koumpouras C, Telesco SE, Monast C, Das A, Brodmerkel C, Schloss PD. Fecal Microbiota Signatures Are Associated with Response to Ustekinumab Therapy among Crohn’s Disease Patients. mBio. 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 121] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 79. | Pinton P. Computational models in inflammatory bowel disease. Clin Transl Sci. 2022;15:824-830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 80. | Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, Li B, Madabhushi A, Shah P, Spitzer M, Zhao S. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 1198] [Article Influence: 199.7] [Reference Citation Analysis (0)] |
| 81. | Gupta R, Srivastava D, Sahu M, Tiwari S, Ambasta RK, Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers. 2021;25:1315-1360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 574] [Cited by in RCA: 453] [Article Influence: 113.3] [Reference Citation Analysis (0)] |
| 82. | Nuzzo A, Saha S, Berg E, Jayawickreme C, Tocker J, Brown JR. Expanding the drug discovery space with predicted metabolite-target interactions. Commun Biol. 2021;4:288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 83. | Tan X, Li C, Yang R, Zhao S, Li F, Li X, Chen L, Wan X, Liu X, Yang T, Tong X, Xu T, Cui R, Jiang H, Zhang S, Liu H, Zheng M. Discovery of Pyrazolo[3,4-d]pyridazinone Derivatives as Selective DDR1 Inhibitors via Deep Learning Based Design, Synthesis, and Biological Evaluation. J Med Chem. 2022;65:103-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 84. | Con D, van Langenberg DR, Vasudevan A. Deep learning vs conventional learning algorithms for clinical prediction in Crohn’s disease: A proof-of-concept study. World J Gastroenterol. 2021;27:6476-6488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (2)] |