Published online Jan 14, 2023. doi: 10.3748/wjg.v29.i2.272
Peer-review started: September 23, 2022
First decision: November 18, 2022
Revised: December 1, 2022
Accepted: December 21, 2022
Article in press: December 21, 2022
Published online: January 14, 2023
Processing time: 104 Days and 21.2 Hours
There is great heterogeneity among inflammatory bowel disease (IBD) patients in terms of pathogenesis, clinical manifestation, response to treatment, and prognosis, which requires the individualized and precision management of patients. Many studies have focused on prediction biomarkers and models for assessing IBD disease type, activity, severity, and prognosis. During the era of biologics, how to predict the response and side effects of patients to different treatments and how to quickly recognize the loss of response have also become important topics. Multiomics is a promising area for investigating the complex network of IBD pathogenesis. Integrating numerous amounts of data requires the use of artificial intelligence.
Core Tip: Inflammatory bowel diseases (IBDs) exhibit different pathogeneses and clinical manifestations. Making precise and appropriate therapeutic decisions according to the condition of each patient remains challenging. We summarize the clustering strategies, the approaches used to apply multiomics and artificial intelligence to IBD precision management.
- Citation: Liu XY, Tang H, Zhou QY, Zeng YL, Chen D, Xu H, Li Y, Tan B, Qian JM. Advancing the precision management of inflammatory bowel disease in the era of omics approaches and new technology. World J Gastroenterol 2023; 29(2): 272-285
- URL: https://www.wjgnet.com/1007-9327/full/v29/i2/272.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i2.272
Since it was first proposed in 2011[1], the concept of precision medicine has become increasingly popular and attracted much attention. Great progress has been made, especially in the treatment of cancer. Precision medicine typically refers to the use of targeted therapy based on etiology and mechanism. The essence of the idea involves classifying individuals with common characteristics into the same subgroup using specific clinical features, treatment features and prognoses. Thus, this strategy should actually combine a wide array of data, including clinical, genetic and environmental infor
Inflammatory bowel disease (IBD) is a group of intestinal disorders of unknown etiology characterized by inflammation that arises from a complex interaction between genetic and environmental factors and immune responses[3]. An increasing number of studies have reported on the great heterogeneity of IBD patients in terms of pathogenesis, clinical manifestation, response to treatment and prognosis, and IBD is currently regarded as a continuous spectrum of diseases[4]. The introduction of biologics has greatly improved the quality of life of IBD patients, which also embodies precision medicine to some extent. However, due to the complexity of pathogenesis, targeting only immune pathways without addressing the genome or microbiome may result in limited success[5]. The treatment strategies used are largely based on evidence from clinical trials, which typically do not stratify patients with enough precision. Additionally, the frequency of treatment may be indicated for a certain population, but this approach might not be the most suitable for an individual. Compared to the oncology field, there is still much room for precision medicine development in IBD.
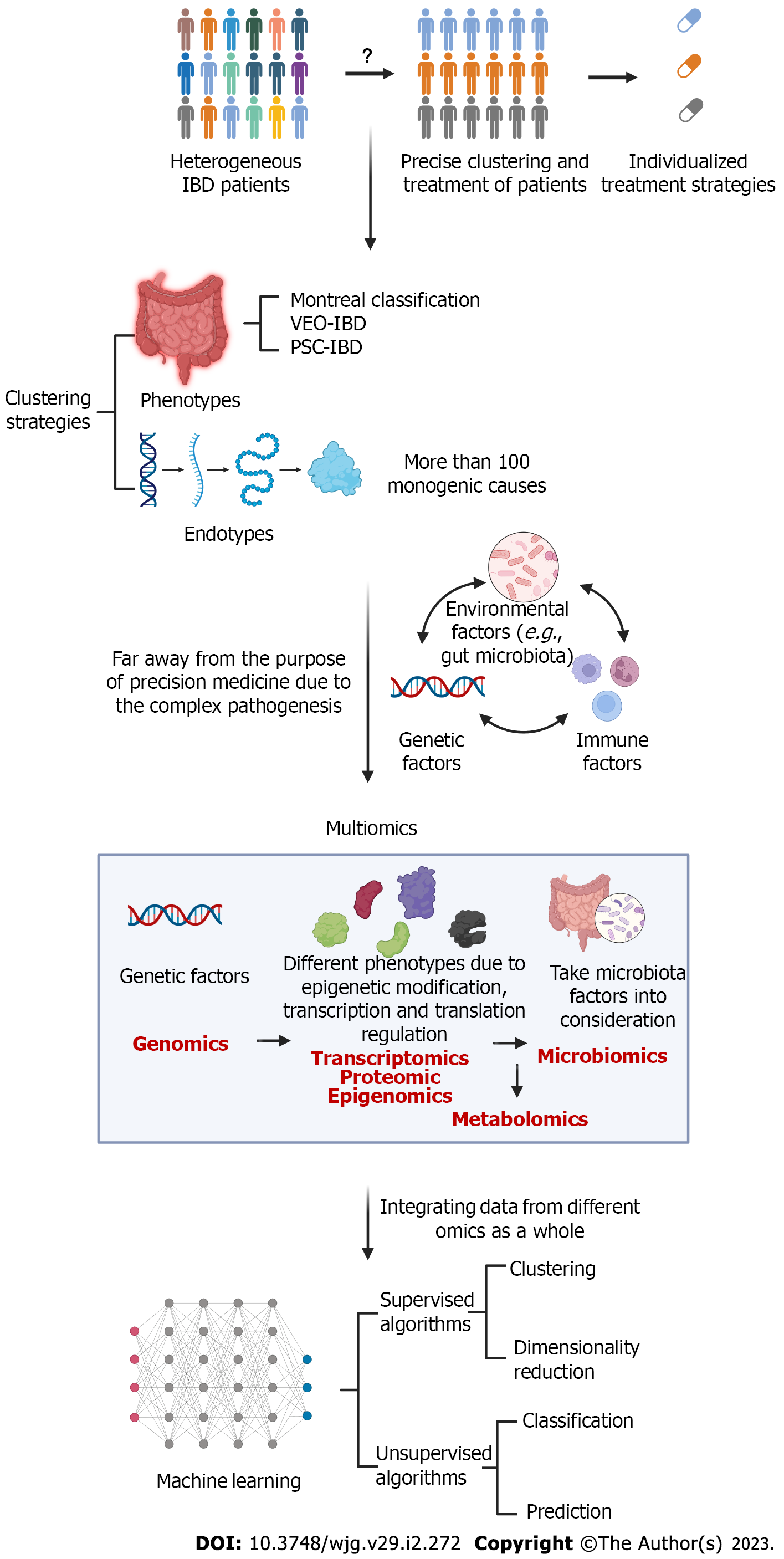
In this review, we discuss the strategies used to categorize IBD patients and biomarkers for identifying these subgroups. We suggest that applications of multiomics and artificial intelligence (AI) approaches could facilitate the precision management of IBD patients (Figure 1).
Phenotype refers to the traits that can be observed in patients, and deep phenotyping plays a key role in the progress of precision medicine[2]. In other disease contexts, there is also the concept of endotype, which is defined as the molecular mechanism underlying the visible phenotype[6]. However, clustering phenotypes and endotypes remains difficult in the context of IBD due to heterogeneity.
The Montreal classification is the most widely used clinical classification of IBD and considers age at diagnosis, location and behavioral factors[7]. The characteristics and natural history of IBD seemed to vary depending on the age of onset[8]. Very early onset IBD (VEO-IBD), defined as IBD occurring in those who are diagnosed under the age of 6 years and sometimes exhibiting a more aggressive disease pattern, seemed to have stronger genetic triggers with less environmental influence[9]. In addition, complications or extraintestinal organ involvement can also be used to identify some unique subsets of IBD patients. For example, IBD patients who experienced complications with primary sclerosing cholangitis (PSC) have been shown to exhibit higher rates of colectomy, cancer and death than non-PSC-IBD patients[10]. However, current clinical classification is far from the precise identification of IBD phenotypes.
Some specific genetic factors have been found to determine disease progression, which is difficult to assess by clinical manifestations. Next-generation sequencing has been used to identify more than 100 monogenic causes that could manifest as IBD-like phenotypes. The genes involved in monogenic IBD disorders are generally classified into six categories according to the mechanisms: Epithelial barrier defects; T-cell and B-cell defects; hyperinflammatory and autoinflammatory disorders; phagocytic defects; immunoregulation defects; and others[11]. For example, mutations in the IL-10 pathway could lead to neonatal or infantile VEO-IBD with severe enterocolitis and crissum disease by impairing IL-10-mediated control of inflammatory responses involving IL-1 and IL-23. Mutations in CYBB could lead to chronic granulomatous disease characterized by intestinal inflammation and autoimmune disease due to impaired antimicrobial activity caused by defects in NADPH oxidase[12,13]. Conventional treatment in patients with the subsets of IBD that are largely driven by genetic factors often exhibit unsatisfactory efficacy, and these patients have poor prognosis. These various mechanisms that underlie the effects of monogenic mutations also have some crossover with the mechanisms involved in sporadic and multifactorial IBD, which reflects the divergence and convergence of the mechanisms. For precision treatment of IBD, strategies should not be limited to therapies targeting upstream etiology; therapy based on more superficial mechanisms should also be pursued.
The etiology of monogenic causes, which account for only a small percentage of IBD cases, is complex, but sporadic IBD involves many more factors. More than 260 risk loci have been identified to be associated with sporadic IBD by genome-wide association studies (GWASs)[14], yet these loci only explained approximately 20% of the genetic heritability in complex adult-onset IBD[15]. This finding is easy to understand because there are also environmental, microbiota or other factors involved in the pathogenesis of IBD. IBD cannot be classified by a single factor, but the application of biomarkers can aid in the advancement of the precision management of IBD to some extent.
Due to the rising incidence of IBD and the inconvenience of endoscopy, there is an urgent need for noninvasive, accessible and cost-efficient biomarkers. Precision medicine should cover the whole management process of IBD patients, including the early identification of patients at potential risk for disease progression and enabling appropriate adjustments in response to ongoing assessments of treatment efficacy. Such a strategy should be a highly sophisticated process, not just the endpoint of a single stratification approach[16]. Accordingly, we reviewed two categories of biomarkers (mainly from serum and feces) for IBD: Those used to identify disease progression risk and activity and those used to predict treatment responses.
C-reactive protein (CRP) is the most widely used serum biomarker for inflammation in IBD[17]. It reflects both clinical disease activity and endoscopic inflammation in IBD patients[18]. Additionally, the level of CRP is not influenced by treatments and thus is also suitable for monitoring treatment response[19]. However, it is not a specific biomarker, and its levels might be elevated in other diseases, including noninflammatory conditions. Additionally, up to 25% of Crohn’s disease (CD) patients with endoscopically proven activity could not be identified by CRP[20]. Fecal calprotectin (FC) is another important noninvasive biomarker widely used in clinical practice. In the assessment of endoscopically defined disease activity in IBD, FC analysis exhibits higher sensitivity in the context of both ulcerative colitis (UC) and CD, especially in UC[21,22]. In particular, FC analysis could be used in the early prediction of relapse risk 6 and even 12 mo in advance[23]. A meta-analysis reported 78% sensitivity and 73% specificity when using FC at remission to predict IBD relapse, with cutoffs varying between 120 μg/g and 340 μg/g[24]. However, this biomarker also faces the problem of limited specificity; inflammation in the gut that is not associated with IBD, such as during infection, necrotizing enterocolitis and drug-induced enteropathy, could confound the results[25]. For patients with borderline FC levels, combining FC analysis with other metrics, such as clinical activity indices or CRP levels, could help the assessment[26]. The levels of serum calprotectin (SC), as an indirect marker of inflammatory activity in UC[27], can indicate the involvement of other extraintestinal organs. Another well-accepted fecal biomarker is fecal lactoferrin (FL), the levels of which could also reflect IBD activity and be used to predict disease relapse. Unlike the analysis of FC levels, the advantage of FL is its specificity[28], and combined analysis of these biomarkers might result in better assessment. In addition, some secondary biomarkers measured using simple laboratory tests, such as the CRP-albumin ratio, neutrophil-lymphocyte ratio (NLR), platelet-lymphocyte ratio (PLR) and lymphocyte-monocyte ratio (LMR), can also be used to infer the activity of IBD[29,30].
In recent years, an increasing number of novel serum and fecal biomarkers with application potential have been discovered. The levels of leucine-rich glycoprotein, a glycoprotein that is also related to IBD activity, could be elevated in patients with normal CRP levels during the active period of UC[31]. Some serum antibodies resulting from autoimmunity and loss of immune tolerance to microbial antigens have been considered in the diagnosis and assessment of IBD[32]. For example, anti-Saccharomyces cerevisiae antibody (ASCA) and perinuclear anti-neutrophil cytoplasmic antibody (pANCA), which are antibodies of microbial antigens and autoantibodies, respectively[33], are two extensively studied antibodies with high specificity for IBD[34]. They could help identify potential CD patients five years before diagnosis when combined with the analysis of other protein markers[35]. In addition to enabling diagnosis, a higher ASCA titer was related to more aggressive fibrosis and stenosis and internal penetrating disease behaviors[36], while the pANCA titer changed with the activity of UC[37]. In addition, cytokines such as granulocyte colony-stimulating factor were associated with endoscopically active disease[38], while IL-6 and IL-2 Levels could also be used to predict the course of disease relapse 12 mo in advance in quiescent CD patients[39]. Circulating noncoding RNAs, including microRNAs (miRNAs) and long ncRNAs (lncRNAs), also play a role in IBD, and the analysis of miRNAs might help monitor disease activity and stricture phenotypes[32]. Other newly emerging biomarkers for disease progression risk and activity include cathelicidin[40], trefoil factor 3[41], and 25-hydroxyvitamin D3[42]. Several extracellular matrix (ECM) components and growth factors are important biomarkers indicating intestinal fibrosis and stenosis[32]. The analysis of fecal biomarkers, cytokines and other indicators of inflammation could also help with the identification of IBD activity[43]. Fecal myeloperoxidase, another biomarker related to neutrophil inflammation in addition to FC, was recently reported to accurately indicate endoscopic activity in IBD and predict the disease course during follow-up[44]. However, all these novel biomarkers are mainly used in research and remain far from clinical use (Table 1).
| Sample | Biomarker | Outcome | Characteristic |
| Serum | CRP[17-20] | Monitor disease activity and mucosal healing | Widely used and low-cost; lack of specificity for intestinal inflammation; relatively low sensitivity |
| SC[27] | Disease burden, prognosis, and relapse | More representative of systemic inflammation | |
| LRG[31] | Monitor disease activity and mucosal healing | More correlated to activity in UC than CRP | |
| Serum antibodies | |||
| ASCA[35,36] | More aggressive fibro stenosing and internal penetrating disease behaviors | CD specificity | |
| pANCA[35,37] | UC disease activity | UC specificity | |
| Cytokines | |||
| G-CSF, IL-1Ra, PDGF-BB[38] | Endoscopically active disease | - | |
| IL-6, IL-2[39] | Predict disease relapse in quiescent CD | - | |
| Noncoding RNAs[32] | Monitor disease activity and stricture phenotypes | - | |
| ECM components[32] | Intestinal fibrosis and stenosis | - | |
| Growth factors[32] | Intestinal fibrosis and stenosis | - | |
| Cathelicidin[40] | Mucosal disease activity in UC, risk of intestinal stricture in CD, and clinical prognosis in IBD | - | |
| Trefoil Factor 3[41] | Monitor disease activity | - | |
| Vitamin D[42] | Disease activity | - | |
| Secondary biomarkers (CRP-albumin ratio, NLR, PLR, LMR)[29,30] | IBD activity | Easy to obtain; fluctuates greatly | |
| Feces | FC[21-26] | Monitor disease activity and mucosal healing; early prediction of relapse risk | Higher sensitivity than CRP; confounding of non-IBD gut inflammation |
| FL[28] | Monitor disease activity and predict disease relapse | Higher specificity than fecal calprotectin | |
| MPO[44] | Endoscopic activity in IBD and predict the disease course during follow-up | - |
The mainstream therapeutic drugs for IBD include aminosalicylates (ASAs), glucocorticoids (GCs) and immunosuppressive agents[45]. The treatment of IBD has greatly advanced since the recent introduction of biologics, including tumour necrosis factor-α (TNF-α) inhibitors (such as infliximab and adali
For 5-ASA, a multicenter prospective cohort study in pediatric UC patients developed a predictive model with initial clinical activity and early treatment response to 5-ASA to predict long-term corticosteroid-free remission[46]. The baseline FC level and UC endoscopic index of severity could be used to predict the early outcome of GCs treatment[47].
Clinical responses to biologics are even more varied. However, the application of biologics is usually tried in a certain order by experienced physicians without effective biomarkers used to influence the selection of different kinds of biologics, which is of concern in research. Some laboratory test results are taken into consideration. For example, a clinical trial revealed that using CRP and FC levels in combination with clinical symptoms could result in better clinical and endoscopic outcomes than considering only clinical symptoms[48]. These downstream indicators of active inflammation could suggest a response to anti-inflammatory TNF-α inhibitors, but they are still not enough for accurate prediction of the likelihood of remission in a given patient in a real clinical environment[49]. Thus, we need to explore more biomarkers that could reveal the molecular heterogeneity of patients treated with different biologics.
Existing biologics can be briefly classified into two groups according to underlying mechanisms: inhibitors of cytokines and inhibitors of lymphocyte migration. TNF-α is considered to be a downstream inflammatory pathway effector in multiple immune-related diseases[50]. It is rational to speculate that IBD patients with increased TNF-α levels might have a good response to anti-TNF-α agents. Detecting membrane-bound TNF (mTNF) by endoscopy with the aid of fluorescence labeling has been used to successfully predict the efficacy of anti-TNF-α treatment[51]. Another study also reached this goal by measuring the TNF production capacity of peripheral blood mononuclear cells (PBMCs)[52]. A lack of response to anti-TNF-α therapy might indicate the activation of other inflammatory pathways. Baseline levels of serum oncostatin M, a member of the IL-6 family that might mediate inflammation in another manner, have been reported to be elevated in anti-TNF-α nonresponders and could be used to predict the efficacy of this treatment[53,54]. In addition, low triggering receptor expressed on myeloid cells 1 (TREM1) expression in both whole peripheral blood samples and intestinal biopsy samples, which indicated a complete macrophage autophagy pathway, could be used to predict a good anti-TNF response in IBD patients[55]. Antibodies including anti-drug antibodies (ADA), pANCA and anti-OmpC (Escherichia coli outer membrane porin) were also found to be associated with the response to infliximab[56]. In the aspect of relapse after discontinuation of anti-TNF therapy, mucosal TNF gene expression and IL1RL1-transcripts might play a role[57].
The IL23/Th17 pathway is also a central cytokine pathway involved in IBD in addition to TNF-α. The levels of IL-22 and IL-17, the downstream factors involved in this pathway, are potential molecular predictors of the response to IL-23 blockers[58]. Another category of biologics for IBD is integrin inhibitors, which act by inhibiting gut-selective lymphocyte homing. The most widely used integrin inhibitor, vedolizumab, works by blocking the binding of the α4β7 integrin heterodimer on lymphocytes to MAdCAM1 on the gut[59]. Higher expression levels of α4β7 on T, B, and NK cells as well as the presence of α4β7+ intestinal mucosa cells could be used to predict responses to vedolizumab[60,61]. However, the predictive role of serum α4β7, VCAM-1 and ICAM-1 remains controversial[62,63]. In addition, higher IL-6 and IL-8 Levels have been reported to be associated with the response to vedolizumab[64] (Table 2).
| Type of agent | Biomarker | Sample | Outcome |
| 5-ASA | Initial clinical activity, early treatment response of 5-ASA[46] | - | Predict long-term corticosteroid-free remission |
| GCs | FC, UCEIS[47] | Feces | Predict the early outcome of GCs treatment |
| Biologics | |||
| TNF-α inhibitors | mTNF[51] | Endoscopy | Predict anti-TNF-α efficacy |
| TNF production capacity of PBMCs[52] | Blood | Predict anti-TNF-α efficacy | |
| OSM[53,54] | Serum | Upregulate in anti-TNF-α non-responders | |
| TREM1 expression[55] | Blood and intestinal biopsies | Predict anti-TNF-α efficacy | |
| Antibodies: ADA, pANCA, anti-OmpC[56] | Serum | Associate with the response to infliximab | |
| Mucosal TNF gene expression and IL1RL1- transcripts[57] | Intestinal biopsies | Predict long-term remission after discontinuation of anti-TNF-α therapy | |
| IL-23 inhibitors | IL-22, IL-17[58] | Serum | Predict anti-IL-23 efficacy |
| Integrin inhibitors | α4β7 on T, B, and NK cells[60,61] | Blood and endoscopy | Predict responses to vedolizumab |
| α4β7, VCAM-1, ICAM-1[62,63] | Serum | Remain controversial | |
| IL-6, IL-8[64] | Serum | Associate with the response to vedolizumab |
Sometimes, simply by assessing early responses to biologics, we can judge the potential future efficacy to some extent[65]. Therapeutic drug monitoring (TDM) is another tool for the assessment of biologic therapeutic outcomes based on the findings that drug concentrations correlate with biologic efficacy. However, due to the long time required for detection and the lack of an instructive reference range, there is still no consensus for the use of TDM[66,67].
In addition to simple serum and fecal biomarkers, emerging high-throughput analytical technologies offer opportunities for the improved management of IBD. Omics strategies, often including genomics, transcriptomics, proteomics, metabolomics, epigenomics and microbiomics, have completely transformed the trajectory of medicine. These different omics approaches might also provide some insights into IBD from different perspectives.
Genomics aims to characterize and quantify all the genetic information of an organism. Numerous variations in factors of genetic susceptibility involved in many complex diseases have been identified. Different genes have been reported to associate with IBD severity and activity. NOD2/CARD15 is the most classic CD-related gene found in Western countries[68,69] and is also associated with stricturing behaviors and the need for operation[70,71]. However, it was not found to be related to CD development in East Asian cohorts[72]. Regarding UC, a GWAS developed a risk score based on 46 single nucleotide polymorphisms to identify medically refractory UC that needed colectomy[73]. Regarding therapeutic efficacy, HLA-DQA1*05 carriage was reported to be associated with ADAs for TNF-α inhibitors in CD and suggested the need for combination therapy[74]. In addition, polymor
In exploring the pathogenesis of complex diseases such as IBD, the limitations of using a single genomics approach are becoming increasingly apparent. The same gene variation might lead to distinct phenotypes by epigenetic modification and transcriptional and translational regulation. Transcriptomics, proteomics and epigenomics could better reflect the gene expression profiles, which combine genetic and environmental factors and thus perform better than simple genomics.
In particular, numerous transcriptomic studies have provided insights into the prediction of IBD progression. Researchers found that the expression of ECM accumulation-associated genes in the ileum was associated with stricturing behaviors in pediatric CD patients. After combining age, race, disease location, and antimicrobial serology factors, they established a competing-risk model that reached a specificity of 71%[78]. However, ileum samples are difficult to access, which poses a barrier to the utility of this approach. Studies on blood samples are thus warranted. Transcriptional profiling of circulating CD8+ T cells successfully distinguished patients with a risk of aggressive disease mainly based on the expression of genes involved in T-cell responses[79]. Furthermore, this classification was also feasible for use in whole blood samples with transcriptional signatures based on 17 genes[80]. The transcriptional risk score, which represented the summation of risk alleles for CD from ileum or blood samples, could be used to identify patients who would progress to complicated disease[81]. Regarding treatment, UC patients could be clustered into different groups with distinct transcriptomic profiles of the rectum and showed different responses to anti-TNF therapy[82].
Unlike transcriptomics, the use of proteomics in the context of IBD is still in its infancy. Some studies have sought to detect proteins involved in early inflammatory mechanisms of IBD, and some proteome analyses have been performed in studies with small sample sizes to investigate the differentiation of disease behavior as well as the prediction of response to biological treatment[83,84]. In the Proteomic Evaluation and Discovery in an IBD Cohort of Tri-service Subjects (PREDICTS) study, a series of protein biomarkers involved in the complement cascade, lysosomes, innate immune response, and glycosaminoglycan metabolism along with some antibodies were able to be used to predict potential CD patients 5 years in advance[35]. Another study revealed that MMP10, CXCL9, CXCL11, and MCP1 were upregulated in UC patients before disease onset[85]. However, there is still a long way to go before these approaches are applied clinically.
Epigenetic mechanisms mainly include DNA methylation, histone modifications, and noncoding RNAs. The complement of methylated DNA in the genome is called the methylome. Epigenetic alterations have been detected when IBD patients were compared with healthy individuals[86]. The number of epigenomic studies investigating IBD subgroup identification is still limited. The earliest finding observed in this area involved the assessment of cancer risk in the context of UC[87]. In future research, epigenomics studies might provide useful biomarkers for the early detection of cancer development in UC patients.
All of the above strategies provide omics analysis of the host. As mentioned previously, the gut microbiota, as an environmental factor, also plays an important role in the pathogenesis of IBD. Due to the convenience of fecal sample collection, microbiomics is promising for monitoring and managing IBD patients. The microbiota might also be able to be used to predict relapse risk. For example, a deficit in some bacterial groups or species, such as Faecalibacterium prausnitzii and Bacteroides, could be a predictive factor for relapse of CD after ileal resection or infliximab cessation[88,89]. Another study revealed that Ruminococcus and Veillonella were associated with stricturing and penetrating complications, respectively[78]. A recent prospective study classified CD patients into different subgroups with different clinical relapse risk based on microbiota[90]. Additionally, microbial analysis revealed distinct microbiota compositions between patients with different responses to anti-TNF-α therapy[84,91] as well as anti-integrin therapy[92]. Furthermore, manipulation of the microbiota might be a direction for IBD treatment.
However, this method is easily influenced by environmental factors such as diet and confounded by the causal relationship between microbiota and IBD; thus, its reliability is questionable. Recent findings are still at a superficial stage of providing simple differences in microbial abundance, and there has been a lack of in-depth analysis of microbial networks and microbiota-host interactions, as well as solid and effective prediction models.
Metabolomics generally includes serum and fecal metabolomics. As a combination of host metabolic factors and environmental gut microbiota factors, it is also a potential technique for use in future IBD research and clinical practice. Several studies have applied metabolic profiling for the diagnosis and classification of IBD[32]. A serum metabolomics study identified altered lipid and amino acid metabolism in parallel with CD activity[93]. Short-chain fatty acids (SCFAs) have been widely validated to be beneficial metabolites[94], among which butyrate is one of the most important. Studies have confirmed that fecal SCFA levels were reduced during active IBD[95]. Butyrate levels were associated with the efficacy of azathioprine, TNF-α inhibitors and integrin antibodies[92,96,97]. Other metabolites, such as bile acids and tryptophan, are also worth studying for future use[94].
Due to the complexity of IBD pathogenesis, interpretation of a single set of omics data often fails to provide insight into complex biological phenomena; thus, the omics approaches discussed above must be considered as a whole. Integrating multiple omics strategies into a network would contribute to the elucidation of the pathway involved in pathogenesis and facilitate the identification of different subgroups and the optimization of therapy regimens in IBD. The analysis of different molecules, including at the genomic, transcriptomic, proteomic, microbiome, epigenetic and metabolomic levels, could be performed simultaneously, and the results could be further integrated into multiomics models[98]. By this approach, we could obtain more insight into disease pathogenesis, identify more promising predictive biomarkers and facilitate early diagnosis[99]. Some multiomics projects are ongoing and are investigating IBD heterogeneity to improve precision management[53].
These high-throughput data need to be modeled by AI algorithms with the aid of advanced computational techniques. Machine learning is a subset of AI where machines can learn from experience provided by the data without the need for programming. Machine learning includes supervised and unsupervised algorithms. Supervised algorithms are often used for classification or prediction using example data, while unsupervised algorithms are often used for clustering according to similarity[100]. These approaches could be well applied to address the need for patient clustering and predictions and the detection of novel biomarkers. Progress made in machine learning has benefited the integrated analysis of multiomics data; these strategies mainly include concatenation-, model- and transformation-based methods[101]. In addition, deep neural networks have been used in the integration of multiomics data for the prediction of drug efficacy in cancer therapy[102], which indicates progress may be made in the context of IBD.
However, due to the obscure nature of machine learning, the robustness of the models established is sometimes uncertain. Thus, testing in independent cohorts and even clinical trials are needed before this approach is employed in a clinical setting. Additionally, products that are easy to implement in clinical settings need to be developed from research.
The pathogenesis of IBD remains uncertain, which challenges the clustering and precision management of patients. Genetic, environmental and immune factors are all involved in the complex process of IBD development. Thus, the future direction of IBD management may largely rely on the development of multiomics analysis. Numerous data processing workflows require the help of AI.
The authors are grateful to the BioRender for aiding the creation of the figure.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kai K, Japan; Mankotia DS, India; Ulasoglu C, Turkey S-Editor: Chen YL L-Editor: A P-Editor: Chen YL
| 1. | Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington (DC): National Academies Press (US); 2011. [PubMed] |
| 2. | Robinson PN. Deep phenotyping for precision medicine. Hum Mutat. 2012;33:777-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 286] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 3. | Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1967] [Cited by in RCA: 2201] [Article Influence: 137.6] [Reference Citation Analysis (6)] |
| 4. | Subramanian S, Ekbom A, Rhodes JM. Recent advances in clinical practice: a systematic review of isolated colonic Crohn's disease: the third IBD? Gut. 2017;66:362-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 5. | Fiocchi C, Iliopoulos D. What's new in IBD therapy: An "omics network" approach. Pharmacol Res. 2020;159:104886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 6. | Kaur R, Chupp G. Phenotypes and endotypes of adult asthma: Moving toward precision medicine. J Allergy Clin Immunol. 2019;144:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 296] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 7. | Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1970] [Cited by in RCA: 2349] [Article Influence: 123.6] [Reference Citation Analysis (2)] |
| 8. | Ruel J, Ruane D, Mehandru S, Gower-Rousseau C, Colombel JF. IBD across the age spectrum: is it the same disease? Nat Rev Gastroenterol Hepatol. 2014;11:88-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 181] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 9. | Moran CJ. Very early onset inflammatory bowel disease. Semin Pediatr Surg. 2017;26:356-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Sørensen JØ, Nielsen OH, Andersson M, Ainsworth MA, Ytting H, Bélard E, Jess T. Inflammatory bowel disease with primary sclerosing cholangitis: A Danish population-based cohort study 1977-2011. Liver Int. 2018;38:532-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 11. | Uhlig HH, Schwerd T, Koletzko S, Shah N, Kammermeier J, Elkadri A, Ouahed J, Wilson DC, Travis SP, Turner D, Klein C, Snapper SB, Muise AM; COLORS in IBD Study Group and NEOPICS. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology. 2014;147:990-1007.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 452] [Cited by in RCA: 475] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 12. | Crowley E, Warner N, Pan J, Khalouei S, Elkadri A, Fiedler K, Foong J, Turinsky AL, Bronte-Tinkew D, Zhang S, Hu J, Tian D, Li D, Horowitz J, Siddiqui I, Upton J, Roifman CM, Church PC, Wall DA, Ramani AK, Kotlarz D, Klein C, Uhlig H, Snapper SB, Gonzaga-Jauregui C, Paterson AD, McGovern DPB, Brudno M, Walters TD, Griffiths AM, Muise AM. Prevalence and Clinical Features of Inflammatory Bowel Diseases Associated With Monogenic Variants, Identified by Whole-Exome Sequencing in 1000 Children at a Single Center. Gastroenterology. 2020;158:2208-2220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 13. | Bolton C, Smillie CS, Pandey S, Elmentaite R, Wei G, Argmann C, Aschenbrenner D, James KR, McGovern DPB, Macchi M, Cho J, Shouval DS, Kammermeier J, Koletzko S, Bagalopal K, Capitani M, Cavounidis A, Pires E, Weidinger C, McCullagh J, Arkwright PD, Haller W, Siegmund B, Peters L, Jostins L, Travis SPL, Anderson CA, Snapper S, Klein C, Schadt E, Zilbauer M, Xavier R, Teichmann S, Muise AM, Regev A, Uhlig HH. An Integrated Taxonomy for Monogenic Inflammatory Bowel Disease. Gastroenterology. 2022;162:859-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 14. | Selin KA, Hedin CRH, Villablanca EJ. Immunological Networks Defining the Heterogeneity of Inflammatory Bowel Diseases. J Crohns Colitis. 2021;15:1959-1973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570-575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 520] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 16. | König IR, Fuchs O, Hansen G, von Mutius E, Kopp MV. What is precision medicine? Eur Respir J. 2017;50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 299] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 17. | Wang C, Baer HM, Gaya DR, Nibbs RJB, Milling S. Can molecular stratification improve the treatment of inflammatory bowel disease? Pharmacol Res. 2019;148:104442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Solem CA, Loftus EV Jr, Tremaine WJ, Harmsen WS, Zinsmeister AR, Sandborn WJ. Correlation of C-reactive protein with clinical, endoscopic, histologic, and radiographic activity in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:707-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 404] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 19. | Plevris N, Lees CW. Disease Monitoring in Inflammatory Bowel Disease: Evolving Principles and Possibilities. Gastroenterology. 2022;162:1456-1475.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 87] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 20. | Vermeire S, Van Assche G, Rutgeerts P. C-reactive protein as a marker for inflammatory bowel disease. Inflamm Bowel Dis. 2004;10:661-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 341] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 21. | Mosli MH, Zou G, Garg SK, Feagan SG, MacDonald JK, Chande N, Sandborn WJ, Feagan BG. C-Reactive Protein, Fecal Calprotectin, and Stool Lactoferrin for Detection of Endoscopic Activity in Symptomatic Inflammatory Bowel Disease Patients: A Systematic Review and Meta-Analysis. Am J Gastroenterol. 2015;110:802-19; quiz 820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 481] [Article Influence: 48.1] [Reference Citation Analysis (1)] |
| 22. | Lin JF, Chen JM, Zuo JH, Yu A, Xiao ZJ, Deng FH, Nie B, Jiang B. Meta-analysis: fecal calprotectin for assessment of inflammatory bowel disease activity. Inflamm Bowel Dis. 2014;20:1407-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 247] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 23. | Theede K, Holck S, Ibsen P, Kallemose T, Nordgaard-Lassen I, Nielsen AM. Fecal Calprotectin Predicts Relapse and Histological Mucosal Healing in Ulcerative Colitis. Inflamm Bowel Dis. 2016;22:1042-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 115] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 24. | Mao R, Xiao YL, Gao X, Chen BL, He Y, Yang L, Hu PJ, Chen MH. Fecal calprotectin in predicting relapse of inflammatory bowel diseases: a meta-analysis of prospective studies. Inflamm Bowel Dis. 2012;18:1894-1899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 25. | Jukic A, Bakiri L, Wagner EF, Tilg H, Adolph TE. Calprotectin: from biomarker to biological function. Gut. 2021;70:1978-1988. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 281] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 26. | Bodelier AG, Jonkers D, van den Heuvel T, de Boer E, Hameeteman W, Masclee AA, Pierik MJ. High Percentage of IBD Patients with Indefinite Fecal Calprotectin Levels: Additional Value of a Combination Score. Dig Dis Sci. 2017;62:465-472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Azramezani Kopi T, Shahrokh S, Mirzaei S, Asadzadeh Aghdaei H, Amini Kadijani A. The role of serum calprotectin as a novel biomarker in inflammatory bowel diseases: a review study. Gastroenterol Hepatol Bed Bench. 2019;12:183-189. [PubMed] |
| 28. | Shi JT, Zhang Y, She Y, Goyal H, Wu ZQ, Xu HG. Diagnostic Utility of Non-invasive Tests for Inflammatory Bowel Disease: An Umbrella Review. Front Med (Lausanne). 2022;9:920732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 29. | Langley BO, Guedry SE, Goldenberg JZ, Hanes DA, Beardsley JA, Ryan JJ. Inflammatory Bowel Disease and Neutrophil-Lymphocyte Ratio: A Systematic Scoping Review. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 30. | Chen YH, Wang L, Feng SY, Cai WM, Chen XF, Huang ZM. The Relationship between C-Reactive Protein/Albumin Ratio and Disease Activity in Patients with Inflammatory Bowel Disease. Gastroenterol Res Pract. 2020;2020:3467419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 31. | Serada S, Fujimoto M, Terabe F, Iijima H, Shinzaki S, Matsuzaki S, Ohkawara T, Nezu R, Nakajima S, Kobayashi T, Plevy SE, Takehara T, Naka T. Serum leucine-rich alpha-2 glycoprotein is a disease activity biomarker in ulcerative colitis. Inflamm Bowel Dis. 2012;18:2169-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 165] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 32. | Chen P, Zhou G, Lin J, Li L, Zeng Z, Chen M, Zhang S. Serum Biomarkers for Inflammatory Bowel Disease. Front Med (Lausanne). 2020;7:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 118] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 33. | Kuna AT. Serological markers of inflammatory bowel disease. Biochem Med (Zagreb). 2013;23:28-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 34. | Prideaux L, De Cruz P, Ng SC, Kamm MA. Serological antibodies in inflammatory bowel disease: a systematic review. Inflamm Bowel Dis. 2012;18:1340-1355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 137] [Article Influence: 10.5] [Reference Citation Analysis (2)] |
| 35. | Torres J, Petralia F, Sato T, Wang P, Telesco SE, Choung RS, Strauss R, Li XJ, Laird RM, Gutierrez RL, Porter CK, Plevy S, Princen F, Murray JA, Riddle MS, Colombel JF. Serum Biomarkers Identify Patients Who Will Develop Inflammatory Bowel Diseases Up to 5 Years Before Diagnosis. Gastroenterology. 2020;159:96-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 171] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 36. | Vasiliauskas EA, Kam LY, Karp LC, Gaiennie J, Yang H, Targan SR. Marker antibody expression stratifies Crohn's disease into immunologically homogeneous subgroups with distinct clinical characteristics. Gut. 2000;47:487-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 231] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 37. | Smids C, Horjus Talabur Horje CS, Groenen MJM, van Koolwijk EHM, Wahab PJ, van Lochem EG. The value of serum antibodies in differentiating inflammatory bowel disease, predicting disease activity and disease course in the newly diagnosed patient. Scand J Gastroenterol. 2017;52:1104-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 38. | Martinez-Fierro ML, Garza-Veloz I, Rocha-Pizaña MR, Cardenas-Vargas E, Cid-Baez MA, Trejo-Vazquez F, Flores-Morales V, Villela-Ramirez GA, Delgado-Enciso I, Rodriguez-Sanchez IP, Ortiz-Castro Y. Serum cytokine, chemokine, and growth factor profiles and their modulation in inflammatory bowel disease. Medicine (Baltimore). 2019;98:e17208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 39. | Louis E, Belaiche J, van Kemseke C, Franchimont D, de Groote D, Gueenen V, Mary JY. A high serum concentration of interleukin-6 is predictive of relapse in quiescent Crohn's disease. Eur J Gastroenterol Hepatol. 1997;9:939-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 94] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 40. | Tran DH, Wang J, Ha C, Ho W, Mattai SA, Oikonomopoulos A, Weiss G, Lacey P, Cheng M, Shieh C, Mussatto CC, Ho S, Hommes D, Koon HW. Circulating cathelicidin levels correlate with mucosal disease activity in ulcerative colitis, risk of intestinal stricture in Crohn's disease, and clinical prognosis in inflammatory bowel disease. BMC Gastroenterol. 2017;17:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 41. | Nakov R, Velikova T, Nakov V, Ianiro G, Gerova V, Tankova L. Serum trefoil factor 3 predicts disease activity in patients with ulcerative colitis. Eur Rev Med Pharmacol Sci. 2019;23:788-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 42. | Lin S, Wang Y, Li L, Chen P, Mao R, Feng R, Qiu Y, He Y, Chen B, Zeng Z, Chen M, Zhang S. A New Model Based on 25-Hydroxyvitamin D3 for Predicting Active Crohn's Disease in Chinese Patients. Mediators Inflamm. 2018;2018:3275025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 43. | Sands BE. Biomarkers of Inflammation in Inflammatory Bowel Disease. Gastroenterology. 2015;149:1275-1285.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 288] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 44. | Swaminathan A, Borichevsky GM, Edwards TS, Hirschfeld E, Mules TC, Frampton CMA, Day AS, Hampton MB, Kettle AJ, Gearry RB. Faecal Myeloperoxidase as a Biomarker of Endoscopic Activity in Inflammatory Bowel Disease. J Crohns Colitis. 2022;16:1862-1873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 45. | Torres J, Bonovas S, Doherty G, Kucharzik T, Gisbert JP, Raine T, Adamina M, Armuzzi A, Bachmann O, Bager P, Biancone L, Bokemeyer B, Bossuyt P, Burisch J, Collins P, El-Hussuna A, Ellul P, Frei-Lanter C, Furfaro F, Gingert C, Gionchetti P, Gomollon F, González-Lorenzo M, Gordon H, Hlavaty T, Juillerat P, Katsanos K, Kopylov U, Krustins E, Lytras T, Maaser C, Magro F, Marshall JK, Myrelid P, Pellino G, Rosa I, Sabino J, Savarino E, Spinelli A, Stassen L, Uzzan M, Vavricka S, Verstockt B, Warusavitarne J, Zmora O, Fiorino G. ECCO Guidelines on Therapeutics in Crohn's Disease: Medical Treatment. J Crohns Colitis. 2020;14:4-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 991] [Cited by in RCA: 903] [Article Influence: 180.6] [Reference Citation Analysis (2)] |
| 46. | Hyams JS, Davis Thomas S, Gotman N, Haberman Y, Karns R, Schirmer M, Mo A, Mack DR, Boyle B, Griffiths AM, LeLeiko NS, Sauer CG, Keljo DJ, Markowitz J, Baker SS, Rosh J, Baldassano RN, Patel A, Pfefferkorn M, Otley A, Heyman M, Noe J, Oliva-Hemker M, Rufo PA, Strople J, Ziring D, Guthery SL, Sudel B, Benkov K, Wali P, Moulton D, Evans J, Kappelman MD, Marquis MA, Sylvester FA, Collins MH, Venkateswaran S, Dubinsky M, Tangpricha V, Spada KL, Saul B, Wang J, Serrano J, Hommel K, Marigorta UM, Gibson G, Xavier RJ, Kugathasan S, Walters T, Denson LA. Clinical and biological predictors of response to standardised paediatric colitis therapy (PROTECT): a multicentre inception cohort study. Lancet. 2019;393:1708-1720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 128] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 47. | Xie T, Zhao C, Ding C, Zhang T, Dai X, Lv T, Li Y, Guo Z, Gong J, Zhu W. Fecal calprotectin as an alternative to ulcerative colitis endoscopic index of severity to predict the response to corticosteroids of acute severe ulcerative colitis: A prospective observational study. Dig Liver Dis. 2017;49:984-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 48. | Colombel JF, Panaccione R, Bossuyt P, Lukas M, Baert F, Vaňásek T, Danalioglu A, Novacek G, Armuzzi A, Hébuterne X, Travis S, Danese S, Reinisch W, Sandborn WJ, Rutgeerts P, Hommes D, Schreiber S, Neimark E, Huang B, Zhou Q, Mendez P, Petersson J, Wallace K, Robinson AM, Thakkar RB, D'Haens G. Effect of tight control management on Crohn's disease (CALM): a multicentre, randomised, controlled phase 3 trial. Lancet. 2017;390:2779-2789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 671] [Article Influence: 83.9] [Reference Citation Analysis (1)] |
| 49. | Kopylov U, Seidman E. Predicting durable response or resistance to antitumor necrosis factor therapy in inflammatory bowel disease. Therap Adv Gastroenterol. 2016;9:513-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 50. | Schett G, McInnes IB, Neurath MF. Reframing Immune-Mediated Inflammatory Diseases through Signature Cytokine Hubs. N Engl J Med. 2021;385:628-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 213] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 51. | Atreya R, Neumann H, Neufert C, Waldner MJ, Billmeier U, Zopf Y, Willma M, App C, Münster T, Kessler H, Maas S, Gebhardt B, Heimke-Brinck R, Reuter E, Dörje F, Rau TT, Uter W, Wang TD, Kiesslich R, Vieth M, Hannappel E, Neurath MF. In vivo imaging using fluorescent antibodies to tumor necrosis factor predicts therapeutic response in Crohn's disease. Nat Med. 2014;20:313-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 294] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 52. | Jessen B, Rodriguez-Sillke Y, Sonnenberg E, Schumann M, Kruglov A, Freise I, Schmidt F, Maul J, Kühl AA, Glauben R, Lissner D, Siegmund B. Level of Tumor Necrosis Factor Production by Stimulated Blood Mononuclear Cells Can Be Used to Predict Response of Patients With Inflammatory Bowel Diseases to Infliximab. Clin Gastroenterol Hepatol. 2021;19:721-731.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 53. | Verstockt B, Parkes M, Lee JC. How Do We Predict a Patient's Disease Course and Whether They Will Respond to Specific Treatments? Gastroenterology. 2022;162:1383-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 54. | Bertani L, Fornai M, Fornili M, Antonioli L, Benvenuti L, Tapete G, Baiano Svizzero G, Ceccarelli L, Mumolo MG, Baglietto L, de Bortoli N, Bellini M, Marchi S, Costa F, Blandizzi C. Serum oncostatin M at baseline predicts mucosal healing in Crohn's disease patients treated with infliximab. Aliment Pharmacol Ther. 2020;52:284-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 55. | Verstockt B, Verstockt S, Dehairs J, Ballet V, Blevi H, Wollants WJ, Breynaert C, Van Assche G, Vermeire S, Ferrante M. Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. EBioMedicine. 2019;40:733-742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 128] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 56. | Elhag DA, Kumar M, Saadaoui M, Akobeng AK, Al-Mudahka F, Elawad M, Al Khodor S. Inflammatory Bowel Disease Treatments and Predictive Biomarkers of Therapeutic Response. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 63] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 57. | Johnsen KM, Florholmen J, Moe ØK, Gundersen M, Beilfuss J, Kileng H, Sørbye SW, Goll R. Prediction of long-term remission in patients following discontinuation of anti-TNF therapy in ulcerative colitis: a 10 year follow up study. BMC Gastroenterol. 2022;22:459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 58. | Gottlieb ZS, Sands BE. Personalised Medicine with IL-23 Blockers: Myth or Reality? J Crohns Colitis. 2022;16:ii73-ii94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 59. | Tong X, Zheng Y, Li Y, Xiong Y, Chen D. Soluble ligands as drug targets for treatment of inflammatory bowel disease. Pharmacol Ther. 2021;226:107859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 60. | Boden EK, Shows DM, Chiorean MV, Lord JD. Identification of Candidate Biomarkers Associated with Response to Vedolizumab in Inflammatory Bowel Disease. Dig Dis Sci. 2018;63:2419-2429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 61. | Rath T, Bojarski C, Neurath MF, Atreya R. Molecular imaging of mucosal α4β7 integrin expression with the fluorescent anti-adhesion antibody vedolizumab in Crohn's disease. Gastrointest Endosc. 2017;86:406-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 62. | Battat R, Dulai PS, Vande Casteele N, Evans E, Hester KD, Webster E, Jain A, Proudfoot JA, Mairalles A, Neill J, Singh S, Chang JT, Rivera-Nieves J, Sandborn WJ, Boland BS. Biomarkers Are Associated With Clinical and Endoscopic Outcomes With Vedolizumab Treatment in Ulcerative Colitis. Inflamm Bowel Dis. 2019;25:410-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 63. | Holmer AK, Battat R, Dulai PS, Vande Casteele N, Nguyen N, Jain A, Miralles A, Neill J, Le H, Singh S, Rivera-Nieves J, Sandborn WJ, Boland BS. Biomarkers are associated with clinical and endoscopic outcomes with vedolizumab treatment in Crohn's disease. Therap Adv Gastroenterol. 2020;13:1756284820971214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 64. | Bertani L, Caviglia GP, Antonioli L, Pellicano R, Fagoonee S, Astegiano M, Saracco GM, Bugianesi E, Blandizzi C, Costa F, Ribaldone DG. Serum Interleukin-6 and -8 as Predictors of Response to Vedolizumab in Inflammatory Bowel Diseases. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 65. | Lopetuso LR, Gerardi V, Papa V, Scaldaferri F, Rapaccini GL, Gasbarrini A, Papa A. Can We Predict the Efficacy of Anti-TNF-α Agents? Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 66. | Papamichael K, Cheifetz AS, Melmed GY, Irving PM, Vande Casteele N, Kozuch PL, Raffals LE, Baidoo L, Bressler B, Devlin SM, Jones J, Kaplan GG, Sparrow MP, Velayos FS, Ullman T, Siegel CA. Appropriate Therapeutic Drug Monitoring of Biologic Agents for Patients With Inflammatory Bowel Diseases. Clin Gastroenterol Hepatol. 2019;17:1655-1668.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 240] [Article Influence: 40.0] [Reference Citation Analysis (2)] |
| 67. | Cheifetz AS, Abreu MT, Afif W, Cross RK, Dubinsky MC, Loftus EV Jr, Osterman MT, Saroufim A, Siegel CA, Yarur AJ, Melmed GY, Papamichael K. A Comprehensive Literature Review and Expert Consensus Statement on Therapeutic Drug Monitoring of Biologics in Inflammatory Bowel Disease. Am J Gastroenterol. 2021;116:2014-2025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 132] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 68. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4223] [Cited by in RCA: 3903] [Article Influence: 162.6] [Reference Citation Analysis (0)] |
| 69. | Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nuñez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3555] [Cited by in RCA: 3476] [Article Influence: 144.8] [Reference Citation Analysis (1)] |
| 70. | Lesage S, Zouali H, Cézard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O'Morain C, Gassull M, Binder V, Finkel Y, Modigliani R, Gower-Rousseau C, Macry J, Merlin F, Chamaillard M, Jannot AS, Thomas G, Hugot JP; EPWG-IBD Group; EPIMAD Group; GETAID Group. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 716] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 71. | Weersma RK, Stokkers PC, van Bodegraven AA, van Hogezand RA, Verspaget HW, de Jong DJ, van der Woude CJ, Oldenburg B, Linskens RK, Festen EA, van der Steege G, Hommes DW, Crusius JB, Wijmenga C, Nolte IM, Dijkstra G; Dutch Initiative on Crohn and Colitis (ICC). Molecular prediction of disease risk and severity in a large Dutch Crohn's disease cohort. Gut. 2009;58:388-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 133] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 72. | Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Dayani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JY, Malekzadeh R, Westra HJ, Yamazaki K, Yang SK; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium, Barrett JC, Alizadeh BZ, Parkes M, Bk T, Daly MJ, Kubo M, Anderson CA, Weersma RK. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1898] [Cited by in RCA: 1862] [Article Influence: 186.2] [Reference Citation Analysis (0)] |
| 73. | Haritunians T, Taylor KD, Targan SR, Dubinsky M, Ippoliti A, Kwon S, Guo X, Melmed GY, Berel D, Mengesha E, Psaty BM, Glazer NL, Vasiliauskas EA, Rotter JI, Fleshner PR, McGovern DP. Genetic predictors of medically refractory ulcerative colitis. Inflamm Bowel Dis. 2010;16:1830-1840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 74. | Sazonovs A, Kennedy NA, Moutsianas L, Heap GA, Rice DL, Reppell M, Bewshea CM, Chanchlani N, Walker GJ, Perry MH, McDonald TJ, Lees CW, Cummings JRF, Parkes M, Mansfield JC, Irving PM, Barrett JC, McGovern D, Goodhand JR, Anderson CA, Ahmad T; PANTS Consortium. HLA-DQA1*05 Carriage Associated With Development of Anti-Drug Antibodies to Infliximab and Adalimumab in Patients With Crohn's Disease. Gastroenterology. 2020;158:189-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 279] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 75. | Bek S, Nielsen JV, Bojesen AB, Franke A, Bank S, Vogel U, Andersen V. Systematic review: genetic biomarkers associated with anti-TNF treatment response in inflammatory bowel diseases. Aliment Pharmacol Ther. 2016;44:554-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 76. | Hlavaty T, Pierik M, Henckaerts L, Ferrante M, Joossens S, van Schuerbeek N, Noman M, Rutgeerts P, Vermeire S. Polymorphisms in apoptosis genes predict response to infliximab therapy in luminal and fistulizing Crohn's disease. Aliment Pharmacol Ther. 2005;22:613-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 144] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 77. | Tong Q, Zhao L, Qian XD, Zhang LL, Xu X, Dai SM, Cai Q, Zhao DB. Association of TNF-α polymorphism with prediction of response to TNF blockers in spondyloarthritis and inflammatory bowel disease: a meta-analysis. Pharmacogenomics. 2013;14:1691-1700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 78. | Kugathasan S, Denson LA, Walters TD, Kim MO, Marigorta UM, Schirmer M, Mondal K, Liu C, Griffiths A, Noe JD, Crandall WV, Snapper S, Rabizadeh S, Rosh JR, Shapiro JM, Guthery S, Mack DR, Kellermayer R, Kappelman MD, Steiner S, Moulton DE, Keljo D, Cohen S, Oliva-Hemker M, Heyman MB, Otley AR, Baker SS, Evans JS, Kirschner BS, Patel AS, Ziring D, Trapnell BC, Sylvester FA, Stephens MC, Baldassano RN, Markowitz JF, Cho J, Xavier RJ, Huttenhower C, Aronow BJ, Gibson G, Hyams JS, Dubinsky MC. Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet. 2017;389:1710-1718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 488] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 79. | Lee JC, Lyons PA, McKinney EF, Sowerby JM, Carr EJ, Bredin F, Rickman HM, Ratlamwala H, Hatton A, Rayner TF, Parkes M, Smith KG. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest. 2011;121:4170-4179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 245] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 80. | Biasci D, Lee JC, Noor NM, Pombal DR, Hou M, Lewis N, Ahmad T, Hart A, Parkes M, McKinney EF, Lyons PA, Smith KGC. A blood-based prognostic biomarker in IBD. Gut. 2019;68:1386-1395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 147] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 81. | Marigorta UM, Denson LA, Hyams JS, Mondal K, Prince J, Walters TD, Griffiths A, Noe JD, Crandall WV, Rosh JR, Mack DR, Kellermayer R, Heyman MB, Baker SS, Stephens MC, Baldassano RN, Markowitz JF, Kim MO, Dubinsky MC, Cho J, Aronow BJ, Kugathasan S, Gibson G. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn's disease. Nat Genet. 2017;49:1517-1521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 127] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 82. | Czarnewski P, Parigi SM, Sorini C, Diaz OE, Das S, Gagliani N, Villablanca EJ. Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat Commun. 2019;10:2892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 83. | Gisbert JP, Chaparro M. Clinical Usefulness of Proteomics in Inflammatory Bowel Disease: A Comprehensive Review. J Crohns Colitis. 2019;13:374-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 84. | Magnusson MK, Strid H, Sapnara M, Lasson A, Bajor A, Ung KA, Öhman L. Anti-TNF Therapy Response in Patients with Ulcerative Colitis Is Associated with Colonic Antimicrobial Peptide Expression and Microbiota Composition. J Crohns Colitis. 2016;10:943-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 128] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 85. | Bergemalm D, Andersson E, Hultdin J, Eriksson C, Rush ST, Kalla R, Adams AT, Keita ÅV, D'Amato M, Gomollon F, Jahnsen J; IBD Character Consortium, Ricanek P, Satsangi J, Repsilber D, Karling P, Halfvarson J. Systemic Inflammation in Preclinical Ulcerative Colitis. Gastroenterology. 2021;161:1526-1539.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 80] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 86. | Agrawal M, Allin KH, Petralia F, Colombel JF, Jess T. Multiomics to elucidate inflammatory bowel disease risk factors and pathways. Nat Rev Gastroenterol Hepatol. 2022;19:399-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 115] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 87. | Rajamäki K, Taira A, Katainen R, Välimäki N, Kuosmanen A, Plaketti RM, Seppälä TT, Ahtiainen M, Wirta EV, Vartiainen E, Sulo P, Ravantti J, Lehtipuro S, Granberg KJ, Nykter M, Tanskanen T, Ristimäki A, Koskensalo S, Renkonen-Sinisalo L, Lepistö A, Böhm J, Taipale J, Mecklin JP, Aavikko M, Palin K, Aaltonen LA. Genetic and Epigenetic Characteristics of Inflammatory Bowel Disease-Associated Colorectal Cancer. Gastroenterology. 2021;161:592-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 88. | Rajca S, Grondin V, Louis E, Vernier-Massouille G, Grimaud JC, Bouhnik Y, Laharie D, Dupas JL, Pillant H, Picon L, Veyrac M, Flamant M, Savoye G, Jian R, Devos M, Paintaud G, Piver E, Allez M, Mary JY, Sokol H, Colombel JF, Seksik P. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn's disease. Inflamm Bowel Dis. 2014;20:978-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 89. | Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731-16736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2747] [Cited by in RCA: 3199] [Article Influence: 188.2] [Reference Citation Analysis (0)] |
| 90. | Buffet-Bataillon S, Bouguen G, Fleury F, Cattoir V, Le Cunff Y. Gut microbiota analysis for prediction of clinical relapse in Crohn's disease. Sci Rep. 2022;12:19929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 91. | Yilmaz B, Juillerat P, Øyås O, Ramon C, Bravo FD, Franc Y, Fournier N, Michetti P, Mueller C, Geuking M, Pittet VEH, Maillard MH, Rogler G; Swiss IBD Cohort Investigators, Wiest R, Stelling J, Macpherson AJ. Microbial network disturbances in relapsing refractory Crohn's disease. Nat Med. 2019;25:323-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 284] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 92. | Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, Xavier RJ. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe. 2017;21:603-610.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 327] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 93. | Lai Y, Xue J, Liu CW, Gao B, Chi L, Tu P, Lu K, Ru H. Serum Metabolomics Identifies Altered Bioenergetics, Signaling Cascades in Parallel with Exposome Markers in Crohn's Disease. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 94. | Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 1197] [Article Influence: 239.4] [Reference Citation Analysis (0)] |
| 95. | Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, Hermoso MA. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol. 2019;10:277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 970] [Cited by in RCA: 2178] [Article Influence: 363.0] [Reference Citation Analysis (0)] |
| 96. | Aden K, Rehman A, Waschina S, Pan WH, Walker A, Lucio M, Nunez AM, Bharti R, Zimmerman J, Bethge J, Schulte B, Schulte D, Franke A, Nikolaus S, Schroeder JO, Vandeputte D, Raes J, Szymczak S, Waetzig GH, Zeuner R, Schmitt-Kopplin P, Kaleta C, Schreiber S, Rosenstiel P. Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology. 2019;157:1279-1292.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 191] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 97. | Effenberger M, Reider S, Waschina S, Bronowski C, Enrich B, Adolph TE, Koch R, Moschen AR, Rosenstiel P, Aden K, Tilg H. Microbial Butyrate Synthesis Indicates Therapeutic Efficacy of Azathioprine in IBD Patients. J Crohns Colitis. 2021;15:88-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 98. | Peters LA, Perrigoue J, Mortha A, Iuga A, Song WM, Neiman EM, Llewellyn SR, Di Narzo A, Kidd BA, Telesco SE, Zhao Y, Stojmirovic A, Sendecki J, Shameer K, Miotto R, Losic B, Shah H, Lee E, Wang M, Faith JJ, Kasarskis A, Brodmerkel C, Curran M, Das A, Friedman JR, Fukui Y, Humphrey MB, Iritani BM, Sibinga N, Tarrant TK, Argmann C, Hao K, Roussos P, Zhu J, Zhang B, Dobrin R, Mayer LF, Schadt EE. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet. 2017;49:1437-1449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 179] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 99. | Li R, Li L, Xu Y, Yang J. Machine learning meets omics: applications and perspectives. Brief Bioinform. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 100. | Stankovic B, Kotur N, Nikcevic G, Gasic V, Zukic B, Pavlovic S. Machine Learning Modeling from Omics Data as Prospective Tool for Improvement of Inflammatory Bowel Disease Diagnosis and Clinical Classifications. Genes (Basel). 2021;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 101. | Reel PS, Reel S, Pearson E, Trucco E, Jefferson E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol Adv. 2021;49:107739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 404] [Article Influence: 101.0] [Reference Citation Analysis (0)] |
| 102. | Sharifi-Noghabi H, Zolotareva O, Collins CC, Ester M. MOLI: multi-omics late integration with deep neural networks for drug response prediction. Bioinformatics. 2019;35:i501-i509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 201] [Article Influence: 33.5] [Reference Citation Analysis (0)] |