Published online Apr 14, 2023. doi: 10.3748/wjg.v29.i14.2172
Peer-review started: December 21, 2022
First decision: January 3, 2023
Revised: January 13, 2023
Accepted: March 9, 2023
Article in press: March 9, 2023
Published online: April 14, 2023
Processing time: 112 Days and 22.7 Hours
Numerous studies have shown that in Crohn’s disease (CD), the gut microbiota is of great importance in the induction and maintenance of inflammation in the gastrointestinal tract. Until recently, studies have focused almost exclusively on bacteria in the gut. Lately, more attention has been paid to the role of intestinal fungi.
To study the gut mycobiome analysis of pediatric patients with CD (in different stages of disease activity) compared to healthy children.
Fecal samples were collected from patients: With active, newly diagnosed CD (n = 50); active but previously diagnosed and treated CD (n = 16); non-active CD and who were in clinical remission (n = 39) and from healthy volunteers (n = 40). Fungal DNA was isolated from the samples. Next, next generation sequencing (MiSeq, Illumina) was performed. The composition of mycobiota was correlated with clinical and blood parameters.
Candida spp. were overrepresented in CD patients, while in the control group, the most abundant genus was Saccharomyces. In CD patients, the percentage of Malassezia was almost twice that of the control (P < 0.05). In active CD patients, we documented a higher abundance of Debaryomyces hansenii (D. hansenii) compared to the non-active CD and control (P < 0.05) groups. Moreover, statistically significant changes in the abundance of Mycosphaerella, Rhodotorula, and Microidium were observed. The analyses at the species level and linear discriminant analysis showed that in each group it was possible to distinguish a specific species characteristic of a given patient population. Moreover, we have documented statistically significant correlations between: D. hansenii and patient age (negative); C. zeylanoides and patient age (positive); C. dubliniensis and calprotectin (positive); C. sake and calprotectin (positive); and C. tropicalis and pediatric CD activity index (PCDAI) (positive).
Mycobiome changes in CD patients, and the positive correlation of some species with calprotectin or PCDAI, give strong evidence that fungi may be of key importance in the development of CD.
Core Tip: There is growing evidence that intestinal microorganisms are associated with pathogenesis of Crohn’s disease (CD). Until recently, studies have focused almost exclusively on bacteria. In this study we showed alterations in the fungal composition in pediatric patients with CD. Changes within the specific species of fungi depending on disease activity, and the positive correlation of some species with calprotectin or pediatric CD activity index, give strong evidence that these microorganisms may be of key importance in the development and course of CD. Some fungal species can be helpful in predicting an exacerbation of the disease or even predicting the diagnosis of CD.
- Citation: Krawczyk A, Salamon D, Kowalska-Duplaga K, Zapała B, Książek T, Drażniuk-Warchoł M, Gosiewski T. Changes in the gut mycobiome in pediatric patients in relation to the clinical activity of Crohn's disease. World J Gastroenterol 2023; 29(14): 2172-2187
- URL: https://www.wjgnet.com/1007-9327/full/v29/i14/2172.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i14.2172
Crohn's disease (CD) belongs to the group of chronic gastrointestinal tract disorders known as inflammatory bowel diseases (IBD), which are characterized by a relapsing and remitting course. The pathogenesis of CD is not fully understood, but it is supposed that its development and severity are influenced by a combination of genetic, immune and microbiological disorders[1,2]. Intestinal microorganisms are increasingly indicated as one of the key factors in the etiology of IBD[1,3-5].
The human gut is colonized by a stable and abundant community of microorganisms, collectively referred to as the gut microbiota, consisting of bacteria, archaea, fungi, protozoans, and viruses[6]. These microbes have physiological functions associated with nutrition, regulation of immune homeostasis, and protection of the host against pathogenic bacteria[3,6]. Recent advances in molecular techniques have recognized alterations in the composition, abundance, and function of the microbiota in CD, which is known as dysbiosis[1,6]. To date, it has not been established whether the changes in the microbiota are a direct cause of the disease or only a secondary effect of the chronic inflammatory process and the treatment applied. However, numerous studies in animal and human models[7-10] have shown that the gut microbiota in CD is of great importance in the induction and maintenance of inflammation in the gastrointestinal tract.
Until recently, studies have focused almost exclusively on bacteria in the gut[4,11-13]. Lately, more attention has been paid to the role of intestinal fungi. These microorganisms, their metabolites, and their interaction with bacterial populations may directly or indirectly influence the host’s immune response[1]. Fungi interact with the immune system among others via Dectin-1, which is one of the most important pattern recognition receptors[7,14]. This receptor has an influence on the activation of macrophages, neutrophils, and dendritic cells. Dectin-1 recognizes β-1.3 glucans that are components of the fungal cell wall and thus activates the intracellular signal that leads to the production of inflammatory cytokines[15]. It has been suggested that Dectin-1 polymorphisms are strongly associated with an excessive immune response to commensal fungi and thus may contribute to the development of CD[7]. In addition, the induction of anti-Saccharomyces cerevisiae antibodies (ASCA) that recognize yeast cell wall mannans is a serological biomarker frequently present in patients with CD, which may suggest that the disease may be associated with an unusual interaction of the immune system with fungi.
Taking into account the likely relationship between the mycobiota and CD, the aim of this study was a detailed taxonomic analysis of the fungal composition in children with CD. In our study, we analyzed the differences between the mycobiomes of CD patients and healthy children, as well as the changes that occur in patients' mycobiomes depending on disease activity.
Pediatric patients with CD, aged 2 to 18 years, hospitalized at the University Children’s Hospital in Krakow in the years 2016–2019, were recruited for the study. Diagnosis was made on the basis of the clinical picture, as well as endoscopic, histopathological, and radiological examinations, according to the revised Porto criteria[16]. The study protocol was approved by the Jagiellonian University Bioethics Committee (No. 1072.6120.21.2020). Disease phenotype was evaluated according to the Paris criteria[17]. The patients were recruited into two research groups on the basis of clinical disease activity according to the pediatric CD activity index (PCDAI).
Patients with active CD, PCDAI > 10 points.
In this group, two subgroups were distinguished: Ia–patients with newly diagnosed CD (before the implementation of any treatment); and Ib–patients previously diagnosed and treated with aminosalicylates (5-ASAs) and/or azathioprine.
Patients with non-active CD, PCDAI ≤ 10 points.
This group (II) included patients who were previously diagnosed with CD and treated with 5-ASAs and/or azathioprine and were in clinical remission at the time of inclusion in the study.
The exclusion criteria were: Lack of consent to participate in the study; patients under 2 or over 18 years of age; use of antibiotics and/or probiotics and/or antifungal drugs within 30 d before collecting stool samples; confirmed infections of the gastrointestinal tract; isolated perianal fistula.
The control group included healthy non-hospitalized children and adolescents without antibiotic, antifungal, or probiotic treatment during the 30-d period before stool sample collection.
At inclusion, routine laboratory tests evaluating biochemical parameters and calprotectin were performed and the PCDAI score was evaluated in all patients. Stool samples from all participants were obtained at the University Children’s Hospital in Krakow and immediately frozen at -80 °C, then delivered under deep freeze conditions to the Department of Microbiology of Jagiellonian University Medical College in Krakow, where DNA was isolated and further analyses were performed.
The stool samples were thawed and 150 mg of each material was used for fungal DNA isolation using the Genomic Mini AX Stool Kit (A&A Biotechnology, Gdansk, Poland), according to our previously described modification[18,19].
The isolates were measured with a NanoDrop spectrophotometer (Thermo Fisher, Waltham, MA, United States) in the A260 nm and A260 nm/280 nm ratios to determine the purity and concentration of DNA.
The DNA extracted was used to perform polymerase chain reaction (PCR) amplification (T100 Thermal Cycler, BioRad, California, United States), with primers targeting the ITS-1 regions of the fungal rDNA gene[20] to prepare ITS libraries.
The primer sequences were the following: [Adapter sequences for MiSeq sequencer (Illumina, San Diego, United States) are written in bold][21]:
Forward primer:
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTAAAAGTCGTAACAAGGTTTC;
Reverse primer:
GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGTTCAAAGAyTCGATGATTCAC.
The composition of the reaction mixture was as follows: 5 μL of DNA, 12.5 μL of Kapa Biosystems (Roche, Basel, Switzerland), 0.5 μL of each primer (20 mmol/L, Genomed, Warsaw, Poland), 6.5 μL of water (A&A Biotechnology, Gdansk, Poland). Thermal cycling conditions included an initial denaturation at 95 °C for 5 min followed by 50 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s.
The volume of 5 μL of each amplicon was subjected to electrophoretic separation on 1.5% agarose gel (Prona, Basica Le, Burgos, Spain) diluted 10 times with TBE buffer (Sigma-Aldrich, Saint Louis, United States). The PCR products were visualized in the FastGene FAS–Digi Pro (Nippon Genetics, Duren, Germany) in the presence of UV light, to check library quality. Subsequent steps (purification, sample indexing, sample quantification, and pooling) were prepared according to the Illumina library preparation protocols[21].
The library concentration was measured using PicoGreen fluorescent dye (Thermo Fisher, Waltham, MA, United States) and pooled with 30% spike-in PhiX control DNA (Illumina, San Diego, United States). Next, the pooled libraries were applied to the Reagent Kit V3 cartridge (600 cycles) (Illumina, San Diego, United States) and the sequencing was carried out using MiSeq (Illumina, San Diego, United States) at the Department of Clinical Immunology of the University Children’s Hospital in Krakow.
Negative control (containing water instead of DNA) and positive control (MSA 1010 Mycobiome Genomic DNA mix, ATCC, Manassas, United States) were included throughout the genomic library procedure and next generation sequencing sequencing.
The ITS-targeted amplicon pair reads were demultiplexed on the basis of the unique molecular barcodes. The short read of raw data was collected as FASTQ files. Then, the per-sample raw taxonomic classification was performed using the Illumina 16S Metagenomics workflow, on the basis of the UNITE Fungal ITS Database v7.2.
Sequences were clustered into operational taxonomic units (OTUs) at a similarity cutoff value of 97%-98%. This classification was performed on the basis of the algorithm described by Wang et al[20]. Next, the metagenomic reads were aligned to the UNITE Fungal ITS Database v7.2 reference taxonomy database. During this step, the candidate fungal reads were identified, and the sequences which did not match the reference fungal ITS database were filtered out (these were features with counts below 4 counts, 20% of prevalence filter). In the normalization step, sequences were trimmed for low-quality scores (less than 3). Finally, filtered and normalized reads were matched again to the fungi, and a definitive taxonomic assignment was performed. Abundance profiling was characterized on the basis of the OTU data, which were compared at different taxonomic levels based on the annotations. The diversity within samples was measured on the basis of alpha and beta diversity indices. To calculate the diversity within samples (richness and evenness), Observed, ACE, Chao1, Shannon, Simpson, and Fisher matrices were applied, and analysis of variance (ANOVA) statistics were performed. Beta diversity analysis was performed on the basis of Principal Coordinate Analysis using Bray–Curtis and the Jaccard index. The statistical significance of sample grouping was tested using the Permutational MANOVA (PERMANOVA) statistical method. The linear discriminant analysis (LDA) Effect Size (LEfSe) algorithm was used to discover statistically significant biomarkers. The Kruskal–Wallis rank sum test was performed to describe features with significant abundance at different taxonomic levels. A 0.05 P value cutoff and 2.0 LDA score cutoff were established to determine the most significant features.
One hundred and five pediatric patients were included in the study. The control group consisted of 40 children. The characteristics of the study groups are presented in Table 1.
| Group Ia (n = 50) | Group Ib (n = 16) | Group II (n = 39) | Control (n = 40) | |
| General characteristics | ||||
| Male:Female (ratio) | 29:21 (1.38) | 8:8 (1) | 22:17 (1.29) | 15:25 (0.6) |
| Age (years), mean ± SD | 13 ± 2.9 | 15 ± 2.8 | 12.5 ± 4.3 | 11 ± 4.1 |
| BMI (kg/m2), mean ± SD | 17.0 ± 3.1 | 18.1 ± 2.7 | 18.1 ± 3.2 | 18.3 ± 3.8 |
| PCDAI, mean ± SD | 34.4 ± 12.7 | 38.2 ± 18.6 | 4.1 ± 3.9 | N/A |
| Disease distribution by Paris classification (number of patients) | ||||
| A1a | 4 | 2 | 7 | |
| A1b | 46 | 14 | 30 | |
| A2 | 0 | 2 | 2 | |
| L1 | 2 | 1 | 2 | |
| L2 | 6 | 2 | 7 | |
| L3 | 16 | 4 | 12 | |
| L4aL1 | 8 | 2 | 6 | |
| L4aL2 | 8 | 3 | 5 | |
| L4aL3 | 10 | 4 | 7 | |
| B1 | 39 | 11 | 27 | |
| B2 | 11 | 5 | 12 |
A DNA purity of ≥ 1.7 was assumed. All samples examined met this criterion.
The data obtained by sequencing consisted of 4859784 reads (1680 minimum reads per sample, 79543 maximum) with an average number of reads of 33748. All samples were included in the analysis.
Alpha diversity at the phylum level (L2) was different and statistically significant between: Ia vs control (expressed as the Shannon, Chao, and Fisher indices; Ia vs II (Fisher index); and II vs control (Chao index) (Table 2). Moreover, we observed statistically significant differences in alpha diversity at the species level (L7) between groups: Ia vs II (expressed as both Chao and Fisher indices); Ia vs control (Chao and Fisher indices); Ib vs II (Fisher index); Ib vs control (Fisher index) (Table 2).
| Index/group | Shannon | Chao | Fisher | |||
| L2 level | L7 level | L2 level | L7 level | L2 level | L7 level | |
| Ia vs Ib | 0.713 | 0.251 | 0.836 | 0.121 | 0.533 | 0.312 |
| Ia vs II | 0.606 | 0.584 | 0.228 | 0.001a | 0.000a | 0.011a |
| Ia vs control | 0.000a | 0.313 | 0.006a | 0.000a | 0.000a | 0.000a |
| Ib vs II | 0.492 | 0.128 | 0.356 | 0.185 | 0.132 | 0.041a |
| Ib vs control | 0.072 | 0.053 | 0.103 | 0.077 | 0.072 | 0.027a |
| II vs control | 0.086 | 0.710 | 0.044a | 0.631 | 0.998 | 0.938 |
Furthermore, statistically significant differences were observed at the phylum and species level for beta diversity between groups Ia vs control for three indices. Additionally, differences at the species level were observed between groups II and control (expressed as the Jensen–Shannon index) (Table 3).
| Index/group | Bray-Curtis | Jensen-Shannon | Jaccard | |||
| L2 level | L7 level | L2 level | L7 level | L2 level | L7 level | |
| Ia vs Ib | 0.753 | 0.727 | 0.746 | 0.92 | 0.813 | 0.632 |
| Ia vs II | 0.409 | 0.291 | 0.404 | 0.176 | 0.469 | 0.389 |
| Ia vs control | 0.038a | 0.006a | 0.009a | 0.001a | 0.035a | 0.008a |
| Ib vs II | 0.532 | 0.374 | 0.543 | 0.468 | 0.517 | 0.332 |
| Ib vs control | 0.172 | 0.086 | 0.125 | 0.064 | 0.15 | 0.094 |
| II vs control | 0.313 | 0.057 | 0.206 | 0.026a | 0.393 | 0.087 |
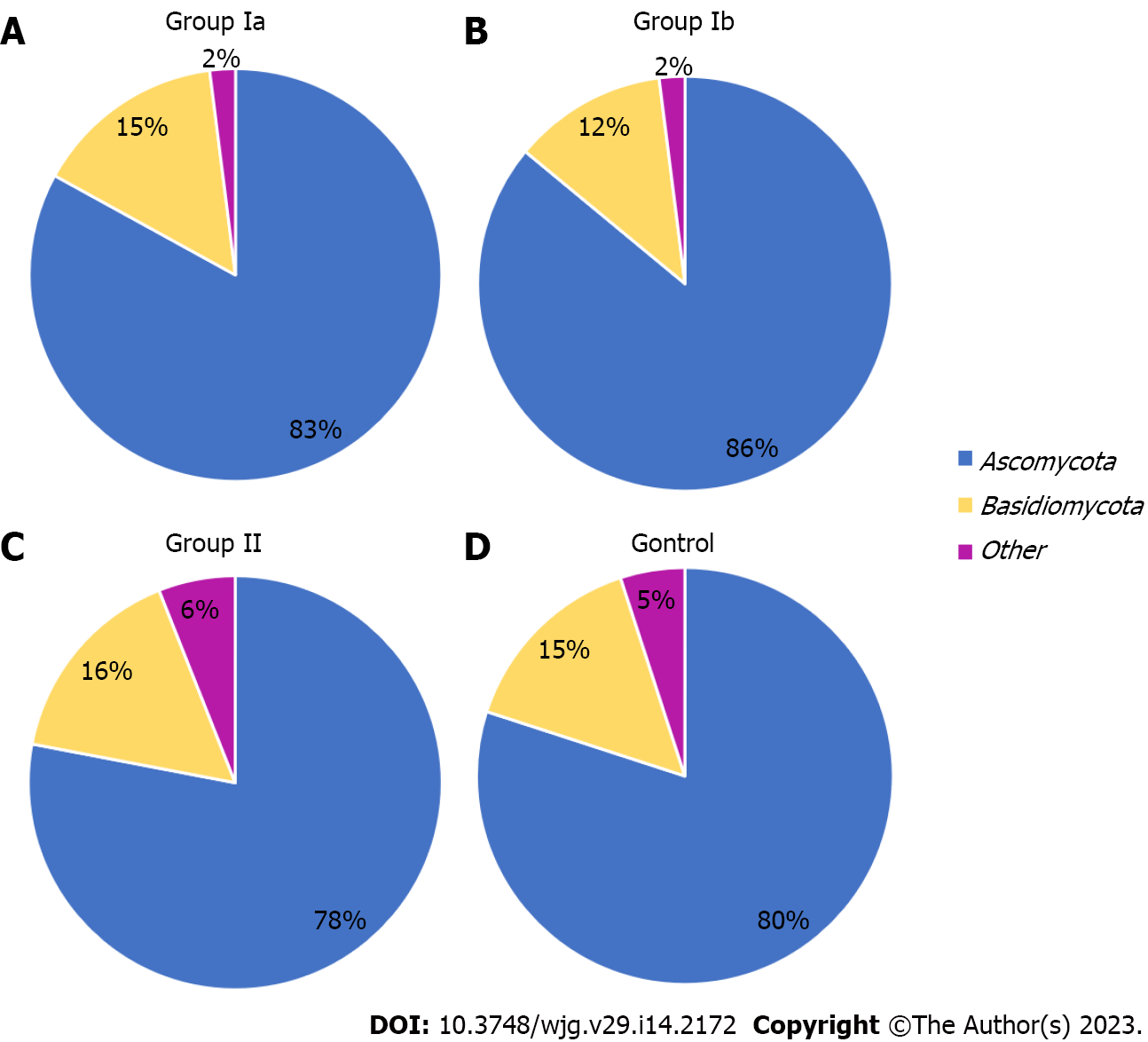
The gut mycobiota was evaluated at six taxonomic levels (phylum, class, order, family, genus, species). Three of these, L2 (phylum), L6 (genus), and L7 (species) were described in this study to include the most representative taxa. At the phylum level (L2), all OTUs obtained were assigned to two main phyla: Ascomycota and Basidiomycota. The others were non-fungal and belonged mainly to plant taxa. The Ascomycota phylum was clearly predominant in all groups, respectively: 83% in group Ia; 86% in group Ib; 78% in group II; 80% in the control group (Figure 1). In groups Ia and Ib, the percentage of these fungi was significantly higher compared to the control group (P = 0.039; P = 0.046, respectively).
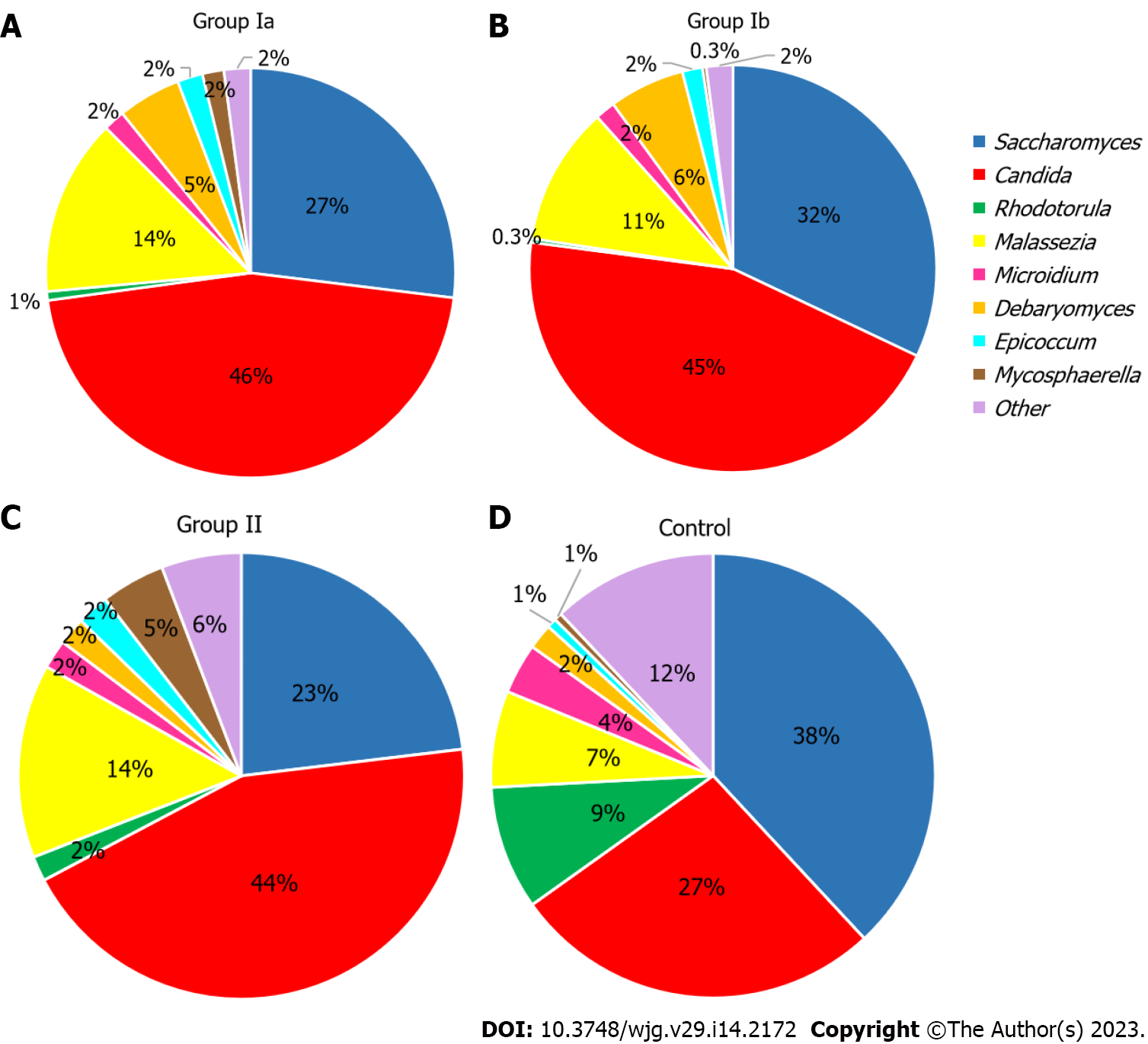
At the genus level (L6) (Figure 2), Candida dominated in all groups including CD patients (Ia–46%, Ib–45%, II–44%), and in groups Ia and II the abundance was statistically higher compared to the control group (26%; respectively: P < 0.001; P = 0.035). On the other hand, in the control group, the dominant genus was Saccharomyces (38%), and the abundance of this fungus was significantly higher in comparison to Ia (27%; P < 0.001). Interestingly, in all groups that included CD patients, the prevalence of Malassezia was almost twice as high (11%–14%) compared to the control group (7%), although statistically significant differences were only documented between group Ia and control (14% vs 7%, P = 0.015). Furthermore, in the group of patients with active CD (Ia and Ib), Debaryomyces constituted a higher percentage (respectively: 5%; 6%) compared to the other groups (2%), but significant differences were noted only between group Ia and the control (P = 0.024). Additionally, a significantly higher level of Rhodotorula was observed in the control group (9%) compared to the groups Ia (1%, P < 0.001), Ib (0.3%, P = 0.001), and II (2%, P = 0.039), and in group group Ib compared to group II (0.3% vs 2%, P = 0.014). Moreover, a significantly lower level of Mycosphaerella was observed in group Ib (0.3%) compared to groups II (5%, P = 0.015) and control (1%, P = 0.016). Statistical differences were also observed between group II (5%) and control (1%, P = 0.027) and for group Ia and II (2% vs 5%, P = 0.029). Statistical differences for Microidium were also found between groups Ib and control (2% vs 4%, P < 0.001).
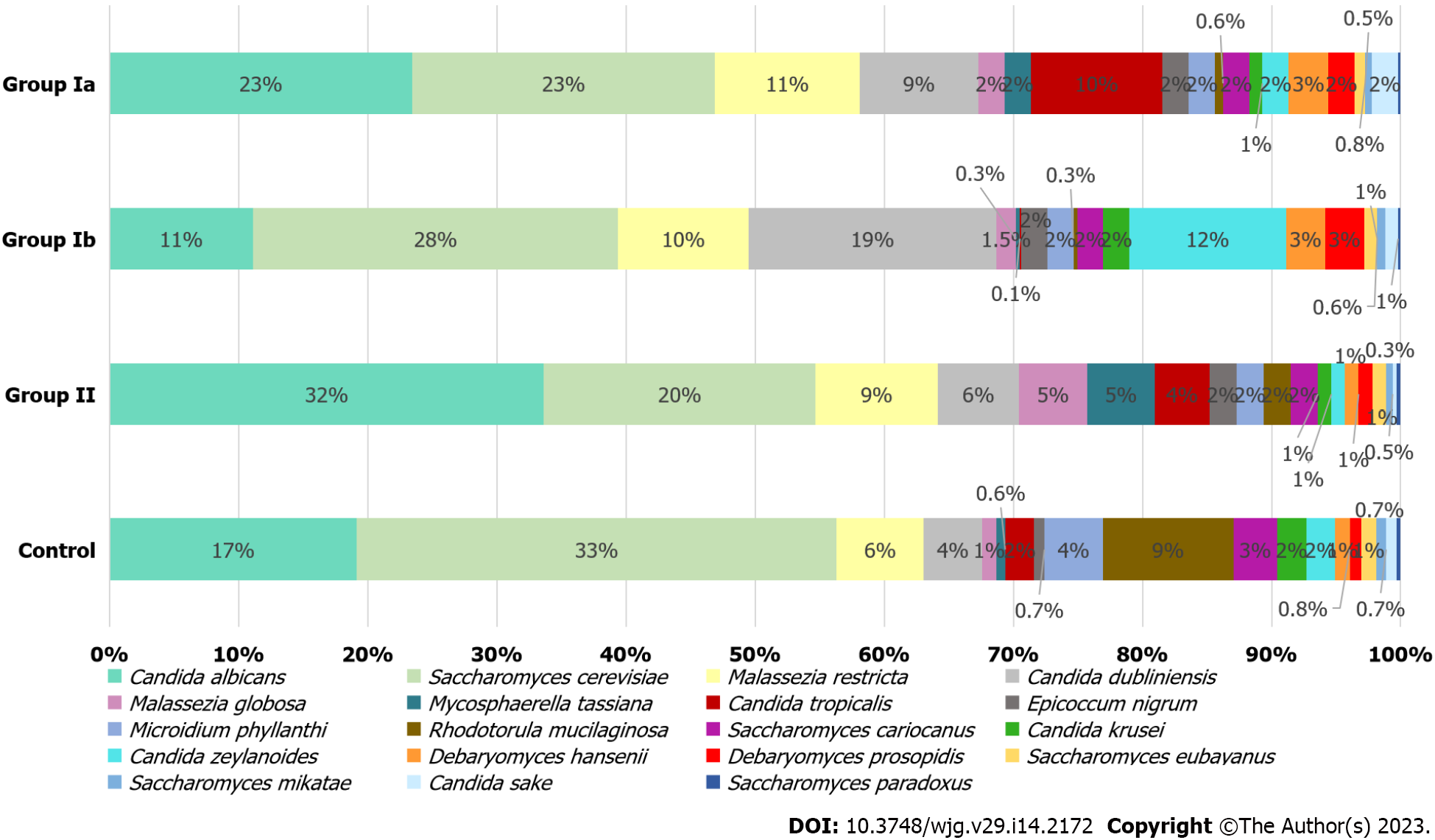
At the species level (L7), S. cerevisiae and Candida albicans (C. albicans) were dominant in all investigated groups and quantitatively constituted nearly 50% of all fungi. In the control group, S. cerevisiae was the most abundant yeast (38%), in contrast to the group of patients with CD (23% in Ia; 28% in Ib; 20% in II), in whom an increased percentage of individual Candida species was observed (Figure 3). In groups Ia, II, and control, the dominant species of the genus Candida was C. albicans (respectively 23%; 32%; 17%), and within this species, statistically significant differences were observed between groups Ia vs Ib (23% vs 11%, P = 0.008) and Ib vs control (11% vs 17%, P = 0.008). Differences close to statistical significance were documented between group II and the control (32% vs 17%, P = 0.054). The mycobiome of the patients in group Ib was overrepresented by Candida dubliniensis (19%) and Candida zeylanoides (12%) and the percentage of these fungi was significantly higher than in group II (respectively: 6%, P = 0.001; 1%, P = 0.003). A higher percentage of Candida tropicalis (10%) was observed in group Ia compared to the other groups (0.1%–4%), although statistically significant changes were found between groups Ia and Ib (10% vs 0.1%, P = 0.008) and Ib and control (0.1% vs 2%, P = 0.021). Furthermore, among the control group, a statistically higher percentage of Candida krusei (C. krusei) was observed compared to group II (2% vs 1%, P = 0.005).
Patients with CD (especially those newly diagnosed with active disease) were characterized by a higher prevalence of Malassezia restricta compared to the control group (11% vs 6%, Figure 3); however, these changes were not statistically significant. Statistical differences were observed between groups Ib and II (10% vs 9%, P = 0.001). Among controls, a significantly higher percentage (9%) of Rhodotorula mucilaginosa (R. mucilaginosa) was found in comparison to Ia (0.7%, P = 0.001); Ib (0.3%, P = 0.002) and II (2%, P = 0.027). Statistically significant differences were also found for groups Ib and II (P = 0.015). In the control group, a slight increase in Saccharomyces cariocanus was observed in comparison to group II (3% vs 2%, P = 0.046) and for Microidium phyllanthi (M. phyllanthi) compared to all CD groups (4% vs 2%, P < 0.05). There were also statistically significant differences in Mycosphaerella tassiana (M. tassiana) abundance between groups Ia vs II (2% vs 5%, P = 0.029) and Ib vs control (0.3% vs 0.6%, P = 0.016). In addition, the changes in abundance of Debaryomyces hansenii (D. hansenii) (3% vs 1%, P = 0.020) and Malassezia globosa (M. globosa) (2% vs 5%, P = 0.037) were documented between groups Ia and II, and in M. globosa between groups II vs control (5% vs 1%, P = 0.030), and also in Epicoccum nigrum (E. nigrum) between Ia vs control (2% vs 0.7%, P = 0.015).
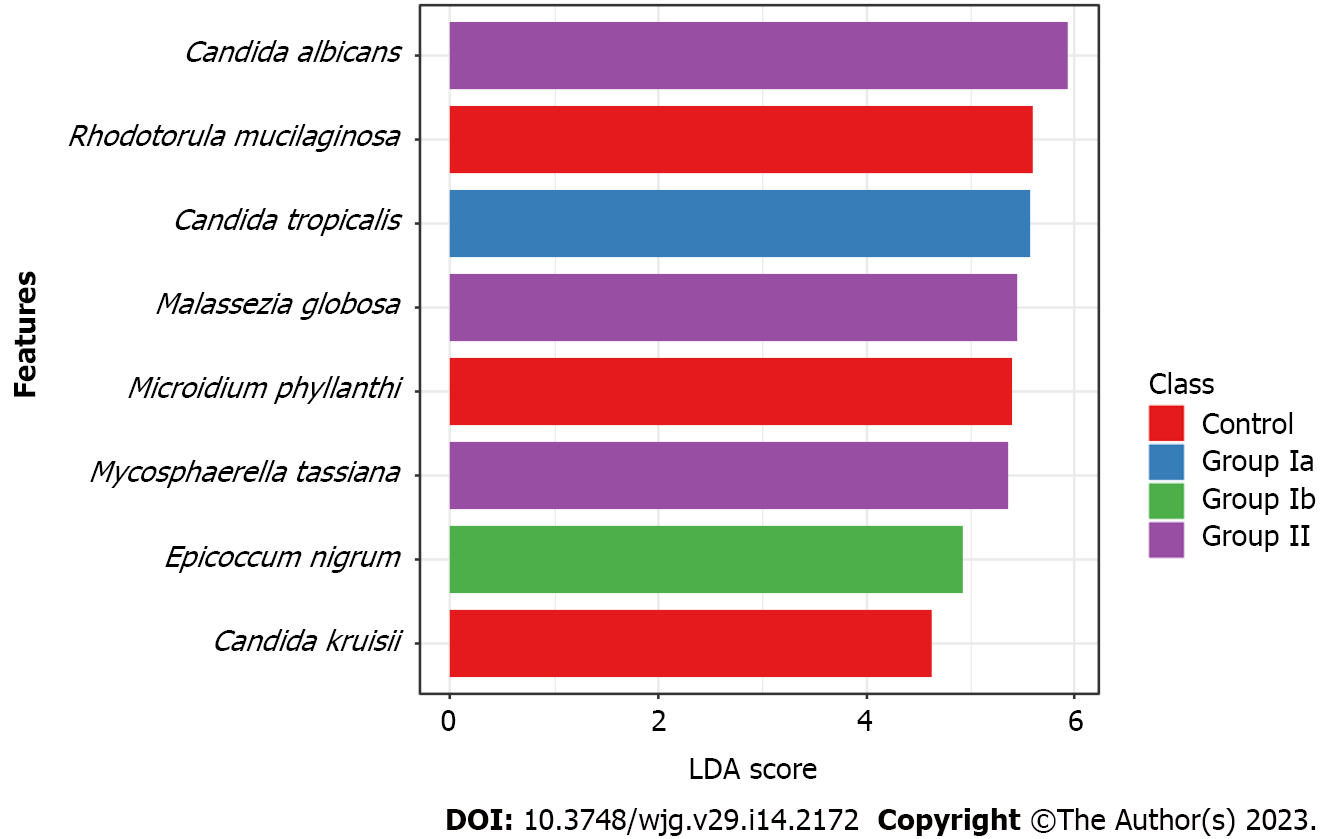
Owing to LDA we have selected species of fungi that could be biomarkers characterizing and distinguishing a given group of patients (Figure 4). We have shown that C. tropicalis was overrepresented in group Ia compared to the other groups (P = 0.026). Whereas increased richness of C. albicans (P = 0.005), M. globosa (P = 0.011) and M. tassiana (P = 0.024) was documented in group II compared to the other groups. The fungal biomarkers that differentiated the control group from the CD patient groups were R. mucilaginosa (P < 0.001), C. krusei (P = 0.013), and M. phyllanthi (P < 0.001). Group Ib was characterized by a higher abundance of E. nigrum compared to the other groups (P = 0.011).
The analysis of the abundance of fungal species showed significant correlations between: D. hansenii and C. zeylanoides and patient age (negative and positive, respectively); C. dubliniensis and C. sake and calprotectin (both positive) and a positive correlation between C. tropicalis and PCDAI (Table 4).
| Variables/age | Variables/calprotectin | Variables/PCDAI | |||
| Debaryomyces hansenii | P < 0.008 | Candida dubliniensis | P < 0.003 | Candida tropicalis | P < 0.006 |
| r = -0.08 | r = 0.29 | r = 0.58 | |||
| Candida zeylanoides | P < 0.004 | Candida sake | P < 0.04 | ||
| r = 0.08 | r = 0.19 | ||||
Numerous studies suggest that changes in the intestinal microbiota and the interaction between microorganisms and the host may lead to the development of IBD. The main limitation of the available studies[12,22-24] is the predominance of experiments in adult patients who have been treated for a long time and who have been sick for many years, which could significantly affect the results obtained. Moreover, the studies of the intestinal microbiota in IBD conducted so far have focused primarily on disorders within the bacteriobiome. There is a lack of more comprehensive research, including fungi, which constitute a very important component of the intestinal microbiome[25]. In our study, we recruited children and adolescents with both newly and previously diagnosed CD and we compared their mycobiome in the exacerbation and remission periods. In addition, we recruited healthy children into a control group. Such a study allowed a more reliable determination of the relationship between fungi in the digestive tract and the course of CD. Moreover, the main exclusion criterion in all groups was the use of antibiotics, probiotics, and antifungal drugs that could significantly change the composition of the mycobiome. Thus, the impact of confounding factors associated with antimicrobial therapy was eliminated. Our data have shown that the mycobiota of pediatric CD patients was different in comparison to healthy controls, and also depended on the stage of disease activity.
The alpha diversity (which is a measure of biodiversity within the study groups) of the fungal microbiota was significantly different (both at the phylum and species levels) in group Ia compared to the control expressed as the Chao and Fisher indices (Table 2). Additionally, biodiversity was higher in control compared to the Ia group for the three analyzed indices. Lower alpha diversity in the samples from patients with active, newly diagnosed CD indicated a reduction in fungal biodiversity, i.e., lower number of taxons in a single sample. These data are consistent with previous reports[24,26,27]. Interestingly, we have shown that patients in the non-active phase of CD (II) were characterized by significantly higher alpha biodiversity indices at the level L7 (species) compared to patients with newly diagnosed, active disease (Ia). This provides the basis for the conclusion that while patients achieve remission, normalization of mycobiome occurs, an indicator of which is the increase in the measure of biodiversity[28].
The analysis of beta diversity (which is a measure of differences between the study groups) has shown that there were statistically significant differences between group Ia and the control, both at the L2 and L7 levels (expressed by the three indices) (Table 3). These results indicate a significantly different taxonomic composition at the phylum and species levels between these groups. The different composition of mycobiota seen in the patients of group Ia compared to healthy children indicates that dysbiosis appears at the very beginning of the disease. However, the treatment (groups Ib and II) likely reduces the differences in mycobiome composition compared to controls.
At the phylum level, Ascomycota was predominant in all groups (Figure 1), but in patients with active CD (irrespective of whether it was a newly diagnosed or previously diagnosed disease), the percentage of this phylum was significantly higher (83%–86%) compared to the control group (80%). These observations are consistent with studies by other researchers showing an increased percentage of Ascomycota in adult patients with active CD[22,29].
At the genus level (Figure 2), we observed that among patients with CD (irrespective of disease activity), Candida was the predominant genus in contrast to the control group (significant differences between Ia vs control and II vs control; P < 0.05). Analyses at the species level (Figure 3), showed that in each of the studied groups it was possible to distinguish a specific species within the genus Candida, characteristic of a given patient population. Thus, an excessive abundance of C. tropicalis has been observed in newly diagnosed patients in active phase of CD (group Ia) (Figure 3). Interestingly, in patients in remission (group II), the abundance of this species decreased more than double. Moreover, in the control group, C. tropicalis was 5 times lower than in group Ia. This may suggest that C. tropicalis may be a biomarker of inflammation in the gut in the course of CD, which was supported by LDA analysis (Figure 4). It has previously been shown that overrepresentation of C. tropicalis is commonly found among patients with active CD[12,23,30] and possibly influences the initiation and maintenance of inflammation[23]. Hoarau et al[23] showed that a higher prevalence of C. tropicalis fungi was positively correlated with levels of ASCA and with the abundance of Serratia marcescens and Escherichia coli. Researchers demonstrated that these microorganisms interact with each other in the gut, forming a biofilm that can promote an excessive immune response and cause intestinal mucosal barrier dysfunction, contributing to the formation of inflammatory lesions[23]. Additionally, studies in animal models of IBD confirmed that C. tropicalis has strong immunogenic potential. Supplementation in mice with C. tropicalis resulted in excessive secretion of pro-inflammatory cytokines, such as tumor necrosis factor alpha, interferon gamma and, interleukin-17, and then intensification of inflammation in the gut[7]. In addition, in other independent studies, C. tropicalis has induced dysbiosis that involved changes in the presence of mucin-degrading bacteria, leading to altered tight junction protein expression with increased intestinal permeability. Then, it induced a strong Th1/Th17 response, leading to an accelerated pro-inflammatory phenotype in experimental colitis mice[31]. Interestingly, in our research, a positive correlation between C. tropicalis and PCDAI was observed, which strengthens the above suggestions that these fungi may be associated with the exacerbation of the disease.
Analysis at the species level has shown that C. albicans constituted the highest percentage in the group of patients in remission (II) compared to the other groups (Figure 3), and LDA analysis has documented that this species may be a fungal biomarker for patients in non-active CD (P = 0.005, Figure 4). Previous studies showed that these yeasts were more abundant in CD patients, and it was suggested that C. albicans may promote IBD by increasing the inflammatory response[27,32,33]. However, it should be noted that these studies did not take into account patients in remission. Patients in the active phase or all CD patients together (undifferentiated according to the phase of disease activity) were compared with healthy people. In our research, an additional comparison of the exacerbation and remission groups has provided evidence that there are more C. albicans in non-active CD compared to active disease. It is likely that the treatment used to lead the patient into remission somehow promotes the growth of C. albicans; however, the effect of anti-inflammatory treatment commonly used in CD (like 5-ASAs; azathioprine) on the mycobiome has not been proven so far. Moreover, the growth of fungi in the gut is closely related to changes in the bacterial microbiota, so it is also possible that changes in the bacterial community that occur during remission contribute to the increased abundance of C. albicans[12,23,34-36].
The results of our study show an increased number of C. dubliniensis and C. zeylanoides in active but already treated patients (Ib) compared to other groups (Figure 3). To our knowledge, this is the first such report. Perhaps the increase in the colonization by these species of fungi was related to the treatment or a long-term inflammatory process. However, it should be noted that among treated patients in remission (II), the abundance of C. dubliniensis and C. zeylanoides significantly decreased (P = 0.001, P = 0.003; respectively) compared to treated patients in the active phase (Ib). It is possible that colonization by these fungi may depend on the disease activity and long-term inflammatory process that promotes their multiplication. Although C. dubliniensis is not an epidemiologically significant species, some reports indicate that it may cause opportunistic infections that may be associated with a higher degree of intestinal colonization[37,38]. Thus, its involvement in the maintenance of inflammation in CD cannot be ruled out, especially since a positive significant correlation of this species with calprotectin (which is a marker of intestinal inflammation) was observed (Table 4).
C. zeylanoides are fungi found in food products[39-41], so their presence may be associated with different diets of patients. On the other hand, we documented that these fungi were positively significantly correlated with age (Table 4). Perhaps, the higher abundance of C. zeylanoides in group Ib was related to the fact that we had slightly elderly patients in this group compared to the other one (Table 1). There are several reported cases in which C. zeylanoides caused fungemia[42-44]; however, there is no evidence that this microorganism has a connection with chronic or autoimmune diseases.
An interesting subject of study is D. hansenii. The abundance of these fungi was higher among patients with active CD, however, statistically significant changes were only observed between groups Ia and II (P = 0.020). The fact that the prevalence of this species was three times higher in patients with active CD compared to patients in remission and control (Figure 3) indicates that this fungus may be involved in maintaining intestinal inflammation or even inducing disease. Jain et al[45] came to similar conclusions. The researchers showed that damaged intestinal tissue (in a mouse model of CD) was over-colonized with D. hansenii, which consequently impeded the healing and regeneration process of intestinal crypts. Interestingly, when mice received an antifungal drug (amphotericin), the wounds began to heal[45]. In the same study, the authors conducted an analysis with the participation of adult people with CD and showed that D. hansenii was detected in all samples taken from patients (100%), but only in one healthy person (10%). Further detailed observation of tissue biopsies showed that this fungus was present only in the inflamed mucosa, whereas it was absent from the non-inflamed mucosa sampled from the same patient[45]. Our study is the first report documenting the presence of D. hansenii in children and adolescents with CD. Furthermore, our data indicate that the abundance of these fungi depends on the phase of disease activity. These observations lead to the conclusion that D. hansenii may be involved in maintaining inflammation.
In this study, we have documented that CD patients were over-colonized with Malassezia species compared to the control group (Figures 2 and 3). This is consistent with the observations of other researchers[12,27,46]. Patients with CD often suffer from malabsorption syndrome, manifested by fatty diarrhea. Malassezia requires lipids to multiply; therefore, it is possible that increased detectability of these yeasts in CD patients is related to the fact that excess fat in the stool creates an optimal environment for Malassezia over-colonization. However, the participation of these yeasts in maintaining inflammation cannot be excluded because there are a few studies that demonstrate the immunogenic properties of Malassezia in the context of IBD[46,47]. The species analysis provided interesting observations regarding the changes in individual species of Malassezia depending on the activity of the disease (Figure 3). Thus, among children in remission (II), M. globosa colonization increased and these changes were statistically significant compared to group Ia (P = 0.037) and the control group (P = 0.030). On the other hand, patients in the exacerbation phase had a higher percentage of M. restricta (P = 0.001). The participation of M. restricta was previously reported in individual studies. Limon et al[46] demonstrated that these fungi were significantly more abundant in CD patients than in healthy subjects, and in addition, their presence was associated with exacerbation in a mouse model of IBD[46]. Furthermore, the team showed that these yeasts were especially present in patients with IBD caspase recruitment domain-containing protein 9 (CARD9) risk. CARD9 is a signaling adapter protein that is essential for antifungal innate immunity in mice and humans. Single nucleotide polymorphism in the gene for CARD9 is the strongest genetic risk factor linked to IBD. A certain variant of this gene caused human immune cells to produce more potent inflammatory cytokines in response to M. restricta[46]. Additionally, Li et al[12] showed that the biopsies collected from inflamed mucosa were over-colonized by M. restricta in relation to tissue without inflammation from the same patient[12]. A study on the immunogenicity of fungi of the genus Malassezia in the context of IBD confirmed that both M. globosa and M. restricta have strong pro-inflammatory properties. Both yeast species induced the production of inflammatory cytokines by dendritic cells in large part mediated through Dectin2 and CARD9 signaling[47]. Furthermore, M. restricta exacerbated colitis in germ-free mouse models of IBD triggered by inflammatory reactions mediated by CARD9[46]. The above data indicate that fungi of the genus Malassezia significantly influence the host's immune response, so their role in the pathogenesis of IBD cannot be ruled out. Understanding the species changes that occur in the various phases of disease activity, which were documented in our analysis, forms the basis for further research and gives an insight into the role of these yeasts in the course of CD.
Another species whose participation in CD is worth considering, is Epicoccum nigrum. These fungi have been overlooked so far, but this is probably due to the difficulties in culturing and detecting these yeasts. In this study, we have shown that the abundance of these fungi was significantly higher in CD patients (Ia) compared to healthy children (P = 0.015, Figure 3). Wheeler et al[48] documented the first reports on E. nigrum in CD and suggested its association with the treatment applied. The researchers showed that extended pretreatment with antifungal drugs restructured the intestinal fungal population. Increased colonization by the E. nigrum fungi, among others, has been observed. It is unknown whether the observed changes were due to alterations in the microbial community that promoted E. nigrum overgrowth, or whether these yeasts were resistant to the applied treatment. However, these changes were correlated with exacerbated disease in chemically induced models of experimental colitis[48]. In our study, the exclusion criterion was the use of antifungal drugs, so the presence of E. nigrum was not related to treatment. However, it cannot be ruled out that the alterations observed in the microbial community in CD patients promote the overgrowth of E. nigrum. Fungal populations of the intestinal tract directly or indirectly help to maintain healthy intestinal homeostasis. Any changes in the composition of the mycobiome may cause serious disruption, especially in genetically susceptible people to diseases (e.g. predisposed to CD). On the other hand, the LDA analysis (Figure 4) has shown that E. nigrum may be a fungal biomarker for patients with active, previously diagnosed disease (Ib). Perhaps the presence of these fungi is associated with the active long-term inflammation. It cannot be ruled out that chronic and long-term inflammatory lesions in the intestine favor the growth of this species. The overrepresentation of E. nigrum found in this report in CD patients requires further research.
In this study, we also demonstrated an increase in colonization level by R. mucilaginosa and M. phyllanthi among healthy subjects compared to CD patients (P < 0.05, Figure 3). LDA analysis has shown that these species may be a fungal biomarker for healthy children (Figure 4). R. mucilaginosa are fungi commonly found in soil, plants, and animals; however, they are also food contaminants. They are often isolated from fruit juices and dairy products[49]. The increase in the occurrence of these fungi in healthy children could be related to a varied and rich diet used by healthy people. In contrast, patients with CD usually are on a dairy-free diet and exclude many products from their meals, so they carry fewer yeasts associated with foods in the digestive tract. On the other hand, M. phyllanthi are plant pathogens that can be delivered to the gastrointestinal tract through the consumption of vegetables and fruits. Patients with CD significantly reduce their consumption of legumes, and stone fruits, and thus, they have a lower abundance of plant-related fungi than healthy people following a varied diet[50,51].
In conclusion, this study confirms alterations in intestinal fungal composition in pediatric patients with CD and shows that some species of fungi may be a microbiological marker related to the activity of the disease. In patients with CD, we have documented an increased load of fungi with potential pro-inflammatory effects (e.g., Candida spp., Malassezia spp.)[7,14,23,46], while fungi with potential anti-inflammatory effects (such as Saccharomyces)[23,52-55] were found in a lower percentage. Moreover, changes within the specific species of fungi depending on disease activity, and the positive correlation of some species with calprotectin or PCDAI, give strong evidence that these microorganisms may be of key importance in the development and course of CD.
Furthermore, we have shown that some fungal species can be helpful in predicting an exacerbation of the disease or even predicting the diagnosis of CD. Further research should focus on selecting fungal species that could be biomarkers to help predict disease exacerbation. Moreover, future research should assess whether the mycobiota could be a therapeutic target.
The recurrent and chronic course of Crohn’s disease (CD), its systemic after effects, and intestinal complications constitute a serious clinical problem, and since the pathogenesis of the disease is unknown, causal treatment is currently not used. As CD incidence age keeps falling and there is a growing number of cases, we are led to undertake intensive studies to determine the possible causes of this disease. Recently, due to the growing interest in the topic of intestinal microbiome, a hypothesis has emerged that the initiation of CD is associated with dysbiosis within the gut microbiota. And while the importance of bacteria in the pathogenesis has been, up to now, a common subject of research, the involvement of fungi has usually been overlooked. The few available studies including mycobiome analysis concern adults and not children, previously treated patients, or those with long-term disease. These shortcomings distort the results due to the impact of confounding variables (such as treatment, age, or the long-term disease process) on changes in the fungal composition.
Undoubtedly, the composition of the microbiome has a significant influence on maintaining internal balance and health and microbial changes constitute an important factor inducing pathological processes. Due to the fact that fungi are an important component of the gut microbiota, it is possible that alterations in the composition of the gut mycobiome may have an impact on the induction of CD.
Taking into account the likely relationship between the mycobiota and the host, the aim of this study was to perform a detailed taxonomic analysis of the fungal composition in pediatric patients with CD. In our study, we recruited children and adolescents with newly and previously diagnosed CD and compared their mycobiome in the exacerbation and remission periods. Additionally, we recruited healthy children into a control group. Such a study allowed for a more reliable determination of the relationship between fungi in the digestive tract and the course of CD.
DNA was isolated from stool samples from patients: With active, newly diagnosed CD (n = 50); active but previously diagnosed and treated CD (n = 16); non-active CD who were in clinical remission (n = 39) and healthy volunteers (n = 40). The next step was to prepare genomic libraries for next generation sequencing (NGS). NGS was performed using a MiSeq sequencer (Illumina). The composition of the gut mycobiota was analyzed using UNITE Fungal ITS Database v7.2, and then correlated with clinical and blood parameters.
Our study confirms alterations in fungal composition in pediatric CD patients and shows that some species of fungi may be a kind of microbiological marker related to the activity of the disease. In CD patients, we have documented an increased load of fungi with potential pro-inflammatory effects (e.g. Candida spp., Malassezia spp.), while fungi with potential anti-inflammatory effects (such as Saccharomyces) were found in a lower percentage. Interestingly, the greatest alterations in mycobiome composition (compared to the control group) were observed among newly diagnosed patients, before implementing any therapeutic approaches. This is strong evidence that fungi may play an important role in the development of CD. This thesis is supported by the fact that a positive correlation of some species with calprotectin or pediatric CD activity index was documented. Furthermore, owing to linear discriminant analysis, we have shown that some fungal species could be biomarkers characterizing and distinguishing a given group of patients (depending on the disease activity) which in the future may be helpful in predicting an exacerbation of the disease or even predicting the diagnosis of CD.
Changes in the composition of the intestinal mycobiome occur already at the beginning of the disease (in newly diagnosed and untreated patients). Furthermore, the composition of fungi changes depending on the activity of CD.
Further research should focus on selecting fungal species that could be biomarkers to help predict disease exacerbation. Furthermore, next research should assess whether the fungal mycobiota could be a therapeutic target.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Poland
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gazouli M, Greece; Yang L, China S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327-339.e4. [PubMed] [DOI] [Full Text] |
| 2. | Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307-317. [PubMed] [DOI] [Full Text] |
| 3. | Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1-10. [PubMed] [DOI] [Full Text] |
| 4. | Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631-637. [PubMed] [DOI] [Full Text] |
| 5. | Salamon D, Gosiewski T, Krawczyk A, Sroka-Oleksiak A, Duplaga M, Fyderek K, Kowalska-Duplaga K. Quantitative changes in selected bacteria in the stool during the treatment of Crohn's disease. Adv Med Sci. 2020;65:348-353. [PubMed] [DOI] [Full Text] |
| 6. | Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759-795. [PubMed] [DOI] [Full Text] |
| 7. | Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314-1317. [PubMed] [DOI] [Full Text] |
| 8. | Håkansson Å, Tormo-Badia N, Baridi A, Xu J, Molin G, Hagslätt ML, Karlsson C, Jeppsson B, Cilio CM, Ahrné S. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin Exp Med. 2015;15:107-120. [PubMed] [DOI] [Full Text] |
| 9. | Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780-13785. [PubMed] [DOI] [Full Text] |
| 10. | Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447-460. [PubMed] [DOI] [Full Text] |
| 11. | Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 2009;15:872-882. [PubMed] [DOI] [Full Text] |
| 12. | Li Q, Wang C, Tang C, He Q, Li N, Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn's disease. J Clin Gastroenterol. 2014;48:513-523. [PubMed] [DOI] [Full Text] |
| 13. | Heidarian F, Alebouyeh M, Shahrokh S, Balaii H, Zali MR. Altered fecal bacterial composition correlates with disease activity in inflammatory bowel disease and the extent of IL8 induction. Curr Res Transl Med. 2019;67:41-50. [PubMed] [DOI] [Full Text] |
| 14. | Wheeler ML, Limon JJ, Underhill DM. Immunity to Commensal Fungi: Detente and Disease. Annu Rev Pathol. 2017;12:359-385. [PubMed] [DOI] [Full Text] |
| 15. | Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, Weiss A, Vasilakos JP, Underhill DM. Activation of the innate immune receptor Dectin-1 upon formation of a 'phagocytic synapse'. Nature. 2011;472:471-475. [PubMed] [DOI] [Full Text] |
| 16. | Levine A, Koletzko S, Turner D, Escher JC, Cucchiara S, de Ridder L, Kolho KL, Veres G, Russell RK, Paerregaard A, Buderus S, Greer ML, Dias JA, Veereman-Wauters G, Lionetti P, Sladek M, Martin de Carpi J, Staiano A, Ruemmele FM, Wilson DC; European Society of Pediatric Gastroenterology, Hepatology, and Nutrition. ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastroenterol Nutr. 2014;58:795-806. [PubMed] [DOI] [Full Text] |
| 17. | Levine A, Griffiths A, Markowitz J, Wilson DC, Turner D, Russell RK, Fell J, Ruemmele FM, Walters T, Sherlock M, Dubinsky M, Hyams JS. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314-1321. [PubMed] [DOI] [Full Text] |
| 18. | Krawczyk A, Sroka-Oleksiak A, Kowalska-Duplaga K, Fyderek K, Gosiewski T, Salamon D. Impact of biological treatment on intestinal microbiom in children with Crohn’s disease. World Sci News. 2018;104:252-263. |
| 19. | Kowalska-Duplaga K, Krawczyk A, Sroka-Oleksiak A, Salamon D, Wędrychowicz A, Fyderek K, Gosiewski T. Dependence of Colonization of the Large Intestine by Candida on the Treatment of Crohn's Disease. Pol J Microbiol. 2019;68:121-126. [PubMed] [DOI] [Full Text] |
| 20. | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261-5267. [PubMed] [DOI] [Full Text] |
| 21. | 16S Metagenomic Sequencing Library Preparation. Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. [cited 10 November 2022]. Available from: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf. |
| 22. | Liguori G, Lamas B, Richard ML, Brandi G, da Costa G, Hoffmann TW, Di Simone MP, Calabrese C, Poggioli G, Langella P, Campieri M, Sokol H. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn's Disease Patients. J Crohns Colitis. 2016;10:296-305. [PubMed] [DOI] [Full Text] |
| 23. | Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn's Disease. mBio. 2016;7. [PubMed] [DOI] [Full Text] |
| 24. | Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039-1048. [PubMed] [DOI] [Full Text] |
| 25. | Ruszkowski J, Kaźmierczak-Siedlecka K, Witkowski JM, Dębska-Ślizień A. Mycobiota of the human gastrointestinal tract. Postepy Hig Med Dosw. 2020;74:301-313. [DOI] [Full Text] |
| 26. | Qiu X, Zhao X, Cui X, Mao X, Tang N, Jiao C, Wang D, Zhang Y, Ye Z, Zhang H. Characterization of fungal and bacterial dysbiosis in young adult Chinese patients with Crohn's disease. Therap Adv Gastroenterol. 2020;13:1756284820971202. [PubMed] [DOI] [Full Text] |
| 27. | Olaisen M, Richard ML, Beisvåg V, Granlund AVB, Røyset ES, Rué O, Martinsen TC, Sandvik AK, Sokol H, Fossmark R. The ileal fungal microbiota is altered in Crohn's disease and is associated with the disease course. Front Med (Lausanne). 2022;9:868812. [PubMed] [DOI] [Full Text] |
| 28. | Manor O, Dai CL, Kornilov SA, Smith B, Price ND, Lovejoy JC, Gibbons SM, Magis AT. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun. 2020;11:5206. [PubMed] [DOI] [Full Text] |
| 29. | Nelson A, Stewart CJ, Kennedy NA, Lodge JK, Tremelling M; UK IBD Genetics Consortium, Probert CS, Parkes M, Mansfield JC, Smith DL, Hold GL, Lees CW, Bridge SH, Lamb CA. The Impact of NOD2 Genetic Variants on the Gut Mycobiota in Crohn's Disease Patients in Remission and in Individuals Without Gastrointestinal Inflammation. J Crohns Colitis. 2021;15:800-812. [PubMed] [DOI] [Full Text] |
| 30. | Krawczyk A, Salamon D, Kowalska-Duplaga K, Bogiel T, Gosiewski T. Association of Fungi and Archaea of the Gut Microbiota with Crohn's Disease in Pediatric Patients-Pilot Study. Pathogens. 2021;10. [PubMed] [DOI] [Full Text] |
| 31. | Di Martino L, De Salvo C, Buela KA, Hager C, Ghannoum M, Osme A, Buttò L, Bamias G, Pizarro TT, Cominelli F. Candida tropicalis Infection Modulates the Gut Microbiome and Confers Enhanced Susceptibility to Colitis in Mice. Cell Mol Gastroenterol Hepatol. 2022;13:901-923. [PubMed] [DOI] [Full Text] |
| 32. | Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2019;16:331-345. [PubMed] [DOI] [Full Text] |
| 33. | Standaert-Vitse A, Sendid B, Joossens M, François N, Vandewalle-El Khoury P, Branche J, Van Kruiningen H, Jouault T, Rutgeerts P, Gower-Rousseau C, Libersa C, Neut C, Broly F, Chamaillard M, Vermeire S, Poulain D, Colombel JF. Candida albicans colonization and ASCA in familial Crohn's disease. Am J Gastroenterol. 2009;104:1745-1753. [PubMed] [DOI] [Full Text] |
| 34. | Jawhara S. How Gut Bacterial Dysbiosis Can Promote Candida albicans Overgrowth during Colonic Inflammation. Microorganisms. 2022;10. [PubMed] [DOI] [Full Text] |
| 35. | Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun. 2012;80:3371-3380. [PubMed] [DOI] [Full Text] |
| 36. | Frau A, Ijaz UZ, Slater R, Jonkers D, Penders J, Campbell BJ, Kenny JG, Hall N, Lenzi L, Burkitt MD, Pierik M, Darby AC, Probert CSJ. Inter-kingdom relationships in Crohn's disease explored using a multi-omics approach. Gut Microbes. 2021;13:1930871. [PubMed] [DOI] [Full Text] |
| 37. | Gutiérrez J, Morales P, González MA, Quindós G. Candida dubliniensis, a new fungal pathogen. J Basic Microbiol. 2002;42:207-227. [PubMed] [DOI] [Full Text] |
| 38. | Sullivan D, Coleman D. Candida dubliniensis: an emerging opportunistic pathogen. Curr Top Med Mycol. 1997;8:15-25. [PubMed] |
| 39. | Seker E. Identification of Candida species isolated from bovine mastitic milk and their in vitro hemolytic activity in Western Turkey. Mycopathologia. 2010;169:303-308. [PubMed] [DOI] [Full Text] |
| 40. | Obasi BC, Whong CMZ, Ado SA, Abdullahi IC. Isolation and Identification Of Yeast Associated With Fermented Orange Juice. Int J Eng Sci (Ghaziabad). 2014;3:64-69. [DOI] [Full Text] |
| 41. | Bintsis T. Yeasts in different types of cheese. AIMS Microbiol. 2021;7:447-470. [PubMed] [DOI] [Full Text] |
| 42. | Pereira GH, Müller PR, Szeszs MW, Levin AS, Melhem MS. Five-year evaluation of bloodstream yeast infections in a tertiary hospital: the predominance of non-C. albicans Candida species. Med Mycol. 2010;48:839-842. [PubMed] [DOI] [Full Text] |
| 43. | Levenson D, Pfaller MA, Smith MA, Hollis R, Gerarden T, Tucci CB, Isenberg HD. Candida zeylanoides: another opportunistic yeast. J Clin Microbiol. 1991;29:1689-1692. [PubMed] [DOI] [Full Text] |
| 44. |
Lee CJ, Shin JH, Kim JP, Kook H, Suh SP, Ryang DW.
A Case of Mixed Fungemia with Cryptococcus laurentii and Candida zeylanoides. |
| 45. | Jain U, Ver Heul AM, Xiong S, Gregory MH, Demers EG, Kern JT, Lai CW, Muegge BD, Barisas DAG, Leal-Ekman JS, Deepak P, Ciorba MA, Liu TC, Hogan DA, Debbas P, Braun J, McGovern DPB, Underhill DM, Stappenbeck TS. Debaryomyces is enriched in Crohn's disease intestinal tissue and impairs healing in mice. Science. 2021;371:1154-1159. [PubMed] [DOI] [Full Text] |
| 46. | Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, Iliev ID, Skalski JH, Brown J, Landers C, Borneman J, Braun J, Targan SR, McGovern DPB, Underhill DM. Malassezia Is Associated with Crohn's Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe. 2019;25:377-388.e6. [PubMed] [DOI] [Full Text] |
| 47. | Wolf AJ, Limon JJ, Nguyen C, Prince A, Castro A, Underhill DM. Malassezia spp. induce inflammatory cytokines and activate NLRP3 inflammasomes in phagocytes. J Leukoc Biol. 2021;109:161-172. [PubMed] [DOI] [Full Text] |
| 48. | Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, Arditi M, Underhill DM, Iliev ID. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe. 2016;19:865-873. [PubMed] [DOI] [Full Text] |
| 49. | Krzysciak P, Halska A, Macura AB. Occurence and pathogenicity of the fungi Rhodotorula spp. Postępy Mikrobiologii. 2007;46:291-300. [DOI] [Full Text] |
| 50. | Cybulska J, Drobek M, Panek J, Cruz-Rubio JM, Kurzyna-Szklarek M, Zdunek A, Frąc M. Changes of pectin structure and microbial community composition in strawberry fruit (Fragaria × ananassa Duch.) during cold storage. Food Chem. 2022;381:132151. [PubMed] [DOI] [Full Text] |
| 51. | Arafat Y, Tayyab M, Khan MU, Chen T, Amjad H, Awais S, Lin X, Lin W, Lin S. Long-Term Monoculture Negatively Regulates Fungal Community Composition and Abundance of Tea Orchards. Agronomy. 2019;9:466. [DOI] [Full Text] |
| 52. | Sivignon A, de Vallée A, Barnich N, Denizot J, Darcha C, Pignède G, Vandekerckove P, Darfeuille-Michaud A. Saccharomyces cerevisiae CNCM I-3856 prevents colitis induced by AIEC bacteria in the transgenic mouse model mimicking Crohn's disease. Inflamm Bowel Dis. 2015;21:276-286. [PubMed] [DOI] [Full Text] |
| 53. | Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:656-665. [PubMed] [DOI] [Full Text] |
| 54. | Jawhara S, Habib K, Maggiotto F, Pignede G, Vandekerckove P, Maes E, Dubuquoy L, Fontaine T, Guerardel Y, Poulain D. Modulation of intestinal inflammation by yeasts and cell wall extracts: strain dependence and unexpected anti-inflammatory role of glucan fractions. PLoS One. 2012;7:e40648. [PubMed] [DOI] [Full Text] |
| 55. | McFarland LV. Systematic review and meta-analysis of Saccharomyces boulardii in adult patients. World J Gastroenterol. 2010;16:2202-2222. [PubMed] [DOI] [Full Text] |