Published online Mar 28, 2023. doi: 10.3748/wjg.v29.i12.1875
Peer-review started: December 18, 2022
First decision: January 22, 2023
Revised: February 2, 2023
Accepted: March 15, 2023
Article in press: March 15, 2023
Published online: March 28, 2023
Processing time: 100 Days and 8.8 Hours
Centipedes have been used to treat tumors for hundreds of years in China. However, current studies focus on antimicrobial and anticoagulation agents rather than tumors. The molecular identities of antihepatoma bioactive components in centipedes have not yet been extensively investigated. It is a challenge to isolate and characterize the effective components of centipedes due to limited peptide purification technologies for animal-derived medicines.
To purify, characterize, and synthesize the bioactive components with the strongest antihepatoma activity from centipedes and determine the antihepatoma mechanism.
An antihepatoma peptide (scolopentide) was isolated and identified from the centipede scolopendra subspinipes mutilans using a combination of enzymatic hydrolysis, a Sephadex G-25 column, and two steps of high-performance liquid chromatography (HPLC). Additionally, the CCK8 assay was used to select the extracted fraction with the strongest antihepatoma activity. The molecular weight of the extracted scolopentide was characterized by quadrupole time of flight mass spectrometry (QTOF MS), and the sequence was matched by using the Mascot search engine. Based on the sequence and molecular weight, scolopentide was synthesized using solid-phase peptide synthesis methods. The synthetic scolopentide was confirmed by MS and HPLC. The antineoplastic effect of extracted scolopentide was confirmed by CCK8 assay and morphological changes again in vitro. The antihepatoma effect of synthetic scolopentide was assessed by the CCK8 assay and Hoechst staining in vitro and tumor volume and tumor weight in vivo. In the tumor xenograft experiments, qualified model mice (male 5-week-old BALB/c nude mice) were randomly divided into 2 groups (n = 6): The scolopentide group (0.15 mL/d, via intraperitoneal injection of synthetic scolopentide, 500 mg/kg/d) and the vehicle group (0.15 mL/d, via intraperitoneal injection of normal saline). The mice were euthanized by cervical dislocation after 14 d of continuous treatment. Mechanistically, flow cytometry was conducted to evaluate the apoptosis rate of HepG2 cells after treatment with extracted scolopentide in vitro. A Hoechst staining assay was also used to observe apoptosis in HepG2 cells after treatment with synthetic scolopentide in vitro. CCK8 assays and morphological changes were used to compare the cytotoxicity of synthetic scolopentide to liver cancer cells and normal liver cells in vitro. Molecular docking was performed to clarify whether scolopentide tightly bound to death receptor 4 (DR4) and DR5. qRT-PCR was used to measure the mRNA expression of DR4, DR5, fas-associated death domain protein (FADD), Caspase-8, Caspase-3, cytochrome c (Cyto-C), B-cell lymphoma-2 (Bcl-2), Bcl-2-associated X protein (Bax), x-chromosome linked inhibitor-of-apoptosis protein and Cellular fas-associated death domain-like interleukin-1β converting enzyme inhibitory protein in hepatocarcinoma subcutaneous xenograft tumors from mice. Western blot assays were used to measure the protein expression of DR4, DR5, FADD, Caspase-8, Caspase-3, and Cyto-C in the tumor tissues. The reactive oxygen species (ROS) of tumor tissues were tested.
In the process of purification, characterization and synthesis of scolopentide, the optimal enzymatic hydrolysis conditions (extract ratio: 5.86%, IC50: 0.310 mg/mL) were as follows: Trypsin at 0.1 g (300 U/g, centipede-trypsin ratio of 20:1), enzymolysis temperature of 46 °C, and enzymolysis time of 4 h, which was superior to freeze-thawing with liquid nitrogen (IC50: 3.07 mg/mL). A peptide with the strongest antihepatoma activity (scolopentide) was further purified through a Sephadex G-25 column (obtained A2) and two steps of HPLC (obtained B5 and C3). The molecular weight of the extracted scolopentide was 1018.997 Da, and the peptide sequence was RAQNHYCK, as characterized by QTOF MS and Mascot. Scolopentide was synthesized in vitro with a qualified molecular weight (1018.8 Da) and purity (98.014%), which was characterized by MS and HPLC. Extracted scolopentide still had an antineoplastic effect in vitro, which inhibited the proliferation of Eca-109 (IC50: 76.27 μg/mL), HepG2 (IC50: 22.06 μg/mL), and A549 (IC50: 35.13 μg/mL) cells, especially HepG2 cells. Synthetic scolopentide inhibited the proliferation of HepG2 cells (treated 6, 12, and 24 h) in a concentration-dependent manner in vitro, and the inhibitory effects were the strongest at 12 h (IC50: 208.11 μg/mL). Synthetic scolopentide also inhibited the tumor volume (Vehicle vs Scolopentide, P = 0.0003) and weight (Vehicle vs Scolopentide, P = 0.0022) in the tumor xenograft experiment. Mechanistically, flow cytometry suggested that the apoptosis ratios of HepG2 cells after treatment with extracted scolopentide were 5.01% (0 μg/mL), 12.13% (10 μg/mL), 16.52% (20 μg/mL), and 23.20% (40 μg/mL). Hoechst staining revealed apoptosis in HepG2 cells after treatment with synthetic scolopentide in vitro. The CCK8 assay and morphological changes indicated that synthetic scolopentide was cytotoxic and was significantly stronger in HepG2 cells than in L02 cells. Molecular docking suggested that scolopentide tightly bound to DR4 and DR5, and the binding free energies were-10.4 kcal/mol and-7.1 kcal/mol, respectively. In subcutaneous xenograft tumors from mice, quantitative real-time polymerase chain reaction and western blotting suggested that scolopentide activated DR4 and DR5 and induced apoptosis in SMMC-7721 Liver cancer cells by promoting the expression of FADD, caspase-8 and caspase-3 through a mitochondria-independent pathway.
Scolopentide, an antihepatoma peptide purified from centipedes, may inspire new antihepatoma agents. Scolopentide activates DR4 and DR5 and induces apoptosis in liver cancer cells through a mitochondria-independent pathway.
Core Tip: Centipedes have been used to treat tumors for hundreds of years in China. However, the molecular identities of their bioactive components have not yet been extensively investigated. We have modified the centipede extraction method and found a polypeptide (scolopentide) with the strongest antihepatoma activity. Mechanistically, scolopentide activated death receptor 4 and death receptor 5 to induce apoptosis through a mitochondria-independent pathway. Scolopentide may be an inspiration for the development of antihepatoma agents. We also aim to provide a reference for the purification and characterization of effective components in animal-derived medicines.
- Citation: Hu YX, Liu Z, Zhang Z, Deng Z, Huang Z, Feng T, Zhou QH, Mei S, Yi C, Zhou Q, Zeng PH, Pei G, Tian S, Tian XF. Antihepatoma peptide, scolopentide, derived from the centipede scolopendra subspinipes mutilans. World J Gastroenterol 2023; 29(12): 1875-1898
- URL: https://www.wjgnet.com/1007-9327/full/v29/i12/1875.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i12.1875
Centipedes have been widely used in traditional Chinese medicine to treat many diseases, including convulsions, strokes, rheumatic joint diseases, snake bites and tumors[1]. It is a major challenge to isolate and characterize the effective components of centipedes due to limited peptide purification technologies for animal-derived medicines. Currently, the main extraction technologies are as follows: Aqueous extraction[2], organic solvent extraction[2-5], enzymatic hydrolysis[6], ultrasonic extraction[2], and freeze-thaw extraction[2]. To obtain low-weight peptides, extracted proteins require further purification. At present, the main purification technologies of animal proteins are as follows: Sephadex column gel chromatography (GC)[3,7,8], HPLC/RP-HPLC[3,5-8], ion exchange chromatography[7,9], and graded ultrafiltration[7]. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) is a traditional separation and identification technique[10], and mass spectrometry (MS) is the most widely used technique for identifying peptides for use in animal-derived medicines[5-7]. To date, there is no single method that can extract a single peptide from complex mixtures (Table 1). Compound extraction plans are mostly used, which are costly and complicated. Consequently, animal-derived medicines have not been developed as herbal medicines. Currently, the main extracts are still crude peptides, and they have some disadvantages, such as a low extraction ratio, undefined composition, unstable properties and susceptibility[11,12]. These disadvantages may be prevented by identifying and characterizing the effective compositions of animal-derived medicines and synthesizing them in vitro.
| Name | Extract methods | Function | Ref. |
| Scolopendrin I | Cation-exchange chromatography and two steps RP-HPLC | Antimicrobial (G+, G-, and fungi) | [53] |
| Scolopin 1 | (1) Sephadex Gel Chromatography; (2) RP-HPLC; and (3) characterized by MALDI-TOF MS | Antimicrobial (G+, G-, fungi, drug-resistant bacteria) | [37] |
| Scolopin 2, Scolopin 2-NH2 | (1) Sephadex Gel Chromatography; (2) RP-HPLC; and (3) Scolopin2-NH2 was amidation modification from scolopin 2 | (1) Antimicrobial (G+, G-, fungi, and drug-resistant bacteria); and (2) inhibit proliferation of cervical cancer HeLa cells | [16,37,45] |
| LBLP | Transcriptomic data analysis, peptide synthesis | Anti-fungi | [54] |
| Scolopendrasin II, V, III, X | Next-generation sequencing | (1) Antimicrobial (G+, G-, drug-resistant bacteria, fungi); and (2) regulating immune system to antimicrobial indirectly | [40,55-57] |
| Scolopendrasin IX | Next-generation sequencing | (1) Regulating immune system to antimicrobial indirectly; and (2) treating autoimmune arthritis | [58] |
| Scolopendrasin VII | Next-generation sequencing | (1) Antimicrobial (G+, G-, drug-resistant bacteria, and fungi); (2) regulating immune system to antimicrobial indirectly; and (3) inhibiting leukemia U937 and jurkat cells | [17,59,60] |
| Scolopendin, 1, 2 | RNA sequencing | Antimicrobial (G+, G-, drug-resistant bacteria, and fungi) | [41-44] |
| SerGln-Leu (SQL) | (1) Ultrafiltration, Sephadex G-50 column, Source 15Q anion exchange Column; (2) RP-HPLC C18 column; and (3) characterized by ESI-MS | Anticoagulation and antithrombotic | [7] |
| Thr-Asn-Gly-Tyr-Thr, (TNGYT) | (1) Sephadex G-50 column; (2) RP-HPLC C18 column; and (3) Characterized by ESI-MS | Anticoagulation and antithrombotic | [38] |
| SsTx-R12A | Red-headed centipede’s venom cDNA arsenal | Treating autoimmune diseases mediated by T cells | [61] |
| SsmTX-I | Red-headed centipede’s venom cDNA arsenal | Analgesic | [62] |
| SsTx | Sephadex G-50 Gel Chromatography and RP-HPLC | Blocking KCNQ potassium channels leads to cardiovascular, respiratory, muscular and nervous system damage | [8] |
| Ssm6a | Sephadex Gel Chromatography and RP-HPLC | Analgesic | [39] |
| Haemolymph and tissue extracts | (1) Enzymatic hydrolysis; (2) HPLC; and (3) characterized by MS | Antimicrobial (G+, G-, fungi, viruses, and parasites) | [6] |
| Lacrain | RP-HPLC and characterized by MS | Antibacterial (G-) | [63] |
| Pinipesin | (1) Organic solvent extraction; (2) HPLC Jupiter® C18 column; and (3) characterized by MS | Antimicrobial | [5] |
| AECS | Organic solvent extraction (ethanol) | Inducing apoptosis of human epidermal carcinoma A431 cells and human melanoma HEK293/EGFR, A375 cells | [4,18] |
| Two isoquinoline alkaloids | (1) Organic solvent extraction (ethanol); (2) sephadex G-25 Gel chromatography; and (3) HPLC | Inducing apoptosis of brain glioma U87 cells | [3] |
| Centipede water extract, alcohol extract | (1) Water extraction; (2) freeze-thawing with liquid nitrogen, ultrasonic extraction; and (3) organic solvent extraction (ethanol) | No tumour inhibition in S180 and H22 Transplanted tumour mice | [2] |
| CE, CA | Organic solvent extraction | Inhibiting cervical tumours in vivo | [64] |
| Centipede chloroform extract | Organic solvent extraction (hexane, chloroform, ethanol) | Reducing central nervous system symptoms of Alzheimer’s disease | [65] |
| Scom 5 | (1) Superdex 75 gel filtration; (2) RESOURCE S ion chromatography; and (3) characterized by LC-MSMS, MALDI-TOF/TOF and protein sequencing techniques | Human allergens | [11] |
Some ancient Chinese medical studies have reported that centipedes can be used to treat tumors. “Compendium of Materia Medica”[13], a famous ancient Chinese pharmaceutical work, recorded that centipedes can be used to treat tumors in the chest and abdomen. It is also recorded in the modern Chinese medical literature “Integrating Chinese and Western Medicine”[14] that centipede liquor can be used to treat esophageal cancer with excellent clinical effects. Moreover, the Shendan Sanjie capsule, a medicine containing centipedes, has already been applied to treat cancers and has been proven to inhibit colitis-associated cancer[15]. Although the antitumor theory and clinical efficacy of centipedes have been verified for hundreds of years in China, current experimental studies still focus on antimicrobial and anticoagulation agents. Only a few studies have observed the antitumor effects of centipedes (Table 1). The potential mechanism is also poorly understood. The tumor types focused on have been gliomas[3], cervical cancer[16], leukemia[17], epidermal cancer[4], and melanoma[18], rather than tumors in the chest and abdomen. Purified centipede peptides can suppress tumor cells by inducing caspase-related apoptosis through a mitochondria-dependent pathway[3,4,16,18], inducing cell cycle arrest[3,18] and necrosis through a specific interaction with phosphatidylserine (Table 1)[17]. Additionally, our previous studies showed that crude centipede peptides could induce apoptosis in HepG2 cells and lung cancer A549 cells by arresting cells at the G2/M phase[19,20].
Apoptosis has been widely used as a target in cancer treatment[21]. There are two main apoptosis signaling pathways: Mitochondria-dependent and mitochondria-independent pathways. The mitochondria-dependent pathway is controlled by Bcl-2 family proteins and is activated by DNA damage caused by chemotherapy and radiotherapy through activation of the tumor suppressor gene p53[22]. However, the loss of p53 is common in the clinic, contributing to the resistance of conventional chemotherapy and the development of most cancers[23,24]. Thus, the mitochondria-independent pathway, which is independent of p53, has become a promising target[25]. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is part of the mitochondria-independent pathway. TRAIL can selectively initiate apoptosis of cancer cells but has no obvious toxic effect on normal cells[26-28], which makes it a promising chemotherapy drug for many cancers. Nonetheless, recent studies have shown that some cancers, including liver cancer and esophageal cancer, are resistant to TRAIL-mediated apoptosis, which greatly reduces its clinical efficacy[29,30]. Current research focuses on finding suitable TRAIL sensitizers and developing TRAIL receptor agonists to maximize the cell-killing effect. However, current studies mainly focus on sensitizers rather than receptor agonists, and the clinical and laboratory results are not ideal[26,28]. Therefore, for research and clinical applications, it would be useful to find stable and efficient TRAIL receptor agonists. Recently, the regulatory effect of Chinese herbal extracts on the TRAIL apoptosis pathway was gradually discovered. Chinese herbal extracts such as luteolin and artemisinin derivatives can induce apoptosis of liver cancer cells by upregulating the expression of death receptor 4 (DR4) or DR5, which are TRAIL receptors on the cell surface[31,32].
In this study, we isolated a novel low-molecular-weight antihepatoma peptide, designated scolopentide, from Scolopendra subspinipes mutilans L. Koch and synthesized it in vitro. We observed that synthetic scolopentide inhibited tumor proliferation and was more toxic to HepG2 human liver cancer cells than to L02 human embryonic hepatocytes. Mechanistically, scolopentide activated TRAIL receptors (DR4 and DR5), upregulated the expression of fas-associated death domain protein (FADD), caspase-8 and caspase-3 through a mitochondria-independent pathway and finally induced tumor cell apoptosis.
Dulbecco’s modified Eagle medium (DMEM) was purchased from Gibco (Thermo Fisher Scientific Inc., United States) and HyClone (Logan, United States). Fetal bovine serum (FBS) was purchased from Gibco and Invitrogen (Thermo Fisher Scientific Inc., United States). DMSO was purchased from Amresco (Solon, United States). PBS was purchased from Boster (Wuhan, Hubei Province, China) and WellBio (Shanghai, China). Trypsin was purchased from Invitrogen. Penicillin-streptomycin was purchased from Beyotime (Shanghai, China). Hoechst was purchased from SolarBio (Beijing, China).
The Bel-7402 Liver cancer cell line, Eca-109 esophageal cancer cell line and A549 Lung cancer cell line were purchased from the Cell Resource Center of Shanghai University of Biology. The HepG2 Liver cancer cell line and L02 human embryonic hepatocyte cell line were purchased from The Cell Center of Xiangya Medical College, Central South University. The SMMC-7721 liver cancer cell line was purchased from the BeNa Culture Collection (Suzhou, Jiangsu Province, China). Cells were placed in culture flasks (Corning Inc., United States) and cultured in a constant temperature incubator (SHEL-LAB, Hitachi, United States) at 37 °C and 5% CO2 with saturated humidity. The medium comprised 10% FBS, low-glucose DMEM and penicillin-streptomycin (100 U/mL). Cells were observed under an inverted microscope (Olympus, Japan).
The viability of HepG2, A549, Eca-109 and Bel7402 cells treated with different centipede extracts was determined by a Cell Counting Kit-8 (Dojindo, Japan). The viability of HepG2 and L02 cells treated with synthetic scolopentide was also determined. Groups and treatments were as follows: (1) Vehicle group: 100 μL cell suspension + 100 μL complete medium; (2) zero group: 200 μL of complete medium; and (3) scolopentide group: Cells were treated with different centipede extracts/synthetic scolopentide. Three replicate wells were used for each group.
Cells at the logarithmic growth stage were collected, and the density was adjusted to 1 × 104 cells/mL. The cell suspension (100 μL) was plated into a 96-well plate (Corning Inc., United States). Four hours later, drugs were added after the cells had adhered to the well. Twelve hours later, 100 μL culture solution containing 10% CCK8 was added to each well. Cells were incubated for 2 h at 37 °C with 5% CO2. Then, the absorbance (OD) at 450 nm was analyzed by a microplate reader, and the mean was calculated. Cell viability (%) = scolopentide group OD/vehicle group OD × 100%. The cell suppression ratio (IR) was determined with the following formula: IR (%) = (vehicle group OD-scolopentide group OD)/vehicle group OD × 100%. The zero group OD was subtracted from the OD of all groups for correction. The 50% maximum inhibitory concentration (IC50) was determined by constructing dose-response curves.
Crude centipede peptides were obtained by enzymolysis combined with acetone precipitation. The “Pharmacopoeia of the People’s Republic of China (2020)” references the use of the whole dried body of Scolopendra subspinipes mutilans[1]. Centipedes were purchased from the pharmacy department of the First Affiliated Hospital of Hunan University of Chinese Medicine. They were identified as Scolopendra subspinipes mutilans L. Koch by Chinese herbal experts. Then, the whole bodies of the centipedes were ground into ultrafine powder by low-temperature ultrafine grinding in the Superfine Powder Engineering Research Center of the Ministry of Education of China. First, 100 g of centipede was dried at 50-60 °C. The superfine powder (particle size 1-75 μm) was crushed by a BFM-6 Bailey superfine powder machine (amplitude 100 Hz, grinding temperature 0-10 °C, grinding time 10-15 min). Microscopic image analysis and laser light scattering were used in combination to assess the particle size, and the particle size of more than 95% of the powder was smaller than 75 μm. Finally, the powder was stored at room temperature until subsequent experiments.
We chose an optimized enzymatic hydrolysis method. The extraction process was as follows: 2 g of centipede superfine powder was added to 10 mL of double distilled water, mixed with trypsin, and incubated in water at 55 °C for 4 h. The trypsin was inactivated in a 99 °C water bath for 10 min. Then, the samples were centrifuged at 3500 r/min for 5 min (low temperature ultracentrifuge, Pharmacia Inc., Sweden), and the supernatant was collected. The samples were centrifuged at 5000 r/min at 4 °C for 10 min. The supernatant was collected again. Two volumes of precooled acetone were added to precipitate the former supernatant, which was centrifuged at 5000 r/min at 4 °C for 10 min. The precipitate was dissolved in double steaming water, and the process was repeated twice. Finally, 2 volumes of precooled acetone and petroleum ether (1:1) were added to precipitate again, which was centrifuged at 5000 r/min at 4 °C for 10 min, and the precipitate was collected. Finally, the precipitate was freeze-dried (FD-1 Freeze Dryer, Rikakika Co., Ltd., Tokyo, Japan), weighed, and stored at -20 °C, and the protein content was determined for subsequent experiments.
In the process of preparing crude centipede peptides, we tested the influence of single factors on the extraction rate of the crude extract. The five factors included protease type (trypsin, pepsin), water-centipede ratio, protease dose, enzymolysis temperature and enzymolysis time. Then, an L9(34) orthogonal design was adopted to investigate the effects of enzymolysis time, enzymolysis temperature and trypsin dose on crude extracts, which were the three most influential factors in single-factor experiments (Table 2). In addition to the extraction ratio, a CCK8 assay was used to detect the suppression effect of different crude extracts on HepG2 cell viability to determine the optimal technological conditions. We also compared the cytotoxicity of the crude extracts obtained from two methods, freeze-thawing with liquid nitrogen and enzymolysis with acetone precipitation, under optimal technological conditions to determine the significance of this improvement.
| Level | Trypsin (g) | Time (h) | Temperature (°C) |
| 1 | 0.1 | 3 | 37 |
| 2 | 0.2 | 4 | 46 |
| 3 | 0.3 | 5 | 55 |
Gel chromatography: For the first step, a Sephadex G-25 column was used to isolate the component with the strongest antitumor effect. With 0.1% trifluoroacetic acid-acetonitrile as the mobile phase, gradient elution was performed using an AKTA protein purification system (Amersham Bioscience, United States). In detail, a crude centipede peptide solution (50 mg/mL) was slowly added to a Sephadex G-25 column. Chromatographic columns were eluted with 0.1% trifluoroacetic acid-acetonitrile (flow rate 0.5 mL/min). The resulting columns (3 mL/tube) were collected by an automatic collector. After isolation, tubes with the same peak are collected together, according to the peaks displayed on the computer ultraviolet spectrum. The protein contents of different peaks were determined by a fluorescence microplate reader (TECAN, Austria). The CCK8 assay was used to assess the antitumor activity of different peaks and to select the strongest peak for subsequent experiments.
High-performance liquid chromatography: For further purification, high-performance liquid chromatography (HPLC) was used as the next step. Initially, we used a conventional C18 column, but the results showed that the sample was hydrophilic. Thus, a hydrophilic XAmide column (5 μm, 100 A, 4.6 mm × 250.0 mm) was chosen. The sample solution (1 mg/mL) was filtered through a 0.45 μm microporous membrane and eluted with acetonitrile-water-100 mmol/L ammonium formate (pH 3.5) (flow rate 4 mL/min, detection wavelength 260 mm, injection volume 20 μL). Different fractions were collected according to the HPLC chromatogram peaks. The CCK8 assay was used to select the fraction with the strongest antitumor activity. Then, the above HPLC and CCK8 assays were repeated. The final centipede peptide was concentrated, purified, freeze dried and designated scolopentide.
Quadrupole time of flight mass spectrometry (UPLC-QTOF MS 6530, SHIMADZU, Japan) was used to measure molecular weight. The ion source parameters were as follows: Electrospray ion source, positive ion mode, temperature 300 °C, gas flow 11 L/min, and spray pressure 15 psi. Scolopentide did not pass through the liquid column but went directly through the mobile phase to the QTOF system. The Mascot search engine from Matrix Science was used to match peptide sequences. Based on the protein sequence library of Mascot, the molecular weight of scolopentide was compared with that of known centipede proteins. The amino acid sequence that best matched the molecular weight was selected.
Scolopentides that were characterized by QTOF MS and Mascot were synthesized using solid-phase peptide synthesis methods. Peptide synthesis was performed by Genscript (Nanjing, China) as follows: (1) A total of 400 mg chloromethyl resin was selected to synthesize the polypeptides, 20% piperidine/N, n-dimethylformamide was added to the reactor, and the reactor was shaken for 20 min; (2) the solvent was removed by filtration, N, n-dimethylformamide was added to the system, and the reactor was shaken for 1 min, followed by filtering to remove the liquid, which was carried out three times; (3) a total of 150 µl ninhydrin and a small amount of resin were added to the detection tube, which was placed at 100 °C for 20 s, after which the resin’s color was checked for change, which indicated that Fmoc was removed successfully; (4) the prepared amino acid solution was added to the reactor, 1 mL of N,N’-diisopropyl carbon diimine/1-hydroxybenzotriazole was added, and the reactor was shaken for 1 h; (5) step 3 was repeated, and if the color did not change, the coupling was considered successful; (6) step 2 was repeated to wash the resin; and (7) the above operation was repeated, and the corresponding amino acids were added until polypeptide synthesis was completed. Then, HPLC was used to test the purity of the synthetic scolopentide, and MS was used to test the molecular weight. Finally, the peptides were freeze-dried and stored at -20 °C until they were used.
The apoptosis of HepG2 cells treated with extracted scolopentide was determined by Annexin V-Alexa Fluor660/7-AAD (BB20121, BestBio, China). The cells were digested and collected by trypsin. Approximately 5 × 105 cells were collected after washing with PBS twice and centrifuged at 2000 rpm for 5 min. Then, 100 μL of Annexin V binding buffer was added to resuspend the cells, and the concentration was adjusted to 3 × 106 cells/mL. Five microliters of annexin V-FITC and 5 μL of propidium iodide were added to the cell suspension, mixed and incubated for 10 min at 4 °C in the dark. Then, 400 μL of PBS was added to the cell suspension and mixed. Within 1 h, flow CytoFlex (A00-1-1102, Beckman, United States) was utilized to detect cell apoptosis.
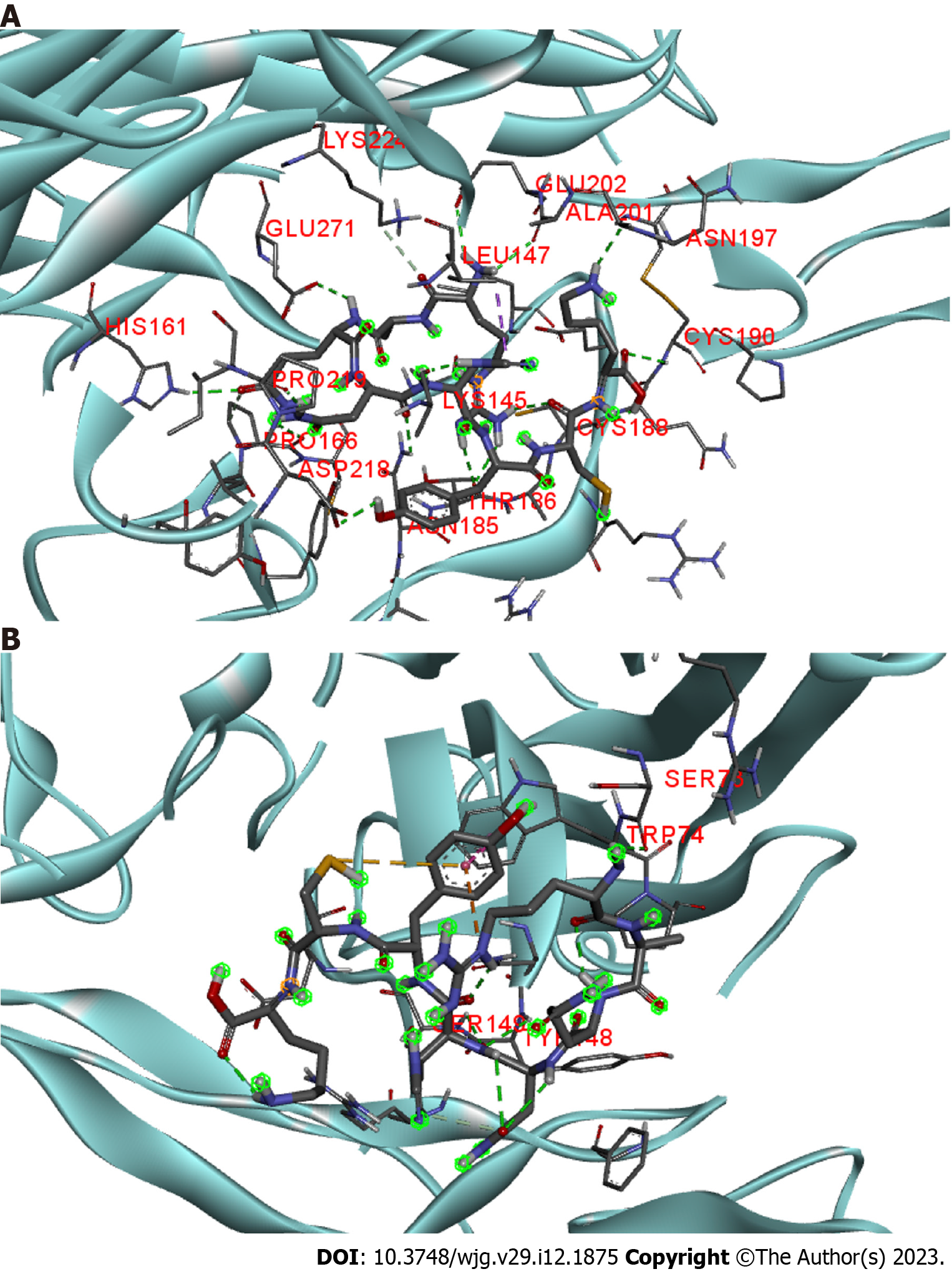
To investigate the antihepatoma mechanism of scolopentide, Discovery Studio 2020 Client and AutoDock-Vina 1.1.2 were used to predict the binding affinity between scolopentide and DR4 and DR5. BIOVIA Discovery Studio 2020 was applied to process and visualize the results. To obtain molecular structure files, scolopentide structures were constructed with Discovery Studio (Small Molecules) according to the sequence RAQNHYCK. Molecular structure files of DR4 (PDB code 5CIR) and DR5 (PDB code 419X) were retrieved from the RCSB PDB database (https://www.rcsb.org/). The DR4 and DR5 structures were modified by Discovery Studio for water deletion, binding pocket prediction, hydrogenation, and conformation optimization. Then, AutoDock-Vina was used to perform molecular docking and calculate the affinity potential energy of the protein molecule potential binding modes between scolopentide and DR4 and DR5.
HepG2 cells were inoculated into 6-well plates (1 × 106 cells per well), and 100 μg/mL synthetic scolopentide was added. The cell slides were removed and washed with PBS 2 or 3 times after incubation at 37 °C and 5% CO2 with saturated humidity for different durations (6 h, 12 h, and 24 h). Then, the slides were fixed with 4% paraformaldehyde for 30 min and rinsed with PBS 3 times. Hoechst (working concentration: 25 μM) was added to the slides, which were incubated for 3 min and rinsed with PBS 3 times. Finally, the slides were sealed with glycerin buffer, stored away from light and observed under a fluorescence microscope (Zeiss, Germany).
Male 5-wk-old BALB/c nude mice (Shanghai SLAC Laboratory Animal Co., Ltd., Shanghai, China) were chosen for the formation of subcutaneous xenograft tumors. SMMC-7721 cells (0.2 mL/mouse, 1 × 107 cells/mL) were subcutaneously injected into the right back. The tumor diameter reached 1 cm at 3 wk. Qualified model mice were randomly divided into 2 groups (n = 6 per group): The scolopentide group (intraperitoneal injection of synthetic scolopentide, 500 mg/kg/d) and the vehicle group (same volume of normal saline). The length and width of tumors were measured every 2 d, and volume was calculated as the maximum tumor length × width2 × 0.5. After 14 d of treatment, all mice were euthanized by cervical dislocation, and tumor weights were measured. All operations followed the United Kingdom Animals (Scientific Procedures) Act, 1986. All laboratory animals were carefully handled, and the study was reviewed and approved by the Ethical Review Committee of Experimental Animal Welfare at Hunan University of Chinese Medicine (Approval No. LL2021040705).
Total RNA was extracted from tumor tissues based on the instructions of the RNA Simple Total RNA Kit (Novoprotein Scientific Inc., Suzhou, Jiangsu Province, China), and the RNA concentration was measured by a microplate reader. Then, NovoScript@Plus (Novoprotein Scientific Inc., Suzhou, China) was utilized to reverse-transcribe the RNA into cDNA, which was stored at -20 °C until subsequent use. According to the instructions of NovoStart@SYBR (Novoprotein Scientific Inc., Suzhou, Jiangsu Province, China), 10 µL SuperMix Plus, 7 μL RNase Freewater, 2 μL cDNA, 0.5 μL forward primers and 0.5 reverse primers (Genecreate Inc., Wuhan, Hubei Province, China) were mixed. GAPDH was used as the internal reference. A LightCycler®96 (Roche, Switzerland) was used for quantitative real-time polymerase chain reaction (qRT-PCR). The primer sequences are shown in Table 3. The qRT-PCR conditions were as follows: Predenaturation for 60 s at 95 °C; 45 cycles of denaturation for 20 s at 95 °C, 20 s at 60 °C, and 30 s at 72 °C; 1 cycle of melting for 10 s at 95 °C, 60 s at 65 °C, and 1 s at 97 °C; and cooling for 30 s at 37 °C. Data were individually normalized to the mean of the relative expression of GAPDH. The fold change was calculated using the 2-ΔΔCT method. The experiment was repeated at least three times, and the results that were the most representative are shown.
| Gene | Sequence (5’ to 3’) | |
| GAPDH | F: AGAAGGCTGGGGCTCATTTG | R: AGGGGCCATCCACAGTCTTC |
| DR4 | F: CCAGGCAGCATTGAAGTCAG | R: GTTTCCAGCATCACCAGGGT |
| DR5 | F: CCCTGGAGTGACATCGAATG | R: CAGCCACAATCAAGACTACGG |
| FADD | F: TGCGGGAGTCACTGAGAATC | R: GGAGGTAGATGCGTCTGAGTTC |
| Caspase-8 | F: GAGAAGCAGCAGCCTTGAAG | R: GACAGTATCCCCGAGGTTTG |
| Caspase-3 | F: CCCTCCTCAGCATCTTATCC | R: TGGTACAGTCAGAGCCAACCT |
| Cyto-C | F: CACAAGACTGGGCCAAATCTC | R: CCAGGGATGTACTTCTTGGGA |
| XIAP | F: CTATGCTCACCTAACCCCAAG | R: TTCTGACCAGGCACGATCAC |
| c-FLIP | F: GGAACCCTCACCTTGTTTCG | R: CTCCTTGCTTATCTTGCCTCG |
| Bcl-2 | F: CCTTCTTTGAGTTCGGTGGG | R: CGGTTCAGGTACTCAGTCATCC |
| Bax | F: CACCAGCTCTGAGCAGATCA | R: TGTTACTGTCCAGTTCGTCCC |
Tumor tissue specimens were prepared and suspended in ice-cold RIPA lysis buffer (CWBIO Co., Ltd, Beijing, China) and incubated for 20 min. The supernatant was removed after centrifugation. The protein levels of tumor tissues were determined by the BCA assay using a BCA Protein Assay Kit (CWBIO Co., Ltd, Beijing, China). Proteins (50 μg) were mixed with pure water and SDS loading buffer and denatured at 100 °C for 10 min. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to PVDF membranes (Merck Millipore Ltd., Germany). Then, PVDF membranes were blocked for 1 h in blocking buffer (5% milk in TBST). The membranes were removed and washed with PBST (PBS + 0.1% Tween 20) 3 times, incubated with primary antibodies overnight at 4 °C, washed with TBST 3 times, incubated with Proteintech HRP Goat Anti-Rabbit IgG (H + L) (SA00001-2, diluted 1:6000) for 90 min at room temperature and washed with TBST 3 times. Protein signals were visualized by enhanced chemiluminescence using an eECL western blot kit (CWBIO Co., Ltd, Beijing, China). Membrane images were collected by an Amersham Imager 600 (GE Healthcare, United States) and qualified by Quantity One software. All experiments were repeated three times. Primary antibodies against DR5 (ab199357, diluted 1:1000), FADD (ab108601, diluted 1:1000), cleaved caspase-3 (ab32042, diluted 1:1000), and cytochrome c (Cyto-C) (ab133504, diluted 1:5000) were obtained from Abcam (Cambridge, England). Antibodies against GAPDH (10494-1-AP, diluted 1:5000) and DR4 (24063-1-AP, diluted 1:2000) were obtained from Proteintech Group, Inc. (Chicago, United States). The antibody against cleaved caspase-8 (AF5267, diluted 1:1000) was obtained from Affinity Biosciences, Inc. (Jiangsu, China).
Reactive oxygen species (ROS) were measured using a ROS Assay Kit (E004-1-1 Jiancheng, Nanjing, China) according to the manufacturer’s instructions. Briefly, the tumor tissue was prepared into a single cell suspension by enzymatic hydrolysis. Three groups of tubes were set up as follows: (1) Negative vehicle tube: cells without any treatment were resuspended in 0.01 M PBS; (2) positive vehicle tube: cells were resuspended in diluted 2,7-dichlorofluorescin diacetate (DCFH-DA), and an ROS hydrogen donor (concentration: 30 μM) was added to induce the cells; and (3) sample tube: cells were resuspended in diluted DCFH-DA (cell density: 1 × 106-2 × 107/mL). Then, all cells were incubated for 50 min at 37 °C. After labeling with the probe, the single-cell suspensions were subsequently collected and centrifuged for 5-10 min. The precipitates were collected and washed with 0.01 M PBS 1-2 times. The precipitates were centrifuged and collected again. Finally, the cell precipitates were resuspended in PBS and measured on a fluorescence microplate reader (optimum excitation wavelength 500 nm, optimum emission wavelength 525 nm). All results are expressed as fluorescence values.
SPSS 25.0 and GraphPad Prism 8.3 software were used for all statistical analyses. Unless otherwise stated, P < 0.05 was considered statistically significant. All data are expressed as the mean ± SD. For the cell viability assay, ANOVA was used to compare the means of multiple groups, Student’s t test was used for pairwise comparisons, and Fisher’s exact test was used for ratio comparisons. For tumor weight, qRT-PCR and WB, Student’s t test was used when normality and ANOVA were satisfied. For ROS assessment, the Mann-Whitney rank-sum test was used due to unsatisfactory normality. Two-way ANOVA was used for tumor volume analysis.
Based on the extract ratio and protein content, we carried out five single-factor experiments and finally obtained the optimum conditions: the protease was trypsin, the water-centipede ratio was 5:1, the protease dose was 0.1 g (300 U/g, with a centipede-trypsin ratio of 20:1), the enzymolysis temperature was 46 °C, and the enzymolysis time was 4 h. The three most influential factors were enzymolysis time, enzymolysis temperature and trypsin dose. Therefore, an L9(34) orthogonal design was adopted to further investigate the optimal group of three factors on the extract ratio, as shown in Table 4. In conclusion, Group 4 (A2B1C2), Group 2 (A1B2C2), and Group 9 (A3B3C2) had higher extract ratios.
| Run | A (mg, trypsin) | B (min, time) | C (°C) (temperature) | D (blank) | Extract ratio (%) |
| 1 | 1 | 1 | 1 | 1 | 4.55 |
| 2 | 1 | 2 | 2 | 2 | 5.86 |
| 3 | 1 | 3 | 3 | 3 | 4.32 |
| 4 | 2 | 1 | 2 | 3 | 5.63 |
| 5 | 2 | 2 | 3 | 1 | 4.21 |
| 6 | 2 | 3 | 1 | 2 | 3.98 |
| 7 | 3 | 1 | 3 | 2 | 3.34 |
| 8 | 3 | 2 | 1 | 3 | 3.68 |
| 9 | 3 | 3 | 2 | 1 | 5.94 |
| K1 | 14.73 | 13.52 | 12.21 | 14.70 | - |
| K2 | 13.82 | 16.01 | 14.83 | 13.18 | - |
| K3 | 12.96 | 11.64 | 11.87 | 13.63 | - |
| R | 1.77 | 4.37 | 2.96 | 1.52 | - |
The L9(34) orthogonal design was adopted to further investigate the optimal group of the three factors for cytotoxicity in HepG2 cells, as shown in Table 5. A CCK8 assay was used to evaluate HepG2 cell viability. The results indicated that the 9 extracts induced cell death in a concentration-dependent manner (P < 0.05). Extracts from Groups 4, 2, and 9 showed stronger cytotoxicity, and Groups 1, 3, 5, 6, 7, and 8 showed moderate cytotoxicity. In the L9(34) orthogonal experiment, Group 2 (5.86%, 0.310 mg/mL), which used the optimal process of the single-factor experiment, showed little difference from the nonoptimal process of Group 9 (5.94% and 0.278 mg/mL). Indeed, Group 2 required less time and a lower trypsin dose. Based on the IC50, extract ratio and suppression ratio shown in Table 5, Group 2 was ultimately determined to have the optimal extraction process. The optimal extraction conditions were as follows: Trypsin at 0.1 g (300 U/g, centipede-trypsin ratio of 20:1), enzymolysis temperature of 46 °C, and enzymolysis time of 4 h.
| Concentration (mg/mL) | Suppression ratio (%) | ||||||||
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | Group 6 | Group 7 | Group 8 | Group 9 | |
| 0.039 | 12.510 | 6.880 | 12.070 | 5.394 | 3.389 | 3.213 | 0.570 | 1.641 | 7.094 |
| 0.078 | 22.270 | 11.240 | 13.060 | 16.430 | 12.890 | 4.709 | 2.280 | 4.540 | 9.883 |
| 0.156 | 45.770 | 36.430 | 45.680 | 52.290 | 18.610 | 16.620 | 26.110 | 11.540 | 33.170 |
| 0.3125 | 85.510 | 85.040 | 88.540 | 94.360 | 68.110 | 63.660 | 58.500 | 64.170 | 76.610 |
| 0.625 | 89.090 | 86.060 | 92.440 | 94.520 | 94.170 | 94.96 | 83.200 | 95.180 | 94.570 |
| 1.250 | 91.840 | 87.490 | 93.050 | 94.770 | 96.780 | 95.730 | 94.660 | 95.300 | 96.000 |
| 2.500 | 92.070 | 89.470 | 93.430 | 94.690 | 96.940 | 96.670 | 95.540 | 95.670 | 96.920 |
| 5.000 | 92.750 | 92.000 | 95.800 | 96.680 | 97.720 | 97.780 | 95.880 | 95.790 | 97.650 |
| IC50 (mg/mL) | 0.457 | 0.310 | 0.480 | 0.360 | 0.584 | 0.681 | 0.812 | 0.757 | 0.278 |
| Extract ratio (%) | 4.550 | 5.860 | 4.320 | 5.630 | 4.210 | 3.980 | 3.340 | 3.680 | 5.940 |
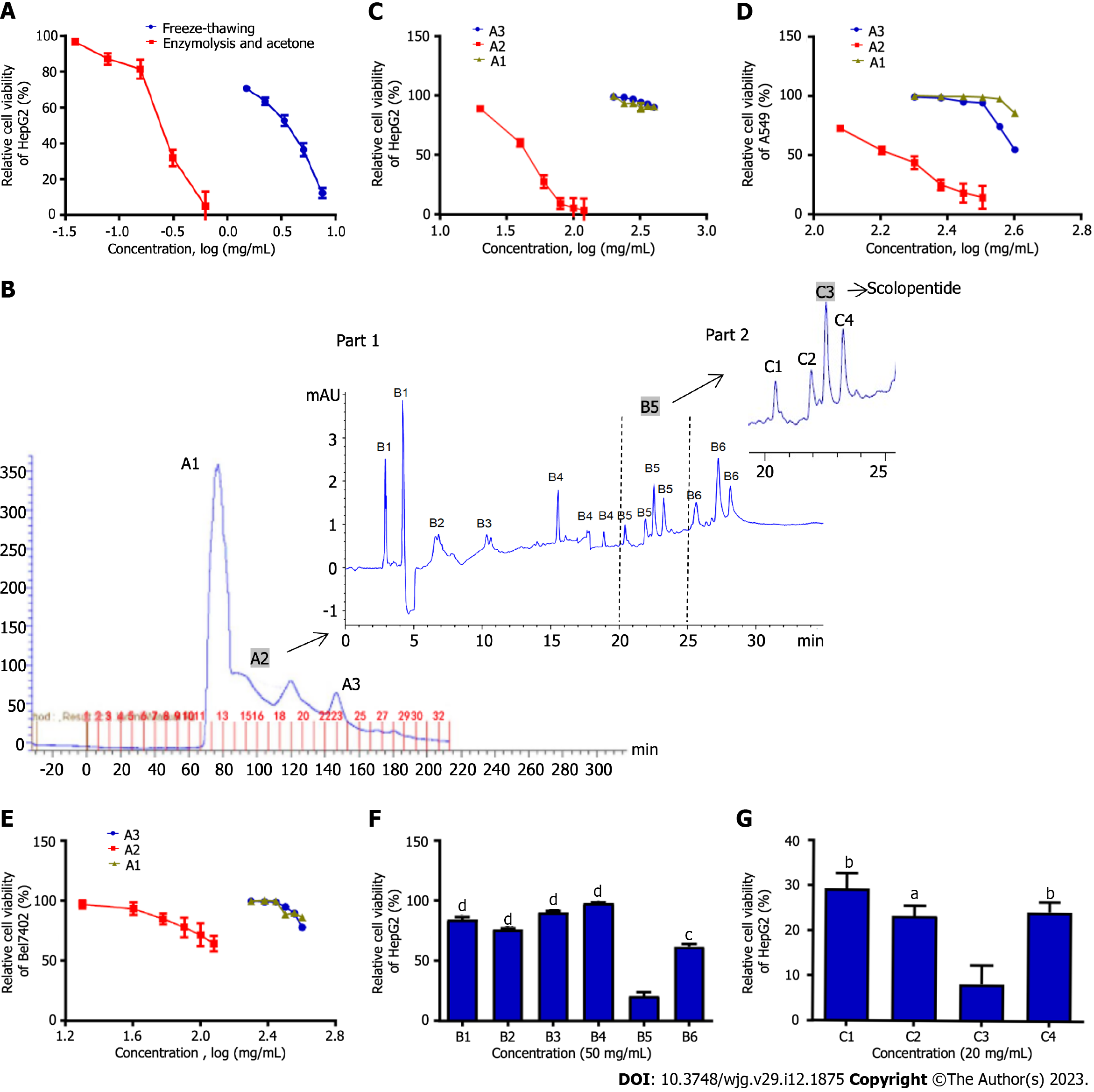
HepG2 cells were treated with crude peptides extracted using two extraction methods. The results showed that extracts obtained from the two methods significantly inhibited HepG2 cells in a concentration-dependent manner. Optimal enzymatic hydrolysis (IC50: 0.31 mg/mL) was superior to freeze-thawing with liquid nitrogen (IC50: 3.07 mg/mL), and the cytotoxicity of extracts increased nearly 10 times (Figure 1A).
We purified and screened crude centipede peptides three times and obtained a low-molecular-weight peptide with the strongest antihepatoma effect, which was designated scolopentide.
Sephadex G-25 gel chromatography was used, and the chromatogram obtained showed 3 peaks (Figure 1B). Part A samples (sample ID nos. A1 to A3) were then collected based on 3 peaks and assessed by a microplate reader. The protein and polypeptide contents of A1, A2 and A3 were 55, 40 and 20 μg/mL, indicating that samples A1-3 were mainly composed of protein. Since the separation range of the Sephadex G-25 column was 1-5 kDa, we speculated that A1 contained proteins > 5 kDa and that A2 and A3 were composed of low-molecular-weight polypeptides (1-5 kDa). Subsequently, a CCK8 assay was used to detect the suppressive effects of A1, A2 and A3 on HepG2, Bel-7402 and A549 cells (Figure 1C-E). The CCK8 assay showed the following: (1) A2 showed the most significant inhibition of proliferation of the three tumor cell lines among samples A1-3 (P < 0.05); (2) A2 showed the strongest inhibitory effect on HepG2 cells: The IC50 values of A2 against the three tumor cell lines were 50.1 mg/mL (HepG2), 132.8 mg/mL (Bel-7402) and 154.5 mg/mL (A549); and (3) A1-3 showed an inhibitory effect on the three tumor cell lines in a concentration-dependent manner.
In Part B, A2, which had the best antineoplastic effect in vitro among samples A1-3, was used. HepG2 cells, which were the most strongly inhibited among the three tumor cell lines, were used. First, tricine-SDS-PAGE[10] was performed to further purify this sample in Part B, but the peptide separation was not obvious, and the band patterns were light and diffuse. The reason may be that the molecular weight of the sample was too low for separation by tricine-SDS-PAGE. However, satisfactory results were obtained from HPLC. Therefore, HPLC was adopted for further purification. The chromatogram indicated that the sample from Part B was composed of a variety of peptides with very similar and low molecular weights in 6 peaks (Figure 1B; part 1). Subsequently, the Part B sample was collected in 6 parts (sample ID nos. B1 to B6) based on 6 peaks. A CCK8 assay was used to assess the suppressive effect of B1-6 on HepG2 cells cultured for 48 h, which were the tumor cells that were most sensitive to the centipede extracts. The CCK8 assay showed that B1-4 hardly inhibited the proliferation of HepG2 cells, while B5 and B6 inhibited proliferation (P < 0.05), with B5 having the strongest effect (Figure 1F).
In Part C, the B5 sample, which had the best antihepatoma effect in vitro among B1-6, was used. HPLC was adopted for purification, and the chromatogram showed 4 peaks (Figure 1B; part 2). Then, the sample from Part C was collected in 4 parts (C1-4) based on the 4 peaks. The CCK8 assay showed that C3 had the strongest inhibitory effect on HepG2 cells cultured for 48 h, while C1, C2, and C4 showed relatively weak inhibition of proliferation (Figure 1G). Finally, we concentrated and purified C3 and designated it scolopentide.
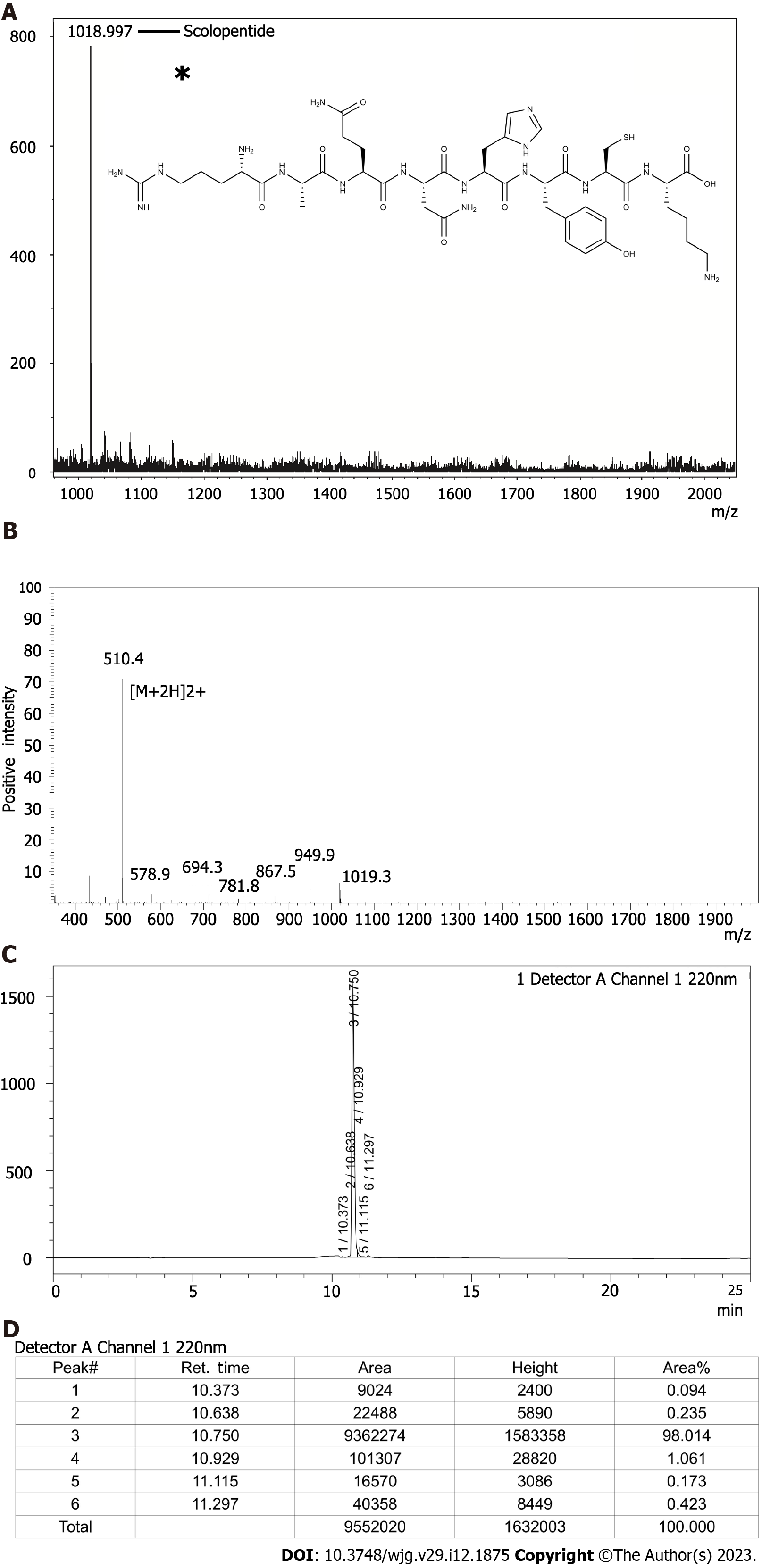
The peptide sequence of the extracted scolopentide is RAQNHYCK, and the mass spectrum (Figure 2A) showed that its molecular weight was 1018.997 Da (the highest peak). Mascot was used to determine the specific peptide sequence of scolopentide, and the final matched sequence was RAQNHYCK, which conformed to the restriction site of the protease. The molecular structure of scolopentide (C42H66N16O12S) is shown in Figure 2A (the asterisk).
Scolopentide was synthesized in vitro according to the peptide sequence RAQNHYCK, the length of which is 8 AA. Then, MS was used to detect the molecular weight. The observed molecular weight of synthetic scolopentide was 1018.8 Da (Figure 2B), which was similar to that of the extract (1018.997 Da). HPLC was used to assess the purity. The chromatogram is shown in Figure 2C, and the peak proportion is shown in Figure 2D. The synthetic scolopentide corresponds to peak 3; thus, its purity was inferred to be 98.014% according to the area fraction of peak 3. Consequently, the purity and molecular weight of synthetic scolopentide met the requirements.
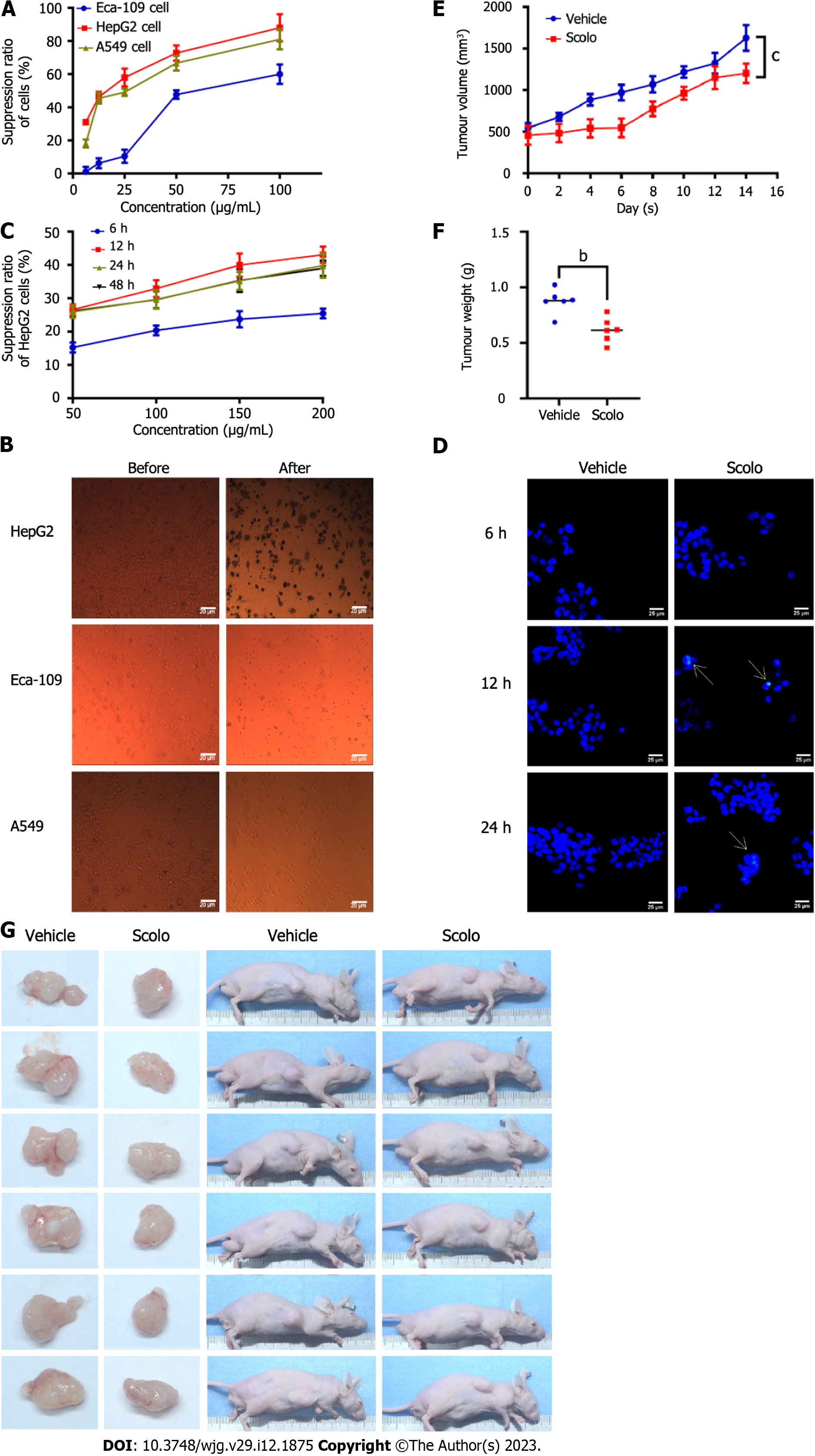
Eca-109, HepG2, and A549 cells were treated with the extracted scolopentide. A CCK8 assay showed that the IC50 values in the three cell types were 76.27 μg/mL (Eca-109), 22.06 μg/mL (HepG2), and 35.13 μg/mL (A549) (Figure 3A). Morphological changes in the three cell lines showed that the A549 and HepG2 cells changed considerably (Figure 3B). Based on the CCK8 assay and morphological changes, the following conclusions were drawn: extracted scolopentide still has antineoplastic effects in vitro, which inhibited the proliferation of three tumor cell lines, especially HepG2 cells.
We wanted to determine whether synthetic scolopentide can exert an antihepatoma effect. Our experiments suggested that synthetic scolopentide had antihepatoma effects in vitro and in vivo, which may be related to apoptosis induction. In vitro, the CCK8 assay suggested that synthetic scolopentide inhibited the proliferation of HepG2 cells in a concentration-dependent manner. The inhibitory effects were the strongest at 12 h, and there was no significant difference between 24 h and 48 h (Figure 3C). Moreover, a Hoechst staining assay was used to observe the morphological changes in HepG2 cells treated with scolopentide (100 μg/mL) (6 h, 12 h, and 24 h). The results showed that cytoplasmic staining and nuclear pyknosis occurred after treatment for 12 h and 24 h, which indicated apoptosis. Apoptosis was most clearly observed at 12 h (IC50: 208.11 μg/mL), consistent with the CCK8 assay (Figure 3D). In vivo, the tumor xenograft experiment suggested that the mean tumor volume of the two groups increased gradually, but that of the scolopentide group grew slower. The tumor weight of the scolopentide group was lower than that of the vehicle group (Figure 3E-G).
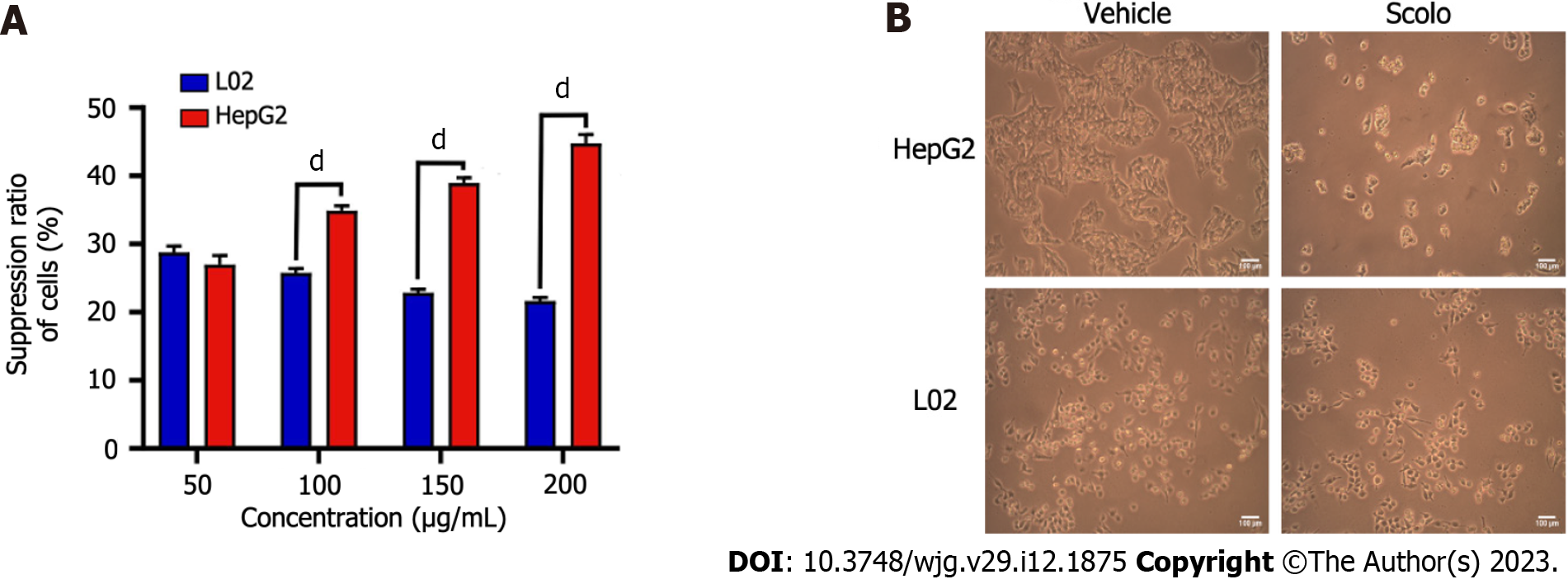
CCK8 assays and morphological changes were used to compare the cytotoxicity of synthetic scolopentide to liver cancer cells and normal liver cells. After treatment with synthetic scolopentide for 12 h, the CCK8 assay suggested that the cytotoxicity in L02 cells was significantly lower than that in HepG2 cells (100, 150, and 200 μg/mL) (Figure 4A). Morphological changes in the scolopentide group (100 μg/mL) and vehicle group (0 μg/mL) are shown in Figure 4B. Compared to the vehicle group, most HepG2 cells in the scolopentide group died, while some L02 cells survived. The CCK8 assay and morphological changes indicated that synthetic scolopentide was cytotoxic and was significantly stronger in HepG2 cells than in L02 cells.
Scolopentide was more cytotoxic in HepG2 cells than in L02 cells (Figure 4), which is similar to the activity of TRAIL, which selectively initiates apoptosis in cancer cells without significant toxicity to normal cells. Among the known TRAIL receptors, only DR4 and DR5 are able to induce apoptosis[33]. In addition, Hoechst staining indicated that HepG2 cells underwent apoptosis after treatment with synthetic scolopentide (Figure 3D). Therefore, we hypothesized that scolopentide could induce apoptosis by activating DR4/DR5. The binding free energies of scolopentide to DR4 and DR5 were-10.4 kcal/mol and-7.1 kcal/mol, respectively. Molecular docking suggested that scolopentide tightly bound to DR4 (Figure 5A) and DR5 (Figure 5B), which supported our hypothesis.
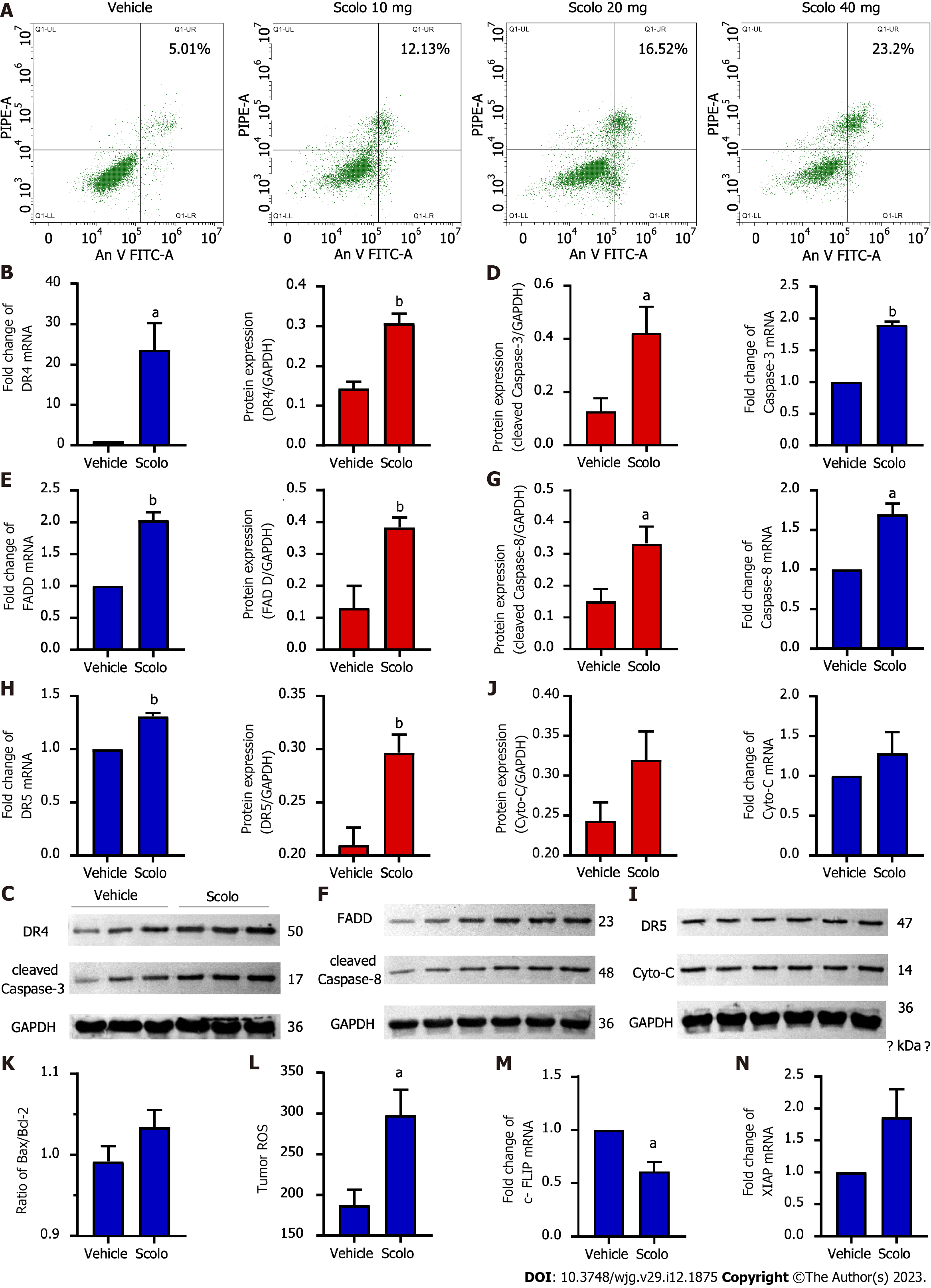
Flow cytometry suggested that apoptosis occurred in HepG2 cells after treatment with extracted scolopentide in vitro. The apoptosis ratios were 5.01% (0 μg/mL), 12.13% (10 μg/mL), 16.52% (20 μg/mL), and 23.2% (40 μg/mL), which indicates concentration-dependence (Figure 6A).
Animal experiments were used to further verify our hypothesis that scolopentide can stimulate the TRAIL pathway to induce apoptosis. The results of qRT-PCR and western blotting suggested that scolopentide activated DR4 (Figure 6B and C) and DR5 (Figure 6H and I), which promoted the expression of FADD (Figure 6E and F), activated the apoptosis promoter caspase-8 (Figure 6F and G) and the executor caspase-3 (Figure 6C and D), and finally induced apoptosis in SMMC-7721 Liver cancer cells. This form of apoptosis appears to be more related to the mitochondria-independent pathway, as Cyto-C and Bcl-2-associated X protein/B-cell lymphoma-2 (Bax/Bcl-2), which are key indicators in the mitochondria-dependent pathway, showed only slight upregulation and insignificant differences (Figure 6I-K). In addition, ROS levels were upregulated (Figure 6L). Cellular fas-associated death domain-like interleukin-1β converting enzyme inhibitory protein (c-FLIP), an inhibitory protein of caspase-8, was downregulated (Figure 6M), and x-chromosome linked inhibitor-of-apoptosis protein (XIAP), an inhibitory protein of caspase-3, was insignificantly upregulated (Figure 6N).
Peptides from centipedes have potential antitumor activity that should not be ignored. We isolated and purified crude centipede peptides 3 times and finally obtained a low molecular weight peptide (scolopentide) with the strongest antihepatoma effect. We further demonstrated that mechanistically, scolopentide induced apoptosis in liver cancer cells by activating DR4 and DR5 and promoting the upregulation of FADD, caspase-8 and caspase-3.
Centipedes, an important part of animal Chinese medicine, have played an important role in clinical treatment[1]. The remarkable activities and novel structure also give bioactive components of centipedes enormous potential to be exploited and modified as biological drugs[34]. However, pharmacological studies of centipedes are far behind those of other animals, such as snakes and scorpions[34,35]. Similar to animal-derived medicines, most centipede extracts are still crude. These crude extracts may contain a large number of histamines, polypeptide toxins and other biologically active substances[34], leading to allergic reactions such as itching or serious adverse reactions such as acute myocardial infarction, arrhythmia, tissue necrosis, respiratory depression and hemolysis, which hinder the clinical use of centipedes[11,36]. Small molecular substances from centipedes may not cause adverse reactions and may play a critical role with precise effects.
We tried a variety of ways to obtain a higher extraction rate and more accurate activity. For protein extraction, the optimal conditions were determined: 0.1 g trypsin (300 U/g, centipede-trypsin ratio of 20:1), enzymolysis temperature of 46 °C and enzymolysis time of 4 h. For peptide purification, GC[37-39] and HPLC[3,5,6], widely used techniques, were adopted to further purify crude centipede peptides. Mass spectrometry[5-7], the most widely used technique for peptide characterization, was used to identify scolopentide. The object of extraction and method used here are also noteworthy. The whole body of the centipede was chosen for extraction as recorded in Chinese works[14], even though most current studies focus on the venom[12,34]. In addition, enzymatic hydrolysis[6], a special extract technique, was chosen for the initial extraction to obtain crude centipede peptides. We aimed to simulate the environment of Chinese medicine being digested by various proteinases in the stomach after oral administration.
Currently, studies on the antihepatoma components of centipedes are rare (Table 1). Thus, we isolated, purified, and screened the crude centipede peptides three times. Finally, we obtained scolopentide (1018.997 Da), which had the strongest antihepatoma activity. Moreover, the peptide sequence RAQNHYCK of scolopentide was matched with the Mascot search engine, which helped to achieve scolopentide synthesis in vitro. Mascot also revealed that scolopentide was supposed to be a polypeptide from centipede venom proteins located in Kappa-scoloptoxin (07)-Ssm2a OS. It is worth noting that the tumor types used for screening were tumors in the chest and abdomen, such as liver cancer and esophageal cancer, as recorded in ancient Chinese works[13]. Liver cancer cells were more sensitive than other tumor cells and were chosen for subsequent mechanistic exploration (Figure 3A).
Antimicrobial activity is the most common pharmacological characteristic known among isolated centipede peptides at present (Table 1). Centipede peptides mainly destroy the integrity of cell membranes, which leads to the death of microbes[40-42]. Inducing apoptosis is the second major mechanism by which centipede peptides induce resistance to microorganisms (Table 1). Two antimicrobial centipede peptides, scolopendin and scolopendin 1, were obtained by RNA sequencing[41,43]. They have been shown to cause ROS accumulation in Candida albicans, which leads to mitochondrial depolarization, thus releasing Cyto-C into the cytoplasm from mitochondria and resulting in an increase in Ca2+ in the cytoplasm and mitochondria, ultimately inducing caspase-related apoptosis[43,44]. Some centipede peptides can also block the cell cycle and affect the expression of genes related to DNA replication and repair[45]. Additionally, some scholars have shown that a few antimicrobial centipede peptides also have antitumor activity[17,26]. Distinct from the antimicrobial mechanism, the antitumor activities are mainly related to apoptosis, especially the mitochondrial-dependent pathway[3,4,18]. To date, there have been no studies related to the mitochondrial-independent pathway of centipede effects (Table 1).
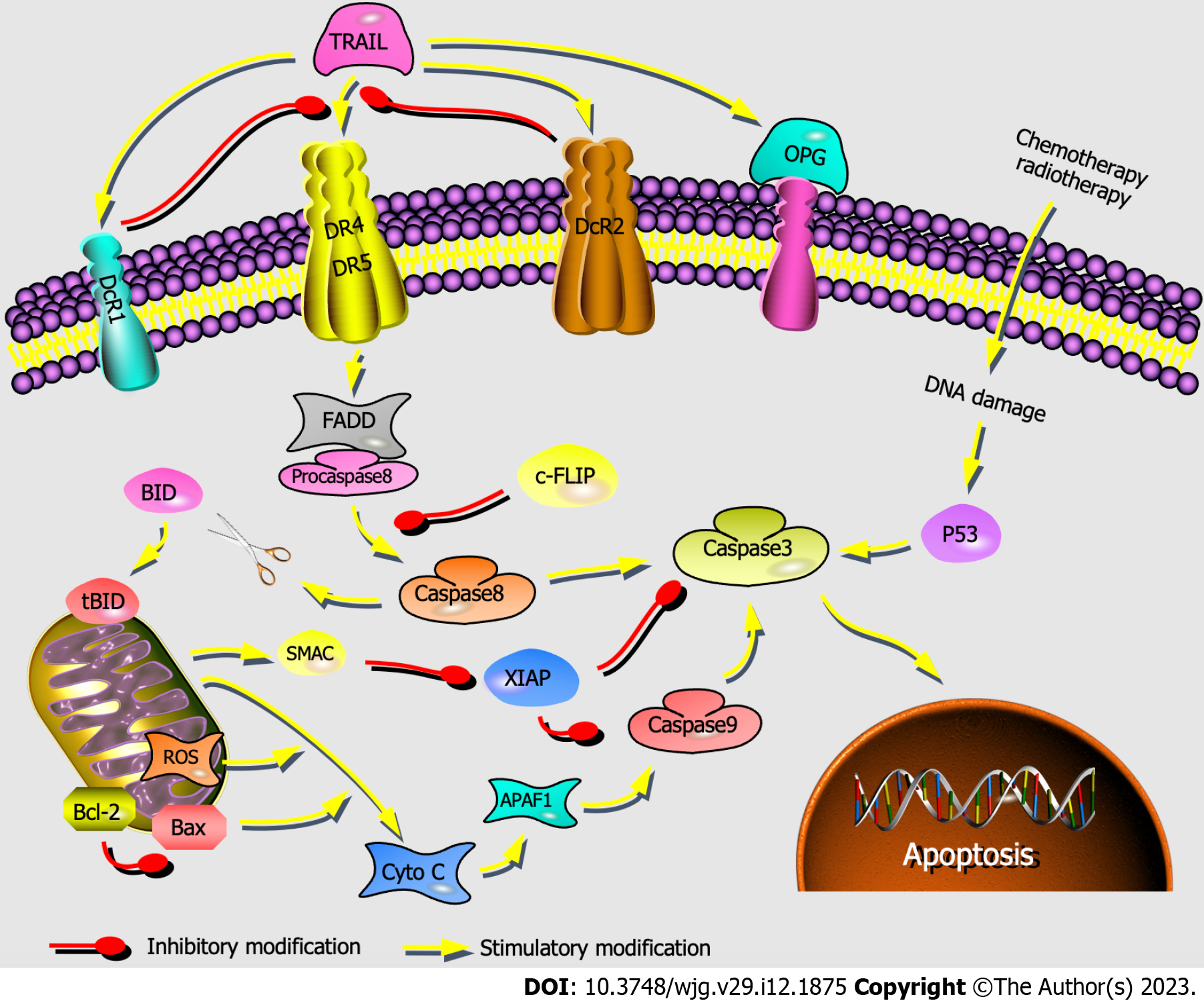
DR4 and DR5 are involved in the mitochondria-independent apoptosis pathway, which can receive extracellular death signals and induce intracellular apoptosis[46]. FADD receives apoptosis signals transmitted from DR4/DR5 and forms the death-inducing signaling complex TRAIL-DR4/DR5-FADD-procaspase. Then, caspase-8 is activated to initiate two apoptotic pathways: (1) The mitochondria-independent pathway, in which caspase-8 directly activates caspase-3; and (2) the mitochondria-dependent pathway, in which caspase-8 cleaves Bid, resulting in tBID that triggers the release of Cyto-C. Then, Cyto-C successively activates the promoter caspase-9 and the executor caspase-3. ROS accumulation can cause mitochondrial permeability transition pore opening, also leading to Cyto-C release. Apoptosis is the common end of these two pathways (Figure 7)[28,47-49].
We found that scolopentide may activate DR4 and DR5 and induce apoptosis. Flow cytometry suggested that apoptosis occurred in HepG2 cells after treatment with extracted scolopentide in vitro (Figure 6A). Hoechst staining showed the occurrence of apoptosis in HepG2 cells after treatment with synthetic scolopentide (Figure 3D). The CCK8 assay showed that scolopentide was significantly more cytotoxic in HepG2 cells than in L02 cells (Figure 4), which was similar to the effect of TRAIL, which selectively induced tumor cell apoptosis. Molecular docking also indicated that scolopentide binds well to TRAIL receptors (DR4 and DR5) (Figure 5). In our previous study, crude centipede peptides were also found to inhibit STAT3 protein phosphorylation[19], while some studies have suggested that dovitinib and sorafenib could overcome TRAIL resistance in HCC by inhibiting STAT3[50,51]. Consequently, animal experiments were used to further verify our hypothesis that scolopentide can stimulate the TRAIL pathway to induce apoptosis. In animal experiments, after treatment with synthetic scolopentide, the expression of DR4 and DR5, which are upstream of the TRAIL pathway, was clearly upregulated, especially the expression of DR4. The expression of FADD, caspase-8, and caspase-3, which are downstream of the TRAIL pathway, was also upregulated (Figure 6B-I). The animal experiments further confirmed the viewpoint that scolopentide induces apoptosis by activating DR4 and DR5.
This process may be more related to the mitochondria-independent pathway, as Cyto-C and Bax/Bcl-2 showed only slight upregulation and insignificant differences (Figure 6I-K), which are key indicators in the mitochondria-dependent pathway[52]. ROS were significantly upregulated but could not cause Cyto-C release (Figure 6I, J and L). Thus, we hypothesized that ROS mainly acted as signal transduction factors of the TRAIL pathway rather than the mitochondrial-dependent pathway[49]. Additionally, c-FLIP was inhibited, but XIAP was slightly upregulated (Figure 6M and N). A negative feedback mechanism of apoptosis may contribute to preventing excessive apoptosis in the body. Overall, our findings suggested that scolopentide may induce apoptosis in tumor cells, especially gastrointestinal tumors, by activating DR4 and DR5 and leading to the caspase cascade.
In summary, we obtained a small molecule polypeptide from Scolopendra subspinipes mutilans L. Koch with the strongest antitumor activity, especially for liver tumors. Mechanistically, scolopentide may induce tumor cell apoptosis by activating DR4 and DR5. Scolopentide is considered to be a promising drug candidate for cancer treatment, especially treatment of gastrointestinal tumors. Our studies are also expected to provide a reference for the extraction, purification and characterization of effective components in animal-derived medicines. Both extracted and synthesized scolopentide had anti-hepatoma activity. When HepG2 cells were cultured with two scolopentides for 48 h, a CCK8 assay showed that the IC50 values were 22.06 μg/mL (extracted scolopentide) and 237.726 μg/mL (synthesized scolopentide), which indicates that the antihepatoma activity of synthesized scolopentide was weaker than that of the extracted scolopentide. This may be due to the lack of dimensional folding configurations in the synthesized peptide during synthesis. Additionally, the TRAIL pathway was activated but decreased incrementally. Key areas of future research include investigating methods of modifying the spatial architecture of synthetic scolopentide, fully activating the TRAIL pathway and improving its antihepatoma activity.
Centipedes have been used to treat tumors for hundreds of years in China, while current studies rarely focus on hepatoma. The molecular identities of antihepatoma bioactive components in centipedes have not yet been extensively investigated. It is a challenge to isolate and characterize the effective components of centipedes due to limited peptide purification technologies for animal-derived medicines.
The antihepatoma components in centipedes remain unclear. We investigated the centipede components with the strongest antihepatoma activity to develop candidates for antihepatoma drugs.
To purify, characterize, and synthesize the bioactive components with the strongest antihepatoma activity from centipedes and determine the antihepatoma mechanism. To provide a reference for the extraction, purification and characterization of effective components for animal-derived medicines.
An antihepatoma peptide (scolopentide) was isolated and identified from the centipede scolopendra subspinipes mutilans using a combination of enzymatic hydrolysis, a Sephadex G-25 column, and two steps of high-performance liquid chromatography. Additionally, the CCK8 assay was used to select the extracted fraction with the strongest antihepatoma activity. The molecular weight of the extracted scolopentide was characterized by quadrupole time-of-flight mass spectrometry, and the sequence was matched by using the Mascot search engine. Scolopentide was then synthesized using solid-phase peptide synthesis methods. The antihepatoma effects of extracted and synthetic scolopentide were confirmed in vitro and in vivo. Mechanistically, flow cytometry and Hoechst staining were conducted to confirm the occurrence of apoptosis. Molecular docking and CCK8 assays were performed to determine the relationship between scolopentide and the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) pathway. Reactive oxygen species assessment, quantitative real-time polymerase chain reaction and Western blot were used to further verify the hypothesis that scolopentide can stimulate the TRAIL pathway to induce apoptosis.
A small molecule polypeptide with the strongest antihepatoma activity was derived from Scolopendra subspinipes mutilans L. Koch. The molecular weight was 1018.997 Da, and the peptide sequence was RAQNHYCK. Both extracted and synthesized scolopentide had antihepatoma activity in a concentration-dependent manner. Mechanistically, scolopentide activated death receptor 4 (DR4) and DR5 and induced apoptosis in liver cancer cells by promoting the expression of Fas-associated death domain protein (FADD), caspase-8 and caspase-3 through a mitochondria-independent pathway.
Scolopentide, an antihepatoma peptide, was isolated and identified from centipedes, which activated DR4 and DR5 and induced apoptosis through a mitochondria-independent pathway.
Scolopentide is considered to be a promising drug candidate for cancer treatment, especially treatment of liver cancer. Ways in which to modify the spatial architecture of synthetic scolopentide, fully activate the TRAIL pathway and improve its antihepatoma activity will be key areas for future research.
The authors would like to thank Professor Feng Li (University of California) for providing suggestion on peptide sequence screening, and Professor Yangu Peng (Hunan University of Chinese Medicine) for providing guidance on extraction technology of centipede.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Integrative and complementary medicine
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lekshmi.R Nath India; Portugal RV, Portugal S-Editor: Chen YL L-Editor: A P-Editor: Zhang XD
| 1. | Commission NP. Pharmacopoeia of the People’s Republic of China. 2020. [cited 3 September 2022]. Available from: https://www.semanticscholar.org/paper/Pharmacopoeia-of-the-People%27s-Republic-of-China-Pharmacopoeia/8726eceef0b79ae17b4f2fd7490728945711b717. |
| 2. | Xu XL, Wang CM, Geng D, Zhang L, Hu B. [Effects of centipede extracts on normal mouse and S180, H22 bearing mouse]. Zhong Yao Cai. 2010;33:499-503. [PubMed] |
| 3. | Ding D, Guo YR, Wu RL, Qi WY, Xu HM. Two new isoquinoline alkaloids from Scolopendra subspinipes mutilans induce cell cycle arrest and apoptosis in human glioma cancer U87 cells. Fitoterapia. 2016;110:103-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Ma W, Zhang D, Zheng L, Zhan Y, Zhang Y. Potential roles of Centipede Scolopendra extracts as a strategy against EGFR-dependent cancers. Am J Transl Res. 2015;7:39-52. [PubMed] |
| 5. | Chaparro-Aguirre E, Segura-Ramírez PJ, Alves FL, Riske KA, Miranda A, Silva Júnior PI. Antimicrobial activity and mechanism of action of a novel peptide present in the ecdysis process of centipede Scolopendra subspinipes subspinipes. Sci Rep. 2019;9:13631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Ali SM, Khan NA, Sagathevan K, Anwar A, Siddiqui R. Biologically active metabolite(s) from haemolymph of red-headed centipede Scolopendra subspinipes possess broad spectrum antibacterial activity. AMB Express. 2019;9:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 7. | Kong Y, Huang SL, Shao Y, Li S, Wei JF. Purification and characterization of a novel antithrombotic peptide from Scolopendra subspinipes mutilans. J Ethnopharmacol. 2013;145:182-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Luo L, Li B, Wang S, Wu F, Wang X, Liang P, Ombati R, Chen J, Lu X, Cui J, Lu Q, Zhang L, Zhou M, Tian C, Yang S, Lai R. Centipedes subdue giant prey by blocking KCNQ channels. Proc Natl Acad Sci U S A. 2018;115:1646-1651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 9. | Kong Y, Xu C, He ZL, Zhou QM, Wang JB, Li ZY, Ming X. A novel peptide inhibitor of platelet aggregation from stiff silkworm, Bombyx batryticatus. Peptides. 2014;53:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8980] [Cited by in RCA: 9032] [Article Influence: 237.7] [Reference Citation Analysis (0)] |
| 11. | Lan XQ, Zhao F, Wang QQ, Li JH, Zeng L, Zhang Y, Lee WH. Isolation and characterization of the major centipede allergen Sco m 5 from Scolopendra subspinipes mutilans. Allergol Int. 2021;70:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 12. | Han Y, Kamau PM, Lai R, Luo L. Bioactive Peptides and Proteins from Centipede Venoms. Molecules. 2022;27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 13. | Li SZ. Compendium of Materia Medica. 21st Century Press. Nanchang: National Top 100 Publishing House, 2017: 273-274. |
| 14. | Zhang X. Integrating Chinese and Western Medicine. Shijiazhuang: Hebei Science and Technology Press, 1985: 135-136. |
| 15. | Xie LS, Huan T, Yang JL, Wu J, Zhao P. [The effect of Shendan Sanjie capsule on angiogenesis in mice with colitis associated cancer and mechanism]. Zhonghua Zhong Liu Za Zhi. 2021;43:1170-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 16. | Yan W, Lu J, Li G, Wei H, Ren WH. Amidated Scolopin-2 inhibits proliferation and induces apoptosis of Hela cells in vitro and in vivo. Biotechnol Appl Biochem. 2018;65:672-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Lee JH, Kim IW, Kim SH, Kim MA, Yun EY, Nam SH, Ahn MY, Kang D, Hwang JS. Anticancer Activity of the Antimicrobial Peptide Scolopendrasin VII Derived from the Centipede, Scolopendra subspinipes mutilans. J Microbiol Biotechnol. 2015;25:1275-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Ma W, Liu R, Qi J, Zhang Y. Extracts of centipede Scolopendra subspinipes mutilans induce cell cycle arrest and apoptosis in A375 human melanoma cells. Oncol Lett. 2014;8:414-420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 19. | Yong-Jiea T, Zhuob L, Liuc L, Yuanc C, Xiao-Did H, Xue-Feic T. STAT3 Inhibition by Centipede Scolopendra Extract in Liver Cancer HepG2 Cells and Orthotopic Mouse Models of Hepatocellular Carcinoma. Dig Chin Med. 2020;3:67-79. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Chen Y, Ai XJ, Wang ZQ, Tian S, Zhou Q, Pei G, Tian XF. Study on Anti-lung Cancer Efficiency of Centipede Extracts in Vitro and Vivo Experiments. Zhongguo Zhongyiyao Xinxi Zazhi. 2016;23:61-63. [DOI] [Full Text] |
| 21. | Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 832] [Cited by in RCA: 834] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 22. | Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol Ther. 2005;4:139-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 730] [Cited by in RCA: 887] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 23. | Labuschagne CF, Zani F, Vousden KH. Control of metabolism by p53 - Cancer and beyond. Biochim Biophys Acta Rev Cancer. 2018;1870:32-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 134] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 24. | Marei HE, Althani A, Afifi N, Hasan A, Caceci T, Pozzoli G, Morrione A, Giordano A, Cenciarelli C. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021;21:703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 289] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 25. | Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 509] [Cited by in RCA: 929] [Article Influence: 116.1] [Reference Citation Analysis (0)] |
| 26. | Holland PM. Death receptor agonist therapies for cancer, which is the right TRAIL? Cytokine Growth Factor Rev. 2014;25:185-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 27. | Yuan X, Gajan A, Chu Q, Xiong H, Wu K, Wu GS. Developing TRAIL/TRAIL death receptor-based cancer therapies. Cancer Metastasis Rev. 2018;37:733-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 191] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 28. | von Karstedt S, Montinaro A, Walczak H. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat Rev Cancer. 2017;17:352-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 426] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 29. | Chen CY, Yiin SJ, Hsu JL, Wang WC, Lin SC, Chern CL. Isoobtusilactone A sensitizes human hepatoma Hep G2 cells to TRAIL-induced apoptosis via ROS and CHOP-mediated up-regulation of DR5. J Agric Food Chem. 2012;60:3533-3539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Kondo K, Yamasaki S, Sugie T, Teratani N, Kan T, Imamura M, Shimada Y. Cisplatin-dependent upregulation of death receptors 4 and 5 augments induction of apoptosis by TNF-related apoptosis-inducing ligand against esophageal squamous cell carcinoma. Int J Cancer. 2006;118:230-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Zhou X, Zijlstra SN, Soto-Gamez A, Setroikromo R, Quax WJ. Artemisinin Derivatives Stimulate DR5-Specific TRAIL-Induced Apoptosis by Regulating Wildtype P53. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Nazim UM, Park SY. Luteolin sensitizes human liver cancer cells to TRAILinduced apoptosis via autophagy and JNKmediated death receptor 5 upregulation. Int J Oncol. 2019;54:665-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Pavet V, Beyrath J, Pardin C, Morizot A, Lechner MC, Briand JP, Wendland M, Maison W, Fournel S, Micheau O, Guichard G, Gronemeyer H. Multivalent DR5 peptides activate the TRAIL death pathway and exert tumoricidal activity. Cancer Res. 2010;70:1101-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 34. | Luo A, Wang A, Kamau PM, Lai R, Luo L. Centipede Venom: A Potential Source of Ion Channel Modulators. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 35. | Undheim EA, Jenner RA, King GF. Centipede venoms as a source of drug leads. Expert Opin Drug Discov. 2016;11:1139-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Ombati R, Luo L, Yang S, Lai R. Centipede envenomation: Clinical importance and the underlying molecular mechanisms. Toxicon. 2018;154:60-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 37. | Peng K, Kong Y, Zhai L, Wu X, Jia P, Liu J, Yu H. Two novel antimicrobial peptides from centipede venoms. Toxicon. 2010;55:274-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Kong Y, Shao Y, Chen H, Ming X, Wang JB, Li ZY, Wei JF. A Novel Factor Xa-Inhibiting Peptide from Centipedes Venom. Int J Pept Res Ther. 2013;19:303-311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 39. | Yang S, Xiao Y, Kang D, Liu J, Li Y, Undheim EA, Klint JK, Rong M, Lai R, King GF. Discovery of a selective NaV1.7 inhibitor from centipede venom with analgesic efficacy exceeding morphine in rodent pain models. Proc Natl Acad Sci U S A. 2013;110:17534-17539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 142] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 40. | Lee JH, Kim IW, Kim MA, Ahn MY, Yun EY, Hwang JS. Antimicrobial Activity of the Scolopendrasin V Peptide Identified from the Centipede Scolopendra subspinipes mutilans. J Microbiol Biotechnol. 2017;27:43-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 41. | Lee W, Hwang JS, Lee DG. A novel antimicrobial peptide, scolopendin, from Scolopendra subspinipes mutilans and its microbicidal mechanism. Biochimie. 2015;118:176-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 42. | Lee H, Hwang JS, Lee J, Kim JI, Lee DG. Scolopendin 2, a cationic antimicrobial peptide from centipede, and its membrane-active mechanism. Biochim Biophys Acta. 2015;1848:634-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 43. | Choi H, Hwang JS, Lee DG. Identification of a novel antimicrobial peptide, scolopendin 1, derived from centipede Scolopendra subspinipes mutilans and its antifungal mechanism. Insect Mol Biol. 2014;23:788-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 44. | Lee H, Hwang JS, Lee DG. Scolopendin, an antimicrobial peptide from centipede, attenuates mitochondrial functions and triggers apoptosis in Candida albicans. Biochem J. 2017;474:635-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 45. | Lu J, Qiu PD, Wen HR. Mechanism of Antimicrobial Peptide Scolopin 2-NH2 Isolated from Scolopendra subspinipes mutilans. Biol Bull. 2018;34:179-190. [DOI] [Full Text] |
| 46. | den Hollander MW, Gietema JA, de Jong S, Walenkamp AM, Reyners AK, Oldenhuis CN, de Vries EG. Translating TRAIL-receptor targeting agents to the clinic. Cancer Lett. 2013;332:194-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 47. | Kelley SK, Ashkenazi A. Targeting death receptors in cancer with Apo2L/TRAIL. Curr Opin Pharmacol. 2004;4:333-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 276] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 48. | Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94:909-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2139] [Cited by in RCA: 3820] [Article Influence: 347.3] [Reference Citation Analysis (0)] |
| 49. | Eberle J. Countering TRAIL Resistance in Melanoma. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 50. | Chen KF, Tai WT, Liu TH, Huang HP, Lin YC, Shiau CW, Li PK, Chen PJ, Cheng AL. Sorafenib overcomes TRAIL resistance of hepatocellular carcinoma cells through the inhibition of STAT3. Clin Cancer Res. 2010;16:5189-5199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 51. | Chen KF, Chen HL, Liu CY, Tai WT, Ichikawa K, Chen PJ, Cheng AL. Dovitinib sensitizes hepatocellular carcinoma cells to TRAIL and tigatuzumab, a novel anti-DR5 antibody, through SHP-1-dependent inhibition of STAT3. Biochem Pharmacol. 2012;83:769-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 52. | Yamaguchi R, Lartigue L, Perkins G. Targeting Mcl-1 and other Bcl-2 family member proteins in cancer therapy. Pharmacol Ther. 2019;195:13-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 53. | Wenhua R, Shuangquan Z, Daxiang S, Kaiya Z, Guang Y. Induction, purification and characterization of an antibacterial peptide scolopendrin I from the venom of centipede Scolopendra subspinipes mutilans. Indian J Biochem Biophys. 2006;43:88-93. [PubMed] |
| 54. | Choi H, Hwang JS, Lee DG. Antifungal effect and pore-forming action of lactoferricin B like peptide derived from centipede Scolopendra subspinipes mutilans. Biochim Biophys Acta. 2013;1828:2745-2750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Park YJ, Kim HS, Lee HY, Hwang JS, Bae YS. A novel antimicrobial peptide isolated from centipede Scolopendra subspinipes mutilans stimulates neutrophil activity through formyl peptide receptor 2. Biochem Biophys Res Commun. 2017;494:352-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 56. | Kwon YN, Lee JH, Kim IW, Kim SH, Yun EY, Nam SH, Ahn MY, Jeong M, Kang DC, Lee IH, Hwang JS. Antimicrobial activity of the synthetic peptide scolopendrasin ii from the centipede Scolopendra subspinipes mutilans. J Microbiol Biotechnol. 2013;23:1381-1385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 57. | Park YJ, Lee SK, Jung YS, Lee M, Lee HY, Kim SD, Park JS, Koo J, Hwang JS, Bae YS. Promotion of formyl peptide receptor 1-mediated neutrophil chemotactic migration by antimicrobial peptides isolated from the centipede Scolopendra subspinipes mutilans. BMB Rep. 2016;49:520-525. [PubMed] |
| 58. | Park YJ, Park B, Lee M, Jeong YS, Lee HY, Sohn DH, Song JJ, Lee JH, Hwang JS, Bae YS. A novel antimicrobial peptide acting via formyl peptide receptor 2 shows therapeutic effects against rheumatoid arthritis. Sci Rep. 2018;8:14664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 59. | Yoo WG, Lee JH, Shin Y, Shim JY, Jung M, Kang BC, Oh J, Seong J, Lee HK, Kong HS, Song KD, Yun EY, Kim IW, Kwon YN, Lee DG, Hwang UW, Park J, Hwang JS. Antimicrobial peptides in the centipede Scolopendra subspinipes mutilans. Funct Integr Genomics. 2014;14:275-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 60. | Park YJ, Lee HY, Jung YS, Park JS, Hwang JS, Bae YS. Antimicrobial peptide scolopendrasin VII, derived from the centipede Scolopendra subspinipes mutilans, stimulates macrophage chemotaxis via formyl peptide receptor 1. BMB Rep. 2015;48:479-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 61. | Du C, Li J, Shao Z, Mwangi J, Xu R, Tian H, Mo G, Lai R, Yang S. Centipede KCNQ Inhibitor SsTx Also Targets K(V)1.3. Toxins (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 62. | Wang Y, Li X, Yang M, Wu C, Zou Z, Tang J, Yang X. Centipede venom peptide SsmTX-I with two intramolecular disulfide bonds shows analgesic activities in animal models. J Pept Sci. 2017;23:384-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 63. | Chaparro E, da Silva PI Junior. Lacrain: the first antimicrobial peptide from the body extract of the Brazilian centipede Scolopendra viridicornis. Int J Antimicrob Agents. 2016;48:277-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 64. | Zhou YQ, Han L, Liu ZQ, Du KC, Li KY. [Effect of centipede extract on cervical tumor of mice and its mechanism]. Zhong Yao Cai. 2011;34:859-864. [PubMed] |
| 65. | Ren Y, Houghton P, Hider RC. Relevant activities of extracts and constituents of animals used in traditional Chinese medicine for central nervous system effects associated with Alzheimer's disease. J Pharm Pharmacol. 2006;58:989-996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |