Published online Mar 21, 2023. doi: 10.3748/wjg.v29.i11.1651
Peer-review started: September 21, 2022
First decision: November 5, 2022
Revised: January 5, 2023
Accepted: March 7, 2023
Article in press: March 7, 2023
Published online: March 21, 2023
Processing time: 177 Days and 4.1 Hours
Liver disease has become a leading cause of death, particularly in the West, where it is attributed to more than two million deaths annually. The correlation between gut microbiota and liver disease is still not fully understood. However, it is well known that gut dysbiosis accompanied by a leaky gut causes an increase in lipopolysaccharides in circulation, which in turn evoke massive hepatic inflammation promoting liver cirrhosis. Microbial dysbiosis also leads to poor bile acid metabolism and low short-chain fatty acids, all of which exacerbate the inflammatory response of liver cells. Gut microbial homeostasis is maintained through intricate processes that ensure that commensal microbes adapt to the low oxygen potential of the gut and that they rapidly occupy all the intestinal niches, thus outcompeting any potential pathogens for available nutrients. The crosstalk between the gut microbiota and its metabolites also guarantee an intact gut barrier. These processes that protect against destabilization of gut microbes by potential entry of pathogenic bacteria are collectively called colonization resistance and are equally essential for liver health. In this review, we shall investigate how the mechanisms of colonization resistance influence the liver in health and disease and the microbial-liver crosstalk potential as therapeutic target areas.
Core Tip: The influence of the gut microbiome on various body systems has important implications for health and disease, such as liver disease. While the exact mechanisms of how the microbiome contributes to liver disease are unknown, there is strong evidence that the translocation of various metabolites across the mucosal barrier plays a strong role, which is precipitated by dysbiotic gut microbiota. Considering the importance of the microbiome in liver disease, powerful therapeutic options that can manipulate the gut microbiome are being explored. These approaches could have the potential for effective treatments for various stages of liver disease. This review will explore how the mechanisms of colonization resistance influence the liver in health and disease and finally examine potential therapeutic targets in the gut-liver axis.
- Citation: Kirundi J, Moghadamrad S, Urbaniak C. Microbiome-liver crosstalk: A multihit therapeutic target for liver disease. World J Gastroenterol 2023; 29(11): 1651-1668
- URL: https://www.wjgnet.com/1007-9327/full/v29/i11/1651.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i11.1651
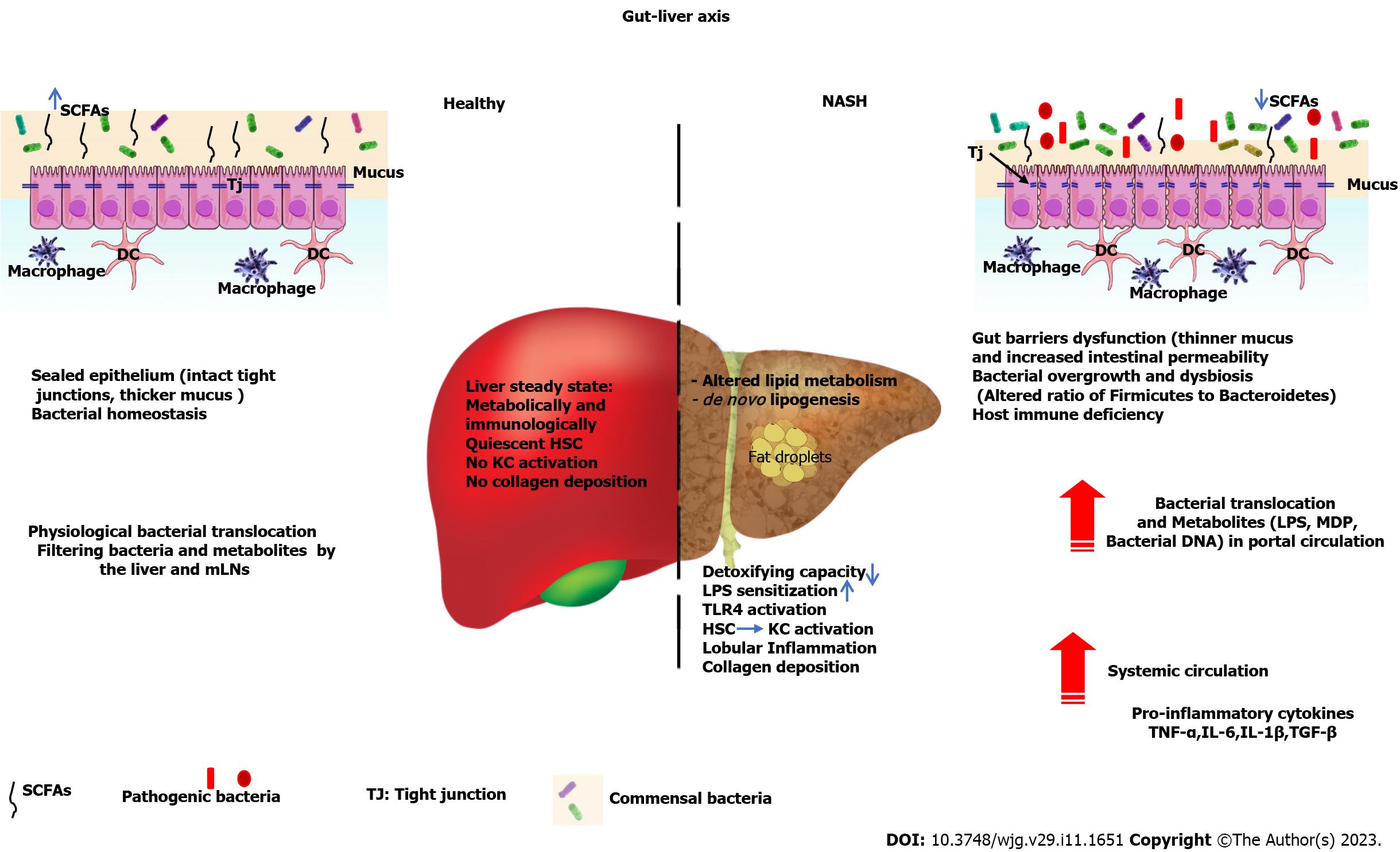
A healthy gut microbiota plays a significant role in maintaining a homeostatic gut environment. One such role is colonization resistance, which is defined as the microbial capacity to resist invasion of exogenous microorganisms (for example, pathogens) and/or prevent uncontrolled overgrowth of endogenous microbes (for example, pathobionts). For gut homeostasis to be achieved, microbial alpha diversity must remain high, gut mucosal integrity must be maintained, and tolerance to the billions of microbial immunogens present in the gut must be established. This is all achieved through intricate microbe-to-microbe and microbe-to-host interactions mediated by microbial metabolites, such as short-chain fatty acids, or microbial cell wall components, such as lipopolysaccharides, lipoteichoic acid, peptidoglycans and flagellin. Homeostasis is also achieved through the production of antimicrobial peptides, resource and oxygen competition, host immunomodulation, and conjugation of bile acids. The mechanisms by which these inter/intramicrobial interactions mediate colonization resistance or how their perturbation leads to disease have not yet been fully elucidated. However, it is known that an imbalance in microbial composition, otherwise known as dysbiosis, which may arise from dietary changes, ingestion of exogenous toxins such as antibiotics or xenobiotics, or through infections that suppress the immune system, has serious and sometimes long-term clinical implications. Diseases such as diabetes, obesity, atherosclerosis, and liver disease are associated with dysbiosis and the translocation of gut microbial products into circulation. As the liver is the first organ to be exposed to the gut bacterial products and digested food delivered through the portal vein, any leakage of microbial products into circulation will lead to hepatocellular immune activation, thereby promoting systemic and hepatic inflammation, which may lead to liver disease[1]. An understanding of the mechanisms involved in colonization resistance and its influencing factors is therefore crucial to establish their link to the etiology of liver disease as well as to identify possible hit points along the gut-liver axis that can be utilized as therapeutic targets for liver disease[2]. This review explores some of the mechanisms of colonization resistance and their importance to the etiology of the different stages of nonalcoholic fatty liver disease (NAFLD) from simple steatosis to liver inflammation, as well as alcohol-associated liver disease (ALD), and highlights potential entry points that may be used as therapeutic targets for liver disease. A summary of the interplay between the microbiome, liver, immune system, and metabolome is presented in Figure 1.
The gut microbiome starts taking shape at birth, where it is initially influenced by the mode of delivery. Vaginally born babies will have a gut microbial composition very close to the maternal vaginal microbiota, while the caesarian born will adopt mainly the skin microbiota[3]. Mammals have five phyla that predominate the gut: Firmicutes (e.g., Lactobacillus, Clostridium, Ruminococcus, Eubacterium, Fecalibacterium and Roseburia), Actinobacteria (with Bifidobacterium as one of its most important members), Bacteroidetes (e.g., Bacteroides, Prevotella, and Xylanibacter), Proteobacteria (e.g., Escherichia and Desulfovibrio) and Verrucomicrobia (e.g., Akkermansia)[4]. The earliest colonizers are mainly facultative aerobes of the phyla Firmicutes and Actinobacteria, which play a significant role in lowering the gut’s oxygen level to allow for the colonization of obligate anaerobes. These aerotolerant microbes reside in the upper gut, where they continue to reduce the amount of oxygen in the gut for life. Escherichia coli and Enterococcus faecalis are the most abundant in the oxygen-high neonatal gut, and they rapidly expand in the early phase, leading to a gradual depression of oxygen levels and allowing growth of the facultative anaerobes Bifidobacterium, Bacteroides and Clostridium, which colonize most of the lower gut[4,5]. The neonatal microbiota is also influenced by the mode of feeding, where breast-fed babies show a more stable microbiota that has a higher copy number of Bacteroides and Bifidobacterium but a lower abundance of Enterococcus and Streptococcus species, while formula-fed babies have a higher abundance of Clostridium, Streptococcus and Enterococcus[6]. The early life microbiota only begins to take a semblance of adult microbiota when solid food is introduced and will remain relatively unstable until 3-5 years after birth[7]. The rapid expansion of early life microbiota and the adaptation to oxygen levels signify the earliest mechanisms for initiating gut microbial homeostasis[8].
The colon has the highest density of microbes in the gastrointestinal tract, harboring approximately 70% of all gut microbes, which are mostly members of the Firmicutes and Bacteroidetes phyla[9]. The Firmicutes to Bacteroidetes axis is important in maintaining gut homeostasis, as members of each phylum have specialized metabolic roles (i.e., metabolism of sugar vs. indigestible fibers) that impact the microbiome and the host. It is believed that the role in homeostasis is optimized when the relative abundance is 80% Firmicutes and 15% Bacteroidetes[8,10,11]. However, the significance of this value and the actual impact it has on the host have been questioned by some researchers[12], emphasizing the importance of more research on the role of Firmicutes and Bacteroidetes in gut microbial homeostasis, health and disease. Nutrients, metabolic byproducts and the competition between exogenous microbes and commensals help prevent colonization of pathogens and maintain homeostasis. Different animal studies have shown that nutrient competition occurs between metabolically related microbiota members. For example, germ-free mice colonized with three human commensal strains of Escherichia coli (E. coli HS, E. coli Nissle 1917, E. coli MG1655) successfully prevented colonization of the cecum by the pathogen enterohaemorrhagic Escherichia coli (EHEC) EDL933, an E. coli 0157:H7 biotype, due to the three precolonized commensal biotypes outcompeting E. coli EDL933 for nutrients[13]. This colonization resistance was further shown to occur using multiple sugars as metabolic substrates for probiotic E. coli Nissle 1917 and commensal subtype E. coli HS, whose rapid growth effectively limited the colonization of EHEC E. coli EDL933 in a mouse model[14]. Competition for a shared nutritional niche of proline was similarly demonstrated in a gnotobiotic mouse model colonized with early life microbiota where early-life E. coli 1 was shown to outcompete E. coli 0157:H7[15]. This colonization resistance was also thought to be attributed to the production of lactate and acetate by bifidobacteria and enterococci, which can suppress the motility of E. coli 0157:H7 under cecal anaerobic conditions[15]. Colonization resistance is also aided by the production of toxic antimicrobial peptides by commensals. For example, many members of the phylum Bacteroidetes produce toxic antimicrobial peptides through their type 6 secretion systems (T6SS)[16], E. coli produces narrow-spectrum antibiotics called microcins that effectively kill competitors within their niche[17,18], and the probiotic Bifidobacterium secretes broad-spectrum bacteriocins[19].
Overall, any extrinsic or intrinsic factors that upset the stable microbial communities will in essence destabilize the colonization resistance mechanisms and lead to disease by allowing colonization of pathogenic microbes and/or leakage of microbes and microbial toxins into circulation.
The gut is lined with a thick mucus layer made of a highly glycosylated mucin 2 protein, which is densely packed and insoluble in the layer closest to the epithelium but loosely packed and soluble on the outer layer[20,21]. This mucus layer prevents direct contact of bacteria with the gut epithelium, thereby reducing the potential for pathogen colonization[20,21]. The development of the mucus layer is enhanced by the gut microbiota and depends on the intestinal microbial composition. It has been shown that germ-free rodents have a much thinner mucus layer than their conventionally colonized counterparts[22]. Petersson and colleagues have shown that a thin colon mucosal layer in a colitis germ-free mouse model can be restored by administering lipopolysaccharides or peptidoglycans to germ-free mice[22]. Bacteria enhance the mucus layer in numerous ways, such as through the production of secondary metabolites. Short chain fatty acids (SCFAs), such as acetate produced by Bifidobacterium or butyrate produced by gram-positive Firmicutes such as Faecalibacterium prausnitzii, Roseburia sp, and Butyricicoccus pullicaecorum[23,24], are known to strengthen gut barrier function, normalize permeability, improve intestinal epithelium defense, protect against pathogenic infections, and reduce inflammation[25-28].
Intestinal epithelial cells are held together by a set of tight junction proteins that are molecules situated at the tight junctions of epithelial cells. The integrity of these tight junctions can be influenced by commensal bacteria and their effects on tight junction proteins. For example, Lactobacillus rhamnosus GG induces claudin-3 expression, L. acidophilus and L. plantarum stimulate the expression of occludin, and Bifidobacterium infantis preserves claudin-4 and occludin deposition at tight junctions[29,30]. In a mouse necrotizing enterocolitis model, Bifidobacterium was found to preserve claudin 4 and occludin localization in tight junctions, thereby preventing gut permeability[31]. In mouse models, probiotics have been shown to improve the integrity of the intestinal barrier, which has also been observed in Crohn’s and colitis patients[28]. In vitro treatment of Caco-2 cells with the probiotic E. coli Nissle 1917 increases the expression and peripheral migration of ZO-2[32]. Treatment of Caco2 cells with the probiotic Lactobacillus plantarum MB452 increased occludin and cingulin gene expression[33]. These results indicate that in vitro, certain probiotics can improve gut barrier function. Maintaining the integrity of the intestinal barrier is essential due to the high levels of microbial lipopolysaccharide (LPS) present within the lumen of the gut, as LPS is a potent immunological signal that can induce an inflammatory cascade if detected systemically, which a healthy intestinal barrier effectively prevents[34]. Leakage of LPS and other microbial polypeptides into circulation due to dysbiosis can lead to inflammation in the liver (among other organs), which can lead to the development of liver disease[34].
Pericentral hepatocytes primarily produce bile acids from cholesterol[35]. In humans, these acids are then transported to the gut, where they are dehydroxylated, epimerized, or dehydrogenated into different secondary bile acids, such as deoxycholic acid (DCA), ursodeoxycholic acid (UDCA), ursocholic acid, or lithocholic acid (LCA)[35]. In mice, murideoxycholic acid and hyodeoxycholic acid are also produced[35]. Secondary bile acids are known to bind to the intestinal farnesoid X receptor (FXR) and G-protein coupled receptor 5 (TGR5)[36]. Some bile acid metabolites have also been shown to have a contradictory effect on gut barrier tight junctions[36]. UDCA and LCA, for example, have opposing effects on the barrier of human colonic T84 cells[36]. Treatment of these cells with primary bile acid-chenodeoxycholic acid (CDCA) combined with LCA leads to an increase in barrier permeability and the inflammatory cytokine IL-8[37]. Using a Caco-2 cell model, it was demonstrated that DCA led to an increase in the phosphorylation of epithelial growth factor receptor, which induced barrier dysfunction[38]. Prematurely weaned piglets treated with CDCA showed an improvement in the gut barrier with higher ZO-1 expression and increased expression of the proinflammatory cytokines tumor necrosis factor (TNF)-α and interleukin (IL)-6 and the anti-inflammatory cytokine IL-10[39]. The authors speculated that the anti-inflammatory effects of both IL-10 and ZO-1 counteracted the inflammatory effects of IL-6 and TNF-α, thus precipitating a net improvement in the intestinal barrier[39]. These examples demonstrate that bile acid metabolism is a significant key player in gut health, and it can be utilized as a therapeutic target for liver disease and other metabolic disorders, as will be discussed later.
Liver disease has been shown through preclinical and clinical trials to be accompanied by gut dysbiosis[40-44]. It has been shown that liver cirrhosis is also correlated with bacteremia, increased gut permeability, and increased circulatory LPS[43]. Dysbiosis has been noted in many mouse models of liver disease, such as secondary biliary fibrosis (common) induced by bile duct ligation, alcoholic liver disease induced by alcohol uptake in drinking water and hepatotoxicity-induced liver cirrhosis using carbon tetrachloride (CCL4) treatment[42,43]. In humans, several gram-positive bacteria, including members of the genera Clostridium XI, Anaerobacter, Streptococcus, and Lactobacillus, were found to be more abundant in the gut in NAFLD patient biopsies than in healthy volunteers[45]. In contrast, Oscillibacter and Flavonifractor of the family Ruminococcaceae were abundant in healthy volunteers relative to NAFLD patients[45]. In severe fibrosis forms of NAFLD, the bacteria Bacteroidetes vulgatus and Escherichia coli were identified as the most abundant[46]. Although there has not yet been a general consensus on what microbial ratios of different strains exist in NAFLD patients, many research findings indicate that a lower Firmicutes to Bacteroidetes ratio is associated with liver disease[11,47]. Dysbiosis may be caused by a reduction in bile acids (which are bacteriostatic) of a cirrhotic liver, which precipitates inflammation and immunosuppression, factors that can positively feedback on cirrhosis[42]. Dysbiosis may also arise from increased saprophytic fungal growth in the alimentary canal. Cirrhotic liver patients who routinely receive antimicrobial treatment have an overgrowth of fungi, especially Candida, leading to fungal-bacterial balance in the gut and worsening dysbiosis[42]. Although cirrhosis is a systemic disease, it is believed to be worsened by dysbiosis both in the gut liver axis and outside this axis, such as in saliva and serum[42,48].
While there is a knowledge gap on the use of microbial interventions for NAFLD therapy, there are data showing that nonalcoholic steatohepatitis (NASH) patients improve following treatment with the antibiotic rifaximin, which is used for the treatment of traveler’s diarrhea caused by Escherichia coli[49]. In a study examining the gut microbiota of stage 4 hepatitis C virus (HCV) patients, Prevotella and Faecalibacterium were found to be more abundant in HCV patients than in healthy controls, while Ruminococcus and some Clostridium species were more abundant in healthy controls than in HCV patients. Bifidobacterium was found only in healthy individuals[50]. Germ-free mice were shown to develop NAFLD following fecal microbial transplantation from donor hyperglycemic mice with systemic inflammation when fed a high-fat diet[51]. On the other hand, germ-free recipients that received fecal transplantation from normal donors (i.e., normoglycemic with negligible systemic inflammation) did not develop NAFLD and were normoglycemic when fed a high-fat diet[51]. Rabot et al[52] also showed that germ-free mice fed a high-fat diet were more resistant to hepatic steatosis than colonized controls. In an experimental mouse model of cholestasis-induced liver fibrosis induced either through bile duct ligation or by CCl4 treatments, colonization with complex microbiota (specific pathogen-free mice) was protective against severe fibrosis when compared to limited colonization (Altered Schaedler Flora)[53]. How the gut microbiota induces a leaky gut, bacteriaemia and an inflammatory flare leading to liver disease has been the subject of intense research. Brown and colleagues fed mice a high carbohydrate diet to induce a leaky gut[54]. This high carbohydrate diet caused a sloughing of the intestinal villi and reduced tight junction integrity, which allowed bacteria to translocate into the circulatory system[54]. In cirrhotic patients, it has been shown that microbial components leaking through the intestinal barrier, such as LPS, lipoteichoic acid, lipopolypeptides, and peptidoglycans, activate Toll-like receptors (TLRs) in hepatic stellate cells, Kupffer cells, and hepatocytes (all of which are differentially populated with TLRs 1-9), inducing severe inflammatory responses and fibrosis in the liver[43,55] Microbial activation of TLR2 in monocytes has especially been identified as significant in liver fibrosis through the production of TNF alpha, which initiates a cascade of reactions leading to increased gut permeability[43].
Gut microbiota-host crosstalk in liver disease remains widely unclear. However, in recent years, many studies have established a correlation between different microbial metabolites and liver disease[56]. LPS are gut microbiota-derived endotoxins that form the major component of the gram-negative bacterial outer cell wall. High plasma levels of LPS have been identified in NAFLD patients and are associated with gram-negative intestinal bacterial overgrowth and compromised gut lining epithelial tight junctions[57,58]. LPS induces an inflammatory response by activating hepatic Kupffer cells through TLR4. Apart from inducing proinflammatory cytokines and chemokines from hepatic Kupffer cells, LPS also activates hepatic stellate cells (HSCs) to differentiate into myofibroblast-like cells by producing extracellular matrix proteins, thus promoting liver fibrosis[49,59-61]. Other important metabolites are SCFAs from the fermentation of indigestible dietary fiber, which are mostly found in the colon, where most of them are produced and absorbed[62]. The major microbial fermentation products following microbial degradation of fiber are the SCFAs butyrate, propionate, and acetate. The body utilizes approximately 10% of the energy supply from microbially derived SCFAs, meaning that 90% is stored in white adipose tissue[63]. Several studies have revealed that gut microbial dysbiosis is associated with chronic liver diseases such as NAFLD or ALD[45,64]. In a metabolomic study in children with NASH, serum levels of 2-butanone and 4-methyl-2-pentanone were found to be elevated compared to those in healthy individuals[65]. Adults with NAFLD were found to have higher levels of fecal propionate and isobutyric acid, which are part of the fecal SCFA family[66]. Obese patients with NAFLD were also found to have high levels of propanoic acid and butanoic acid[67]. SCFAs such as acetate and butyrate modulate the host immune response by dampening the LPS-induced hepatocellular inflammatory response and restoring mucosal and systemic immunologic homeostasis, thus minimizing liver injury[68,69]. SCFAs can act as hormonal molecules by binding to G-protein-coupled receptors (GPCRs), which leads to activation of the GPCR pathway, slowing gut motility and increasing energy harvest[70-72]. Upon activation, glucagon-like peptide-1 is secreted from epithelial L-cells, enters circulation, and induces insulin release from the pancreas[70]. GPCR pathway activation also limits insulin-mediated hepatic and muscular fat accumulation and stimulates energy expenditure[71]. In adipocytes, SCFAs activate G protein-coupled receptor (GPR) 41 and GPR43 to inhibit lipolysis and activate adipocyte differentiation[70]. SCFAs also regulate immune cell functions through GPR43, which is widely expressed in most immune cells[73-75]. SCFAs have also been shown to inhibit histone deacetylases, which downregulate gene expression and reduce the production of inflammatory cytokines, particularly in macrophages and blood mononuclear cells during acute inflammatory hepatitis[69]. Therefore, it can be argued that dysbiosis that reduces microbial SCFA generation will result in a dysregulated inflammatory response and thus contribute to the progression of liver disease”
Indole and its derivatives are microbial metabolites of tryptophan breakdown. Indole upregulates tight junction proteins in the gut and downregulates colonic epithelium inflammatory genes through the aryl hydrocarbon receptor[76]. Indole-3-propionate activates pregnane X receptor to downregulate proinflammatory cytokine production and has been associated with protection against injury through oxidative stress signaling[76,77]. Indole-3-acetate has been shown to modulate hepatocyte lipogenesis, thus playing a protective role against NAFLD[78]. Microbial metabolism of dietary choline and L-carnitine produces trimethylamine (TMA), which is oxidized to trimethylamine N-oxide (TMAO) during hepatic detoxification of the blood through catalysis of the liver enzyme hepatic flavin monooxygenases[79]. TMAO is excreted in urine, and recent findings in animal NAFLD models fed a high-fat diet have shown increased urine levels of TMAO[80]. In a Chinese cohort study, the severity of NAFLD was closely associated with circulatory TMAO[81]. Bacteria are essential for the conversion of dietary choline to TMA, which is oxidized in the liver through the catalysis of hepatic flavin monooxygenase to generate trimethylamine-N-oxide, whose accumulation has been associated with both cardiac and renal disease[82,83]. Phosphatidylcholine is also metabolized by gut microbes to generate TMA, whose oxidation in the liver yields TMAO and, as previously described, may lead to kidney and cardiac disease[84,85]. It is now thought that accumulation of TMAO in the liver causes NASH through the inhibition of FXR and alteration of bile acid homeostasis[86]. SCFAs are significant microbial metabolites in the etiology of liver disease. More studies are required to target SCFAs as diagnostic or therapeutic tools for predicting or treating liver disease.
Liver disease is highly influenced by exposure to different environmental factors, which has recently been referred to as the exposome. It is now known that liver disease is impacted by an interaction between the genetic makeup of the host, exposome, and gut microbiome[87,88]. Certain types of gut microbiota have been associated with endogenous alcohol generation, which may in turn be hepatotoxic, leading to NASH[89]. The gut microbiota is important for the metabolism of bile acids, and in the absence or deficiency of bacteria that can convert primary bile acids to secondary bile acids, there is an accumulation of circulatory bile acids, which in turn activate TGR5, leading to monocyte dysfunction, which may exacerbate the hepatic inflammatory response and lead to liver disease[90]. High circulatory bile acids reflect a dysfunctional FXR, the nuclear receptor responsible for bile acid homeostasis, whose function is to facilitate enterohepatic bile acid circulation[91]. Dysbiosis affecting 7α-dehydroxylation-rich Firmicutes, which convert primary bile acids to FXR-low-binding secondary bile acids, will inevitably affect the function of FXR, leading to liver disease[92]. The liver is a crucial filter for toxins that find their way into the body either accidentally or deliberately. Alcohol is by far the most significant xenobiotic causing liver disease in humans, and it has been identified as the cause of ALD[93]. It can be argued that alcohol consumption causes both destruction of microbial communities and rupture of the barrier wall integrity in the gut and leads to induction of inflammation during detoxification in the liver. A compromised gut barrier leads to leakage of LPS and other microbial ligands into circulation, triggering inflammation of liver cells.
A high-fat diet and environmental pollutants are further risk factors for liver disease, and their effects are exacerbated by microbial metabolites[94,95]. It is likely that most xenobiotics, in addition to being directly toxic to hepatic cells, will cause dysbiosis that favors changes in microbial composition that generate toxic liver disease-causing metabolites. The changes in these microbial metabolites may therefore be used as noninvasive diagnostic biomarkers for liver disease[96] but may also become significant therapeutic targets for the treatment of this disease[96-98]. A high carbohydrate diet has been demonstrated in environmental enteropathy animal models to lead to intestinal wall epithelial brush-border shortening and loosening of tight junctions[54]. Furthermore, small intestinal gram-negative bacterial overgrowth and high plasma LPS levels can lead to liver disease[56]. In the absence of dietary fibers, the gut microbiota cannot produce sufficient SCFAs, which may lead to a dysregulated inflammatory response and liver disease[99,100]. Most liver metabolism occurs through the catalysis of cytochrome P-450 (CYP-450), and it is known that many dietary biproducts can influence the activity of CYP-450[101]. Dietary retinoids, for example, are metabolized by hepatic cells, including hepatic stellate cells. An alteration in the uptake and metabolism of retinoids may influence retinoic acid signaling, which may activate hepatic stellate cells, resulting in loss of retinoid stores, aberrant extracellular matrix generation and the onset of fibrosis, which inevitably precipitate liver disease[102]. Additionally, alcohol consumption affects hepatic retinoid metabolism through inhibition of retinoid oxidation, induction of CYP2E1 enzymes to increase retinoic acid metabolism, or increased peripheral tissue damping of retinoic acid, all of which leads to activation of hepatic stellate cells and development of liver disease[103]. Retinoic acid is a gut microbial metabolite of vitamin A whose intestinal concentration is modulated by suppression of retinol dehydrogenase 7 expression by commensal Clostridia microbes[104]. Retinoic acid not only regulates bile acid homeostasis but also shares with it the receptors retinoid X receptor and FXR and therefore shares the functions of lipid metabolism and insulin sensitivity[105]. In a rat model, a high-fat diet in combination with high glucocorticoid treatment resulted in a fourfold hepatic lipid deposition and an almost threefold increase in circulatory alanine aminotransferase indicative of liver injury[106]. A high-fat diet also caused severe liver damage with high levels of circulatory alanine transaminases (ALT) and aspartate aminotransferases (AST) in a mouse model[107]. Mice fed a high-fat diet developed high intestinal gram-negative microbial growth and an increase in ethanol-producing bacteria when compared to mice fed normal chow[107]. This result is consistent with findings from clinical studies where it has been documented that microbial diversity rapidly changes with a change in diet[108]. Therefore, it can be concluded that diet, food additives, and xenobiotics affect liver disease by influencing gut microbial composition, gut permeability, and microbial metabolites. The liver plays a major role in metabolism and blood detoxification and is thus prone to damage from microbial endotoxins, environmental toxins, and microbial dietary metabolites, all of which work together in cascaded inflammatory responses to cause liver injury. Understanding the individualized microbial signatures and their influence on gut permeability, immunologic inflammatory responses, and the hepatic response to insult will expose multientry avenues to precision liver disease therapy.
The intestine is heavily colonized with microbiota, yet the surrounding tissues remain sterile. This barrier is maintained by intricate crosstalk between gut microbes, the gut wall epithelium, and the innate immune system[109,110]. The expression of intercellular tight junction proteins between the intestinal epithelium is regulated by cytokines such as interferon gamma and TNF and other regulatory cytokines that interact with immunoglobulin A (IgA)-coated gut microbiota to maintain gut and immune homeostasis[111]. A change in diet or intake of xenobiotics such as alcohol, prescription/over-the-counter drugs, or other environmental chemicals may lead to destabilization of the intestinal homeostatic environment either through selective overgrowth or reduction of specific microbial strains or injury to the mucosal lining. A destabilization of the homeostatic environment will give way to a shift in the immunological signaling molecules protective to the gut tight junctions and a sloughing of intestinal villi. This breach in the barrier allows leakage of microbial endotoxins into circulation and microbial translocation into the liver, thus triggering an immunological inflammatory response once the microbial products are detected by the liver’s pathogen recognition receptors, mainly the TLR and nucleotide oligomerization domain-like receptors[109,112]. HSCs are endowed with TLR 2, 4, and 9, which are associated with promoting TLR4 fibrosis[43,60]. Kupffer cells are lined with TLR 2, 3, 4, and 9 and are hepatic macrophages that form the main targets of microbial ligands within the liver[43]. Furthermore, hepatocytes express TLR 1-9 and are the most abundant cells in the liver, playing a critical role in the acute phase of the immunologic response through cytokine-like IL-6[113]. The inflammatory response of the liver to leaked gut-microbial endotoxins is not yet fully understood. However, it is known that upon activation, Kupffer cells release proinflammatory and profibrogenic cytokines, such as TNF-α, Transforming Growth Factor (TGF)-β and IL-1β, and a few more members of the inflammasome whose effect is to induce inflammation and accumulation of lipids in the liver, and if this is not resolved, it leads to fibrosis NAFLD[114]. Therapeutic target efforts are geared toward minimizing the hepatic inflammation seen after proinflammatory cytokine release. Chemokine receptor antagonists such as C-C motif chemochine receptor (CCR) 2 and CCR5 [Cenicrivinoc (CVC)] have been used with some success to decrease leukocyte infiltration, and when used in a diet-induced NASH mouse model and a thioacetamide-induced fibrosis rat model, liver fibrosis was effectively reduced[115,116]. This outcome has since been replicated in phase 2 clinical trials with a remarkable reduction in fibrosis[117]. Several other proinflammatory cytokines, including IL-17, IL-11, and IL-1, are still under investigation. A clinical trial therapy utilizing an IL-1 pathway anti-inflammatory drug, diacerelin, achieved a remarkable reduction in fibrosis in NAFLD patients with diabetes[118].
The dynamics of the gut microbiome could be used as a noninvasive diagnostic tool for liver cirrhosis and hepatocellular carcinoma (HCC)[119]. In a cross-regional prospective validation study in China, human fecal samples analyzed for microbial diversity revealed a significant rise in diversity as the liver condition advanced from cirrhosis to HCC with cirrhosis[119]. There was also a high level of butyrate-producing bacteria in healthy controls relative to early cirrhosis patients and a notable rise in LPS-producing bacteria in HCC patients[119]. In a different experiment, gut microbiota known to originate from the oral cavity were found to be enriched in liver cirrhosis patients relative to healthy volunteers[120]. In an Asian NAFLD cohort, Ruminococcaceae and Veillonellaceae species were found to be more predominant in NAFLD patients relative to healthy individuals[121]. These microbiome changes could not be associated with genetic predispositions known to influence NAFLD and were thought to be environmentally driven[121]. Bacteroides and Escherichia spp. have, on the other hand, been associated with liver fibrosis in NAFLD patients[122]. Overall, these multiregional studies indicate that there is great potential for the gut microbiota as a noninvasive diagnostic biomarker for liver disease with distinct indications of the staging of fibrosis and inflammation[121,123]. There is also great potential for the gut microbiota and associated metabolites to be utilized as therapeutic biomarkers[119-121]. It must, however, be appreciated that as of yet, a single microbial signature indicative of liver disease does not exist mainly because disease outcome is influenced by multiple factors such as diet, genetic background, age, and lifestyle (such as alcohol consumption), all of which must be considered while interpreting data on the predictive value of fecal microbiota on liver disease[124].
As we have discussed above, dysbiosis and a dysfunctional gut barrier promote the leakage of microbial endotoxins and components, as well as bile acid metabolites, into circulation, which can eventually lead to liver injury. Various therapeutic approaches (which are at various stages of testing) could be used to address these different factors for the treatment or prevention of liver disease, which will be highlighted below. Although SCFA supplements could be an attractive therapeutic approach in liver disease, their taste is normally not well tolerated. However, methods such as microencapsulation[125], either as soft gels or liquid capsules, are available that mask the taste of bitter medications and could be used for oral delivery of SCFA, which has the added benefit of being slow release and helps prevent evaporation of some volatile SCFAs, such as butyrate. Butyrate enemas have been used in a rat model, with the treatment group showing improved mucosal repair and reduced colonic damage compared to the untreated control groups[126]. However, butyrate enemas did not show any improvement in clinical studies with ulcerative colitis patients[127]. There is potential for the use of SCFA as a therapeutic approach, but more research is required to develop an optimal approach. Prebiotics such as inulin represent a substitute approach for the supply of SCFAs[98]. Multiple agonists of FXR are under investigation, including GS-9674 and LJN452, in phase 2 trials for NASH[98]. Some fibroblast growth factors (FGFs), such as FGF19 and FGF21, have shown encouraging results for NAFLD therapy[128,129].
Treating dysbiosis and restoring homeostasis is complicated due to the wide range of associated factors that lead to a loss of important microbial populations or diversity in the first place. In most cases, treating dysbiosis with a single approach usually gives discouraging outcomes. However, studies involving probiotics have shown encouraging results in terms of safety, tolerance, and efficacy[130]. In a Phase 1 clinical trial, Lactobacillus rhamnosus GG administered to cirrhotic patients resulted in reduced Enterobacteraceae and increased relative abundance of Clostridiales incertae Sedis XIV and Lachnospiriceae with reduced endotoxemia and decreased pathogenic bacterial growth indicative of improved health[131]. In another study using multiple probiotic strains, a reduction in inflammatory cytokine flares in cirrhotic patients was observed[132]. In obese, sonographically identified NAFLD children, treatment with a probiotic combination of Bifidobacteria (B. bifidum and B. lactis) and two Lactobacilli (L. rhamnosus DSMZ 21690 and L. acidophilus) strains significantly lowered intrahepatic fat content and ALT levels as well as AST relative to the placebo treatment[133]. This reduction in hepatic steatosis was replicated in NAFLD patients treated with a multistrain probiotic[134]. In another study, a twelve-week treatment of 30 NAFLD volunteers with six strains of bacteria containing Bifidobacterium breve and B. lactis, Lactobacillus rhamnosus, L. acidophilus and L. paracasei pacasei and Pediococcus pentosaceus in a randomized, double-blind, placebo-controlled study led to an improvement in proinflammatory cytokines, a reduction in cholesterol and a decrease in body weight[135]. When probiotics are mixed with compatible prebiotics, better outcomes have been achieved in clinical trials, but more studies are needed to determine the most effective combinations[136,137]. Hepatic steatosis has, for example, been reported to decrease in patients with NASH following symbiotic and prebiotic treatment. Serum alkaline phosphatase was decreased following treatment with probiotics, prebiotics and synbiotics[136] However, it is noteworthy that the outcomes are dependent on the composition of probiotics, the exposure time, and the dosage[136]. Studies in animal models have shown similar outcomes as in human studies. In rats fed a high-fat diet, treatment with Bifidobacteria longum or Lactobacillus acidophilus significantly reduced hepatic fat accumulation[138]. There was also a strong negative correlation between fat liver content and probiotic concentration in the stool[138]. In addition, hepatic steatosis was markedly reduced after 12 wk of treatment with B. longum, but this was not the case with L. acidophilus treatment[138]. In a diabetic rat model, treatment with Akkermansia muciniphila led to a decreased inflammatory response and improved liver function[139]. In hepatic encephalopathy, a mixture of Lactobacillus plantarum, L. casei, L. delbrueckii subsp. Bulgaricus, Bifidobacterium infantis, B. longum, B, breve, and Streptococcus salivarius subsp. Thermophilius has been associated with both primary and secondary prophylaxis[140,141]. Yogurts containing L. bulgaricus, S. thermophilus, L. acidopilus La5 and B. lactis Bb12 as well as a prebiotic mixture of fruco-oligosaccharides and L. casei, L. rhamnosus, S. thermophilus, B. breve, L. acidophilus, B. longum, and L. bulgaricus have been shown to improve aminotransferase in NAFLD patients[142-144]. In NASH patients, probiotics containing L. bulgaricus and S. thermophilus have also shown improvement in aminotransferase[145]. A combination of B. longum W11 and fructooligosaccharides, on the other hand, has shown improvement in aminotransferase and the histological score activity of NASH patients[146]
Fecal microbiota transplantation (FMT) is the administration of a solution containing fecal material from a “healthy” donor into the intestinal tract of a recipient to modify that recipient’s gut microbial composition for targeted health benefits[147]. To date, FMT has been successfully used in the treatment of recurrent Clostridium difficile infection, and there is growing evidence that FMT can be used to treat noninfectious diseases such as inflammatory bowel disease, obesity, and other metabolic disorders [147]. FMT has also been tried as a therapeutic option for liver disease. In a diet-induced steatohepatitis mouse model, FMT-treated mice showed increased SCFAs, improved expression of tight junction proteins, reduced proinflammatory cytokines and less intrahepatic lipid deposition compared to controls (i.e., no FMT)[148]. There have also been several human clinical trials but with mixed outcomes, with some achieving a significant reduction in proinflammatory cytokines and improved gut barrier function and others not responding to therapy[149,150]. Future experiments should address the question of who qualifies as a healthy donor, how should we deal with the variation in gut microbial diversity among the recipients, and how best to package the product for better acceptability.
A recent study in mice indicated that during antibiotic-induced dysbiosis, the homeostasis of bile acids was equally destabilized[151,152]. Treatment of these mice with flavanones and total phenolic extracts of citrus aurantium L. (TPE-CA) restored bile acid homeostasis and gut barrier integrity[152]. TPE-CA also regulates the enterohepatic circulation entry of bile acids through the farnesoid X receptor-fibroblast-growth factor 15 pathway[152]. The effects of dysbiosis and increased intestinal unconjugated bile acid that are observed in ALD were reversed through improved FXR activity and gut barrier function following treatment with fexaramine, which is an intestine restricted FXR agonist. These results indicate that modulation of cyp7a1 and lipid metabolism can be achieved in a mouse model and thereby minimize ethanol-derived liver damage by targeting the bile acid-FXR-fibroblast-growth factor 15 signaling pathway[153]. Future experiments to verify these findings in higher mammals and translate the results to therapeutic interventions for human liver disease are warranted.
The mechanisms by which the intestinal microbiota influences the development and/or progression of liver disease are only beginning to unfold, but to fully elucidate the microbiome role in liver disease, a more comprehensive picture of the dynamics of the gut ecosystem is needed. Unfortunately, most of our knowledge about the intestinal microbiota arises from fecal or biopsy sample analysis, which is not representative of the entire gut microbiome. However, novel technologies are being developed to address this knowledge gap. One such innovation is a capsule sampler and drug delivery system that is swallowed and utilizes mechanical gut peristaltic movements to guide the capsule down the entire length of the gut as the capsule collects samples[154]. Recently, a capsule robot was designed from a shape memory alloy spring with a chamber of a storage capacity of 500 µL, which showed enhanced sample preservation[155]. Another approach consists of an inexpensive 3D-printed sampler containing a hydrogel whose swelling ability seal and protects the liquid gut samples[156]. Such strategies that analyze small samples from various sites will provide information on microbiota distribution and will make microbial engineering and microbial targeting more feasible.
One such microbial engineering approach being developed is the use of Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR) Cas-based instructions to precisely cut off targeted genetic sequences of the microbial genome and thus change their function in vivo[157]. A conjugative plasmid, TP114, was recently used as a delivery vehicle for CRISPR-Cas9, targeted at drug-resistant Escherichia coli and Citrobacter rodentium, which led to full clearance of these organisms in a mouse model four days after administration[157]. More recent delivery systems for CRISPR-Cas9 have been designed to utilize probiotics as a genetically engineered conjugative vehicle that are more efficient and practical to use than bacteriophage-based systems[158,157]. The use of CRISPR-Cas9 as antimicrobial therapy is still in its early stages but has the potential to be an effective therapy for targeting specific, undesired microbes in the dysbiotic gut of liver disease. Other approaches to manipulate the gut microbiome are mucosal vaccines. IgA is the predominant antibody in the gut that binds to pathogens and commensals, preventing their translocation across the mucosal barrier. Using a probiotic-based mucosal vaccine with Lactobacillus acidophilus, Fox et al[159] showed that a potent, diverse IgA response could be elicited, which could help with colonization resistance. In another study, Slack and colleagues designed an oral vaccine using genetically modified Salmonella enterica capable of setting evolutionary traps for prophylaxis treatment in a mouse model[160,161]. While this technology was advanced into a pig model and is currently being tested on human neonates to treat neonatal sepsis and necrotizing enterocolitis, it has hallmarks to be equally beneficial as therapeutic approaches for liver disease.
There are many therapeutic options for NAFLD that are being explored, some of which are in advanced levels of clinical trials; however, no treatment is yet available[124]. Diet and lifestyle changes remain the most effective methods of managing liver disease[162]. Low caloric diets, low carbohydrate intake and low protein diets have all been shown to be effective in the management of liver disease[163,162]. It should, however, be noted that dietary changes alone cannot achieve the intended long-term weight loss goals to reduce liver inflammation. It is rather a combination of correct diet and exercise that is most effective against NAFLD[162]. The response to dietary changes and exercise on both gut microbiota that are negatively associated with liver disease and the amount of fat in the liver is different between individuals and between races[164] The amount of Bacteroides, for example, is lower in Chinese NAFLD individuals after diet and exercise compared to people from the West, and this is correlated with lower hepatic fat[164]. It has also been noted that Bacteroides increases in obese volunteers but decreases in lean volunteers following exercise and diet intervention[165]. This is suggestive of personalized intervention approaches of diet and lifestyle changes[164].
The influence of the gut microbiome on various body systems has important implications for health and disease, such as liver disease. While the exact mechanisms by which the microbiome contributes to liver disease are unknown, there is strong evidence that translocation of various metabolites across the mucosal barrier plays a strong role, which is precipitated by a dysbiotic gut microbiota. Considering the importance of the microbiome in liver disease, powerful therapeutic options that can manipulate the gut microbiome are being explored. These approaches could have the potential for effective treatments for various stages of liver disease. More research needs to be done to understand the crosstalk between the microbiome and host as it relates to liver disease so that more effective and targeted preventative and therapeutic options can be developed.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ogino S, United States; Patno N, United States S-Editor: Liu GL L-Editor: A P-Editor: Liu GL
| 1. | Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol. 2020;72:558-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 1266] [Article Influence: 253.2] [Reference Citation Analysis (1)] |
| 2. | Kisseleva T, Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 1172] [Article Influence: 293.0] [Reference Citation Analysis (0)] |
| 3. | Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971-11975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2922] [Cited by in RCA: 3171] [Article Influence: 211.4] [Reference Citation Analysis (0)] |
| 4. | Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 665] [Cited by in RCA: 951] [Article Influence: 105.7] [Reference Citation Analysis (0)] |
| 5. | Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol. 2012;2:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 231] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 6. | Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, Zhuo N, Wang H, Wang L, Wu D. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci Rep. 2020;10:15792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 176] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 7. | Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, Marchesi JR, Collado MC. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis. 2015;26:26050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 455] [Cited by in RCA: 586] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 8. | Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev. 2017;81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1147] [Article Influence: 143.4] [Reference Citation Analysis (0)] |
| 9. | Iebba V, Totino V, Gagliardi A, Santangelo F, Cacciotti F, Trancassini M, Mancini C, Cicerone C, Corazziari E, Pantanella F, Schippa S. Eubiosis and dysbiosis: the two sides of the microbiota. New Microbiol. 2016;39:1-12. [PubMed] |
| 10. | Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, Corthier G, Furet JP. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 997] [Cited by in RCA: 1246] [Article Influence: 77.9] [Reference Citation Analysis (0)] |
| 11. | Manor O, Dai CL, Kornilov SA, Smith B, Price ND, Lovejoy JC, Gibbons SM, Magis AT. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun. 2020;11:5206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 332] [Cited by in RCA: 506] [Article Influence: 101.2] [Reference Citation Analysis (0)] |
| 12. | Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P, Balamurugan R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 1317] [Article Influence: 263.4] [Reference Citation Analysis (0)] |
| 13. | Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun. 2009;77:2876-2886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 160] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 14. | Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One. 2013;8:e53957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 198] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 15. | Momose Y, Hirayama K, Itoh K. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie Van Leeuwenhoek. 2008;94:165-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 267] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 17. | Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology (Reading). 2003;149:2557-2570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 178] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 18. | Baquero F, Lanza VF, Baquero MR, Del Campo R, Bravo-Vázquez DA. Microcins in Enterobacteriaceae: Peptide Antimicrobials in the Eco-Active Intestinal Chemosphere. Front Microbiol. 2019;10:2261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 19. | Javvadi SG, Kujawska M, Papp D, Jordan A, Lawson MAE, Clarke P, Beraza N, Hall LJ, Microbes G, Microbiome I, Kingdom U. A novel bacteriocin produced by Bifidobacterium longum subsp. infantis has dual antimicrobial and immunomodulatory activity. 2022 Preprint. Available from: bioRxiv. [DOI] [Full Text] |
| 20. | Johansson ME, Ambort D, Pelaseyed T, Schütte A, Gustafsson JK, Ermund A, Subramani DB, Holmén-Larsson JM, Thomsson KA, Bergström JH, van der Post S, Rodriguez-Piñeiro AM, Sjövall H, Bäckström M, Hansson GC. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci. 2011;68:3635-3641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 384] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 21. | Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4659-4665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 1019] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 22. | Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, Roos S, Holm L, Phillipson M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327-G333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 275] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 23. | Geirnaert A, Steyaert A, Eeckhaut V, Debruyne B, Arends JB, Van Immerseel F, Boon N, Van de Wiele T. Butyricicoccus pullicaecorum, a butyrate producer with probiotic potential, is intrinsically tolerant to stomach and small intestine conditions. Anaerobe. 2014;30:70-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 24. | Van Immerseel F, Ducatelle R, De Vos M, Boon N, Van De Wiele T, Verbeke K, Rutgeerts P, Sas B, Louis P, Flint HJ. Butyric acid-producing anaerobic bacteria as a novel probiotic treatment approach for inflammatory bowel disease. J Med Microbiol. 2010;59:141-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 25. | Geirnaert A, Calatayud M, Grootaert C, Laukens D, Devriese S, Smagghe G, De Vos M, Boon N, Van de Wiele T. Butyrate-producing bacteria supplemented in vitro to Crohn's disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci Rep. 2017;7:11450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 227] [Cited by in RCA: 320] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 26. | Iacob S, Iacob DG, Luminos LM. Intestinal Microbiota as a Host Defense Mechanism to Infectious Threats. Front Microbiol. 2018;9:3328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 27. | Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, Hermoso MA. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol. 2019;10:277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 970] [Cited by in RCA: 2182] [Article Influence: 363.7] [Reference Citation Analysis (0)] |
| 28. | Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, Looijer-van Langen M, Madsen KL. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1025-G1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 413] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 29. | Rose EC, Odle J, Blikslager AT, Ziegler AL. Probiotics, Prebiotics and Epithelial Tight Junctions: A Promising Approach to Modulate Intestinal Barrier Function. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 126] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 30. | Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141:769-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 855] [Article Influence: 61.1] [Reference Citation Analysis (1)] |
| 31. | Bergmann KR, Liu SX, Tian R, Kushnir A, Turner JR, Li HL, Chou PM, Weber CR, De Plaen IG. Bifidobacteria stabilize claudins at tight junctions and prevent intestinal barrier dysfunction in mouse necrotizing enterocolitis. Am J Pathol. 2013;182:1595-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 191] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 32. | Guo S, Chen S, Ma J, Ma Y, Zhu J, Liu Y, Wang P, Pan Y. Escherichia coli Nissle 1917 Protects Intestinal Barrier Function by Inhibiting NF-κB-Mediated Activation of the MLCK-P-MLC Signaling Pathway. Mediators Inflamm. 2019;2019:5796491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 33. | Bruewer M, Samarin S, Nusrat A. Inflammatory bowel disease and the apical junctional complex. Ann N Y Acad Sci. 2006;1072:242-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 164] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 34. | Mohammad S, Thiemermann C. Role of Metabolic Endotoxemia in Systemic Inflammation and Potential Interventions. Front Immunol. 2020;11:594150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 261] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 35. | Dawson PA, Karpen SJ. Intestinal transport and metabolism of bile acids. J Lipid Res. 2015;56:1085-1099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 413] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 36. | Ghosh S, Whitley CS, Haribabu B, Jala VR. Regulation of Intestinal Barrier Function by Microbial Metabolites. Cell Mol Gastroenterol Hepatol. 2021;11:1463-1482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 402] [Article Influence: 100.5] [Reference Citation Analysis (0)] |
| 37. | Stenman LK, Holma R, Korpela R. High-fat-induced intestinal permeability dysfunction associated with altered fecal bile acids. World J Gastroenterol. 2012;18:923-929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 120] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 38. | Raimondi F, Santoro P, Barone MV, Pappacoda S, Barretta ML, Nanayakkara M, Apicella C, Capasso L, Paludetto R. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am J Physiol Gastrointest Liver Physiol. 2008;294:G906-G913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 213] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 39. | de Diego-Cabero N, Mereu A, Menoyo D, Holst JJ, Ipharraguerre IR. Bile acid mediated effects on gut integrity and performance of early-weaned piglets. BMC Vet Res. 2015;11:111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 40. | Fouts DE, Torralba M, Nelson KE, Brenner DA, Schnabl B. Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J Hepatol. 2012;56:1283-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 252] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 41. | Yan AW, Fouts DE, Brandl J, Stärkel P, Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA, Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53:96-105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 668] [Cited by in RCA: 638] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 42. | Albhaisi SAM, Bajaj JS, Sanyal AJ. Role of gut microbiota in liver disease. Am J Physiol Gastrointest Liver Physiol. 2020;318:G84-G98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 43. | Brenner DA, Paik YH, Schnabl B. Role of Gut Microbiota in Liver Disease. J Clin Gastroenterol. 2015;49 Suppl 1:S25-S27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 44. | Balmer ML, Slack E, de Gottardi A, Lawson MA, Hapfelmeier S, Miele L, Grieco A, Van Vlierberghe H, Fahrner R, Patuto N, Bernsmeier C, Ronchi F, Wyss M, Stroka D, Dickgreber N, Heim MH, McCoy KD, Macpherson AJ. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med. 2014;6:237ra66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 350] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 45. | Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, Hu Y, Li J, Liu Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 473] [Cited by in RCA: 453] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 46. | Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, Jones MB, Sirlin CB, Schnabl B, Brinkac L, Schork N, Chen CH, Brenner DA, Biggs W, Yooseph S, Venter JC, Nelson KE. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25:1054-1062.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 754] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 47. | Wang B, Jiang X, Cao M, Ge J, Bao Q, Tang L, Chen Y, Li L. Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Sci Rep. 2016;6:32002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 274] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 48. | Trebicka J, Macnaughtan J, Schnabl B, Shawcross DL, Bajaj JS. The microbiota in cirrhosis and its role in hepatic decompensation. J Hepatol. 2021;75 Suppl 1:S67-S81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 146] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 49. | Gangarapu V, Ince AT, Baysal B, Kayar Y, Kılıç U, Gök Ö, Uysal Ö, Şenturk H. Efficacy of rifaximin on circulating endotoxins and cytokines in patients with nonalcoholic fatty liver disease. Eur J Gastroenterol Hepatol. 2015;27:840-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 50. | Aly AM, Adel A, El-Gendy AO, Essam TM, Aziz RK. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016;8:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 51. | Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, Martin P, Philippe C, Walker F, Bado A, Perlemuter G, Cassard-Doulcier AM, Gérard P. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787-1794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 601] [Cited by in RCA: 715] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 52. | Rabot S, Membrez M, Bruneau A, Gérard P, Harach T, Moser M, Raymond F, Mansourian R, Chou CJ. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24:4948-4959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 382] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 53. | Moghadamrad S, Hassan M, McCoy KD, Kirundi J, Kellmann P, De Gottardi A. Attenuated fibrosis in specific pathogen-free microbiota in experimental cholestasis- and toxin-induced liver injury. FASEB J. 2019;33:12464-12476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 54. | Brown EM, Wlodarska M, Willing BP, Vonaesch P, Han J, Reynolds LA, Arrieta MC, Uhrig M, Scholz R, Partida O, Borchers CH, Sansonetti PJ, Finlay BB. Diet and specific microbial exposure trigger features of environmental enteropathy in a novel murine model. Nat Commun. 2015;6:7806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 166] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 55. | Oliveira-Nascimento L, Massari P, Wetzler LM. The Role of TLR2 in Infection and Immunity. Front Immunol. 2012;3:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 372] [Cited by in RCA: 547] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 56. | Zhao ZH, Lai JK, Qiao L, Fan JG. Role of gut microbial metabolites in nonalcoholic fatty liver disease. J Dig Dis. 2019;20:181-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 57. | Thuy S, Ladurner R, Volynets V, Wagner S, Strahl S, Königsrainer A, Maier KP, Bischoff SC, Bergheim I. Nonalcoholic fatty liver disease in humans is associated with increased plasma endotoxin and plasminogen activator inhibitor 1 concentrations and with fructose intake. J Nutr. 2008;138:1452-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 309] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 58. | Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, Mascianà R, Forgione A, Gabrieli ML, Perotti G, Vecchio FM, Rapaccini G, Gasbarrini G, Day CP, Grieco A. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49:1877-1887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1133] [Cited by in RCA: 1103] [Article Influence: 68.9] [Reference Citation Analysis (1)] |
| 59. | Seki E, Tsutsui H, Nakano H, Tsuji N, Hoshino K, Adachi O, Adachi K, Futatsugi S, Kuida K, Takeuchi O, Okamura H, Fujimoto J, Akira S, Nakanishi K. Lipopolysaccharide-induced IL-18 secretion from murine Kupffer cells independently of myeloid differentiation factor 88 that is critically involved in induction of production of IL-12 and IL-1beta. J Immunol. 2001;166:2651-2657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 187] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 60. | Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1361] [Cited by in RCA: 1557] [Article Influence: 86.5] [Reference Citation Analysis (1)] |
| 61. | Hritz I, Mandrekar P, Velayudham A, Catalano D, Dolganiuc A, Kodys K, Kurt-Jones E, Szabo G. The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology. 2008;48:1224-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 332] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 62. | Rahat-Rozenbloom S, Fernandes J, Gloor GB, Wolever TM. Evidence for greater production of colonic short-chain fatty acids in overweight than lean humans. Int J Obes (Lond). 2014;38:1525-1531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 216] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 63. | Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1559] [Cited by in RCA: 1679] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 64. | Wieland A, Frank DN, Harnke B, Bambha K. Systematic review: microbial dysbiosis and nonalcoholic fatty liver disease. Aliment Pharmacol Ther. 2015;42:1051-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 164] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 65. | Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D, Furlanello C, Zandonà A, Paci P, Capuani G, Dallapiccola B, Miccheli A, Alisi A, Putignani L. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65:451-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 529] [Article Influence: 66.1] [Reference Citation Analysis (0)] |
| 66. | Rau M, Rehman A, Dittrich M, Groen AK, Hermanns HM, Seyfried F, Beyersdorf N, Dandekar T, Rosenstiel P, Geier A. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J. 2018;6 1496-1507 [PMID:30574320 DOI: 10.1177/2050640618804444. |
| 67. | Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Myers RP, Rioux KP. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:868-75.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 522] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 68. | Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016;5:e73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 596] [Cited by in RCA: 885] [Article Influence: 98.3] [Reference Citation Analysis (0)] |
| 69. | Visekruna A, Luu M. The Role of Short-Chain Fatty Acids and Bile Acids in Intestinal and Liver Function, Inflammation, and Carcinogenesis. Front Cell Dev Biol. 2021;9:703218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 70. | van der Hee B, Wells JM. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021;29:700-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 564] [Article Influence: 141.0] [Reference Citation Analysis (0)] |
| 71. | Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, Terasawa K, Kashihara D, Hirano K, Tani T, Takahashi T, Miyauchi S, Shioi G, Inoue H, Tsujimoto G. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 833] [Cited by in RCA: 1084] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 72. | Towle HC. Glucose and cAMP: adversaries in the regulation of hepatic gene expression. Proc Natl Acad Sci U S A. 2001;98:13476-13478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 73. | Silva YP, Bernardi A, Frozza RL. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol (Lausanne). 2020;11:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 780] [Cited by in RCA: 1611] [Article Influence: 322.2] [Reference Citation Analysis (0)] |
| 74. | Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol. 2019;16:461-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 850] [Cited by in RCA: 1856] [Article Influence: 309.3] [Reference Citation Analysis (1)] |
| 75. | Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1076] [Cited by in RCA: 1616] [Article Influence: 146.9] [Reference Citation Analysis (0)] |
| 76. | Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1512] [Cited by in RCA: 1724] [Article Influence: 143.7] [Reference Citation Analysis (1)] |
| 77. | Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med. 2016;8:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 371] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 78. | Krishnan S, Ding Y, Saeidi N, Choi M, Sridharan GV, Sherr DH, Yarmush ML, Alaniz RC, Jayaraman A, Lee K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2019;28:3285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 79. | Velasquez MT, Ramezani A, Manal A, Raj DS. Trimethylamine N-Oxide: The Good, the Bad and the Unknown. Toxins (Basel). 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 360] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 80. | Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Mitchell SC, Holmes E, McCarthy MI, Scott J, Gauguier D, Nicholson JK. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511-12516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 810] [Cited by in RCA: 804] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 81. | Chen YM, Liu Y, Zhou RF, Chen XL, Wang C, Tan XY, Wang LJ, Zheng RD, Zhang HW, Ling WH, Zhu HL. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci Rep. 2016;6:19076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 254] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 82. | Rath S, Rud T, Pieper DH, Vital M. Potential TMA-Producing Bacteria Are Ubiquitously Found in Mammalia. Front Microbiol. 2019;10:2966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 83. | Tang WH, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B, Li XS, Levison BS, Hazen SL. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116:448-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 918] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 84. | Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio. 2015;6:e02481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 396] [Cited by in RCA: 557] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 85. | Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57-63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4469] [Cited by in RCA: 4061] [Article Influence: 290.1] [Reference Citation Analysis (0)] |
| 86. | Tan X, Liu Y, Long J, Chen S, Liao G, Wu S, Li C, Wang L, Ling W, Zhu H. Trimethylamine N-Oxide Aggravates Liver Steatosis through Modulation of Bile Acid Metabolism and Inhibition of Farnesoid X Receptor Signaling in Nonalcoholic Fatty Liver Disease. Mol Nutr Food Res. 2019;63:e1900257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 154] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 87. | Jones DP. Sequencing the exposome: A call to action. Toxicol Rep. 2016;3:29-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 88. | Wild CP. Complementing the genome with an "exposome": the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847-1850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1198] [Cited by in RCA: 1446] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 89. | Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1283] [Article Influence: 106.9] [Reference Citation Analysis (1)] |
| 90. | Leonhardt J, Haider RS, Sponholz C, Leonhardt S, Drube J, Spengler K, Mihaylov D, Neugebauer S, Kiehntopf M, Lambert NA, Kortgen A, Bruns T, Tacke F, Hoffmann C, Bauer M, Heller R. Circulating Bile Acids in Liver Failure Activate TGR5 and Induce Monocyte Dysfunction. Cell Mol Gastroenterol Hepatol. 2021;12:25-40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 91. | Armstrong LE, Guo GL. Role of FXR in Liver Inflammation during Nonalcoholic Steatohepatitis. Curr Pharmacol Rep. 2017;3:92-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 92. | Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74:2959-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 437] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 93. | Osna NA, Donohue TM Jr, Kharbanda KK. Alcoholic Liver Disease: Pathogenesis and Current Management. Alcohol Res. 2017;38:147-161. [PubMed] |
| 94. | Rosenfeld CS. Gut Dysbiosis in Animals Due to Environmental Chemical Exposures. Front Cell Infect Microbiol. 2017;7:396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 95. | Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016;375:2369-2379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1826] [Cited by in RCA: 2326] [Article Influence: 258.4] [Reference Citation Analysis (0)] |
| 96. | Sharpton SR, Schnabl B, Knight R, Loomba R. Current Concepts, Opportunities, and Challenges of Gut Microbiome-Based Personalized Medicine in Nonalcoholic Fatty Liver Disease. Cell Metab. 2021;33:21-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 97. | Hyun JY, Kim SK, Yoon SJ, Lee SB, Jeong JJ, Gupta H, Sharma SP, Oh KK, Won SM, Kwon GH, Cha MG, Kim DJ, Ganesan R, Suk KT. Microbiome-Based Metabolic Therapeutic Approaches in Alcoholic Liver Disease. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 98. | Dai X, Hou H, Zhang W, Liu T, Li Y, Wang S, Wang B, Cao H. Microbial Metabolites: Critical Regulators in NAFLD. Front Microbiol. 2020;11:567654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 99. | Campo L, Eiseler S, Apfel T, Pyrsopoulos N. Fatty Liver Disease and Gut Microbiota: A Comprehensive Update. J Clin Transl Hepatol. 2019;7:56-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 100. | Kendrick SF, O'Boyle G, Mann J, Zeybel M, Palmer J, Jones DE, Day CP. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis. Hepatology. 2010;51:1988-1997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 136] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 101. | Mega A, Marzi L, Kob M, Piccin A, Floreani A. Food and Nutrition in the Pathogenesis of Liver Damage. Nutrients. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 102. | Shirakami Y, Lee SA, Clugston RD, Blaner WS. Hepatic metabolism of retinoids and disease associations. Biochim Biophys Acta. 2012;1821:124-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 103. | Liu C, Russell RM, Seitz HK, Wang XD. Ethanol enhances retinoic acid metabolism into polar metabolites in rat liver via induction of cytochrome P4502E1. Gastroenterology. 2001;120:179-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 91] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 104. | Grizotte-Lake M, Zhong G, Duncan K, Kirkwood J, Iyer N, Smolenski I, Isoherranen N, Vaishnava S. Commensals Suppress Intestinal Epithelial Cell Retinoic Acid Synthesis to Regulate Interleukin-22 Activity and Prevent Microbial Dysbiosis. Immunity. 2018;49:1103-1115.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 105. | Graham TE, Yang Q, Blüher M, Hammarstedt A, Ciaraldi TP, Henry RR, Wason CJ, Oberbach A, Jansson PA, Smith U, Kahn BB. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med. 2006;354:2552-2563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 971] [Cited by in RCA: 968] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 106. | D'souza AM, Beaudry JL, Szigiato AA, Trumble SJ, Snook LA, Bonen A, Giacca A, Riddell MC. Consumption of a high-fat diet rapidly exacerbates the development of fatty liver disease that occurs with chronically elevated glucocorticoids. Am J Physiol Gastrointest Liver Physiol. 2012;302:G850-G863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (1)] |
| 107. | Singh RP, Halaka DA, Hayouka Z, Tirosh O. High-Fat Diet Induced Alteration of Mice Microbiota and the Functional Ability to Utilize Fructooligosaccharide for Ethanol Production. Front Cell Infect Microbiol. 2020;10:376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 108. | David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5625] [Cited by in RCA: 6839] [Article Influence: 569.9] [Reference Citation Analysis (0)] |
| 109. | König J, Wells J, Cani PD, García-Ródenas CL, MacDonald T, Mercenier A, Whyte J, Troost F, Brummer RJ. Human Intestinal Barrier Function in Health and Disease. Clin Transl Gastroenterol. 2016;7:e196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 431] [Cited by in RCA: 593] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 110. | Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3003] [Cited by in RCA: 2991] [Article Influence: 230.1] [Reference Citation Analysis (0)] |
| 111. | Zhang H, Luo XM. Control of commensal microbiota by the adaptive immune system. Gut Microbes. 2015;6:156-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 112. | Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther. 2021;6:291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 414] [Cited by in RCA: 917] [Article Influence: 229.3] [Reference Citation Analysis (0)] |
| 113. | Crispe IN. Hepatocytes as Immunological Agents. J Immunol. 2016;196:17-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 114. | Mridha AR, Wree A, Robertson AAB, Yeh MM, Johnson CD, Van Rooyen DM, Haczeyni F, Teoh NC, Savard C, Ioannou GN, Masters SL, Schroder K, Cooper MA, Feldstein AE, Farrell GC. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol. 2017;66:1037-1046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 546] [Cited by in RCA: 830] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 115. | Parthasarathy G, Malhi H. Macrophage Heterogeneity in NASH: More Than Just Nomenclature. Hepatology. 2021;74:515-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 116. | Lefebvre E, Moyle G, Reshef R, Richman LP, Thompson M, Hong F, Chou HL, Hashiguchi T, Plato C, Poulin D, Richards T, Yoneyama H, Jenkins H, Wolfgang G, Friedman SL. Antifibrotic Effects of the Dual CCR2/CCR5 Antagonist Cenicriviroc in Animal Models of Liver and Kidney Fibrosis. PLoS One. 2016;11:e0158156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 260] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 117. | Ratziu V, Sanyal A, Harrison SA, Wong VW, Francque S, Goodman Z, Aithal GP, Kowdley KV, Seyedkazemi S, Fischer L, Loomba R, Abdelmalek MF, Tacke F. Cenicriviroc Treatment for Adults With Nonalcoholic Steatohepatitis and Fibrosis: Final Analysis of the Phase 2b CENTAUR Study. Hepatology. 2020;72:892-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 278] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 118. | Leite NC, Viegas BB, Villela-Nogueira CA, Carlos FO, Cardoso CRL, Salles GF. Efficacy of diacerein in reducing liver steatosis and fibrosis in patients with type 2 diabetes and non-alcoholic fatty liver disease: A randomized, placebo-controlled trial. Diabetes Obes Metab. 2019;21:1266-1270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 119. | Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 542] [Cited by in RCA: 509] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 120. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1539] [Article Influence: 139.9] [Reference Citation Analysis (40)] |
| 121. | Lee G, You HJ, Bajaj JS, Joo SK, Yu J, Park S, Kang H, Park JH, Kim JH, Lee DH, Lee S, Kim W, Ko G. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat Commun. 2020;11:4982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 248] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 122. | Sharpton SR, Ajmera V, Loomba R. Emerging Role of the Gut Microbiome in Nonalcoholic Fatty Liver Disease: From Composition to Function. Clin Gastroenterol Hepatol. 2019;17:296-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 123. | Ahmad MI, Khan MU, Kodali S, Shetty A, Bell SM, Victor D. Hepatocellular Carcinoma Due to Nonalcoholic Fatty Liver Disease: Current Concepts and Future Challenges. J Hepatocell Carcinoma. 2022;9:477-496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 124. | Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4054] [Cited by in RCA: 3801] [Article Influence: 543.0] [Reference Citation Analysis (2)] |
| 125. | Weisany W, Yousefi S, Tahir NA, Golestanehzadeh N, McClements DJ, Adhikari B, Ghasemlou M. Targeted delivery and controlled released of essential oils using nanoencapsulation: A review. Adv Colloid Interface Sci. 2022;303:102655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 48] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 126. | Butzner JD, Parmar R, Bell CJ, Dalal V. Butyrate enema therapy stimulates mucosal repair in experimental colitis in the rat. Gut. 1996;38:568-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 155] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 127. | Hamer HM, Jonkers DM, Vanhoutvin SA, Troost FJ, Rijkers G, de Bruïne A, Bast A, Venema K, Brummer RJ. Effect of butyrate enemas on inflammation and antioxidant status in the colonic mucosa of patients with ulcerative colitis in remission. Clin Nutr. 2010;29:738-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 128. | Harrison SA, Rossi SJ, Paredes AH, Trotter JF, Bashir MR, Guy CD, Banerjee R, Jaros MJ, Owers S, Baxter BA, Ling L, DePaoli AM. NGM282 Improves Liver Fibrosis and Histology in 12 Weeks in Patients With Nonalcoholic Steatohepatitis. Hepatology. 2020;71:1198-1212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 208] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 129. | Harrison SA, Ruane PJ, Freilich BL, Neff G, Patil R, Behling CA, Hu C, Fong E, de Temple B, Tillman EJ, Rolph TP, Cheng A, Yale K. Efruxifermin in non-alcoholic steatohepatitis: a randomized, double-blind, placebo-controlled, phase 2a trial. Nat Med. 2021;27:1262-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 238] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 130. | Meroni M, Longo M, Dongiovanni P. The Role of Probiotics in Nonalcoholic Fatty Liver Disease: A New Insight into Therapeutic Strategies. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 93] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 131. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, Puri P, Sterling RK, Luketic V, Stravitz RT, Siddiqui MS, Fuchs M, Thacker LR, Wade JB, Daita K, Sistrun S, White MB, Noble NA, Thorpe C, Kakiyama G, Pandak WM, Sikaroodi M, Gillevet PM. Randomised clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment Pharmacol Ther. 2014;39:1113-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 223] [Article Influence: 20.3] [Reference Citation Analysis (1)] |
| 132. | Román E, Nieto JC, Gely C, Vidal S, Pozuelo M, Poca M, Juárez C, Guarner C, Manichanh C, Soriano G. Effect of a Multistrain Probiotic on Cognitive Function and Risk of Falls in Patients With Cirrhosis: A Randomized Trial. Hepatol Commun. 2019;3:632-645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 133. | Famouri F, Shariat Z, Hashemipour M, Keikha M, Kelishadi R. Effects of Probiotics on Nonalcoholic Fatty Liver Disease in Obese Children and Adolescents. J Pediatr Gastroenterol Nutr. 2017;64:413-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 189] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 134. | Kobyliak N, Abenavoli L, Mykhalchyshyn G, Kononenko L, Boccuto L, Kyriienko D, Dynnyk O. A Multi-strain Probiotic Reduces the Fatty Liver Index, Cytokines and Aminotransferase levels in NAFLD Patients: Evidence from a Randomized Clinical Trial. J Gastrointestin Liver Dis. 2018;27:41-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 177] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 135. | Ahn SB, Jun DW, Kang BK, Lim JH, Lim S, Chung MJ. Randomized, Double-blind, Placebo-controlled Study of a Multispecies Probiotic Mixture in Nonalcoholic Fatty Liver Disease. Sci Rep. 2019;9:5688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 141] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 136. | Castillo V, Figueroa F, González-Pizarro K, Jopia P, Ibacache-Quiroga C. Probiotics and Prebiotics as a Strategy for Non-Alcoholic Fatty Liver Disease, a Narrative Review. Foods. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 137. | Hu D, Yang W, Mao P, Cheng M. Combined Amelioration of Prebiotic Resveratrol and Probiotic Bifidobacteria on Obesity and Nonalcoholic Fatty Liver Disease. Nutr Cancer. 2021;73:652-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 138. | Xu RY, Wan YP, Fang QY, Lu W, Cai W. Supplementation with probiotics modifies gut flora and attenuates liver fat accumulation in rat nonalcoholic fatty liver disease model. J Clin Biochem Nutr. 2012;50:72-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 139. | Zhang L, Qin Q, Liu M, Zhang X, He F, Wang G. Akkermansia muciniphila can reduce the damage of gluco/Lipotoxicity, oxidative stress and inflammation, and normalize intestine microbiota in streptozotocin-induced diabetic rats. Pathog Dis. 2018;76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 140. | Lunia MK, Sharma BC, Sharma P, Sachdeva S, Srivastava S. Probiotics prevent hepatic encephalopathy in patients with cirrhosis: a randomized controlled trial. Clin Gastroenterol Hepatol. 2014;12:1003-8.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 154] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 141. | Zhao LN, Yu T, Lan SY, Hou JT, Zhang ZZ, Wang SS, Liu FB. Probiotics can improve the clinical outcomes of hepatic encephalopathy: An update meta-analysis. Clin Res Hepatol Gastroenterol. 2015;39:674-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 142. | Shavakhi A, Minakari M, Firouzian H, Assali R, Hekmatdoost A, Ferns G. Effect of a Probiotic and Metformin on Liver Aminotransferases in Non-alcoholic Steatohepatitis: A Double Blind Randomized Clinical Trial. Int J Prev Med. 2013;4:531-537. [PubMed] |
| 143. | Nabavi S, Rafraf M, Somi MH, Homayouni-Rad A, Asghari-Jafarabadi M. Effects of probiotic yogurt consumption on metabolic factors in individuals with nonalcoholic fatty liver disease. J Dairy Sci. 2014;97:7386-7393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 144. | Eslamparast T, Poustchi H, Zamani F, Sharafkhah M, Malekzadeh R, Hekmatdoost A. Synbiotic supplementation in nonalcoholic fatty liver disease: a randomized, double-blind, placebo-controlled pilot study. Am J Clin Nutr. 2014;99:535-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 287] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 145. | Aller R, De Luis DA, Izaola O, Conde R, Gonzalez Sagrado M, Primo D, De La Fuente B, Gonzalez J. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: a double blind randomized clinical trial. Eur Rev Med Pharmacol Sci. 2011;15:1090-1095. [PubMed] |
| 146. | Malaguarnera M, Vacante M, Antic T, Giordano M, Chisari G, Acquaviva R, Mastrojeni S, Malaguarnera G, Mistretta A, Li Volti G, Galvano F. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig Dis Sci. 2012;57:545-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 333] [Article Influence: 25.6] [Reference Citation Analysis (1)] |
| 147. | Gupta S, Allen-Vercoe E, Petrof EO. Fecal microbiota transplantation: in perspective. Therap Adv Gastroenterol. 2016;9:229-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 302] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 148. | Zhou D, Pan Q, Shen F, Cao HX, Ding WJ, Chen YW, Fan JG. Total fecal microbiota transplantation alleviates high-fat diet-induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci Rep. 2017;7:1529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 305] [Cited by in RCA: 296] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 149. | Forlano R, Sivakumar M, Mullish BH, Manousou P. Gut Microbiota-A Future Therapeutic Target for People with Non-Alcoholic Fatty Liver Disease: A Systematic Review. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 150. | Craven L, Rahman A, Nair Parvathy S, Beaton M, Silverman J, Qumosani K, Hramiak I, Hegele R, Joy T, Meddings J, Urquhart B, Harvie R, McKenzie C, Summers K, Reid G, Burton JP, Silverman M. Allogenic Fecal Microbiota Transplantation in Patients With Nonalcoholic Fatty Liver Disease Improves Abnormal Small Intestinal Permeability: A Randomized Control Trial. Am J Gastroenterol. 2020;115:1055-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 229] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 151. | Kuno T, Hirayama-Kurogi M, Ito S, Ohtsuki S. Reduction in hepatic secondary bile acids caused by short-term antibiotic-induced dysbiosis decreases mouse serum glucose and triglyceride levels. Sci Rep. 2018;8:1253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 152. | Liu L, Liu Z, Li H, Cao Z, Li W, Song Z, Li X, Lu A, Lu C, Liu Y. Naturally Occurring TPE-CA Maintains Gut Microbiota and Bile Acids Homeostasis via FXR Signaling Modulation of the Liver-Gut Axis. Front Pharmacol. 2020;11:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 153. | Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, Llorente C, Wang L, Alnouti Y, Fouts DE, Stärkel P, Loomba R, Coulter S, Liddle C, Yu RT, Ling L, Rossi SJ, DePaoli AM, Downes M, Evans RM, Brenner DA, Schnabl B. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology. 2018;67:2150-2166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 219] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 154. | Cui J, Zheng X, Hou W, Zhuang Y, Pi X, Yang J. The study of a remote-controlled gastrointestinal drug delivery and sampling system. Telemed J E Health. 2008;14:715-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 155. | Rehan M, Al-Bahadly I, Thomas DG, Avci E. Capsule robot for gut microbiota sampling using shape memory alloy spring. Int J Med Robot. 2020;16:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 156. | Waimin JF, Nejati S, Jiang H, Qiu J, Wang J, Verma MS, Rahimi R. Smart capsule for non-invasive sampling and studying of the gastrointestinal microbiome. RSC Adv. 2020;10:16313-16322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 157. | Neil K, Allard N, Roy P, Grenier F, Menendez A, Burrus V, Rodrigue S. High-efficiency delivery of CRISPR-Cas9 by engineered probiotics enables precise microbiome editing. Mol Syst Biol. 2021;17:e10335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 68] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 158. | Neil K, Allard N, Grenier F, Burrus V, Rodrigue S. Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl(2) conjugative plasmid TP114. Commun Biol. 2020;3:523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 159. | Fox BE, Vilander AC, Gilfillan D, Dean GA, Abdo Z. Oral Vaccination Using a Probiotic Vaccine Platform Combined with Prebiotics Impacts Immune Response and the Microbiome. Vaccines (Basel). 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 160. | Moor K, Slack E. What makes a bacterial oral vaccine a strong inducer of high-affinity IgA responses? Antibodies. 2015;4:295-313. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 161. | Diard M, Bakkeren E, Lentsch V, Rocker A, Bekele NA, Hoces D, Aslani S, Arnoldini M, Böhi F, Schumann-Moor K, Adamcik J, Piccoli L, Lanzavecchia A, Stadtmueller BM, Donohue N, van der Woude MW, Hockenberry A, Viollier PH, Falquet L, Wüthrich D, Bonfiglio F, Loverdo C, Egli A, Zandomeneghi G, Mezzenga R, Holst O, Meier BH, Hardt WD, Slack E. A rationally designed oral vaccine induces immunoglobulin A in the murine gut that directs the evolution of attenuated Salmonella variants. Nat Microbiol. 2021;6:830-841. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 162. | Romero-Gómez M, Zelber-Sagi S, Trenell M. Treatment of NAFLD with diet, physical activity and exercise. J Hepatol. 2017;67:829-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 844] [Cited by in RCA: 926] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 163. | Huang MA, Greenson JK, Chao C, Anderson L, Peterman D, Jacobson J, Emick D, Lok AS, Conjeevaram HS. One-year intense nutritional counseling results in histological improvement in patients with non-alcoholic steatohepatitis: a pilot study. Am J Gastroenterol. 2005;100:1072-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 606] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 164. | Cheng R, Wang L, Le S, Yang Y, Zhao C, Zhang X, Yang X, Xu T, Xu L, Wiklund P, Ge J, Lu D, Zhang C, Chen L, Cheng S. A randomized controlled trial for response of microbiome network to exercise and diet intervention in patients with nonalcoholic fatty liver disease. Nat Commun. 2022;13:2555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 68] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 165. | Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, Holscher HD, Woods JA. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med Sci Sports Exerc. 2018;50:747-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 508] [Article Influence: 72.6] [Reference Citation Analysis (0)] |