Published online Sep 21, 2022. doi: 10.3748/wjg.v28.i35.5141
Peer-review started: November 9, 2021
First decision: April 16, 2022
Revised: April 28, 2022
Accepted: September 1, 2022
Article in press: September 1, 2022
Published online: September 21, 2022
Processing time: 309 Days and 21.2 Hours
Pancreatic ductal cancer (PDAC) has high malignancy and poor prognosis. Long noncoding RNAs (lncRNAs) are associated with high levels of malignancy, including PDAC. However, the biological and clinical significance of negative regulator of antiviral response (NRAV) in PDAC is unclear.
To study the regulatory role of lncRNA NRAV in PDAC.
GEPIA analyzed lncRNA NRAV and miRNA (miR-299-3p) expression levels in PDAC tissues and measured them in PDAC cells by quantitative measurements in real time. The specific role of NRAV and miR-299-3p in cell proliferation and transfer potential was evaluated by cell formation analysis, Cell Counting Kit-8 and Transwell analysis. The relationship between NRAV and miR-299-3p was studied by predictive bioinformatics, RNA immunoassay, and fluorescence enzyme analysis. In vivo experiments included transplantation of simulated tumor cells under naked mice.
The expression level of lncRNA NRAV was higher in both tumor tissues and cell lines of PDAC and was negatively associated with the clinical survival of PDAC patients. Functionally, overexpression of NRAV promoted cell proliferation and metastasis of PDAC cells, while knockdown of NRAV reversed these effects. Finally, NRAV was performed as a molecular sponge of miR-299-3p. Moreover, overexpression of miR-299-3p could reverse the promoting effects of NRAV on cell proliferation and metastasis of PDAC cells.
NRAV facilitates progression of PDAC as a molecular sponge of miR-299-3p and may be a potential molecular marker for diagnosis and treatment of PDAC.
Core Tip: In the present research, the expression level of long noncoding RNA negative regulator of antiviral response (NRAV) in pancreatic ductal adenocarcinoma (PDAC) was detected, and the clinicopathological relationship between NRAV and PDAC was demonstrated. Moreover, cell and animal tests were conducted to assess the concrete roles of NRAV in the progression of PDAC. Finally, the existence of potential specific molecular mechanisms that can provide new ideas for finding new molecular markers for the diagnosis and treatment of PDAC is demonstrated.
- Citation: Wang HQ, Qian CH, Guo ZY, Li PM, Qiu ZJ. Long noncoding RNA negative regulator of antiviral response contributes to pancreatic ductal adenocarcinoma progression via targeting miR-299-3p. World J Gastroenterol 2022; 28(35): 5141-5153
- URL: https://www.wjgnet.com/1007-9327/full/v28/i35/5141.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i35.5141
As an extremely malignant tumor, pancreatic ductal adenocarcinoma (PDAC) has shown a rapidly increasing incidence rate in recent years worldwide[1]. According to the latest cancer report in 2021, there are 495773 new cases reported globally every year and about 466003 people die of PDAC annually[2]. Due to insufficient means of early diagnosis and effective treatment, the 5-year survival rate of PDAC patients is only about 8% and will drop to about 3% in patients with advanced PDAC[1]. Therefore, it is urgent to establish a comprehensive pathological mechanism of PDAC and explore a more effective PDAC diagnosis and treatment center.
Long noncoding RNAs (lncRNAs) are new ncRNAs that are composed of > 200 nucleotides, accounting for the largest proportion of the entire human gene transcriptome[3,4]. lncRNAs mainly regulate gene expression in a variety of ways, including chromatin remodeling and transcriptional and post-transcriptional processing, and participate in regulating a variety of biological processes[5,6]. Recently, a large number of researches showed that lncRNAs are dysregulated in most tumors and function as key regulators in the process of tumor growth, metastasis, drug tolerance, and angiogenesis[7,8]. The lncRNA negative regulator of antiviral response (NRAV) is a newly identified lncRNA and is mainly related to immunity[9]. Xu et al[10] found that NRAV is highly expressed in the cell system of hepatocytes and regulates the course of hepatocellular carcinoma. However, the statements and specific role of NRAV in PDAC are still unclear.
lncRNAs play an important regulatory role in the occurrence and development of various cancers and may be considered a potential molecular marker. Many studies have shown that RNA plays its biological function of competing with endogenous RNAs[11,12]. Hsa-miR-299-3p plays an important role in the development of many cancers. Many studies describe the regulation of the relationship between miR-299-3p and lncRNAs[13,14]. Many experts conducted in-depth research on the regulation of the relationship between the two and stated that miR-299-3p increased the volume and cellular system and prevented proliferation and metastasis of pancreatic cancer cells with exposure to Notch1[15]. The relationship between NRAV and miR-299-3p in PDAC has not yet been investigated.
In this study, we tested the expression of NRAV in PDAC and studied its function, and found that NRAV and PDAC have obvious overlaps. The results of functional experiments showed that the level of NRAV expression was directly associated with cell proliferation and metastasis and was closely related to the growth of tumors in naked mice. In addition, we found that NRAV, as a molecular sponge of miR-299-3p, contributes to PDAC progression. Therefore, the results of this study may provide new insights into the role of NRAV in PDAC. It can be a biomarker for the diagnosis and treatment of PDAC.
PDAC cell lines (PANC-1, AsPC-1, Mia Paca-2 and BxPC-3) and human immortalized normal pancreatic duct epithelial (HPDE) cells were obtained from the American Type Culture Collection (Manassas, VA, United States). The cell lines were cultured at an appropriate concentration in a specific environment in a moistened cell incubator with a CO2 concentration of 5% at 37 °C. Percentage of 10 fetal serum and 1% penicillin–streptomycin were added to the culture medium, including RPMI-1640 and Dulbecco’s modified Eagle’s medium.
Total cellular RNA was extracted from cultured cells using Trizol reagent (Invitrogen, Carlsbad, CA, United States). For lncRNA, FastKing gDNA Dispeeling RT Supermix (TIANGEN, China) was selected for reverse transclncRNAn. The specific cDNAs of miRNAs were obtained with a specific kit (miDETECT A Track RT Reagent Kit; RiboBio, China). The quantitative real-time polymerase chain reaction (qRT-PCR) system wasABI 7300 PCR (Foster City, CA, United States). U6 and ACTB were respectively deemed as standardized internal controls of miRNA and lncRNA. The specific sequences of the primers were designed as follows. For NRAV, 5’-GGAGTTGATGCCTCCGAACA-3’ (forward) and 5’-ATGACCGGAGCTGAAAGGTG-3’ (reverse); for β-actin, 5’-TCCCTGGAGAAGAGCTACGA-3’ (forward) and 5’-AGCACTGTGTTGGCGTACAG-3’ (reverse); for miR-299-3p, 5’-ACACTCCAGCTGGGTATGTGGGATGGTAAAC-3’ (forward) and 5’-GTGCAGGGTCCGAGGT-3’ (reverse); and for U6, 5’-CTCGCTTCGGCAGCACA-3’ (forward) and 5’-AACGCTTCACGAATTTGCGT-3’ (reverse). The relative expression level of each gene was calculated with the 2−ΔΔCt method.
Lipofectamine 2000 kit (Invitrogen) was used to conduct cell transfection. Short hairpin RNAs (shRNAs) targeting NRAV were designed by Genepharma (Shanghai, China). The specific sequences targeting NRAV were designed as follows: For sh-NRAV #1, 5’-CACCTCATCCACAAGTAGGAC-3’; for sh-NRAV #2, 5’-TTGGAGCCAAGGACTGTACTG-3’; and negative control: 5’-TTCTCCGAACGTGTCACGT-3’. NRAV overexpression plasmid and shRNAs against NRAV were inserted into the pcDNA3.1 vector and pGpU6/GFP/Neo vector, respectively. The miR-299-3p mimic was purchased from RiboBio. qRT-PCR was performed to examine the transfection efficiency.
A 96-well plate was selected, and 2000 tumor cells were cultured in each well. The cells were transfected with different vectors and allowed to stand for 48 h. Cell Counting Kit-8 (CCK-8) solution (10 μL; Mashiki, Japan) was added to each well, and incubated for 2 h. A microplate reader (Winooski, VT, United States) was used to check cell viability.
The specific PDAC cells transfected with different plasmids were inoculated into six-well plates and incubated at 37°C for 10 d. Tumor cells were fixed for 20 min in methanol after being washed with phosphate-buffered saline in triplicate. Cell colonies were stained and counted.
In migration and invasion assays, PDAC cells were added to resuspension after setting a concentration of 105 mL in a serum-free environment. Two hundred microliters of cell suspension was added to the upper layer of each Transwell chamber and 600 μL of complete media containing 10% fetal serum was added to the lower layer. To set up the invasion analysis phase, the Matrigel layer (BD Biosciences, United States) was first applied to the chamber membrane, and the cell suspension was placed on top of the transition chamber. A 24-h incubation was followed and the invasive cells were finally fixed and monitored and compared in five random fields.
All animal studies were approved by the Ethics Committee for Animal Studies at Shanghai 10th People’s Hospital. Specific PANC-1 cells (2 × 106, 200/L) were transplanted subcutaneously to the right side of naked male mice aged 4 wk. The condition of the mice was monitored daily and the size of the tumor was measured every 5 d. Tumor volume was calculated according to tumor length and length formula volume = length through × wide2/2. Twenty days after inoculation, mice were killed and the entire tumor removed and weighed.
Mouse tumors were removed for immunohistochemical staining to visualize Ki-67 expression. Paraffin-embedded sections were prepared and wax removed in 100% xylene, followed by rehydration with various gradients of ethanol and distilled water. The fabric is dyed with a protractor for 1 h at room temperature and removed during washing with distilled water. Tissue sections were incubated for 20 min with secondary antibodies bound to horseradish peroxidase. To facilitate visualization, 3,3’-diaminobenzidine tetrahydrochloride Ki-67 was selected for positive staining.
The PARIS Kit (Life Technologies, United States) was used to isolate RNA from the nucleus and cytoplasm. qRT-PCR assays were conducted as described above.
RNA immunoprecipitation (RIP) assays were performed by EZ-Magna RNA Immunosuppression Kit (Millipore, United States). The cells were collected and resuspended in an immunodeposition buffer and kept on ice for 30 min. The cell suspension was incubated with a separate RIP buffer containing magnetic beads. Proteinase K was added to the bead state after buffering for further digestion of proteins. All RNA was extracted with Trizol and measured with qRT-PCR.
According to a special combination of software for network prediction starbes 3.0 and NRAV miR-299-3p (http://starbase.sysu.edu.cn/ b). Other matters according to the V2 specification (enzyme, China), it is a rapid mutation reagent for NRAV, and supplementary DNA and mutant miR-299-3p are added to psicheck2 (US Promega) (NRAV wild and music) satellite. Paycheck-2 and miR-229-3p plasmid samples or miR-229-3p negative control were transferred to PDAC cells. After 48 h, the fluorescent enzyme activity of each well was examined by the Promega reporting system. The relative activity of luciferase is used for normalization.
Statistical analysis was performed using GraphPad Prisma 8.2 (LA Jolla, CA, United States) and SPSS 22.0 (IBM, Armonk, NY, United States). Student t test was used to compare the differences between the two groups and calculate the P value. P < 0.05 was statistically significant.
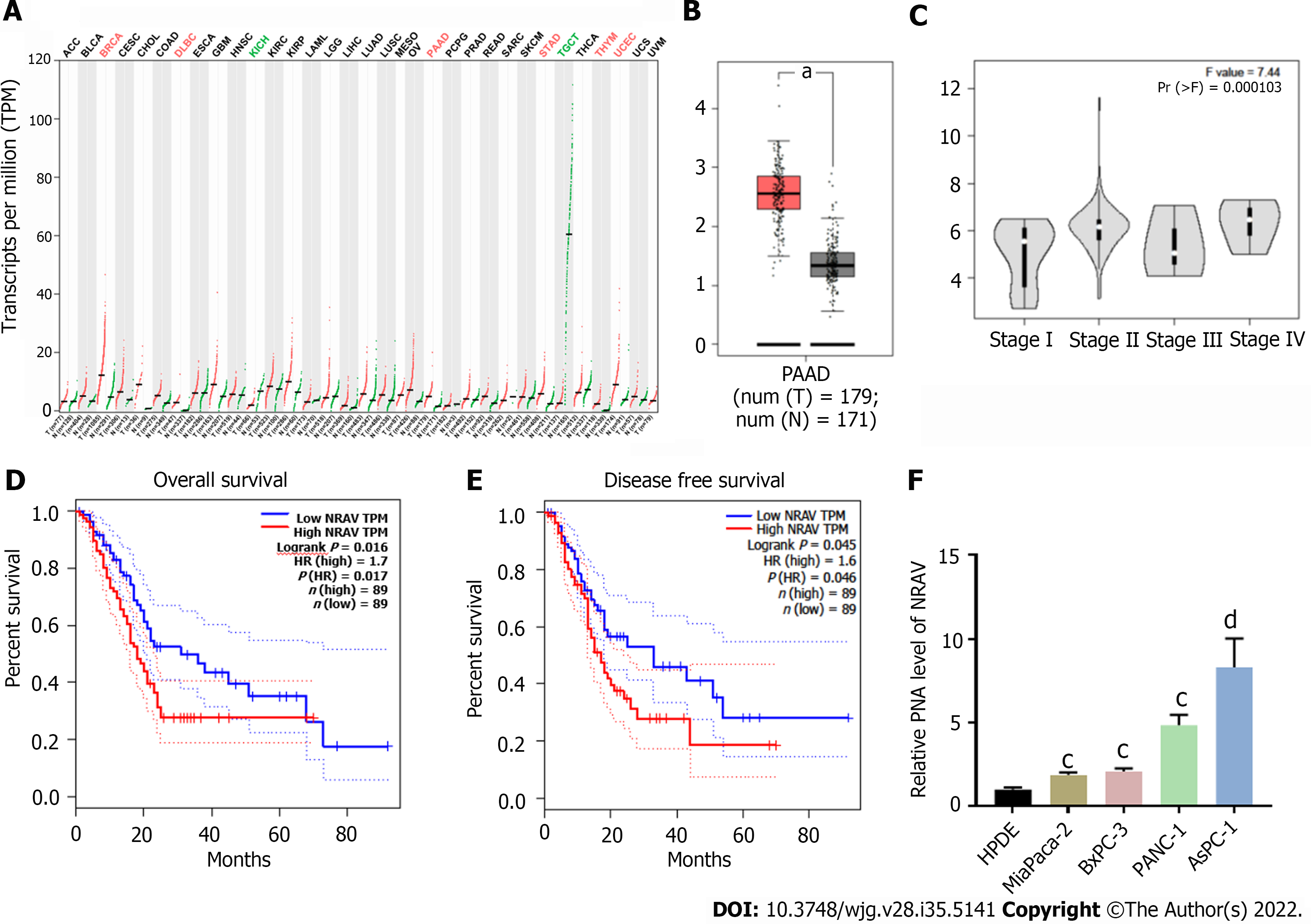
We used GEPIA to determine NRAV expression levels as an online bioinformatics tool for gene expression analysis based on the cancer genome database (TCGA)[16]. NRAV expression of PDAC in tissues was significantly higher than that in nonmedical tissues (Figure 1A and B). The survival curve showed that the total life cycle (P = 0.034; Figure 1D) and disease-free life cycle (P = 0.046; Figure 1E) of PDAC patients were decreasing. The data of the TCGA database showed that the level of emotional expression was negatively correlated with the level of clinical pathology (Figure 1C). The level of NRAV expression in HPDE cells and four PDAC systems was tested. NRAV in the PDAC cell system was significantly higher than that in HPDE cells (Figure 1F). Based on these data, we believe that NRAV may be a potential carcinogen related to lncRNA.
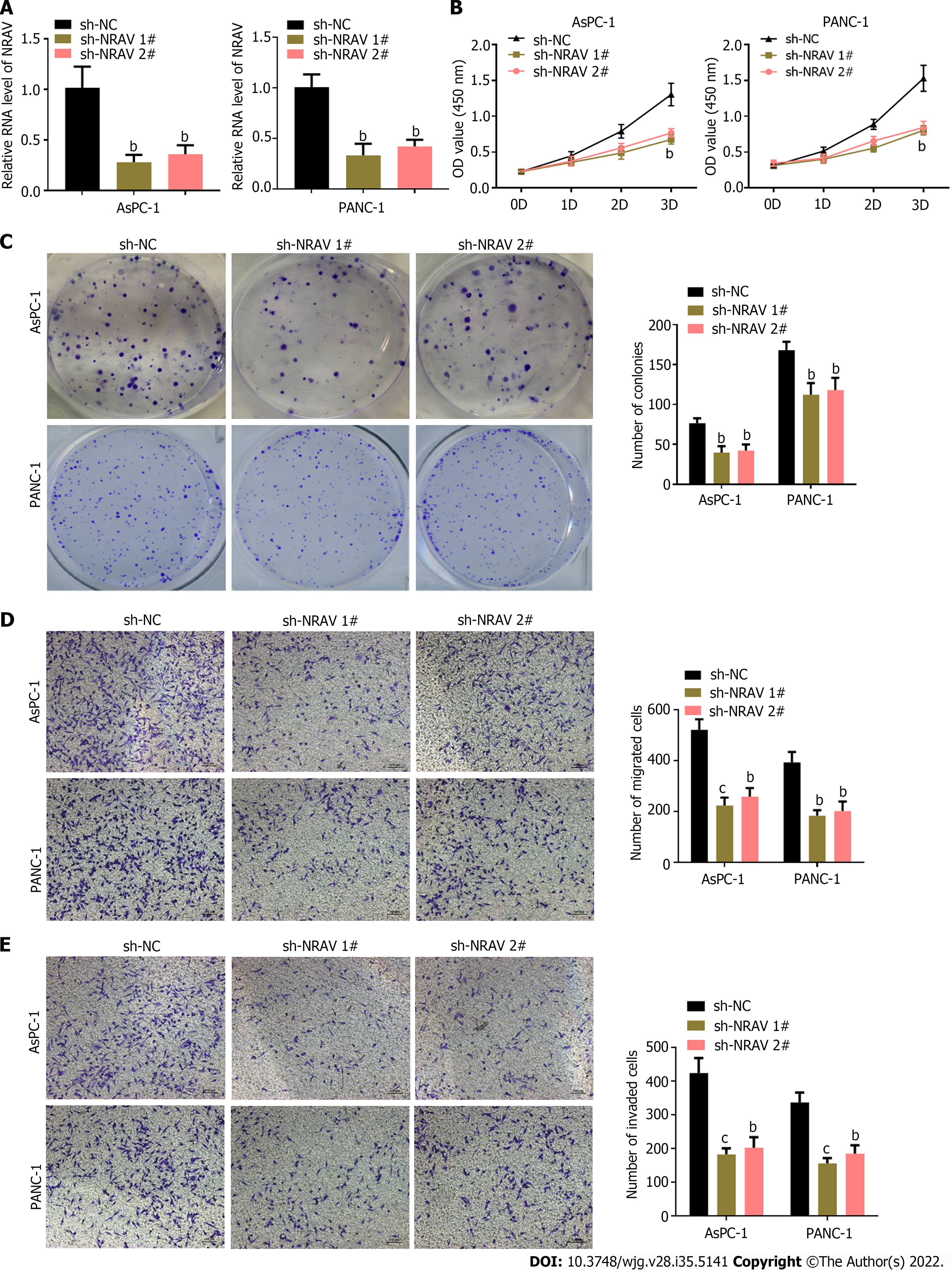
We studied the biological role of NRAV in the PDAC cell system. We used heterogeneous RNA transplantation to reduce NRAV expression in PANC-1 and AsPC-1 cells. qRT-PCR showed a significant decrease in expression after NRAV (Figure 2A). After showing the results of CCK-8 tests and colony formation, there was a significant reduction after removal of signs of proliferation and abscision of PANC-1 and AsPC-1 cells (Figure 2B and C). We performed Transwell analysis to test whether NRAV affected metastasis and damage to police cells. Similarly, after the removal of PANC-1 and AsPC-1 cells, migration and invasion rates slowed significantly (Figure 2D and E). Knockdown of NRAV inhibited proliferation, migration and invasion of PDAC cells.
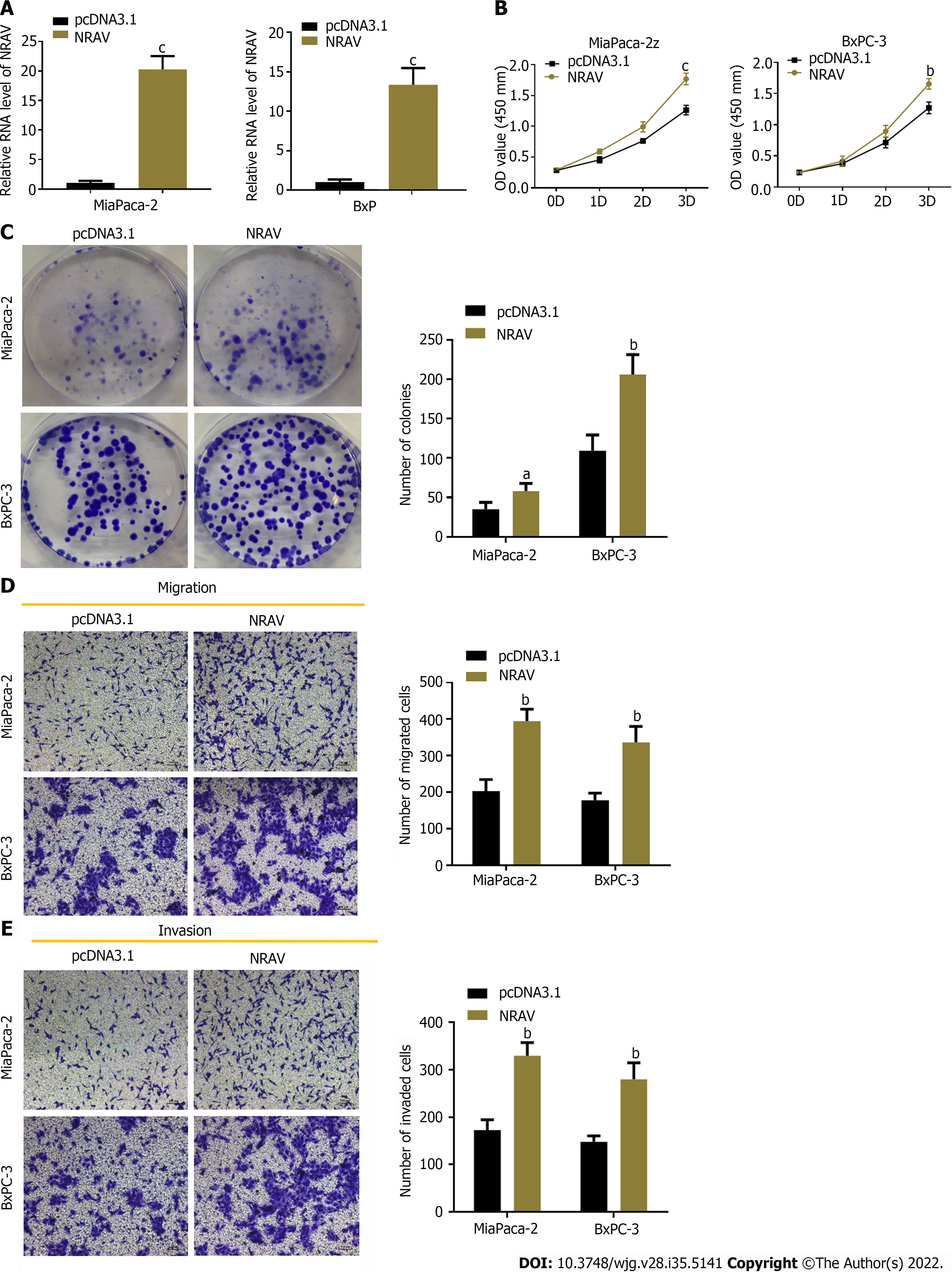
High expression of NRAV in BXPC-3 and Mia PaCa-2 cells used to display exogenous particles confirmed the effectiveness of the expression (Figure 3A). The CCK-8 test and the product reduction test showed that NRAV overexpression significantly supported proliferation and formation of PDAC cells (Figure 3B and C). Transwell analysis found that NRAV overexpression significantly increased displacement and invasion of Mia PaCa-2 and BXPC-3 cells (Figure 3D and E). Overall, the results showed that NRAV promotes proliferation, metastasis and invasion of PDAC cells.
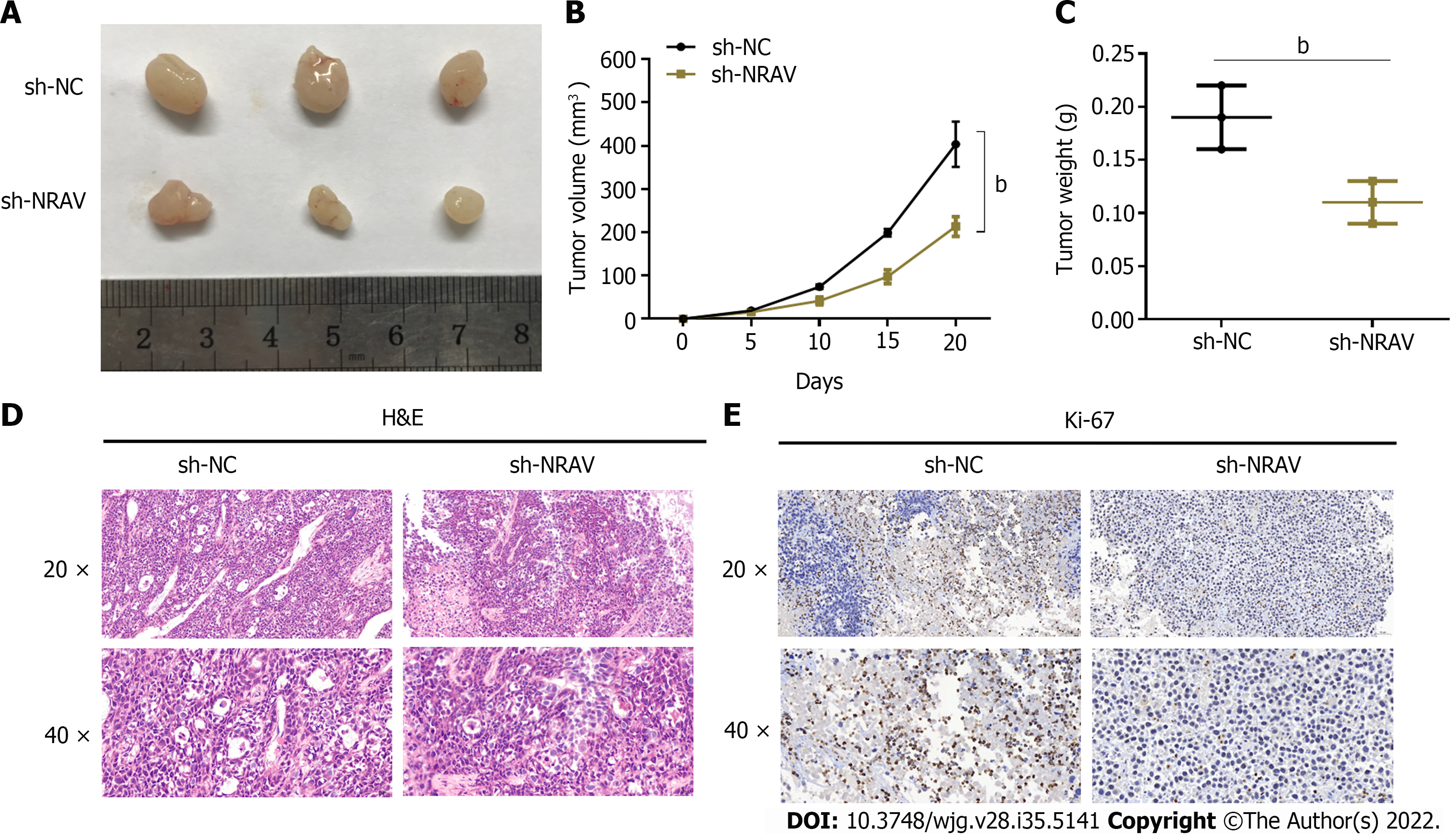
To further investigate whether NRAV supports tumor growth in vivo, PANC-1 cells were injected into naked mice treated with sh-NRAV and internal control cells. The tumor volume of the NRAV genotype was significantly lower than the negative control volume (Figure 4A and B). Similarly, the mass of the tumor in the NRAV group (Figure 4C) was significantly reduced compared to the control group. Ki-67 was analyzed for lignin content and immunochemical staining. Reduction of NRAV led to a significant reduction of Ki-67 and inhibition of tumor growth (Figure 4D and E). Overall, these findings suggest that NRAV does not inhibit tumor growth in vivo.
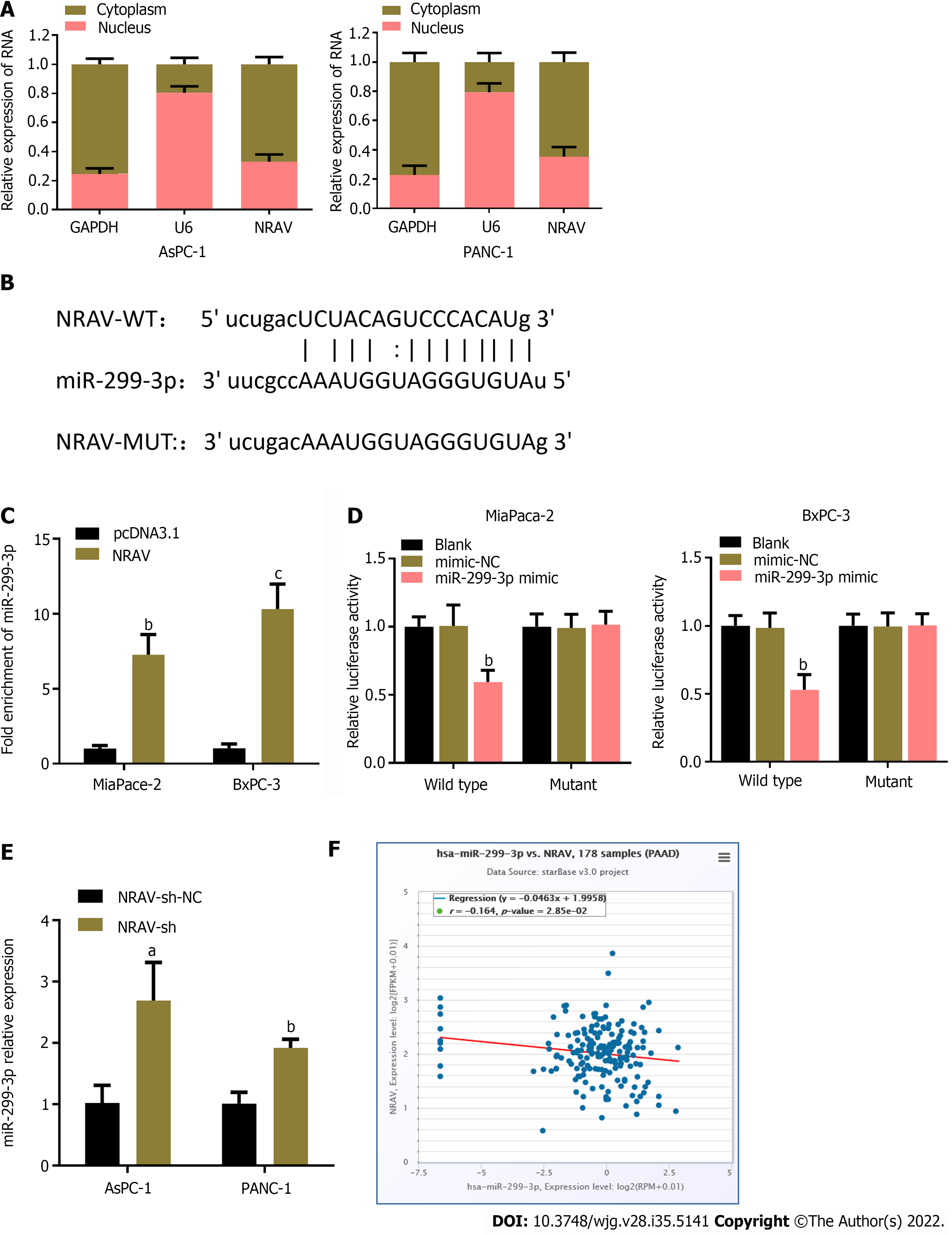
To study how NRAV plays a catalytic role in PDAC, we used the nuclear/cell separation method to determine the location of NRAV in PDAC. Spleen qi was mainly located in cells (Figure 5A), and it can exert its biological function as a competitive endogenous RNA[17,18]. We used starbase V3.0 (http://starbase.sysu.edu.cn/) Interactive Bio information Analysis Software to identify potential miRNAs that may interact with genes. miR-299-3p was selected as the tempered target due to potential point complementarity (Figure 5B). Rip analysis confirms this point[19]. As shown in Overexpression in PDAC cells resulted in significant enrichment of miR-299-3p on ago2 (Figure 5C). The miR-299-3p simulator significantly reduced the fluorescence activity of the wild-type NRAV group, while this change did not occur in the modified NRAV group (Figure 5D). qRT-PCR showed that expression of miR-299-3p in PDAC cells was significantly enhanced after removing the traits (Figure 5E). Spearman’s analysis showed that expression of NRAV and miR-299-3p in PDAC tissue was opposite (Figure 5F). Therefore, these results suggest that temper may play the role of molecular sponge miR-299-3p.
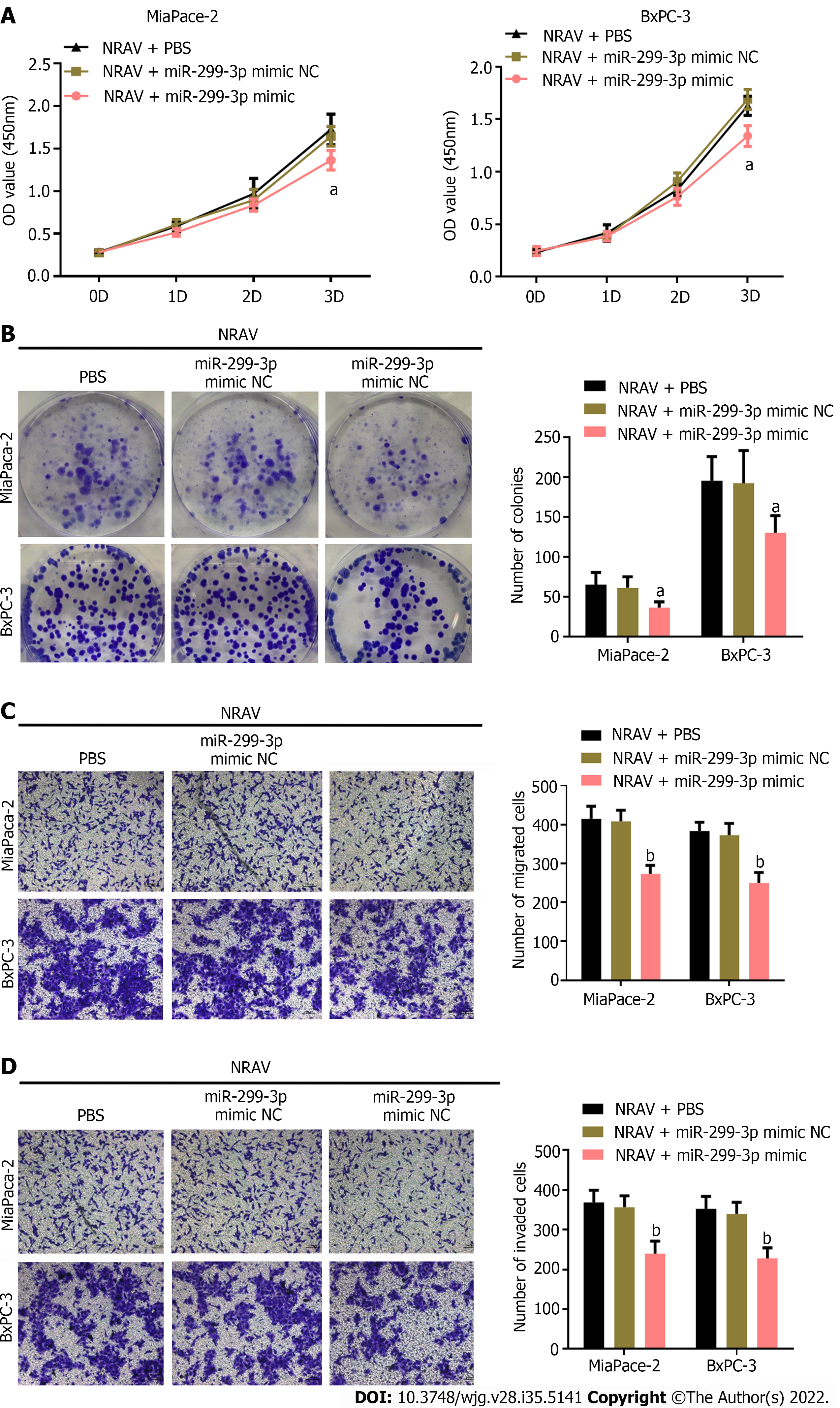
To validate whether NRAV affected proliferation and metastasis of PDAC cells through sponging miR-299-3p, specific miR-299-3p mimic or mimic NC vector was cotransfected into PDAC cells after overexpression of NRAV. Subsequently, we evaluated the cell proliferation, migration and invasion of PDAC cells. miR-299-3p mimic reversed the dramatic promotion by overexpression of NRAV on proliferation (Figure 6A and B). Similarly, miR-299-3p mimic inhibited the promoting effects of NRAV overexpression on migration and invasion of PDAC cells (Figure 6C and D). These results indicated that miR-299-3p mediated the promoting effects of NRAV on PDAC cell proliferation, migration and invasion.
An increasing number of studies have shown that lncRNAs play an important role in the development of various malignant tumors. Recent studies have shown that large amounts of lncRNAs are abnormally expressed in PDAC and participate in the tumor process[20,21]. Guo et al[22] found that under hypoxia, lncRNAs extract of PDAC UCA1 cells increased to promote angiogenesis. We used the TCGA database to identify NRAV in PDAC. We confirmed that the code of good conduct is a key element related to the program of action, and that significant progress has been made in the program of action. In addition, in PDAC patients, high levels of NRAV expression inhibit overall and disease-free survival. It is worth noting that NRAV may participate in PDAC progression.
The specific role of NRAV in oncology is not clear due to increased immunization. Recent studies have shown that NRAV expression is significantly increased in liver cancer and myeloma and can play a key role in the development of these two tumors[10,23]. To verify the biological function of NRAV in PDAC, function loss and acquisition studies were carried out. The results of in vitro experiments showed that temper has a major contribution to the migration, invasion and reproduction of PDAC cells. In addition, ablation temperament significantly reduced the tumor of the nude mouse model overall, these results show that the NRAV plays an important role in promoting cancer and can be used as a new biometric index for diagnosis and prediction of diseases caused by PDAC.
To further study the potential mechanism of NRAV, nuclear/cell division experiments were carried out in PDAC, and NRAV was mainly in the cytoplasm. Further analysis of the rip report analysis showed that NRAV may play the role of miR-299-3p molecular sponge (Figure 7). In addition, the stimulation effect of NRAV on malignant tumors in PDAC cells was also proved in the rescue experiment. The role of NRAV in promoting progression of PDAC mainly depends on miR-299-3p.
At the beginning of the study, it was emphasized that the significant increase in lncRNA was negatively correlated with overall and disease-free survival in PDAC patients. In addition, it also plays a carcinogenic role by promoting the proliferation and metastasis of cells in the sponge world. Our research shows that NRAV/miR-299-3p play a key role in PDAC and can be used as potential biomarkers for PDAC diagnosis and treatment.
However, because no samples from patients with pancreatic cancer were collected, there was a lack of correlation analysis between temperament expression and PDAC clinicopathological features. In addition, it is also of great significance to study the significance of NRAV and miR-299-3p in PDAC organization.
NRAV can act as a molecular sponge of miR-299-3p and significantly promote the proliferation and metastasis of PDAC cells.
Pancreatic ductal adenocarcinoma (PDAC) has high malignancy and poor prognosis. Long noncoding RNAs (lnRNAs) are recognized as crucial factors and associated with progression of PDAC. However, the specific biological role and practical clinical significance of lnRNAs and negative regulator of antiviral response (NRAV) in PDAC remain unclear.
Early and timely diagnosis and treatment of PDAC are still scarce. Therefore, it is a matter of urgency to comprehensively understand the pathogenesis of PDAC and explore more effective targets for its diagnosis and treatment.
To study the role of NRAV in the growth and metastasis of PDAC.
Real-time polymerase chain reaction detected expression of NRAV and miR-299-3p in PDAC cells. The temperament correction and miR-299-3p in the process of cell proliferation, metastasis and invasion were verified by Cell Counting Kit-8, precipitation test, and Transwell assay. RNA and fluorescent enzyme immunoprecipitation test to test the interaction between NRAV and miR-299-3p. Verify the interaction between NRAV and miR-299-3p.
According to our data, NRAV in PDAC was significantly increased, which is related to the negative survival rate of PDAC patients. NRAV overexpression was conducive to the proliferation and metastasis of PDAC cells, and NRAV knockout reversed these effects. Finally, in terms of mechanism, NRAV acts as a miR-299-3p molecular sponge. Overexpression of miR-299-3p significantly changed the role of NRAV in the proliferation, metastasis and invasion of PDAC cells.
NRAV promotes proliferation and metastasis of PDAC by playing the molecule sponge function of miR-299-3p.
NRAV facilitated the progression of PDAC, which provides a potential biological marker for diagnosis and therapeutic target for PDAC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Karamarkovic AR, Serbia; Nagaya M, Japan; Nakano H, Japan; Sheykhhasan M, Iran S-Editor: Chen YL L-Editor: Kerr C P-Editor: Cai YX
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8287] [Cited by in RCA: 11942] [Article Influence: 2985.5] [Reference Citation Analysis (4)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64684] [Article Influence: 16171.0] [Reference Citation Analysis (177)] |
| 3. | Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3151] [Cited by in RCA: 3684] [Article Influence: 263.1] [Reference Citation Analysis (0)] |
| 4. | Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 722] [Cited by in RCA: 739] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 5. | Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1576] [Cited by in RCA: 1818] [Article Influence: 139.8] [Reference Citation Analysis (0)] |
| 6. | Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21:542-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 1041] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 7. | Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965-3981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2088] [Cited by in RCA: 2163] [Article Influence: 270.4] [Reference Citation Analysis (0)] |
| 8. | Arun G, Diermeier SD, Spector DL. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med. 2018;24:257-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 470] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 9. | Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16:616-626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 286] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 10. | Xu Q, Wang Y, Huang W. Identification of immune-related lncRNA signature for predicting immune checkpoint blockade and prognosis in hepatocellular carcinoma. Int Immunopharmacol. 2021;92:107333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 11. | Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 528] [Cited by in RCA: 832] [Article Influence: 118.9] [Reference Citation Analysis (0)] |
| 12. | Chen W, Zhang K, Yang Y, Guo Z, Wang X, Teng B, Zhao Q, Huang C, Qiu Z. MEF2A-mediated lncRNA HCP5 Inhibits Gastric Cancer Progression via MiR-106b-5p/p21 Axis. Int J Biol Sci. 2021;17:623-634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 13. | Duan H, Li X, Chen Y, Wang Y, Li Z. LncRNA RHPN1-AS1 promoted cell proliferation, invasion and migration in cervical cancer via the modulation of miR-299-3p/FGF2 axis. Life Sci. 2019;239:116856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 14. | Bai N, Ma Y, Zhao J, Li B. Knockdown of lncRNA HCP5 Suppresses the Progression of Colorectal Cancer by miR-299-3p/PFN1/AKT Axis. Cancer Manag Res. 2020;12:4747-4758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Xu K, Zhang L. Inhibition of TUG1/miRNA-299-3p Axis Represses Pancreatic Cancer Malignant Progression via Suppression of the Notch1 Pathway. Dig Dis Sci. 2020;65:1748-1760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 16. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7089] [Article Influence: 886.1] [Reference Citation Analysis (0)] |
| 17. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5558] [Article Influence: 397.0] [Reference Citation Analysis (0)] |
| 18. | Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1761] [Cited by in RCA: 2113] [Article Influence: 176.1] [Reference Citation Analysis (0)] |
| 19. | Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1109] [Cited by in RCA: 1108] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 20. | Wang Y, Zhou L, Lu J, Jiang B, Liu C, Guo J, Xiao GG. Research progress on long non-coding RNAs and their roles as potential biomarkers for diagnosis and prognosis in pancreatic cancer. Cancer Cell Int. 2020;20:457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Tasaki Y, Suzuki M, Katsushima K, Shinjo K, Iijima K, Murofushi Y, Naiki-Ito A, Hayashi K, Qiu C, Takahashi A, Tanaka Y, Kawaguchi T, Sugawara M, Kataoka T, Naito M, Miyata K, Kataoka K, Noda T, Gao W, Kataoka H, Takahashi S, Kimura K, Kondo Y. Cancer-Specific Targeting of Taurine-Upregulated Gene 1 Enhances the Effects of Chemotherapy in Pancreatic Cancer. Cancer Res. 2021;81:1654-1666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 22. | Guo Z, Wang X, Yang Y, Chen W, Zhang K, Teng B, Huang C, Zhao Q, Qiu Z. Hypoxic Tumor-Derived Exosomal Long Noncoding RNA UCA1 Promotes Angiogenesis via miR-96-5p/AMOTL2 in Pancreatic Cancer. Mol Ther Nucleic Acids. 2020;22:179-195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 23. | Maimaiti A, Jiang L, Wang X, Shi X, Pei Y, Hao Y, Paerhati H, Zibibula Y, Abudujielili A, Kasimu M. Identification and validation of an individualized prognostic signature of lower-grade glioma based on nine immune related long non-coding RNA. Clin Neurol Neurosurg. 2021;201:106464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |