INTRODUCTION
The latest estimates of global cancer incidence and mortality provided for 2020 show that colorectal cancer (CRC) ranks third in terms of incidence and second in terms of mortality[1]. More than 1.9 million new CRC cases and over 935000 deaths caused by this disease were registered worldwide by GLOBOCAN in 2020[1]. Being the most prevalent type of gastrointestinal (GI) neoplasia, CRC is also the most preventable due to its association with modifiable life-style factors[2]. Besides, colorectal tumors are often curable as they grow slowly and can be detected and treated early if effective screening is applied[2,3]. One important biological feature of CRC is its origination from the colonic epithelium, enormous surface of which constitutes the interface between the human body and potentially carcinogenic gut content. Indeed, the intestinal lumen is inhabited by billions of diverse microorganisms forming a unique microbial ecosystem within the host’s organism[4]. Microbial density inside the human colon reaches 1012 bacterial cells per gram of predominantly anaerobic colonic content that is, nevertheless, indispensable for host development and physiology[5]. This complex and sometimes aggressive ecosystem permanently interacts with the single layer of intestinal epithelial cells that are highly active, being involved in various absorption and secretion processes.
The immense surface of the intestinal mucosa defines the utmost importance of the continuous interplay between its epithelium and gut microbiota. Until the first decade of the XXI century bacteria were presumed to directly contact the epithelial cells[6]. Only in 2008 a research group from Gothenburg (Sweden) convincingly demonstrated the existence of a well-structured system of protective mucus that effectively separates the epithelium from gut content[7]. That seminal study was followed by impressively productive exploration of the gut mucus and its functional significance in health and disease. This review is focused on recent developments in this field, particularly highlighting the emerging evidence of the importance of colon mucus (CM) for the pathogenesis of inflammatory bowel disease (IBD) and colorectal tumors, as well as associated problems of colorectal disease diagnosis, prevention and treatment.
CM COMPOSITION AND STRUCTURAL CHARACTERISTICS
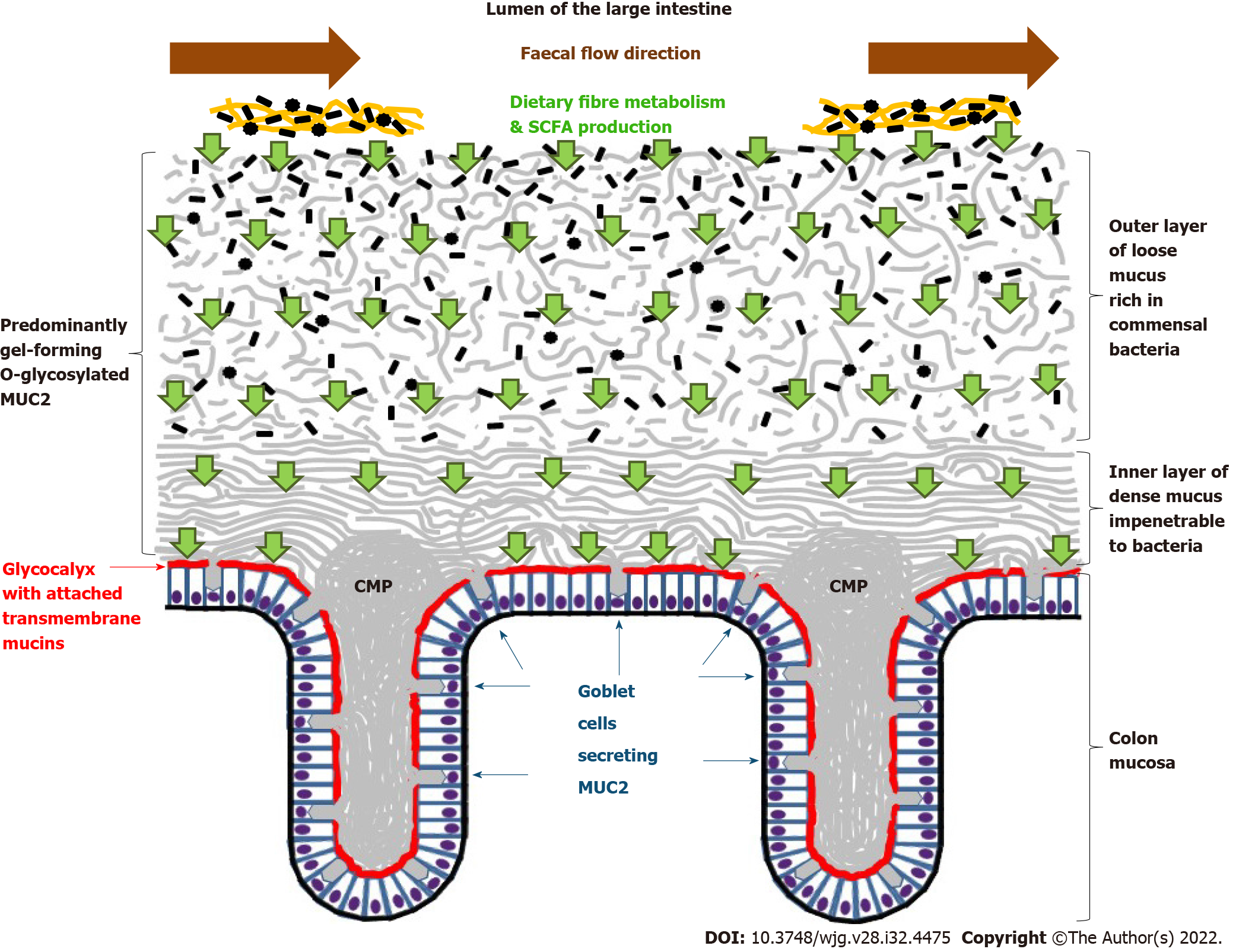
It is now well established that all surfaces of the columnar epithelia lining inner cavities of the human body are overlayed by mucus, which can be generally defined as a complex viscoelastic substance usually secreted by goblet or mucous cells and protecting the underlying epithelium[6,8]. Distinct mucus types naturally adapted for organ-specific functions are described for the GI tract, the respiratory system, the reproductive organs and the ocular surface[6,8,9]. Although mucus composition is organ-specific, its constituents always comprise water (90%-95%), electrolytes, lipids and various proteins[8]. The high water content makes normal CM transparent, which could be one of the reasons of the late discovery of its structure[6,7] schematically presented in Figure 1.
Figure 1 Schematic representation of normal human colonic mucosa and overlaying mucus layers.
Small green arrows show short-chain fatty acid transport through colon mucus layers. Small black shapes show bacteria. CMP: Colon mucus plumes; MUC2: Mucins 2; SCFA: Short-chain fatty acids.
The review is focused on the colon, therefore further discussion is largely devoted to the CM, properties of which are determined by its specific proteins, mucins, defining structural and functional characteristics of this substance[10]. All GI tract mucins are densely decorated with complex carbohydrates or glycans[11]. It is important to stress that the high degree of mucin O-glycosylation protects the peptide bonds, hence rendering these proteins inert to the proteolytic action of the host’s proteases[11,12]. There are 22 genes encoding mucins, the presence of phosphotransferase system (PTS)-rich sequences [tandem repeat units rich in proline, threonine (Thr) and serine (Ser)] being their characteristic feature[8-10]. In these proteins a large proportion of the Ser and Thr residues are O-glycosylated with the creation of a “bottle brush” configuration important for high water-binding capacity and gel-forming properties[8-10,13,14]. GI tract mucins can be subdivided into two classes comprising: (1) Transmembrane mucins that have a transmembrane domain enabling them to be anchored in the apical cell membrane; and (2) Gel-forming mucins that are synthesised and secreted by the goblet cells (GCs), thus being the key CM components[8,10,13,14].
Transmembrane mucins
The transmembrane mucins of epithelial cells are a family of large and extended glycoproteins that are attached to the apical cell membrane through a single-pass transmembrane domain involved in intracellular signaling. This family includes MUC1, MUC3, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17, MUC 21 and MUC22 that vary in length and the presence of several specific domains[14,15]. All transmembrane mucins have PTS domains, which are heavily O-glycosylated, but glycosylation patterns along the GI tract vary, also being different in health and disease[15]. The transmembrane mucins MUC3, MUC12 and MUC17 cover the apical cell membrane of enterocytes and colonocytes, forming the attached protective glycan-rich diffusion barrier, called glycocalyx (Figure 1). It was reported that MUC3 is expressed throughout the whole intestine, MUC12 appears to be colon-specific, and MUC17 is abundant in the small intestine but is also found in the transverse colon[11,13,14]. The transmembrane mucins do not belong to the CM, and their precise role in health and disease remains to be fully elucidated. Detailed discussion on this subject can be found in a recent review by Pelaseyed and Hansson[15].
Gel-forming mucins
The gel-forming mucins of the GI tract are synthesised and secreted by the mucosal GCs. The most abundant and best characterized of the secreted mucins is MUC2, the main element of the mucus overlaying mucosal surfaces of the small intestine and colon. Other gel-forming mucins of the GI tract are MUC5AC (expressed in the stomach), MUC5B (weakly expressed in the colon) and MUC6 (expressed in the stomach and duodenum)[13,14]. The structural organization of the GI mucus was initially determined in rodents, and it was shown that mouse stomach and colon have two distinct layers of dense (inner) and loose (outer) mucus, whereas this pattern is absent in the small intestine, where only a single layer of loose mucus is present[7,11,16]. It is now known that in the human colon the thickness of the dense inner mucus layer is about 200-300 μm, and the loose outer layer is at least twice as thick[6,10,11]. Figure 1 schematically shows the structure of the human CM. It should be stressed that, despite almost identical protein profiles dominated by MUC2, the two CM layers are essentially different[16]. In the normal conditions the dense inner layer is permanently renewed by colonic GCs producing MUC2, which remains anchored to the GCs and attached to the epithelium[10]. Remarkably, the high density of the inner CM layer appears to make it devoid of bacteria[7,11,16]. At a certain distance from the epithelial surface (over 200 μm in humans), the inner mucus is abruptly replaced by the loose outer CM layer, where commensal bacteria live and thrive[16]. The two mucus layers form a sharp border separating them. According to the current paradigm, the dense inner layer is converted into the loose outer mucus by endogenous (host’s) proteases, however it is impossible to exclude that gut bacteria populating the outer layer also contribute to the conversion[7,10,11,16]. It is also worth noting that alongside the mammalian colon there are longitudinal differences in both microbiota composition and MUC2 O-glycan pattern distribution in the CM. It the mouse CM was generally characterized by the abundance of highly charged fucosylated glycans, but glycan sulfation level was higher in the distal colon, whereas sialic acid was more common in the proximal colon[17]. It was suggested that the observed differences in mucin glycan structures might be involved in the bacterial selection process by bacterial adhesin - mucin glycan interactions[17]. All these important basic findings are further discussed below in relation to interactions between gut microbiome, CM and colonic epithelium.
GOBLET CELLS AND THEIR ROLE IN CM FORMATION AND MAINTENANCE
Mucus production
Intestinal GCs are responsible for the continuous synthesis and secretion of gut mucus. The details of the complex process of MUC2 synthesis in the GCs are described in recent reviews[8,12,14], but it is useful to note that MUC2 monomers form C-terminal dimers and N-terminal trimers in the endoplasmic reticulum, and the extensive O-glycosylation of the large PTS domain of MUC2 occurs in the Golgi apparatus. The MUC2 polymer is then densely packed in the secretory granules of the GCs under conditions of the low pH and high calcium concentration[11,14,18]. It is believed that the formation of secretory granules occurs during the maturation of GCs coinciding with their migration from the crypt bottom towards the luminal surface[10]. Finally, the secretory granules undergo exocytosis by fusion with the apical membrane, thus releasing their content to the surface of the epithelium. Upon its release, which is accompanied by an increase in pH and a decrease in calcium concentration, the densely packed mucin expands over than 1000-fold[6,10]. The secreted MUC2 immediately unfolds into large net-like planar sheets that are then assembled in porous lamellar networks forming the dense inner CM layer[12,19,20]. The stability of the inner mucus layer is further increased by the formation of isopeptide bond crosslinks catalyzed by transglutaminase 3 produced by GCs and neighbouring colonocytes[21].
The process of permanent mucus renewal is very intense, inner layer renewal time in the mouse colon being only about 1 h[22]. It was also observed that GCs of the luminal surface epithelium produce mucus faster than their counterparts located in the crypt epithelium[22], and it has later been shown that the properties of mucus generated by GCs of the colonic surface epithelium (intercrypt GCs) differ from those of mucus secreted by crypt-resident GCs. Intercrypt GCs have a specific transcript profile and produce less dense and more penetrable mucus compared to the mucus “plumes” synthesised by crypt-resident GCs and possessing more stringent barrier properties[23] (Figure 1). Single GC analysis revealed that within the colonic epithelium there are several different GC clusters, all originating from stem cells at the bottom of the crypt but forming two distinct “differentiation trajectories”. One of them, termed “canonical” GCs, is characterized by the expression of typical GC markers, whereas the other one expresses enterocyte-associated genes and was designated as “noncanonical”[23]. The intercrypt GC population appears to be predominantly composed of the most differentiated “canonical” GCs continuously secreting mucus at baseline. In contrast, “noncanonical” GCs, which are more abundant in the crypts and contribute to mucus “plume” secretion, are likely to be more responsive to external stimulation[23]. These results are in agreement with the identification of at least five clusters of GCs with distinct gene expression profiles in the human colon[24].
The discovery of colonic GC diversity inevitably leads to questions on functional differences between GC subpopulations and alternative regulatory mechanisms governing CM production in health and disease. The emerging evidence of GC participation in controlling immune responses in the gut is especially important in this context[25]. In 2012, McDole et al[26] reported that small intestinal GCs could endocytose low molecular weight soluble antigens from the gut lumen, being capable of delivering them through GC-associated antigen passages (GAPs) to underlying CD103+ dendritic cells of the intestinal Lamina propria. Further studies of this group revealed the existence of this antigen-presenting phenomenon in the colon as well, when acetylcholine-induced GAP formation in colonic GCs was observed[27]. These GAPs could even translocate live commensal bacteria across the epithelium following antibiotic treatment[28]. It is also believed that GAP formation may be associated with compound exocytosis by GCs, but not primary exocytosis[29]. Hence, the process of GAP formation involves both endocytosis and exocytosis, the two interlinked pathways closely related to autophagy[30], and it was not surprising that autophagy proteins were found to control mucin granule accumulation in colonic GCs[31]. Among autophagy-related factors associated with intestinal GCs, the NOD-like receptor family pyrin domain containing 6 (NLRP6) inflammasome has recently attracted considerable interest[32]. Wlodarska et al[33] first demonstrated that the NLRP6 inflammasome acts as a major orchestrator of mucin granule exocytosis in the colon. This theme was further developed by the identification of sentinel GCs localized at the colonic crypt entrance and capable of endocytosing microbe-derived ligands of toll-like receptors (TLRs) from the lumen. This, in turn, triggers the TLR pathway stimulation and leads to the activation of NLRP6 inflammasome. The response provokes compound exocytosis of MUC2 from the sentinel GCs, entailing their death and inducing enhanced mucus production from adjacent GCs through intercellular gap junction signaling[34]. Nevertheless, the described NLRP6 inflammasome involvement does not seem to be important for either CM formation or its function at baseline[35]. Taken together, these results suggest that NLRP6 modulates GC function and CM secretion only in response to interactions with TLR agonists, being inactive in the normal conditions[33]. The role of GCs in monitoring extracellular environment, interacting with the gut microbiota and communicating with the immune cells of the Lamina propria, is being actively studied and discussed[25,29,36], but further research is required for its comprehensive clarification.
Other products of goblet cells and protective properties of the CM
Although MUC2 is the main product of the GCs in the colon, mucin granules of these cells also contain other mucus components, such as Fc fragment of immunoglobulin (Ig) G-binding protein, calcium-activated chloride channel regulator 1, zymogen granule protein 16 (ZG16), anterior gradient 2, and trefoil factor 3[11,12]. Some of these proteins possess antibacterial properties. In particular, the lectin-like protein ZG16 prevents bacterial penetration into the inner CM layer[37]. Likewise, proteins resistin-like molecule beta, predominantly synthesised by colon GCs[38], and cathelin-related antimicrobial peptide, which is produced by both epithelial cells in colonic crypts and mucosal macrophages[39], were demonstrated to be bactericidal. Moreover, the loose outer mucus layer in the colon is likely to be intermixed with distally moving loose mucus generated in the small intestine and rich in antimicrobial peptides (AMPs) secreted by Paneth cells located in the mucosa of the small intestine[40]. Besides, the process of mucus barrier formation appears to be even more complex as it has recently been shown that in mice mucus abundantly produced in the proximal colon and encapsulating the faecal material considerably differs from that secreted in the distal colon for secondary strengthening the encapsulation process[41]. However, given numerous species-specific differences, direct extrapolation of these results to humans remains impossible until the matter is comprehensively investigated[42]. In any case, the presence of a range of AMPs probably produced at different sites throughout the GI tract was clearly demonstrated in human rectal mucus samples[43]. It can be added that secretory IgA antibodies produced by plasma cells residing in the intestinal Lamina propria were shown to be transported to the mucus by transcytosis[44], and IgA in the colon is concentrated in the outer mucus layer[45]. Finally, it should be mentioned that the steep oxygen gradient keeping the inner CM layer relatively well oxygenated also prevents anaerobic luminal pathogens from reaching the epithelium[46]. The presence of all these factors and the permanent distal movement of the outer CM with the peristaltic waves[10,47], contribute to the protection of the inner CM layer from bacterial invasion.
INTERACTIONS BETWEEN CM AND GUT MICROBIOTA
The human gut harbors a highly diverse community of commensal bacteria usually referred as microbiota, which exhibits both longitudinal and cross-sectional variation in both location and density[48,49]. Microbiota composition in the normal human colon is dominated by Bacteroidetes and Firmicutes phyla, with the presence of members of a few other phyla and strong variability between individuals[48,50]. Notably, dietary fiber digestion is one of the most important functions of the gut microbiota as these plant polysaccharides are largely indigestible by human glycoside hydrolases[51]. The anaerobic fermentation of fiber-derived sugars by gut lumen bacteria produces short-chain fatty acids (SCFAs), including butyrate, acetate and propionate. These SCFAs obviously pass through the intestinal mucus (Figure 1) before being taken up and utilized by the epithelium (butyrate provides up to 70% of colonocyte energy supply). SCFAs are indispensable for both intestinal homeostasis and a variety of effects on tissues and organs beyond the gut (see review by van der Hee and Wells[52]). Remarkably, abundantly glycosylated mucins of the CM can also be targeted by gut bacteria as nutrient sources[50]. Hence, mucin-decorating glycans are thought to constitute a critical resource utilized by commensals to enable them to thrive when diet-derived glycans are limited[53]. This suggestion was experimentally proven by the observations of gut microbiota switch to consuming host-secreted mucus glycoproteins during chronic or intermittent dietary fiber deficiency in mice[54]. The diet-induced microbiota behavior change was, however, unfavorable for the host, leading to CM barrier degradation and lethal colitis development[54].
The described observations demonstrate that both the loose mucus of the small intestine and the loose outer CM layer serve as both energy sources and habitats for human gut microbiota[50]. In the colon of experimental animals (information regarding human microbiota is scarce) the outer mucus layer harbors a mixed population of commensals with the typical presence of Akkermansia muciniphila and bacteria of Bacteroides genus (e.g., Bacteroides thetaiotaomicron, Bacteroides fragilis, Bacteroides vulgatus)[48-50]. Outer CM layer also contains Rumminococcus gnavus, Rumminococcus torques, and Desulfovibrio desulfuricans, as well as probiotic Lactobacillus reuteri, Lactobacillus rhamnosus, Lactobacillus johnsonii, Bifidobacterium breve and Bifidobacterium longum[55]. In addition, bacteria of relatively aerotolerant phyla (Proteobacteria, Actinobacteria) were reported to be occasionally present even closer to the mucosal surface[48,49].
It has been hypothesised that in health CM not only constitutes a physical barrier between bacteria and the epithelium but provides a bioactive environment selectively favorable for beneficial commensals and hostile for intruding pathogens[56]. As mentioned above, mucin glycoproteins provide nutrition for mucus-dwelling bacteria that are able to degrade them, like Akkermansia muciniphila, Bacteroides fragilis and Rumminococcus gnavus[50,53,56]. However, mucin glycans may not be the preferred carbon sources for some host commensals, including Bacteroides thetaiotaomicron[57]. Interestingly, this human gut symbiont was recently shown to possess a unique ability to initiate degradation of the complex sulphated O-glycans of the distal colon[58]. The activities of 12 sulfatases produced by this species were collectively sufficient for degrading all known sulphate linkages in mucin O-glycans, but only a single key sulfatase was disproportionally important for the utilization of sulphated O-glycans[58]. This example illustrates the challenging complexity of mucin degradation pathways that may become relevant for the development of future therapeutic applications.
CM is also responsible for the spatial organization of mucus-dwelling bacteria. Mucin-based networks with varying pore size and adhesiveness provide three-dimensional scaffolds for bacterial settlement, thus governing the process of bacterial colonisation[56]. Adhesiveness modulation by the host can also be employed as a mechanism altering microbiota composition[59]. Heterogenous glycan patterns of mucins affect bacterial motility and aggregation and prevent certain pathogens from aggregating and forming biofilms[56]. In addition, CM is believed to control the traffic of small signaling molecules, thereby regulating microbial behavior, and in some circumstances mucin glycans probably exert signaling functions themselves[56].
The diversity of mucin-decorating glycans constitutes a highly variable element of human CM and influences both gut microbiota behavior and host susceptibility to infectious and metabolic diseases[53,60]. Its importance for microbial community composition can be illustrated by the dependence of the latter on the presence of a functional copy of the galactoside 2-alpha-L-fucosyltransferase 2 (FUT2) gene encoding FUT2 that facilitates the attachment of the L-fucose monosaccharide to O-glycans, producing α(1,2)-fucosylated glycans[61]. The FUT2 is one of the genes responsible for the expression of ABO histo-blood group antigen precursors in gut mucus[53,60-62]. Individuals with at least one functional FUT2 allele are termed “secretors”, although the gene does not actually regulate the secretion of proteins bearing α(1,2)-fucosylated glycans[60]. The homozygosity for the loss-of-function FUT2 mutations (found in about 20% of Caucasians) defines “non-secretors”[53,60,62]. Interestingly, considerable differences in CM bacterial profiles were repeatedly detected between “secretors” and “non-secretors”. Rausch et al[62] found that in healthy “secretors” the probiotic Lactobacillus genus was more prominent, whereas a clear association of the “non-secretor” status with the genus Prevotella was observed. Other FUT2 genotype-associated phylotype changes, including the decrease of Roseburia and Faecalibacterium in endoscopic lavage samples from “non-secretors” or probiotic Bifidobacterium decrease in faeces from “non-secretors” were also reported[63,64]. In contrast, a few relatively large studies failed to detect any associations between FUT2 genotype and gut microbiome shifts[65-67]. However, an even larger genome-wide association study (GWAS) comprising 8956 German individuals from five independent cohorts confirmed the previously identified associations of ABO histo-blood groups and FUT2 secretor status with Bacteroides and Faecalibacterium species[68]. These discrepancies are not surprising since GWAS studies assessing gut microbiome changes are usually based upon using homogenised stool samples. The use of this crude approach means that it is impossible to find out which bacteria are where, how their gene expression and functions are related to the local environment and how their spatial organization changes in disease[48]. It can be argued that samples obtained by mucosal biopsy, colonic lavage or CM collection are much more reliable and informative for analysing the microbiota populating CM. However, it is obvious that O-glycans associated with MUC2 strongly influence the composition of the commensal microbial population confined to the outer mucus layer in the colon.
It was already stated before that there are longitudinal differences in both microbiota composition and MUC2 O-glycan pattern distribution in the mammalian gut[17]. The distribution of O-glycosyltransferases produced by the epithelial cells along the colon correlated well with the pattern of O-glycans[69]. These regional-specific characteristics of the gut glycan pattern are certainly acquired after birth, as neither sialic acid nor fucose gradients exist along the foetal intestine[70]. The differences developing in the postnatal life are thought to be related to the establishment of luminal microbiota acting as a potent environmental factor that provokes dramatic gene expression shifts in the host epithelium[70]. This view is supported by mouse experiments demonstrating that commensal colonisation promotes structural and physiological adaptations of mucus barrier properties, thus contributing to intestinal homeostasis[71,72]. The complex interplay between gut microbiota and colonic mucosa continues into the maturity of the host and becomes even more important during ageing. In mice ageing causes a progressive decrease of CM thickness that is accompanied by considerable changes in faecal microbiota composition and more frequent contacts of luminal bacteria with the epithelium[73]. A similar reduction of CM layer thickness, primarily attributed to the reduced number of GCs, was observed in ageing rats[74]. Interestingly, a recent report shows that indoles produced by commensal bacteria can act via the aryl hydrocarbon receptor and interleukin 10 to restore the depleted population of colonic GCs in aged animals, hence supporting homeostasis[75]. Although age-related changes in the human CM system are poorly studied, it is notable that the intestinal microbiota composition in elderly subjects substantially differed from that in young adults, with a greater proportion of Bacteroides species and distinct abundance of Clostridium groups in the elderly[76].
To conclude this section of the review it is important to clarify that the gut mucus barrier, albeit essential, is just an element of the complex mechanism responsible for protecting the host’s organism from unwanted risks. The immune system is the key driving force of this mechanism playing a central role in shaping the composition of the microbiota and defining its spatial distribution in close proximity to host tissues. Nevertheless, resident microorganisms interact with the immune system and influence the development of immune responses. Disruption of this complex and dynamic cross-talk can have deleterious consequences for host health contributing to the pathogenesis of many diseases including IBD and cancer[77].
CONSEQUENCES OF GUT MUCUS DETERIORATION EXPOSING COLONIC EPITHELIUM
CM and underlying epithelium: Host cells are released into mucus in the normal physiological conditions
It is well established that the intestinal epithelium is renewed every 4-5 d[78], and, before the discovery of CM structure and recognition of its importance[6], it was presumed that terminally differentiated enterocytes or colonocytes undergo spontaneous apoptosis and are finally “shed into the gut lumen”[78]. This simplistic notion needed to be revised, given the complexity of the two-layered CM structure and high density of the inner mucus layer[6,10,11]. Our group previously demonstrated that in healthy individuals exfoliated colonocytes are rarely found in CM samples collected either from the surface of the rectal mucosa or non-invasively[79,80]. Similarly, only small numbers of normal exfoliated colonocytes in mucus-containing surface washes of stool samples obtained from healthy volunteers were observed in earlier studies[81-83]. In view of the proven existence of the two-layered CM structure (Figure 1), an immediate question emerges: How exfoliated colonocytes and occasional neutrophils released from the epithelial surface manage to penetrate the dense inner mucus layer impenetrable for much smaller bacteria? No satisfactory answer has hitherto been found, but it was previously assumed that shed cells could be “trapped in the mucus” and degraded there[7].
Remarkably, well-preserved colonocytes were identified in all quoted studies that analysed CM[47,79-83]. One could argue that intrarectal mucus collection might mechanically detach epithelial cells from the mucosa[79], however this explanation is not valid when CM is obtained non-invasively or from the surface of freshly excreted stool[80-83]. Although it is possible to hypothesise that the transport of the released cells might be assisted by the rapid CM layer renewal[22], or that focal partial cleavage of MUC2 may occur at sites of colonocyte exfoliation or neutrophil migration[16], mechanisms of this phenomenon remain obscure. In contrast, abundant presence of both exfoliated colonocytes and migrating inflammatory cells in CM samples obtained from patients with IBD and CRC is easy to explain by the structural damage of the CM barrier observed during these conditions and considered below. Despite the existing uncertainties on the mechanisms involved in cell accumulation in the mucus, it is indisputable that this easily accessible substance presents a highly informative material for multiple diagnostic applications.
CM barrier damage and its consequences in gut inflammation predisposing to CRC
CM presents the first defensive barrier against the luminal microbiota, its dense inner layer making bacterial contact with the colonic epithelium hardly possible in the healthy gut[7]. Although loose mucus of the small intestine contains commensal bacteria, it is rich in antimicrobial substances, and its permanent production by GCs in the normal conditions creates a continuous mucus flow preventing bacteria from reaching the epithelium[11,84]. Therefore, only severe CM impairment can lead to exposing the epithelial surface to direct contacts with the microbiota. The unique importance of MUC2-rich mucus for colonic epithelium protection was graphically demonstrated by the development of spontaneous colitis in MUC2-knockout mice unable to produce MUC2[85]. Likewise, microbiota-induced mucus layer defects were observed in genetically obese mice[86]. Experimentally modelled dietary fiber deficiency also led to CM barrier degradation, bacterial invasion of the mucosa and lethal colitis development[54]. The composition of intestinal microbiota is now recognised as a key factor for defining properties of the inner mucus layer[71,72], and it was clearly demonstrated that dysbiosis in the mucus preceded experimental colitis development[87]. Results of all these experimental studies clearly demonstrate that the loss of CM barrier integrity combined with dysbiosis usually results in opportunistic invasion of the colonic mucosa by resident bacteria, inevitably leading to inflammation[77].
In patients with IBD, comprising ulcerative colitis (UC) and Crohn’s disease (CD), CM deterioration and the abundance of mucolytic bacteria were observed[88,89]. Aforementioned mucus microbiota shifts related to FUT2 “non-secretor” genotype entailed an increased risk of developing CD[62]. The gut bacteria in IBD patients are often confined to mucosa-adhering biofilms that can be defined as matrix-enclosed multispecies bacterial communities forming higher-order structures[90,91]. Mucosal biofilms were frequently found in IBD patients and patients with irritable bowel syndrome (IBS), and, sometimes, in clinically healthy individuals[88,91,92]. These findings indicate that bacterial biofilm formation does not immediately lead to inflammation development, being, at least to some extent, compatible with the preservation of homeostasis control by the immune system. Probable involvement of biofilms in IBS pathogenesis may open new ways for devising diagnostic approaches and therapeutic strategies addressing this highly prevalent condition[92].
Patterns of alterations in the colonic mucosa, its mucus layers and gut microbiota composition tend to differ between CD and UC[84]. For example, it was shown that a global expression of mucin genes was reduced in CD patients[93], whereas it was elevated in patients with UC[94]. Conversely, UC was also found to be associated with decreased numbers of mucosal GCs[95,96], as well as signs of CM layer reduced thickness and disruption[95,97,98]. In addition, the secretory response of colonic GCs to microbial challenge in active UC seems to be impaired, with the number of sentinel GCs significantly reduced and protective mucin sulphation decreased[99,100]. Intestinal mucosa dysfunction in IBD was further confirmed by signs of transcriptomic dysregulation of some key genes involved in colonic barrier maintenance, especially those encoding transmembrane mucins MUC1, MUC4 and MUC22[101]. Consequently, the inner mucus layer of UC patients could be easily penetrated by luminal bacteria[98]. The observed dysregulation of transmembrane mucin synthesis might also indicate possible alterations of colonocyte glycocalyx[101].
The extreme complexity of IBD pathogenesis is generally admitted[102], and this fascinating subject is beyond the scope of this review. Nevertheless, a few more points related to IBD need to be addressed here, especially given the clearly elevated probability of CRC development in IBD patients[103]. In particular, modern Western diet characterized by limited dietary fiber intake is now regarded as a major risk factor for both IBD and CRC[104-106]. Consequently, diet correction may constitute an effective approach to disease prevention. Experimental studies demonstrate that fiber addition to the diet and administration of probiotic microbiota, especially Bifidobacterium species, can restore CM layer functionality and provide anti-inflammatory effects[107,108]. Likewise, tea polyphenols and citrus flavonoids were shown to protect CM integrity[109,110]. These findings look promising, however further research is required for designing dietary interventions targeting CM and suitable for treating IBD patients and preventing CRC development in this risk group.
Another consequence of CM barrier deterioration in IBD patients is related to the phenomenon of massive inflammatory cell migration from the Lamina propria of colonic mucosa to the impaired CM layer. Neutrophil and eosinophil biomarkers are abundantly present in stool samples from IBD patients, and stool calprotectin quantification is widely used for diagnosing UC and CD[111,112]. However, the scale of inflammatory cell migration towards the gut lumen became evident only recently. We were able to demonstrate that numerous immune cells can be found in CM samples obtained from IBD patients either intrarectally[79,113], or using non-invasive collection from the anal area following defaecation[80,114,115]. Cytological analysis of the collected CM revealed that neutrophils were the most abundant cell type, but macrophages, eosinophils (especially in UC patients), plasma cells and lymphocytes were frequently detected as well[114]. The collected cells were very well preserved and looked viable, as phagocytosis and erythrophagocytosis by neutrophils and macrophages could be seen[114]. It was also hypothesised that neutrophils and eosinophils migrating to the loosened CM of IBD patients can easily undergo extracellular DNA trap formation (ET-osis), cell death producing the formation of antibacterial “extracellular traps” composed of released DNA and globular proteins [also called ET-osis exerted by neutrophils (NET-osis)][116]. This hypothesis, also assuming that CM in IBD serves as a supporting milieu for immune responses expanding from the mucosa, is discussed in detail elsewhere[116]. Taken together, recent advances in CM research suggest that even partial loss of this protective barrier compromises its function at different levels, including impaired control of the relationship between gut microbiota and mucus, dysregulation of CM layer maintenance by the epithelium and immune cells of the Lamina propria, insufficient production of AMPs and defects in the process of autophagy[117,118]. Initial signs of intestinal barrier impairment may precede the onset of clinical IBD manifestations by years[119], this interval probably constituting a good window of opportunity for applying preventive interventions and early therapeutic measures.
CM CHANGES ASSOCIATED WITH COLORECTAL NEOPLASIA
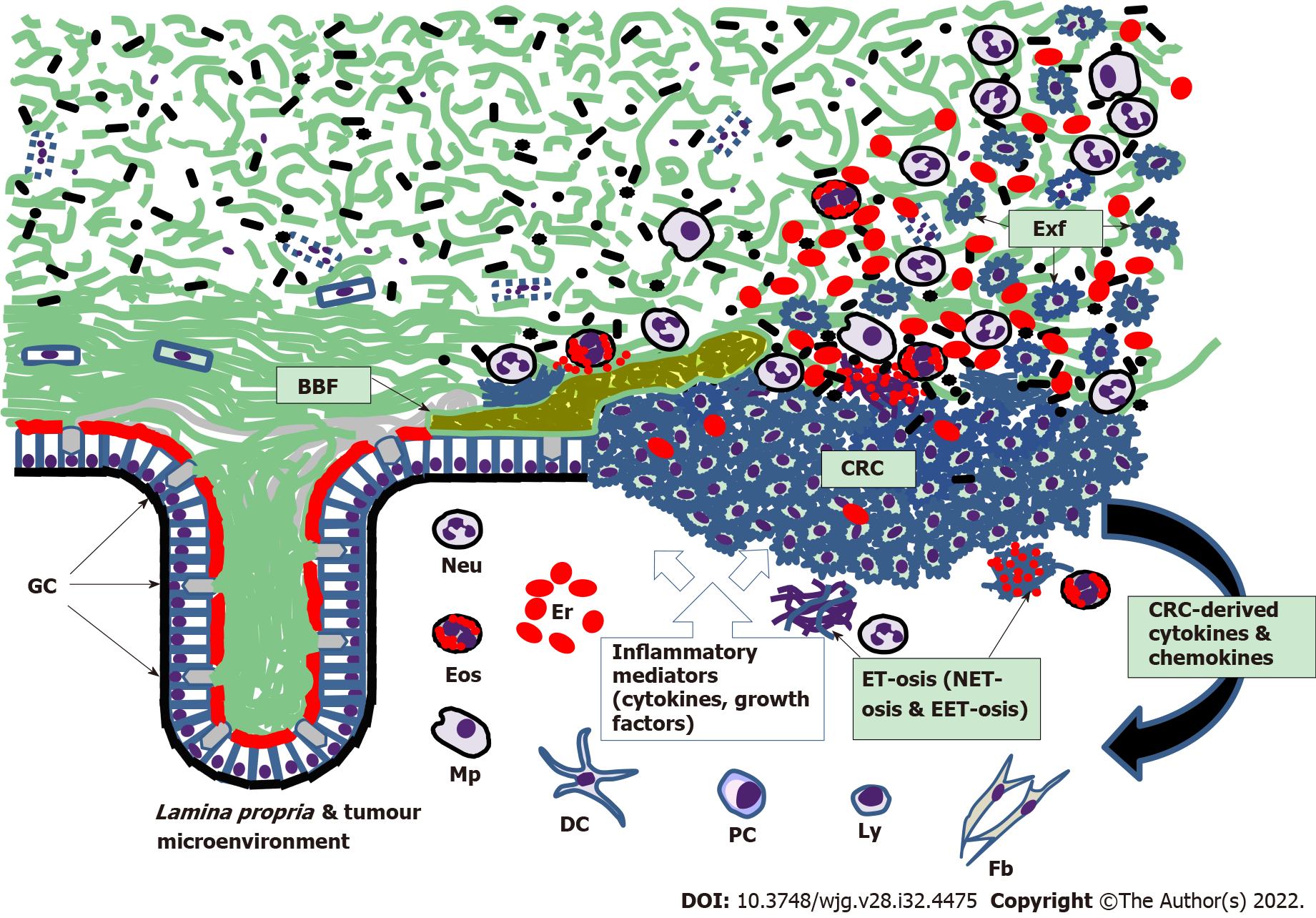
Although inflammation-related changes involving CM can contribute to colorectal tumor development[120], sporadic CRC grows slowly, early stages of this neoplasia being confined exclusively to colorectal mucosa[3]. However, it is evident that, like in active IBD, CM from CRC patients is very rich in cells shed from tumor surface (Figure 2). It is well established that numerous CRC markers can be detected in stool samples obtained from CRC patients[121,122], and the presence of cancer cells on the stool surface is well documented[79-81,123-125]. CM overlaying tumor surface, which was initially defined as “mucocellular layer”[126], serves as a medium accepting and preserving all cells and biomolecules released by the neoplastic tissue. This diagnostically informative material could be obtained from CRC patients by intrarectal collection with an inflatable device designed for this purpose[79]. Human DNA measurements in the mucus collected from tumor surface and at equal distances proximally and distally from tumor margins (resected colon segments were examined) revealed significantly higher DNA levels in the samples collected distally, hence confirming distal movement of the CM[79]. The latter finding of our group resulted in devising a completely non-invasive technique for CM sampling by swabbing the anal area immediately following defaecation[80]. The new method has recently been successfully applied for non-invasive CRC detection[127,128]. These findings confirm that CM constitutes a uniquely informative biological material possessing an enormous diagnostic potential owing to abundant presence of various biomarkers, comprising a wide range of proteins and nucleic acids[116,127,128]. While the described diagnostic importance of CM for CRC detection becomes obvious, this biological substance also emerges among major pathogenetic factors contributing to colorectal carcinogenesis. The key mechanisms of CM involvement in this process are probably related to the loss of homeostatic balance between CM layers and gut microbiota, which exposes the mucosa to both commensal and pathogenic bacteria and triggers cascades of unfavorable host responses that may eventually lead to neoplastic growth[120,129-131]. CM depletion effect was graphically demonstrated experimentally, when mice genetically deficient in the MUC2 were shown to develop colorectal tumors[132,133]. Furthermore, colon tumor development was observed in mice with colon-specific loss of Atonal homolog 1 gene, which determines normal differentiation of secretory cells, comprising GCs[134]. GC depletion during 1,2-dimethylhydrazine-induced colon carcinogenesis in rats was reported as well[135]. In human colorectal tumors, MUC2 expression was decreased, and the degree of expression inhibition correlated with the progression from adenomas to advanced carcinomas[136]. In terms of prognostic significance, reduced MUC2 expression in CRC patients corresponded to poorer prognosis[137,138]. However, different types of CRC display different mucin expression patterns, as several mucin-encoding genes are overexpressed in mucinous carcinomas[139-141]. Mucus synthesis in the colonic GCs involves MUC2 O-glycosylation, and this process was found to be impaired in genetically modified mice predisposed to colitis-associated carcinogenesis[142,143]. Likewise, aberrant O-glycosylation was observed in malignant tumors removed from CRC patients[144]. All these findings indicate that CRC-associated changes in colon mucin gene expression and protein synthesis may occur either because of intrinsic genetic and immune disturbances or due to interactions of the preneoplastic or neoplastic epithelium with the microbial populations of the gut, impact of which needs to be considered as well.
Figure 2 Scheme of colorectal mucus-associated events involved in colorectal cancer development.
BBF: Bacterial biofilm; Exf: Exfoliated malignant cells of colorectal cancer; GC: Goblet cells; Neu: Neutrophils; Eos: Eosinophils; Mp: Macrophages; Er: Erythrocytes; DC: Dendritic cells; PC: Plasma cells; Ly: Lymphocytes; Fb: Fibroblasts; ET-osis: Extracellular DNA trap formation; NET-osis: ET-osis exerted by neutrophils; EET-osis: ET-osis exerted by eosinophils. Small black shapes show bacteria; CRC: Colorectal cancer.
It is now recognized that colorectal tumor development strongly depends on compositional and ecological changes of the microbiota[145]. Recent metagenomic studies revealed differences between gut bacterial communities in CRC patients and healthy individuals, the latter category being characterized by a lower abundance of potentially protective taxa (e.g., Roseburia) combined with an increased presence of pro-carcinogenic taxa, such as Bacteroides, Escherichia, Fusobacterium, and Porphyromonas[145-147]. However, the existing body of evidence on CRC-related gut microbiome alterations was generated mostly by molecular analyses of faecal material that may not reliably reflect changes in CM-dwelling bacteria. For this reason, this theme will not be further discussed here, and a comprehensive review by Wong and Yu[145] can be recommended to interested readers.
In contrast, recent reports suggesting that bacterial biofilms formed on the surface of the colonic epithelium may be implicated in carcinogenesis deserve close attention[90,148]. This association is especially intriguing since bacterial biofilms can be easily identified endoscopically[92], thus potentially presenting a very good early indicator of neoplastic transformation risk. Biofilms in the colon of CRC patients are typically confined to the CM overlaying tumor surface or margins and seem to replace the inner mucus layer[148-150], which is apparently damaged during tumor growth. There is no clarity regarding mechanisms of biofilm carcinogenicity, but it was suggested that certain individuals are predisposed to form these bacterial structures, which may be capable of driving neoplastic transformation[149,151]. Proximal CRC and, especially, mucinous carcinomas were strongly associated with the presence of bacterial biofilms, whereas they were observed relatively rarely in distal CRC cases[149,150,152]. This pattern suggests that biofilms tend to specifically contribute to the serrated pathway of colorectal carcinogenesis[153].
The analysis of biofilm bacterial composition in CRC patients has revealed that human gut commensal Bacteroides fragilis capable of generating enterotoxigenic strains[154], as well as oral pathogens Fusobacterium nucleatum, Parvimonas micra and Peptostreptococcus stomatis were the main species detected in these biofilms[150]. Further work of the same group demonstrated the presence of patchy bacterial biofilms dominated by enterotoxigenic Bacteroides fragilis and colibactin-expressing Escherichia coli on the surface of the colonic mucosa of patients with familial adenomatous polyposis[155]. In these patients the biofilms were not confined to polyp surface, and no association with proximal tumor location could be observed[155]. Interestingly, co-colonisation of tumor-prone mice with the bacterial strains of these biofilms caused DNA damage in the colonic epithelium of the animals and accelerated carcinogenesis[155]. These findings were later confirmed in three murine models, where bacterial biofilm homogenates obtained from either CRC patients or healthy individuals manifestly promoted colon carcinogenesis[156]. Notably, Fusobacterium nucleatum did not seem to be required for carcinogenesis in these experiments, which might indicate that it is possibly involved at later stages of CRC development[156]. Overall, these results strongly suggest that polymicrobial biofilms may now be regarded as a colon carcinogen.
The reviewed advances in defining CM role in CRC development highlight a previously obscure area of pathogenetically important interplay between multiple elements. These elements include CM, gut microbiota (both mucus-dwelling and luminal), colonic epithelial cells (normal, malignant and especially mucus-producing GCs) and immune cells (both belonging to the tumor microenvironment or adjacent Lamina propria and actively migrating to CM through the epithelium). Figure 2 schematically depicts some of these interactions. Progressive CM deterioration results in its loosening that initially exposes colonic epithelium to occasional contacts with gut commensals and can later lead to possible pathogen invasion to the mucosa. CM deficiency probably triggers: (1) Epithelial homeostasis dysregulation accompanied by the loss of control over cell renewal process; and (2) Cascades of inflammatory responses exerted by both immune cells of the tumor microenvironment and adjacent Lamina propria and free immune cells migrating through colonic epithelium to the loosened CM. Although these events may potentially be reversible, one needs to be aware of less favorable scenarios, such as the development of IBS, IBD and colorectal tumors. The reported formation of bacterial biofilms in healthy individuals and patients with IBD, polyps and CRC illustrates this range of scenarios[92,149,155]. However, biofilm formation may depend on the genetic background of the host[153], and appears to be just one of at least a few possible pathways eventually leading to CRC.
Of course, there is no chance to properly discuss here multiple molecular pathways of CRC development[120], or complex impacts of immune responses affecting this process[157]. Nevertheless, a few more points directly related to the CM deserve to be addressed at the end. One of them concerns the process of antibacterial extracellular DNA trap formation or ET-osis by effector immune cells (termed NET-osis when exerted by neutrophils) that massively migrate to the loosened CM in both IBD and CRC[116]. It is apparent that this process basically constitutes an element of a protective inflammatory response aiming to eliminate bacteria contacting colonic epithelium, but arriving neutrophils and eosinophils inevitably undergo degranulation and ET-osis, thus releasing cytotoxic factors (Figure 2) that damage host cells and probably induce poorly regulated compensatory proliferation of normal or malignant epithelial cells, which can stimulate neoplastic growth. The role of the ET-osis in CRC is being discussed but remains largely obscure and needs further exploration[116,158]. The other important point is related to possible ways of preserving CM integrity and avoiding its deterioration in view of CRC prevention. This goal can be achieved by establishing proper communication between the host and gut microbiota via balanced dietary patterns preventing dysbiosis development[131]. Indeed, diet is a major determinant of CRC risk, with red meat consumption increasing the risk and fiber uptake being protective[159]. Interestingly, both effects can be mediated by the CM. SCFAs (especially butyrate) produced by luminal microbiota during dietary fiber fermentation stimulates MUC2 expression in the colon and promotes proliferation of normal colonocytes[160], whereas cancer cell proliferation tends to be inhibited by butyrate because of the low differentiation of these cells[161]. There is no doubt that fiber-rich diets and SCFAs are beneficial for CM preservation and CRC prevention, but additional investigations are indispensable for designing effective dietary intervention schemes. On the other hand, it was shown that CM damage inflicted by sulphide-producing and mucin-degrading bacteria can faciliate epithelial hyperproliferation in the colon induced by heme-rich diet modelling high red meat consumption[162]. In that experimental model antibiotic treatment allowed normalizing gut microbiota composition, reinforcing the mucus barrier and eliminating the abnormal proliferation[162]. These examples demonstrate that both dietary corrections and drug therapy can be considered for CM protection and restoration with the purpose of preventing CRC.
The on-going progress in exploring the role of the CM in the pathogenesis of major colorectal diseases, especially CRC, is impressive, however many important points remain obscure. Therefore, thoroughly designed further studies are needed for elucidating fine mechanisms governing structural and functional CM changes in disease. It can be expected that numerous innovative practical applications addressing colorectal disease diagnosis, prevention and treatment will be developed once this challenging goal is achieved.
CONCLUSION
The presented analysis of the existing information regarding mammalian and human CM shows that this area was actively explored only since the beginning of the 21st century. Considerable progress has been made in basic research of CM structure and function, especially in experimental models. It is now evident that in the normal mammalian colon the CM is formed by two distinct layers composed of gel-forming glycosylated mucins that are permanently secreted by goblet cells of the colonic epithelium. The inner layer is dense and impenetrable for bacteria, whereas the loose outer layer provides a habitat for abundant commensal microbiota. Mucus barrier integrity is essential for preventing bacterial contact with the mucosal epithelium and maintaining homeostasis within the GI tract, but it can be impaired by a variety of factors, including CM-damaging switch of commensal bacteria to mucin glycan consumption due to dietary fiber deficiency. It is already proven that impairments in CM structure and function can lead to colonic barrier deterioration that opens direct bacterial access to the epithelium. Bacteria-induced damage dysregulates epithelial proliferation and causes inflammatory responses that may expand to the loosened CM and eventually result in severe disorders, including colitis and colorectal neoplasia. Recently described formation of bacterial biofilms within the inner CM layer was shown to be associated with both inflammation and cancer. Although obvious gaps in our knowledge of human CM remain, its importance for the pathogenesis of major colorectal diseases, comprising IBD and CRC, is generally recognized. Continuing progress in the field of CM exploration promises considerable future achievements and is likely to result in the development of a range of new useful clinical applications addressing colorectal disease diagnosis, prevention and therapy.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: ECCO, 38848.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Almeida C, Portugal; M’Koma AE, United States S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ