Published online Aug 7, 2022. doi: 10.3748/wjg.v28.i29.3886
Peer-review started: January 29, 2022
First decision: April 10, 2022
Revised: April 26, 2022
Accepted: July 11, 2022
Article in press: July 11, 2022
Published online: August 7, 2022
Processing time: 186 Days and 6.2 Hours
The high prevalence and persistence of Helicobacter pylori (H. pylori) infection, as well as the diversity of pathologies related to it, suggest that the virulence factors used by this microorganism are varied. Moreover, as its proteome contains 340 hypothetical proteins, it is important to investigate them to completely understand the mechanisms of its virulence and survival. We have previously reported that the hypothetical protein HP0953 is overexpressed during the first hours of adhesion to inert surfaces, under stress conditions, suggesting its role in the environmental survival of this bacterium and perhaps as a virulence factor.
To investigate the expression and localization of HP0953 during adhesion to an inert surface and against gastric (AGS) cells.
Expression analysis was performed for HP0953 during H. pylori adhesion. HP0953 expression at 0, 3, 12, 24, and 48 h was evaluated and compared using the Kruskal-Wallis equality-of-populations rank test. Recombinant protein was produced and used to obtain polyclonal antibodies for immunolocalization. Immunogold technique was performed on bacterial sections during adherence to inert surfaces and AGS cells, which was analyzed by transmission electron microscopy. HP0953 protein sequence was analyzed to predict the presence of a signal peptide and transmembrane helices, both provided by the ExPASy platform, and using the GLYCOPP platform for glycosylation sites. Different programs, via, I-TASSER, RaptorX, and HHalign-Kbest, were used to perform three-dimensional modeling.
HP0953 exhibited its maximum expression at 12 h of infection in gastric epithelium cells. Immunogold technique revealed HP0953 localization in the cytoplasm and accumulation in some peripheral areas of the bacterial body, with greater expression when it is close to AGS cells. Bioinformatics analysis revealed the presence of a signal peptide that interacts with the transmembrane region and then allows the release of the protein to the external environment. The programs also showed a similarity with the Tip-alpha protein of H. pylori. Tip-alpha is an exotoxin that penetrates cells and induces tumor necrosis factor alpha production, and HP0953 could have a similar function as posttranslational modification sites were found; modifications in turn require enzymes located in eukaryotic cells. Thus, to be functional, HP0953 may necessarily need to be translocated inside the cell where it can trigger different mechanisms producing cellular damage.
The location of HP0953 around infected cells, the probable posttranslational modifications, and its similarity to an exotoxin suggest that this protein is a virulence factor.
Core Tip: The high prevalence and persistence of Helicobacter pylori infection and the diverse pathologies associated with it suggest that the virulence factors of this microorganism are varied. Moreover, its proteome contains 340 hypothetical proteins, so it is crucial to investigate them to elucidate the mechanisms of its virulence and survival. We studied the hypothetical protein HP0953, its location around infected cells, and the probable posttranslational modifications. Its similarity to an exotoxin suggests that HP0953 is a virulence factor.
- Citation: Arteaga-Resendiz NK, Rodea GE, Ribas-Aparicio RM, Olivares-Cervantes AL, Castelán-Vega JA, Olivares-Trejo JJ, Mendoza-Elizalde S, López-Villegas EO, Colín C, Aguilar-Rodea P, Reyes-López A, Salazar García M, Velázquez-Guadarrama N. HP0953 - hypothetical virulence factor overexpresion and localization during Helicobacter pylori infection of gastric epithelium. World J Gastroenterol 2022; 28(29): 3886-3902
- URL: https://www.wjgnet.com/1007-9327/full/v28/i29/3886.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i29.3886
The estimated prevalence of Helicobacter pylori (H. pylori) infection is 70% in developing countries and 30%-40% in the United States and other industrialized countries[1]. However, not all people infected with this bacterium have symptoms. H. pylori has been associated with gastroduodenal diseases and it is considered the major risk factor for the development of gastric cancer[2]. H. pylori colonizes the gastric mucosa, adhering to the mucous epithelial cells and to the mucous layer that covers the gastric epithelium. Adherence to gastric cells is the first step in establishing an infection and for successful colonization, the bacterium releases various effector proteins/toxins[3-5].
Upon colonization, the pathogenic mechanism of H. pylori is mediated by virulence factors such as flagella and the ability to form a biofilm (both in vitro and in vivo)[6]. Biofilm formation enhances the ability of microorganisms to survive in a hostile environment, which would cause and exacerbate infections, rather than extend them, resulting in chronicity[7]. The glycocalyx participates in adhesion to inert surfaces, allowing biofilm formation but also participates in cell attachment; the major components of the glycocalyx are glycans and may contain digestive proteins or secreted virulence factors[8,9]. H. pylori secretes various virulence factors, including urease, cag pathogenicity island proteins, hemagglutinin, Lewis antigens (Lex and Ley), BabA CagA, and VacA[10-12]. All these virulence factors have been extensively investigated elsewhere.
Furthermore, H. pylori contains hypothetical proteins, some of which are secreted particularly in the context of interactions with the host. The bacterium uses a set of secreted and translocated proteins to adapt itself to the mucosal environment[13]. H. pylori possesses a relatively small genome of < 1.6 Mb[14,15] compared with other gram-negative prokaryotes, such as Escherichia coli, whose genome is > 4.5 Mb[16]. Despite its small size, an ineligible fraction of H. pylori proteins, possibly 30%-40%, are annotated as “hypothetical proteins”. Using bioinformatics tools, Naqvi et al[17] and Zanotti and Cendron[18] respectively analyzed 340 hypothetical proteins of H. pylori 26695. The function of some of these proteins can be hypothesized based on a weak homology with proteins of other bacteria, whereas most proteins do not exhibit similarities with others, and their function cannot be predicted[17,18].
HP0953 is a hypothetical and uncharacterized protein of H. pylori secretome; it lacks conserved domains but, interestingly, is present in all H. pylori strains whose genomes have been sequenced, as well as in H. acinonychis that colonizes the gastric mucosa of large felines. The closely related H. hepaticus and Campylobacter jejuni lack HP0953 homologs, suggesting that HP0953 is unique to the gastric Helicobacter species[11]. Moreover, we previously observed that this protein is overexpressed during adhesion to inert surfaces under stress conditions. Most of the possible effects of secreted proteins on the host have yet to be discovered. In addition, secreted factors represent eligible proteins for identifying promising targets for the development of new antimicrobial drugs[17,18].
In recent years, there has been an increasing importance on H. pylori; on the one hand when it is present, it is associated with gastric disease, and on the other hand, it is associated with gastroduodenal reflux, food allergy, and asthma when it is absent[19]. This dual role in the human microbiota emphasizes the need for further characterization of the hypothetical proteins already described. In this study, the location of the protein is determined for the first time, inside and outside the bacterium, and we predict some biochemical characteristics, through in silico analysis that contribute toward understanding the importance of this hypothetical protein to the microorganism in the establishment of infection, its role in such infection, or colonization of the host.
We obtained the amino acid sequence of HP0953 (accession number NP_207745) from the National Center for Biotechnology Information database[20]. Then, we used the Pfam database to analyze the HP0953 sequence and search for conserved domains or homologous regions present in other proteins[21].
The amino acid sequence of HP0953 was analyzed using the SignalP-4.1 tool provided by the ExPASy platform to predict the presence of a signal peptide[22].
The amino acid sequence previously obtained was analyzed to predict transmembrane helices using the TM pred tool provided by the ExPASy platform[23,24]. The glycosylation sites were investigated using the GLYCOPP platform[25].
To explore the remote homologs that could be used as a template for modeling, we used the HHpred tool that uses the HMM comparison[26]. Different programs such as I-TASSER[27], RaptorX[28], and HHalign-Kbest[29] were used to perform three-dimensional (3D) modeling.
H. pylori strain 26695 was cultured on Casman agar (Dibico; Mexico) supplemented with 5% sheep erythrocytes and incubated in an atmosphere of 50 mL/L CO2 and 37 °C for 48 h. The culture was harvested for infected human against gastric (AGS) cell-line assay and genomic DNA extraction. Genomic DNA extraction was performed according to the protocol established for the Wizard kit (Promega; United States) by its manufacturer.
HP0953 was amplified by polymerase chain reaction (PCR) using H. pylori genomic DNA as the template. To amplify the complete gene, 50 μL of reaction mixture was prepared as follows: 1.25 U polymerase GoTaq®, 0.2 mmol/L dATP, 0.2 mmol/L dGTP, 0.2 mmol/L dCTP, 0.2 mmol/L dTTP, 1.5 mmol/L MgCl2 (Promega), HP0953F oligonucleotide (25 pmol) (5’-GGGGGATCCATGGTTTTAATCGCTCTTTTAGGGGTG-3’), HP0953R oligonucleotide (25 pmol) (5’-GGGGTCGACCCTTAACGCACAAACGCTACC-3’), and H. pylori 26695 DNA (250 ng). For amplification of the gene without the signal peptide, the same PCR was performed substituting the oligonucleotide HP0953F by HP095WSPF (5’-GGGGGATCCGTCTCTGATTTAAAGGGCATG-3’). The amplification cycle consisted of 94 °C for 1 min, followed by 30 cycles at 94 °C for 30 s, 68.4 °C for 30 s, 72 °C for 1 min, and a final step at 72 °C for 7 min. Both PCR products (555 and 474 bp) were purified using the protocol established by Promega for using the Wizard® SV gel and the PCR Clean-Up System (Promega).
Complete HP0953 and its version without the signal peptide were inserted into the vector pJET1.2 (Thermo Scientific; Lithuania) according to the manufacturer’s instructions to obtain the recombinant pJET-HP0953 and pJET-HP0953WSP plasmids, respectively. Chemically competent DH5α cells of Escherichia coli (E. coli) were transformed with each plasmid by thermal shock[30,31]. The recombinant colonies were selected in LB agar (Sigma-Aldrich; MO) containing 100 μg/mL ampicillin. The plasmids were extracted using the illustraTM plasmidPrep Mini Spin Kit (GE Healthcare; United Kingdom).
The pJET-HP0953 and pJET-HP0953WSP plasmids were digested with the fast restriction enzymes BamHI and Sal I according to the manufacturer’s instructions (Thermo Scientific) to free both versions of HP0953. Restriction products were purified using the protocol established by Promega for the Wizard® SV gel and the PCR Clean-Up System. DNA fragments were ligated into the expression vectors pET28a(+) (Novagen, United States) for the complete HP0953 and pGEX-6p-2 (GE Healthcare) for the version without the signal peptide. The vectors were digested with the same enzymes and treated with alkaline phosphatase (Thermo Scientific). The Rapid DNA Ligation Kit (Thermo Scientific) was used to obtain the recombinant pET-HP0953 and pGEX-HP0953WSP plasmids. Chemically competent DH5α cells of E. coli were transformed with each plasmid by thermal shock. The recombinant colonies were selected in LB agar containing 50 μg/mL kanamycin for pET-HP0953 or 100 μg/mL ampicillin for pGEX-HP0953WSP. The plasmids were extracted using the illustraTM plasmidPrep Mini Spin Kit (GE Healthcare) and analyzed by PCR. DNA inserts in the recombinant plasmids were characterized by sequencing; for this purpose, PCRs were performed as described earlier using the following primers: T7F (5’-TAATACGACTCACTATAGGG-3’) and T7R (5’-GCTAGTTATTGCTCAGCG-3’) for pET-HP0953 and HP095WSPF (5’-GGGGGATCCGTCTCTGATTTAAAGGGCATG-3’) and HP0953R (5’-GGGGTCGACCCTTAACGCACAAACGCTACC-3’) for pGEX-HP0953WSP. The annealing temperature was 68.4 °C for pET-HP0953 and 60 °C for pGEX-HP0953WSP. The PCR products were purified as described earlier and sequenced by Synthesis and Sequencing Unit, Institute of Biotechnology, Universidad Nacional Autónoma de México. Chemically competent BL21 pLysS cells of E. coli were transformed with each recombinant plasmid and the empty plasmids by thermal shock[31]. The recombinant colonies were selected in LB agar containing the corresponding antibiotic.
The HP0953 protein was expressed according to the manufacturer’s instructions for pET28a(+) and pGEX-6p-2 vectors using 1 mmol/L IPTG (Sigma-Aldrich) and incubating the E. coli pET28a(+)/pET-HP0953-transformed cells for 16 h at 30 °C to obtain the recombinant protein His-HP0953 and the E. coli pGEX-6p-2/pGEX-HP0953WSP-transformed cells for 3 h at 37 °C to obtain the recombinant protein GST-HP0953WSP. The inclusion bodies were extracted as described in the inclusion body preparation method without solubilizing the inclusion bodies in a denaturing agent[32]. The cell pellet was resuspended in 1 mL of 4 × sample buffer. Aliquots were collected throughout the process and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot assays. The inclusion bodies were electrophoresed on SDS-PAGE, and the recombinant protein band was purified by electroelution. The glutathione-S-transferase (GST)-HP0953WSP recombinant protein was used to produce polyclonal antibodies, as described later, and the His-HP0953 protein was used to purify antibodies.
Anti-GST-HP0953WSP antibodies were obtained by immunization of a New Zealand rabbit (2 mo age) according to the Howard and Bethell method[33] (approved by the Bioethics Committee of the Hospital Infantil de México Federico Gómez).
Purification of polyclonal antibody was performed by immunoadsorption. The His-HP0953 protein was subjected to SDS-PAGE on preparative gels. The bands were transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore; United States), which was blocked with 0.5% bovine serum albumin (BSA) and washed with phosphate buffer solution (PBS)-Tween. Serum was diluted 1:100 in PBS-Tween, incubated with the membrane for 2 h under stirring at room temperature, and washed with PBS-Tween; then, the membrane was incubated with 0.02 M glycine-HCl (pH 2.2) for 10 min at room temperature. Finally, 1 M Tris base (pH 9.1) was added. The antibody was recovered and stored at -20 °C. Nonpurified antisera and purified antibodies were tested against a total extract of H. pylori, and the two purified recombinant proteins were evaluated by western blotting using the same PVDF membrane, which was stripped between each test[34].
The chloramphenicol resistance gene was obtained from the pLysS plasmid of the E. coli BL21 (DE3) pLysS strain by PCR, for which 50 μL of reaction mix was prepared, consisting of 1 × HF Fusion buffer (Thermo Scientific); 0.2 mmol/L each of dATP, dGTP, dCTP, and dTTP (Promega); 6 pmol each of primer CMF (5’-GGG AGATCTTTACGCCCCGCCCTG-3’) and CMR (5’-GGGCATATGATGGAGAAAAAAATCACTGG-3’); plasmid pLysS (250 ng); and Phusion DNA Polymerase (1 U) (Thermo Scientific). The CMF and CMR primers contain restriction sites for BglII and NdeI, respectively. The amplification program consisted of initial denaturation at 98 °C for 30 s; 35 cycles of denaturation at 98 °C for 10 s, primer annealing at 65 °C for 30 s, and extension at 72 °C for 1 min; and a final extension at 72 °C for 7 min.
The 5’ and 3’ fragments of the HP0953 gene were obtained from the plasmid pJET-HP0953, constructed above by PCR. To amplify the 5’ fragment, 50 μL of the reaction mixture was prepared, consisting of GoTaq DNA polymerase (Promega) (1.25 U); 0.2 mmol/L each of dATP, dGTP, dCTP, and dTTP; 1.5 mmol/L MgCl2 (Promega); primer HP0953F (25 pmol) (5’-GGGGGATCCATGGTTTTAATCGCTCTTTTAGGGGTG-3’); primer HP0953R (5’-GGGAGATCTCATGGAATTGCTCCATGAAGCG-3’); and plasmid DNA from pJET-HP0953 (250 ng). The amplification program consisted of initial denaturation at 94 °C for 1 min; 35 cycles of denaturation at 94 °C for 30 s, primer annealing at 68.4 °C for 30 s, and extension at 72 °C for 30 s; and a final extension at 72 °C for 7 min. To amplify the 3’ fragment, PCR was performed under the same conditions but using the primers HP0953F (5’-GGGCATATGAGTCCGAGCAGACGACTACG-3’) and HP0953R (5’-GGGGTCGACCTACCTTAACGCACAAACGCTACC-3’). Amplicons of 304 and 248 bp were obtained, respectively.
We subjected 5 μL of each amplification product to 1.5% agarose gel electrophoresis at 85 volts for 75 min. Then, we stained the gel with ethidium bromide (Sigma-Aldrich) and observed the bands under ultraviolet light. Upon confirmation of the amplified product, the remainder was purified using a Wizard SV Gel and PCR Clean-up System, as per the manufacturer’s protocol. Each fragment was inserted into a pJET1.2 vector. The ligation reaction of each fragment in the pJET1.2 vector consisted of 1 × reaction buffer, 0.15 pmol purified PCR product (5’ fragment, 3’ fragment, or chloramphenicol resistance gene; 0.15 pmol), pJET1.2/Blunt Cloning Vector (0.05 pmol), and T4 DNA ligase (5 U). The reaction mixture was incubated at 22 °C for 20 min.
After ligation, we used heat shock treatment to transform 50 μL of competent E. coli DH5α cells with each plasmid[31]. Then, they were centrifuged at 5000 × g for 1 min, the supernatant was decanted, and the pellet was resuspended in the residual medium. Subsequently, the bacterial suspension was seeded on LB agar plates containing ampicillin (100 μg/mL; Sigma-Aldrich) and incubated at 37 °C for 18-24 h to select the recombinant clones. Following incubation, we selected five colonies from the plate and resuspended them in 2 mL of LB broth (Sigma-Aldrich) containing ampicillin (100 μg/mL), and the plasmid was extracted using the alkaline lysis method[35]. We quantified the plasmid DNA using an Epoch microplate spectrophotometer (Biotek Instruments, Inc.; United States) and stored it at -20 °C.
We performed three PCR assays to verify the sequence of the cloned fragments. Each PCR reaction was performed in a final volume of 20 μL, consisting of 1 × HF Fusion buffer (Thermo Scientific); 0.2 mmol/L each of dATP, dGTP, dCTP, and dTTP (Thermo Scientific); primer pJET1.2F (0.2 μM) (5’-CGACTCACTATAGGGAGAGCGGC-3’) (Thermo Scientific); primer pJET1.2R (0.2 μM) (5’-AAGAACATCGATTTTCCATGGCAG-3’) (Thermo Scientific); plasmid DNA (100 ng/μL) (pJET-CM, pJET-5’ or pJET-3’); and Phusion DNA polymerase (Thermo Scientific) (0.4 U). The amplification program consisted of initial denaturation at 98 °C for 30 s; 35 cycles of 98 °C for 10 s, 68 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 7 min. The PCR products were sequenced by Macrogen (Seoul, Korea).
The 5’ fragments, 3’ fragments, and the chloramphenicol resistance gene were obtained from the plasmids previously constructed by enzymatic digestion. BamHI and BglII were used for the 5’ fragment, NdeI and SalI for the 3’ fragment, and BglII and NdeI for the chloramphenicol resistance gene, according to the above-described digestion protocols. The fragments were inserted into the pBluescript II KS+ vector (Agilent Technologies, Inc.; United States), which allows for the synthesis of ssDNA and has an ampicillin resistance gene to facilitate selection following the above-described protocols.
E. coli XL1-blue MRF’ cells were transformed with the constructed vector (pBlue-5CM3) using heat shock treatment and selected following culture on LB agar plates (Sigma-Aldrich) with 100 μg/mL of ampicillin[31]. The plasmid was extracted using alkaline lysis[35], and fragment insertion was verified by automated sequencing. H. pylori was transformed with the plasmid using electroporation[36]. The mutant H. pylori strain was cultured on Casman agar supplemented with 5% sheep erythrocytes and incubated in an atmosphere of 50 mL/L CO2 at 37 °C for 48 h.
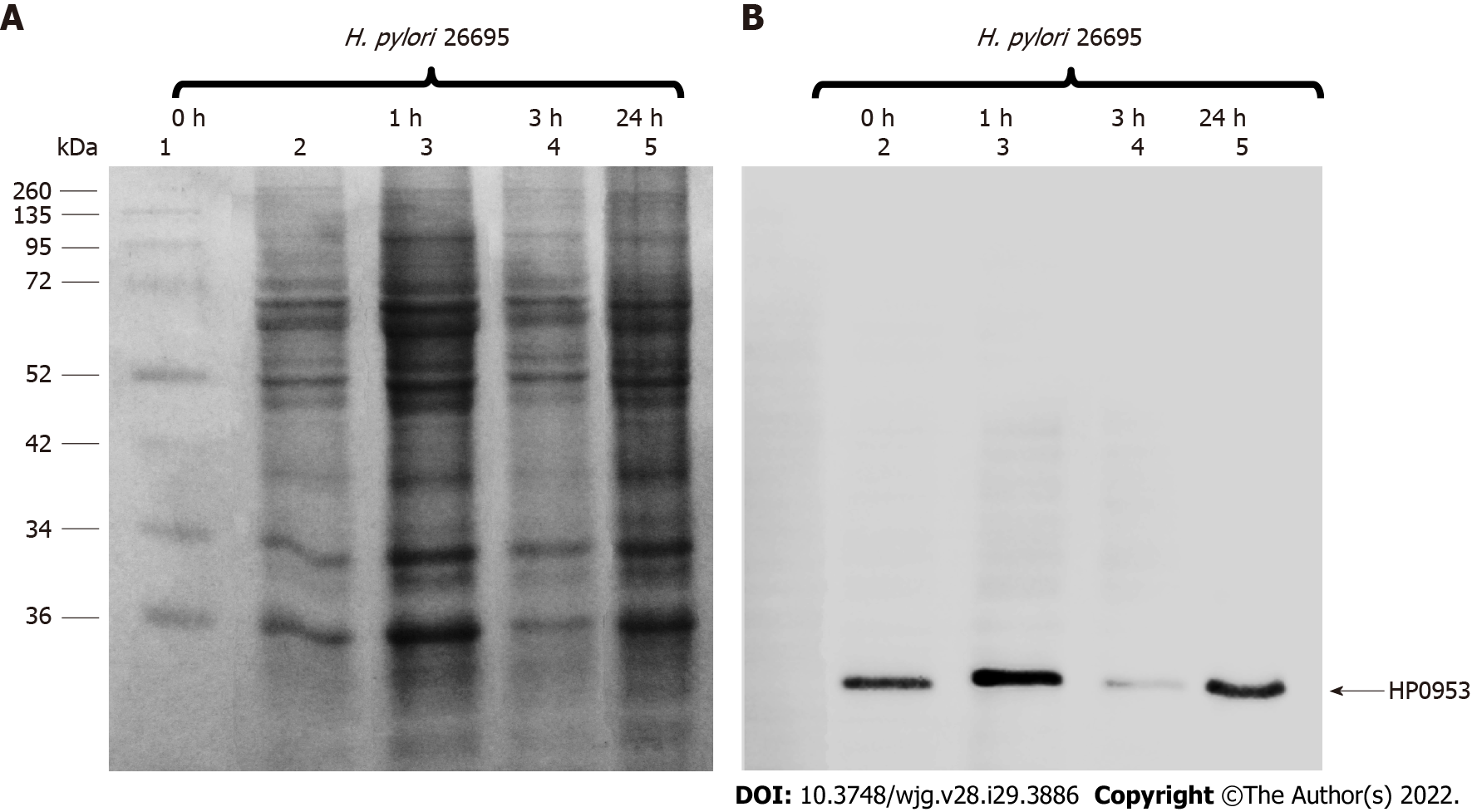
The purified antibody was used for identifying the protein HP0953 in H. pylori 26695 by western blotting. A total extract obtained from H. pylori adhesion test (see below) was used, and soluble and insoluble fractions were separated according to the inclusion body preparation method[32]. These fractions were subjected to SDS-PAGE (50 mg of protein) and transferred onto a PVDF membrane, where western blotting was performed using the purified antibody diluted 1:2 in PBS-Tween and incubated overnight under stirring at 4 °C and the secondary antibody diluted 1:10000 [anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP)-conjugated antibody] (Santa Cruz Biotechnology; United States)[34]. Washing was done with TBST (2 mmol/L Tris, 15 mmol/L NaCl, 0.1% Tween 20, pH 7.6). Finally, the membrane was visualized using Immobilon Western Chemiluminescent HRP substrate (Millipore).
A suspension (6 × 108) of H. pylori 26695 culture in Brucella broth (Dibico) supplemented with 1% yeast extract was prepared. Next, 50 mL of the suspension was placed in cell culture bottles and incubated at 37 °C in an atmosphere of 50 mL/L CO2 for 0, 1, 3, 12, and 24 h; these time points are the first hours of biofilm formation where adherence occurs. After the incubation period, the supernatant was removed, and the bottle walls were scraped to obtain adherent cells, which were resuspended in 1 mL of sterile 1 × PBS.
First, 1 mL of AGS cell suspension was placed in a 15 mL conical tube. Subsequently, 2 mL of DMEM-F12 (Gibco, Life Technologies; United States) medium was added for washing by centrifugation at 1500 rpm for 5 min. A second washing step was performed, and the cells were resuspended again in DMEM-F12 medium. Next, 1 mL of the cell suspension was taken to seed into cell culture bottles. The bottles were incubated in a CO2 chamber for 24 h at 37 °C.
The gene expression assay during biofilm formation was performed in duplicate in ten cell culture bottles of 225 mL each. A suspension of H. pylori (strain 26695, previously cultured for 24 h in a plate with Casman medium) was prepared in DMEM-F12 culture medium. The cells were washed by centrifugation at 3000 rpm for 15 min. After washing, the cell pellet was used to prepare a bacterial suspension in 300 mL of DMEM-F12 culture medium inside a flask until the growth level reached similar to that in tube number 2 on the McFarland scale. To each bottle, 30 mL of the bacterial suspension similar to McFarland tube 2 suspension was added, and then 20 mL of DMEM-F12 medium without the bacteria was added to complete 50 mL. One bottle was not infected with the bacterial strain and was used as the control for the transmission electron microscopy (TEM) assay. The bottles were incubated at 3, 12, 24, and 48 h post-infection. Time 0 was taken as the culture of the bacterial strain with no contact with AGS cells for the expression assay. After each time point of incubation, the supernatant was discarded, and 1 mL of sterile 1 × PBS was added for washing the cells. Subsequently, the wall of the bottle was scraped with a gendarme to detach the cells and remove the adhered biofilm. Each sample was processed for RNA extraction assay. An aliquot was taken for use in TEM, and the remainder was centrifuged at 7000 rpm for 15 min. The test was conducted in triplicate.
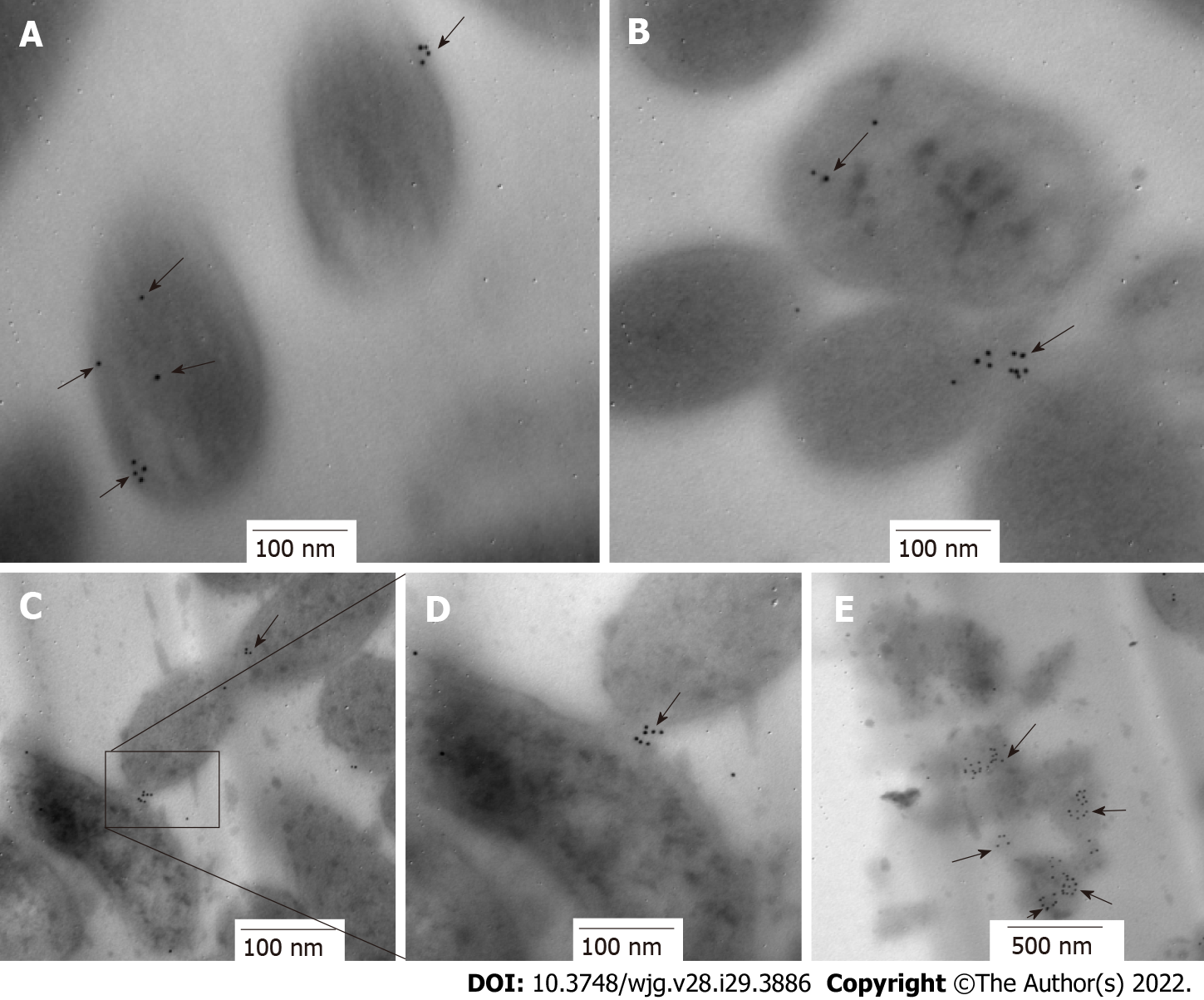
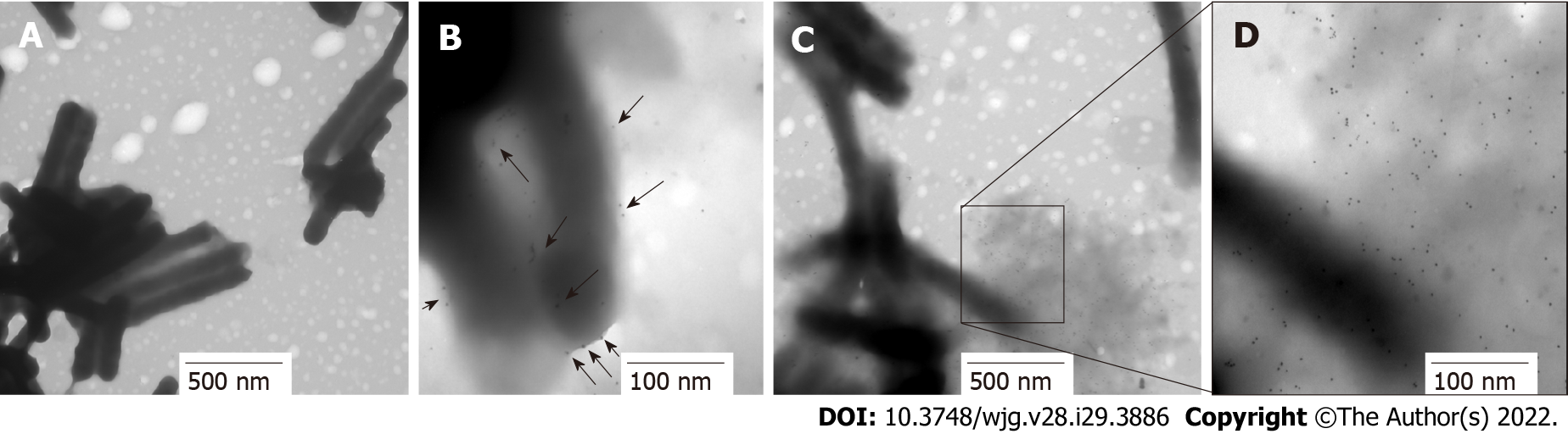
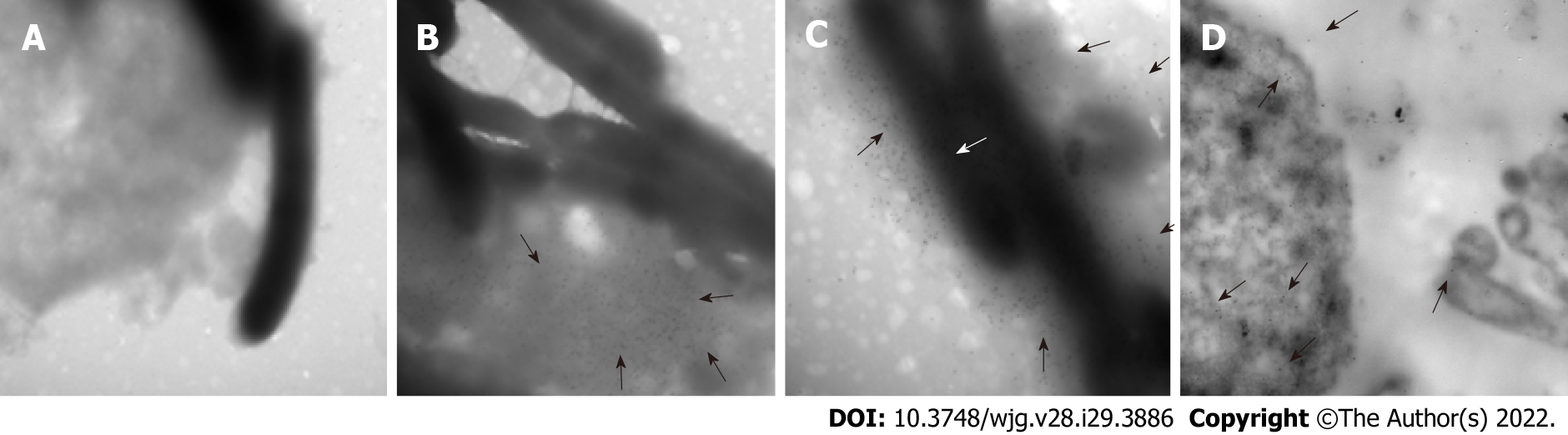
First, 500 μL of the samples obtained from the adhesion test was centrifuged at 2500 rpm for 10 min and fixed with 4% paraformaldehyde (MP Biomedicals; United States) in 1 × PBS (pH 7.4) for 1 h at 4 °C. Then, three washes were performed with 1 × PBS (pH 7.4), and the sample was dehydrated by adding 50% ethanol (Merck; IRL) for 10 min, followed by two washes with 70% ethanol for 15 min at room temperature. The inclusion was done using the LR-White method as described by Vázquez and Echeverría[37], and ultrathin sections of 60-90 nm were obtained. The sections were placed on nickel grids, blocked with 5% BSA for 20 min, and incubated with the purified polyclonal antibody overnight at 4 °C. The sample was then washed with TBST and incubated with the secondary antibody (goat anti-rabbit IgG coupled to 10-nm colloidal gold particles, 1:5, Sigma-Aldrich) diluted in deionized water for 1 h at room temperature. The grids were rinsed with TBST. Finally, they were observed under a transmission electron microscope (JEM 1010 JEOL Ltd. Japan) equipped with a TEM Imaging Systems AMT (Woburn; United States).
Expression analysis was performed by real time-PCR (RT-qPCR). The glutamate racemase (glmM) gene was used as an internal control for the bacterial cell, and the β-globin gene was used as an internal control for human AGS cells. HP0953 expression was evaluated at different time points (0, 3, 12, 24, and 48 h). Time 0 was taken as the culture of the bacterial strain with no contact with AGS cells.
Gene-specific primers were developed for HP0953 and glmM using the Primer3 software, v.0.4.0[38,39]. The sequences of all the primers were as follows: HP0953 (5’-CATATGCCGAGCAGACGACTACG-3’; 5’-GTCGACCTACCTTAACGCACAAACGCTACC-3’), glmM (5’-ACCGACGCTCTCACCCACTT-3’; 5’-AGCGCGAGCCACAACCCTTT-3’). Total RNA was isolated from the AGS cell cultures with and without H. pylori at a multiplicity of infection of 1:100. RNA was extracted using TRIzol® Reagent (Ambion RNA, Life Technologies; United States), followed by phenol:chloroform extraction. The quality and amount of the resulting RNA were evaluated using a Nano-Drop spectrophotometer (The Epoch™ Multi-Volume Spectrophotometer System; United States) at 260 and 280 nm, respectively.
Reverse transcription reaction and SYBR-Green PCR were performed as described below. To initiate the first-strand cDNA synthesis, 1 mg of total RNA was treated with 500 μL of DNAse I (10 U/mL; Invitrogen; United States). Then, it was incubated for 30 min at 37 °C and 5 min at 75 °C with 4 μL of 5 × RT buffer, 2 μL of 25 mmol/L MgCl2, 2 μL of 10 mmol/L dNTP mix, 2 μL of 50 mmol/L random hexamer primers, 1 μL of RNAse inhibitor (20 U/μL), and sufficient H2O for a final volume of 20 μL. The mixture was then incubated for 10 min at 25 °C and 30 min at 48 °C with 2 μL of MultiScribe reverse transcriptase (50 U/mL; Applied Biosystems; United States). The reaction was terminated by heating the sample at 95 °C for 10 min. Aliquots of 200 ng of cDNA were used in the SYBR-Green PCR analysis according to the manufacturer’s protocol. RT-PCR amplification was performed in triplicate under the following conditions: 2 min at 50 °C (for UNG activation) and 10 min at 95 °C (for polymerase activation), followed by 40 cycles of denaturation and alignment/extension at 95 °C for 15 s and 60 °C for 1 min, respectively, using the Stratagene Mx3005p qPCR System and MxPro-Mx3005p software (Santa Clara; United States). Relative quantification of HP0953 RNA expression was performed using the comparative CT method. The ΔCt (difference in CT values) between HP0953 and the glmM internal control and the ΔΔCt were calculated to normalize the differences in cDNA concentrations for each reaction. The RNA expression level was calculated using the equation 2-(ΔΔCt)[40]. The Kruskal-Wallis equality-of-populations rank test was used to compare the expression of the gene at all study time points. Differences were considered to be significant when the P value was < 0.05.
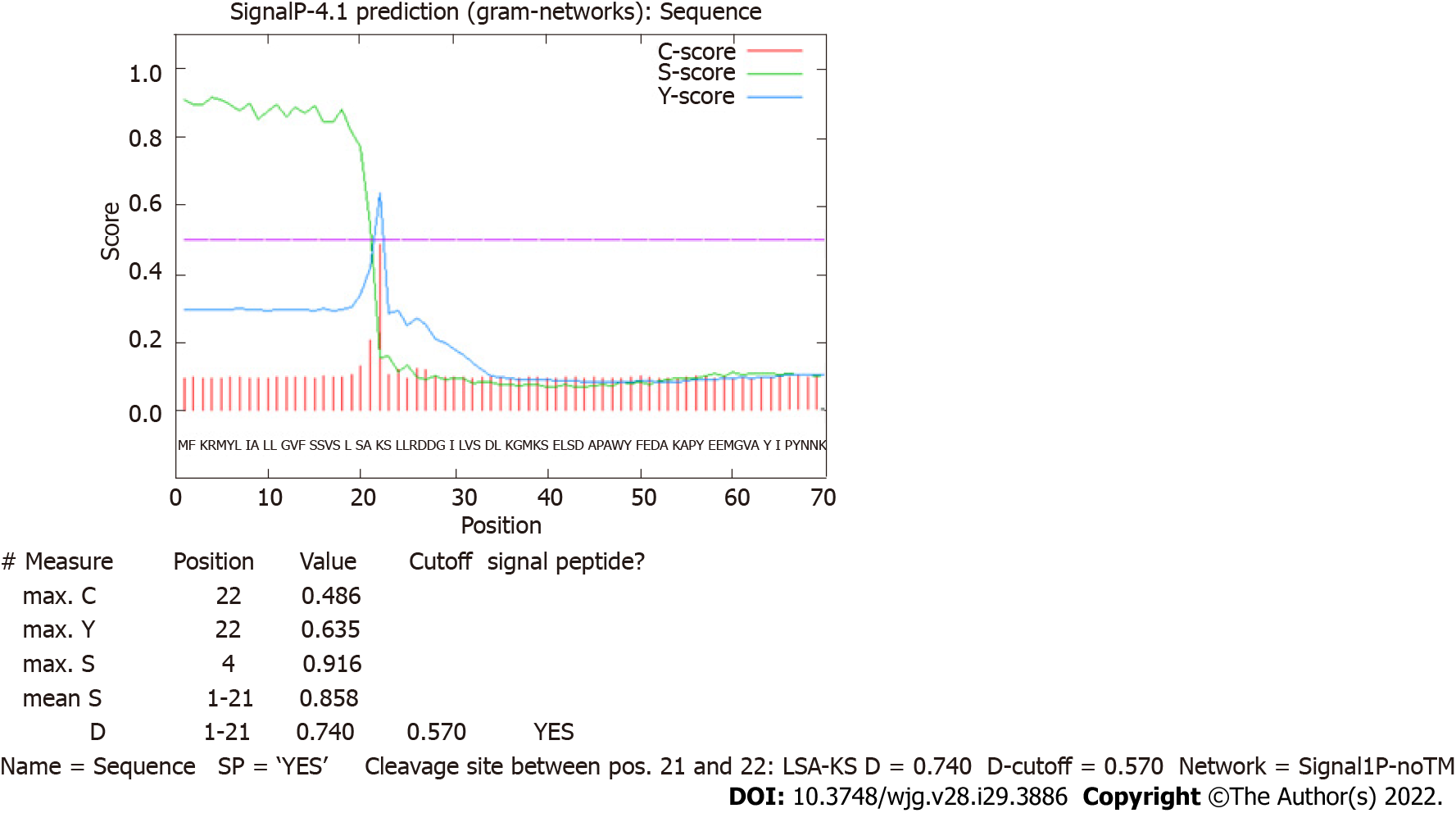
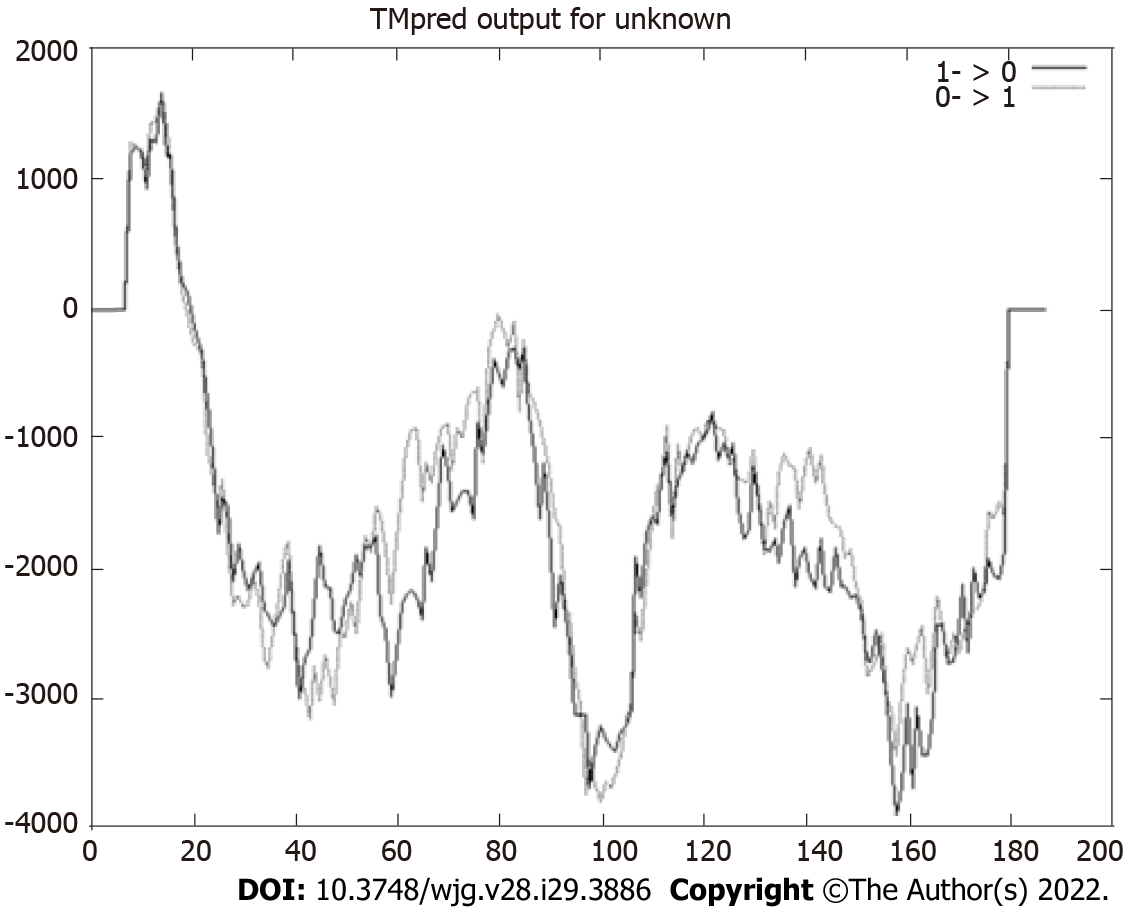
In silico analyses allow us to foresee probable protein maturation. The analysis conducted using the SignalP 4.1 tool revealed that HP0953 protein loses a signal peptide after excision between amino acids 21 and 22 which could lead to extracellular release (Figure 1). This region is also believed to present a transmembrane helix (Figure 2). The protein modifications provided us an insight into their probable function. Tables 1 and 2 show the predicted modification sites for HP0953. There are three predicted N-linked glycosylation sites in amino acid positions 113, 164, and 168 and six predicted O-linked glycosylation sites in positions 15, 23, 43, 111, 115, and 183. Moreover, there are four predicted myristoylation sites in positions 12-17, 29-34, 73-78, and 182-187 and one predicted prenylation site in the position 185-188.
| Position | Residue score | Prediction | |
| Probable N-linked glycosylation sites | |||
| 68 | NNK | -0.20374993 | Non-glycosylated |
| 69 | NKY | -0.11718906 | Non-glycosylated |
| 80 | NAK | -0.46398761 | Non-glycosylated |
| 113 | NIS | 0.031759513 | Potentially glycosylated |
| 120 | NYL | -0.12578944 | Non-glycosylated |
| 130 | NTY | -0.42088241 | Non-glycosylated |
| 134 | NLK | -0.028092183 | Non-glycosylated |
| 164 | NLN | 0.45527702 | Potentially glycosylated |
| 166 | NDN | -0.14904342 | Non-glycosylated |
| 168 | NEI | 0.2906731 | Potentially glycosylated |
| Probable O-linked glycosylation sites | |||
| 15 | S | 0.3493683 | Potentially glycosylated |
| 16 | S | -0.20711247 | Non-glycosylated |
| 18 | S | -0.033164353 | Non-glycosylated |
| 20 | S | -0.066886402 | Non-glycosylated |
| 23 | S | 0.33910492 | Potentially glycosylated |
| 33 | S | -0.58418843 | Non-glycosylated |
| 40 | S | -0.76207501 | Non-glycosylated |
| 43 | S | 0.16899016 | Potentially glycosylated |
| 78 | T | -0.75721234 | Non-glycosylated |
| 84 | S | -0.20144186 | Non-glycosylated |
| 107 | S | -0.2527357 | Non-glycosylated |
| Position | Residue | Prediction |
| PS00008 MYRISTYL N-myristoylation site | ||
| 12-17 | GVfsSV | N-myristoylation site |
| 29-34 | GilvSD | N-myristoylation site |
| 73-78 | GIeqAT | N-myristoylation site |
| 182-187 | GSvcAL | N-myristoylation site |
| PS00005 PKC_PHOSPHO_SITE Protein Kinase C phosphorylation site | ||
| 20-22 | SaK | MOD_RES 20 phosphoserine [condition: S] |
| PS00001 ASN_GLYCOSYLATION N-glycosylation site | ||
| 113-116 | NISY | CARBOHYD 113 N-linked (GlcNAc) asparagine [condition: N] |
| PS00294 PRENYLATION Prenyl group binding site (CAAX box) | ||
| 185-188 | CALr | Prenyl group binding site (CAAX box) |
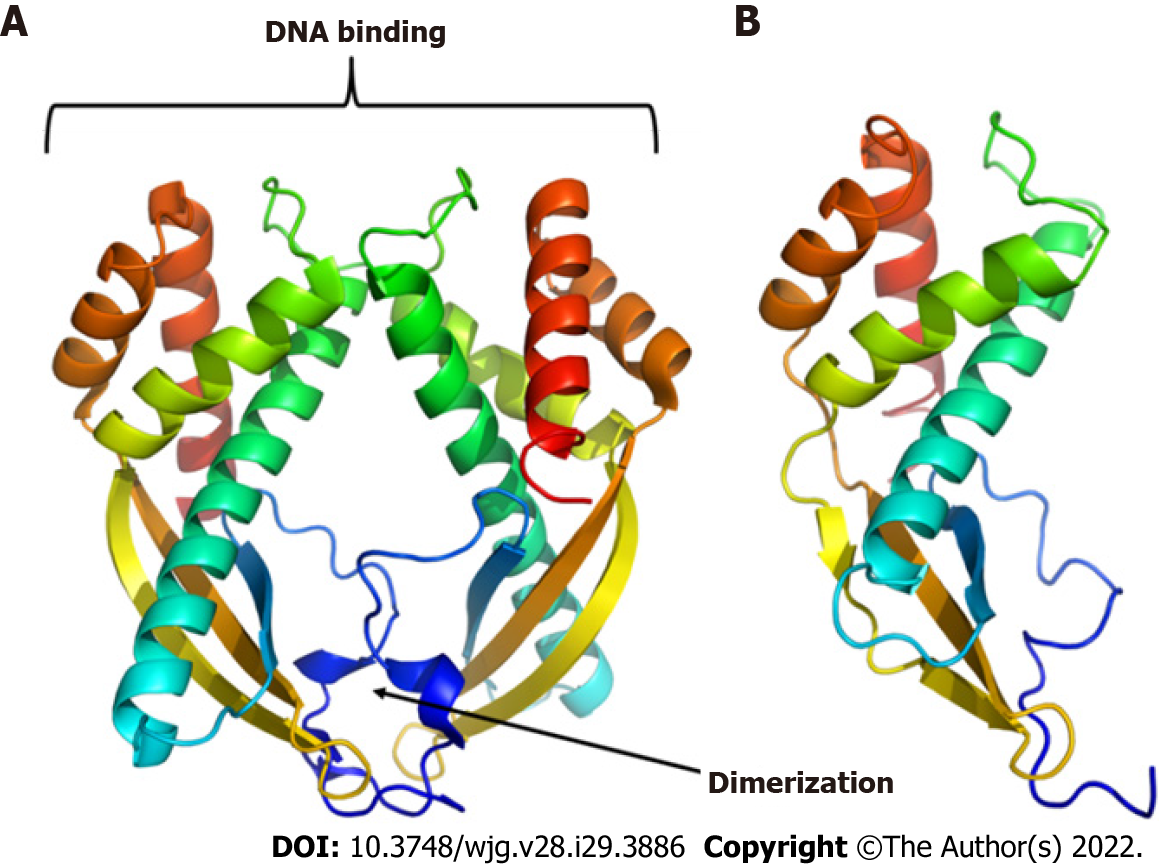
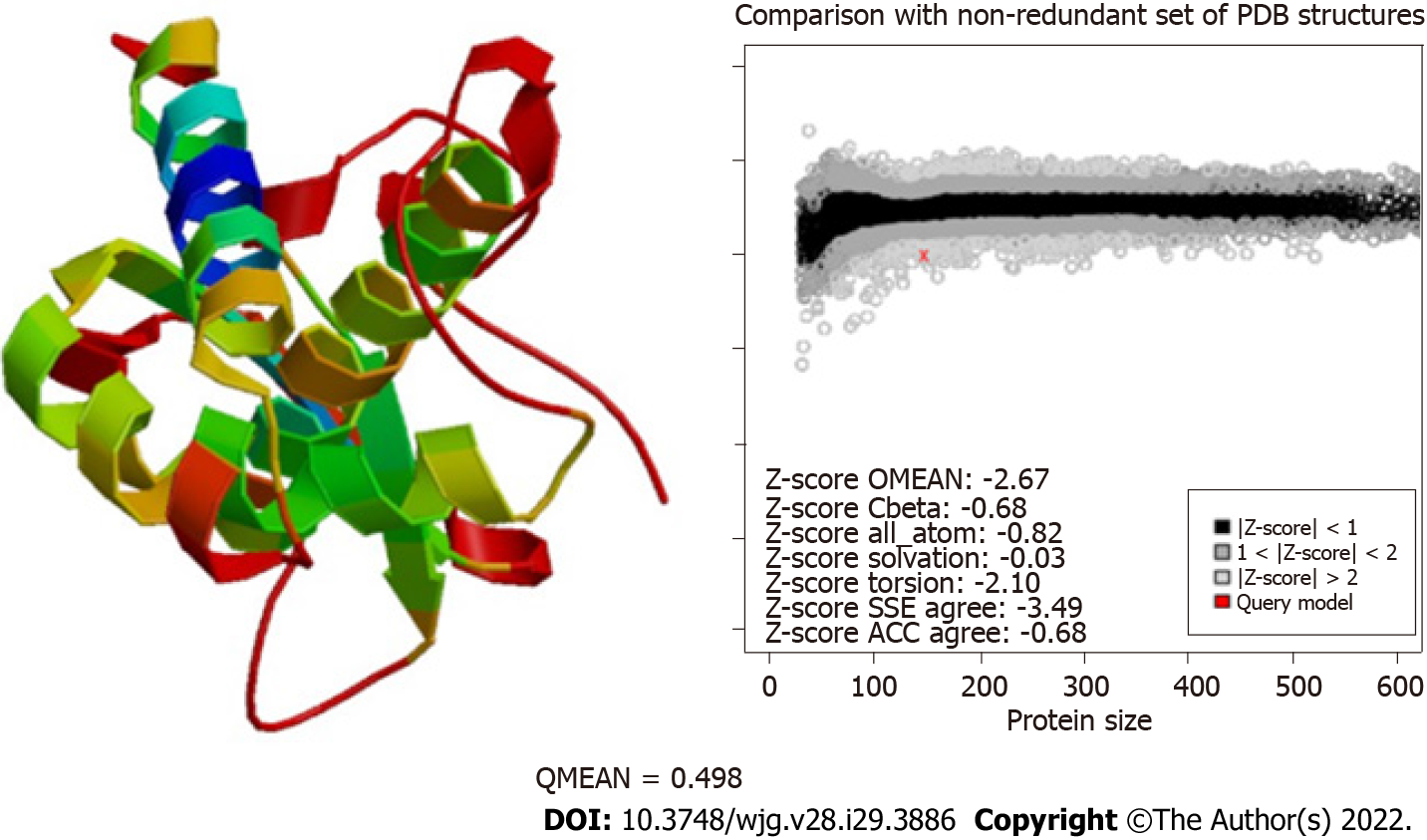
To predict the structural model, we used different programs such as I-TASSER, RaptorX, and HHalign-Kbest, the latter being the one that generated a better quality model (Figures 3 and 4). The HHpred program disclosed a similarity with the Tip-alpha protein of H. pylori; thus, it was used as a template for modeling. Additionally, no conserved domains were found or matches with other proteins.
Plasmid pBlue-5CM3 was constructed for HP0953 site-directed mutagenesis (Supplementary Figure 1). Sequencing the Dhp0953::Cm Blue-5CM3 construction multiple cloning site corresponded to inserted HP0953 and Cm cassette. However, we were unable to propagate the H. pylori-transformed strain.
For H. pylori HP0953 protein identification, insoluble and soluble fractions were obtained and resolved by SDS-PAGE assay. HP0953 protein was not detected in the soluble fraction, and its total concentration was very low in the insoluble fraction (data not shown). Interestingly, HP0953 was expressed constitutively at all the tested time points (Figure 5).
Protein location is indicative of its function. HP0953 was identified in the insoluble fraction of H. pylori lysates, which probably implies that the function of HP0953 is elicited in the vicinity of the outer membrane. To examine this hypothesis, we performed the immunogold technique on bacterial sections that were analyzed by TEM. The micrographs showed that HP0953 protein is located in the cytoplasm and accumulated in some peripheral areas of the bacterial body (Figure 6A), as well as in bacteria-bacteria contact areas (Figures 6B, 6C and 6D). Furthermore, there was a large accumulation of the protein in the bacterial debris (Figure 6E). Interestingly, the presence of this protein was also observed in the area of the glycocalyx that covers H. pylori (Figure 7), and accumulated where said coating spreads, producing a type of net, perhaps indicating the formation of a biofilm. Interestingly, a large amount of HP0953 protein was secreted during the infection of gastric cells because it was found in the H. pylori-AGS cell contact area (Figure 8).
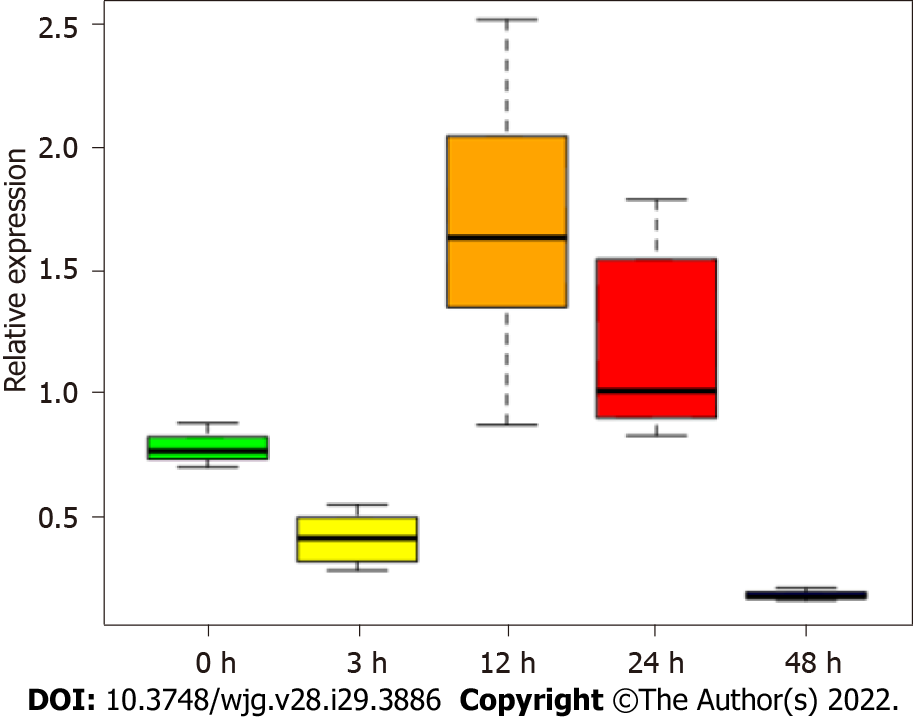
RT-qPCR was performed to elucidate the behavior of HP0953 during the contact of H. pylori with AGS cells. Figure 9 shows a comparison of the expression ratios of the treated and untreated cultures. HP0953 expression was observed at all the tested time points; however, higher expression values were observed after 12 h of infection. glmM was the prokaryotic constitutive gene that was used as an internal control (Figure 9).
The high prevalence of H. pylori infection and its significant persistence in the gastrointestinal tract in the population with limited economic resources, as well as the diversity of related pathologies, the environment, and the genetics of the host, suggest that the pathogenicity mechanisms used by this microorganism are numerous. The different bacterial strains express variations in their virulence factors, including those associated with the colonization process (e.g., urease, flagella, a chemotaxis system, and adhesins), immune response evaders (e.g., lipopolysaccharide and flagella; CagA and the type IV secretion system, VacA; gamma-glutamyl transpeptidase, and catalase superoxide dismutase), and disease inducers (e.g., VacA, BabA, IceA, HtrA, CagA, and the type IV secretion system, DupA, and OipA)[41]. Furthermore, the H. pylori proteome consists of 340 proteins that are described as hypothetical, implying that their structure or function is unknown, thus indicating that some proteins may be related to pathogenicity mechanisms or are important to adapt to the surrounding environment. The HP0953 protein exhibited its maximum expression at the first 12 h of infection in a culture of gastric epithelium cells; this finding along with its location near the infected cells and the predicted prenylation and myristoylation sites suggest that it is an important protein during the bacterium’s colonization phase of the gastric epithelium.
Although various studies, such as those conducted by Naqvi et al[17] in 2016 and Park et al[42] in 2012, have performed in silico analyses to determine the probable structures and functions of the hypothetical proteins of H. pylori, none of them evaluated the probable function of HP0953[17,42]. Furthermore, the bioinformatics analysis performed in this study highlighted the presence of a signal peptide that may anchor at the transmembrane region and then allow the release of the protein to the external environment after its excision. The analyses also revealed some probable glycosylation sites, which suggests that HP0953 is a protein from the glycocalyx. The glycocalyx is defined as any glycoprotein-associated polysaccharide that contains bacterial surface structures and is distant from the outer membrane or cell wall surface[43]. Bacterial glycocalyx generally has protection functions that allow bacterial survival and persistence in the natural environment and cell attachment[44].
Different modification patterns are found within the HP0953 sequence, such as the myristoylation pattern. This modification is essential for the removal of membrane proteins[45]; the existence of these putative sites provides an idea about its pathogenicity. Importantly, these bacteria lack the enzyme N-myristoyl transferase (NMT) required for producing this modification; hence, the proteins that are myristoylated are processed by the NMT of their eukaryotic hosts[46]. NMT is a suitable therapeutic target in opportunistic infections in humans; in fact, it has been associated with carcinogenesis, in particular colon cancer[47]. Myristoylation has been suggested to occur as part of the secretory machinery[48,49]. Hypothetical patterns of prenylation were also detected, which is a class of modification involving the addition of isoprenoids or geranylgeranyl, and it is involved in membrane interactions due to the hydrophobicity of these lipids[50]. This modification is often a step to target the affected proteins to specific membranes[51]. As this modification was predicted in HP0953 by the ProtComB tool, it could be relevant for its function.
Extracellular proteins can play a vital role in bacterial pathogenesis[11], which correlates with our findings of the similarity between the HP0953 protein and the Tip-alpha protein, which is described as the only virulence factor secreted by all H. pylori strains[52]. Moreover, its overexpression has been related to the development of gastric cancer. Tip-alpha is an exotoxin that penetrates cells and induces the production of tumor necrosis factor alpha[53]. HP053 protein could have a similar function because probable posttranslational modification sites related to bacterial pathogenesis were also found. It has been reported that some bacteria secrete effectors through the type III or type IV secretion systems, mimicking the functions of eukaryotic proteins to evade the defense systems. Subsequently, they undergo a posttranslational modification mediated by the same host enzymes to become biologically active[54], stimulating or inhibiting the immune response, interfering signaling pathways, affecting T-cell activation, or whatever their function may be[55]. Among these posttranslational modifications are myristoylation and prenylation[56,57], which were also detected in the in silico analysis conducted in this study. These modifications added lipid residues that allow the protein to anchor to the cell membrane to exert some unknown function.
HP0953 lacks homologous proteins outside H. pylori, and this absence limits comparisons and the elucidation of its potential role. X-ray crystallography analysis can be performed to determine its 3D structure, which could then be used in comparisons with other proteins in the database. However, it was observed that HP0953 is an insoluble protein, which makes it difficult to purify in its native form.
The persistence of H. pylori in an environment where there are peristaltic movements and cell shedding is mediated by a variety of adhesins present on the bacterial surface[10,58], and the adhesion of H. pylori can be achieved by a connection of the plasma membrane through filamentous materials. However, it is known that glycocalyx preferentially adheres to the surfaces of biomaterials and compromised tissues, forming biofilms[55]. Research on biofilm composition has shown that a pericellular matrix, also known as the glycocalyx, develops to preserve and concentrate the lytic enzymes secreted by bacteria, which, in turn, increases their metabolic efficiency[8]. This suggests that the hypothetical protein HP0953 could be involved in lytic processes by being present in the glycocalyx and concentrated in the bacterial debris; however, functionality studies are still required to elucidate the role of this protein in the persistence of H. pylori in the human stomach. Smith et al[11] suggested that this protein could be involved in the survival of the bacterium in the gastric environment based on their finding that it is also present in H. acinonychis, which colonizes the gastric mucosa of felines. Nevertheless, other closely related ε-proteobacteria, such as H. hepaticus and C. jejuni, lack sequences that are homologous to HP0953, thus suggesting that this protein is unique to the gastric Helicobacter species[11], suggesting an important role of HP0953 in H. pylori gastric pathogenesis.
In our previous study, we observed the overexpression of HP0953 protein during the first hours of adherence to inert surfaces and AGS cells[59], suggesting its involvement in the survival of H. pylori in the environment or in its pathogenic mechanisms. In the present study, we found that initial levels of HP0953 expression become diminished, perhaps as a result of the adaptation of H. pylori to its environment. The levels of HP0953 rise after 3 h, indicating that this protein is required for the interaction of H. pylori with AGS cells, and the maximum level of HP0953 expression occurred 12 h after the infection of AGS cells. Genetic regulation in pathogens allows for fluctuation in the expression of constitutive genes following initial contact with the target cells[60,61]. The immunogold assay demonstrated that HP0953 protein was located in the cytoplasm, accumulated in some peripheral areas of the bacterial body, and exhibited a greater expression when it is close to AGS cells. These data suggest that HP0953 protein is synthesized in the cytoplasm and subsequently exported to the outer membrane, where it is involved in adhesion functions as its agglomeration was detected at the binding sites between the bacterium and in the space between the bacterium and gastric cells. Similarly, the immunogold assay of HP0953 protein in the whole bacterium revealed that the protein was primarily located in the bacterial coat or glycocalyx, which is consistent with the finding reported by Smith et al[11], who investigated the proteins secreted by H. pylori and found that HP0953 protein was a component of this set of proteins.
Furthermore, we altered HP0953 to obtain a mutant strain. Unfortunately, we were unable to propagate the resultant HP0953 mutant clone, but we are continuing with our efforts to modify the methodology to obtain a mutant strain that can be successfully propagated. Further studies are necessary to determine the structure and function of hypothetical proteins of H. pylori, such as HP0953, which may be associated with the disease pathology.
HP0953 protein exhibited its maximum expression during the first 12 h of infection in a culture of gastric epithelium cells. This finding indicated its location surrounding the infected cells, and the predicted presence of prenylation and myristoylation sites suggests that HP0953 is a relevant protein during the bacterium’s colonization phase of the gastric epithelium. HP0953 was also found to be strikingly similar to an exotoxin, possibly associating it with the development of gastric cancer.
Helicobacter pylori (H. pylori) is a bacterium of clinical relevance. Approximately 50% of the worldwide human population is colonized by this bacterium, where up to 3% can develop gastric cancer. Several environmental conditions such as the genetic predisposition of the host, and bacterial virulence factors are directly associated.
H. pylori is a bacterium associated with gastric diseases. This microorganism owns more than 200 hypothetical proteins with functions yet unknown. Some of them could be related to the disease or adaptation to gastric microenvironment. However, in a short-term future, they can be considered as therapeutic targets.
The objective was to characterize the hypothetical protein HP0953, such as structure, location, and expression, to determine if it is a virulence factor employed by H. pylori during infection of against gastric (AGS) cells.
Several bioinformatic platforms (SignalP, Expasy, GLYCOPP, I-Tasser) were employed to predict some features of the hypothetical protein HP0953, such as peptide-signaling, three-dimensional structure, and possible post-translational modifications. HP0953 was located during H. pylori infection to AGS cells by transmission electron microscopy, employing a specific antibody for said protein and immunogold technique. The expression of HP0953 during the infection of AGS cell cultures by the bacterium was evaluated by real-time polymerase chain reaction.
The hypothetical protein HP0953 was observed in the cytoplasm and embedded in the glycocalyx secreted by the bacterium, as well as in the space between the bacterium and the AGS cell. The overexpression of the protein was observed at 12 h of infection. HP0953 was predicted to possess a signal peptide, which can interact with transmembranal regions for its secretion. Additionally, the HP0953 protein acts as like an exotoxin, which can then undergo post-translational modifications, which might then require the eukaryotic cell to be carried out.
The hypothetical protein HP0953 is a protein located in the periphery of infected cells. It is overexpressed during infection. The in silico analysis revealed prenylation and myristoylation sites. These findings suggest that the protein has a relevant function during the colonization or infection of H. pylori into gastric epithelium.
Propagate a H. pylori modified strain, which lacks the HP0953 protein. The study will define its functions and establish a new precedent for the study of the pathogenesis of the disease, and its relevance in the adaptation process in the gastric microenvironment.
The authors would like to acknowledge Alejandra S Rodríguez for her skillful technical assistance.
Provenance and peer review: Invited article; Externally peer reviewed
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Hospital Infantil de México Federico Gómez, 11217.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Keikha M, Iran; Kirkik D, Turkey; Mi Y, China S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Connor BA. Helicobacter pylori. In: Centers for Disease Control and Prevention. CDC Yellow Book 2020: Health Information for International Travel. New York: Oxford University Press, 2017. |
| 2. | Cheok YY, Lee CYQ, Cheong HC, Vadivelu J, Looi CY, Abdullah S, Wong WF. An Overview of Helicobacter pylori Survival Tactics in the Hostile Human Stomach Environment. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 3. | Huang Y, Wang QL, Cheng DD, Xu WT, Lu NH. Adhesion and Invasion of Gastric Mucosa Epithelial Cells by Helicobacter pylori. Front Cell Infect Microbiol. 2016;6:159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 103] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 4. | Wizenty J, Tacke F, Sigal M. Responses of gastric epithelial stem cells and their niche to Helicobacter pylori infection. Ann Transl Med. 2020;8:568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Sharndama HC, Mba IE. Helicobacter pylori: an up-to-date overview on the virulence and pathogenesis mechanisms. Braz J Microbiol. 2022;53:33-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 112] [Article Influence: 37.3] [Reference Citation Analysis (1)] |
| 6. | Yonezawa H, Osaki T, Kamiya S. Biofilm Formation by Helicobacter pylori and Its Involvement for Antibiotic Resistance. Biomed Res Int. 2015;2015:914791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Percival SL, Suleman L. Biofilms and Helicobacter pylori: Dissemination and persistence within the environment and host. World J Gastrointest Pathophysiol. 2014;5:122-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 8. | Gristina AG, Costerton JW. Bacterial adherence and the glycocalyx and their role in musculoskeletal infection. Orthop Clin North Am. 1984;15:517-535. [PubMed] |
| 9. | Argüeso P, Woodward AM, AbuSamra DB. The Epithelial Cell Glycocalyx in Ocular Surface Infection. Front Immunol. 2021;12:729260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Liu Y, Hidaka E, Kaneko Y, Akamatsu T, Ota H. Ultrastructure of Helicobacter pylori in human gastric mucosa and H. pylori-infected human gastric mucosa using transmission electron microscopy and the high-pressure freezing-freeze substitution technique. J Gastroenterol. 2006;41:569-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Smith TG, Lim JM, Weinberg MV, Wells L, Hoover TR. Direct analysis of the extracellular proteome from two strains of Helicobacter pylori. Proteomics. 2007;7:2240-2245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Chmiela M, Kupcinskas J. Review: Pathogenesis of Helicobacter pylori infection. Helicobacter. 2019;24 Suppl 1:e12638. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 13. | Alzahrani S, Lina TT, Gonzalez J, Pinchuk IV, Beswick EJ, Reyes VE. Effect of Helicobacter pylori on gastric epithelial cells. World J Gastroenterol. 2014;20:12767-12780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 105] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (6)] |
| 14. | Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2635] [Cited by in RCA: 2586] [Article Influence: 92.4] [Reference Citation Analysis (0)] |
| 15. | Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191:447-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 167] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 16. | Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5555] [Cited by in RCA: 5384] [Article Influence: 192.3] [Reference Citation Analysis (0)] |
| 17. | Naqvi AA, Anjum F, Khan FI, Islam A, Ahmad F, Hassan MI. Sequence Analysis of Hypothetical Proteins from Helicobacter pylori 26695 to Identify Potential Virulence Factors. Genomics Inform. 2016;14:125-135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Zanotti G, Cendron L. Structural and functional aspects of the Helicobacter pylori secretome. World J Gastroenterol. 2014;20:1402-1423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 19. | Otero LL, Ruiz VE, Perez Perez GI. Helicobacter pylori: the balance between a role as colonizer and pathogen. Best Pract Res Clin Gastroenterol. 2014;28:1017-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | National Center of Biotechnology Information. Protein. [cited 8 July 2021]. Available from: https://www.ncbi.nlm.nih.gov/protein. |
| 21. | Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412-D419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1180] [Cited by in RCA: 3543] [Article Influence: 885.8] [Reference Citation Analysis (0)] |
| 22. | Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7001] [Cited by in RCA: 6999] [Article Influence: 499.9] [Reference Citation Analysis (0)] |
| 23. | Siss Institute of Bioinformatics. ExPASy. [cited 8 July 2021]. Available from: https://www.expasy.org/. |
| 24. | Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem. 1993;374:166. |
| 25. | GlycoPP V2.0 A webserver for glycosites prediction in procaryotes. [cited 8 July 2021]. Available from: https://datascience.imtech.res.in/alkarao/glycopp2/. |
| 26. | Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430:2237-2243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1367] [Cited by in RCA: 1708] [Article Influence: 213.5] [Reference Citation Analysis (0)] |
| 27. | Zhang Lab. I-TASSER Protein Structure α Function Predctions. [cited 9 May 2021]. Available from: https://zhanggroup.org/I-TASSER/. |
| 28. | RaptorX. RaptorX: Protein Structure and Function Prediction Powered by Deep Learning. [cited 4 November 2016]. Available from: http://raptorx.uchicago.edu/. |
| 29. | HHalign-Kbest: A service for optimized model generation through sub optimal HMM-HMM sequence alignment. [cited 4 November 2016]. Available from: https://bioserv.rpbs.univ-paris-diderot.fr/services/HHalign-Kbest/. |
| 30. | Vosshall L. Hanahan competent cell protocol. [cited 8 July 2021]. Available from: https://www.rockefeller.edu/research/uploads/www.rockefeller.edu/sites/8/2018/10/Hanahan.pdf. |
| 31. | Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp. 2007;253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 186] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 32. | Hyvonen M. Inclusion body preparation. [cited 8 July 2021]. Available from: https://hyvonen.bioc.cam.ac.uk/wp-content/uploads/2017/09/ib.pdf. |
| 33. | Howard GC, Bethell DR. Polyclonal antibodies, In: Basic Methods in Antibody Production and Characterization. Florida, USA: CRC Press, 2000: 31-48. |
| 34. | Abcam. Western blot membrane stripping for restaining protocol. [cited 8 July 2021]. Available from: https://docs.abcam.com/pdf/protocols/western-blot-membrane-stripping-for-restaining-protocol.pdf. |
| 35. | Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10464] [Cited by in RCA: 11251] [Article Influence: 244.6] [Reference Citation Analysis (0)] |
| 36. | Ge Z, Taylor DE. H. pylori DNA Transformation by Natural Competence and Electroporation. In: Clayton CL, Mobley HLT. Helicobacter pylori Protocols. New Jersey: Humana Press, 1997: 145-152. |
| 37. | Vázquez NG, Echeverría O. Introducción a la microscopía Electrónica Aplicada a las Ciencias Biológicas. Facultad de Ciencias de la Universidad Nacional Autónoma de México. México: Fondo de Cultura Económica, 2000: 148-151. |
| 38. | Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5674] [Cited by in RCA: 6214] [Article Influence: 478.0] [Reference Citation Analysis (0)] |
| 39. | Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1799] [Cited by in RCA: 1835] [Article Influence: 101.9] [Reference Citation Analysis (0)] |
| 40. | Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24144] [Cited by in RCA: 25992] [Article Influence: 1083.0] [Reference Citation Analysis (0)] |
| 41. | Chang WL, Yeh YC, Sheu BS. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci. 2018;25:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 42. | Park SJ, Son WS, Lee BJ. Structural analysis of hypothetical proteins from Helicobacter pylori: an approach to estimate functions of unknown or hypothetical proteins. Int J Mol Sci. 2012;13:7109-7137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 43. | Luong P, Dube DH. Dismantling the bacterial glycocalyx: Chemical tools to probe, perturb, and image bacterial glycans. Bioorg Med Chem. 2021;42:116268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 44. | Ogata M, Araki K, Ogata T. An electron microscopic study of Helicobacter pylori in the surface mucous gel layer. Histol Histopathol. 1998;13:347-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 45. | Hayashi N, Titani K. N-myristoylated proteins, key components in intracellular signal transduction systems enabling rapid and flexible cell responses. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:494-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 46. | Maurer-Stroh S, Eisenhaber F. Myristoylation of viral and bacterial proteins. Trends Microbiol. 2004;12:178-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 118] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 47. | Wright MH, Heal WP, Mann DJ, Tate EW. Protein myristoylation in health and disease. J Chem Biol. 2010;3:19-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 194] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 48. | Simon SM, Aderem A. Myristoylation of proteins in the yeast secretory pathway. J Biol Chem. 1992;267:3922-3931. [PubMed] |
| 49. | Yuan M, Song ZH, Ying MD, Zhu H, He QJ, Yang B, Cao J. N-myristoylation: from cell biology to translational medicine. Acta Pharmacol Sin. 2020;41:1005-1015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 50. | Palsuledesai CC, Distefano MD. Protein prenylation: enzymes, therapeutics, and biotechnology applications. ACS Chem Biol. 2015;10:51-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 182] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 51. | Hildebrandt D. Heterogeneous Prenyl Processing of the Heterotrimeric G protein Gamma Subunits. The Enzymes. 2011;29:97-124. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 52. | Tosi T, Cioci G, Jouravleva K, Dian C, Terradot L. Structures of the tumor necrosis factor alpha inducing protein Tipalpha: a novel virulence factor from Helicobacter pylori. FEBS Lett. 2009;583:1581-1585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 53. | Mahant S, Chakraborty A, Som A, Mehra S, Das K, Mukhopadhyay AK, Gehlot V, Bose S, Das R. The Synergistic Role of Tip α, Nucleolin and Ras in Helicobacter pylori Infection Regulates the Cell Fate Towards Inflammation or Apoptosis. Curr Microbiol. 2021;78:3720-3732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 54. | Al-Quadan T, Price CT, London N, Schueler-Furman O, AbuKwaik Y. Anchoring of bacterial effectors to host membranes through host-mediated lipidation by prenylation: a common paradigm. Trends Microbiol. 2011;19:573-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 55. | Ismail S, Hampton MB, Keenan JI. Helicobacter pylori outer membrane vesicles modulate proliferation and interleukin-8 production by gastric epithelial cells. Infect Immun. 2003;71:5670-5675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 133] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 56. | Popa CM, Tabuchi M, Valls M. Modification of Bacterial Effector Proteins Inside Eukaryotic Host Cells. Front Cell Infect Microbiol. 2016;6:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 57. | Hicks SW, Galán JE. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol. 2013;11:316-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 58. | Vázquez-Jiménez FE, Torres J, Flores-Luna L, Cerezo SG, Camorlinga-Ponce M. Patterns of Adherence of Helicobacter pylori Clinical Isolates to Epithelial Cells, and its Association with Disease and with Virulence Factors. Helicobacter. 2016;21:60-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 59. | Arteaga-Resendiz NK, Velázquez-Guadarrama N, Rivera-Gutiérrez S, Olivares Trejo J J, Méndez-Tenorio A, Valencia Mayoral P, López-Villegas E O, Rodríguez-Leviz A, Vigueras J C, Arellano-Galindo J, Girón J, Torres-López J. In silico analysis of proteins potentially involved in fimbrial biogenesis in Helicobacter pylori. Bol Med Hosp Infant Mex. 2013;70:78-88. |
| 60. | De la Cruz MA, Ares MA, von Bargen K, Panunzi LG, Martínez-Cruz J, Valdez-Salazar HA, Jiménez-Galicia C, Torres J. Gene Expression Profiling of Transcription Factors of Helicobacter pylori under Different Environmental Conditions. Front Microbiol. 2017;8:615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 61. | Kitamoto S, Nagao-Kitamoto H, Kuffa P, Kamada N. Regulation of virulence: the rise and fall of gastrointestinal pathogens. J Gastroenterol. 2016;51:195-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |