Published online Mar 28, 2022. doi: 10.3748/wjg.v28.i12.1257
Peer-review started: September 29, 2021
First decision: December 4, 2021
Revised: December 10, 2021
Accepted: February 15, 2022
Article in press: February 15, 2022
Published online: March 28, 2022
Processing time: 176 Days and 16.3 Hours
Choledocholithiasis is a severe disorder that affects a significant portion of the world’s population. Treatment using endoscopic sphincterotomy (EST) has become widespread; however, recurrence post-EST is relatively common. The bile microbiome has a profound influence on the recurrence of choledocholithiasis in patients after EST; however, the key pathogens and their functions in the biliary tract remain unclear.
To investigate the biliary microbial characteristics of patients with recurrent choledocholithiasis post-EST, using next-generation sequencing.
This cohort study included 43 patients, who presented with choledocholithiasis at the Guangdong Second Provincial General Hospital between May and June 2020. The patients had undergone EST or endoscopic papillary balloon dilation and were followed up for over a year. They were divided into either the stable or recurrent groups. We collected bile samples and extracted microbial DNA for analysis through next-generation sequencing. Resulting sequences were analyzed for core microbiome and statistical differences between the diagnosis groups; they were examined using the Kyoto Encyclopedia of Genes and Genomes pathway hierarchy level using analysis of variance. Correlation between the key genera and metabolic pathways in bile, were analyzed using Pearson’s correlation test.
The results revealed distinct clustering of biliary microbiota in recurrent choledocholithiasis. Higher relative abundances (RAs) of Fusobacterium and Neisseria (56.61% ± 14.81% vs 3.47% ± 1.10%, 8.95% ± 3.42% vs 0.69% ± 0.32%, respectively) and the absence of Lactobacillus were observed in the bile of patients with recurrent disease, compared to that in stable patients. Construction of a microbiological co-occurrence network revealed a mutual relationship among Fusobacterium, Neisseria, and Leptotrichia, and an antagonistic relationship among Lactobacillales, Fusobacteriales, and Clostridiales. Functional prediction of biliary microbiome revealed that the loss of transcription and metabolic abilities may lead to recurrent choledocholithiasis. Furthermore, the prediction model based on the RA of Lactobacillales in the bile was effective in identifying the risk of recurrent choledocholithiasis (P = 0.03).
We demonstrated differences in the bile microbiome of patients with recurrent choledocholithiasis compared to that in patients with stable disease, thereby adding to the current knowledge on its microbiologic etiology.
Core Tip: Treatment of choledocholithiasis by endoscopic sphincterotomy (EST) has become widespread, but recurrence post-EST is relatively common. In this study, we analyzed the bile microbiome of patients with recurrent choledocholithiasis. Increase in Fusobacterium and Neisseria, and the absence of Lactobacillus in bile were the key microbiologic features of recurrent choledocholithiasis. Bile microbiome imbalance might cause poor metabolism of carbohydrates and amino acids and increased glycan biosynthesis in the biliary tract, leading to disease recurrence. The microbiological features in bile could be an effective predictor for choledocholithiasis recurrence post-EST. The findings of our study will help develop new prevention strategies for post-surgery recurrence of choledocholithiasis.
- Citation: Li Y, Tan WH, Wu JC, Huang ZX, Shang YY, Liang B, Chen JH, Pang R, Xie XQ, Zhang JM, Ding Y, Xue L, Chen MT, Wang J, Wu QP. Microbiologic risk factors of recurrent choledocholithiasis post-endoscopic sphincterotomy. World J Gastroenterol 2022; 28(12): 1257-1271
- URL: https://www.wjgnet.com/1007-9327/full/v28/i12/1257.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i12.1257
Cholelithiasis is a common and socially significant health problem worldwide, occurring in approximately 5%-22% of adults, with synchronous common bile duct stone (CBDS) in 20% of these patients[1-4]. In western countries, it is one of the leading gastrointestinal conditions that results in hospitalization[4]. Cholelithiasis with CBDS can lead to biliary obstruction, secondary cholangitis, and obstructive jaundice, endangering lives in some severe cases and often requiring surgical interventions[5]. The introduction of endoscopic treatment started a new era in the treatment of choledocholithiasis[6-9], and management by endoscopic sphincterotomy (EST) or endoscopic papillary balloon dilation (EPBD) has become widespread, replacing open laparoscopic cholecystectomy or open common bile duct exploration with choledochoscopy[10,11].
However, long-term surveys have revealed up to 39% recurrence of choledocholithiasis post-EST, and life-long follow-ups are still needed after surgery[12-14]. Recurrent choledocholithiasis post-EST involves complicated factors, including infections and biliary anatomical abnormalities[15,16]. The elimination of certain pathogens in the bile duct can significantly reduce the recurrence rate[17,18]. Therefore, further investigations into the microbiological etiology and underlying mechanisms of recurrent choledocholithiasis post-EST are crucial for its prediction and prevention in clinical practice.
Complex microbiomes in the biliary system have been observed using next-generation sequencing (NGS)[19]. In these systems, the microbiota metabolize and secrete cholesterol and bile acids; their dysfunction may cause pathophysiological defects and result in stone formation[20,21]. Unlike primary stones, secondary stones in recurrent choledocholithiasis predominately consist of more cholesterol than calcium bilirubinate[22], and their microbiological etiology remains unclear.
In this study, we investigated the microbiological etiology of recurrent choledocholithiasis using NGS to find the key pathogens associated with recurrence post-EST and their metabolic characteristics in disease relapse.
Ethical compliance: Consecutive recruitment of eligible patients was carried out in the Department of Endoscopy at Guangdong Second Provincial General Hospital from May to June 2020. All experimental protocols were approved by this hospital’s ethics committee (project 2019-QNJJ-14-02). The study design complied with all relevant ethical regulations in accordance with the Declaration of Helsinki and Belgian Privacy Commission. Written consent was obtained from all patients in the study.
Study cohort: In this study, we included 43 choledocholithiasis participants diagnosed using computed tomography (CT) or magnetic resonance cholangiopancreatography (MRCP). All patients were assessed by experienced doctors, without risk of EST-related complications[10]. Patients with a history of malignant diseases, autoimmune diseases, diabetes, structure abnormality of the biliary tract, or any exposure to antibiotics within one month were excluded from the study. All patients accepted laparoscopic cholecystectomy (LC) treatment following an EST or endoscopic papillary balloon dilation (EPBD), during the same hospitalization episode. The components of the stones were recorded according to the method of Dosch[23]. All patients received CT or MRCP examinations one week after the treatment to ensure the complete removal of stone in the biliary tract. All participants underwent at least one-year follow-up with transabdominal ultrasonography every three months, and CT or MRCP was performed once recurrence of choledocholithiasis was indicated through clinical presentations or imaging examinations. Patients were divided into stable and recurrent groups according to their disease evaluation at the end of the follow-up period.
Bile sample collection: Bile samples were collected during endoscopic treatment. The ERCP was performed to confirm the diagnosis of choledocholithiasis, followed by the EST or EPBD treatment, and the bile sample was collected through suction during the treatment. The bile samples were immediately transported to the laboratory and stored at -80 °C until extraction.
Bile sample (3 mL) was centrifuged at 16000 × g at 4 °C for 10 min, and the pellet was washed twice with phosphate-buffered saline before DNA extraction. Microbiome DNA was then extracted using the QIAamp PowerFecal DNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
The 16S rRNA gene obtained from each bile sample was amplified by targeting the V3-V4 hypervariable regions using the following primers: 341F 5′-CCTACGGGNGGCWGCAG-3′ and 806R 5′-GGACTACHVGGGTWTCTAAT-3′ using the UCP Multiplex PCR Kit (Qiagen)[24]. The amplicon library was prepared using the QIAseq Ultralow Input Library Kit (Qiagen). An Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States) and Qubit dsDNA HS Assay Kit (Invitrogen Life Technologies, Carlsbad, CA, United States) were used to validate the library pooling. Paired-end sequencing was conducted using the MiSeq platform (Illumina, San Diego, CA, United States) with MiSeq Reagent Kit version V3 (Illumina).
Trimming and quality filtering of the data were performed using the CLC Genomic Workbench version 20.0, with the Microbial Genomics Module (Qiagen). Sequences were matched to the Greengenes database version 13.5.
The amplicon sequencing, and the taxonomic and statistical analyses were performed using Calypso version 8.84[25]. Alpha diversity was determined based on Fisher’s alpha index, which was assessed using the analysis of variance test. Microbial diversity was visualized using the canonical correspondence analysis based on the prognosis groups. Key taxonomic discovery analysis related to prognosis was performed using linear discriminant analysis effect size (LEfSe) at the genus level[26]. The relative abundance (RA) measurements of the genera, with biomarker significance were compared using the Wilcoxon rank test.
The core microbiome was identified as described by Ainsworth[27]. Network analysis was performed to identify the co-occurring and exclusive bacteria using Calypso[25]. Genera and orders of the bile microbiome were represented as nodes, taxa RAs as node size, and edges as positive and negative associations. Networks were generated based on the associations between both genera and orders using Pearson’s correlation; nodes were colored based on their association with different prognosis groups. Only relationships with statistical significance (P < 0.05) were visualized in the network.
Metagenome prediction of the bile microbiome was performed using the amplicon sequencing approach in the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)[28]. The statistical differences between the diagnosis groups were examined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway hierarchy level using analysis of variance. Correlation analysis was carried out between the key genera and metabolic pathways in bile using Pearson’s correlation test. Survival analysis of the identical microbiological risk factors was carried out using Kaplan-Meier analysis.
Except for those analyzed using Calypso, data were analyzed using GraphPad Prism v7.00 software (GraphPad, La Jolla, CA, United States). All analyses in the study were statistically significant at P < 0.05, and P values were adjusted using false discovery rate, Bonferroni, or area under curve correction.
Forty-three choledocholithiasis patients, who underwent LC following EST were recruited in this study and received a one-year follow-up survey. Thirteen patients had co-occurrence of cholelithiasis; the baseline clinical characteristics of the 43 choledocholithiasis patients were shown in Table 1. The stone components were recorded according to the methods of Dosch[23]; they were classified as brown pigmented stones, black pigmented stones, cholesterol stones, and mixed stones. Four recurrent cases without other complications were observed using routine ultrasonography as well as CT during the follow-up period. No significant differences were found in the clinical features or the stone components between patients with and without recurrent choledocholithiasis.
| Stable (n = 39) | Relapse (n = 4) | P value | |
| Age (yr) (range) | 47 (38-64) | 44 (38-46) | 0.142 |
| Sex | |||
| Male (cases) (%) | 24 (61.54) | 3 (75.00) | 0.626 |
| Female (cases) (%) | 15 (38.46) | 1 (25.00) | |
| History of smoking (cases) (%) | 10 (25.64) | 1 (25.00) | 0.978 |
| Comorbidities | |||
| Type 2 diabetes mellitus | 4 (10.26) | 1 (25.00) | 0.381 |
| Hypertension | 6 (15.38) | 1 (25.00) | 0.620 |
| Hyperlipoidemia | 13 (33.33) | 2 (50.00) | 0.505 |
| Accompanied diagnosis | |||
| Cholelithiasis (cases) (%) | 11 (28.21) | 2 (50.00) | 0.366 |
| Acute cholangitis (cases) (%) | 4 (10.26) | 1 (25.00) | 0.381 |
| Pancreatitis (cases) (%) | 1 (2.56) | 0 (0.00) | - |
| Serum biochemical indexes | |||
| ALT (U/L) | 161.50 ± 159.11 | 67.75 ± 75.61 | 0.210 |
| AST (U/L) | 128.51 ± 151.74 | 34.75 ± 38.20 | 0.063 |
| Total Bilirubin (μmol/L) | 92.19 ± 82.97 | 24.23 ± 23.50 | 0.057 |
| Direct Bilirubin (μmol/L) | 84.92 ± 91.99 | 18.73 ± 23.69 | 0. 060 |
| Amylase (U/L) | 118.69 ± 192.30 | 77.50 ± 30.39 | 0.544 |
| Follow-up time (d) | 369.80 ± 2.67 | 372.00 ± 4.00 | 0.101 |
| Recurrent time from EST (d) | - | 208.80 ± 87.97 | - |
| Stone components | |||
| Brown pigment (cases) (%) | 29 (74.36) | 2 (50.00) | 0.303 |
| Black pigment (cases) (%) | 8 (20.51) | 1 (25.00) | |
| Cholesterol (cases) (%) | 0 (0.00) | 0 (0.00) | |
| Mixed component (cases) (%) | 2 (5.13) | 1 (25.00) |
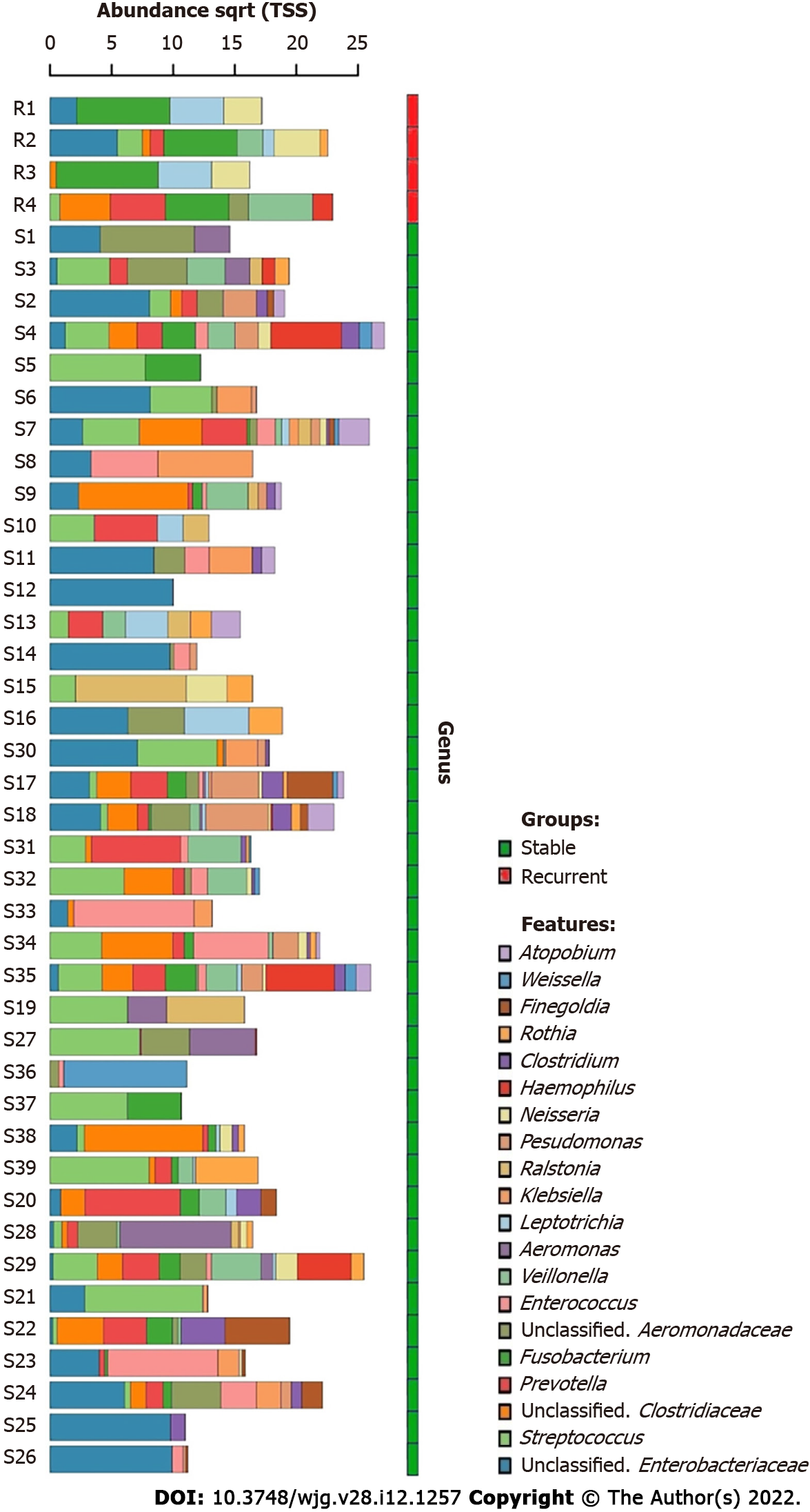
A total of 702 unique operational taxonomic units were identified in the bile of all patients with choledocholithiasis, indicating the diversity of the microbiome in bile (Figure 1). Streptococcus and an unclassified genus of Enterobacteriaceae were the most dominant genera; they were detected in the bile of 28 and 29 patients, respectively. The average RAs of Streptococcus and Fusobacterium in bile were 13.59% and 19.91%, respectively.
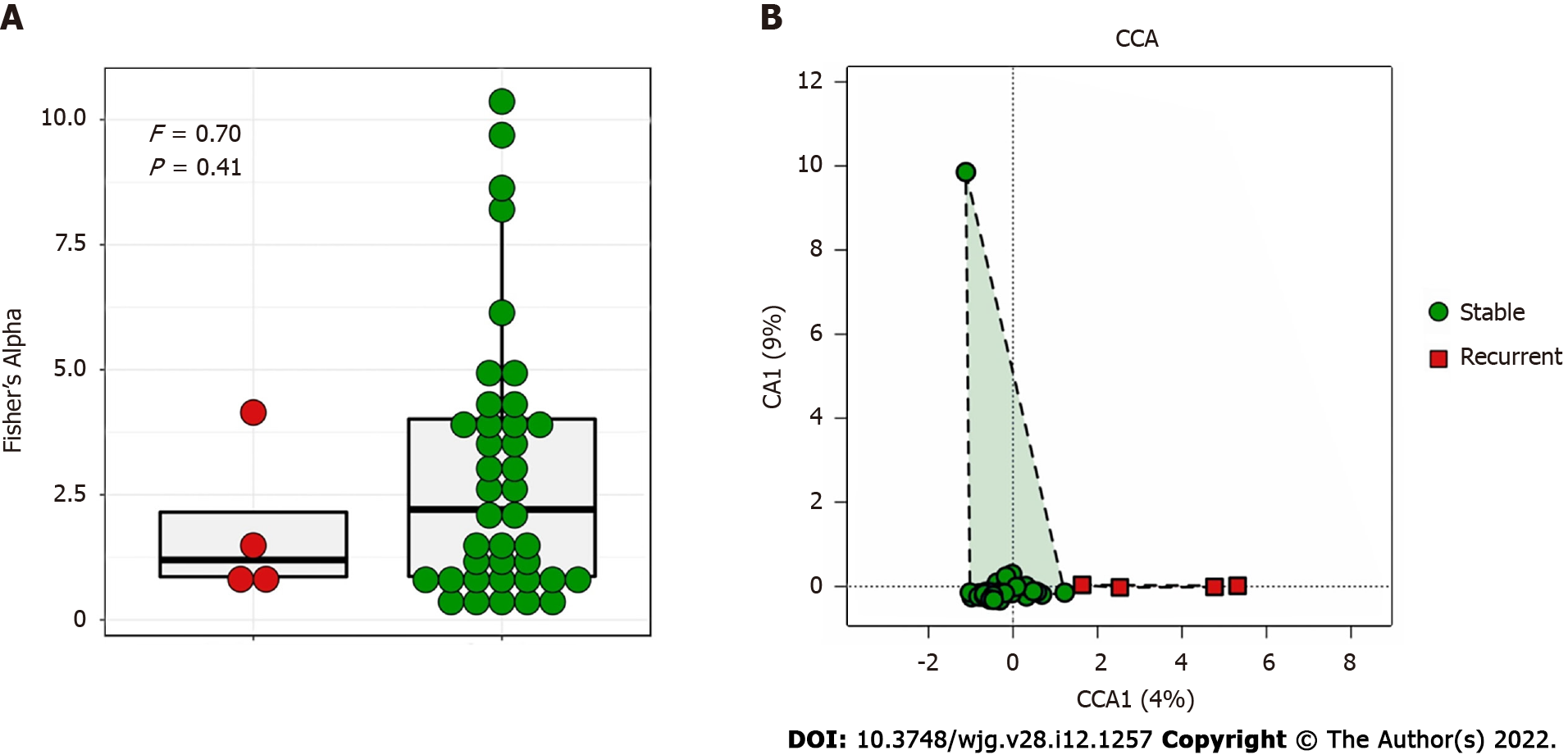
The bile microbial structure in patients with recurrent choledocholithiasis was different from that in patients without recurrence, with lower alpha diversity (P = 0.41) and distinct beta diversity (P = 0.03; Figure 2).
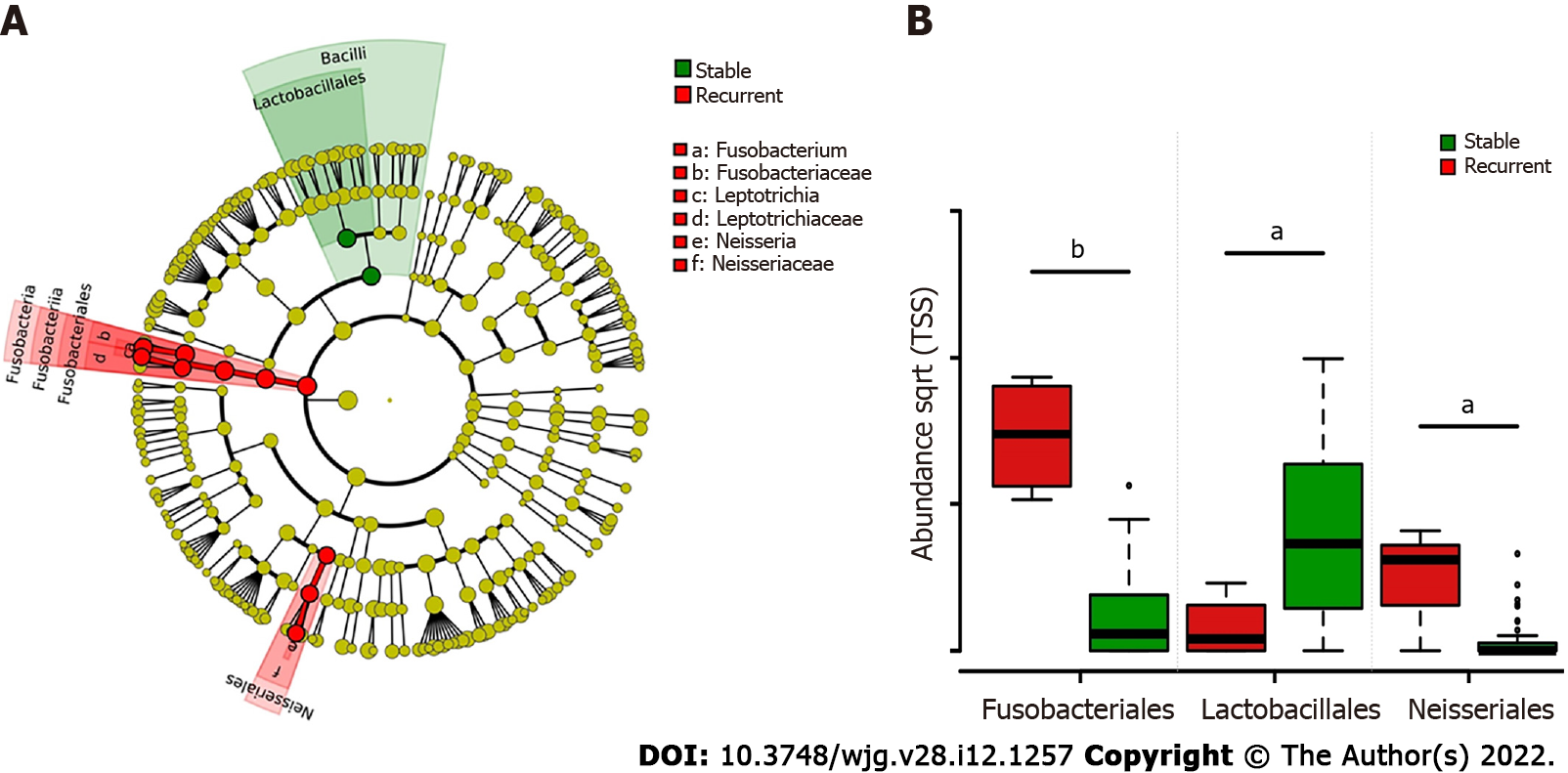
LEfSe biomarker discovery analysis identified Fusobacteriales and Neisseriales as biomarkers in the recurrent group and Lactobacillales in the stable group at the order level (Figure 3A). The RAs of Fusobacteriales (56.61% ± 14.81% vs 3.47% ± 1.10%) and Neisseriales (8.95% ± 3.42% vs 0.69% ± 0.32%) were higher in patients with recurrent choledocholithiasis than that in stable patients post-EST (P < 0.05), while the RA of Lactobacillales was significantly lower in the recurrent group (1.48% ± 1.28%) than that in the stable group (25.04% ± 4.76%; P < 0.05; Figure 3B).
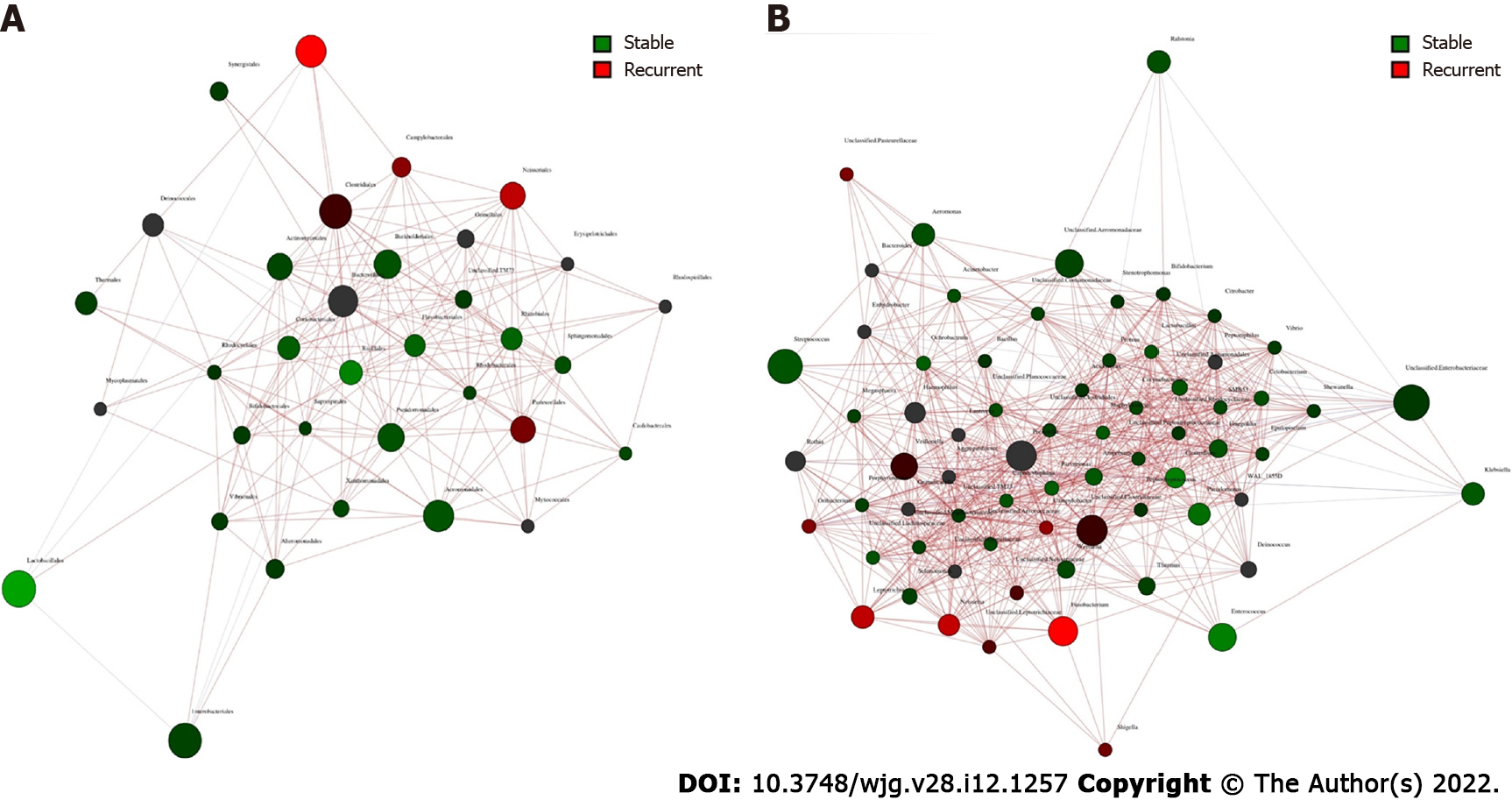
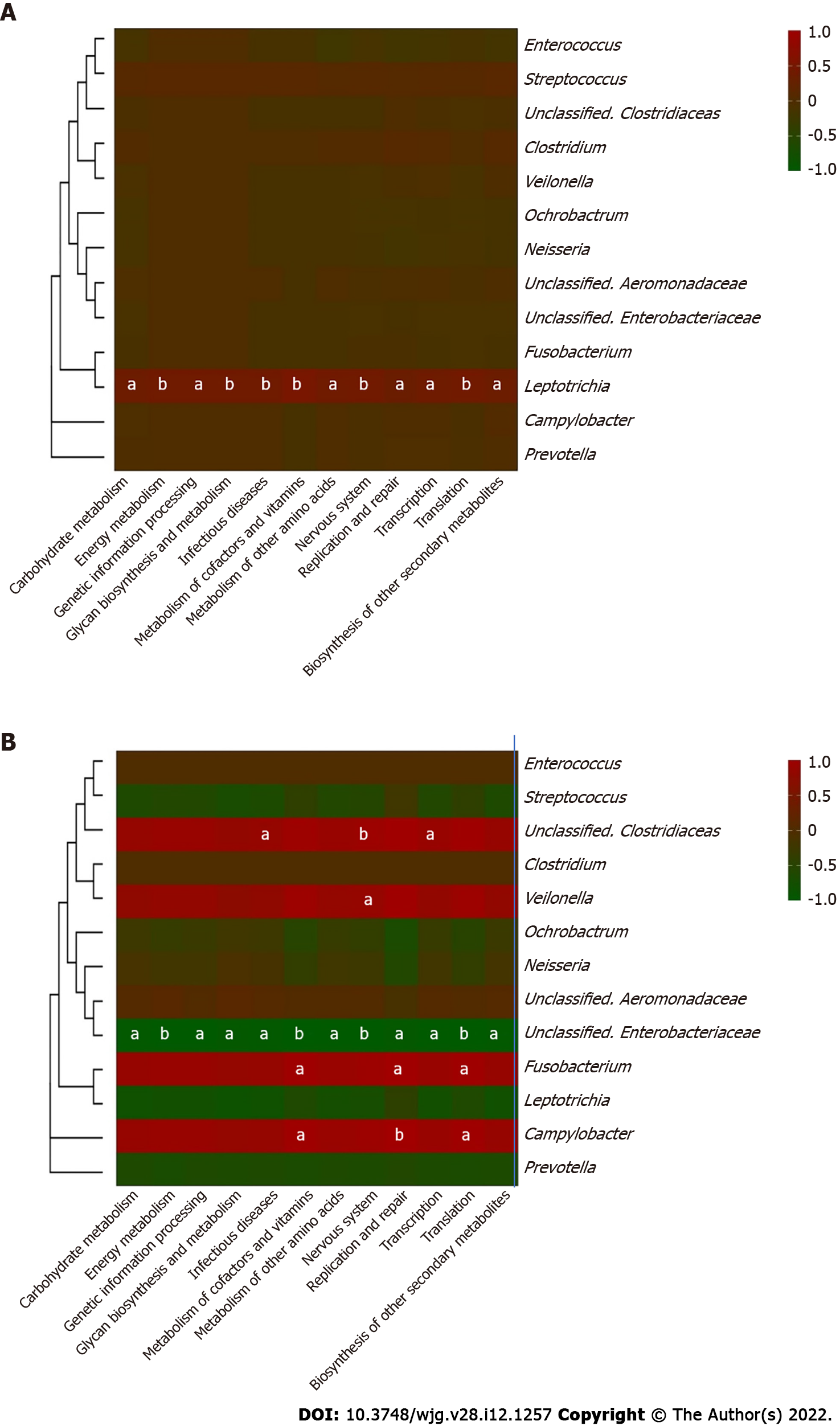
Core microbiome analyses showed that Streptococcus, Prevotella, Fusobacterium, an unclassified genus of Enterobacteriaceae, and an unclassified genus of Clostridiaceae were the shared core genera in both the stable and the recurrent group. Veillonella, Oribacterium, Neisseria, Leptotrichia, and Campylobacter were the specific core genera in the recurrent group, while Enterococcus, Clostridium, and an unclassified genus of Aeromonadaceae were the unique genera in the stable group (Table 2). Construction of a microbiological co-occurrence network revealed a mutual relationship among Fusobacterium, Neisseria, and Leptotrichia (Figure 4A).
| Core microbiome | Type | Group | Recurrent. Occ | Stable. Occ |
| Clostridium | Unique | Stable | 0 | 0.44 |
| Enterococcus | Unique | 0 | 0.44 | |
| Unclassified genus of Aeromonadaceae | Unique | 0.25 | 0.49 | |
| Fusobacterium | Core | Recurrent&Stable | 1 | 0.41 |
| Prevotella | Core | 0.5 | 0.56 | |
| Streptococcus | Core | 0.5 | 0.67 | |
| Unclassified genus of Clostridiaceae | Core | 0.75 | 0.49 | |
| Unclassified genus of Enterobacteriaceae | Core | 0.5 | 0.69 | |
| Campylobacter | Unique | Recurrent | 0.5 | 0.18 |
| Leptotrichia | Unique | 0.75 | 0.28 | |
| Neisseria | Unique | 0.75 | 0.31 | |
| Oribacterium | Unique | 0.5 | 0.18 | |
| Veillonella | Unique | 0.5 | 0.38 |
Additionally, Lactobacillales, Fusobacteriales, Enterobacteriales, Clostridiales, and Bacteroidales were the shared core orders in both the stable and the recurrent group. Pasteurellales, Neisseriales, and Campylobacterales were the unique core orders in the recurrent group, while Pseudomonadales, Burkholderiales, Bacillales, Aeromonadales, and Actinomycetales were the unique orders in the stable group. Co-occurrence network analyses suggested mutual enhancement among the key recurrence-related pathogens in bile and antagonistic relationships among Lactobacillales, Fusobacteriales, and Clostridiales in the ecosystem, indicating the role of probiotics in the prevention of recurrence (Figure 4B).
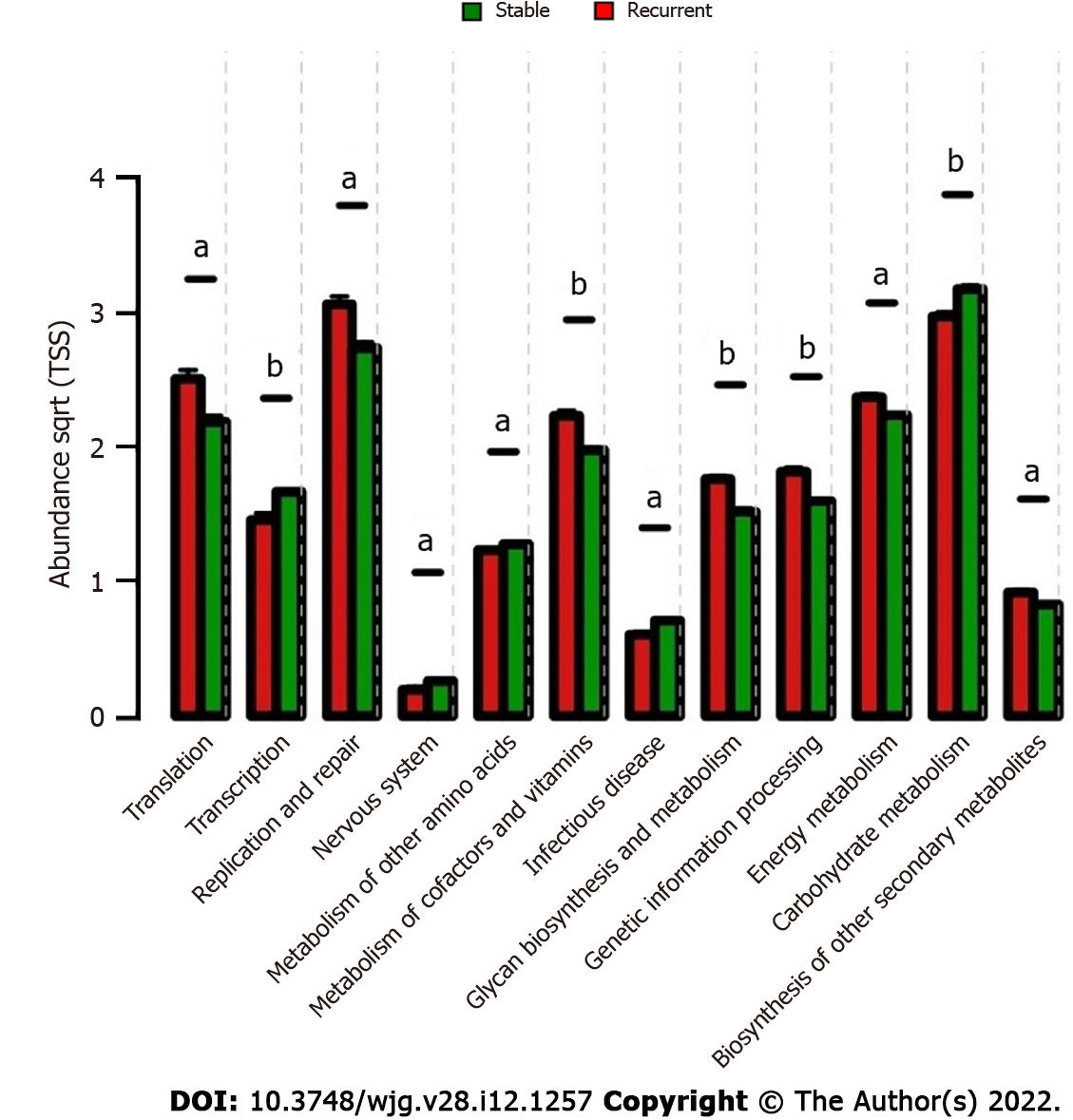
The metabolites from microorganisms are the key pathogenic factors for the host; therefore, the characteristics of the metabolic pathways in bile were analyzed for deeper insight into the microbiologic etiology of recurrent choledocholithiasis post-EST. Comparative analyses of microbiological functions were carried out at the 2nd hierarchy level of the KEGG pathway. In the stable group, the bile microorganisms were active in the transcription and metabolism related to the nervous system, infectious diseases, biosynthesis of carbohydrates and amino acids; while, in the recurrent group, the microbes were active in translation, replication and repair, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism, genetic information processing, energy metabolism, and biosynthesis of secondary metabolites (Figure 5).
Furthermore, correlations between the key genera in the two groups and the different metabolic pathways were analyzed to identify the influence of certain microbes on the host (Figure 6). In the bile ecosystem of the patients with recurrent disease, Fusobacterium and Campylobacter had positive correlations with the metabolism of amino acids, replication and repair, and translation (P < 0.05), while the unclassified genus of Enterobacteriaceae had a negative correlation with all the discrepant metabolic pathways between the two groups (P < 0.05). Leptotrichia had a positive correlation with all the discrepant metabolic pathways in the bile of the stable group (P < 0.05). Correlation analyses indicated that in bile of the recurrent group, increased Fusobacterium could alter the metabolism of amino acids, replication and repair, and translation functions, leading to the formation of secondary bile stones.
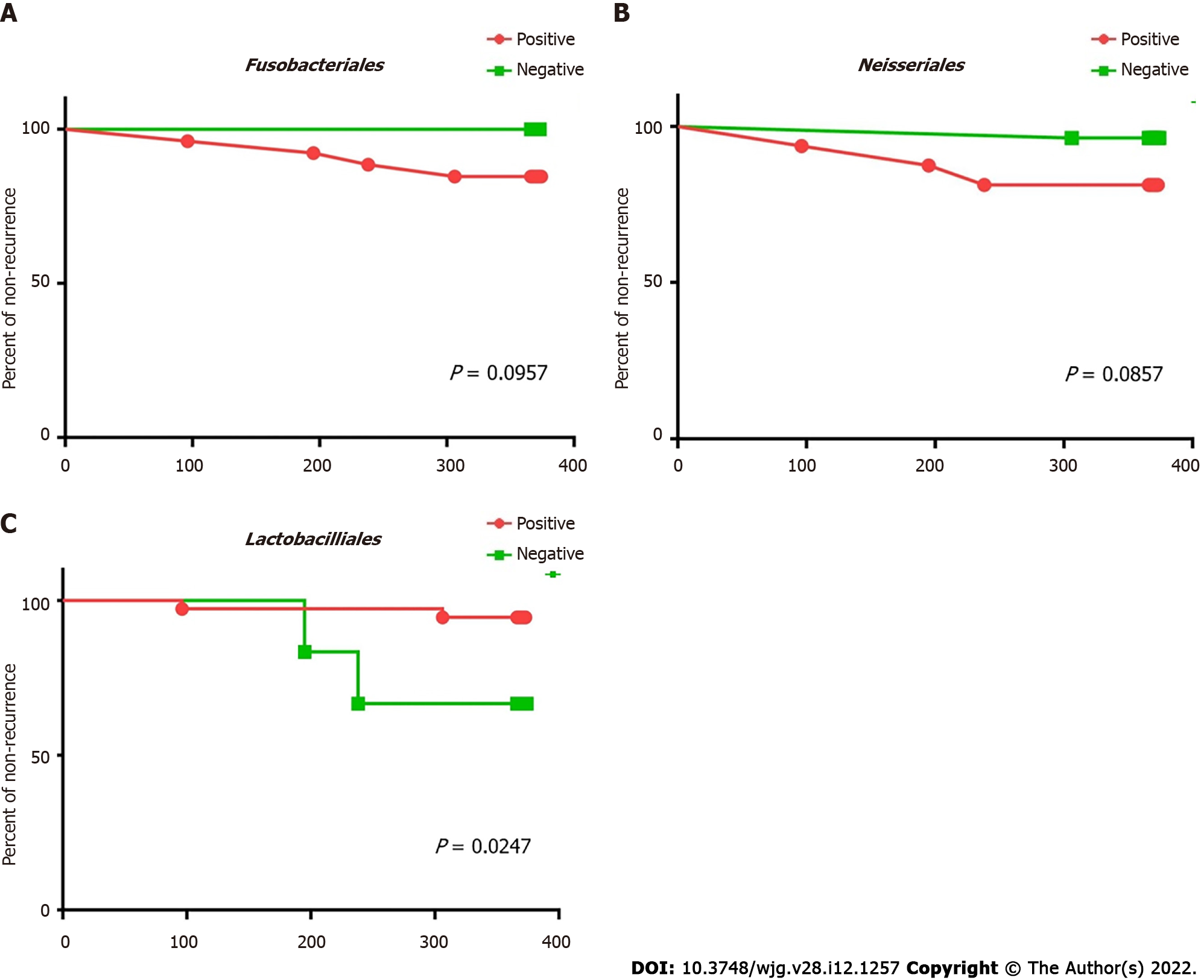
Fusobacteriales and Neisseriales were identified as the bile biomarkers in the recurrent group and Lactobacillales in the stable group. Kaplan-Meier analysis was carried out to confirm whether these biomarkers can be used as independent predictive factors for recurrence post-EST (Figure 7). The statistical results revealed that patients with Lactobacillales in the bile were at a lower risk of recurrence post-EST (P = 0.03) than patients, who lacked this order in their bile.
It was assumed that the biliary system is sterile in healthy people; however, an increasing amount of NGS-supported evidence shows that bile supports a complex and abundant microbiome in healthy individuals[19,29]. The frequently identified microorganisms using traditional culture techniques are Enterococcus, Klebsiella, and Pseudomonas; these bacteria are active in reducing the bile acid pool and regulating bile acid metabolism[30-33]. However, the contribution of microbes to the biliary system is still unclear. NGS techniques revealed that the most common inhabitants of the biliary tract are Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Synergistetes, and candidate phylum Saccharibacteria (TM7)[34]. Some of these microorganisms regulate the hydrolysis of bile acids to constituent components, cleavage of exogenous aromatic rings, deconjugation of bile acid complexes by hydrolytic enzymes, and the formation of free bile acids[35]. The disturbance of the microbiologic ecosystems in bile may lead to dysfunctional bile acid metabolism, resulting in a series of bile duct diseases[20,21]; however, the most disease-specific pathogens and their unique functions remain unknown.
Similar to the results from previous studies[29,34,36], this study revealed that the biliary tract was composed of a diversity of bacteria, and the majority of microorganisms in the bile were Streptococcus, Prevotella, Fusobacterium, Enterococcus, Veillonella, and Clostridium. Lactobacillus and Lactococcus were reported as the major genera in bile[36]; however, these two genera could only be detected in 11 patients in this study. These differences could be attributed to the differences in study designs; we included only patients with severe choledocholithiasis, who needed surgical intervention, for the analysis of microbial risk factors for disease recurrence. Another factor could be the difference in bile sampling; we chose the endoscopic route over open surgery for the collection of bile.
Endoscopic treatment such as EST can provide definitive relief to choledocholithiasis; however, the formation of gallstones will not stop unless the etiologic factors are eliminated[12,13,37]. Among all the risk factors for choledocholithiasis recurrence, only biliary infections are correctable; microbiological treatment is the most potential therapy against the recurrence of choledocholithiasis[5,15,16,38,39]. Therefore, investigation into the biliary microbiology characteristics of recurrent choledocholithiasis is crucial to both etiology and prevention studies. To the best of our knowledge, this is the first pilot study to investigate the microbiological risk factors in recurrent choledocholithiasis post-EST. Increased Fusobacterium and Neisseria were recurrence-related biomarkers in the bile microbiome. Furthermore, we discovered the antagonistic potentials of Lactobacillus and an unclassified genus of Enterobacteriales against Fusobacterium and Neisseria, indicating the potential use of probiotics in the prevention of recurrence post-EST.
Bacteria in bile play an active role in gallstone formation[35]. Escherichia coli and Klebsiella in bile can produce hydrolytic enzymes such as β-glucuronidase, phospholipase A[40], and conjugated bile acid hydrolase; in addition, they can cause deconjugation of bilirubin diglucuronide and precipitation of calcium bilirubinate, which ultimately leads to biliary stone formation[41,42]. We identified Clostridium as one of the key microorganisms in the bile microbiome, which, according to Leung et al[43], is a more important microorganism in the deconjugation of bilirubin diglucuronide than E. coli, because it exhibits a 34-fold higher β-glucuronidase enzyme activity in the biliary tract. A lack of Lactobacillus in the bile could be a probable risk factor for choledocholithiasis, because Lactobacillus in bile can absorb cholesterol and reduce total serum cholesterol[44,45]. The core microbiome pattern in the bile of patients with choledocholithiasis in this study offers a more comprehensive understanding of the influence of the bile microbiome on biliary stone formation.
Furthermore, functional analysis indicated that the loss of transcription and metabolic abilities, and increased function of translation, replication and repair, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism, genetic information processing, energy metabolism, and biosynthesis of other secondary metabolites could lead to recurrent choledocholithiasis. Most of these microbiologic functions were caused by the increased abundance of Fusobacterium and Leptotrichia and the loss of an unclassified genus of Enterobacteriales. However, little is known about the specific health-related functions of the metabolites of these microbes in the bile, and these important metabolic pathways require further research.
Certain microorganisms in bile could predict the time taken before disease recurrence post-EST, and this was evaluated. The existence of Lactobacillales is crucial for predicting recurrence time post-EST, because patients with Lactobacillales in their bile had a longer progression-free time post-EST than patients without Lactobacillales. Therefore, the examination of Lactobacillales existence in bile at the time of endoscopic examination could help doctors identify high-risk patients, who are likely to have early choledocholithiasis recurrence post-EST.
The limited number of early recurrent choledocholithiasis patients could have introduced a bias in statistical analysis and could have limited the generalizability of the prediction model in this study. Furthermore, the diagnosis of choledocholithiasis recurrence relied mainly on the imaging examinations; we could have missed some stones which were invisible in the CT, underestimating the rate of choledocholithiasis recurrence. The molecular mechanisms of microorganisms underlying the recurrence post-EST was based on the PICRUSt model. Verification experiments such as analyzing the correlation between the bile microbiome and the stone composition, and animal experiments to ascertain the preventive effects of Lactobacillus in choledocholithiasis recurrence are warranted.
The microbiological characteristics of bile from patients with recurrent choledocholithiasis post-EST indicated that an increase in Fusobacterium and Neisseria are potential biomarkers for the identification of high-risk patients in the first EST. It elucidated the role of microbial metabolites in the underlying etiology of choledocholithiasis. A co-occurrent network of the biliary bacterial community was constructed. Potential preventive therapy against recurrent choledocholithiasis through supplementation with Lactobacillus and maintenance of the balance of the microbial systems could be promising. These findings could help doctors better understand the etiology of recurrent choledocholithiasis and develop better monitoring and treatment strategies against recurrence post-EST.
Choledocholithiasis is a common and socially significant health problem worldwide, and endoscopic sphincterotomy (EST) has become widespread in treating choledocholithiasis; however, recurrence post-EST is relatively common. The bile microbiome has a profound influence on the recurrence of choledocholithiasis; however, the key pathogens and their functions are not fully elucidated.
To determine the microbiologic risk factors of recurrent choledocholithiasis post EST.
To investigate the biliary microbial characteristics of the recurrent choledocholithiasis post-EST, using next-generation sequencing.
This cohort study included 43 choledocholithiasis patients who had undergone EST were followed up for over a year. They were divided into either the stable or recurrent groups and comparison of their bile microbiome was carried out through next-generation sequencing. Resulting sequences were analyzed for core microbiome and statistical differences between the microbiologic compositions and functions. Correlation between the key genera and metabolic pathways in bile, were analyzed using Pearson’s correlation test.
The results revealed distinct clustering of biliary microbiota in recurrent choledocholithiasis, in which higher relative abundances (RAs) of Fusobacterium and Neisseria and the absence of Lactobacillus were observed in the bile of the recurrent patients. Microbiological co-occurrence network revealed a mutual relationship among Fusobacterium, Neisseria, and Leptotrichia, and an antagonistic relationship among Lactobacillales, Fusobacteriales, and Clostridiales. Functional analysis revealed that the loss of microbiologic transcription and metabolic abilities may lead to the choledocholithiasis recurrence. Furthermore, the prediction model based on the RA of Lactobacillales in the bile was effective in identifying the risk of recurrent choledocholithiasis.
We concluded the microbiologic differences in the bile of recurrent choledocholithiasis patients post EST, thereby adding to the current knowledge on its microbiologic etiology.
The findings of our study will help develop new prevention strategies for post-surgery recurrence of choledocholithiasis.
The authors sincerely thank Wang Z for his expert technical advices in the amplicon analysis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Chinese Academy of Engineering, Academician of Chinese Academy of Engineering; Chinese Institute of Food Science and Technology, vice president; China Edible Fungus Association, vice chairman.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fukui T, Lee Y S-Editor: Wang LL L-Editor: A P-Editor: Li X
| 1. | Tazuma S. Gallstone disease: Epidemiology, pathogenesis, and classification of biliary stones (common bile duct and intrahepatic). Best Pract Res Clin Gastroenterol. 2006;20:1075-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 311] [Article Influence: 16.4] [Reference Citation Analysis (1)] |
| 2. | Kratzer W, Mason RA, Kächele V. Prevalence of gallstones in sonographic surveys worldwide. J Clin Ultrasound. 1999;27:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 3. | Everhart JE, Ruhl CE. Burden of digestive diseases in the United States part I: overall and upper gastrointestinal diseases. Gastroenterology. 2009;136:376-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 418] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 4. | European Association for the Study of the Liver (EASL). EASL Clinical Practice Guidelines on the prevention, diagnosis and treatment of gallstones. J Hepatol. 2016;65:146-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 346] [Article Influence: 38.4] [Reference Citation Analysis (2)] |
| 5. | Cai JS, Qiang S, Bao-Bing Y. Advances of recurrent risk factors and management of choledocholithiasis. Scand J Gastroenterol. 2017;52:34-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 6. | Rhodes M, Sussman L, Cohen L, Lewis MP. Randomised trial of laparoscopic exploration of common bile duct vs postoperative endoscopic retrograde cholangiography for common bile duct stones. Lancet. 1998;351:159-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 288] [Article Influence: 10.7] [Reference Citation Analysis (1)] |
| 7. | Geron N, Reshef R, Shiller M, Kniaz D, Eitan A. The role of endoscopic retrograde cholangiopancreatography in the laparoscopic era. Surg Endosc. 1999;13:452-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Perissat J, Huibregtse K, Keane FB, Russell RC, Neoptolemos JP. Management of bile duct stones in the era of laparoscopic cholecystectomy. Br J Surg. 1994;81:799-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 74] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Arregui ME, Davis CJ, Arkush AM, Nagan RF. Laparoscopic cholecystectomy combined with endoscopic sphincterotomy and stone extraction or laparoscopic choledochoscopy and electrohydraulic lithotripsy for management of cholelithiasis with choledocholithiasis. Surg Endosc. 1992;6:10-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 58] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Williams EJ, Green J, Beckingham I, Parks R, Martin D, Lombard M; British Society of Gastroenterology. Guidelines on the management of common bile duct stones (CBDS). Gut. 2008;57:1004-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 353] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 11. | Qiu W, Sun XD, Wang GY, Zhang P, Du XH, Lv GY. The clinical efficacy of laparoscopy combined with choledochoscopy for cholelithiasis and choledocholithiasis. Eur Rev Med Pharmacol Sci. 2015;19:3649-3654. [PubMed] |
| 12. | Suc B, Escat J, Cherqui D, Fourtanier G, Hay JM, Fingerhut A, Millat B. Surgery vs endoscopy as primary treatment in symptomatic patients with suspected common bile duct stones: a multicenter randomized trial. French Associations for Surgical Research. Arch Surg. 1998;133:702-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 79] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 13. | Hammarström LE, Stridbeck H, Ihse I. Endoscopic sphincterotomy for bile duct calculi-factors influencing the success rate. Hepatogastroenterology. 1996;43:127-133. [PubMed] |
| 14. | Lai KH, Lo GH, Lin CK, Hsu PI, Chan HH, Cheng JS, Wang EM. Do patients with recurrent choledocholithiasis after endoscopic sphincterotomy benefit from regular follow-up? Gastrointest Endosc. 2002;55:523-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 15. | Kaufman HS, Magnuson TH, Lillemoe KD, Frasca P, Pitt HA. The role of bacteria in gallbladder and common duct stone formation. Ann Surg. 1989;209:584-591; discussion 591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 91] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Song ME, Chung MJ, Lee DJ, Oh TG, Park JY, Bang S, Park SW, Song SY, Chung JB. Cholecystectomy for Prevention of Recurrence after Endoscopic Clearance of Bile Duct Stones in Korea. Yonsei Med J. 2016;57:132-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 17. | Yamamoto R, Tazuma S, Kanno K, Igarashi Y, Inui K, Ohara H, Tsuyuguchi T, Ryozawa S. Ursodeoxycholic acid after bile duct stone removal and risk factors for recurrence: a randomized trial. J Hepato-Bil Pancreat Sci. 2016;23:132-136. [RCA] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 18. | Park CH. The Management of Common Bile Duct Stones. Korean J Gastroenterol. 2018;71:260-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Verdier J, Luedde T, Sellge G. Biliary Mucosal Barrier and Microbiome. Viszeralmedizin. 2015;31:156-161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 20. | Wang DQ, Cohen DE, Carey MC. Biliary lipids and cholesterol gallstone disease. J Lipid Res. 2009;50 Suppl:S406-S411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 146] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 21. | Van Erpecum KJ, Van Berge-Henegouwen GP. Gallstones: an intestinal disease? Gut. 1999;44:435-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Kim MH, Myung SJ, Seo DW, Lee SK, Kim YS, Lee MH, Yoo BM, Min MI. Association of periampullary diverticula with primary choledocholithiasis but not with secondary choledocholithiasis. Endoscopy. 1998;30:601-604. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 23. | Dosch AR, Imagawa DK, Jutric Z. Bile Metabolism and Lithogenesis: An Update. Surg Clin North Am. 2019;99:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 24. | Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1103] [Cited by in RCA: 1118] [Article Influence: 101.6] [Reference Citation Analysis (0)] |
| 25. | Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion MJ, Berger B, Krause L. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33:782-783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 348] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 26. | Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11842] [Cited by in RCA: 10292] [Article Influence: 735.1] [Reference Citation Analysis (0)] |
| 27. | Da T, Krause L, Bridge T, Torda G, Raina JB, Zakrzewski M, Gates RD, Padilla-Gamiño JL, Spalding HL, Smith C, Woolsey ES, Bourne DG, Bongaerts P, Hoegh-Guldberg O, Leggat W. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015;9:2261-2274. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 339] [Cited by in RCA: 342] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 28. | Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814-821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5694] [Cited by in RCA: 6295] [Article Influence: 524.6] [Reference Citation Analysis (0)] |
| 29. | Shen H, Ye F, Xie L, Yang J, Li Z, Xu P, Meng F, Li L, Chen Y, Bo X, Ni M, Zhang X. Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci Rep. 2015;5:17450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 30. | Hazrah P, Oahn KT, Tewari M, Pandey AK, Kumar K, Mohapatra TM, Shukla HS. The frequency of live bacteria in gallstones. HPB (Oxf). 2004;6:28-32. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 36] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 31. | Moazeni-Bistgani M, Imani R. Bile bacteria of patients with cholelithiasis and theirs antibiogram. Acta Med Iran. 2013;51:779-783. [PubMed] |
| 32. | Abeysuriya V, Deen KI, Wijesuriya T, Salgado SS. Microbiology of gallbladder bile in uncomplicated symptomatic cholelithiasis. Hepatobiliary Pancreat Dis Int. 2008;7:633-637. [PubMed] |
| 33. | Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1367] [Cited by in RCA: 1678] [Article Influence: 139.8] [Reference Citation Analysis (0)] |
| 34. | Ye F, Shen H, Li Z, Meng F, Li L, Yang J, Chen Y, Bo X, Zhang X, Ni M. Influence of the Biliary System on Biliary Bacteria Revealed by Bacterial Communities of the Human Biliary and Upper Digestive Tracts. PLoS One. 2016;11:e0150519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 35. | Molinero N, Ruiz L, Sánchez B, Margolles A, Delgado S. Intestinal Bacteria Interplay With Bile and Cholesterol Metabolism: Implications on Host Physiology. Front Physiol. 2019;10:185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 159] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 36. | Wu T, Zhang Z, Liu B, Hou D, Liang Y, Zhang J, Shi P. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics. 2013;14:669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 37. | Tazuma S, Unno M, Igarashi Y, Inui K, Uchiyama K, Kai M, Tsuyuguchi T, Maguchi H, Mori T, Yamaguchi K, Ryozawa S, Nimura Y, Fujita N, Kubota K, Shoda J, Tabata M, Mine T, Sugano K, Watanabe M, Shimosegawa T. Evidence-based clinical practice guidelines for cholelithiasis 2016. J Gastroenterol. 2017;52:276-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (1)] |
| 38. | Caddy GR, Kirby J, Kirk SJ, Allen MJ, Moorehead RJ, Tham TC. Natural history of asymptomatic bile duct stones at time of cholecystectomy. Ulster Med J. 2005;74:108-112. [PubMed] |
| 39. | Lopez AJ, O'Keefe P, Morrissey M, Pickleman J. Ceftriaxone-induced cholelithiasis. Ann Intern Med. 1991;115:712-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Trotman BW. Pigment gallstone disease. Gastroenterol Clin North Am. 1991;20:111-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 55] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Cetta F. The role of bacteria in pigment gallstone disease. Ann Surg. 1991;213:315-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 88] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Feretis CB, Contou CT, Manouras AJ, Apostolidis NS, Golematis BC. Long term consequences of bacterial colonization of the biliary tract after choledochostomy. Surg Gynecol Obstet. 1984;159:363-366. [PubMed] |
| 43. | Leung JW, Liu YL, Leung PS, Chan RC, Inciardi JF, Cheng AF. Expression of bacterial beta-glucuronidase in human bile: an in vitro study. Gastrointest Endosc. 2001;54:346-350. [RCA] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 44. | Grigor'eva IN, Romanova TI. Gallstone Disease and Microbiome. Microorganisms. 2020;8:835. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 45. | Wang L, Guo MJ, Gao Q, Yang JF, Yang L, Pang XL, Jiang XJ. The effects of probiotics on total cholesterol: A meta-analysis of randomized controlled trials. Med (Baltim). 2018;97:e9679. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |