Published online Nov 28, 2021. doi: 10.3748/wjg.v27.i44.7705
Peer-review started: May 31, 2021
First decision: July 1, 2021
Revised: July 9, 2021
Accepted: November 9, 2021
Article in press: November 9, 2021
Published online: November 28, 2021
Processing time: 177 Days and 19.1 Hours
Interleukin 10 receptor alpha subunit (IL10RA) dysfunction is the main cause of very early-onset inflammatory bowel disease (VEO-IBD) in East Asians.
To identify disease-causing gene mutations in four patients with VEO-IBD and verify functional changes related to the disease-causing mutations.
From May 2016 to September 2020, four young patients with clinically diagnosed VEO-IBD were recruited. Before hospitalization, using targeted gene panel sequencing and trio-whole-exome sequencing (WES), three patients were found to harbor a IL10RA mutation (c.301C>T, p.R101W in one patient; c.537G>A, p.T179T in two patients), but WES results of the fourth patient were not conclusive. We performed whole-genome sequencing (WGS) on patients A and B and reanalyzed the data from patients C and D. Peripheral blood mononuclear cells (PBMCs) from patient D were isolated and stimulated with lipopolysaccharide (LPS), interleukin 10 (IL-10), and LPS + IL-10. Serum IL-10 levels in four patients and tumor necrosis factor-α (TNF-α) in the cell supernatant were determined by enzyme-linked immunosorbent assay. Phosphorylation of signal transducer and activator of transcription 3 (STAT3) at Tyr705 and Ser727 in PBMCs was deter
The four children in our study consisted of two males and two females. The age at disease onset ranged from 18 d to 9 mo. After hospitalization, a novel 333-bp deletion encompassing exon 1 of IL10RA was found in patients A and B using WGS and was found in patients C and D after reanalysis of their WES data. Patient D was homozygous for the 333 bp deletion. All four patients had elevated serum IL-10 levels. In vitro, IL-10-stimulated PBMCs from patient D failed to induce STAT3 phosphorylation at Tyr705 and only minimally suppressed TNF-α production induced by LPS. Phosphorylation at Ser727 in PBMCs was not affected by LPS or LPS + IL-10 in both healthy subjects and in patient D.
WGS revealed a novel 333-bp deletion of IL10RA in four patients with VEO-IBD, whereas the WES results were inconclusive.
Core Tip: Children less than 6 years old with very early-onset inflammatory bowel disease (VEO-IBD) exhibit severe and refractory disease phenotypes, which indicate a monogenic type disease. Here, we report four cases clinically diagnosed with VEO Crohn’s disease, of which three were compound heterozygous carriers for a 333-bp deletion and an additional single-nucleotide variant, and one was homozygous for the 333-bp deletion in IL10RA. Based on these cases with heterozygous pathogenic variants in IL10RA, the possibility of another large fragment deletion that can be missed by whole-exon sequencing or gene panels should be considered, particularly when serum IL-10 is increased in patients with VEO-IBD.
- Citation: Lv JJ, Su W, Chen XY, Yu Y, Xu X, Xu CD, Deng X, Huang JB, Wang XQ, Xiao Y. Autosomal recessive 333 base pair interleukin 10 receptor alpha subunit deletion in very early-onset inflammatory bowel disease. World J Gastroenterol 2021; 27(44): 7705-7715
- URL: https://www.wjgnet.com/1007-9327/full/v27/i44/7705.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i44.7705
Inflammatory bowel disease (IBD) in children < 6 years of age is known as very early-onset IBD (VEO-IBD)[1] and represents a specific disease course with a distinct phenotype that can be more severe and refractory than classic IBD[2-3]. Recent studies suggested that patients with VEO-IBD, particularly those with symptoms such as perianal disease soon after birth, suffer from failed treatment, indicating a monogenic type of disease[4-6].
By utilizing next-generation sequencing (NGS), many genetic disorders associated with epithelial defects or immunodeficiencies have been found in patients[7-9]. Notably, interleukin 10 receptor alpha subunit (IL10RA) dysfunction is the most common cause of the disease in East Asians, particularly in the Chinese, Japanese, and Korean populations[10-12].
According to our previous retrospective study, increased serum ferritin levels in VEO-IBD patients are indicative of monogenic disease, and very high serum levels of IL-10 suggest that patients with VEO-IBD are more likely to have IL10RA mutations[13].
We report four cases clinically diagnosed with VEO-Crohn's disease with high serum IL-10 levels, indicating IL10RA dysfunction. However, neither results of targeted gene panel sequencing (TGPS) nor those of whole-exome sequencing (WES) in the probands were conclusive. Whole-genome sequencing (WGS) was performed in two patients, and a novel 333-bp deletion in IL10RA was identified. The results of trio-WES for the other two patients were subsequently reanalyzed, and the same novel 333-bp deletion was found.
Four patients with VEO-IBD, including two boys and two girls, were enrolled in our study. The medical history and clinical characteristics of the patients are summarized in Table 1. All patients were of Chinese Han ethnicity and were born to parents who were non-consanguineous, presented disease at the age of less than 1 year (range: 11 d to 8 mo), and experienced severe diarrhea with fistulas in the perianal region; blood samples were collected from three healthy volunteers.
| Patient A | Patient B | Patient C | Patient D | Normal range | |
| Sex | Male | Female | Male | Female | |
| Consanguinity | - | - | - | - | |
| Disease onset | 3 mo | 9 mo | 11 d | 18 d | |
| Diarrhea (times/d) | 1 | 8 | 20 | 6 | |
| Bloody stool | + | - | + | + | |
| Weight (kg) SDS | 7.65 (-1.475) | 6.2 (-1.4) | 8.2 (-1.05) | 6.3 (-1.15) | |
| Height (cm) SDS | 76 (-1.7) | 65.2 (-3.65) | 78 (0.25) | 67 (1) | |
| Perianal disease | + | + | + | + | |
| Extragastro-intestinal manifestations | Fever, UTI, Sepsis | Recurrent otitis media | Fever, respiratory infetion | Fever, UTI, eczama | |
| WBC (× 109) | 12.4 | 19.44 | 15.34 | 24.5 | 3.69-9.16 |
| Hemoglobin (g/L) | 101 | 77 | 107 | 96 | 113-151 |
| Platelet (× 109) | 624 | 621 | 424 | 387 | 101-320 |
| IL-10 (pg/mL) | 87.3 | 106 | 43.4 | 80.4 | < 12.9 |
| TNF-α (pg/mL) | 20.7 | 30.5 | 3.9 | 19.8 | < 16.5 |
| ESR (mm/h) | 6 | 55 | 19 | 23 | F: 0-20; M: 0-15 |
| Albumin (g/L) | 28 | 29 | 33 | 23 | 35-55 |
| Ferritin | 17 | 47.3 | 46.5 | 68.2 | 11-306.8 |
| Identified mutation | IL10RA (c.301C>T, p.R101W): Exon 1 del | IL10RA (c.537G>A, p.T179T): Exon 1 del | IL10RA (c.537G>A, p.T179T): Exon1 del | IL10RA (exon.1 del): Exon 1 del |
Written informed consent was obtained from the parents of the four patients who participated in the study. This study was approved by the Institutional Review Board of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (No. 2019-15).
Sample preparation and WGS were carried out by Beijing Berry Genomics Co., Ltd. (Beijing, China). The quality of the isolated genomic DNA was verified using the following two methods: (1) DNA degradation and contamination were monitored by electrophoresis on 1% agarose gels; and (2) DNA concentration was measured using the Qubit DNA Assay Kit and Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States).
A total of 1 μg DNA per sample was used as the input material for DNA library preparation. The DNA sequencing library was generated using the CLEANNGS DNA kit following the manufacturer’s recommendations, and indexing codes were added to each sample. Briefly, genomic DNA samples were enzymatically disrupted to a size of 350 bp. The DNA fragments were then end-polished, a-tailed, and ligated with a full-length adapter for Illumina sequencing, followed by further polymerase chain reaction (PCR) amplification. After the PCR products were purified (AMPure XP system, Beckman, Brea, CA, United States), libraries were analyzed to determine theirsize distribution using an Agilent 2100 Bioanalyzer (Santa Clara, CA, United States) and quantified by qPCR.
Clustering of the index-coded samples was performed on a cBot Cluster Generation System using the NovaSeq 5000/6000 S4 Reagent Kit (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions. After cluster generation, the DNA libraries were sequenced on an Illumina NovaSeq 6000 platform, and 150-bp paired-end reads were generated.
The pathogenicity of all mutations was further evaluated according to the American College of Medical Genetics and Genomics guidelines.
Peripheral blood mononuclear cells (PBMCs) were isolated according to a previous study, with minor modifications[14]. Briefly, blood was drawn from patient D and healthy controls by standard venipuncture in our pediatric ward and collected into a tube containing ethylenediamine tetraacetic acid. Blood (4 mL) was diluted 1:1 with sterile RPMI 1640 medium (Hyclone, Logan, UT, United States) at room temperature (RT) and carefully dropped into a 15-mL tube (Corning, Inc., Corning, NY, United States) containing 4 mL Ficoll-Paque Plus (GE Healthcare, Little Chalfont, United Kingdom). Notably, diluted blood was present on the surface of the Ficoll gradient. The 15-mL tube was centrifuged at 800 × g at RT for 20 min (brake off), after which the buffy coat was carefully aspirated and transferred to another sterile 15-mL tube. After washing the cells with 5 mL RPMI 1640 medium three times by centrifugation at 400 × g for 15 min at RT, most of the supernatant, as much as possible, was pipetted off.
Cells were aspirated with complete RPMI 1640 (10% fetal bovine serum and 1% penicillin-streptomycin) and cultured in 6-well plates at a density of 2 × 106 cells/well. Four groups of PBMCs from patients or healthy controls were established as follows: Unstimulated phosphate buffered saline (PBS), lipopolysaccharide (LPS) (100 ng/mL), LPS (100 ng/mL) + IL-10 (20 ng/mL), and IL-10 (20 ng/mL)[15]. Cells were cultured in the indicated milieu for 12 h at 37 °C. Proteins in PBS-, LPS-, and IL-10-stimulated PBMCs were collected in radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors for western blot analysis. The supernatants of the PBS-, LPS-, and LPS + IL-10-stimulated PBMCs were collected to determine tumor necrosis factor-α (TNF-α) level.
Western blotting was performed as described previously[16]. Polyvinylidene fluoride membranes were blotted with monoclonal antibodies against phos-signal transducer and activator of transcription 3 (STAT3) (Tyr705), phos-STAT3 (Ser727), STAT3 (Cell Signaling Technology, Danvers, MA, United States), and glyceraldehyde-3-phosphate dehydrogenase (Servicebio, Wuhan, China). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit (Cell Signaling Technology) secondary antibodies were detected using a chemiluinescent substrate (Millipore, Billerica, MA, United States). Images were captured using an automatic chemiluminescence image analysis system (Tanon, Shanghai, China).
The supernatant of PBMCs after stimulation with PBS, LPS or LPS + IL-10 was collected. IL-10 and TNF-α levels were determined using sandwich ELISA kits (DAKEWE, Shenzhen, China) according to the manufacturer’s instructions.
Continuous variables are presented as the mean ± SEM, and the unpaired two-tailed Student’s t-test or analysis of variance was used to compare the differences between groups as appropriate. Bonferroni correction was used for pairwise comparisons (GraphPad Prism v.5.0 software; GraphPad, Inc., La Jolla, CA, United States). Statis
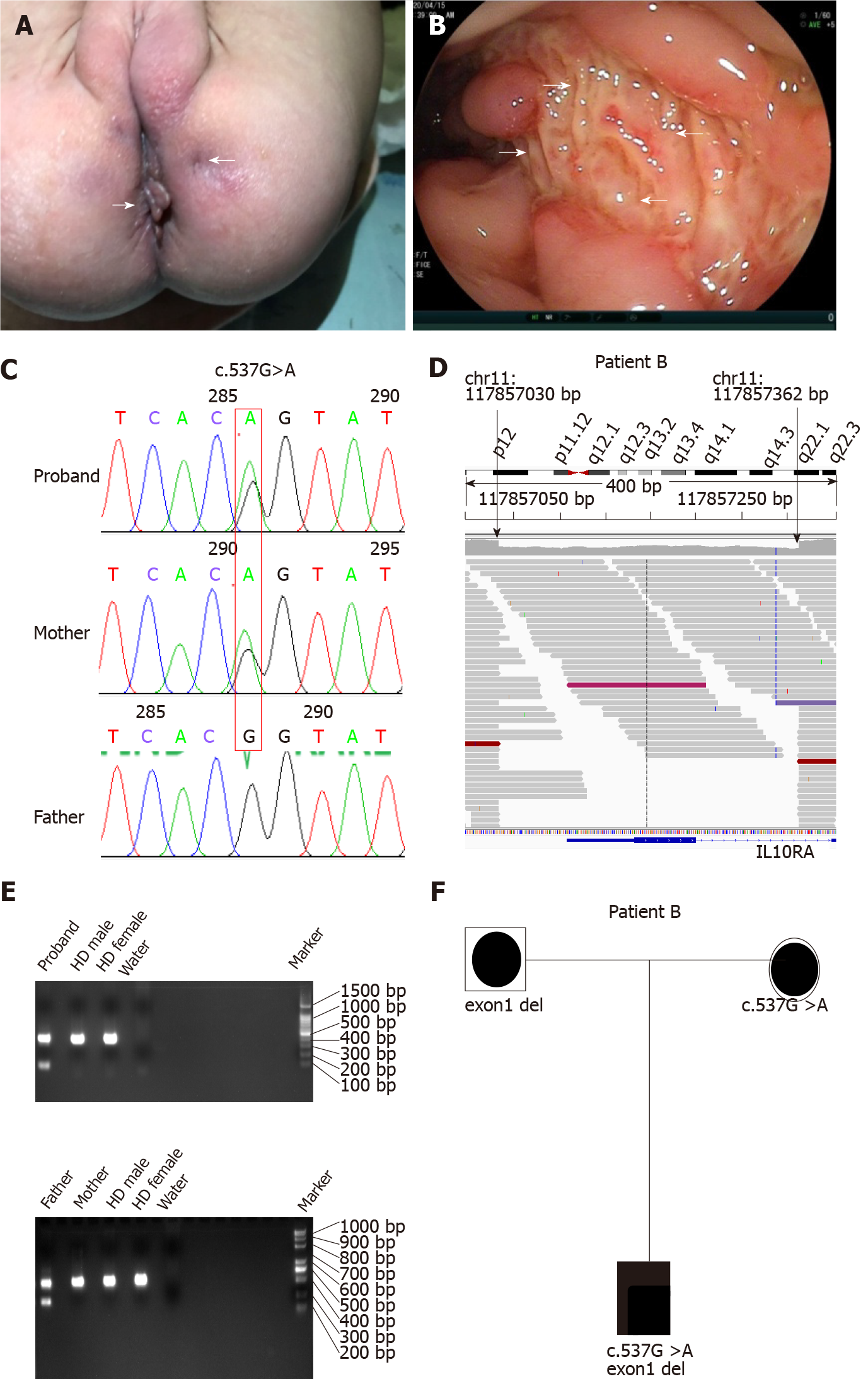
All four patients had severe diarrhea (> 6 times/d) and hematochezia during the first year of life. Patients C and D suffered from the disease during the newborn period. In addition to gastrointestinal symptoms, all cases exhibited extraintestinal manifestations, such as perianal abscesses, skin tags (Figure 1A), rectoperineal fistula, failure to thrive, recurrent otitis, urinary tract or respiratory infection, folliculitis, and even sepsis (Table 1). Elevated numbers of peripheral white blood cells and platelets and decreased hemoglobin and albumin levels were found in each patient. Remarkably, immune-related investigations showed that all patients had high serum levels of IL-10 (Table 1). All patients underwent colonoscopy and intestinal biopsy under general anesthesia, revealing erosive lesions (Figure 1B), and were diagnosed with Crohn’s disease.
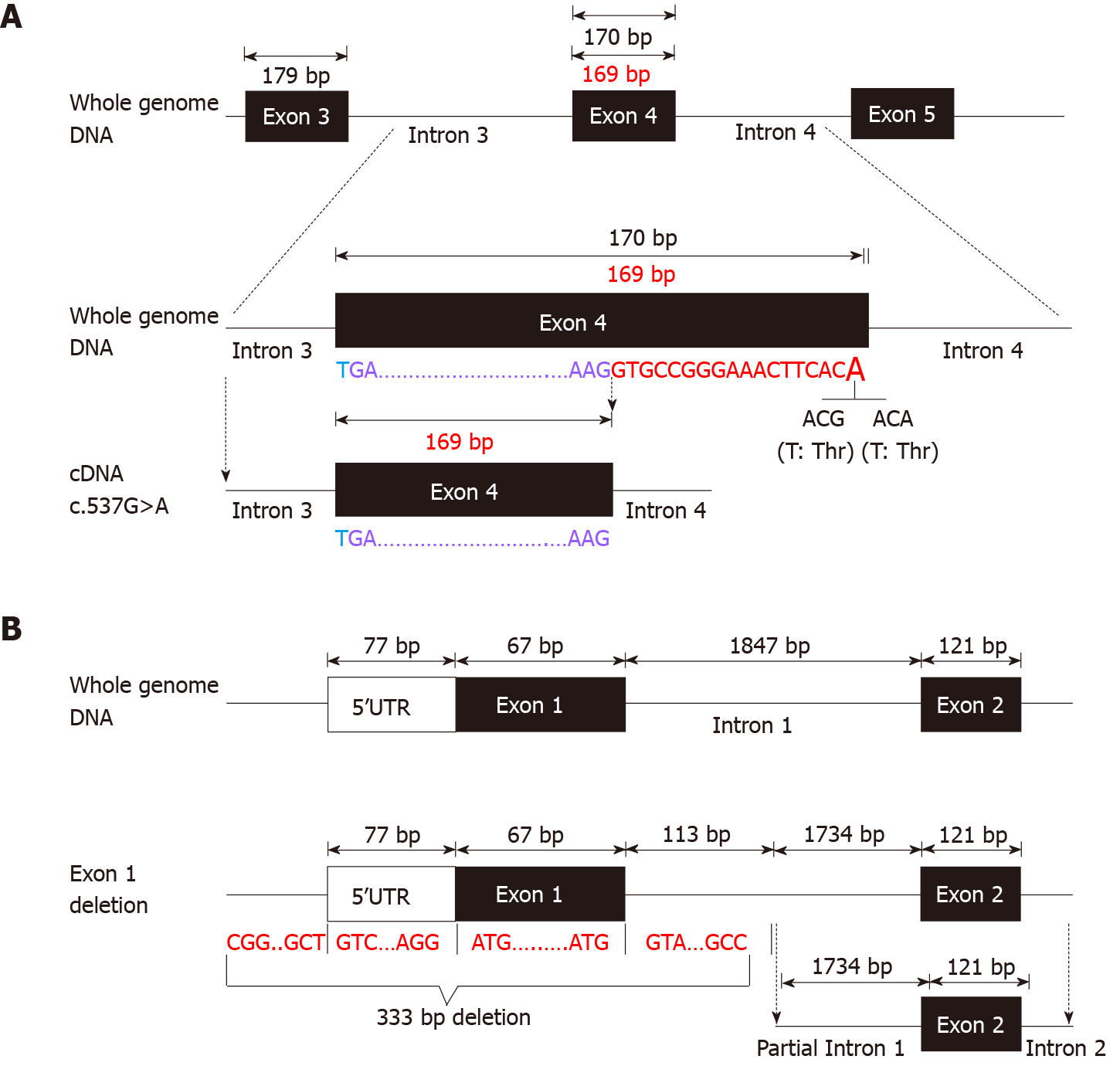
Before admission, all patients underwent TGPS or trio-WES. Two heterozygous pathogenic variants of IL10RA were detected in three patients (patient A: c.301C>T, p.R101W; patients B and C: c.537G>A, p.T179T) (Figure 1C and Supplementary Figures 1 and 2). Mutation of c.537G>A occurred at the exon-intron boundary of exon 4, which is a variant hotspot and disrupts RNA splicing (Figure 2A). No pathogenic or likely pathogenic variants were found in patient D.
As IL10RA mutation causes infantile IBD in an autosomal recessive manner and serum levels of IL-10 were very high in the four infantile patients with IBD, which is a valuable clinical indicator for identifying infantile IBD as a monogenic disease as we demonstrated previously[13], we suspected that mutations had been overlooked in WES owing to the techniques’ limitations. After performing WGS in patients A and B, the breakpoints of the novel deletion were identified by manual review and correction. The deletion was located at chr11:117857030 upstream of exon 1 and chr11:117857362 in intron 1, which contains the 5′-untranslated region (UTR), all of exon 1, and part of intron 1 in IL10RA (Figure 1D and Figure 2B). PCR revealed a paternally-inherited 333-bp deletion in addition to the point mutations mentioned above (Figure 1E). The deletion and point mutations were inherited from both parents and eventually constituted compound heterozygotes in patients B (Figure 1F) and A (Supplementary Figure 1).
After identifying the 333-bp deletion in the gene in patients A and B, we reanalyzed the trio-WES data for patients C and D, specifically in the region from 117857030 and 117857362 on chromosome 11 and detected the same deletion. Patient C was a compound heterozygous carrier for c.537G>A, p.T179T (maternal), and the 333-bp deletion (paternal) (Supplementary Figure 2), and patient D was homozygous for the 333-bp deletion.
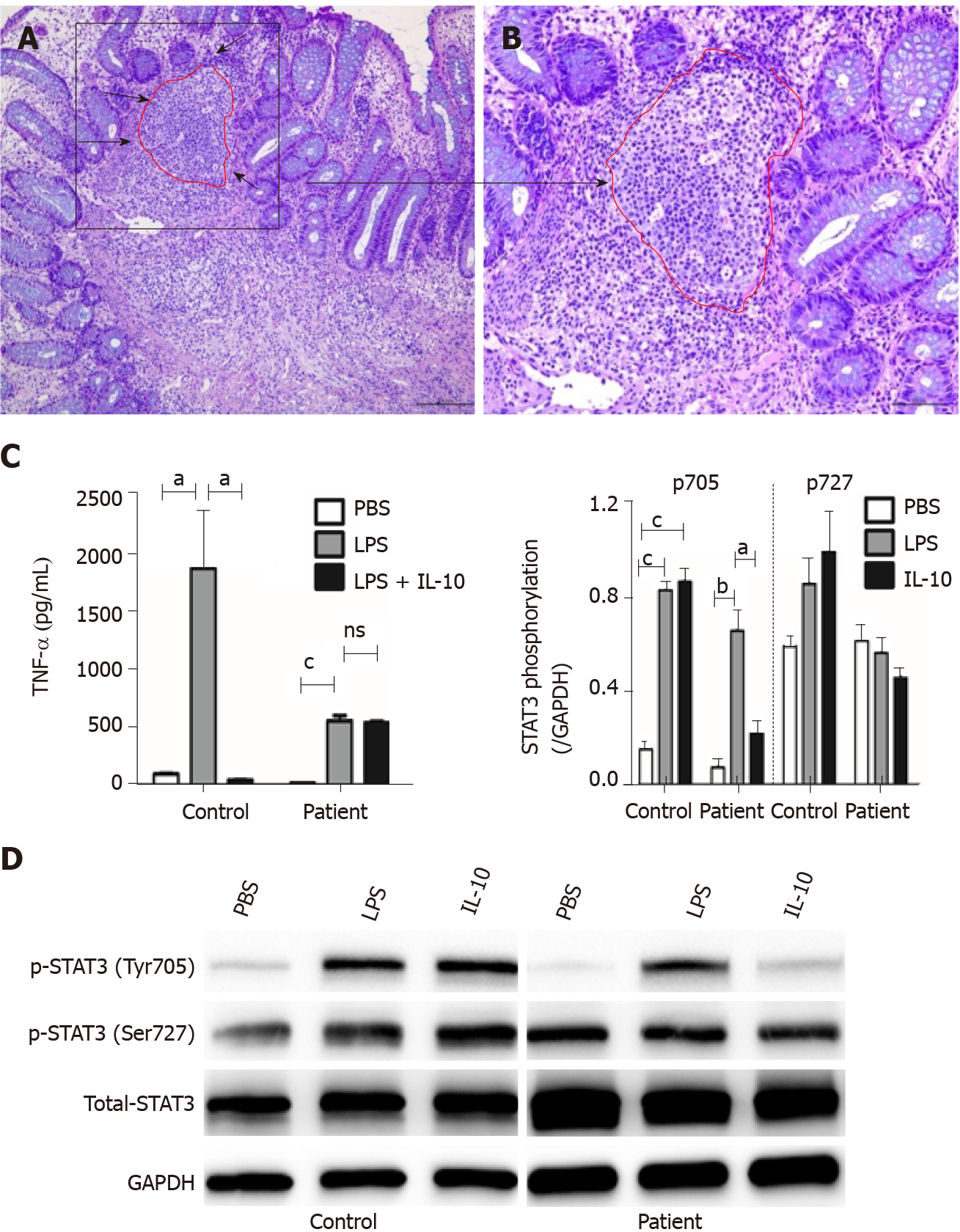
Histological findings in a specimen from the colon obtained during colonoscopy revealed oval-shaped intramural abscesses in the submucosa (Figure 3A). Figure 3B shows a higher magnification of inset 1 in Figure 3A, depicting intramural micro-abscesses.
To determine whether the novel 333-bp deletion in IL10RA caused IL-10R dysfunction and subsequently inhibited TNF-α production, supernatants of cultured PBMCs were collected and used to determine TNF-α levels. In healthy controls, LPS stimulation caused a remarkable increase in TNF-α production, whereas addition of IL-10 significantly decreased its abundance. Although LPS led to increased TNF-α production in patient D, this phenomenon was not reversed by the addition of IL-10, as observed in the healthy controls (Figure 3C).
To clarify the exact mechanisms involved, PBMCs were isolated from patient D because the patient was homozygous for the IL10RA deletion. PBMCs were stimulated with LPS in the presence or absence of IL-10. The results of western blot analysis showed that in PBMCs from healthy controls, both LPS and IL-10 stimulation caused an increase in the phosphorylation of STAT3 at Tyr705 but not at Ser727 (Figure 3D). In PBMCs from patient D, LPS stimulation also induced increased phosphorylation of STAT3 at Tyr705 but not at Ser727. However, IL-10 stimulation failed to significantly increase phosphorylation of STAT3 at Tyr705 in PBMCs of patient D compared with that in PBS-stimulated PBMCs. No significant differences in STAT3 phosphorylation at Ser727 were observed among the PBS-, LPS-, and IL-10-stimulated PBMCs from patient D (Figure 3D).
VEO-IBD is challenging to diagnose and treat because the patients are critically ill and exhibit numerous potential mono
We evaluated four patients who had Crohn’s disease from early infancy and exhibited failure to thrive, severe perianal disease, and resistance to medication. These characteristics indicate the presence of underlying genetic conditions[7]. Although the patients underwent NGS in a local hospital, neither TGPS nor WES revealed conclusive results. However, all patients showed very high serum IL-10 levels, and three patients had a disease-causing heterozygous mutation in IL10RA. According to our previous research, serum IL-10 levels > 33.05 pg/mL in patients with VEO-IBD strongly indicates the presence of IL10RA dysfunction[13]. Thus, we predicted that additional IL10RA mutations were missed during TGPS or WES. We did not detect the 333-bp deletion in IL10RA in two patients until WGS was performed. We then analyzed the same deletion in the other two patients by reanalyzing the trio-WES data. Finally, all patients were precisely diagnosed with VEO-IBD owing to compound heterozygous mutations in IL10RA in three patients and homozygous deletion involving IL10RA in one patient.
NGS, including TGPS and WES, is a powerful tool for identifying Mendelian genetic diseases in patients with VEO-IBD. The position paper on VEO-IBD by NASPGHAN/ESPGHAN suggests that NGS combined with the patient clinical history represents a vital component of the diagnostic approach[1]. A previous multicenter study showed that molecular diagnosis was achieved in 32% of patients with VEO-IBD when NGS was employed[18]. However, clinical NGS applications have limitations such as short read lengths, relatively high error rates, and incomplete coverage. Non-coding, yet potentially functional regions, and approximately 5% of exons are poorly covered in WES[19]. It is difficult to detect variants involving extensive deletions/insertions or short tandem repeats[20]. Charbit-Henrion et al[18] reported three WES-negative cases harboring large deletions in LRBA and NCF1. Compared to WES, WGS can detect all single-nucleotide variants, small indels, large indels, and copy number variants. In our cases, the 333-bp IL10RA deletion contained a 5′-UTR, exon 1, and part of intron 1. This large deletion was easy to overlook when WES was used because of its technical limitations and insufficient bioinformatics analysis. After detecting this deletion, we requested re-analysis of the WES data for patients C and D, specifically for the 333-bp region spanning IL10RA exon 1. As expected, these two patients harbored the deletion. These results indicate that WES can detect the 333-bp deletion, which was easily overlooked in bioinformatics analysis because of algorithm defects and insufficient experience. WGS compensates for the limitations of WES. Our results indicated that WGS should be performed in VEO-IBD cases with negative WES results, particularly in those with infantile-onset IBD and treatment failure.
A comprehensive range of defects may be associated with VEO-IBD. It is difficult to differentiate every patient based on these defects and their underlying genetic disorders. A small number of patients show distinct phenotypes associated with specific functions, such as IL-10/IL-10R defects, IPEX, CGD, and XIAP[1]. IL10RA mutation-induced VEO-IBD shows specific characteristics such as refractory pan-colitis, perianal defects, fistulas, and growth failure, which occur during the neonatal period. An assay that can detect the lack of IL-10 inhibition in LPS can confirm receptor mutations[1]. Nevertheless, this assay is complicated and not routinely available in most hospitals in China. In a retrospective study, we found that the assay was useful for diagnosing IL10RA defects when the serum level of IL-10 was > 33.05 pg/mL. The assay sensitivity was very close to 100%, and the specificity was approximately 84%[13]. Elevated serum levels of IL-10 in patients with VEO-IBD indicated that even such high level of IL-10 could not inhibit TNF-α release and alleviate inflammation. Thus, serum IL-10 level may be a substitute for determining IL-10 inhibition in LPS functional testing. Therefore, when classic symptoms and laboratory results indicate that patients may have IL-10/IL-10R dysfunction but WES is inconclusive, IL-10/IL10RA/IL-10RB must be investigated and analyzed. It is recommended to use WGS or specific PCR to detect whether a large deletion involving these genes has occurred.
Interestingly, TNF-α production in LPS-stimulated PBMCs was not as robust in patient D as in control subjects; in our study, patient D was administered with anti-TNF-α antibody (Infliximab) before blood collection, which may have led to relatively low TNF-α production in the supernatant of LPS-stimulated PBMCs compared to that in healthy controls. Another possible reason is that increased TNF-α in the blood of patient D led to activation of the TNF-α receptor, resulting in JNK signaling-dependent inhibition of Bcl-2 expression, which acts as a major anti-apoptosis protein[21].The lack of a comprehensive functional test for IL10RA mutation is the major limitation of our study, although we conducted western blotting to determine the possible mechanism of IL10RA mutation-induced dysfunction of IL10RA signaling. Further studies are needed to explore other potential differences in IL10RA signaling.
Using WGS, we identified a novel 333-bp deletion in IL10RA that contributed to four cases of clinically diagnosed VEO-IBD with inconclusive IL10RA mutations. Most importantly, we confirmed that typical clinical manifestations and increased serum levels of IL-10 strongly indicate the existence of IL-10R dysfunction even when WES results are negative.
Interleukin 10 receptor alpha (IL10RA) gene mutations constitute the most common monogenic disease in East Asia, affecting the health of children. However, identifying disease-causing mutant sites or copy number variants remains challenging in the clinic.
According to the results of our previous study, severe clinical symptoms as well as significantly increased serum IL-10 indicate IL10RA dysfunction, a monogenic phenotype of very early-onset inflammatory bowel disease (VEO-IBD). In addition, such very early-onset IBD showed a heterozygous IL10RA gene mutation by whole exon sequencing, leading to the employment of WGS and subsequent identification of 333-bp deletions in IL10RA.
We investigated the potential disease-causing gene mutations missed by WES and target gene panel sequencing (TGPS). Our results may contribute to monogenic disease diagnosis.
Four patients clinically diagnosed with VEO-IBD during the past 5 years were recruited for this study. Based on their severe clinical phenotypes and the fact that before hospitalization, three patients harbored an IL10RA mutation (c.301C>T, p.R101W in one patient; c.537G>A, p.T179T in two patients), as detected by TGPS and trio-WES, and because WES did not show conclusive results in the fourth patient, we performed whole-genome sequencing (WGS) on patients A and B and reanalyzed the trio-WES data from patients C and D. To verify the functional change caused by the novel mutation, peripheral blood mononuclear cells (PBMCs) from patient D were isolated and stimulated in vitro with lipopolysaccharide (LPS), IL-10, and LPS + IL-10. Serum IL-10 levels in four patients and tumor necrosis factor-α (TNF-α) in the cell supernatant were determined by ELISA. Phosphorylation of signal transducer and activator of transcription 3 (STAT3) at Tyr705 and Ser727 in PBMCs was determined by western blot analysis.
Results of WGS revealed a novel 333-bp deletion encompassing exon 1 of IL10RA in patients A and B, which was also found in patients C and D after reanalyzing their WES data. Patient D was homozygous for the 333-bp deletion. All four patients showed elevated serum IL-10 levels. In vitro, IL-10-stimulated PBMCs from patient D failed to induce STAT3 phosphorylation at Tyr705 and minimally suppressed TNF-α production induced by LPS. Phosphorylation at Ser727 in PBMCs was not affected by LPS or LPS + IL-10 in both healthy subjects and patient D.
Genome-wide uniformity of coverage of WGS identified a novel 333-bp deletion in IL10RA in four patients with VEO-IBD, whereas the results of initially performed WES were inconclusive. WGS, which was more informative than WES, is the most important comprehensive second-tier genomic test for monogenic diseases in the clinic.
We will customize a multiplex ligation-dependent amplification probe of the 333-bp deletion in IL10RA to help diagnose IL10RA mutation-related monogenic diseases.
The authors would like to thank the members of the Department of Pediatrics and Pediatric Laboratory.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nayudu SK S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Wang JJ
| 1. | Kelsen JR, Sullivan KE, Rabizadeh S, Singh N, Snapper S, Elkadri A, Grossman AB. North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition Position Paper on the Evaluation and Management for Patients With Very Early-onset Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr. 2020;70:389-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 2. | Cannioto Z, Berti I, Martelossi S, Bruno I, Giurici N, Crovella S, Ventura A. IBD and IBD mimicking enterocolitis in children younger than 2 years of age. Eur J Pediatr. 2009;168:149-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 3. | Gupta N, Bostrom AG, Kirschner BS, Cohen SA, Abramson O, Ferry GD, Gold BD, Winter HS, Baldassano RN, Smith T, Heyman MB. Presentation and disease course in early- compared to later-onset pediatric Crohn's disease. Am J Gastroenterol. 2008;103:2092-2098. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 113] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 4. | Fried K, Vure E. A lethal autosomal recessive entero-colitis of early infancy. Clin Genet. 1974;6:195-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 5. | Mégarbané A, Sayad R. Early lethal autosomal recessive enterocolitis: report of a second family. Clin Genet. 2007;71:89-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 6. | Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1970] [Cited by in RCA: 2353] [Article Influence: 123.8] [Reference Citation Analysis (2)] |
| 7. | Uhlig HH, Schwerd T, Koletzko S, Shah N, Kammermeier J, Elkadri A, Ouahed J, Wilson DC, Travis SP, Turner D, Klein C, Snapper SB, Muise AM; COLORS in IBD Study Group and NEOPICS. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology. 2014;147:990-1007.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 452] [Cited by in RCA: 476] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 8. | Crowley E, Muise A. Inflammatory Bowel Disease: What Very Early Onset Disease Teaches Us. Gastroenterol Clin North Am. 2018;47:755-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Leung G, Muise AM. Monogenic Intestinal Epithelium Defects and the Development of Inflammatory Bowel Disease. Physiology (Bethesda). 2018;33:360-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Shim JO, Seo JK. Very early-onset inflammatory bowel disease (IBD) in infancy is a different disease entity from adult-onset IBD; one form of interleukin-10 receptor mutations. J Hum Genet. 2014;59:337-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 11. | Suzuki T, Sasahara Y, Kikuchi A, Kakuta H, Kashiwabara T, Ishige T, Nakayama Y, Tanaka M, Hoshino A, Kanegane H, Abukawa D, Kure S. Targeted Sequencing and Immunological Analysis Reveal the Involvement of Primary Immunodeficiency Genes in Pediatric IBD: a Japanese Multicenter Study. J Clin Immunol. 2017;37:67-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Huang Z, Peng K, Li X, Zhao R, You J, Cheng X, Wang Z, Wang Y, Wu B, Wang H, Zeng H, Yu Z, Zheng C, Huang Y. Mutations in Interleukin-10 Receptor and Clinical Phenotypes in Patients with Very Early Onset Inflammatory Bowel Disease: A Chinese VEO-IBD Collaboration Group Survey. Inflamm Bowel Dis. 2017;23:578-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 13. | Su W, Yu Y, Xu X, Wang XQ, Huang JB, Xu CD, Xiao Y. Valuable clinical indicators for identifying infantile-onset inflammatory bowel disease patients with monogenic diseases. World J Gastroenterol. 2021;27:92-106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (2)] |
| 14. | Luotti S, Pasetto L, Porcu L, Torri V, Elezgarai SR, Pantalone S, Filareti M, Corbo M, Lunetta C, Mora G, Bonetto V. Diagnostic and prognostic values of PBMC proteins in amyotrophic lateral sclerosis. Neurobiol Dis. 2020;139:104815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hätscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1149] [Cited by in RCA: 1092] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 16. | Lin XL, Lv JJ, Lv J, Di CX, Zhang YJ, Zhou T, Liu JL, Xia ZW. Heme oxygenase-1 directly binds STAT3 to control the generation of pathogenic Th17 cells during neutrophilic airway inflammation. Allergy. 2017;72:1972-1987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 17. | Crowley E, Warner N, Pan J, Khalouei S, Elkadri A, Fiedler K, Foong J, Turinsky AL, Bronte-Tinkew D, Zhang S, Hu J, Tian D, Li D, Horowitz J, Siddiqui I, Upton J, Roifman CM, Church PC, Wall DA, Ramani AK, Kotlarz D, Klein C, Uhlig H, Snapper SB, Gonzaga-Jauregui C, Paterson AD, McGovern DPB, Brudno M, Walters TD, Griffiths AM, Muise AM. Prevalence and Clinical Features of Inflammatory Bowel Diseases Associated With Monogenic Variants, Identified by Whole-Exome Sequencing in 1000 Children at a Single Center. Gastroenterology. 2020;158:2208-2220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 18. | Charbit-Henrion F, Parlato M, Hanein S, Duclaux-Loras R, Nowak J, Begue B, Rakotobe S, Bruneau J, Fourrage C, Alibeu O, Rieux-Laucat F, Lévy E, Stolzenberg MC, Mazerolles F, Latour S, Lenoir C, Fischer A, Picard C, Aloi M, Dias JA, Hariz MB, Bourrier A, Breuer C, Breton A, Bronsky J, Buderus S, Cananzi M, Coopman S, Crémilleux C, Dabadie A, Dumant-Forest C, Gurkan OE, Fabre A, Diaz MG, Gonzalez-Lama Y, Goulet O, Guariso G, Gurcan N, Homan M, Hugot JP, Jeziorski E, Karanika E, Lachaux A, Lewindon P, Lima R, Magro F, Major J, Malamut G, Mas E, Mattyus I, Mearin LM, Melek J, Navas-Lopez VM, Paerregaard A, Pelatan C, Pigneur B, Pais IP, Rebeuh J, Romano C, Siala N, Strisciuglio C, Tempia-Caliera M, Tounian P, Turner D, Urbonas V, Willot S, Ruemmele FM, Cerf-Bensussan N. Diagnostic Yield of Next-generation Sequencing in Very Early-onset Inflammatory Bowel Diseases: A Multicentre Study. J Crohns Colitis. 2018;12:1104-1112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 19. | Adams DR, Eng CM. Next-Generation Sequencing to Diagnose Suspected Genetic Disorders. N Engl J Med. 2019;380:201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Płoski R. Next Generation Sequencing—General Information about the Technology, Possibilities, and Limitations. In: Płoski R. Clinical Applications for Next-Generation Sequencing 2016: 1-18. [DOI] [Full Text] |
| 21. | Lin YT, Wang CT, Lee JH, Chu CY, Tsao WC, Yang YH, Chiang BL. Higher Bcl-2 Levels decrease staphylococcal superantigen-induced apoptosis of CD4+ T cells in atopic dermatitis. Allergy. 2007;62:520-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |