Published online Nov 21, 2021. doi: 10.3748/wjg.v27.i43.7509
Peer-review started: April 21, 2021
First decision: July 14, 2021
Revised: July 21, 2021
Accepted: September 14, 2021
Article in press: September 14, 2021
Published online: November 21, 2021
Processing time: 212 Days and 7.6 Hours
Serum small extracellular vesicles (sEVs) and their small RNA (sRNA) cargoes could be promising biomarkers for the diagnosis of liver injury. However, the dynamic changes in serum sEVs and their sRNA components during liver injury have not been well characterized. Given that hepatic macrophages can quickly clear intravenously injected sEVs, the effect of liver injury-related serum sEVs on hepatic macrophages deserves to be explored.
To identify the characteristics of serum sEVs and the sRNAs during liver injury and explore their effects on hepatic macrophages.
To identify serum sEV biomarkers for liver injury, we established a CCL4-induced mouse liver injury model in C57BL/6 mice to simulate acute liver injury (ALI), chronic liver injury (CLI) and recovery. Serum sEVs were obtained and characterized by transmission electron microscopy and nanoparticle tracking analysis. Serum sEV sRNAs were profiled by sRNA sequencing. Differentially expressed microRNAs (miRNAs) were compared to mouse liver-enriched miRNAs and previously reported circulating miRNAs related to human liver diseases. The biological significance was evaluated by Ingenuity Pathway Analysis of altered sEV miRNAs and conditioned cultures of ALI serum sEVs with primary hepatic macrophages.
We found that both ALI and CLI changed the concentration and morphology of serum sEVs. The proportion of serum sEV miRNAs increased upon liver injury, with the liver as the primary contributor. The altered serum sEV miRNAs based on mouse studies were consistent with human liver disease-related circulating miRNAs. We established serum sEV miRNA signatures for ALI and CLI and a panel of miRNAs (miR-122-5p, miR-192-5p, and miR-22-3p) as a common marker for liver injury. The differential serum sEV miRNAs in ALI contributed mainly to liver steatosis and inflammation, while those in CLI contributed primarily to hepatocellular carcinoma and hyperplasia. ALI serum sEVs decreased both CD86 and CD206 expression in monocyte-derived macrophages but increased CD206 expression in resident macrophages in vitro.
Serum sEVs acquired different concentrations, sizes, morphologies and sRNA contents upon liver injury and could change the phenotype of liver macrophages. Serum sEVs therefore have good diagnostic and therapeutic potential for liver injury.
Core Tip: The liver injury changed the concentration, morphology and small RNA contents of serum small extracellular vesicles (sEVs). Altered serum sEV microRNAs (miRNAs) based on mouse studies were highly consistent with the circulating miRNAs reported in human liver diseases. Serum sEV miRNA signatures for acute liver injury and chronic liver injury and a panel of miRNAs that can be used as a common marker for liver injury were established. Acute liver injury serum sEVs depolarized monocyte-derived macrophages and educated resident liver macrophages to transform into M2-like cells. Serum sEVs have good diagnostic and therapeutic potential for liver injury.
- Citation: Lv XF, Zhang AQ, Liu WQ, Zhao M, Li J, He L, Cheng L, Sun YF, Qin G, Lu P, Ji YH, Ji JL. Liver injury changes the biological characters of serum small extracellular vesicles and reprograms hepatic macrophages in mice. World J Gastroenterol 2021; 27(43): 7509-7529
- URL: https://www.wjgnet.com/1007-9327/full/v27/i43/7509.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i43.7509
Because of its strategic location and biological functions, the liver is particularly susceptible to various pathogenic factors, including drugs, alcohol and viruses. The initial insult induces acute liver injury (ALI) or even liver failure. Repeated or persistent insults will cause chronic liver injury (CLI), resulting in liver fibrosis and finally fatal cirrhosis[1,2]. Therefore, it is important to identify individuals with liver injury. However, liver injury does not always cause noticeable signs and symptoms. Aside from the widely used blood liver function tests on serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST), which are not always restricted to liver injury[3], there is still a need to explore specific and sensitive biomarkers. With the rapid progress in medical research, it is now possible and necessary to search for new biomarkers from serum small extracellular vesicles (sEVs).
Recently, serum sEVs have attracted tremendous interest due to their essential roles in intercellular communication and to their diagnostic and therapeutic potential[4]. The term sEVs refers to extracellular vesicles released by cells that are of relatively small size (< 200 nm) and were previously regarded as exosomes[5]. The cargoes carried by sEVs represent a snapshot of the parental cells at the time of release and change depending on the physiological and pathological states[6,7]. In the liver, sEVs are released from both hepatocytes and nonparenchymal cells into the extracellular space and circulation. Several studies have reported that circulating sEV RNAs or proteins are abnormally expressed in the contexts of drug-induced liver injury (DILI), steatohepatitis, viral hepatitis and hepatocellular carcinoma (HCC)[6,8].
MicroRNAs (miRNAs) are 22-24 nt small noncoding RNAs involved in posttranscriptional regulation and various biological processes[9]. Tissue-specific distribution is a key feature of miRNAs, making miRNAs good candidates as biomarkers or therapeutic targets for particular types of tissue injury[10-12]. Serum miRNAs have been studied in a variety of liver diseases[13]. However, compared to serum miRNAs, serum sEV miRNAs are well protected from RNA enzymes. Thus, serum sEV can serve as a more reliable miRNA pool[14]. We hypothesized that serum sEVs and their miRNA cargoes might reflect liver damage upon injury and could be promising biomarkers.
In the present study, we tried to determine the effects of liver injury on serum sEVs and the small RNAs (sRNAs) they transport; we were also interested in determining if there is any difference between acute and chronic injury. A study in this regard will aid in identification of potential serum sEV miRNA biomarkers. The dynamic changes in the number and morphology of serum sEVs and the sRNA components of serum sEVs were examined. The profiles of deregulated serum sEV miRNAs were obtained and compared to those of mouse liver enriched miRNAs and previously reported circulating miRNAs related to human liver diseases (HLD). To further evaluate the biological significance of serum sEVs upon liver injury, conditioned cultures of ALI serum sEVs and primary hepatic macrophages were carried out.
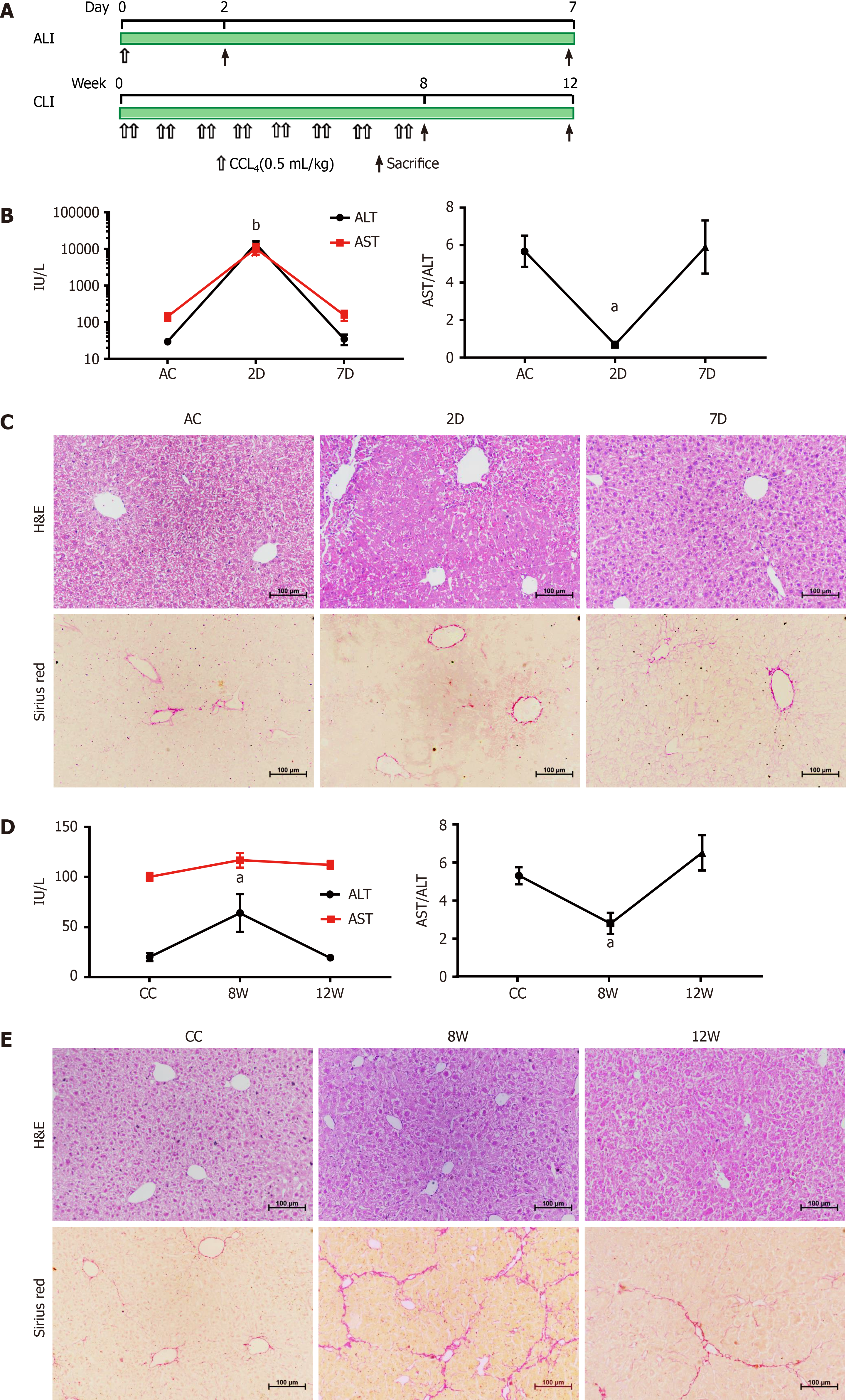
Male C57BL/6 mice (8 wk old) were purchased from the Shanghai Medical Laboratory Animal Center (Shanghai, China) and housed in the animal facility of Nantong University with temperature 25 ± 2 °C and 12 h light/dark cycle controls. All experimental protocols were approved by the Animal Ethics Committee of Nantong University. The animal care and experiments were performed in accordance with the relevant guidelines and regulations. For ALI, mice were treated with a single dose of CCL4 (0.5 mL/kg intraperitoneal injection) dissolved in olive oil (1:9). The mice were sacrificed at 2 d or 7 d. For CLI, mice were treated with CCL4 (0.5 mL/kg) or vehicle twice a week for 8 wk[15]. The mice were sacrificed 48 h after the last injection at 8 wk or at 12 wk (Figure 1A). Mice treated with the same volume of olive oil served as the controls for the ALI and CLI models, and 10-12 mice were used in each group. Blood or livers were collected from each group for further analyses.
Blood was collected by left ventricular puncture from mice and was left undisturbed for 1 h at 37 °C and 2 h at 4 °C. Afterward, the samples were centrifuged at 1000 × g for 10 min at 4 °C; the clear upper fractions were aliquoted and stored at -80 °C. Serum ALT and AST levels were measured on an ADVIA 1800 autoanalyzer (Siemens Healthcare Diagnostics, Deerfield, IL, United States). The livers were preserved in 4% paraformaldehyde, paraffin-embedded and sectioned. The liver tissue sections were stained with hematoxylin and eosin (Beyotime Biotechnology, Shanghai, China) for routine histology and 0.1% Sirius Red (Sigma-Aldrich, St. Louis, MO, United States) for collagen evaluation.
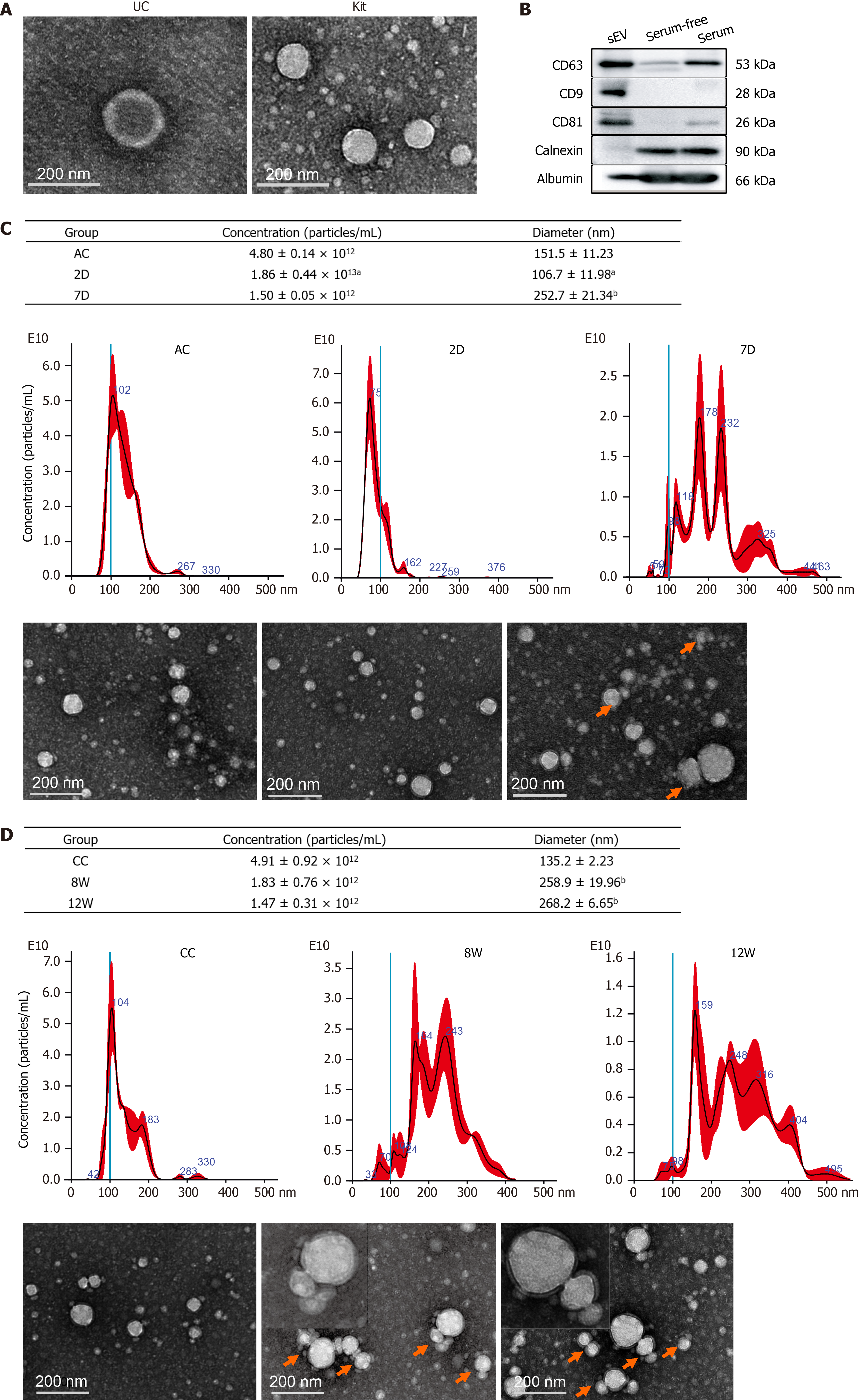
Exosome-enriched serum sEV fractions were precipitated using ultracentrifugation and an ExoQuick precipitation kit (System Biosciences Inc., Mountain View, CA, United States)[16]. The sizes and particle concentrations of the isolated serum sEVs were measured by nanoparticle tracking analysis (NTA, NanoSight NS300, Malvern, United Kingdom). Serum sEVs were visualized using transmission electron microscopy (TEM, HT7700, Hitachi Ltd., Tokyo, Japan). The expression of exosomal protein markers was determined by Western blot analysis. The details are provided in the Supplementary material, Supporting Information.
Serum sEV sRNA sequencing (RNA-seq) was conducted by BMK Biotech Co., Ltd. (Beijing, China) with biological replicates for each group.
The raw data were processed as described previously[16]. The trimmed sequencing reads were deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9462). Bioinformatic analysis of the differentially expressed serum sEV miRNAs was performed using Ingenuity Pathways Analysis (Qiagen, Valencia, CA, United States). The significance of enrichment for genes with particular biologically relevant functions was determined with a one-sided Fisher’s exact test.
The detailed procedure is provided in the Supplementary material, Supporting Information.
Primary mouse hepatic macrophages were isolated from male C57BL/6 mouse livers by Percoll (GE Healthcare, Princeton, NJ, United States) density gradient centrifugation. Incubation of liver macrophages with mouse serum sEVs and subsequent multiple-color flow cytometric analysis were carried out. The details are provided in the Supplementary material, Supporting Information.
Statistical analyses were performed with GraphPad Prism 7.0 software (GraphPad Software, Inc., La Jolla, CA, United States). Quantitative data were reported as the mean ± standard deviation. Comparisons between groups were made by Student’s t-test or one-way analysis of variance. All P values were two-sided, and statistical significance was accepted for a value less than 0.05. Except for the data from sRNA-seq experiments, which included two biological replicates for each group, the data provided in the present study were from three or more independent experiments.
Additional methods and details are provided in the Supplementary material, Supporting Information.
The mouse CCL4-induced ALI and recovery model and the mouse CCL4-induced CLI and recovery model were established and validated (Figure 1).
In acutely injured mice (at 2 d; 2D group), serum ALT and AST levels were increased, and hepatocyte necrosis and inflammatory cell infiltration were observed around the lobular central vein. After 5 d of recovery (at 7 d; 7D group), the elevated ALT and AST levels had returned to the baseline of the ALI control group, and the histological changes were also reversed (Figure 1B and C). For CLI, repeated CCL4 treatment induced a slight elevation in serum ALT (at 8 wk; 8W group), but the level returned to the baseline of the CLI control (CC) group by 4 wk after cessation of CCL4 treatment (at 12 wk; 12W group) (Figure 1D). Although the ALT level change in CLI at 8W was not as prominent as those in ALI at 2D, the change was comparable to the ALT level changes reported by other study groups using the same mice CLI model[17,18]. Damaged hepatocytes and centrilobular contracture were observed in the livers of the CLI mice (8W), with mild inflammatory cell infiltration. Sirius Red staining showed obvious collagen deposition and pseudolobule formation in 8W CLI mice. These morphological changes were alleviated in recovered mice (12W) (Figure 1E).
The isolated particles were spherical or cup-shaped, as observed by TEM (Figure 2A). Exosomal protein markers, including CD63, CD81 and CD9, were all highly expressed (Figure 2B), as determined by Western blot analysis.
NTA showed that the mean diameter of the particles ranged from 90.2 nm to 127.8 nm. The number of particles was higher in the 2D group but lower in the 7D group than in the control group, and the particle diameters were smaller in the 2D group but larger in the 7D group than in the control group. We also noticed that the size distribution of sEVs widened and that multiple peaks were present in the 7D group (Figure 2C). For CLI mice, the particle concentrations in the 8W and 12W groups tended to be lower than those in the control group, although there were no significant differences. The size distribution of sEVs was expanded with multiple peaks in both the 8W and 12W groups, and the particle diameters were larger in both the 8W and 12W groups (Figure 2D). TEM examination revealed that the multiple peaks reflected the aggregation or fusion of mouse serum sEVs present in 7D, 8W and 12W samples (Figure 2C and D).
These findings suggested that both ALI and CLI changed the number and morphology of mouse serum sEVs, and even when the visible histological changes of the liver had recovered in the 7D and 12W groups, the changes in particle number and morphology of serum sEVs persisted.
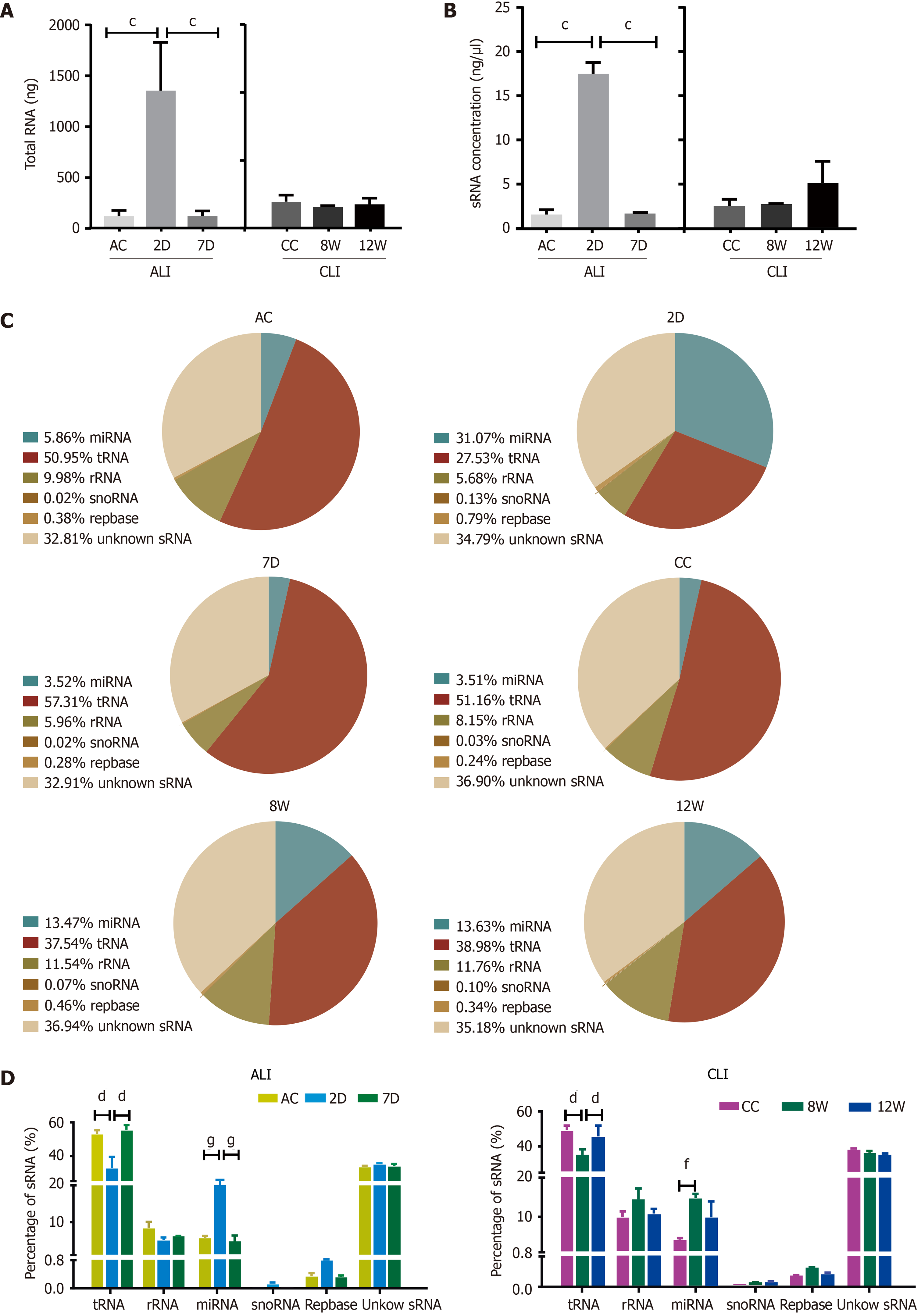
Dramatic increases in both total RNA and sRNA levels in serum sEVs were observed upon ALI (2D) (Figure 3A and B). For CLI, there was no significant difference in either sEV RNA or sRNA content among the groups. RNA-seq and annotation revealed that each pair of biological repeats had at least 94.97% common sequences in clean reads (Supplementary Figure 1). In the control groups (ALI control and CC), tRNA was the dominant sRNA species in serum sEVs, followed by rRNA and miRNA (Figure 3C). The most remarkable change in serum sEV sRNAs was the increase in miRNA proportion in both ALI and CLI mice. Compared to the control condition, ALI increased the proportion of miRNAs by more than four-fold, but the proportion returned to baseline by 7 d; the proportion of miRNAs increased by almost three-fold in CLI mice but was partially restored by 12 wk. With the increase in miRNA, the proportion of tRNA decreased (Figure 3D).
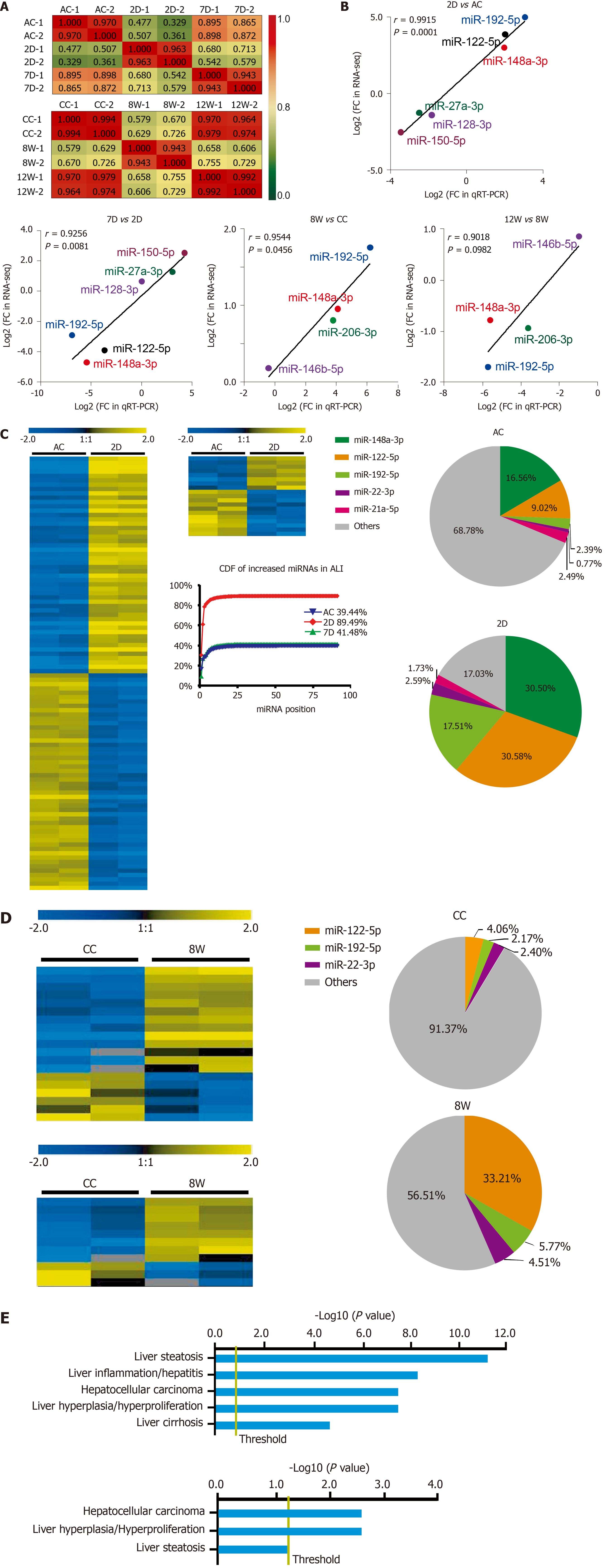
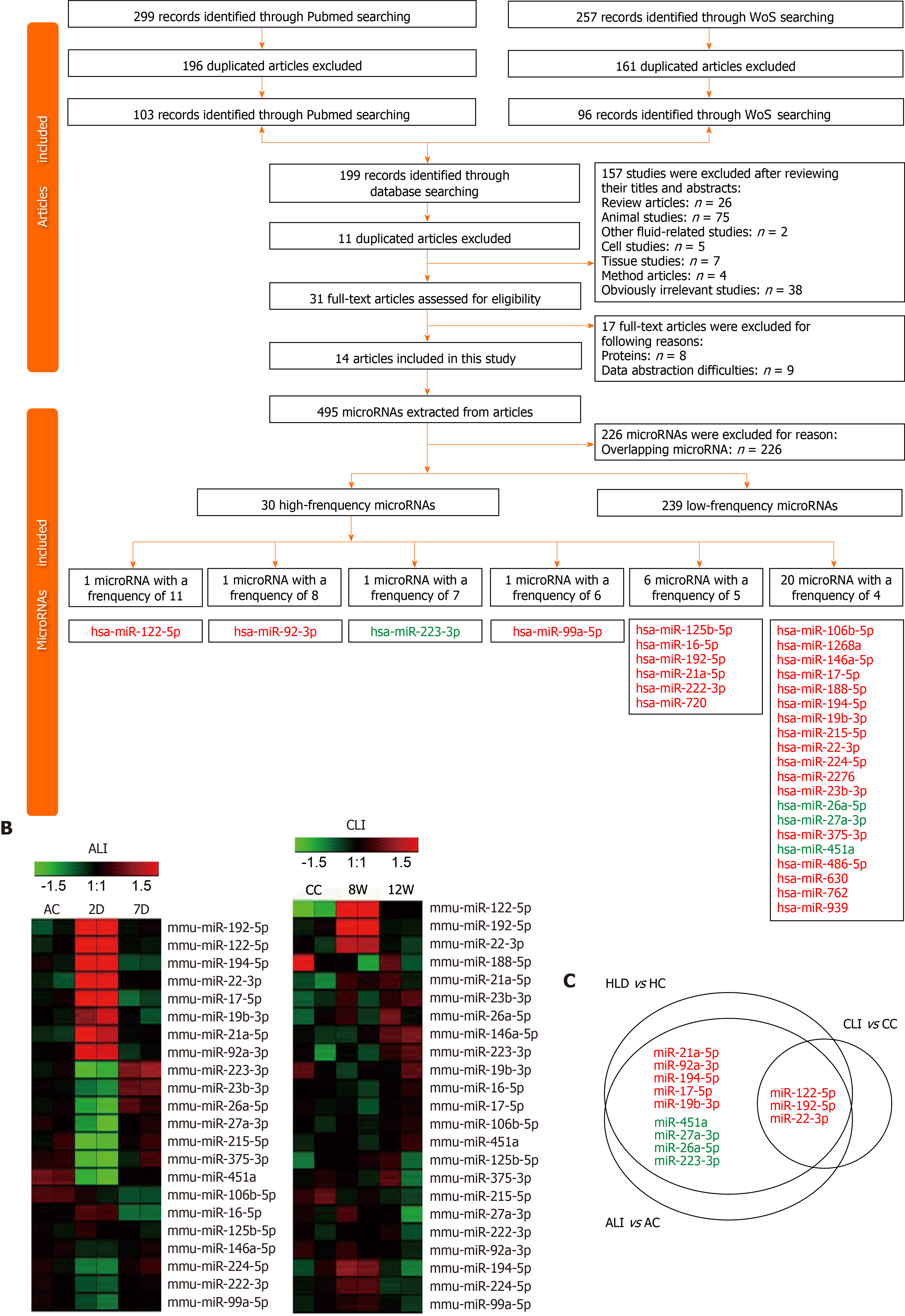
For the ALI and CLI groups, 467 and 488 detectable sEVs (transcripts per million reads ≥ 5.0) miRNAs were obtained, respectively. The biological replications were highly correlated in each group (Figure 4A). The RNA-seq data were further validated by quantitative real-time polymerase chain reaction. Differentially expressed miRNAs with different abundances were selected for validation (Figure 4B).
In total, 91 miRNAs were upregulated and 85 miRNAs were downregulated (fold change ≥ 2.0, P < 0.05) in the 2D group compared with the ALI control group (Figure 4C and Supplementary Table 1). The levels of most of these miRNAs had recovered to baseline levels in the 7D group, in which only 8 upregulated miRNAs and 11 downregulated miRNAs were detected (fold change ≥ 2.0, P < 0.05) (Figure 4C and Supplementary Table 2). The cumulative distribution frequency was calculated by adding each proportion of miRNAs from most to least abundant. The plot showed that the top five upregulated miRNAs (miR-148a-3p, miR-122-5p, miR-192-5p, miR-22-3p and miR-21a-5p) in the 2D group accounted for up to 84.27% of all detectable miRNAs (Figure 4C).
Only 13 miRNAs were upregulated and six miRNAs were downregulated (fold change ≥ 2.0, P < 0.05) in the 8W group compared with the CC group (Figure 4D and Supplementary Table 3); in addition, 8 miRNAs were upregulated and 3 miRNAs were downregulated (fold change ≥ 2.0, P < 0.05) in the 12W group compared with the CC group (Figure 4D and Supplementary Table 4). The cumulative distribution frequency analysis showed that the top three upregulated miRNAs (miR-122-5p, miR-192-5p, and miR-22-3p) in the 8W group constituted up to 43.48% of all detectable miRNAs (Supplementary Figure 2). These findings suggested that both ALI and CLI induced changes in serum sEV miRNA composition. The changes were caused by the differential expression of a small number of miRNAs with high abundance.
The biological significance of these differentially expressed serum sEV miRNAs in liver injury was explored by Ingenuity Pathway Analysis (www.qiagen.com/ingenuity). For the 176 differentially expressed serum sEV miRNAs upon ALI, hepatic steatosis was the most significant hepatotoxic effect, followed by liver inflammation. For the 19 differentially expressed serum sEV miRNAs upon CLI, HCC was the most significant, followed by liver hyperplasia (Figure 4E).
We were interested in determining the contribution of liver cells to the changes in serum sEV miRNAs upon ALI and CLI. First, the liver miRNA expression profile for wild-type male C57BL/6 mice was established using BRB-Array Tool v4.6.0 (https://brb.nci.nih.gov) based on the RNA-seq data from GSE78792 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78792)[19] (Supplementary Table 5). Differentially expressed serum sEV miRNAs from ALI and CLI mice were compared to the liver miRNA expression profile. Among the top ten most abundant liver miRNAs, seven were also among the top ten increased miRNAs in serum sEVs upon ALI (Table 1) and constituted up to 84.70% of the increased serum sEV miRNAs. For CLI, the levels of three were raised in serum sEVs. These three miRNAs were the top three miRNAs that increased upon CLI (Table 1) and constituted up to 60.56% of the total increased serum sEV miRNAs. These findings suggested the liver as the primary contributor to the upregulated serum sEV miRNAs during ALI and CLI and confirmed that the serum sEV miRNA test could be a reliable and sensitive way to monitor either ALI or CLI.
| GSE78792 liver | Rank | ALI | ALI rank | CLI | CLI rank |
| miR-192 | 1 | miR-192-5p | 3 | miR-192-5p | 2 |
| miR-22 | 2 | miR-22-3p | 4 | miR-22-3p | 3 |
| miR-30a | 3 | miR-30a-5p | 7 | N/A | N/A |
| miR-148a | 4 | miR-148a-3p | 1 | N/A | N/A |
| miR-21a | 5 | miR-21a-5p | 5 | N/A | N/A |
| miR-26a-2 | 6 | miR-26a-5p | 105 | N/A | N/A |
| miR-122 | 7 | miR-122-5p | 2 | miR-122-5p | 1 |
| miR-10a | 8 | N/A | N/A | N/A | N/A |
| miR-143 | 9 | miR-143-3p | 92 | N/A | N/A |
| miR-27b | 10 | miR-27b-3p | 10 | N/A | N/A |
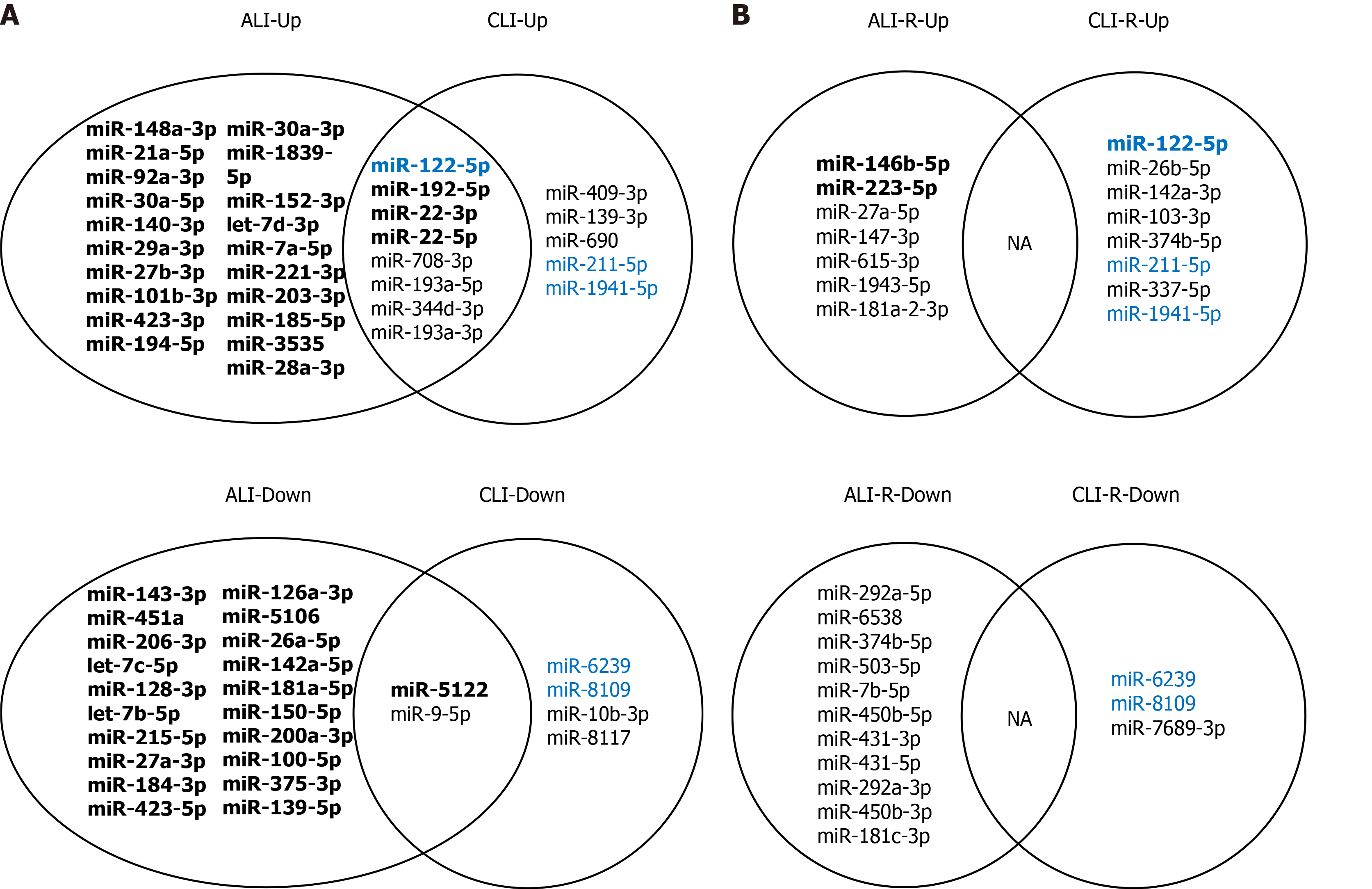
To identify serum sEV miRNA signatures for liver injury, we compared the differentially expressed serum sEV miRNAs in various stages of ALI and CLI. Compared to the levels in the vehicle control samples, eight miRNAs were upregulated and two miRNAs were downregulated significantly during the acute injury stage, and these changes were sustained through the chronic phase (fold change ≥ 2.0, P < 0.05); thus, they can serve as common liver injury signatures (Figure 5A). In addition, the levels of 166 miRNAs changed significantly during acute injury, while those of nine miRNAs changed dramatically during chronic injury. These miRNAs with high abundance (transcripts per million reads _mean > 1000) have the potential to be ALI or CLI signatures. The complete lists of these potential ALI and CLI serum sEV miRNA signatures are provided in Supplementary Table 6. According to their abundance, up to the top 20 miRNAs are listed in Figure 5. For the recovery stage, 18 miRNAs were changed significantly in the ALI group, and 11 miRNAs were changed significantly in the CLI group (Figure 5B). Some of these miRNAs overlapped with differentially expressed serum sEV miRNAs in corresponding acute or chronic injury stages.
To explore the biological significance of the ALI and CLI serum sEV miRNA signatures in HLDs, we performed a systematic review of abnormally expressed circulating miRNAs reported in various HLDs. In total, 299 and 257 studies were identified from PubMed (https://pubmed.ncbi.nlm.nih.gov) and Web of Science (http://apps.webofknowledge.com/) databases, respectively (Figure 6A). Data were retrieved from 14 studies, including drug-induced liver injury[20], chronic hepatitis B[21-25], chronic hepatitis C[21,25-27], nonalcoholic fatty liver disease[28], nonalcoholic steatohepatitis[23], liver cirrhosis[24,29,30] and HCC[21-24,28-33] studies. Details on the 14 articles are summarized in Supplementary Table 7.
In total, 269 nonredundant abnormally expressed circulating miRNAs related to HLDs were extracted, and those that appeared ≥ four times were defined as high-frequency miRNAs (Figure 6A). Of the 30 high-frequency miRNAs (SupplementaryTable 7), 23 miRNAs were detected in ALI and CLI serum sEVs (Figure 6B), 14 miRNAs overlapped with ALI and CLI serum sEV signatures, and 12 miRNAs showed the same expression trend. Of the 12 miRNAs, three miRNAs (miR-122-5p, miR-192-5p, and miR-22-3p) were identified as being increased in both ALI and CLI mice and thus have the potential to serve as common signatures for either ALI or CLI. The other 9 miRNAs were identified as ALI signatures (miR-21a-5p, miR-92a-3p, miR-194-5p, miR-17-5p and miR-19b-3p were increased; miR-451a, miR-27a-3p, miR-26a-5p, and miR-223-3p were decreased) and may reflect acute or active liver injury (Figure 6C). In addition, it was noteworthy that the four high-frequency circulating miRNAs reported in HLDs with decreased levels all exhibited decreased levels in serum sEVs upon ALI.
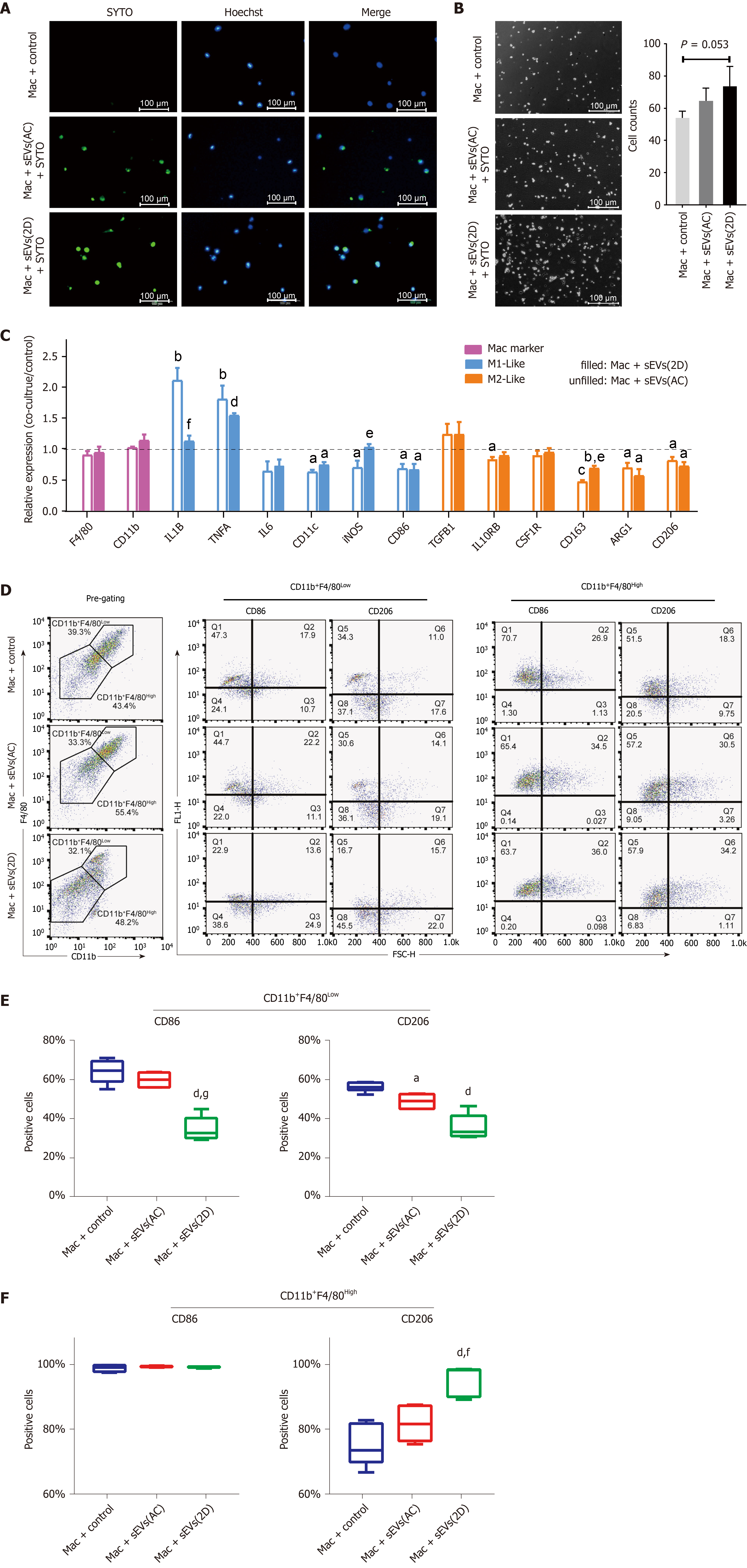
Primary mouse hepatic macrophages were isolated, purified and incubated with SYTO-labeled serum sEVs. After 24 h, green fluorescence was observed in most macrophages in both the control and ALI (2D) serum sEV incubation groups (Figure 7A). These observations indicated that serum sEVs could be taken up by hepatic macrophages. Serum sEVs from ALI mice (2D) accelerated the adhesion of hepatic macrophages (Figure 7B). As determined by quantitative real-time polymerase chain reaction, normal serum sEVs increased M1-like gene (IL-1B and TNFA) expression and decreased the expression of some M2-like genes (IL-10RB, CD163, ARG1 and CD206); ALI serum sEVs showed similar effects. However, compared to normal serum sEVs, ALI serum sEVs tended to decrease M1-like IL-1B but increase M2-like CD163 gene expression (Figure 7C).
In order to further dissect the effects of ALI serum sEVs on monocyte-derived and resident hepatic macrophage subgroups, multiple-color flow cytometric analyses were performed to assess the expression of M1-like CD86 and M2-like CD206 in CD11b+F4/80Low monocyte-derived and CD11b+F4/80High resident macrophages (Tacke and Zimmermann[34] and our unpublished data). ALI serum sEVs decreased both CD86 and CD206 expression in the CD11b+F4/80Low subgroup but increased CD206 expression in the CD11b+F4/80High subgroup (Figure 7D and E). These findings indicated that ALI serum sEVs might induce depolarization of CD11b+F4/80Low monocyte-derived macrophages but M2 differentiation of CD11b+F4/80High resident macrophages.
A growing number of studies have suggested the diagnostic value of serum sEV content for liver injury, especially miRNAs[6,8]. To explore potential serum sEV miRNA biomarkers for liver injury, we simulated the complex processes of liver injury and recovery in CCL4-induced ALI and CLI mouse models. The 2D group and 8W group represented ALI and CLI, respectively, while the 7D group and 12W group represented the recovery stage of ALI and CLI, respectively. The ALT and AST levels elevated in the 2D group and 8W group and returned to the baseline of the control groups (Figure 1B and D). Moreover, the histological changes also reversed in the 7D and 12W recovery groups (Figure 1C and E), which indicated that the ALI and CLI and recovery models were well established.
Unexpectedly, aside from differentially expressed miRNAs, we found that the concentration, size and morphology of serum sEVs might be essential features in liver injury (Figure 2). The number of serum sEVs increased upon ALI, which has been reported in human alcoholic hepatitis and alcoholic liver injury mouse models[35-37]. Furthermore, we found sustained decreases in serum sEV number during the chronic injury stage and the recovery stage for both ALI and CLI. Moreover, serum sEVs became smaller upon ALI but enlarged during ALI recovery and the CLI stage, and the increased size persisted through the CLI recovery stage, at which time there were multiple peaks, as observed by NTA. Interestingly, aggregation of serum sEV particles was observed in samples from the ALI recovery stage, CLI stage and CLI recovery stage by TEM, which explained the multiple peaks and increased particle size found by NTA. We propose that the aggregation of sEVs might reflect membrane damage of extracellular vesicles following liver injury. Thus, changes in serum sEV concentration, size, and morphology are well connected to a particular stage of liver injury and could provide diagnostic clues. In addition, the changes in serum sEVs persisted even when liver function and visible histopathological changes were restored; thus, they could be useful to trace recent liver injury.
Then, we showed that ALI and CLI altered the sRNA levels and components in serum sEVs. For ALI, both the total RNA and sRNA levels of serum sEVs increased significantly, and the proportion of miRNAs in sRNA also increased. Although there were no significant increases in RNA or sRNA levels for CLI, the proportion of miRNAs increased significantly. Compared to CLI, ALI significantly changed more miRNA species (176 vs 19, fold change ≥ 2.0, P < 0.05). The increased proportion of miRNAs upon liver injury was mainly attributable to a few highly abundant miRNAs. To traceback the primary source of the increased serum sEV miRNAs, the highly abundant miRNAs detected in serum sEVs from ALI and CLI mice were evaluated in the livers of normal male C57BL/6 mice. The miRNAs with the highest abundance were all liver-enriched miRNAs (Table 1). We propose that the liver is the main contributor to the elevations in miRNAs in serum sEVs for both ALI and CLI. Thus, serum sEVs carry the miRNA messages released from the injured liver, and examining the serum sEV miRNAs could be a reliable way to monitor either ALI or CLI.
By comparing the expression profiles of serum sEV miRNAs in various stages of ALI and CLI, we obtained a list of miRNAs that can be used as common liver injury signatures as well as the miRNA signatures for ALI, CLI and the recovery stages. However, these signatures were obtained from mouse models and need to be validated in human patients. Hence, we carried out a systematic review of previously published studies and obtained 30 miRNAs that were highly correlated with HLD, including drug-induced liver injury, chronic hepatitis B, chronic hepatitis C, nonalcoholic fatty liver disease, nonalcoholic steatohepatitis and liver cirrhosis (Figure 6). Of these 30 miRNAs, 3 miRNAs (miR-122-5p, miR-192-5p and miR-22-3p) were identified as common injury signatures that were increased in both ALI and CLI mice. Although these three miRNAs have been studied separately[24,36,38,39], here, for the first time, they were combined as a universal signature for either ALI or CLI. Nine miRNAs were identified as ALI signatures. Along with the five increased miRNAs (miR-21a-5p, miR-92a-3p, miR-194-5p, miR-17-5p and miR-19b-3p), four miRNAs were decreased (miR-451a, miR-27a-3p, miR-26a-5p and miR-223-3p): the same four miRNAs that are frequently reported to be decreased in HLD (Figure 6). Although the human data were mostly from patients with CLI, the overlapping ALI signatures might reflect active lesions. Based on serum sEV miRNAs, we established common signatures for liver injury and specific signatures for acute/active liver injury.
The biological significance of the alterations in sEV miRNAs upon liver injury was explored by Ingenuity Pathway Analysis. The top hepatotoxicity-related functions of the altered serum sEV miRNAs in ALI were hepatic steatosis and liver inflammation, while the altered miRNAs carried by serum sEVs from CLI were linked primarily to HCC, followed by liver hyperplasia. These findings are consistent with the clinical findings that ALI mainly causes inflammation and that sustained damage increases the risk of carcinoma[40,41]. Hepatic steatosis reflects fatty degeneration that is typically caused by CCL4[42]. These findings reveal the pathological importance of serum sEVs during the processes of ALI and CLI.
It has been reported that hepatic macrophages can take up serum sEVs and play essential roles in the clearance of intravenously injected sEVs from the systemic circulation[43,44]. We were interested in determining the effects of liver injury-related serum sEVs on hepatic macrophages and whether these effects could aggravate liver damage or play a protective role. We found that in vitro, ALI serum sEVs could be taken up by hepatic macrophages and promote macrophage adhesion. Furthermore, ALI serum sEVs tended to decrease M1-like gene expression, such as IL-1B and TNFA expression, and increase M2-like gene expression, including CD163 expression. However, the trends were not consistent. Considering that hepatic macrophages are heterogeneous populations composed of two subgroups, including resident macrophages and circulating monocyte-derived macrophages[34], we propose that macrophages of different origins might react inconsistently to ALI serum sEVs. The expression of M1 (CD86) and M2 (CD206) polarization signatures on the two subgroups of hepatic macrophages was examined by flow cytometry. ALI serum sEVs induced depolarization of CD11b+F4/80Low monocyte-derived macrophages but induced M2 differentiation of CD11b+F4/80High resident macrophages. We propose that the changes in serum sEVs upon ALI might alleviate liver damage by depolarizing monocyte-derived macrophages and educating resident hepatic macrophages to transform into M2-like cells.
In conclusion, we found that the concentration, size and morphology of serum sEV particles were essential features for liver injury. We established specific serum sEV miRNA signatures for different liver injury stages and created a list of miRNAs that can be used as common liver injury biomarkers. The altered ALI and CLI serum sEV miRNAs were connected to diverse liver pathological processes. ALI serum sEVs reprogrammed hepatic macrophage subgroups differently. Serum sEVs not only have good diagnostic potential but also could be used to ameliorate liver injury. However, the diagnostic and therapeutic potential of these altered serum sEVs upon liver injury deserves further study.
Both acute liver injury (ALI) and chronic liver injury (CLI) do not always cause noticeable signs and symptoms. Serum small extracellular vesicles (sEVs) have attracted tremendous interest due to their essential roles in intercellular communication and their diagnostic and therapeutic potential. The cargoes carried by sEVs represent a snapshot of the parental cells and change depending on the physiological and pathological states.
Serum sEVs and their small RNA (sRNA) cargoes could be promising biomarkers for the diagnosis of liver injury.
The present study aimed to characterize the dynamic changes of serum sEVs and their sRNA components during liver injury and to explore the effect of liver injury-related serum sEVs on hepatic macrophages.
Male C57BL/6 mice were treated with CCL4 to establish a mouse liver injury model for simulating ALI, CLI and recovery. Serum sEVs were obtained and characterized by transmission electron microscopy and nanoparticle tracking analysis. Serum sEV sRNAs were profiled by sRNA sequencing. Differentially expressed microRNAs (miRNAs) were compared to mouse liver-enriched miRNAs and previously reported circulating miRNAs related to human liver diseases. The biological significance was evaluated by Ingenuity Pathway Analysis of the altered sEV miRNAs and conditioned cultures of ALI serum sEVs with primary hepatic macrophages.
Both ALI and CLI changed the concentration and morphology of serum sEVs. The proportion of serum sEV miRNAs increased upon liver injury, with the liver as the primary contributor. The altered serum sEV miRNAs based on mouse study were consistent with human liver disease-related circulating miRNAs. We established serum sEV miRNA signatures for ALI and CLI and a panel of miRNAs (miR-122-5p, miR-192-5p, and miR-22-3p) as a common marker for liver injury. ALI serum sEVs decreased both CD86 and CD206 expression in monocyte-derived macrophages but increased CD206 expression in resident macrophages in vitro.
Serum sEVs acquired different concentrations, sizes, morphologies and sRNA contents upon diverse liver injured pathological processes. ALI serum sEVs reprogrammed hepatic macrophage subgroups differently.
Serum sEVs have good diagnostic and therapeutic potential for liver injury.
The authors would like to acknowledge Shen Y for the statistical review.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Li WJ S-Editor: Gao CC L-Editor: Filipodia P-Editor: Li JH
| 1. | Reuben A, Koch DG, Lee WM; Acute Liver Failure Study Group. Drug-induced acute liver failure: results of a U.S. multicenter, prospective study. Hepatology. 2010;52:2065-2076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 526] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 2. | Kullak-Ublick GA, Andrade RJ, Merz M, End P, Benesic A, Gerbes AL, Aithal GP. Drug-induced liver injury: recent advances in diagnosis and risk assessment. Gut. 2017;66:1154-1164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 274] [Cited by in RCA: 334] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 3. | Nathwani RA, Pais S, Reynolds TB, Kaplowitz N. Serum alanine aminotransferase in skeletal muscle diseases. Hepatology. 2005;41:380-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 261] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 4. | Hubbell CL, Mankes RF, Reid LD. A small dose of morphine leads rats to drink more alcohol and achieve higher blood alcohol concentrations. Alcohol Clin Exp Res. 1993;17:1040-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D'Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr, Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-'t Hoen EN, Noren Hooten N, O'Driscoll L, O'Grady T, O'Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Østergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr, Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėkas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6453] [Cited by in RCA: 7698] [Article Influence: 1099.7] [Reference Citation Analysis (1)] |
| 6. | Szabo G, Momen-Heravi F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat Rev Gastroenterol Hepatol. 2017;14:455-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 219] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 7. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8246] [Cited by in RCA: 9798] [Article Influence: 544.3] [Reference Citation Analysis (0)] |
| 8. | Urban SK, Mocan T, Sänger H, Lukacs-Kornek V, Kornek M. Extracellular Vesicles in Liver Diseases: Diagnostic, Prognostic, and Therapeutic Application. Semin Liver Dis. 2019;39:70-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 9. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27845] [Article Influence: 1326.0] [Reference Citation Analysis (0)] |
| 10. | Erdos Z, Barnum JE, Wang E, DeMaula C, Dey PM, Forest T, Bailey WJ, Glaab WE. Evaluation of the Relative Performance of Pancreas-Specific MicroRNAs in Rat Plasma as Biomarkers of Pancreas Injury. Toxicol Sci. 2020;173:5-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL, Sistare FD, Glaab WE. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977-1983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 484] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 12. | Sun P, Liu DZ, Jickling GC, Sharp FR, Yin KJ. MicroRNA-based therapeutics in central nervous system injuries. J Cereb Blood Flow Metab. 2018;38:1125-1148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 13. | Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431-1443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 229] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 14. | Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 465] [Cited by in RCA: 630] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 15. | Gu L, Deng WS, Sun XF, Zhou H, Xu Q. Rapamycin ameliorates CCl4-induced liver fibrosis in mice through reciprocal regulation of the Th17/Treg cell balance. Mol Med Rep. 2016;14:1153-1161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Zhao F, Cheng L, Shao Q, Chen Z, Lv X, Li J, He L, Sun Y, Ji Q, Lu P, Ji Y, Ji J. Characterization of serum small extracellular vesicles and their small RNA contents across humans, rats, and mice. Sci Rep. 2020;10:4197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 17. | Kwon HJ, Won YS, Park O, Chang B, Duryee MJ, Thiele GE, Matsumoto A, Singh S, Abdelmegeed MA, Song BJ, Kawamoto T, Vasiliou V, Thiele GM, Gao B. Aldehyde dehydrogenase 2 deficiency ameliorates alcoholic fatty liver but worsens liver inflammation and fibrosis in mice. Hepatology. 2014;60:146-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 153] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 18. | Ma X, Luo Q, Zhu H, Liu X, Dong Z, Zhang K, Zou Y, Wu J, Ge J, Sun A. Aldehyde dehydrogenase 2 activation ameliorates CCl4 -induced chronic liver fibrosis in mice by up-regulating Nrf2/HO-1 antioxidant pathway. J Cell Mol Med. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Langfelder P, Gao F, Wang N, Howland D, Kwak S, Vogt TF, Aaronson JS, Rosinski J, Coppola G, Horvath S, Yang XW. MicroRNA signatures of endogenous Huntingtin CAG repeat expansion in mice. PLoS One. 2018;13:e0190550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 20. | Russo MW, Steuerwald N, Norton HJ, Anderson WE, Foureau D, Chalasani N, Fontana RJ, Watkins PB, Serrano J, Bonkovsky HL. Profiles of miRNAs in serum in severe acute drug induced liver injury and their prognostic significance. Liver Int. 2017;37:757-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 21. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 22. | Wang Y, Gao Y, Shi W, Zhai D, Rao Q, Jia X, Liu J, Jiao X, Du Z. Profiles of differential expression of circulating microRNAs in hepatitis B virus-positive small hepatocellular carcinoma. Cancer Biomark. 2015;15:171-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, Shim SG, Paik YH. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 343] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 24. | Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X, Wang Z, Qiu S, Wang X, Yang G, Sun H, Tang Z, Wu Y, Zhu H, Fan J. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 498] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 25. | Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, Kosaka N, Ochiya T, Taguchi YH. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS One. 2012;7:e48366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 26. | Murakami Y, Tanahashi T. Analysis of circulating microRNA by microarray in liver disease. Methods Mol Biol. 2013;1024:173-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 27. | Santangelo L, Bordoni V, Montaldo C, Cimini E, Zingoni A, Battistelli C, D'Offizi G, Capobianchi MR, Santoni A, Tripodi M, Agrati C. Hepatitis C virus direct-acting antivirals therapy impacts on extracellular vesicles microRNAs content and on their immunomodulating properties. Liver Int. 2018;38:1741-1750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 28. | Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 2014;9:e105192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 29. | Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, Run W, Tian L, Jia X, Gao Y. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond). 2011;120:183-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 183] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 30. | Fornari F, Ferracin M, Trerè D, Milazzo M, Marinelli S, Galassi M, Venerandi L, Pollutri D, Patrizi C, Borghi A, Foschi FG, Stefanini GF, Negrini M, Bolondi L, Gramantieri L. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, Identify Cirrhotic Patients with HCC. PLoS One. 2015;10:e0141448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 116] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 31. | Chiu LY, Kishnani PS, Chuang TP, Tang CY, Liu CY, Bali D, Koeberl D, Austin S, Boyette K, Weinstein DA, Murphy E, Yao A, Chen YT, Li LH. Identification of differentially expressed microRNAs in human hepatocellular adenoma associated with type I glycogen storage disease: a potential utility as biomarkers. J Gastroenterol. 2014;49:1274-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 32. | Lu L, Guo D, Chen X, Xiong W, Jie S, Li H. Abnormal miRNAs Targeting Chromosome Open Reading Frame Genes were Enriched in Microvesicles Derived from the Circulation of HCC. Biochem Genet. 2016;54:120-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 33. | Xue X, Zhao Y, Wang X, Qin L, Hu R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 34. | Tacke F, Zimmermann HW. Macrophage heterogeneity in liver injury and fibrosis. J Hepatol. 2014;60:1090-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 600] [Cited by in RCA: 813] [Article Influence: 73.9] [Reference Citation Analysis (0)] |
| 35. | Momen-Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med. 2015;13:261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 252] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 36. | Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep. 2015;5:9991. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 37. | Eguchi A, Lazaro RG, Wang J, Kim J, Povero D, Willliams B, Ho SB, Stärkel P, Schnabl B, Ohno-Machado L, Tsukamoto H, Feldstein AE. Extracellular vesicles released by hepatocytes from gastric infusion model of alcoholic liver disease contain a MicroRNA barcode that can be detected in blood. Hepatology. 2017;65:475-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 38. | Motawi TK, Mohamed MR, Shahin NN, Ali MAM, Azzam MA. Time-course expression profile and diagnostic potential of a miRNA panel in exosomes and total serum in acute liver injury. Int J Biochem Cell Biol. 2018;100:11-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 39. | Arataki K, Hayes CN, Akamatsu S, Akiyama R, Abe H, Tsuge M, Miki D, Ochi H, Hiraga N, Imamura M, Takahashi S, Aikata H, Kawaoka T, Kawakami Y, Ohishi W, Chayama K. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J Med Virol. 2013;85:789-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 40. | Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10:656-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 772] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 41. | Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat. 2009;16:453-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 232] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 42. | Recknagel RO, Glende EA Jr, Dolak JA, Waller RL. Mechanisms of carbon tetrachloride toxicity. Pharmacol Ther. 1989;43:139-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 834] [Cited by in RCA: 801] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 43. | Imai T, Takahashi Y, Nishikawa M, Kato K, Morishita M, Yamashita T, Matsumoto A, Charoenviriyakul C, Takakura Y. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J Extracell Vesicles. 2015;4:26238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 281] [Cited by in RCA: 436] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 44. | Matsumoto A, Takahashi Y, Chang HY, Wu YW, Yamamoto A, Ishihama Y, Takakura Y. Blood concentrations of small extracellular vesicles are determined by a balance between abundant secretion and rapid clearance. J Extracell Vesicles. 2020;9:1696517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (0)] |