Published online Nov 21, 2021. doi: 10.3748/wjg.v27.i43.7480
Peer-review started: April 3, 2021
First decision: July 3, 2021
Revised: August 2, 2021
Accepted: November 15, 2021
Article in press: November 15, 2021
Published online: November 21, 2021
Processing time: 230 Days and 3.6 Hours
Pancreatic ductal adenocarcinoma (PDAC) remains the most lethal type of cancer. The 5-year survival rate for patients with early-stage diagnosis can be as high as 20%, suggesting that early diagnosis plays a pivotal role in the prognostic improvement of PDAC cases. In the medical field, the broad availability of biomedical data has led to the advent of the “big data” era. To overcome this deadly disease, how to fully exploit big data is a new challenge in the era of precision medicine. Artificial intelligence (AI) is the ability of a machine to learn and display intelligence to solve problems. AI can help to transform big data into clinically actionable insights more efficiently, reduce inevitable errors to improve diagnostic accuracy, and make real-time predictions. AI-based omics analyses will become the next alterative approach to overcome this poor-prognostic disease by discovering biomarkers for early detection, providing molecular/genomic subtyping, offering treatment guidance, and predicting recurrence and survival. Advances in AI may therefore improve PDAC survival outcomes in the near future. The present review mainly focuses on recent advances of AI in PDAC for clinicians. We believe that breakthroughs will soon emerge to fight this deadly disease using AI-navigated precision medicine.
Core Tip: Pancreatic ductal adenocarcinoma (PDAC) remains the most lethal type of cancer. Artificial intelligence (AI) is the ability of a machine to learn and display intelligence to solve problems. AI can help to transform big data into clinically actionable insights more efficiently, reduce inevitable errors to improve diagnostic accuracy, and make real-time predictions. AI-based omics analyses should be the next alternative approach to improve survival outcomes in PDAC by discovering biomarkers for early detection, molecular/genomic subtyping, treatment guidance, and predicting recurrence and survival. The present review mainly focuses on recent advances of AI in PDAC for clinicians.
- Citation: Hayashi H, Uemura N, Matsumura K, Zhao L, Sato H, Shiraishi Y, Yamashita YI, Baba H. Recent advances in artificial intelligence for pancreatic ductal adenocarcinoma. World J Gastroenterol 2021; 27(43): 7480-7496
- URL: https://www.wjgnet.com/1007-9327/full/v27/i43/7480.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i43.7480
Pancreatic ductal adenocarcinoma (PDAC) stands the most life-threatenning type of cancer[1]. The recent 5-year survival rate for PDAC in all stages is 8.5% according to American Cancer Society statistics 2017. In patients with early-stage diagnosis, the 5-year survival rate for can be as high as 20%. During the past ten years, median overall survival (OS) has improved from 22.1 mo to 35 mo in resectable PDAC, considerably owing to improvements in adjuvant therapies[2-5]. These findings suggest that early diagnosis plays a pivotal role in the prognostic improvement of PDAC cases. Furthermore, the high recurrence rate, even in patients who have undergone curative resection, and chemoresistance to the current systemic chemotherapies (FOLFIRINOX: 5-fluorouracil, folinic acid, irinotecan, and oxaliplatin; and GnP: Gemcitabine plus nab-paclitaxel)[6,7] are major issues. Based on recent advances in genetic analysis, PDACs have been divided into several molecular subtypes[8-11], which is a prelude of precision medicine. Genetic and molecular profiling researches have revealed that up to 25% (range 12%-25%) of PDACs maintained actionable molecular alterations. Actually, matching to relevant molecular-specific treatments improves the OS compared to that of those without actionable mutations or those who do not receive molecular-specific therapy[12]. The comprehensive biomedical data has led to the dawn of the “big data” era in the medical field[13].
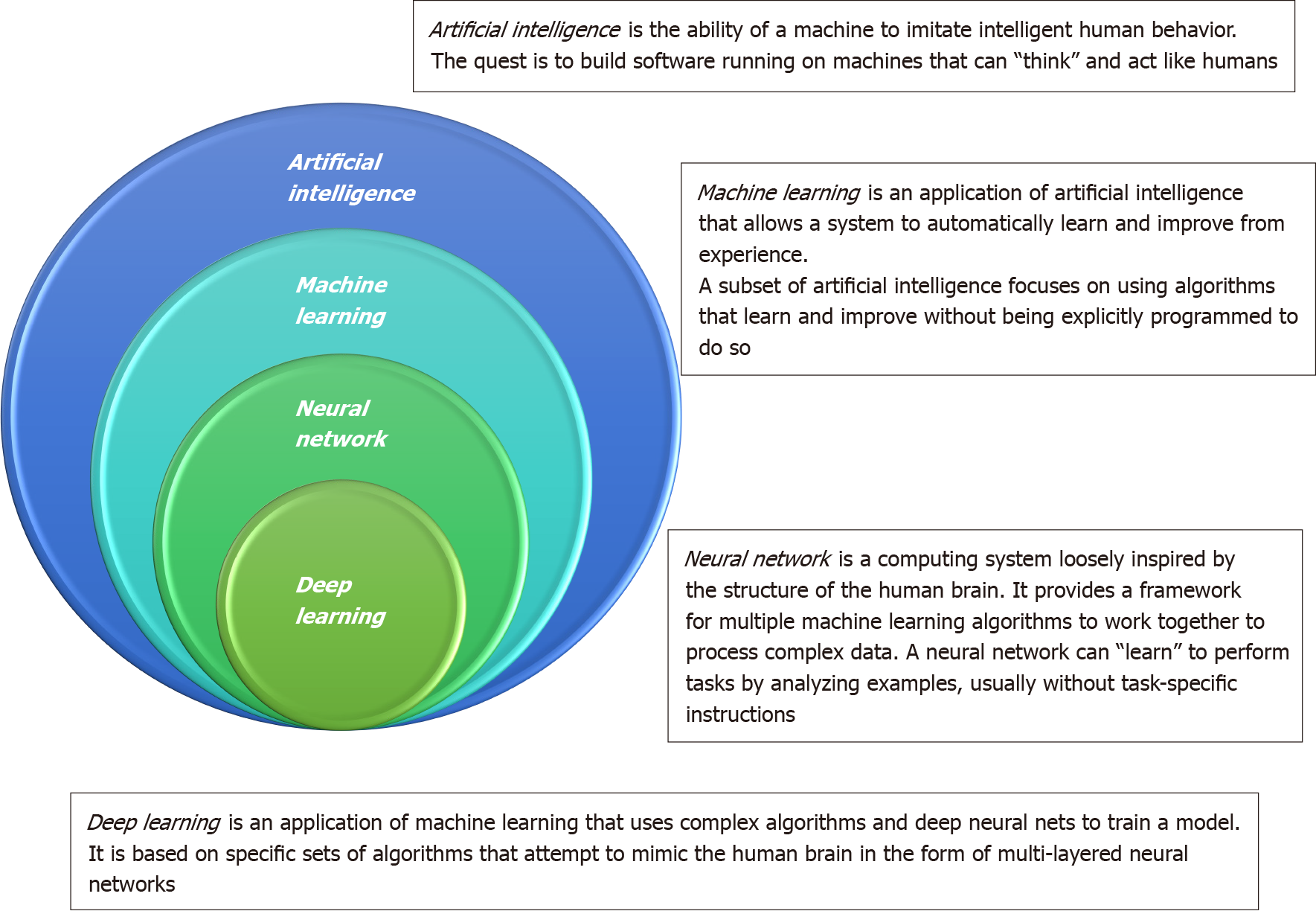
To overcome this deadly disease, how to well utilize big data is a next step for physicians and researchers physicians in the era of precision medicine. The main issue for physicians has shifted from gathering data to competently analyzing it. Artificial intelligence (AI) is the ability of a machine to learn and display intelligence to solve problems (Figure 1)[14]. An artificial neural network (ANN) can imitate the human neural meridian system. It is divided into three parts: Input layer, hidden layer, and output layer. “Deep learning” refers to an ANN with multiple hidden layers. Machine learning helps researchers spend less time on data processing. The processess for employing machine learning generally contain the following: Gathering the basic data, separating the data into an experimental group and a verification group, buildinging a screening and processing model, inputting the experimental group data into the model, accounting the output results, and confirming the model’s workability using the verification group. The verification group can be employed to examine the sensitivity and specificity of the experimental group, while the experimental group can fabricate more intelligent model. An overview of types in AI is provided in the Supplementary material. Chen et al[15] developed a survival prediction model of non-small cell lung cancer patients through the use of ANN. AI has also been applied to tackle and manage the recent coronavirus disease 2019 crisis in many areas, including screening, diagnosis, severity stratification, mortality prediction, and epidemiology controls[16].
In the age of precision medicine, AI can support to convert big data into clinically actionable perception more conveniently, reduce the inevitable errors to improve diagnostic accuracy, and make real-time predictions[17,18]. Due to latest break
| Ref. | Modality | Type of algorithm | Sensitivity (%) | Specificity (%) | ROC-AUC (or accuracy %) |
| PDAC risk prediction | |||||
| Boursi et al[25], 2021 | 7 clinical variables | Logistic regression | 66.53 | 54.91 | 0.71 |
| Appelbaum et al[29], 2021 | 18 risk factors | Logistic regression | NA | NA | 0.71 |
| Muhammad et al[30], 2018 | Personal health data (18 features) | ANN | 80.7 | 80.7 | 0.85 |
| Hsieh et al[28], 2018 | ICD-9 code | Logistic regression | NA | NA | 0.727 |
| Boursi et al[26], 2017 | 10 clinical variables | Logistic regression | 44.7 | 94 | 0.82 |
| Cai et al[27], 2011 | 5 clinical variables | Logistic regression | NA | NA | 0.72 |
| Early diagnosis of PDAC | |||||
| Zhang et al[34], 2020 | Nine-gene signature | Support vector machine | 98.65 | 100 | 93.3 |
| Zhang et al[83], 2020 | CT | DCNN | 83.76 | 91.79 | 0.9455 |
| Si et al[42], 2021 | CT | Fully end-to-end deep learning | 86.8 | 69.5 | 0.871 |
| Liu et al[54], 2020 | CT | CNN | 79 (United States) | 97.6 (United States) | 0.920 (United States) |
| Ma et al[84], 2020 | CT | CNN | 98.2 | 91.6 | 95 |
| Chu et al[85], 2019 | CT | Deep learning (details are NA) | 94.1 | 98.5 | NA |
| Liu et al[53], 2019 | CT | CNN | NA | NA | 0.9632 |
| Tonozuka et al[86], 2021 | EUS | CNN | 90.2 | 74.9 | 0.924 |
| Ozkan et al[87], 2016 | EUS | ANN | 83.3 | 93.3 | 87.5 |
| Săftoiu et al[88], 2015 | EUS | ANN | 94.64 | 94.44 | NA |
| Zhu et al[63], 2013 | EUS | Support vector machine | 92.52 | 93.03 | NA |
| Zhang et al[62], 2010 | EUS | Support vector machine | 94.32 | 99.45 | NA |
| Das et al[61], 2008 | EUS | ANN | 93 | 92 | 0.93 |
| Săftoiu et al[89] 2008 | EUS elastography | NN | 91.4 | 87.9 | 89.7 |
| Norton et al[60], 2001 | EUS | NN | 73 | NA | 83 |
| Alizadeh Savareh et al[40], 2020 | Circulating microRNA signatures | PSO + ANN + NCA | 93 | 92 | 93 |
| Urman et al[90], 2020 | Bile juice | NN | 88 | 100 | 0.98 |
| Pancreatic fistula after pancreaticoduodenectomy | |||||
| Kambakamba et al[71], 2020 | CT | k-NN, random forest classifier, etc | 96 | 98 | 0.95 |
| Mu et al[72], 2020 | CT | CNN | 86.7 | 87.3 | 0.89 |
| Pathological tumor response to neoadjuvant chemotherapy | |||||
| Watson et al[80], 2020 | CT and CA19-9 | CNN | NA | NA | 0.785 |
| Survival model | |||||
| Zhang et al[77], 2020 | CT | CNN | NA | NA | 11.81% in IPA |
| Alizadeh Savareh et al[40], 2020 | Circulating microRNA signatures | PSO + ANN + NCA | NA | NA | NA |
| Kaissis et al[66], 2019 | MRI | Random forest | 87 | 80 | 0.90 |
| Walczak et al[79], 2017 | 14 clinical variables | ANN | 91 | 38 | 0.6576 |
| Molecular subtype | |||||
| Kaissis et al[68], 2020 | CT | Random forest | 84 | 92 | 0.93 |
| Tumor subtype (QM vs non-QM) | |||||
| Kaissis et al[67], 2019 | MRI | Gradient boosting decision tree | 90 | 92 | 0.93 |
| Molecular subtype (KRT81 positive vs negative) | |||||
| Microsatellite instability status | |||||
| Li et al[19], 2020 | PreMSIm (15-gene signature) | k-NN | 85 | 97 | 95 |
The radiographic traits of unoperability and the appearance of symptoms of PDAC occur concurrently[22]. At the time of diagnosis, only a small part of patients (< 15%) have surgically resectable state[22]. In addition, identification of individuals at high risk for PDAC or with early stage is hard due to the absence of trusty screening tools, the lack of clinically relevant biomarkers, and low prevalence[22]. No established screening strategy has been introduced for sporadic PDAC. It is estimated that symptoms manifest about 6 mo after PDAC becomes unresectable[22]. Identifying individuals at high risk but asymptomatic is crucial for finding PDAC while it is still resectable.
Approximately 50% of all patients with PDAC develop diabetes mellitus prior to their diagnosis[23,24]. Screening patients with new-onset diabetes may enable earlier diagnosis of PDAC. In pre-diabetic and new-onset diabetic patients, an AI-based prediction model of PDAC risk has been developed[25,26]. In a pre-diabetic study, 245 of 138232 patients with impaired fasting glucose were thereafter diagnosed as having PDAC within 3 years of impaired fasting glucose detection. The AI (logistic regression model)-based prediction model consisted of age, body mass index, PPIs, total cholesterol, low-density lipoprotein, alanine transaminase, and alkaline phosphatase[25]. This model achieved an area under the curve (AUC) of 0.71. Furthermore, by analyzing 109,385 new-onset diabetic patients including 390 PDAC cases, a multivariable prediction (logistic regression) model that included age, smoking, body mass index, change in body mass index, usage of proton pump inhibitors and anti-diabetic medications (insulin, oral hypoglycemic except metformin, and metformin), as well as levels of hemoglobin, hemoglobin A1C, creatinine, cholesterol, and alkaline phosphatase, was established (AUC, 0.82)[26]. Among these diabetic patients, 390 (0.4%) were diagnosed with PDAC within 3 years. If the predicted risk threshold for definitive PDAC screening was set at 1% over 3 years, only 6.19% of the new-onset diabetes cases would undergo definitive screening, which could identify PDAC cases with 94.0% specificity, 44.7% sensitivity, and a positive predictive value of 2.6%[26].
Cai et al[27] established a PDAC risk prediction model by analyzing 138 chronic pancreatitis patients with focal mass lesions. A scoring method based logistic regression was employed to build the prediction model, which included five variables: sex, mass number, mass location, bilirubin, and carbohydrate antigen 19-9 (CA19-9) (AUC, 0.72). Hsieh et al[28] predicted PDAC in patients with type 2 diabetes using ICD-9 code data by logistic regression and ANN models. The AUCs achieved by these models were 0.72[27] and 0.73[28], respectively.
Appelbaum et al[29] used a logistic regression model and developed a prediction model of PDAC using electronic health record data. A total of 18 risk factors (i.e., age, gender, race, abdominal pain, angina pectoris, asthma, atherosclerotic heart disease, calculus gallbladder, chest pain, chronic pancreatitis, coronary heart disease, diabetes mellitus, emphysema, essential hypertension, family history pancreatic cancer, jaundice, stroke, and ulcer) were used to weigh the risk factors, and their prediction model displayed an AUC of 0.71. Their risk model based on patients’ prior diagnoses derived from electronic health record data would predict PDAC 6-12 mo before an eventual diagnosis date. Such a risk score could be employed as an initial screening prior to additional biomarkers or genetic testing, to pick out individuals from the general population for closer surveillance.
Muhammad et al[30] used the ANN model to focus on the early prediction and stratification of PDAC risk based on personal health data (800114 answers in the National Health Interview Survey and Pancreatic, Lung, Colorectal, and Ovarian cancer datasets, including 898 cases diagnosed with pancreatic cancer) before symptoms appear. The prediction model using 18 personal health features produced a specificity of 80.7%, a sensitivity of 80.7%, and an AUC of 0.85 to predict PDAC[30]. Furthermore, the model based solely on personal health data was able to divide individuals into low, medium, and high cancer risk. Identification of high-risk individuals who would benefit from tailored screening may increase the probability of detecting early PDAC. While logistic regression was used to develop risk prediction models in many previous studies, Muhammad et al[30] employed an ANN model based on personal health big data and produced the highest AUC in the prediction model of PDAC risk.
Such prediction models using AI will be beneficial for clinicians to estimate the PDAC risk of their patients easily after inputting their data. These models can be combined into an electronic medical record system or be available on portable devices such as tablets and mobile phones. They may also be useful for primary care physicians to stratify individuals into various risk categories. By such PDAC risk stratification, higher-risk individuals can be referred to a diagnostic department for more intensive and tailored assessments. More data and testing will be required to refine the performance of the AI-based prediction model of PDAC in order to facilitate its application in the clinical setting. An AI-based prediction model using clinical variables is non-invasive, cost-effective, and easy for early diagnosis of PDAC. Using AI to recognize signs in early PDAC and precancerous lesions is one of the key strategies to improving survival.
It is highly desirable to identify an effective PDAC diagnostic biomarker. Currently, the most widely employed biomarker for early PDAC detection is CA19-9, however it is not an perfect because of its comparatively low level of specificity and sensitivity (70% with a 5% error rate, for diagnosis of PDAC)[31,32]. Several molecular elements such as CA19-9, CEA, DUPAN, and Span-1 have been employed as biomarkers for diagnosis of pancreatic tumors[33], but none of them are sufficiently specific and sensitive to clearly distinguish cancer from healthy or benign diseases. Therefore, a solid tool with sufficient specificity and sensitivity is required to enable early PDAC diagnosis. Zhang et al[34] designed a novel AI (support vector machine) method based on relative gene expression ranking within tissue samples using the microarray gene expression data and RNA-seq data collected from two databases, GEO and TCGA. Zhang et al[34] then identified a qualitative diagnostic signature comprising 9 gene pairs (16 genes), that could distinguish PDAC (using expression profiles from PDAC and adjacent normal tissues) patients from non-PDAC (pancreatitis and normal tissues) and was a useful biomarker for early detection of PDAC. Seven genes in the nine-gene-pair signature, namely CTSE, HOXB7, LAMC2, ONECUT1, RRM2, SERPINB5, and UBE2C, had previously been known to be associated with PDAC. Thus, AI-based tissue biomarker analysis identified a multiple-gene expression signature for detection of early PDAC.
MicroRNAs (miRNAs) have been proposed as promising biomarkers for diagnosis of PDAC[35]. miRNAs are a group of short non-coding RNA molecules with 19-25 nucleotides that have been considered as candidate biomarkers for early cancer diagnosis and precise prognosis[36]. Recently, miRNA-used liquid biopsy has become a promising approach for early detection of cancers. Several miRNAs in plasma of PDAC patients are abundantly expressed, supporting that circulating miRNAs could be helpful for PDAC detection[37]. Ganepola et al[38] employed three circulating miRNAs (miR-22, miR-642b-3p, and miR-885-5) for detection of PDAC, and the AUC value was 0.97 for discrimination of the PDAC cases. Liu et al[39] utilized a serum panel including miR-20a, miR-21, miR-24, miR-25, miR-99a, miR-185, and miR-191 for diagnosis of PDAC at different stages, and the AUC value was 0.99. Alizadeh Savareh et al[40] assessed the value of top miRNAs using a machine learning method (particle swarm optimization + ANN + neighborhood components analysis) to assist early diagnosis of PDAC. They identified a number of serum miRNAs that were significantly differentially expressed in 671 microarray PDAC expression profiles, using bioinformatics techniques Their final model comprised the most promising miRNAs (miR-92a-2-5p, miR-125b-3p, miR-532e5p, miR-663a, and miR-1469) with the high performance (accuracy, 0.93; sensitivity, 0.93; and specificity, 0.92) in differentiation of PDAC from controls.
Early detection of PDAC using tissue and/or blood biomarkers in conjunction with AI is an alternative approach to improving survival.
Nowadays, computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound, endoscopic ultrasound are the most popular imaging modalities for PDAC detection. However, these modalities are often employed in patients with symptoms, which results in delayed detection of PDACs in most cases. A promising application of AI technology is in the earlier detection of PDAC from radiological findings. CT is the most frequently used modality for the initial assessment of suspected PDAC, and its sensitivity of detection ranges from 76%-96%[41]. CT imaging can collect information about tumor location, size, and morphology. The pancreas is considerably different in size, shape, and location among the individuals and possesses only a very small part of the entire CT image, or about 1.3% of each CT image in a CT dataset[42]. Further
The features of early PDAC can be delicate and retrospectively ascertained up to 34 mo before the diagnosis of PDAC[44]. In a tertiary medical center, 7.1% of PDACs were missed even by radiologist assessment. This fact emphasizes the limitations in the conventional CT approach for PDAC. Prognostic outcome in patients with PDACs considerably deteriorates when tumor size exceeds 2 cm[45], however tumors smaller than 2 cm are frequently invisible on CT images and so about 40% of small PDACs are missed[46]. An AI-based diagnostic tool might minimize such oversight. Therefore, there is a growing need to develop AI-based algorithms for accurate pancreatic tumor detection. Although deep learning has been investigated for the diagnosis of pancreatic cystic neoplasms[47], neuroendocrine tumors[48] and segmentation of the pancreas[49-52], the usefulness of AI in the detection of PDAC has not yet been widely explored. AI can analyze thousands of images on a pixel-by-pixel level and is not susceptible to mistakes due to human error. Another strength of AI is automatic diagnosis, which takes no more than approximately 20 s per case from inputting the original CT image to obtaining a diagnosis.
Liu et al[53] reported that the AUC of an AI [convolutional neural network (CNN)] platform for CT-assisted diagnosis of PDAC was 0.963. Furthermore, the time of the CT-assisted diagnosis was 20 s/case, which is remarkably shorter than the duration required for diagnosis by radiologists, indicating AI has good clinical feasibility. In a deep learning (fully end-to-end deep learning) model for diagnosing pancreatic tumors, Si et al[42] reported that the average test time per case was 18.6 s, compared with at least 8 min for manual reviewing. Thus, the AI diagnosis system was more efficient than the conventional diagnostic approach.
Liu et al[54] showed that CNN-based analysis could precisely discriminate cases with and without PDAC in portal venous CT. The CNN-based analysis achieved an accuracy approaching 99% and missed fewer tumors than did radiologists. In this study, CNN-based analysis provided higher sensitivity compared to radiologists (0.983 vs 0.929, respectively)[54]. CNN missed three (1.7%) of 176 PDACs (1.1-1.2 cm). Radiologists missed 12 (7%) of 168 PDACs (1.0-3.3 cm), of which 11 (92%) were correctly classified by CNN. The sensitivity of CNN for tumors smaller than 2 cm was 92.1% in local test sets and 63.1% in an external (US) test set. Although the latter sensitivity for tumors smaller than 2 cm initially seemed unsatisfactory, DeWitt et al[46] reported that the sensitivity of CT by radiologist assessment was 53% for PDACs smaller than 2.5 cm[46]. Consequently, the sensitivity of the CNN-based analysis was equivalent to radiologist assessment. The lower sensitivity of the CNN in the external test set compared with local test sets might be attributed to differences in patients' ethnicity and race, and protocols or scanners, between the training and external test sets, which could present greater challenges for small tumors. An important factor that affects the imaging features of the pancreas is fat content. Higher fat content decreases the density of the pancreas on CT images, and several studies reported marked differences in pancreatic fat content between ethnicities and races[55,56].
Radiologists were given with important clinical information from the clinicians when they assessed the CT images, whereas the CNN was provided with no information except CT images. Therefore, the major utility of the CNN was to support radiologists in judging whether a lesion or suspicious area in the pancreas harbored pancreatic cancer. For example, patients present with obstructive jaundice which is a typical sign of pancreatic cancer in the pancreatic head. Nevertheless, the CT findings are negative or equivocal. In such a situation, occult pancreatic cancer should be highly suspected even if no apparent mass is noted on CT image, given that about 40% of PDACs smaller than 2 cm are missed on CT image due to undefined borders with surrounding tissue[46,57].
With the wide application of endoscopic ultrasonography (EUS) and EUS-fine needle aspiration (FNA) have become the important diagnostic modalities for PDAC; these modalities provide diagnostic accuracies up to 85%, which are remarkably greater than the 50% accuracy in CT-assisted diagnosis. The sensitivity of diagnosis of pancreatic tumors 3 cm in diameter was reported to be 93% for EUS, which was greater than that of CT (53%) and MRI (67%)[58]. A meta-analysis revealed that CT and EUS were comparable in determining the resectability of PDAC, with high sensitivity and specificity[59].
However, based on EUS for early diagnosis of PDAC, the experience and subjective factors affect on the accuracy, especially in the presence of chronic pancreatitis. Additionally, the availability of the EUS-FNA is restricted in community hospitals. Even when the EUS-FNA is utilized, the diagnosis can be also influenced by the operator's experience and the location of the needle insertion. In 2001, Norton et al[60] reported the usefulness of neural network analysis of EUS images to distinguish between PDAC and chronic pancreatitis using 4 different image parameters. Although they provided a high sensitivity, the specificity was only 50%. In 2008, Das et al[61] applied techniques of digital image analysis to EUS images of the pancreas to develop a classification model that could differentiate PDAC from non-neoplastic tissue using ANN. The model accurately classified PDAC, with an AUC of 0.93 and a 93% sensitivity rate[61]. Digital analysis of EUS images is useful in differentiating PDAC from normal tissue and chronic inflammation. Given the possibility of real-time application, digital image analysis may become a helpful diagnostic modality in pancreatic diseases and may sometimes evade EUS-guided FNA. In another study, Zhang et al[62] differentiated between PDAC and normal tissue on EUS images. Regions of interest were selected from 216 images obtained from 153 cancer and 63 non-cancer patients, and a 97.98% sensitivity rate was obtained from the 29 features that were identified[62]. Zhu et al[63] conducted a computer-aided diagnosis utilizing EUS images of 262 PDAC and 126 chronic pancreatitis patients, from which 105 features were extracted. Sixteen of these features were selected for classification by a support vector machine and a 94% sensitivity rate was obtained[63].
EUS imaging is a common imaging method for diagnosing PDAC, and is often applied with FNA in distinguishing benign and malignant tumors. However, FNA is not available in all health centers. AI-assisted diagnosis via EUS images should guide physicians toward more accurate and easier diagnosis.
Collectively, AI can supplement radiologists to reduce miss rates, rather than replace them. The AI stands as a diagnostic tool to assist clinicians and radiologists in diagnosing PDAC. The application of AI in the diagnosis of PDAC has made subs
Recent advances in biotechnology enable us to execute comprehensive genomic, transcriptomic, proteomic, and metabolomic analyses rapidly and cheaply. Such inclusive gene expression studies have uncovered subtypes of PDAC with biological and prognostic relevance. Collisson et al[9] proposed the categorization of PDACs into three subtypes: classical, quasi-mesenchymal (QM), and exocrine-like. The prognostic outcome of PDAC patients following operation and conventional medical treatment was notably better in the classical subtype than in patients with the QM subtype; patientss with the exocrine-like subtype displayed intermediate prognostic outcome between the two other subtypes[9]. Muckenhuber et al[64] subsequently reported that the most of PDAC can be categorized into two distinct subtypes based on transcriptome profiling and on immunohistochemical staining of cytokeratin-81 (KRT81) and hepatocyte nuclear factor-1A (HNF1a). The epithelial KRT81-/HNF1a- (double-negative) subtype (the so-called classical subtype) showed better survival and response to chemotherapy, notably to the FOLFIRINOX regimen, but not to a gemcitabine-based regimen. On the other hand, the epithelial KRT81+/HNF1a- subtype (the so-called QM subtype) has worse OS. But, the QM subtype displays a better response to the gemcitabine-based regimen compared to the non-QM subtype[65]. These features encourage precision medicine based on individual molecular features.
Recent developments in AI using medical image analysis such as radiomics reveal promising models of molecular phenotyping from imaging data. The radiomics approach can perform whole-tumor analytics without invasiveness. Kaissis et al[66,67] have reported on machine learning algorithms to preoperativelly predict molecular subtypes and survival risk in PDAC patients from MRI. However, the restricted availability of MRI data, overall decreased image quality, and the less-quantitative and unstandardized nature of MRI render obstacles to algorithm development and generalization. To reinfoce clinical application, Kaissis et al[68] extended their previous results to CT by training and validating an algorithm (random forest) capable of discriminating between the QM and the non-QM subtypes of PDAC. The advantages of CT comprise broad availability, fewer motion artifacts, and high standardization. Their retrospective study assessed baseline CT from 207 PDAC cases. By immunohistochemical staining for KRT81 and HNF1a, the molecular subtype was determined as QM vs non-QM. The random forest algorithm was used to predict the molecular subtype from the radiomic features. Then, the algorithm was applied to an independent cohort of histopathologically unclassifiable tumors. The classification algorithm achieved sensitivity, specificity, and AUC of 0.84, 0.92, and 0.93, respectively. The median OS for predicted QM and non-QM tumors was 16.1 and 20.9 mo, respectively. The application of the algorithm to histopathologically unclassifiable tumors showed two groups with remarkably different survival (8.9 and 39.8 mo). Thus, the machine learning-based analysis of CT imaging provided the possibility of the prediction of molecular subtypes that is clinically relevant for prognostic outcome, permitting pre-operative stratification for precision medicine. This approach is encouraged by the fact that histopathological approachs are by default a significant underrepresentation of the tumor, since they are derived from a small sub-section of the tissue, and regions of differing molecular subtype are likely to coexist within the same tumor[69]. On the other hand, the radiomic approach enables whole-tumor assessment, providing better information required for precision therapy.
Microsatellite instability (MSI) is a genomic property of cancers with defective DNA mismatch repair. Notably, MSI has been recognized as a biomarker for the favorable immune checkpoint blockade therapy response in cancer[70]. Most standard methods for examing MSI are based on DNA sequencing data and a few are based on mRNA expression data. Using RNA-Seq pan-cancer datasets for three cancer cohorts (colon, endometrial, and gastric cancers) from TCGA program, Li et al[19] established an algorithm called PreMSIm (Predicting MSI from mRNA) to predict MSI in cancer from the expression profiling of a 15-gene panel. A benefit of mRNA-based over DNA-based MSI prediction algorithms is that mRNA data are closer to protein and phenotype than DNA data. Pathway analysis revealed that these genes were mainly involved in DNA damage repair (MLH1 and MSH4), gene expression (MLH1, HENMT1, and RPL22L1), cell cycle regulation (MLH1, MSH4, and HENMT1), and metabolism (NHLRC1 and RPL22L1). Gene ontology analysis showed that these genes were involved in the biological processes of DNA repair (MLH1, MSH4, and RTF2), gene expression regulation (NHLRC1 and HENMT1), cell cycle (MLH1, MSH4, RPL22L1, and RTF2), biogenesis (DDX27, EPM2AIP1, NHLRC1, and RNLS), metabolic process (HENMT1, LYG1, NHLRC1, and SMAP1), and cell and organism development (SMAP1, SHROOM4, and TTC30A). The PreMSIm algorithm provided high performance in predicting MSI using both RNA-Seq and microarray gene expression datasets[19]. Furthermore, PreMSIm showed superior or comparable performance vs other DNA- or mRNA-based methods. Li et al[19] comment that PreMSIm can be an alternative approach for identifying MSI. The introduction of machine learning algorithms such as this as a clinical decision support tool should be beneficial to predict molecular/genetic signatures that may help to stratify patients in clinical routines.
In the pancreatic fields, the availability of AI in surgery is still very limited. An AI-based risk prediction model of postoperative complication has been reported[71,72]. Postoperative pancreatic fistula (POPF) is a serious complication after pancreatoduodenectomy (PD). The fistula risk score (FRS), which consists of four variables — soft pancreas, small main pancreatic duct, high-risk pathology (PDAC or chronic pancreatitis), and massive intraoperative blood loss — is useful to predict clinically relevant POPF development after PD[73,74]. However, the score contains subjective factors related to surgeons. Therefore, an accurate and easy-to-use preoperative index is desired. Kambakamba et al[71] examined whether quantitative analysis of plain CT with five types of machine learning algorithms (k-nearest neighbors, sequential minimum optimization, multilayer perceptron, random forest, and C5.0) could predict clinically relevant POPF in 110 patients from a single institution, and found that machine learning-based CT analysis provided an magnificent AUC of 0.95 in predicting clinically relevant POPF[71]. Mu et al[72] tried to predict clinically relevant POPF after PD using a deep learning (CNN model) score derived from preoperative CT. The deep learning score offered significantly greater predictability compared to FRS in training (0.85 vs 0.78 in AUC, respectively), validation (0.81 vs 0.76 in AUC, respectively) and test (0.89 vs 0.73 in AUC, respectively) cohorts. In particular, in patients of intermediate risk (FRS 3-6), the deep learning score achieved remarkably higher accuracy compared to FRS (test: 92.1% vs 65.8%, respectively). Interestingly, the deep learning score was independently associated with pancreatic fibrosis, diameter of main pancreatic duct and remnant volume in multivariate linear regression analysis. The automated scores reflected histomorphological features related to pancreatic duct, parenchymal fibrosis, and remnant pancreatic tissue volume. Thus, an AI model using preoperative CT represents a novel tool to predict clinically relevant POPF after PD, especially at intermediate risk levels. Such an AI system helps surgeons to optimize preoperative strategy.
The potential of radiomics in prediction of clinically relevant conditions, such as expected OS or response to a specific therapy, has been reported in recent studies[75,76]: For instance, CT-derived radiomic features were useful to predict local disease control and OS in PDAC[75,76]. Entropy-related and cluster tendency features were described as predictive of OS in PDAC[76]. Zhang et al[77] proposed that a CNN-based survival model outperforms a Cox proportional hazard model-based radiomics pipeline in PDAC prognosis. This model provides a better fit for survival patterns based on CT images and overcomes the limitations of conventional survival models. Kaissis et al[66] reported that a machine learning algorithm (random forest) using MRI achieved 87% sensitivity, 80% specificity, and AUC 0.9 for the prediction of above- vs below-median OS in the independent validation cohort. Alizadeh Savareh et al[40] have identified several circulating miRNAs as a diagnosis model in PDAC patients by analyzing microarray miRNA expression profiles from the Gene Expression Omnibus database. Three (hsa-mir-1469, hsa-mir-663a and hsa-mir-532) of five miRNAs with a high rank in the final model were comprehensively associated with the OS of patients with PDAC based on their up- or down-regulated expression patterns[40].
Late diagnosis of PDAC can cause to lose the chance of surgical treatment and lead to a high mortality rate[78]. On the other hand, surgical treatments for PDAC can have a high morbidity and mortality rate. Therefore, the clinicians must weigh the potential survival advantage of the invasive treatment, the complications due to invasive treatment, and the impacts on the patient’s quality of life with and without treatment.
Walczak and Velanovich[79] established ANN models that could accurately predict the 7-mo survival of PDAC patients using 14 clinical variables including eight SF-36 domain values, both with and without surgical resection, at 91% sensitivity and 38% specificity. The ANN model to predict 7-mo survival consisted of age, sex, the eight domains of quality of life measurements from the SF-36, the stage of the cancer, whether or not a resection had taken place, if any adjuvant therapy had been given, and time in months since diagnosis. The quality of life domains from the SF-36 are bodily pain, vitality, physical functioning, social functioning, role-physical, role-emotional, general health, and mental health. Such an ANN model for predicting the survival of PDAC patients helps physicians and patients to reduce anticipated regret from treatment decisions including observation. This information may be useful for patients and surgeons in determining invasive treatment plans to minimize regret and improve the patients' quality of life.
Neoadjuvant therapy may provide improved survival of PDAC patients; but, determining the efficacy is difficult. Watson et al[80] hypothesized that a deep learning (CNN) model could predict the tumor response to neoadjuvant therapy using CT and CA19-9. A total of 81 cases were divided between partial responder (333 images) and non-responder (443 images). The model using only the deep learning model had an AUC of 0.738, whereas a hybrid model incorporating a decrease in CA19-9 of 10% in addition to the deep learning model had an AUC of 0.785. CA19-9 reduction alone was not an effective predictor of the response to neoadjuvant therapy, with an AUC of 0.564. A deep learning model can predict the pathological response to neoadjuvant therapy for PDAC patients, and the model is amended with the incorporation of decreases in serum CA19-9. Abraham et al[81] investigated the clinical relevance of a machine learning-derived signature in predicting the responses from first-line oxaliplatin-based chemotherapy in PDAC and advanced colorectal cancer. The machine learning-derived signature was effective for metastatic colorectal cancer, but not for PDAC. AI has already been applied to match biological information with chemical properties of specific drugs to predict the response to these specific agents in cancers[82].
In the near future, the combined analysis of clinical variables, less-invasive biological samples, and radiological features through machine learning should be able to simulate responses to chemotherapy and patient survival. On the other hand, radiological features and biological tissue can be variable in response to the treatment including chemotherapy and radiotherapy. In particular, PDAC has a high potential for acquired drug resistance. During sequential treatment, good communication and the accumulation of knowledge from various fields such as gastroenterology, radiology, oncology, computer science, and pathology must will be required to fight this deadly disease.
A major limitation is the lack of adequate standardization. Universal and uniform protocols for data collection, data quality, storage, processing, reproduction, and analysis must be established. For instance, ANNs can be trained to appropriately categorize histologic slides of pancreatic biopsies. However, the trained ANNs may underperform, or not perform at all, when the prepared slides are fixed and stained in a different manner. Development of universal and uniform protocols during data and sample processing will be required for medical AI to be feasible. Further improvement of the technology is also essential for medical AI in clinical practice.
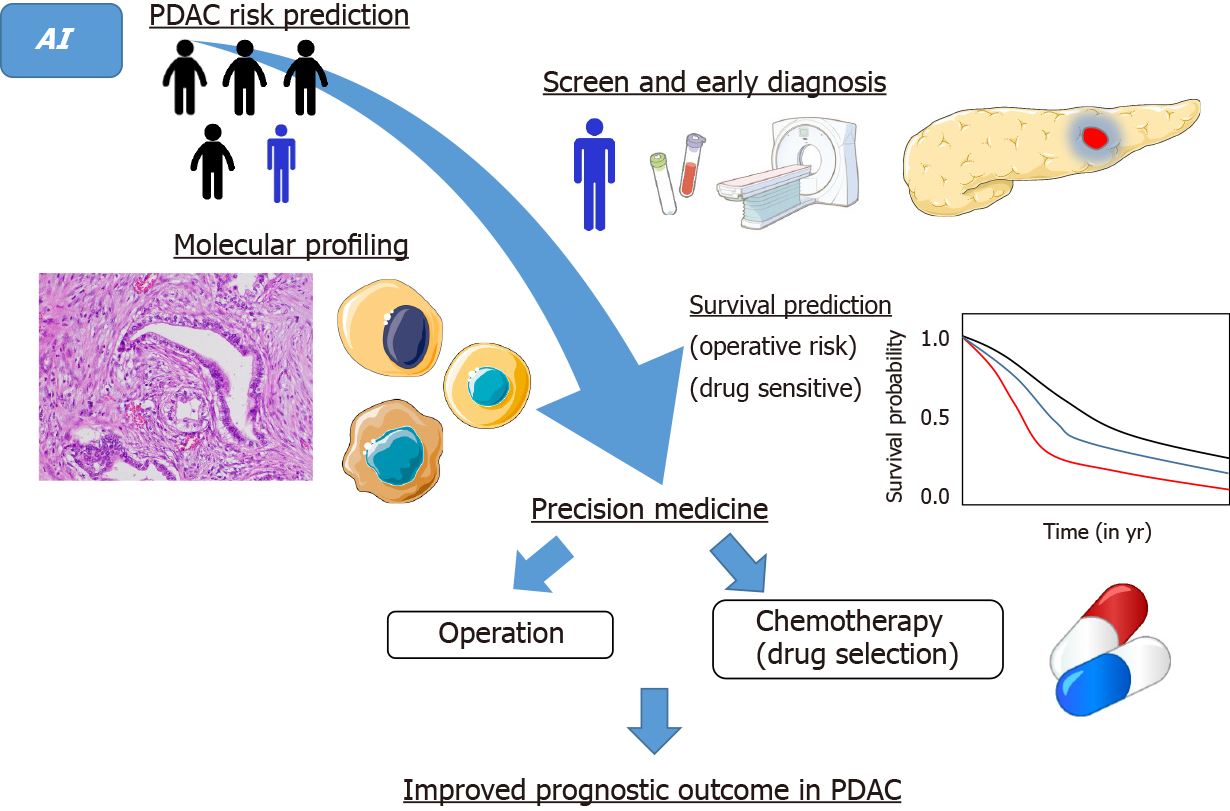
AI in the medical field should become an indispensable tool to reduce human error. Because of human limitations, we cannot achieve zero errors. Furthermore, it is time-consuming to train professional radiologists, gastroenterologists, oncologists, and pathologists. Combined work by experts from multiple fields will be needed to establish feasible medical AI systems in clinical practice. With further research, AI must have a great impact on the diagnosis and treatment of PDAC in near future. Ultimately, a sequential approach involving risk prediction, diagnosis, treatment, and survival prediction using IA will realize timely and consecutive precision medicine and lead to improved prognosis in PDAC (Figure 2). Dr. William Osler stated: “Medicine is a science of uncertainty and an art of probability.” This is still true in medical AI, which is also “a science of uncertainty and an art of probability.” However, the degree of uncertainty and probability will consistently shrink with the advance of AI technology and cooperation among various experts. While AI applications in PDAC are still in the early stage of development, further research must lead to great advances in screening, early diagnosis, and treatment.
Here we summarize the current advances of AI in PDAC. AI-based omics analyses are likely to be the next alternative approach to overcome this poor-prognostic disease by the discovery of biomarkers for early detection, molecular/genomic subtyping, and treatment guidance, and by the improved prediction of recurrence and survival. How to entirely utilize “big data” is a new challenge for physicians and researchers in the era of precision medicine. On the other hand, AI will not entirely act for doctors — human beings and machines working harmoniously together is the ideal state that results in excellent performance. Although AI data reveal that the diagnostic accuracy of deep learning models is better than that of radiologists, the aim in this field is to develop a helpful tool to aid radiologists in making effective and accurate diagnoses, not to be a replacement for doctors. To facilitate AI-based omics analyses, multidisciplinary collaboration between physicians, basic scientists, radiologists, statisticians, and engineers is mandatory. To further validate the clinical relevance of AI systems, next step is to conduct a prospective study based on multicenter clinical data. We believe that breakthroughs will soon emerge to fight this deadly disease using AI-navigated precision medicine.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hameed MM, Moldogazieva NT, Santos-García G, Surlin VM S-Editor: Gao CC L-Editor: A P-Editor: Gao CC
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15468] [Article Influence: 2578.0] [Reference Citation Analysis (2)] |
| 2. | Oettle H, Post S, Neuhaus P, Gellert K, Langrehr J, Ridwelski K, Schramm H, Fahlke J, Zuelke C, Burkart C, Gutberlet K, Kettner E, Schmalenberg H, Weigang-Koehler K, Bechstein WO, Niedergethmann M, Schmidt-Wolf I, Roll L, Doerken B, Riess H. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1779] [Cited by in RCA: 1763] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 3. | Neoptolemos JP, Stocken DD, Bassi C, Ghaneh P, Cunningham D, Goldstein D, Padbury R, Moore MJ, Gallinger S, Mariette C, Wente MN, Izbicki JR, Friess H, Lerch MM, Dervenis C, Oláh A, Butturini G, Doi R, Lind PA, Smith D, Valle JW, Palmer DH, Buckels JA, Thompson J, McKay CJ, Rawcliffe CL, Büchler MW; European Study Group for Pancreatic Cancer. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010;304:1073-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1144] [Cited by in RCA: 997] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 4. | Uesaka K, Boku N, Fukutomi A, Okamura Y, Konishi M, Matsumoto I, Kaneoka Y, Shimizu Y, Nakamori S, Sakamoto H, Morinaga S, Kainuma O, Imai K, Sata N, Hishinuma S, Ojima H, Yamaguchi R, Hirano S, Sudo T, Ohashi Y; JASPAC 01 Study Group. Adjuvant chemotherapy of S-1 vs gemcitabine for resected pancreatic cancer: a phase 3, open-label, randomised, non-inferiority trial (JASPAC 01). Lancet. 2016;388:248-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 780] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 5. | Neoptolemos JP, Palmer DH, Ghaneh P, Psarelli EE, Valle JW, Halloran CM, Faluyi O, O'Reilly DA, Cunningham D, Wadsley J, Darby S, Meyer T, Gillmore R, Anthoney A, Lind P, Glimelius B, Falk S, Izbicki JR, Middleton GW, Cummins S, Ross PJ, Wasan H, McDonald A, Crosby T, Ma YT, Patel K, Sherriff D, Soomal R, Borg D, Sothi S, Hammel P, Hackert T, Jackson R, Büchler MW; European Study Group for Pancreatic Cancer. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet. 2017;389:1011-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1327] [Cited by in RCA: 1392] [Article Influence: 174.0] [Reference Citation Analysis (0)] |
| 6. | Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardière C, Bennouna J, Bachet JB, Khemissa-Akouz F, Péré-Vergé D, Delbaldo C, Assenat E, Chauffert B, Michel P, Montoto-Grillot C, Ducreux M; Groupe Tumeurs Digestives of Unicancer; PRODIGE Intergroup. FOLFIRINOX vs gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817-1825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4838] [Cited by in RCA: 5628] [Article Influence: 402.0] [Reference Citation Analysis (1)] |
| 7. | Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, Harris M, Reni M, Dowden S, Laheru D, Bahary N, Ramanathan RK, Tabernero J, Hidalgo M, Goldstein D, Van Cutsem E, Wei X, Iglesias J, Renschler MF. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691-1703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4035] [Cited by in RCA: 4877] [Article Influence: 406.4] [Reference Citation Analysis (0)] |
| 8. | Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM; Australian Pancreatic Cancer Genome Initiative, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2848] [Cited by in RCA: 2548] [Article Influence: 283.1] [Reference Citation Analysis (0)] |
| 9. | Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, Feiler HS, Ko AH, Olshen AB, Danenberg KL, Tempero MA, Spellman PT, Hanahan D, Gray JW. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500-503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1536] [Cited by in RCA: 1358] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 10. | Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK, Anderson JM, Kim HJ, Bentrem DJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Yeh JJ. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168-1178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1041] [Cited by in RCA: 1474] [Article Influence: 147.4] [Reference Citation Analysis (0)] |
| 11. | Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grützmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA; Australian Pancreatic Cancer Genome Initiative, Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2130] [Cited by in RCA: 1987] [Article Influence: 198.7] [Reference Citation Analysis (1)] |
| 12. | Pishvaian MJ, Blais EM, Brody JR, Lyons E, DeArbeloa P, Hendifar A, Mikhail S, Chung V, Sahai V, Sohal DPS, Bellakbira S, Thach D, Rahib L, Madhavan S, Matrisian LM, Petricoin EF 3rd. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 2020;21:508-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 365] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 13. | Andreu-Perez J, Poon CC, Merrifield RD, Wong ST, Yang GZ. Big data for health. IEEE J Biomed Health Inform. 2015;19:1193-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 261] [Article Influence: 26.1] [Reference Citation Analysis (1)] |
| 14. | Colom R, Karama S, Jung RE, Haier RJ. Human intelligence and brain networks. Dialogues Clin Neurosci. 2010;12:489-501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 156] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 15. | Chen YC, Ke WC, Chiu HW. Risk classification of cancer survival using ANN with gene expression data from multiple laboratories. Comput Biol Med. 2014;48:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 16. | Islam MM, Poly TN, Alsinglawi B, Lin MC, Hsu MH, Li YJ. A State-of-the-Art Survey on Artificial Intelligence to Fight COVID-19. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Koumakis L, Kanterakis A, Kartsaki E, Chatzimina M, Zervakis M, Tsiknakis M, Vassou D, Kafetzopoulos D, Marias K, Moustakis V, Potamias G. MinePath: Mining for Phenotype Differential Sub-paths in Molecular Pathways. PLoS Comput Biol. 2016;12:e1005187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Zhu W, Xie L, Han J, Guo X. The Application of Deep Learning in Cancer Prognosis Prediction. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 19. | Li L, Feng Q, Wang X. PreMSIm: An R package for predicting microsatellite instability from the expression profiling of a gene panel in cancer. Comput Struct Biotechnol J. 2020;18:668-675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 20. | Hashimoto DA, Rosman G, Rus D, Meireles OR. Artificial Intelligence in Surgery: Promises and Perils. Ann Surg. 2018;268:70-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 594] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 21. | Fard MJ, Ameri S, Darin Ellis R, Chinnam RB, Pandya AK, Klein MD. Automated robot-assisted surgical skill evaluation: Predictive analytics approach. Int J Med Robot. 2018;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 22. | Pannala R, Basu A, Petersen GM, Chari ST. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 2009;10:88-95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 428] [Cited by in RCA: 403] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 23. | Pannala R, Leirness JB, Bamlet WR, Basu A, Petersen GM, Chari ST. Prevalence and clinical profile of pancreatic cancer-associated diabetes mellitus. Gastroenterology. 2008;134:981-987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 469] [Cited by in RCA: 422] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 24. | Aggarwal G, Rabe KG, Petersen GM, Chari ST. New-onset diabetes in pancreatic cancer: a study in the primary care setting. Pancreatology. 2012;12:156-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 25. | Boursi B, Finkelman B, Giantonio BJ, Haynes K, Rustgi AK, Rhim AD, Mamtani R, Yang YX. A clinical prediction model to assess risk for pancreatic cancer among patients with prediabetes. Eur J Gastroenterol Hepatol. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 26. | Boursi B, Finkelman B, Giantonio BJ, Haynes K, Rustgi AK, Rhim AD, Mamtani R, Yang YX. A Clinical Prediction Model to Assess Risk for Pancreatic Cancer Among Patients With New-Onset Diabetes. Gastroenterology. 2017;152:840-850.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 138] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 27. | Cai QC, Chen Y, Xiao Y, Zhu W, Xu QF, Zhong L, Chen SY, Zhang MM, Wang LW, Li ZS. A prediction rule for estimating pancreatic cancer risk in chronic pancreatitis patients with focal pancreatic mass lesions with prior negative EUS-FNA cytology. Scand J Gastroenterol. 2011;46:464-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Hsieh MH, Sun LM, Lin CL, Hsieh MJ, Hsu CY, Kao CH. Development of a prediction model for pancreatic cancer in patients with type 2 diabetes using logistic regression and artificial neural network models. Cancer Manag Res. 2018;10:6317-6324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 29. | Appelbaum L, Cambronero JP, Stevens JP, Horng S, Pollick K, Silva G, Haneuse S, Piatkowski G, Benhaga N, Duey S, Stevenson MA, Mamon H, Kaplan ID, Rinard MC. Development and validation of a pancreatic cancer risk model for the general population using electronic health records: An observational study. Eur J Cancer. 2021;143:19-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 30. | Muhammad W, Hart G, Johung K, Liang Y, Nartowt B, Ali I, Deng J. Pancreatic Cancer Risk Prediction Through an Artificial Neural Network. Med Phys. 2018;45:E146. |
| 31. | Datta J, Vollmer CM Jr. Investigational biomarkers for pancreatic adenocarcinoma: where do we stand? South Med J. 2014;107:256-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33:266-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 611] [Article Influence: 32.2] [Reference Citation Analysis (1)] |
| 33. | Miyata T, Hayashi H, Yamashita YI, Matsumura K, Nakao Y, Itoyama R, Yamao T, Tsukamoto M, Okabe H, Imai K, Chikamoto A, Ishiko T, Baba H. Prognostic Value of the Preoperative Tumor Marker Index in Resected Pancreatic Ductal Adenocarcinoma: A Retrospective Single-Institution Study. Ann Surg Oncol. 2021;28:1572-1580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 34. | Zhang ZM, Wang JS, Zulfiqar H, Lv H, Dao FY, Lin H. Early Diagnosis of Pancreatic Ductal Adenocarcinoma by Combining Relative Expression Orderings With Machine-Learning Method. Front Cell Dev Biol. 2020;8:582864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 35. | Du Y, Liu M, Gao J, Li Z. Aberrant microRNAs expression patterns in pancreatic cancer and their clinical translation. Cancer Biother Radiopharm. 2013;28:361-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 36. | Costello E, Greenhalf W, Neoptolemos JP. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat Rev Gastroenterol Hepatol. 2012;9:435-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 37. | Duell EJ, Lujan-Barroso L, Sala N, Deitz McElyea S, Overvad K, Tjonneland A, Olsen A, Weiderpass E, Busund LT, Moi L, Muller D, Vineis P, Aune D, Matullo G, Naccarati A, Panico S, Tagliabue G, Tumino R, Palli D, Kaaks R, Katzke VA, Boeing H, Bueno-de-Mesquita HBA, Peeters PH, Trichopoulou A, Lagiou P, Kotanidou A, Travis RC, Wareham N, Khaw KT, Ramon Quiros J, Rodríguez-Barranco M, Dorronsoro M, Chirlaque MD, Ardanaz E, Severi G, Boutron-Ruault MC, Rebours V, Brennan P, Gunter M, Scelo G, Cote G, Sherman S, Korc M. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int J Cancer. 2017;141:905-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 38. | Ganepola GA, Rutledge JR, Suman P, Yiengpruksawan A, Chang DH. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J Gastrointest Oncol. 2014;6:22-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 88] [Cited by in RCA: 99] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 39. | Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, Yuan Y, Li Z, Zen K, Ba Y, Zhang CY. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 313] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 40. | Alizadeh Savareh B, Asadzadeh Aghdaie H, Behmanesh A, Bashiri A, Sadeghi A, Zali M, Shams R. A machine learning approach identified a diagnostic model for pancreatic cancer through using circulating microRNA signatures. Pancreatology. 2020;20:1195-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 41. | Chu LC, Goggins MG, Fishman EK. Diagnosis and Detection of Pancreatic Cancer. Cancer J. 2017;23:333-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 209] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 42. | Si K, Xue Y, Yu X, Zhu X, Li Q, Gong W, Liang T, Duan S. Fully end-to-end deep-learning-based diagnosis of pancreatic tumors. Theranostics. 2021;11:1982-1990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 43. | Pawlik TM, Laheru D, Hruban RH, Coleman J, Wolfgang CL, Campbell K, Ali S, Fishman EK, Schulick RD, Herman JM; Johns Hopkins Multidisciplinary Pancreas Clinic Team. Evaluating the impact of a single-day multidisciplinary clinic on the management of pancreatic cancer. Ann Surg Oncol. 2008;15:2081-2088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 205] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 44. | Gonoi W, Hayashi TY, Okuma H, Akahane M, Nakai Y, Mizuno S, Tateishi R, Isayama H, Koike K, Ohtomo K. Development of pancreatic cancer is predictable well in advance using contrast-enhanced CT: a case-cohort study. Eur Radiol. 2017;27:4941-4950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 45. | Agarwal B, Correa AM, Ho L. Survival in pancreatic carcinoma based on tumor size. Pancreas. 2008;36:e15-e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 46. | Dewitt J, Devereaux BM, Lehman GA, Sherman S, Imperiale TF. Comparison of endoscopic ultrasound and computed tomography for the preoperative evaluation of pancreatic cancer: a systematic review. Clin Gastroenterol Hepatol. 2006;4:717-25; quiz 664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 168] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 47. | Chen WX, Ji HC, Feng JJ, Liu R, Yu Y, Zhou RQ, Zhou J. Classification of Pancreatic Cystic Neoplasms Based on Multimodality Images. Lect Notes Comput Sc. 2018;11046:161-169. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 48. | Li J, Lu J, Liang P, Li A, Hu Y, Shen Y, Hu D, Li Z. Differentiation of atypical pancreatic neuroendocrine tumors from pancreatic ductal adenocarcinomas: Using whole-tumor CT texture analysis as quantitative biomarkers. Cancer Med. 2018;7:4924-4931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 49. | Fu M, Wu W, Hong X, Liu Q, Jiang J, Ou Y, Zhao Y, Gong X. Hierarchical combinatorial deep learning architecture for pancreas segmentation of medical computed tomography cancer images. BMC Syst Biol. 2018;12:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 50. | Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, Davidson B, Pereira SP, Clarkson MJ, Barratt DC. Automatic Multi-Organ Segmentation on Abdominal CT With Dense V-Networks. IEEE Trans Med Imaging. 2018;37:1822-1834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 325] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 51. | Roth HR, Lu L, Lay N, Harrison AP, Farag A, Sohn A, Summers RM. Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med Image Anal. 2018;45:94-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 144] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 52. | Xie L, Yu Q, Zhou Y, Wang Y, Fishman EK, Yuille AL. Recurrent Saliency Transformation Network for Tiny Target Segmentation in Abdominal CT Scans. IEEE Trans Med Imaging. 2020;39:514-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 53. | Liu SL, Li S, Guo YT, Zhou YP, Zhang ZD, Lu Y. Establishment and application of an artificial intelligence diagnosis system for pancreatic cancer with a faster region-based convolutional neural network. Chin Med J (Engl). 2019;132:2795-2803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 54. | Liu KL, Wu T, Chen PT, Tsai YM, Roth H, Wu MS, Liao WC, Wang W. Deep learning to distinguish pancreatic cancer tissue from non-cancerous pancreatic tissue: a retrospective study with cross-racial external validation. Lancet Digit Health. 2020;2:e303-e313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 55. | Szczepaniak LS, Victor RG, Mathur R, Nelson MD, Szczepaniak EW, Tyer N, Chen I, Unger RH, Bergman RN, Lingvay I. Pancreatic steatosis and its relationship to β-cell dysfunction in humans: racial and ethnic variations. Diabetes Care. 2012;35:2377-2383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 56. | Lê KA, Ventura EE, Fisher JQ, Davis JN, Weigensberg MJ, Punyanitya M, Hu HH, Nayak KS, Goran MI. Ethnic differences in pancreatic fat accumulation and its relationship with other fat depots and inflammatory markers. Diabetes Care. 2011;34:485-490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 57. | Frampas E, David A, Regenet N, Touchefeu Y, Meyer J, Morla O. Pancreatic carcinoma: Key-points from diagnosis to treatment. Diagn Interv Imaging. 2016;97:1207-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 58. | Müller MF, Meyenberger C, Bertschinger P, Schaer R, Marincek B. Pancreatic tumors: evaluation with endoscopic US, CT, and MR imaging. Radiology. 1994;190:745-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 304] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 59. | Rahman MIO, Chan BPH, Far PM, Mbuagbaw L, Thabane L, Yaghoobi M. Endoscopic ultrasound vs computed tomography in determining the resectability of pancreatic cancer: A diagnostic test accuracy meta-analysis. Saudi J Gastroenterol. 2020;26:113-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 60. | Norton ID, Zheng Y, Wiersema MS, Greenleaf J, Clain JE, Dimagno EP. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest Endosc. 2001;54:625-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 61. | Das A, Nguyen CC, Li F, Li B. Digital image analysis of EUS images accurately differentiates pancreatic cancer from chronic pancreatitis and normal tissue. Gastrointest Endosc. 2008;67:861-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 62. | Zhang MM, Yang H, Jin ZD, Yu JG, Cai ZY, Li ZS. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointest Endosc. 2010;72:978-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 63. | Zhu M, Xu C, Yu J, Wu Y, Li C, Zhang M, Jin Z, Li Z. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: a diagnostic test. PLoS One. 2013;8:e63820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (2)] |
| 64. | Muckenhuber A, Berger AK, Schlitter AM, Steiger K, Konukiewitz B, Trumpp A, Eils R, Werner J, Friess H, Esposito I, Klöppel G, Ceyhan GO, Jesinghaus M, Denkert C, Bahra M, Stenzinger A, Sprick MR, Jäger D, Springfeld C, Weichert W. Pancreatic Ductal Adenocarcinoma Subtyping Using the Biomarkers Hepatocyte Nuclear Factor-1A and Cytokeratin-81 Correlates with Outcome and Treatment Response. Clin Cancer Res. 2018;24:351-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 65. | Aung KL, Fischer SE, Denroche RE, Jang GH, Dodd A, Creighton S, Southwood B, Liang SB, Chadwick D, Zhang A, O'Kane GM, Albaba H, Moura S, Grant RC, Miller JK, Mbabaali F, Pasternack D, Lungu IM, Bartlett JMS, Ghai S, Lemire M, Holter S, Connor AA, Moffitt RA, Yeh JJ, Timms L, Krzyzanowski PM, Dhani N, Hedley D, Notta F, Wilson JM, Moore MJ, Gallinger S, Knox JJ. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin Cancer Res. 2018;24:1344-1354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 430] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 66. | Kaissis G, Ziegelmayer S, Lohöfer F, Algül H, Eiber M, Weichert W, Schmid R, Friess H, Rummeny E, Ankerst D, Siveke J, Braren R. A machine learning model for the prediction of survival and tumor subtype in pancreatic ductal adenocarcinoma from preoperative diffusion-weighted imaging. Eur Radiol Exp. 2019;3:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 67. | Kaissis G, Ziegelmayer S, Lohöfer F, Steiger K, Algül H, Muckenhuber A, Yen HY, Rummeny E, Friess H, Schmid R, Weichert W, Siveke JT, Braren R. A machine learning algorithm predicts molecular subtypes in pancreatic ductal adenocarcinoma with differential response to gemcitabine-based vs FOLFIRINOX chemotherapy. PLoS One. 2019;14:e0218642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 68. | Kaissis GA, Ziegelmayer S, Lohöfer FK, Harder FN, Jungmann F, Sasse D, Muckenhuber A, Yen HY, Steiger K, Siveke J, Friess H, Schmid R, Weichert W, Makowski MR, Braren RF. Image-Based Molecular Phenotyping of Pancreatic Ductal Adenocarcinoma. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 69. | Chan-Seng-Yue M, Kim JC, Wilson GW, Ng K, Figueroa EF, O'Kane GM, Connor AA, Denroche RE, Grant RC, McLeod J, Wilson JM, Jang GH, Zhang A, Dodd A, Liang SB, Borgida A, Chadwick D, Kalimuthu S, Lungu I, Bartlett JMS, Krzyzanowski PM, Sandhu V, Tiriac H, Froeling FEM, Karasinska JM, Topham JT, Renouf DJ, Schaeffer DF, Jones SJM, Marra MA, Laskin J, Chetty R, Stein LD, Zogopoulos G, Haibe-Kains B, Campbell PJ, Tuveson DA, Knox JJ, Fischer SE, Gallinger S, Notta F. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52:231-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 414] [Article Influence: 82.8] [Reference Citation Analysis (0)] |
| 70. | Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-2520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6096] [Cited by in RCA: 7227] [Article Influence: 722.7] [Reference Citation Analysis (0)] |
| 71. | Kambakamba P, Mannil M, Herrera PE, Müller PC, Kuemmerli C, Linecker M, von Spiczak J, Hüllner MW, Raptis DA, Petrowsky H, Clavien PA, Alkadhi H. The potential of machine learning to predict postoperative pancreatic fistula based on preoperative, non-contrast-enhanced CT: A proof-of-principle study. Surgery. 2020;167:448-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 72. | Mu W, Liu C, Gao F, Qi Y, Lu H, Liu Z, Zhang X, Cai X, Ji RY, Hou Y, Tian J, Shi Y. Prediction of clinically relevant Pancreatico-enteric Anastomotic Fistulas after Pancreatoduodenectomy using deep learning of Preoperative Computed Tomography. Theranostics. 2020;10:9779-9788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 73. | Callery MP, Pratt WB, Kent TS, Chaikof EL, Vollmer CM Jr. A prospectively validated clinical risk score accurately predicts pancreatic fistula after pancreatoduodenectomy. J Am Coll Surg. 2013;216:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 918] [Article Influence: 70.6] [Reference Citation Analysis (2)] |
| 74. | McMillan MT, Soi S, Asbun HJ, Ball CG, Bassi C, Beane JD, Behrman SW, Berger AC, Bloomston M, Callery MP, Christein JD, Dixon E, Drebin JA, Castillo CF, Fisher WE, Fong ZV, House MG, Hughes SJ, Kent TS, Kunstman JW, Malleo G, Miller BC, Salem RR, Soares K, Valero V, Wolfgang CL, Vollmer CM Jr. Risk-adjusted Outcomes of Clinically Relevant Pancreatic Fistula Following Pancreatoduodenectomy: A Model for Performance Evaluation. Ann Surg. 2016;264:344-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 75. | Cozzi L, Comito T, Fogliata A, Franzese C, Franceschini D, Bonifacio C, Tozzi A, Di Brina L, Clerici E, Tomatis S, Reggiori G, Lobefalo F, Stravato A, Mancosu P, Zerbi A, Sollini M, Kirienko M, Chiti A, Scorsetti M. Computed tomography based radiomic signature as predictive of survival and local control after stereotactic body radiation therapy in pancreatic carcinoma. PLoS One. 2019;14:e0210758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 76. | Eilaghi A, Baig S, Zhang Y, Zhang J, Karanicolas P, Gallinger S, Khalvati F, Haider MA. CT texture features are associated with overall survival in pancreatic ductal adenocarcinoma - a quantitative analysis. BMC Med Imaging. 2017;17:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 77. | Zhang Y, Lobo-Mueller EM, Karanicolas P, Gallinger S, Haider MA, Khalvati F. CNN-based survival model for pancreatic ductal adenocarcinoma in medical imaging. BMC Med Imaging. 2020;20:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 78. | Hayashi H, Baba H. Current statement and safe implementation of minimally invasive surgery in the pancreas. Ann Gastroenterol Surg. 2020;4:505-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 79. | Walczak S, Velanovich V. An Evaluation of Artificial Neural Networks in Predicting Pancreatic Cancer Survival. J Gastrointest Surg. 2017;21:1606-1612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 80. | Watson MD, Baimas-George MR, Murphy KJ, Pickens RC, Iannitti DA, Martinie JB, Baker EH, Vrochides D, Ocuin LM. Pure and Hybrid Deep Learning Models can Predict Pathologic Tumor Response to Neoadjuvant Therapy in Pancreatic Adenocarcinoma: A Pilot Study. Am Surg. 2020;3134820982557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 81. | Abraham JP, Magee D, Cremolini C, Antoniotti C, Halbert DD, Xiu J, Stafford P, Berry DA, Oberley MJ, Shields AF, Marshall JL, Salem ME, Falcone A, Grothey A, Hall MJ, Venook AP, Lenz HJ, Helmstetter A, Korn WM, Spetzler DB. Clinical Validation of a Machine-learning-derived Signature Predictive of Outcomes from First-line Oxaliplatin-based Chemotherapy in Advanced Colorectal Cancer. Clin Cancer Res. 2021;27:1174-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 82. | Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603-607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5947] [Cited by in RCA: 5805] [Article Influence: 446.5] [Reference Citation Analysis (0)] |
| 83. | Zhang Z, Li S, Wang Z, Lu Y. A Novel and Efficient Tumor Detection Framework for Pancreatic Cancer via CT Images. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:1160-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 84. | Ma H, Liu ZX, Zhang JJ, Wu FT, Xu CF, Shen Z, Yu CH, Li YM. Construction of a convolutional neural network classifier developed by computed tomography images for pancreatic cancer diagnosis. World J Gastroenterol. 2020;26:5156-5168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 56] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 85. | Chu LC, Park S, Kawamoto S, Wang Y, Zhou Y, Shen W, Zhu Z, Xia Y, Xie L, Liu F, Yu Q, Fouladi DF, Shayesteh S, Zinreich E, Graves JS, Horton KM, Yuille AL, Hruban RH, Kinzler KW, Vogelstein B, Fishman EK. Application of Deep Learning to Pancreatic Cancer Detection: Lessons Learned From Our Initial Experience. J Am Coll Radiol. 2019;16:1338-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 86. | Tonozuka R, Itoi T, Nagata N, Kojima H, Sofuni A, Tsuchiya T, Ishii K, Tanaka R, Nagakawa Y, Mukai S. Deep learning analysis for the detection of pancreatic cancer on endosonographic images: a pilot study. J Hepatobiliary Pancreat Sci. 2021;28:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 87. | Ozkan M, Cakiroglu M, Kocaman O, Kurt M, Yilmaz B, Can G, Korkmaz U, Dandil E, Eksi Z. Age-based computer-aided diagnosis approach for pancreatic cancer on endoscopic ultrasound images. Endosc Ultrasound. 2016;5:101-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 88. | Săftoiu A, Vilmann P, Dietrich CF, Iglesias-Garcia J, Hocke M, Seicean A, Ignee A, Hassan H, Streba CT, Ioncică AM, Gheonea DI, Ciurea T. Quantitative contrast-enhanced harmonic EUS in differential diagnosis of focal pancreatic masses (with videos). Gastrointest Endosc. 2015;82:59-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 89. | Săftoiu A, Vilmann P, Gorunescu F, Gheonea DI, Gorunescu M, Ciurea T, Popescu GL, Iordache A, Hassan H, Iordache S. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest Endosc. 2008;68:1086-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 173] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 90. | Urman JM, Herranz JM, Uriarte I, Rullán M, Oyón D, González B, Fernandez-Urién I, Carrascosa J, Bolado F, Zabalza L, Arechederra M, Alvarez-Sola G, Colyn L, Latasa MU, Puchades-Carrasco L, Pineda-Lucena A, Iraburu MJ, Iruarrizaga-Lejarreta M, Alonso C, Sangro B, Purroy A, Gil I, Carmona L, Cubero FJ, Martínez-Chantar ML, Banales JM, Romero MR, Macias RIR, Monte MJ, Marín JJG, Vila JJ, Corrales FJ, Berasain C, Fernández-Barrena MG, Avila MA. Pilot Multi-Omic Analysis of Human Bile from Benign and Malignant Biliary Strictures: A Machine-Learning Approach. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |