Published online Nov 14, 2021. doi: 10.3748/wjg.v27.i42.7247
Peer-review started: March 16, 2021
First decision: August 9, 2021
Revised: August 17, 2021
Accepted: October 27, 2021
Article in press: October 27, 2021
Published online: November 14, 2021
Processing time: 238 Days and 15.8 Hours
Bacteria are known to communicate with each other and regulate their activities in social networks by secreting and sensing signaling molecules called autoinducers, a process known as quorum sensing (QS). This is a growing area of research in which we are expanding our understanding of how bacteria collectively modify their behavior but are also involved in the crosstalk between the host and gut microbiome. This is particularly relevant in the case of pathologies associated with dysbiosis or disorders of the intestinal ecosystem. This review will examine the different QS systems and the evidence for their presence in the intestinal ecosystem. We will also provide clues on the role of QS molecules that may exert, directly or indirectly through their bacterial gossip, an influence on intestinal epithelial barrier function, intestinal inflammation, and intestinal carcinogenesis. This review aims to provide evidence on the role of QS molecules in gut physiology and the potential shared by this new player. Better understanding the impact of intestinal bacterial social networks and ultimately developing new therapeutic strategies to control intestinal disorders remains a challenge that needs to be addressed in the future.
Core Tip: Host-microbiota interactions play a crucial role in the pathophysiology of many intestinal diseases. While biological components have been repeatedly described, a largely overlooked component is quorum sensing (QS), a density-dependent system able to coordinate bacterial responses and interact with host cells constantly exposed to bacteria. This review intends to describe the different QS systems to show evidence that QS is part of the intestinal ecosystem and highlight its impact on intestinal epithelial barrier function, inflammation, and intestinal carcinogenesis. From this report, we open up a new area of intestinal physiology.
- Citation: Coquant G, Aguanno D, Pham S, Grellier N, Thenet S, Carrière V, Grill JP, Seksik P. Gossip in the gut: Quorum sensing, a new player in the host-microbiota interactions. World J Gastroenterol 2021; 27(42): 7247-7270
- URL: https://www.wjgnet.com/1007-9327/full/v27/i42/7247.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i42.7247
Gut microbiota mutually interacts with coevolved host epithelial and immune cells in a beneficial reciprocal relationship[1]. The advent of multi-omics sequencing in the past decade has allowed researchers to investigate the complexity of the intestinal microbiota in various human disorders[2]. Many lines of evidence support a role for alteration of gut microbiota (dysbiosis) in the development or perpetuation of inflammatory and metabolic disorders; recent data pointed out the consequences of dysbiosis on host-microbiota interactions in this setting[3,4]. Currently, gut microbiota metabolites recognized as the main drivers of the impact of gut microbiota on hosts are short-chain fatty acids (SFCAs), branched-chain amino acids, trimethylamine N-oxide, bile acids, tryptophan (Trp), and indole derivatives[3,5]. A largely overlooked component is diffusible signaling molecules, which modulate the physiological response in the three domains of life[6]. A particular class of these signaling compounds is represented by bacterial quorum sensing (QS) molecules called autoinducers (AIs). QS is a density-dependent mechanism allowing bacterial populations to coordinate gene expression and physiology by modulating, for example, metabolic pathways, secretion of virulence factors, or biofilm formation in response to AIs[7]. Drawing on its density-dependent nature, it can be hypothesized that the production of bacterial signaling molecules is abundant in the highly densely populated environment of the mammalian intestinal tract.
Moreover, since several eukaryotic systems from fungi to plants and animals are known to recognize and respond to bacterial signaling compounds, it seems likely that human intestinal cells constantly exposed to bacterial compounds might also have developed response mechanisms to AIs with consequences on intestinal physiology[8]. The purpose of this current review is to provide clues to consider bacterial QS as a new actor of host-microbiota interactions. We will start by presenting the bacterial QS systems and evidence of QS in the gut. We will then provide an overview of the impact of QS molecules on host cell functions within the gut. Finally, we will investigate how modulation of the QS could be thought of as a therapeutic option, determine the key challenges, and suggest directions for future QS research.
In the conventional view of prokaryotic existence, bacteria live as unicellular orga
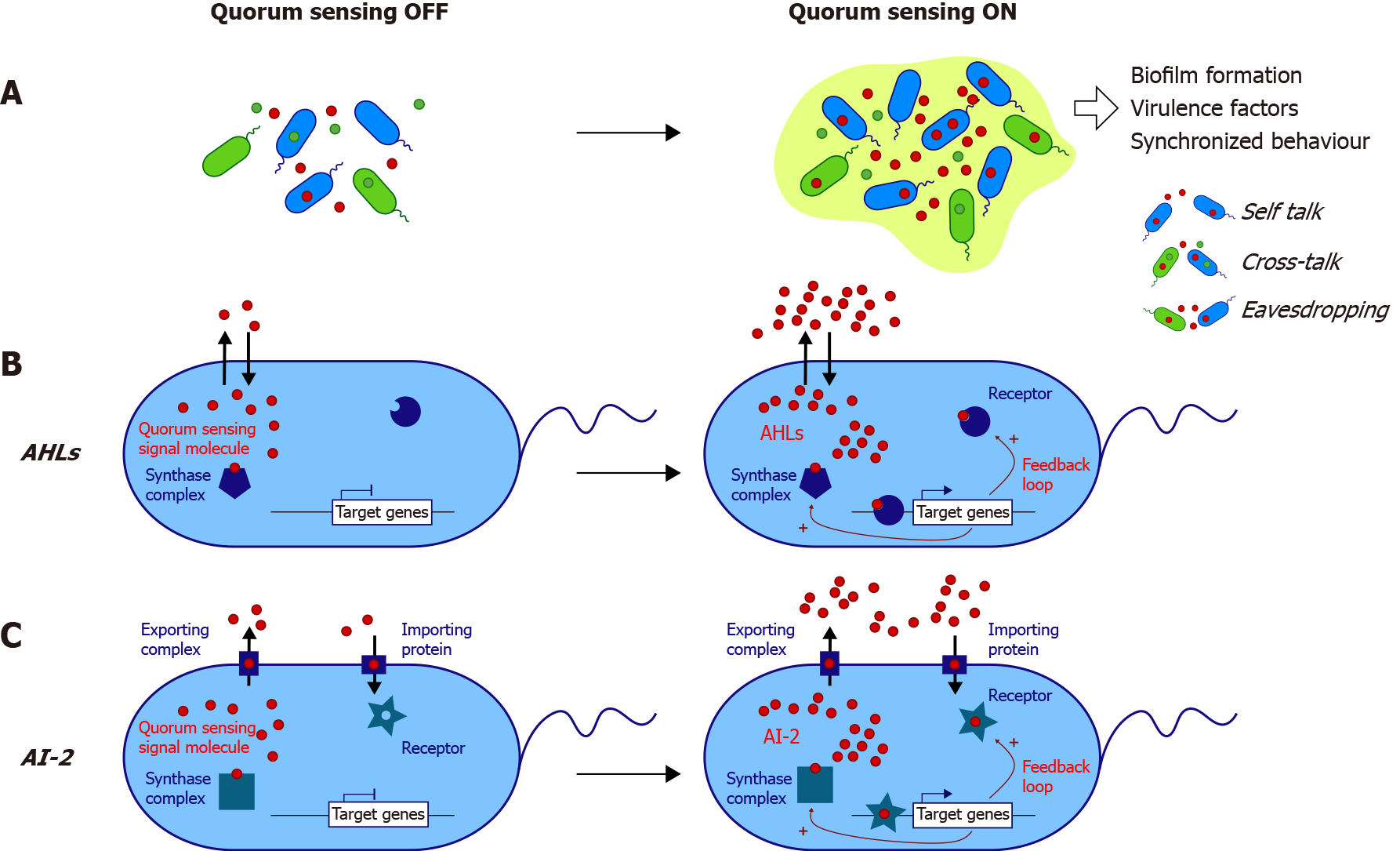
Bacterial QS is highly complex and mediates communications thanks to the diversity of its different systems. QS systems can be divided into systems specific to species and mediating communication between Gram-positive bacteria, Gram-negative bacteria, and interspecies systems (Table 1 and Figure 1).
| AI | Example of producing bacteria | QS system | Bacterial QS-regulated processes | Ref. | |
| Gram + | AI peptide | Staphylococcus aureus | agr | Virulence | Novick et al[155] |
| Listeria monocytogenes | agr | Virulence | Autret et al[156] | ||
| Clostridium perfringens | agr | Virulence | Ohtani et al[157] | ||
| Enterococcus faecalis | FsR | Virulence | Sifri et al[158] | ||
| Bacillus subtilis | com | Competence | Magnuson et al[159] | ||
| γ-butyrolactone | Streptomyces genus | scb | Antibiotics | Takano et al[13] | |
| scg | Metabolism | Du et al[14] | |||
| Gram - | AI-1 (acyl-homoserine lactones) | Vibrio fischeri | LuxI/LuxR | Luminescence | Engebrecht et al[16] |
| Vibrio harveyi | LuxLM/LuxN | Luminescence | Mok et al[160] | ||
| Virulence | Waters and Bassler[161] | ||||
| Pseudomonas aeruginosa | LasI/LasR | Virulence and biofilm | Gambello and Iglewski[162], Gambello et al[163], Winson et al[164], and Chapon-Hervé et al[165] | ||
| RhlI/RhlR | |||||
| PQS | Pseudomonas aeruginosa | PqsABCD/PqsR | QS regulation | Pesci et al[166] | |
| Pyocyanin | Gallagher et al[167] | ||||
| Iron homeostasis | Bredenbruch et al[168] and Diggle et al[169] | ||||
| Virulence | Gallagher et al[167] and Cao et al[170] | ||||
| Biofilm | Diggle et al[171] | ||||
| IQS | Pseudomonas aeruginosa | AmbBCDE/IqsR | Response to stress | Lee et al[172] | |
| CAI | Vibrio (cholerae) | CqsA/CqsS | Virulence | Ng et al[173] | |
| AI-3 | EHEC O157:H7 | Qse/QseBC | Attachment-effacement | Sperandio et al[19], Walters et al[21], and Kim et al[22] | |
| EPEC O26:H11 | Qse/unknown | Unknown | Kim et al[22], and Kaper and Sperandio[40] | ||
| AIEC LF82 | Qse/unknown | Unknown | Kim et al[22] | ||
| Escherichia coli MG1655 | Unknown | Unknown | Kim et al[22] | ||
| Escherichia coli BW25113 | Unknown | Unknown | Kim et al[22] | ||
| Salmonella enterica | Qse/unknown | Unknown | Kim et al[22], Kaper and Sperandio[40], and Walters and Sperandio[174] | ||
| Shigella flexneri | Qse/unknown | Unknown | Kim et al[22], Kaper and Sperandio[40], and Walters and Sperandio[174] | ||
| Yersinia sp. | Qse/unknown | Unknown | Kim et al[22], Kaper and Sperandio[40], and Walters and Sperandio[174] | ||
| Gram + and - | AI-2 | Vibrio harveyi | LuxS/LuxPQ | Bioluminescence, TSS, protease | Surette et al[24], Mok et al[160], and Schauder et al[175] |
| Vibrio cholerae | LuxS/LuxPQ | Virulence and Biofilm | Schauder et al[175], Zhu et al[176], and Hammer and Bassler[177] | ||
| Enterococcus faecalis | LuxS/LuxPQ | Unknown | Surette et al[24], and Schauder et al[175] | ||
| EHEC | LuxS/LsrB (?) | Attachment-effacement | Schauder et al[175], and Bansal et al[178] | ||
| Salmonella enterica | LuxS/LsrB | Pathogenicity and invasion | Miller et al[26], Schauder et al[175], and Choi et al[179] |
QS in Gram-positive species is driven in most cases by 5-17 amino acid oligopeptides (AIPs for AutoInducer Peptides), which are detected by membrane receptors belonging to the histidine kinase family and are involved in virulence or competence[12]. In addition, γ-butyrolactones are produced and integrated by Streptomyces sp. as signals controlling antibiotic production[13] or metabolism[14].
QS in Gram-negative bacteria relies on a high diversity of different systems, with some bacteria, such as Pseudomonas aeruginosa (P. aeruginosa), possessing several QS systems (Table 1). The expression of over 300 genes is regulated by QS. The most common system is driven by AI-1 molecules belonging to the AHL family, which are constituted by a homoserine lactone ring carrying a 4-18 carbon acyl chain. The lengths and modifications of the acyl chain give each AHL its species specificity[15]. The first described model is the Vibrio fischeri system, in which N-3-oxohexanoyl-homoserine lactone (3-oxo-C6) is synthesized by LuxI synthase, passively diffuses out of the cell and enters another bacterium in which it binds its receptor LuxR[16] (Figure 1B). Above a threshold, the AHL-receptor complex binds a consensus DNA sequence, thus triggering luciferase expression[17]. This model applies to all AHL systems (Table 1 and Figure 1B). The system involves a positive feedback loop, thus promoting QS activation at the population scale (Figure 1B). To date, numerous homologous systems (i.e., genes coding synthases and receptors) have been described in many Gram-negative bacteria, including over 70 Proteobacteria species[18] (Table 1).
Other Gram-negative QS systems involve the AI-3 molecule, initially identified in enterohemorrhagic Escherichia coli (E. coli) (EHEC) serotype O157:H7[19]. AI-3 regulates flagellar genes and pathogenicity[20,21] and is thought to be present in other enteropathogens (Table 1). A recent study[22] uncovered the structure of AI-3 and its natural analogs, including the prominent analog in mouse feces in vivo, which belongs to the pyrazinone family. The authors showed that various gram-negative and Gram-positive bacteria produce AI-3 analogs, thus redefining the specificity of AI-3 molecules.
Last, the third type of QS system has been identified in Gram-negative bacteria such as EHEC or Vibrio species and Gram-positive bacteria such as Salmonella enterica[23,24]. It relies on AI-2 molecules such as S-THMF-borate [for (2S,4S)-2-methyl-2,3,3’,4-tetrahydroxy-tetrahydrofurane-borate][25] and R-THMF [for (2R,4S)-2-methyl-2,3,3’,4-tetrahydroxy-tetrahydro furane][26]. AI-2 has now been found in various bacterial species in which it regulates many processes[27] and is proposed to mediate poly-species communication (Figure 1C).
In addition, indole is produced from Trp by Gram-negative and Gram-positive commensal and pathogenic bacteria displaying tryptophanase activity[28-31]. As the source of Trp is supplied by the diet and cannot be synthesized endogenously, either by bacteria or by the host, indole is a bacterial byproduct of Trp metabolism. However, in recent years, some authors have considered indole to be a QS molecule, as it is produced in a density-dependent manner and regulates several bacterial physiological processes, such as the formation of spores or biofilms, virulence traits, bacterial motility, and drug resistance[29,32,33].
The versatility of QS systems and their AI molecules highlights the complexity of communication and thus emphasizes the key role QS could play in a diverse ecosystem: the intestinal microbiota.
The study of QS in the gut is still a relatively recent matter of interest, as QS is generally addressed from the pathogenic bacterium P. aeruginosa point of view in the lung ecosystem. However, many arguments suggest that QS is a new player in the gut ecosystem.
AHLs in the gut: As part of the eavesdropping mechanism, some bacteria from the human gut can sense AHLs from other species (Figure 1A). Gram-negative bacilli such as E. coli, Enterobacter, or Klebsiella express the receptor SdiA, which can sense AHL, without producing such a signal[34]. The opportunistic pathogen P. aeruginosa, which targets the digestive tract in severely immune-compromised patients[35,36], and the more common enteropathogen Yersinia enterolitica are known to produce AHLs[37]. Analyses from sequencing databases have shown the presence of LuxI/LuxR homologs in a few commensals: Hafnia alvei, Edwardsiella tarda, and Ralstonia sp. strain 5_7_47FAA[37]. However, the latter article did not demonstrate the presence of AHLs but only homologs of the genes encoding the synthase complex and the receptor. A cohort low sample size pediatric study (n = 4) demonstrated, thanks to bacterial reporter systems, the presence of AHLs in the feces of patients without identifying them[38]. Our team investigated the question of AHLs in the human gut in the context of inflammatory bowel disease (IBD). With high-resolution mass spectrometry, we identified approximately ten AHLs in the feces of healthy patients, IBD patients in remission, and flare[39]. We also found a never-described AHL, 3-oxo-C12:2-HSL, that was less represented in IBD patients, especially in flares, than in healthy subjects[39].
AI-2 in the gut: AI-2 presence in the gut has been reported by several articles[19,40,41] but is mainly linked to pathogenic bacteria such as enterohemorrhagic E. coli[19]. As AI-2 is considered a “universal language”, it is not surprising to find this AI in an ecosystem as diverse as the gut microbiota.
Thompson et al[42] showed that most Firmicutes contain LuxS protein orthologs, an important enzyme that allows AI-2 production. In contrast, its presence is less represented in Bacteroidetes. Mutant E. coli engineered to regulate AI-2 levels in the mouse gut counteract antibiotic-induced dysbiosis[42]. AI-2 produced by E. coli benefits Firmicutes while restraining Bacteroidetes representation[42], suggesting an important role of AI-2 in the gut ecosystem.
Other QS signals: Concerning other QS signals, there is less evidence of their implications in gut microbiota. A recent study showed a correlation between indole and Clostridioides difficile (C. difficile) infection (CDI) with higher indole concentrations for patients with CDI than for CDI-negative patients with diarrhea[43]. C. difficile induces indole production through overexpression of the tryptophanase gene tnaA in enterotoxigenic E. coli and other indole-producing anaerobes. This increased indole level has been shown to be detrimental to some of the beneficial bacteria and favors C. difficile colonization.
These mechanisms collectively suggest a complex bacteria-bacteria QS network in the gut ecosystem. The key issue is now to decode every language to fully understand its potential in host-microbiota interactions.
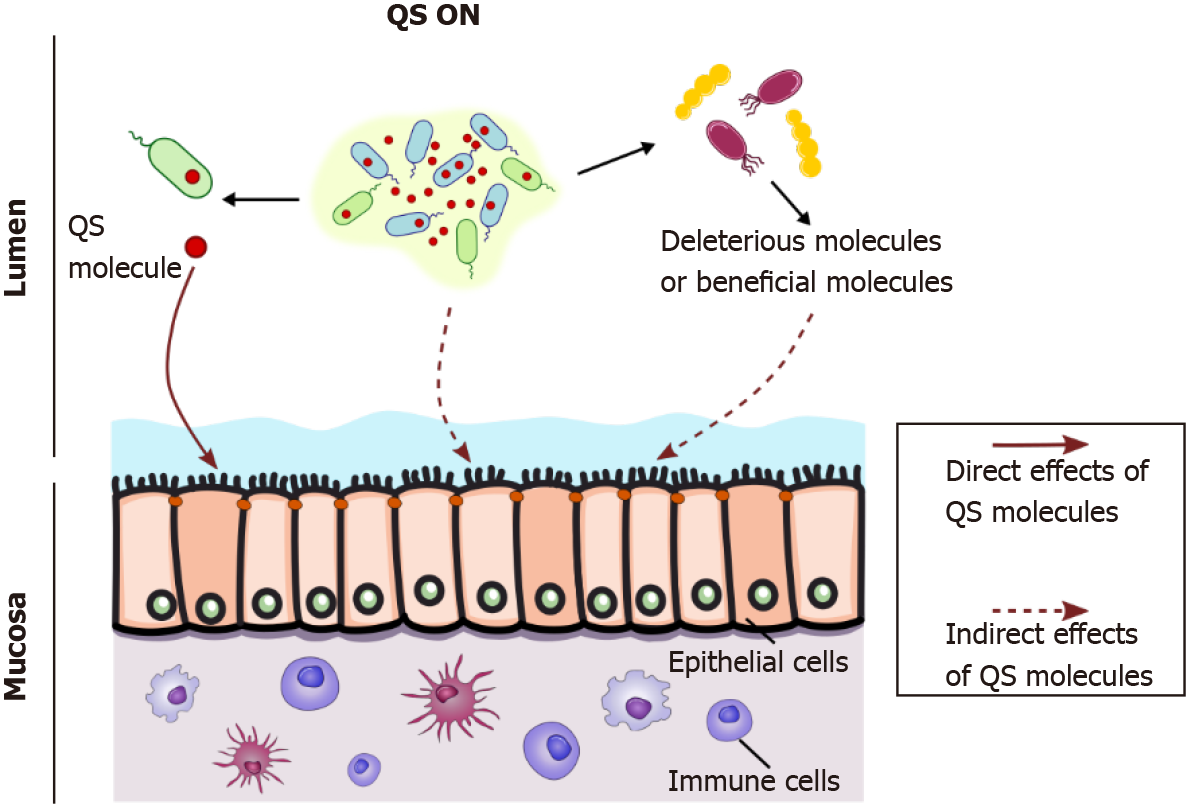
In an ecosystem as networked and complex as that in the gut, bacterial communication has to be seen from a large perspective, with multiple bacterial populations crosstalking to each other through eavesdropping or crosstalking between species (Figure 1A). Therefore, the question of how QS affects the host can be addressed in two ways (Figure 2).
QS modulates microorganism metabolism, which in turn can affect the host’s physiology; one could consider this to be an indirect effect of QS molecules on the host (Figure 2). Bacterial metabolism modifies beneficial byproducts such as SFCAs and bile acids[42]. By modulating intestinal microbiota composition, QS can indirectly influence gut physiology by promoting deleterious or beneficial bacteria (Figure 2). Thompson et al[42] demonstrated that AI-2 modulates dysbiotic microbiota composi
The question addressed in this review is how quorum-sensing molecules can directly affect the host, independent of the producing bacteria (Figure 2).
As discussed above, it remains largely unknown how the microbial communities hosted in the gut lumen use QS communication systems. However, there is evidence that at least several bacterial species commonly found in the gastrointestinal tract have the capacity to synthesize QS molecules[6,8,39].
Studies on the impact of QS molecules on the biology of intestinal host cells have focused on key actors of barrier function and the immune response. Intestinal barrier function includes the ability of epithelial cells to form: (1) A selective barrier whose permeability is controlled by cell-cell junctions[47], (2) Synthesize a protective mucus layer and antimicrobial peptides[48,49], and (3) Secrete cytokines and chemokines allowing appropriate crosstalk with the underlying immune compartment. The intestinal immune system is involved in tolerogenic or inflammatory responses to the commensal microbiota or pathobionts/pathogens[50,51], and it represents the largest immune organ in the body. Intestinal epithelial cells, intraepithelial lymphocytes, and immune cells located in the lamina propria are involved in the modulation of immunity and inflammation by microbiota[52].
The impact of QS molecules on barrier function and the immune response has been mainly studied in the context of host-pathogen interactions, probably because most of the data rely on AHLs produced by P. aeruginosa. However, evidence of the presence of QS molecules in the healthy intestinal lumen has led to further study on their effects on the host compartment, including barrier function, inflammatory process, and carcinogenesis.
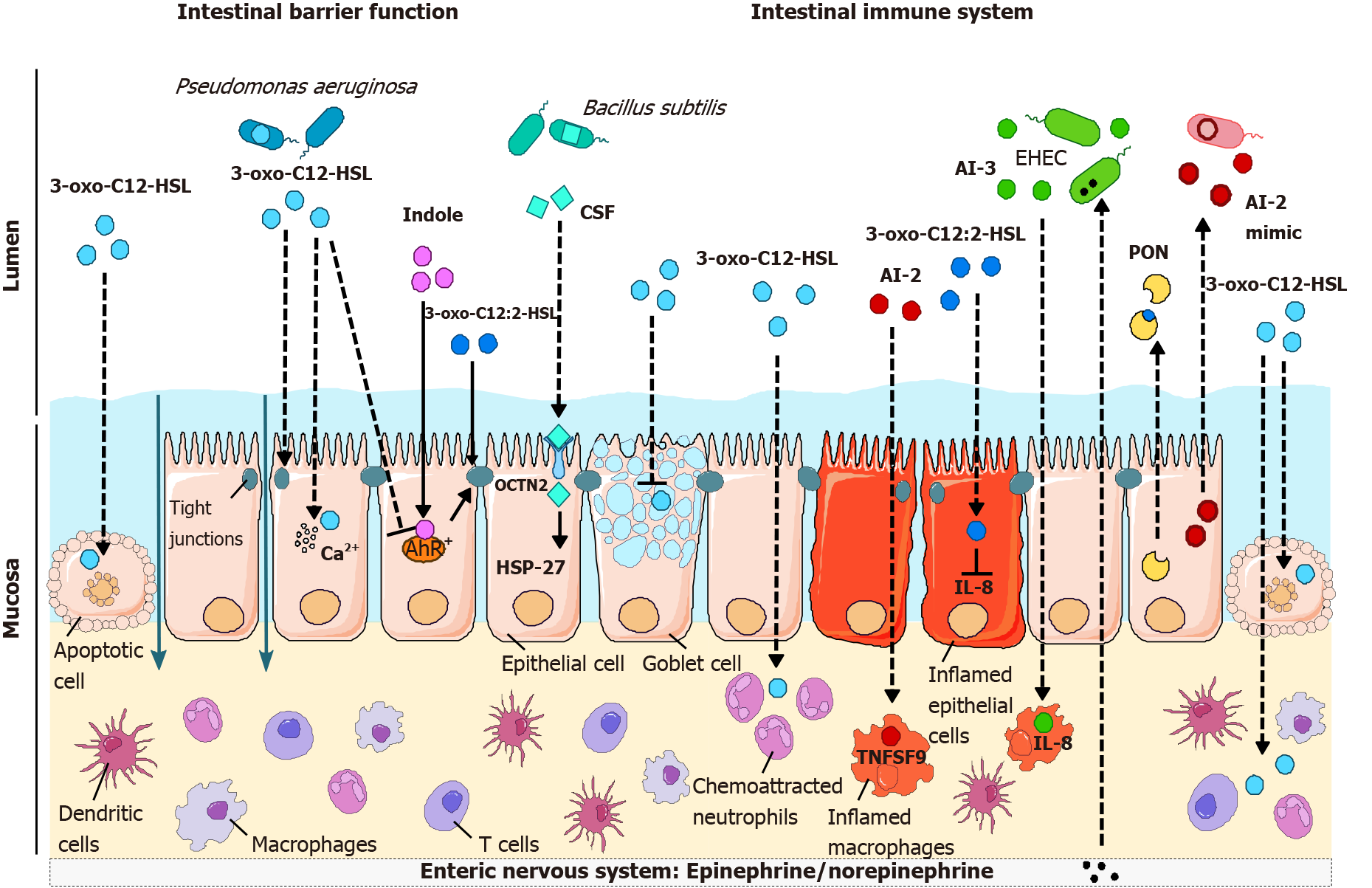
AHLs: 3-oxo-C12-HSL produced by P. aeruginosa is probably the QS molecule whose effects on the barrier function of epithelial cells have been the most studied during the last two decades[53]. P. aeruginosa synthesizes various virulence factors, which act synergistically with QS molecules to destabilize cell-cell junctions and promote bacterial transmigration across epithelial and endothelial barriers[54].
3-oxo-C12-HSL induces an increase in paracellular permeability to ions and macromolecules[55-60] (Table 2). This deleterious effect of 3-oxo-C12-HSL on barrier function is accompanied by an alteration of tight junctions (TJs) (Figure 3). In the Caco-2 intestinal epithelial cell line, 3-oxo-C12-HSL induced a decrease in the expression, as well as mislocalization, of the TJ proteins occludin, tricellulin, ZO- 1, ZO-3, JAM-A, and of the adherent junction proteins E-cadherin and β-catenin[55,58-61] (Table 2). Loss of occludin/ZO-1 and tricellulin/ZO-1 interaction at the plasma membrane suggested the dismantling of TJ protein complexes[61]. In addition, hyperphosphorylation of occludin, ZO-1, ZO-3, JAM-A, E-cadherin, and β-catenin on tyrosine residues (as well as serine and threonine for E-cadherin and ZO-1) was reported in the presence of 3-oxo-C12-HSL, whereas the serine and threonine residues of occludin, JAM-A and β-catenin were less phosphorylated[58,59].
| QS molecule | Effects | Ref. |
| Effects on the intestinal epithelial migration | ||
| 3-oxo-C12-HSL | Increased migration at low concentrations (1.5-12 μmol/L) vs inhibition at 200 μmol/L | Karlsson et al[72] |
| Interaction with IQGAP1 and increase in Rac1/Cdc42 (1.5-200 μmol/L) | Karlsson et al[72] | |
| Effects on the intestinal epithelial permeability and intercellular junctions | ||
| 3-oxo-C12-HSL | Increased permeability to ions and macromolecules (100-400 μmol/L) | Eum et al[55], Vikström et al[58-60], and Aguanno et al[61] |
| Activation of p38 and p42/44 and calcium signaling (100-200 μmol/L) | Vikström et al[58-60] | |
| Decreased expression levels of tight junction genes (100-400 μmol/L); Disassembly of tight and adherens junctions (modification of their phosphorylation status and involvement of MMP-2 and -3) | Eum et al[55], Vikström et al[58-60], and Aguanno et al[61] | |
| Decreased levels of tight junction proteins occludin and tricellulin (100-400 μmol/L) | Eum et al[55] | |
| Decreased protein levels of extracellular matrix and tight junction proteins (400 μmol/L) | Tao et al[62] | |
| 3-oxo-C12:2-HSL | No deleterious effects on permeabilityProtection of tight junction integrity and maintenance of junctional complexes at the plasma membrane under pro-inflammatory conditions | Landman et al[39] and Aguanno et al[61] |
| 3-oxo-C14-HSL | Decreased protein levels of extracellular matrix and tight junction proteins (400 μmol/L) | Tao et al[62] |
| Indole and indole derivatives | Decreased permeability to ions and increased expression of genes coding tight junction and cytoskeleton proteins | Bansal et al[76] and Shimada et al[77] |
| Decreased permeability to macromolecules | Venkatesh et al[79] | |
| Increased transcripts levels of genes coding tight junction proteins | Shin et al[78] | |
| Effects on the mucus layer components | ||
| 3-oxo-C12-HSL | Decreased MUC3 mRNA levels (30 μmol/L) | Taguchi et al[70] |
| Decrease in Muc2 production in goblet cell-like cell line (100 μmol/L) vs increase in colonic cell line (400 μmol/L) | Tao et al[67] | |
| Indole | Increased expression of genes involved in the production of mucins | Bansal et al[76] |
| Effects on intestinal epithelial cell viability | ||
| 3-oxo-C12-HSL | Mitochondrial dysfunction and induction of apoptosis in goblet cell-like cell line (100 μmol/L) and in colonic cell line (30-100 μmol/L) | Tao et al[67-69], and Taguchi et al[70] |
| Induction of apoptosis, mitochondrial dysfunction, oxidative stress and blocking of cell cycle (400 μmol/L) | Tao et al[62] | |
| 3-oxo-C14-HSL | Induction of apoptosis, mitochondrial dysfunction, oxidative stress and blocking of cell cycle (400 μmol/L) | Eum et al[55], Vikström et al[58-60], Aguanno et al[61], and Tao et al[62] |
| CSF | Reduction of oxidative stress-induced cell death and loss of the epithelial barrier (involving HSP27 and p38/MAPK pathway) | Fujiya et al[74] |
Several signaling mechanisms, including p38 and p42/44 MAP kinases[59,60], Ca2+ release[57,58], matrix metalloproteinases MMP-2 and MMP-3 via protease-activated receptor (PAR) signaling[55] and oxidative stress[62], have been implicated in 3-oxo-C12-HSL effects on junctional proteins and on the concomitant increase in permeability (Table 2).
As discussed above, the most prominent AHL detected in the human intestinal ecosystem is unsaturated 3-oxo-C12:2-HSL[39]. Despite a high structural homology with the P. aeruginosa 3-oxo-C12-HSL, this intestinal AHL has recently been found to have opposite properties regarding the barrier function (Table 2). In contrast to 3-oxo-C12-HSL, 3-oxo-C12:2-HSL does not increase paracellular permeability in Caco-2/TC7 enterocytic cells[39,61]. Most importantly, 3-oxo-C12:2-HSL can limit TJ disruption induced by the proinflammatory cytokines interferon-gamma (IFN-γ) and tumor necrosis factor-α (TNF-α)[61] (Figure 3). In these conditions mimicking intestinal inflammation encountered, for example, in IBD, 3-oxo-C12:2-HSL maintains the interaction of the TJ transmembrane proteins occludin and tricellulin with their main cytoplasmic partner ZO-1. It limits cytokine-induced occludin and tricellulin ubiquitination and the interaction of these TJ proteins with the E3 ubiquitin ligase itch, suggesting stabilization of TJ complexes at the plasma membrane in inflammatory conditions. Altogether, these results show that “commensal” intestinal 3-oxo-C12:2-HSL mitigates the deleterious effects of the inflammatory environment on TJs, which are key actors in epithelial barrier function[61].
Epithelial barrier disruption may combine TJ alteration and an unrestricted passage, which occurs following epithelial damage, generated, for example, by cell apoptosis upon exposure to harmful molecules such as high doses of proinflammatory cytokines[63-65]. This TJ-independent breaking of the barrier allows translocation of large particles such as large proteins, entire bacteria, and viruses, which a priori cannot cross the epithelium through the paracellular route even in conditions where TJs are “open”[66]. The 3-oxo-C12-HSL produced by P. aeruginosa exerts cytotoxic effects, particularly through apoptosis induction, in numerous cell types, including the intestinal and colonic epithelial cell lines LS174T[67-69], Caco-2[70], and CT26[62] (Table 2 and Figure 3). Apoptosis triggered by 3-oxo-C12-HSL relies on oxidative stress and caspase-dependent processes[62,69], whereas short-chain C4-HSL does not exert any apoptotic effects[67]. Interestingly, the increase in paracellular permeability to macromolecules induced by 3-oxo-C12-HSL was dramatically exacerbated in Caco-2/TC7 cells cultured in the presence of IFN-γ and TNF-α or cocultured with THP-1 activated monocytic cells[61]. These synergistic effects on barrier disruption probably rely on epithelial cell apoptosis, as they are abolished by a caspase inhibitor (unpublished results). In contrast, intestinal AHL 3-oxo-C12:2-HSL neither exerts cytotoxic effects nor synergizes with proinflammatory cytokines to disrupt the epithelial barrier[61].
Epithelial injury accompanying acute inflammatory conditions is followed by a re-epithelialization phase, during which cell migration plays an important role[71]. Interestingly, 3-oxo-C12-HSL has been shown to dose-dependently modulate Caco-2 cell migration in a wound-healing assay and interact directly with the GTPase activating protein IQGAP1, stressing a potential role of AHL in cytoskeletal reorganization[72] (Table 2).
Another key actor of the intestinal physical barrier is the mucus layer, which is essential to maintain segregation between luminal microorganisms and the epithelium[49]. 3-oxo-C12-HSL induces reduced expression and production of Mucin2 in LS174T cells[67], as well as a decrease in the levels of MUC3 mRNA in the Caco-2 cell line cultivated in an undifferentiated state[70] (Table 2 and Figure 3). Interestingly, differentiated Caco-2 cells, which express higher levels of mucin 3, showed less sensitivity to 3-oxo-C12-HSL-induced apoptosis in the latter study, and the addition of mucin dose-dependently protected cells from apoptosis induced by this AHL[70].
It must be specified that all these studies on barrier function were carried out with high concentrations of AHLs (100-400 μmol/L) (Table 2), knowing that the concentration of 3-oxo-C12-HSL has been estimated to reach 600 μmol/L in biofilms of P. aeruginosa[73].
Gram-positive QS peptides: The effects of AIP (found in Gram-positive bacteria) on intestinal barrier function are much less documented than those of Gram-negative QS molecules. Whereas most of the studies on the effects of AIP on host inflammation describe the indirect effects of AI through the modulation of bacterial metabolism, one article reported the direct effects of AIP. Fujiya et al[74] reported that Bacillus subtilis AIP, named competence and sporulation factor (CSF), induces HSP27 expression and the p38/MAPK pathway and reduces cell death and the loss of the epithelial barrier induced by oxidative stress (Table 2 and Figure 3). Inducible HSPs are needed under stress and help stabilize proteins to prevent denaturation[75]. B. subtilis is part of the normal microbiota; it is also used as a commercial probiotic and is beneficial to the host. Moreover, CSF seems to signal through a receptor named OCTN2 (organic cation/carnitine transporter 2), and polymorphisms of the gene encoding this receptor are part of the susceptibility locus of Crohn’s disease[74].
Indole: Indole exerts a beneficial role on TJ protein expression in several intestinal epithelial cells[76-79] (Table 2). Oral administration of indole to germ-free mice, which display very low indole fecal levels, increased the expression of TJ and adherens junction-associated proteins in the colonic epithelium and improved their resistance to dextran sulfate sodium (DSS)-induced colitis[77]. Indole has been identified as an endogenous agonist of aryl hydrocarbon receptor (AhR), which can compete for receptor binding with well-known AhR ligands[80]; several studies have also stressed the key role of the AhR pathway in indole derivative protective effects[81-83]. Accordingly, several studies have shown that AhR activation strengthens the epithelial barrier by protecting TJs[82,84-87] (Figure 3) or by stimulating antimicrobial peptide production via interleukin (IL)-22[88,89].
In addition to their effects on the intestinal barrier, QS molecules were analyzed on different actors of the immune compartment of the intestine, which is involved in a complex crosstalk with the epithelial compartment to maintain an appropriate immune response toward the content of the intestinal lumen.
AHLs: Our group described that 3-oxo-C12:2-HSL, an AHL recently discovered in the human gut[39], exerts anti-inflammatory effects on intestinal epithelial cells. During inflammation, intestinal epithelial cells can secrete some cytokines, among which the chemokine IL-8 promotes the recruitment of neutrophils in the mucosa and participates in the acute-phase response[90,91]. In a study comparing the effect of 3-oxo-C12:2-HSL to 3-oxo-C12-HSL produced by P. aeruginosa, our group demonstrated in the human enterocytic Caco-2/TC7 cell line that 3-oxo-C12:2-HSL, but not 3-oxo-C12-HSL, attenuated the induction of IL-8 secretion induced by the proinflammatory cytokine IL-1β[39,92] (Table 3 and Figure 3). This potential anti-inflammatory effect of 3-oxo-C12:2-HSL is consistent with the hypothesis of a beneficial role of this AHL in gut ecosystems[39], as are its protective effects on TJ integrity. The impact of intestinal 3-oxo-C12:2-HSL on immune cells remains largely unknown.
| Cell type | QS molecule | Effects | Ref. |
| Effects on innate immune cells | |||
| Macrophages | 3-oxo-C12-HSL | Anti-inflammatory effects on IL-12 and TNF-α (0.1-100 μmol/L) | Telford et al[94] |
| Increased TLR2 and TLR4 expression and decreased TNF-α production (1-100 μmol/L) | Bao et al[180] | ||
| Pro-apoptotic effects (12-50 μmol/L) | Tateda et al[102] | ||
| Increased phagocytosis (100 μmol/L) | Vikström et al[107] | ||
| NF-κB inhibition (4.7 μmol/L) | Kravchenko et al[104] | ||
| Dose-dependent anti-inflammatory effects (1-50 μmol/L) | Kravchenko et al[105] | ||
| Involvement in p38/MAPK signaling (1-100 μmol/L) | Kravchenko et al[105], Vikström et al[107], Glucksam-Galnoy et al[181] | ||
| Activation of the Unfolded Protein Response (6.25-100 μmol/L) | Zhang et al[182] | ||
| Change in cell volume and shape (10-50 μmol/L) | Holm et al[183] | ||
| Indole derivatives | Prevents the induction of pro-inflammatory cytokines | Krishnan et al[184] | |
| AI-2 | Induction of the expression of cytokines, chemokines and TNFSF9 | Li et al[41] | |
| Monocytes | AI-3 and analogues | Increase in IL-8 secretion | Kim et al[22] |
| Dendritic cells | 3-oxo-C12-HSL | Pro-apoptotic effects (100 μmol/L) | Boontham et al[185] |
| No effect on IL-10 secretion (5-30 μmol/L) | Skindersoe et al[100] | ||
| Increased IL-10 production (5-100 μmol/L) | Li et al[99] | ||
| Decreased IL-12 secretion (5-100 μmol/L) | Li et al[99] and Skindersoe et al[100] | ||
| Increased induction of Treg (5-100 μmol/L) | Li et al[99] | ||
| Neutrophils | 3-oxo-C12-HSL | Chemoattraction (0.01-100 μmol/L) | Karlsson et al[186] and Zimmermann et al[187] |
| Activation of MAPK signaling (12-50 μmol/L) | Tateda et al[102] and Singh et al[188] | ||
| Increased phagocytosis (10 μmol/L) | Wagner et al[189] | ||
| Pro-apoptotic effects (12-50 μmol/L) | Tateda et al[102] | ||
| Effects on adaptive immune cells | |||
| T cells | 3-oxo-C12-HSL | Inhibition of proliferation and activation (0.1-100 μmol/L) | Telford et al[94], Boontham et al[185], Gupta et al[190], and Hooi et al[191] |
| Activation of naïve T cells towards Th1 phenotype (5 μmol/L) | Smith et al[95] | ||
| Decreased secretion of IL-4 and IFN-γ (5 μmol/L) | Ritchie et al[96] | ||
| Induction of apoptosis via the mitochondria pathway (100 μmol/L) | Jacobi et al[101] | ||
| Induction of Treg (1-50 μmol/L) | Li et al[99] | ||
| Indole derivatives | Re-programming into tolerogenic T cells | Cervantes-Barragan et al[192] | |
| Promotion of differentiation towards a regulatory type 1 phenotype | Aoki et al[193] | ||
| B cells | 3-oxo-C12-HSL | Modulation of immunoglobulin production (0.1-100 μmol/L) | Telford et al[94] and Ritchie et al[194] |
| ILC | Indole derivatives | Promotion of IL-22 production | Zelante et al[83] |
| Effects on epithelial cells | |||
| Pulmonary tract epithelial cells | 3-oxo-C12-HSL | Induction of IL-8 production and NF-B activation (100 μmol/L) | Smith et al[195] |
| Increased expression levels of pro-inflammatory cytokines | Jahoor et al[115] | ||
| Intestinal epithelial cells | 3-oxo-C12-HSL | Mitigation (1-10 μmol/L) or aggravation (> 50 μmol/L) of IL-8 expression induction | Peyrottes et al[92] |
| 3-oxo-C12:2-HSL | Attenuation of the induction of IL-8 expression (5-50 μmol/L) | Landman et al[39] | |
The effects of 3-oxo-C12-HSL depend on the concentration and cell type studied[6,93]. Telford et al[94] showed that 3-oxo-C12-HSL inhibits the production of TNF-α and IL-12 [a cytokine involved in the T helper cell-1 type response (Th1-type response)] by lipopolysaccharide-activated macrophages at high concentrations and stimulates the production of antibodies, particularly immunoglobulin G1, which is an indicator of a Th2-type response at lower concentrations (Table 3). Conversely, Smith et al[95] showed that 3-oxo-C12-HSL activates and promotes the differentiation of naive T lymphocytes toward a Th1-like phenotype, while Ritchie et al[96] observed that 3-oxo-C12-HSL inhibits the differentiation of both Th1 and Th2 T lymphocytes (Table 3). Altogether, these results demonstrated that 3-oxo-C12-HSL is an immunomodulator of the Th1/Th2 response. 3-oxo-C12-HSL and two other QS molecules from P. aeruginosa, PQS (Pseudomonas quinolone signal), and HHQ (4-hydroxy-2-heptylquinoline), suppress both innate and adaptive immune responses acting on lymphoid cells, dendritic cells, and neutrophil monocytes/macrophages[22,97,98]. 3-oxo-C12-HSL and PQS decreased the production of the cytokines IL-12 and IFN-γ by activated dendritic cells, which in turn decreased T-cell proliferation and activity[98-100] while promoting the induction of regulatory T-cells[99] (Table 3). 3-oxo-C12-HSL provoked apoptosis of macrophages, neutrophils, and T lymphocytes through activation of caspases and the mitochondrial apoptosis pathway[101,102]. Several reports described inhibition of the nuclear factor-kappa B (NF-κB) pathway by QS molecules from P. aeruginosa[93,103-106] and/or the activation of signaling pathways such as p38 MAPK[105,107] (Table 3). It has been recently demonstrated that only long-chain AHLs such as 3-oxo-C12-HSL modulate the phenotype of dendritic cells and the type 2 immune response through mechanisms involving retinoic acid signaling and the protein kinase AKT[106].
The molecular mechanisms involved in the effects of QS molecules from P. aeruginosa on immune cells are independent of the Toll-like receptor pathway[105], which is a classical cell process involved in recognizing pathogen fragments. Some reports have indicated that the perception of AHL by mammalian cells involves the bitter taste receptor T2R38[108-110], which is widely expressed in the human digestive tract from the tongue to the colon[111]. Polymorphisms in the TAS2R38 gene may increase susceptibility to infections and colorectal cancer (CRC)[112]. It has been shown that these receptors use inflammatory pathways, which differ according to the cell type and their localization[113]. 3-oxo-C12-HSL binds to the transcription factor peroxisome proliferator-activated receptor γ[114], which has been proposed as a potential receptor for AHL and seems to be involved in AHL proinflammatory effects[115]. Recently, Moura-Alves et al[116] showed that QS molecules produced by P. aeruginosa modulated the activity of the transcription factor AhR, which plays an important role in regulating innate and adaptive immunity[117,118].
Overall, it has been demonstrated that QS molecules from P. aeruginosa have an immunosuppressive effect, allowing the pathogen to evade the immune system during infection. It remains to be determined whether endogenous intestinal 3-oxo-C12:2-HSL participates in controlling intestinal immunity in health and diseases and to decipher the underlying mechanisms.
AI-2 and AI-3: AI-2 is produced by both Gram-negative and Gram-positive bacteria and is mainly studied for its role in bacteria-bacteria communication and the virulence of pathogenic strains[8]. However, little is known about the effect of AI-2 on immune cells. In mice, AI-2 administration has no effect by itself on cytokine expression but aggravates lung inflammation during P. aeruginosa infection by interfering with QS molecules produced by this pathogen[119]. In cultured macrophages, AI-2 induces the expression of several cytokines and chemokines as well as the expression of TNF superfamily member 9 (TNFSF9), a protein involved in the immune response[41] (Figure 3).
The AI-3 system is mainly described in enterohemorrhagic E. coli and is therefore linked to the development of intestinal epithelial lesions, suggesting its proinflammatory activity[120] (Table 3 and Figure 3). Indeed, AI-3 and its analogs increase IL-8 secretion by THP-1 monocytes[22]. Given that the AI-3 structure has only been uncovered recently[22], the direct effect of this molecule on the host is poorly known so far. In addition, since the AI-3 bacterial receptor can recognize host-synthesized epinephrine/norepinephrine (Figure 3), one could suggest that adrenergic receptors could recognize AI-3[19]. However, it has been shown that AI-3 and its analogs do not activate or modulate adrenergic signaling[22].
Gram-positive AIP: The effects of Gram-positive AIP bacteria on inflammation are far less documented than those of Gram-negative QS. Moreover, most of the studies on the effects of AIP on host inflammation describe the indirect effects of AIs through the modulation of bacterial metabolism.
A study described that AIP could selectively cross intestinal epithelial cell Caco-2 cell monolayers[121]. Additionally, it has been reported that AIP can cross the highly selective blood-brain barrier in vivo[122]. These processes seem to be peptide-specific: Clostridium acetobutylicum AIP easily penetrates the blood-brain barrier, while Streptococcus pneumonia’s AIP crosses it poorly[122]. This shows that small molecules such as AIs affect the host's physiology beyond the gastrointestinal tract. For instance, it has been described that AIP has various effects on muscle inflammation as part of the gut-muscle axis. De Spiegeleer et al[121] performed an extensive screening of 75 QS molecules on muscle cells. They demonstrated both pro- and anti-inflammatory effects of four peptides from the genera Staphylococcus, Streptococcus, Lactobacillus, and Bacillus, and some of those peptides have been described in the gut.
Indole: Several studies have reported that indole exerts anti-inflammatory effects in the intestine and protects against pathogenic infection[123] (Table 3). AhR, an important contributor to the maintenance of innate and adaptive immunity, drives most of these effects, particularly in the intestinal mucosa[117,118,124].
Several reports have shown altered Trp metabolism in gut inflammation in humans and mice [81,125-127]. A decrease in endogenous indole was observed in human feces from subjects with celiac disease or IBD[127]. This was associated with a decrease in AhR activity in the intestinal mucosa. In parallel, an increase in Trp levels in the same samples suggested that the gut microbiota-dependent metabolism of Trp was altered[127]. In mouse models of celiac disease and IBD, the implantation of indole-producing bacteria increases AhR activity and protects them from gut inflammation[81]. Interestingly, Moura-Alves et al[116] showed that 3-oxo-C12-HSL and HHQ had an inhibitory effect on AhR activity and could compete with well-known activators of AhR. This observation raises the question of potential competition between several QS molecules for AhR-dependent modulation of innate and adaptive immunity.
There is growing evidence that gut microbiota dysbiosis plays a major role in CRC development[128]. Indeed, modifications of commensal gut microbiota in favor of opportunist bacteria promote intestinal inflammation, which is well known as a driver event in CRC onset[129,130]. Thus, the concept of the “bacterial driver-passenger model” highlights the crosstalk between host immunity and colonic microbiota[131]. For example, some driver pathogens, such as Bacteroides fragilis, have been proposed to promote a strong Th17 inflammatory response[132]. This proinflammatory microenvironment might favor colonization by opportunist pathogens such as Fusobacterium spp. Accordingly, Fusobacteria-dominant biofilms were associated with human CRC[133-135]. Altogether, these findings support that polymicrobial interactions and intercellular communications might play an important role in CRC development[136]. Nevertheless, how bacteria communicate with themselves and with the host during CRC remains poorly understood.
Recent findings provide new insights into the role of the QS molecule AI-2 in intercellular communication during CRC. First, the AI-2 concentration is increased in tumors compared to the surrounding normal tissue in human CRC[41]. These levels also correlate with the progression of the disease according to the CRC TNM (tumor node and metastasis) score[41]. Regarding the tumor immune microenvironment, the AI-2 concentration positively correlates with TNFSF9 expression, which is mainly expressed by tumor-associated macrophages, and negatively correlates with the CD4/CD8 ratio, suggesting that AI-2 associates with the antitumor response[41]. At the molecular level, it was demonstrated that AI-2 induces in vitro M1 polarization of U987-derived macrophages through the TNFSF9 signaling pathway[137]. These findings reveal that AI-2 could be an important factor linked to the immune tumor microenvironment and shed light on the role of the quorum-sensing system during CRC development and progression. Interestingly, mammalian epithelial cells are able to produce AI-2 analog molecules that mimic AI-2 effects (Figure 3), illustrating the complexity of bacteria-host crosstalk[45]. Thus, a better characterization of QS molecules involved in tumorigenesis might be an opportunity to improve our knowledge of the mechanisms underlying CRC development.
Interkingdom signaling works in two ways, as host cells are able to counterattack the QS system using several strategies.
As described above, as part of AI-3 signaling, host hormones such as epinephrine and norepinephrine can be recognized by EHEC and lead to the expression of virulence genes[19]. This AI-3/epinephrine/norepinephrine signaling is not restricted to EHEC, and the receptor QseC is also expressed, for example, by the intestinal pathogenic Salmonella enterica serovar Typhimurium[138] (Table 1). Recently, in silico analysis suggested that another catecholamine neurotransmitter, dopamine, can bind to QseC. However, no effect in vitro was measured[139]. This study of interkingdom signaling through hormones has been named “microbial endocrinology”[140].
Interestingly, there is evidence that human epithelial cells can produce AI-2 mimicking molecules. The study was conducted on Caco-2 intestinal epithelial cells, and the authors showed that an AI-2 mimic is produced not only when cells are in contact with bacteria but also after TJ disruption by calcium deprivation or DSS treatment[45]. This emphasizes how much AI-2 is a universal language between Gram-positive and Gram-negative bacteria and the host (Figure 3).
Hosts have also developed defense tools against QS, leading to a mechanism named quorum quenching. Mammals can synthesize enzymes named paraoxonases (PONs) that hydrolyze the lactone ring of long-chain AHLs[141] (Figure 3). There are three types of PONs (PON 1, PON2, PON3) that are highly conserved across species, and PON2 has greater activity on AHLs[142]. It has been demonstrated that PON2 is more highly expressed in the human jejunum than in other parts of the intestine[143]. Interestingly, PON1 and PON3 are expressed at lower levels in patients with Crohn’s disease and ulcerative colitis than in healthy subjects[144]. A case-control study has also shown that carriage of the PON1 R192 allele in Ashkenazi Jewish may confer protection against the development of IBD. This allele was significantly less common among IBD Ashkenazi patients, with a significant odds ratio of 0.61[145].
Gut dysbiosis is an imbalance in the composition of microorganisms inside the digestive tract, especially described with bacteria. This dysregulation has been shown to be a preponderant risk factor in several digestive and extra digestive diseases[146-149]. For example, in IBD and recurrent C. difficile infections, it is well known that the over- and underrepresentation of certain phyla can lead to a pathologic state[150,151]. Modulating the gut microbiota may be the key to treating or even preventing such diseases by restoring normobiosis. Fecal microbiota transplantation is now commonly used in the setting of C. difficile infections[152,153]. However, the lack of standardization and the safety and quality issues of this procedure call for the development of new strategies.
Theoretically, AHLs remain good candidates in this approach using natural molecules from QS to modulate microbiota composition and gut inflammation. As seen above, AHL signaling may involve different pathways that contribute to controlling intestinal inflammation, such as inhibition of NF-κB, modulation, inhibition of MAPK activation, increase in regulatory T cell induction, decrease in proinflammatory cytokines, and modulation of junctional complexes in the epithelial barrier. Indeed, using QS molecules could play a role in both components (gut microbiota and host responses) of gut ecosystem disorders observed in metabolic and inflammatory diseases. AHL-based QS devices already exist as therapeutic applications for the dynamic control of Gram-negative bacterial populations, especially in infectious diseases. Other QS molecules could be extended as potential clinical therapies for diseases related to the gut microbiota that involve biofilm formation and antibiotic resistance[154]. Research efforts must investigate the potential of this new trial.
In addition to therapeutic applications, one could consider QS molecules as reliable biomarkers for dysbiosis-related chronic diseases such as IBD or CRC. Indeed, it has been shown that the presence of some AI-1 QS molecules in the gut ecosystem directly correlates with bacterial group size[39]. AHLs could represent a biomarker of the bacterial level population acting as a magnifying glass for dysbiosis. In addition, AI-2 concentration increased during adenomas to colorectal transition and CRC progression[41]. This opens the perspective for using the QS system as a biomarker for the prevention and follow-up of chronic diseases.
Knowing which commensal bacteria carry QS systems, their site of production, their ability to be mobilized during dysbiosis, and their effect on the luminal or mucosal microenvironment are as many unresolved questions. The scientific community, together with gastroenterologists, needs to tackle these issues to pave the way for translation into clinical use. Future directions also involve designing dedicated QS derivatives targeting either the host cells or the bacterial compartment. Such QS derivatives have already been reported to control the epithelial cell inflammation pathway with a wider effect than natural 3-oxoC12:2 without bacterial-activating properties[92]. Using QS molecules as an approach to tackle the gut microbiota compartment has already been proven to be a successful strategy[42], thus leading to interesting perspectives. Considering the QS system as a new player in the gut ecosystem, it represents a control platform to shape the host's gut microbiota population and/or major physiological pathways.
In conclusion, the intestinal microbiota interacts mutually with epithelial and immune cells of the coevolved host in a beneficial, reciprocal relationship. The QS signaling of bacteria probably contributes substantially to establishing symbiotic interactions in certain dynamics of interaction between the different kingdoms. A better understanding of QS systems by researchers and gastroenterologists involved in describing and managing ecological disorders of the intestinal ecosystem is a new approach that opens up fascinating therapeutic opportunities.
We thank Rainteau D for his support and knowledge on quorum sensing. We acknowledge the Association François Aupetit for its unwavering support.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: French National Society of Gastroenterology; European Crohn's and Colitis Organisation.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dai YC S-Editor: Gao CC L-Editor: A P-Editor: Gao CC
| 1. | Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T; MetaHIT consortium, Bork P, Wang J, Ehrlich SD, Pedersen O. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2727] [Cited by in RCA: 3215] [Article Influence: 267.9] [Reference Citation Analysis (2)] |
| 2. | Sokol H, Seksik P. The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr Opin Gastroenterol. 2010;26:327-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 3. | Agus A, Clément K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 2021;70:1174-1182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 230] [Cited by in RCA: 725] [Article Influence: 181.3] [Reference Citation Analysis (0)] |
| 4. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J; MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9101] [Cited by in RCA: 7845] [Article Influence: 523.0] [Reference Citation Analysis (4)] |
| 5. | Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787-8803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1421] [Cited by in RCA: 1839] [Article Influence: 183.9] [Reference Citation Analysis (58)] |
| 6. | Coquant G, Grill JP, Seksik P. Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Front Immunol. 2020;11:1827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 7. | Prescott RD, Decho AW. Flexibility and Adaptability of Quorum Sensing in Nature. Trends Microbiol. 2020;28:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 8. | Wu L, Luo Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front Microbiol. 2021;12:611413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 9. | Nealson KH, Hastings JW. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979;43:496-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 274] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 10. | Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2025] [Cited by in RCA: 1851] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 11. | Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 683] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 12. | Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1301] [Cited by in RCA: 1108] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 13. | Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol. 2001;41:1015-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 168] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Du YL, Shen XL, Yu P, Bai LQ, Li YQ. Gamma-butyrolactone regulatory system of Streptomyces chattanoogensis links nutrient utilization, metabolism, and development. Appl Environ Microbiol. 2011;77:8415-8426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Galloway WR, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev. 2011;111:28-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 455] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 16. | Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A. 1984;81:4154-4158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 452] [Cited by in RCA: 418] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 17. | Stevens AM, Dolan KM, Greenberg EP. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc Natl Acad Sci U S A. 1994;91:12619-12623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 142] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 18. | Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2008;2:345-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 208] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 19. | Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A. 2003;100:8951-8956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 620] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 20. | Carlson-Banning KM, Sperandio V. Enterohemorrhagic Escherichia coli outwits hosts through sensing small molecules. Curr Opin Microbiol. 2018;41:83-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Walters M, Sircili MP, Sperandio V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668-5681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 142] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 22. | Kim CS, Gatsios A, Cuesta S, Lam YC, Wei Z, Chen H, Russell RM, Shine EE, Wang R, Wyche TP, Piizzi G, Flavell RA, Palm NW, Sperandio V, Crawford JM. Characterization of Autoinducer-3 Structure and Biosynthesis in E. coli. ACS Cent Sci. 2020;6:197-206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 23. | Schauder S, Bassler BL. The languages of bacteria. Genes Dev. 2001;15:1468-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 306] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 24. | Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999;96:1639-1644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 686] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 25. | Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1159] [Cited by in RCA: 1051] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 26. | Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 401] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 27. | Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 394] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 28. | Defoirdt T. Amino acid-derived quorum sensing molecules controlling the virulence of vibrios (and beyond). PLoS Pathog. 2019;15:e1007815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34:426-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 537] [Cited by in RCA: 720] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 30. | Pena RT, Blasco L, Ambroa A, González-Pedrajo B, Fernández-García L, López M, Bleriot I, Bou G, García-Contreras R, Wood TK, Tomás M. Relationship Between Quorum Sensing and Secretion Systems. Front Microbiol. 2019;10:1100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 31. | Rumbaugh KP, Kaufmann GF. Exploitation of host signaling pathways by microbial quorum sensing signals. Curr Opin Microbiol. 2012;15:162-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 32. | Wang G, Huang S, Wang Y, Cai S, Yu H, Liu H, Zeng X, Zhang G, Qiao S. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci. 2019;76:3917-3937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 195] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 33. | Defoirdt T. Quorum-Sensing Systems as Targets for Antivirulence Therapy. Trends Microbiol. 2018;26:313-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 308] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 34. | Tobias NJ, Brehm J, Kresovic D, Brameyer S, Bode HB, Heermann R. New Vocabulary for Bacterial Communication. Chembiochem. 2020;21:759-768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 35. | Agodi A, Barchitta M, Cipresso R, Giaquinta L, Romeo MA, Denaro C. Pseudomonas aeruginosa carriage, colonization, and infection in ICU patients. Intensive Care Med. 2007;33:1155-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 80] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 36. | Bertrand X, Thouverez M, Talon D, Boillot A, Capellier G, Floriot C, Hélias JP. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med. 2001;27:1263-1268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 138] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 37. | Swearingen MC, Sabag-Daigle A, Ahmer BM. Are there acyl-homoserine lactones within mammalian intestines? J Bacteriol. 2013;195:173-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Kumari A, Pasini P, Deo SK, Flomenhoft D, Shashidhar H, Daunert S. Biosensing systems for the detection of bacterial quorum signaling molecules. Anal Chem. 2006;78:7603-7609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 39. | Landman C, Grill JP, Mallet JM, Marteau P, Humbert L, Le Balc'h E, Maubert MA, Perez K, Chaara W, Brot L, Beaugerie L, Sokol H, Thenet S, Rainteau D, Seksik P, Quévrain E; Saint Antoine IBD Network. Inter-kingdom effect on epithelial cells of the N-Acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS One. 2018;13:e0202587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 40. | Kaper JB, Sperandio V. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun. 2005;73:3197-3209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 120] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 41. | Li Q, Peng W, Wu J, Wang X, Ren Y, Li H, Peng Y, Tang X, Fu X. Autoinducer-2 of gut microbiota, a potential novel marker for human colorectal cancer, is associated with the activation of TNFSF9 signaling in macrophages. Oncoimmunology. 2019;8:e1626192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 42. | Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861-1871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 274] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 43. | Darkoh C, Plants-Paris K, Bishoff D, DuPont HL. Clostridium difficile Modulates the Gut Microbiota by Inducing the Production of Indole, an Interkingdom Signaling and Antimicrobial Molecule. mSystems. 2019;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 44. | Chen J, Ma M, Uzal FA, McClane BA. Host cell-induced signaling causes Clostridium perfringens to upregulate production of toxins important for intestinal infections. Gut Microbes. 2014;5:96-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 45. | Ismail AS, Valastyan JS, Bassler BL. A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe. 2016;19:470-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 112] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 46. | Pinheiro J, Lisboa J, Pombinho R, Carvalho F, Carreaux A, Brito C, Pöntinen A, Korkeala H, Dos Santos NMS, Morais-Cabral JH, Sousa S, Cabanes D. MouR controls the expression of the Listeria monocytogenes Agr system and mediates virulence. Nucleic Acids Res. 2018;46:9338-9352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 47. | Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2121] [Cited by in RCA: 2711] [Article Influence: 169.4] [Reference Citation Analysis (0)] |
| 48. | Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 880] [Article Influence: 62.9] [Reference Citation Analysis (0)] |
| 49. | Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 647] [Article Influence: 71.9] [Reference Citation Analysis (0)] |
| 50. | Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 1013] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 51. | Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1682] [Cited by in RCA: 2143] [Article Influence: 194.8] [Reference Citation Analysis (0)] |
| 52. | Wang L, Zhu L, Qin S. Gut Microbiota Modulation on Intestinal Mucosal Adaptive Immunity. J Immunol Res. 2019;2019:4735040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 53. | Holm A, Vikström E. Quorum sensing communication between bacteria and human cells: signals, targets, and functions. Front Plant Sci. 2014;5:309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 54. | Golovkine G, Reboud E, Huber P. Pseudomonas aeruginosa Takes a Multi-Target Approach to Achieve Junction Breach. Front Cell Infect Microbiol. 2017;7:532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Eum SY, Jaraki D, Bertrand L, András IE, Toborek M. Disruption of epithelial barrier by quorum-sensing N-3-(oxododecanoyl)-homoserine lactone is mediated by matrix metalloproteinases. Am J Physiol Gastrointest Liver Physiol. 2014;306:G992-G1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 56. | Halldorsson S, Gudjonsson T, Gottfredsson M, Singh PK, Gudmundsson GH, Baldursson O. Azithromycin maintains airway epithelial integrity during Pseudomonas aeruginosa infection. Am J Respir Cell Mol Biol. 2010;42:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 57. | Schwarzer C, Ravishankar B, Patanwala M, Shuai S, Fu Z, Illek B, Fischer H, Machen TE. Thapsigargin blocks Pseudomonas aeruginosa homoserine lactone-induced apoptosis in airway epithelia. Am J Physiol Cell Physiol. 2014;306:C844-C855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 58. | Vikström E, Bui L, Konradsson P, Magnusson KE. Role of calcium signalling and phosphorylations in disruption of the epithelial junctions by Pseudomonas aeruginosa quorum sensing molecule. Eur J Cell Biol. 2010;89:584-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 59. | Vikström E, Bui L, Konradsson P, Magnusson KE. The junctional integrity of epithelial cells is modulated by Pseudomonas aeruginosa quorum sensing molecule through phosphorylation-dependent mechanisms. Exp Cell Res. 2009;315:313-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 60. | Vikström E, Tafazoli F, Magnusson KE. Pseudomonas aeruginosa quorum sensing molecule N-(3 oxododecanoyl)-l-homoserine lactone disrupts epithelial barrier integrity of Caco-2 cells. FEBS Lett. 2006;580:6921-6928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 61. | Aguanno D, Coquant G, Postal BG, Osinski C, Wieckowski M, Stockholm D, Grill JP, Carrière V, Seksik P, Thenet S. The intestinal quorum sensing 3-oxo-C12:2 Acyl homoserine lactone limits cytokine-induced tight junction disruption. Tissue Barriers. 2020;8:1832877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 62. | Tao S, Xiong Y, Han D, Pi Y, Zhang H, Wang J. N-(3-oxododecanoyl)-l-homoserine lactone disrupts intestinal epithelial barrier through triggering apoptosis and collapsing extracellular matrix and tight junction. J Cell Physiol. 2021;236:5771-5784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 63. | Buckley A, Turner JR. Cell Biology of Tight Junction Barrier Regulation and Mucosal Disease. Cold Spring Harb Perspect Biol. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 479] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 64. | Su L, Nalle SC, Shen L, Turner ES, Singh G, Breskin LA, Khramtsova EA, Khramtsova G, Tsai PY, Fu YX, Abraham C, Turner JR. TNFR2 activates MLCK-dependent tight junction dysregulation to cause apoptosis-mediated barrier loss and experimental colitis. Gastroenterology. 2013;145:407-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 297] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 65. | Thoo L, Noti M, Krebs P. Keep calm: the intestinal barrier at the interface of peace and war. Cell Death Dis. 2019;10:849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 109] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 66. | France MM, Turner JR. The mucosal barrier at a glance. J Cell Sci. 2017;130:307-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 67. | Tao S, Luo Y, Bin He, Liu J, Qian X, Ni Y, Zhao R. Paraoxonase 2 modulates a proapoptotic function in LS174T cells in response to quorum sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone. Sci Rep. 2016;6:28778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 68. | Tao S, Niu L, Cai L, Geng Y, Hua C, Ni Y, Zhao R. N-(3-oxododecanoyl)-l-homoserine lactone modulates mitochondrial function and suppresses proliferation in intestinal goblet cells. Life Sci. 2018;201:81-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 69. | Tao S, Sun Q, Cai L, Geng Y, Hua C, Ni Y, Zhao R. Caspase-1-dependent mechanism mediating the harmful impacts of the quorum-sensing molecule N-(3-oxo-dodecanoyl)-l-homoserine lactone on the intestinal cells. J Cell Physiol. 2019;234:3621-3633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 70. | Taguchi R, Tanaka S, Joe GH, Maseda H, Nomura N, Ohnishi J, Ishizuka S, Shimizu H, Miyazaki H. Mucin 3 is involved in intestinal epithelial cell apoptosis via N-(3-oxododecanoyl)-L-homoserine lactone-induced suppression of Akt phosphorylation. Am J Physiol Cell Physiol. 2014;307:C162-C168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 71. | Sturm A, Dignass AU. Epithelial restitution and wound healing in inflammatory bowel disease. World J Gastroenterol. 2008;14:348-353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 237] [Cited by in RCA: 262] [Article Influence: 15.4] [Reference Citation Analysis (1)] |
| 72. | Karlsson T, Turkina MV, Yakymenko O, Magnusson KE, Vikström E. The Pseudomonas aeruginosa N-acylhomoserine lactone quorum sensing molecules target IQGAP1 and modulate epithelial cell migration. PLoS Pathog. 2012;8:e1002953. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 73. | Charlton TS, de Nys R, Netting A, Kumar N, Hentzer M, Givskov M, Kjelleberg S. A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography-mass spectrometry: application to a model bacterial biofilm. Environ Microbiol. 2000;2:530-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 236] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 74. | Fujiya M, Musch MW, Nakagawa Y, Hu S, Alverdy J, Kohgo Y, Schneewind O, Jabri B, Chang EB. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe. 2007;1:299-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 188] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 75. | Petrof EO, Ciancio MJ, Chang EB. Role and regulation of intestinal epithelial heat shock proteins in health and disease. Chin J Dig Dis. 2004;5:45-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 76. | Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A. 2010;107:228-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 640] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 77. | Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One. 2013;8:e80604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 284] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 78. | Shin JH, Lee YK, Shon WJ, Kim B, Jeon CO, Cho JY, Morse HC 3rd, Choi EY, Shin DM. Gut microorganisms and their metabolites modulate the severity of acute colitis in a tryptophan metabolism-dependent manner. Eur J Nutr. 2020;59:3591-3601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 79. | Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, Fleet JC, Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson JK, Dumas ME, Khanna KM, Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 734] [Article Influence: 66.7] [Reference Citation Analysis (0)] |
| 80. | Vyhlídalová B, Krasulová K, Pečinková P, Marcalíková A, Vrzal R, Zemánková L, Vančo J, Trávníček Z, Vondráček J, Karasová M, Mani S, Dvořák Z. Gut Microbial Catabolites of Tryptophan Are Ligands and Agonists of the Aryl Hydrocarbon Receptor: A Detailed Characterization. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 81. | Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay JM, Langella P, Xavier RJ, Sokol H. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1083] [Cited by in RCA: 1081] [Article Influence: 120.1] [Reference Citation Analysis (0)] |
| 82. | Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, Michel ML, Chong-Nguyen C, Roussel R, Straube M, Jegou S, McQuitty C, Le Gall M, da Costa G, Lecornet E, Michaudel C, Modoux M, Glodt J, Bridonneau C, Sovran B, Dupraz L, Bado A, Richard ML, Langella P, Hansel B, Launay JM, Xavier RJ, Duboc H, Sokol H. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018;28:737-749.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 401] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 83. | Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1512] [Cited by in RCA: 1724] [Article Influence: 143.7] [Reference Citation Analysis (1)] |
| 84. | Han B, Sheng B, Zhang Z, Pu A, Yin J, Wang Q, Yang K, Sun L, Yu M, Qiu Y, Xiao W, Yang H. Aryl Hydrocarbon Receptor Activation in Intestinal Obstruction Ameliorates Intestinal Barrier Dysfunction Via Suppression of MLCK-MLC Phosphorylation Pathway. Shock. 2016;46:319-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 85. | Postal BG, Ghezzal S, Aguanno D, André S, Garbin K, Genser L, Brot-Laroche E, Poitou C, Soula H, Leturque A, Clément K, Carrière V. AhR activation defends gut barrier integrity against damage occurring in obesity. Mol Metab. 2020;39:101007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 86. | Yu M, Wang Q, Ma Y, Li L, Yu K, Zhang Z, Chen G, Li X, Xiao W, Xu P, Yang H. Aryl Hydrocarbon Receptor Activation Modulates Intestinal Epithelial Barrier Function by Maintaining Tight Junction Integrity. Int J Biol Sci. 2018;14:69-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 180] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 87. | Ziegler N, Awwad K, Fisslthaler B, Reis M, Devraj K, Corada M, Minardi SP, Dejana E, Plate KH, Fleming I, Liebner S. β-Catenin Is Required for Endothelial Cyp1b1 Regulation Influencing Metabolic Barrier Function. J Neurosci. 2016;36:8921-8935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 88. | Taleb S. Tryptophan Dietary Impacts Gut Barrier and Metabolic Diseases. Front Immunol. 2019;10:2113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 159] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 89. | Wang J, Wang P, Tian H, Tian F, Zhang Y, Zhang L, Gao X, Wang X. Aryl hydrocarbon receptor/IL-22/Stat3 signaling pathway is involved in the modulation of intestinal mucosa antimicrobial molecules by commensal microbiota in mice. Innate Immun. 2018;24:297-306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 90. | Cotton JA, Platnich JM, Muruve DA, Jijon HB, Buret AG, Beck PL. Interleukin-8 in gastrointestinal inflammation and malignancy: induction and clinical consequences. Int J Interferon Cytokine Mediat Res. 2016;8:13-34. [RCA] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 91. | Ghezzal S, Postal BG, Quevrain E, Brot L, Seksik P, Leturque A, Thenet S, Carrière V. Palmitic acid damages gut epithelium integrity and initiates inflammatory cytokine production. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865:158530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 92. | Peyrottes A, Coquant G, Brot L, Rainteau D, Seksik P, Grill JP, Mallet JM. Anti-Inflammatory Effects of Analogues of N-Acyl Homoserine Lactones on Eukaryotic Cells. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 93. | Turkina MV, Vikström E. Bacteria-Host Crosstalk: Sensing of the Quorum in the Context of Pseudomonas aeruginosa Infections. J Innate Immun. 2019;11:263-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 94. | Telford G, Wheeler D, Williams P, Tomkins PT, Appleby P, Sewell H, Stewart GS, Bycroft BW, Pritchard DI. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-L-homoserine lactone has immunomodulatory activity. Infect Immun. 1998;66:36-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 297] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 95. | Smith RS, Harris SG, Phipps R, Iglewski B. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol. 2002;184:1132-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 243] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 96. | Ritchie AJ, Jansson A, Stallberg J, Nilsson P, Lysaght P, Cooley MA. The Pseudomonas aeruginosa quorum-sensing molecule N-3-(oxododecanoyl)-L-homoserine lactone inhibits T-cell differentiation and cytokine production by a mechanism involving an early step in T-cell activation. Infect Immun. 2005;73:1648-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 97. | Kim K, Kim SH, Lépine F, Cho YH, Lee GR. Global gene expression analysis on the target genes of PQS and HHQ in J774A.1 monocyte/macrophage cells. Microb Pathog. 2010;49:174-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 98. | Lin J, Cheng J, Wang Y, Shen X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front Cell Infect Microbiol. 2018;8:230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 99. | Li Y, Zhou H, Zhang Y, Chen C, Huang B, Qu P, Zeng J, Shunmei E, Zhang X, Liu J. N-3-(oxododecanoyl)-L-homoserine lactone promotes the induction of regulatory T-cells by preventing human dendritic cell maturation. Exp Biol Med (Maywood). 2015;240:896-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 100. | Skindersoe ME, Zeuthen LH, Brix S, Fink LN, Lazenby J, Whittall C, Williams P, Diggle SP, Froekiaer H, Cooley M, Givskov M. Pseudomonas aeruginosa quorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol Med Microbiol. 2009;55:335-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 79] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 101. | Jacobi CA, Schiffner F, Henkel M, Waibel M, Stork B, Daubrawa M, Eberl L, Gregor M, Wesselborg S. Effects of bacterial N-acyl homoserine lactones on human Jurkat T lymphocytes-OdDHL induces apoptosis via the mitochondrial pathway. Int J Med Microbiol. 2009;299:509-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 102. | Tateda K, Ishii Y, Horikawa M, Matsumoto T, Miyairi S, Pechere JC, Standiford TJ, Ishiguro M, Yamaguchi K. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect Immun. 2003;71:5785-5793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 266] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 103. | Kim K, Kim YU, Koh BH, Hwang SS, Kim SH, Lépine F, Cho YH, Lee GR. HHQ and PQS, two Pseudomonas aeruginosa quorum-sensing molecules, down-regulate the innate immune responses through the nuclear factor-kappaB pathway. Immunology. 2010;129:578-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 93] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 104. | Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Grauer DC, Lehmann M, Meijler MM, Janda KD, Ulevitch RJ. Modulation of gene expression via disruption of NF-kappaB signaling by a bacterial small molecule. Science. 2008;321:259-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 176] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 105. | Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Wood MR, Brogan AP, Lehmann M, Mee JM, Iwata K, Pan Q, Fearns C, Knaus UG, Meijler MM, Janda KD, Ulevitch RJ. N-(3-oxo-acyl)homoserine lactones signal cell activation through a mechanism distinct from the canonical pathogen-associated molecular pattern recognition receptor pathways. J Biol Chem. 2006;281:28822-28830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 107] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 106. | Wu R, Li X, Ma N, Jin X, Yuan X, Qu C, Tang H, Liu Z, Zhang Z. Bacterial Quorum Sensing Molecules Promote Allergic Airway Inflammation by Activating the Retinoic Acid Response. iScience. 2020;23:101288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 107. | Vikström E, Magnusson KE, Pivoriūnas A. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone stimulates phagocytic activity in human macrophages through the p38 MAPK pathway. Microbes Infect. 2005;7:1512-1518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 108. | Gaida MM, Dapunt U, Hänsch GM. Sensing developing biofilms: the bitter receptor T2R38 on myeloid cells. Pathog Dis. 2016;74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 109. | Lee RJ, Xiong G, Kofonow JM, Chen B, Lysenko A, Jiang P, Abraham V, Doghramji L, Adappa ND, Palmer JN, Kennedy DW, Beauchamp GK, Doulias PT, Ischiropoulos H, Kreindler JL, Reed DR, Cohen NA. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 2012;122:4145-4159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 428] [Article Influence: 32.9] [Reference Citation Analysis (1)] |
| 110. | Tizzano M, Gulbransen BD, Vandenbeuch A, Clapp TR, Herman JP, Sibhatu HM, Churchill ME, Silver WL, Kinnamon SC, Finger TE. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc Natl Acad Sci U S A. 2010;107:3210-3215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 321] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 111. | Rozengurt E. Taste receptors in the gastrointestinal tract. I. Bitter taste receptors and alpha-gustducin in the mammalian gut. Am J Physiol Gastrointest Liver Physiol. 2006;291:G171-G177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 162] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 112. | Carrai M, Steinke V, Vodicka P, Pardini B, Rahner N, Holinski-Feder E, Morak M, Schackert HK, Görgens H, Stemmler S, Betz B, Kloor M, Engel C, Büttner R, Naccarati A, Vodickova L, Novotny J, Stein A, Hemminki K, Propping P, Försti A, Canzian F, Barale R, Campa D. Association between TAS2R38 gene polymorphisms and colorectal cancer risk: a case-control study in two independent populations of Caucasian origin. PLoS One. 2011;6:e20464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 113. | Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, Chu J, Vila-Farres X, Kaplitt J, Rogoz A, Calle PY, Hunter C, Bitok JK, Brady SF. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 2017;549:48-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 353] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 114. | Cooley M, Chhabra SR, Williams P. N-Acylhomoserine lactone-mediated quorum sensing: a twist in the tail and a blow for host immunity. Chem Biol. 2008;15:1141-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 115. | Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, Wahli W, Li G, Williams SC, Rumbaugh KP. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J Bacteriol. 2008;190:4408-4415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 116. | Moura-Alves P, Puyskens A, Stinn A, Klemm M, Guhlich-Bornhof U, Dorhoi A, Furkert J, Kreuchwig A, Protze J, Lozza L, Pei G, Saikali P, Perdomo C, Mollenkopf HJ, Hurwitz R, Kirschhoefer F, Brenner-Weiss G, Weiner J 3rd, Oschkinat H, Kolbe M, Krause G, Kaufmann SHE. Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science. 2019;366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 115] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 117. | Lamas B, Natividad JM, Sokol H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol. 2018;11:1024-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 372] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 118. | Trikha P, Lee DA. The role of AhR in transcriptional regulation of immune cell development and function. Biochim Biophys Acta Rev Cancer. 2020;1873:188335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 119. | Li H, Li X, Song C, Zhang Y, Wang Z, Liu Z, Wei H, Yu J. Autoinducer-2 Facilitates Pseudomonas aeruginosa PAO1 Pathogenicity in Vitro and in Vivo. Front Microbiol. 2017;8:1944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 120. | Walters M, Sperandio V. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun. 2006;74:5445-5455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 113] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 121. | De Spiegeleer A, Elewaut D, Van Den Noortgate N, Janssens Y, Debunne N, Van Langenhove S, Govindarajan S, De Spiegeleer B, Wynendaele E. Quorum sensing molecules as a novel microbial factor impacting muscle cells. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 122. | Wynendaele E, Verbeke F, Stalmans S, Gevaert B, Janssens Y, Van De Wiele C, Peremans K, Burvenich C, De Spiegeleer B. Quorum Sensing Peptides Selectively Penetrate the Blood-Brain Barrier. PLoS One. 2015;10:e0142071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 123. | Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, Li T, Yin Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front Cell Infect Microbiol. 2018;8:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 752] [Cited by in RCA: 854] [Article Influence: 122.0] [Reference Citation Analysis (0)] |
| 124. | Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9:3294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1202] [Cited by in RCA: 1319] [Article Influence: 188.4] [Reference Citation Analysis (0)] |
| 125. | Alexeev EE, Lanis JM, Kao DJ, Campbell EL, Kelly CJ, Battista KD, Gerich ME, Jenkins BR, Walk ST, Kominsky DJ, Colgan SP. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am J Pathol. 2018;188:1183-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 362] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 126. | Dong F, Perdew GH. The aryl hydrocarbon receptor as a mediator of host-microbiota interplay. Gut Microbes. 2020;12:1859812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 127. | Lamas B, Hernandez-Galan L, Galipeau HJ, Constante M, Clarizio A, Jury J, Breyner NM, Caminero A, Rueda G, Hayes CL, McCarville JL, Bermudez Brito M, Planchais J, Rolhion N, Murray JA, Langella P, Loonen LMP, Wells JM, Bercik P, Sokol H, Verdu EF. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci Transl Med. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (1)] |
| 128. | Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 608] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 129. | Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 2019;51:27-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 2435] [Article Influence: 405.8] [Reference Citation Analysis (0)] |
| 130. | Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1054] [Cited by in RCA: 1235] [Article Influence: 102.9] [Reference Citation Analysis (2)] |
| 131. | Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 668] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 132. | Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1113] [Cited by in RCA: 1310] [Article Influence: 81.9] [Reference Citation Analysis (0)] |
| 133. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1498] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 134. | Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, Stein E, Vadivelu J, Roslani AC, Malik AA, Wanyiri JW, Goh KL, Thevambiga I, Fu K, Wan F, Llosa N, Housseau F, Romans K, Wu X, McAllister FM, Wu S, Vogelstein B, Kinzler KW, Pardoll DM, Sears CL. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014;111:18321-18326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 533] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 135. | Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1499] [Article Influence: 107.1] [Reference Citation Analysis (0)] |
| 136. | Li S, Konstantinov SR, Smits R, Peppelenbosch MP. Bacterial Biofilms in Colorectal Cancer Initiation and Progression. Trends Mol Med. 2017;23:18-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 112] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 137. | Wu J, Li K, Peng W, Li H, Li Q, Wang X, Peng Y, Tang X, Fu X. Autoinducer-2 of Fusobacterium nucleatum promotes macrophage M1 polarization via TNFSF9/IL-1β signaling. Int Immunopharmacol. 2019;74:105724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 138. | Bearson BL, Bearson SM. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog. 2008;44:271-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 141] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 139. | Luqman A, Kharisma VD, Ruiz RA, Götz F. In Silico and in Vitro Study of Trace Amines (TA) and Dopamine (DOP) Interaction with Human Alpha 1-Adrenergic Receptor and the Bacterial Adrenergic Receptor QseC. Cell Physiol Biochem. 2020;54:888-898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 140. | Lyte M. The role of microbial endocrinology in infectious disease. J Endocrinol. 1993;137:343-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 111] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 141. | Grandclément C, Tannières M, Moréra S, Dessaux Y, Faure D. Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev. 2016;40:86-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 384] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 142. | Teiber JF, Horke S, Haines DC, Chowdhary PK, Xiao J, Kramer GL, Haley RW, Draganov DI. Dominant role of paraoxonases in inactivation of the Pseudomonas aeruginosa quorum-sensing signal N-(3-oxododecanoyl)-L-homoserine lactone. Infect Immun. 2008;76:2512-2519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 122] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 143. | Levy E, Trudel K, Bendayan M, Seidman E, Delvin E, Elchebly M, Lavoie JC, Precourt LP, Amre D, Sinnett D. Biological role, protein expression, subcellular localization, and oxidative stress response of paraoxonase 2 in the intestine of humans and rats. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1252-G1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 144. | Rothem L, Hartman C, Dahan A, Lachter J, Eliakim R, Shamir R. Paraoxonases are associated with intestinal inflammatory diseases and intracellularly localized to the endoplasmic reticulum. Free Radic Biol Med. 2007;43:730-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 145. | Karban A, Hartman C, Eliakim R, Waterman M, Nesher S, Barnett-Griness O, Shamir R. Paraoxonase (PON)1 192R allele carriage is associated with reduced risk of inflammatory bowel disease. Dig Dis Sci. 2007;52:2707-2715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 146. | Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology. 2014;146:1547-1553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 329] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 147. | Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, Hirota K, Matsushita M, Furuta Y, Narazaki M, Sakaguchi N, Kayama H, Nakamura S, Iida T, Saeki Y, Kumanogoh A, Sakaguchi S, Takeda K. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016;68:2646-2661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 489] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 148. | Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B, Lequin O, Kharrat P, Thomas G, Rainteau D, Aubry C, Breyner N, Afonso C, Lavielle S, Grill JP, Chassaing G, Chatel JM, Trugnan G, Xavier R, Langella P, Sokol H, Seksik P. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut. 2016;65:415-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 573] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 149. | Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 912] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 150. | Sokol H, Jegou S, McQuitty C, Straub M, Leducq V, Landman C, Kirchgesner J, Le Gall G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Richard ML, Beaugerie L. Specificities of the intestinal microbiota in patients with inflammatory bowel disease and Clostridium difficile infection. Gut Microbes. 2018;9:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 151. | Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, Dekens J, Peters V, Voskuil MD, Visschedijk MC, van Dullemen HM, Keszthelyi D, Swertz MA, Franke L, Alberts R, Festen EAM, Dijkstra G, Masclee AAM, Hofker MH, Xavier RJ, Alm EJ, Fu J, Wijmenga C, Jonkers DMAE, Zhernakova A, Weersma RK. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 379] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 152. | Cammarota G, Ianiro G, Kelly CR, Mullish BH, Allegretti JR, Kassam Z, Putignani L, Fischer M, Keller JJ, Costello SP, Sokol H, Kump P, Satokari R, Kahn SA, Kao D, Arkkila P, Kuijper EJ, Vehreschild MJG, Pintus C, Lopetuso L, Masucci L, Scaldaferri F, Terveer EM, Nieuwdorp M, López-Sanromán A, Kupcinskas J, Hart A, Tilg H, Gasbarrini A. International consensus conference on stool banking for faecal microbiota transplantation in clinical practice. Gut. 2019;68:2111-2121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 319] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 153. | van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2582] [Cited by in RCA: 2678] [Article Influence: 223.2] [Reference Citation Analysis (0)] |
| 154. | Wu S, Liu J, Liu C, Yang A, Qiao J. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell Mol Life Sci. 2020;77:1319-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 155. | Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 311] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 156. | Autret N, Raynaud C, Dubail I, Berche P, Charbit A. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect Immun. 2003;71:4463-4471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 120] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 157. | Ohtani K, Yuan Y, Hassan S, Wang R, Wang Y, Shimizu T. Virulence gene regulation by the agr system in Clostridium perfringens. J Bacteriol. 2009;191:3919-3927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 158. | Sifri CD, Mylonakis E, Singh KV, Qin X, Garsin DA, Murray BE, Ausubel FM, Calderwood SB. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect Immun. 2002;70:5647-5650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 160] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 159. | Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 329] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 160. | Mok KC, Wingreen NS, Bassler BL. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 2003;22:870-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 192] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 161. | Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20:2754-2767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 172] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 162. | Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000-3009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 511] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 163. | Gambello MJ, Kaye S, Iglewski BH. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 251] [Article Influence: 7.8] [Reference Citation Analysis (1)] |
| 164. | Winson MK, Camara M, Latifi A, Foglino M, Chhabra SR, Daykin M, Bally M, Chapon V, Salmond GP, Bycroft BW. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:9427-9431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 371] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 165. | Chapon-Hervé V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:1169-1178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 98] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 166. | Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1999;96:11229-11234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 852] [Cited by in RCA: 809] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 167. | Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472-6480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 436] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 168. | Bredenbruch F, Geffers R, Nimtz M, Buer J, Häussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol. 2006;8:1318-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 211] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 169. | Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Cámara M, Williams P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14:87-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 366] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 170. | Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A. 2001;98:14613-14618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 289] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 171. | Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 445] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 172. | Lee J, Wu J, Deng Y, Wang J, Wang C, Chang C, Dong Y, Williams P, Zhang LH. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol. 2013;9:339-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 294] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 173. | Ng WL, Perez LJ, Wei Y, Kraml C, Semmelhack MF, Bassler BL. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol Microbiol. 2011;79:1407-1417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 106] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 174. | Walters M, Sperandio V. Quorum sensing in Escherichia coli and Salmonella. Int J Med Microbiol. 2006;296:125-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 152] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 175. | Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 764] [Cited by in RCA: 717] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 176. | Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 2002;99:3129-3134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 672] [Cited by in RCA: 631] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 177. | Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 618] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 178. | Bansal T, Jesudhasan P, Pillai S, Wood TK, Jayaraman A. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol. 2008;78:811-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 179. | Choi J, Shin D, Kim M, Park J, Lim S, Ryu S. LsrR-mediated quorum sensing controls invasiveness of Salmonella typhimurium by regulating SPI-1 and flagella genes. PLoS One. 2012;7:e37059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 180. | Bao L, Yu J, Zhong H, Huang D, Lu Q. Expression of toll-like receptors in T lymphocytes stimulated with N-(3-oxododecanoyl)-L-homoserine lactone from Pseudomonas aeruginosa. APMIS. 2017;125:553-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 181. | Glucksam-Galnoy Y, Sananes R, Silberstein N, Krief P, Kravchenko VV, Meijler MM, Zor T. The bacterial quorum-sensing signal molecule N-3-oxo-dodecanoyl-L-homoserine lactone reciprocally modulates pro- and anti-inflammatory cytokines in activated macrophages. J Immunol. 2013;191:337-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 182. | Zhang J, Gong F, Li L, Zhao M, Song J. Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl) homoserine lactone attenuates lipopolysaccharide-induced inflammation by activating the unfolded protein response. Biomed Rep. 2014;2:233-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 183. | Holm A, Magnusson KE, Vikström E. Pseudomonas aeruginosa N-3-oxo-dodecanoyl-homoserine Lactone Elicits Changes in Cell Volume, Morphology, and AQP9 Characteristics in Macrophages. Front Cell Infect Microbiol. 2016;6:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 184. | Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, Yarmush ML, Alaniz RC, Jayaraman A, Lee K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018;23:1099-1111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 434] [Article Influence: 72.3] [Reference Citation Analysis (1)] |
| 185. | Boontham P, Robins A, Chandran P, Pritchard D, Cámara M, Williams P, Chuthapisith S, McKechnie A, Rowlands BJ, Eremin O. Significant immunomodulatory effects of Pseudomonas aeruginosa quorum-sensing signal molecules: possible link in human sepsis. Clin Sci (Lond). 2008;115:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 186. | Karlsson T, Musse F, Magnusson KE, Vikström E. N-Acylhomoserine lactones are potent neutrophil chemoattractants that act via calcium mobilization and actin remodeling. J Leukoc Biol. 2012;91:15-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 187. | Zimmermann S, Wagner C, Müller W, Brenner-Weiss G, Hug F, Prior B, Obst U, Hänsch GM. Induction of neutrophil chemotaxis by the quorum-sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone. Infect Immun. 2006;74:5687-5692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 94] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 188. | Singh PK, Yadav VK, Kalia M, Sharma D, Pandey D, Agarwal V. Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxo-dodecanoyl)-L-homoserine lactone triggers mitochondrial dysfunction and apoptosis in neutrophils through calcium signaling. Med Microbiol Immunol. 2019;208:855-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 189. | Wagner C, Zimmermann S, Brenner-Weiss G, Hug F, Prior B, Obst U, Hänsch GM. The quorum-sensing molecule N-3-oxododecanoyl homoserine lactone (3OC12-HSL) enhances the host defence by activating human polymorphonuclear neutrophils (PMN). Anal Bioanal Chem. 2007;387:481-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 190. | Gupta RK, Chhibber S, Harjai K. Acyl homoserine lactones from culture supernatants of Pseudomonas aeruginosa accelerate host immunomodulation. PLoS One. 2011;6:e20860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 191. | Hooi DS, Bycroft BW, Chhabra SR, Williams P, Pritchard DI. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect Immun. 2004;72:6463-6470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 138] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 192. | Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, Cella M, Gordon JI, Hsieh CS, Colonna M. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science. 2017;357:806-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 639] [Article Influence: 79.9] [Reference Citation Analysis (0)] |
| 193. | Aoki R, Aoki-Yoshida A, Suzuki C, Takayama Y. Indole-3-Pyruvic Acid, an Aryl Hydrocarbon Receptor Activator, Suppresses Experimental Colitis in Mice. J Immunol. 2018;201:3683-3693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 194. | Ritchie AJ, Yam AO, Tanabe KM, Rice SA, Cooley MA. Modification of in vivo and in vitro T- and B-cell-mediated immune responses by the Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone. Infect Immun. 2003;71:4421-4431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 75] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 195. | Smith RS, Fedyk ER, Springer TA, Mukaida N, Iglewski BH, Phipps RP. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappa B and activator protein-2. J Immunol. 2001;167:366-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 210] [Article Influence: 8.8] [Reference Citation Analysis (0)] |