Published online Nov 7, 2021. doi: 10.3748/wjg.v27.i41.7041
Peer-review started: April 29, 2021
First decision: June 17, 2021
Revised: July 2, 2021
Accepted: October 14, 2021
Article in press: October 14, 2021
Published online: November 7, 2021
Processing time: 190 Days and 14.8 Hours
The human gut microbiome has gained increasing attention over the past two decades. Several findings have shown that this complex and dynamic microbial ecosystem can contribute to the maintenance of host health or, when subject to imbalances, to the pathogenesis of various enteric and non-enteric diseases. This scoping review summarizes the current knowledge on how the gut microbiota and microbially-derived compounds affect host metabolism, especially in the context of obesity and related disorders. Examples of microbiome-based targeted intervention strategies that aim to restore and maintain an eubiotic layout are then discussed. Adjuvant therapeutic interventions to alleviate obesity and associated comorbidities are traditionally based on diet modulation and the supplementation of prebiotics, probiotics and synbiotics. However, these approaches have shown only moderate ability to induce sustained changes in the gut microbial ecosystem, making the development of innovative and tailored microbiome-based intervention strategies of utmost importance in clinical practice. In this regard, the administration of next-generation probiotics and engineered microbiomes has shown promising results, together with more radical intervention strategies based on the replacement of the dysbiotic ecosystem by means of fecal microbiota transplantation from healthy donors or with the introduction of synthetic communities specifically designed to achieve the desired therapeutic outcome. Finally, we provide a perspective for future translational investigations through the implementation of bioinformatics approaches, including machine and deep learning, to predict health risks and therapeutic outcomes.
Core Tip: The gut microbiome (GM) has gained increasing attention in recent years due to its key role in contributing to host health, potentially serving as a target for personalized precision medicine. This review summarizes the current evidence for the involvement of the GM in the regulation of various pathophysiological aspects, particularly in obesity and related comorbidities. The influence of diet and the molecules produced by commensal microorganisms is discussed, together with traditional and innovative microbiome-based strategies in the prevention and treatment of obesity, up to the development of machine and deep learning bioinformatics tools for the prediction of health risks and therapeutic outcomes.
- Citation: Barone M, D'Amico F, Fabbrini M, Rampelli S, Brigidi P, Turroni S. Over-feeding the gut microbiome: A scoping review on health implications and therapeutic perspectives. World J Gastroenterol 2021; 27(41): 7041-7064
- URL: https://www.wjgnet.com/1007-9327/full/v27/i41/7041.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i41.7041
Over the past two decades, the trillion-member community that resides in the human gastrointestinal tract (i.e., the gut microbiome-GM) has emerged as a key regulator of host physiology, supporting overall host health or, vice versa, contributing to triggering and sustaining pathological conditions when altered. The wide range of metabolic activities and the multiple levels of bidirectional interaction with the host strongly support GM as a strategic therapeutic target, laying the foundations for the development of innovative microbiome-tailored intervention strategies aimed at restoring an eubiotic layout. In parallel, advances in sequencing techniques and bioinformatics tools are proving crucial to deepen our understanding of the complex interactions established by the GM with the host, as well as to rationally fine-tune and successfully translate personalized microbiome-based interventions into clinical practice.
Our scoping review aims to discuss the state of the art on research and application aspects related to the role of GM in obesity, a complex and multifactorial disease that represents a major health risk factor. In particular, we first discuss the influence of GM on human physiology and its contribution to the pathogenesis of numerous enteric and non-enteric diseases when imbalances occur. Next, we focus on evidence for a link between GM and obesity and discuss the growing literature on the impact of diet on GM structure, and the key role played by GM-produced or derived bioactive compounds [e.g., fatty acids, amines, bile acids (BAs) and neuroactive metabolites] in affecting host physiology and metabolism, at both local and systemic level. We then review the adjuvant interventions currently available to manipulate GM and alleviate the obesity phenotype, which are based on diet modulation and the supplementation of prebiotics, probiotics and synbiotics. Since these traditional approaches have shown only moderate ability to induce sustained change in the complex and dynamic GM ecosystem, we stress the need to develop innovative and tailored microbiome-based intervention strategies, such as the administration of next-generation probiotics (NGPs) and engineered microbiomes, to be organically integrated into clinical practice. More radical approaches involving replacement of the dysbiotic microbial ecosystem through fecal microbiota transplantation (FMT) or the infusion of synthetic microbial communities, rationally designed to meet the desired therapeutic outcome and patient needs, are also discussed as promising alternatives for personalized clinical applications. Finally, we provide a perspective for future translational investigations through the implementation of bioinformatics approaches, including machine and deep learning, to predict health risks and therapeutic outcomes. Current clinical practice is increasingly leveraging new artificial intelligence technologies and refined bioinformatics approaches, but still little has been done in relation to GM. Therefore, we conclude by discussing the possibilities offered by machine and deep learning for the development of microbiome-based strategies, especially in the context of obesity and related morbidities. All articles included in this review were identified through the PubMed platform, the full-text archive of biomedical and life sciences journal literature at the United States National Institutes of Health’s National Library of Medicine. The most pertinent and relevant articles for each aforementioned topic were selected and commented on.
From the development of next-generation sequencing approaches and their application to the microbiome field, the analysis of the intestinal microbial community has had a strong burst, as evidenced by > 3 million hits returned by typing “gut microbiota” on the NCBI database (as accessed on April 26, 2021). Of course, this is not surprising when one considers the plethora of actors and functions involved in the whole GM field[1,2]. Indeed, GM is composed of a multitude (over trillions) of microbial entities from all domains of life (i.e., bacteria, archaeabacteria and micro-eukaryotes), which encode a number of genes probably more than 500 times greater than the human genome. This genetic heritage, still largely underestimated, allows them to perform various functions recognized as instrumental to maintaining host homeostasis[3,4]. With specific regard to the bacterial counterpart, indisputably the most explored to date, despite the thousands of different species identified so far, most of them belong to 2 phyla, Firmicutes and Bacteroidetes, which together account for approximately 90% of an adult-like community, with Actinobacteria, Proteobacteria, Fusobacteria and Verrucomicrobia representing the most commonly found subdominant taxa[3]. This ecosystem is characterized by essential ecological features, such as stability, resistance and resilience, associated with high diversity and functional redundancy, the loss of which can lead to unhealthy microbe-microbe and microbe-host interactions[5]. Established at birth, GM develops structurally and functionally over time, in relation to the personal exposome (i.e., the totality of exposures that individuals experience in their lives), while always providing the host with a series of fundamental immunological and metabolic ecosystem services for a mutualistic GM-host relationship[6,7]. In particular, GM is known to act as a protective barrier against infectious threats (the so-called “colonization resistance” by occupying niches, taking up resources and producing antimicrobials) and play an active role in the development and modulation of immune responses[8,9]. On the other hand, GM is also called “metabolic organ” as it provides numerous bioactive molecules from the degradation of dietary compounds, which are the main actors of the well-known local and systemic functions attributed to GM, just to name a few: (1) Nutritional support for the intestinal epithelium; (2) Synthesis of vitamins and balance of energy intake; (3) Lipid and carbohydrate metabolism-related effects; (4) Immune system modulation; and (5) Enteric and central nervous system regulation through the gut-brain axis[10]. Moreover, recent findings on GM-drug interactions have indicated its role in influencing the response to treatments, including the occurrence of side effects, with potentially groundbreaking implications in precision medicine[11].
That said, it is not hard to imagine how strategic it is for our health to maintain this intricate and complex balance with our microbial inhabitants. Indeed, when this balance fails, the so-called dysbiosis is established, i.e., a disease-promoting or associated GM alteration[12]. Several endogenous (e.g., immune dysregulation and inflammation) and exogenous (unbalanced diet, antibiotic intake, pathogen infection, etc.) factors are capable of promoting GM dysbiosis, and several studies have tried to explain the mechanisms underlying these events[13-15]. Generally speaking, dysbiosis may present with one or more of the following characteristics: (1) Loss of biodiversity (a recognized hallmark of healthy gut); (2) Depletion of beneficial, health-associated taxa, typically short-chain fatty acid (SCFA) producers from the Lachnospiraceae and Ruminococcaceae families; and (3) Enrichment in pathogens or pathobionts[16]. To date, dysbiotic profiles have been associated with a plethora of enteric and non-enteric disorders, including metabolic, hepatic, respiratory, cardiovascular, immunological and oncological disorders, and are supposed to cause some of these[12]. However, it is of the utmost importance to corroborate claims on GM-related causality in human diseases, as only a few of them have been validated[17]. Regardless, a very hot topic in the field now is the development of GM modulatory strategies, to increase the resilience of healthy states (prevention) or overcome that of unhealthy states (treatment) to alleviate the disease phenotype and restore eubiosis.
The prevalence of overweight and obesity has dramatically risen over the past four decades[18]. Combined with polygenic host susceptibility, increased food consumption and sedentary lifestyles lay the foundation for a widespread obesity epidemic[19]. Recognized as a multifactorial disease, obesity represents a major risk factor for health, with dramatic consequences on quality of life and healthcare costs[20-22]. Comorbidities linked to obesity (e.g., diabetes, cardiovascular disease, cancer) are indeed among the leading causes of premature death, and researchers are striving to find more effective treatments for these conditions[23]. Over the past 15 years, pioneering studies have proposed GM as a key factor involved in energy storage and fat mass gain. Among the first, Bäckhed et al[24,25] demonstrated that germ-free mice gained less body weight and fat mass than conventional mice harboring a GM[24], as well as providing proof of concept by showing that the lack of a microbial ecosystem confers resistance to obesity induced by a high-fat diet[25]. In addition, a pivotal study by Turnbaugh et al[26] showed that the obese phenotype can be induced in lean mice by transferring the GM of obese animals[26], thus suggesting a causality between unbalanced GM and the development of obesity. Subsequently, the GM of obese individuals was shown to have reduced microbial richness and biodiversity, combined with an altered representation of the two main phyla, namely Bacteroidetes and Firmicutes, compared to lean subjects[27]. These findings have paved the way for several epidemiological studies focused on showing differences in the GM of obese and lean individuals, which have led to accumulating evidence of the GM role in mediating the impact of environmental factors on obesity pathogenesis[28-31]. As expected, given the high taxonomic level, not all studies have been successful in validating the association between the Firmicutes/Bacteroidetes ratio and obesity, and the biological relevance of this ratio is now highly controversial. On the other hand, greater resolution was gradually provided in the description of the bacterial composition, as well as in the understanding of the underlying mechanisms through which a dysbiotic layout might contribute to triggering host metabolic imbalances. For instance, species-level characterization of GM in twins highlighted a positive association of SCFA producers Eubacterium ventriosum and Roseburia intestinalis with obesity[32]. Similar correlations were observed for Collinsella spp.[33], which were also found to be overrepresented in type 2 diabetes (T2D) and atherosclerosis[34,35]. The hypothesized mechanisms include reduced expression of tight junction proteins, possibly leading to gut leakage and metabolic endotoxemia, as well as impaired cholesterol absorption, decreased hepatic glycogenesis and increased triglyceride synthesis[36,37]. It is therefore not surprising that Collinsella has been proposed as a target in future GM intervention studies for the improvement of metabolic parameters[38]. On the other hand, the most common methanogenic archaeon found in GM, Methanobrevibacter smithii, as well as butyrate producers, such as potentially heritable Oscillospira spp., were found to be more represented in lean individuals[39]. Bacteroides thetaiotaomicron was depleted as well in obesity-related GM configurations and inversely correlated with serum concentration of glutamate, a common food additive able to induce obesity and insulin resistance[40]. In vivo studies have highlighted the ability of this glutamate-fermenting commensal to protect against adiposity[40], suggesting its potential application in probiotic-based intervention strategies in obese individuals. Similarly, Parabacteroides goldsteinii, whose levels are reduced in high-fat diet-fed mice, has been proposed as an anti-obesogenic probiotic, capable of promoting adipose tissue thermogenesis and intestinal integrity, and reducing inflammation and insulin resistance[41]. Finally yet importantly, the mucin-degrading bacterium Akkermansia muciniphila has been repeatedly and consistently found to be inversely correlated with body fat mass, fasting blood glucose levels and subcutaneous adiposity in mice and humans[42,43], potentially representing a NGP or live biotherapeutic candidate for obesity treatment. In human studies, A. muciniphila has shown protective effects against gut permeability and endotoxemia, as well as improving glucose homeostasis and promoting better overall health[43]. Very recently, in a proof-of-concept exploratory study, Depommier et al[44] demonstrated that its daily oral administration, as live or pasteurized bacteria, for three months to overweight/obese insulin-resistant volunteers, was safe and well tolerated, and led to the improvement of multiple metabolic parameters[44]. However, it should also be remembered that microorganisms interact with each other in complex syntrophic networks, the understanding of which could help guide more rational preventive and therapeutic strategies. In an attempt to take a step forward in this direction, Tavella et al[45] recently identified a distinct GM compositional structure (with elevated proportions of Christensenellaceae, Porphyromonadaceae and Rikenellaceae) associated with reduced visceral adipose tissue and healthier metabolic profile in elderly Italians[45]. It has also been hypothesized that peculiar “steady states” of GM, combined with long-term dietary habits, may predict the development of childhood obesity[31].
Diet is a major driver of GM variation and undoubtedly plays a central role in promoting and maintaining GM diversity, a strategic element to ensure eubiosis and resilience. Indeed, GM can rapidly shift its composition and functionality in response to dietary changes, contributing to the generation of health-relevant metabolic outputs[46]. In recent years, interest in understanding the relationship between dietary habits, GM and host physiology has grown remarkably. Different dietary patterns, such as Western-type diets, vegetarian or vegan diets and Mediterranean-style diets have been explored and each has been found to be associated with quite distinct GM profiles, which obviously affect host metabolism in a distinct way[47]. In particular, the advent of a Westernized lifestyle, with the fast-paced globalization of food, excessive sanitation and modern medicines, has led to the introduction of a dietary pattern mostly based on saturated fat, sugar and salt, and dramatically low in fiber, otherwise known as “microbiota-accessible carbohydrates”[13]. Several studies have shown that this type of diet is associated with alterations in the GM structure and functionality, in particular enrichment in mucus degraders, bile-tolerant and antibiotic-resistant species (“BloSSUM”, i.e., bloom or selected in societies of urbanization/modernization) and the loss of diversity, ancestral fiber-degrading taxa and related functions (“VANISH”, i.e., volatile and/or associated negatively with industrialized societies of humans)[48]. This has collectively been referred to as the “microbiota insufficiency syndrome” and is supposed to result in broad dysfunctions, including obesity and chronic inflammation, thus contributing to the emergence of non-communicable chronic diseases. In contrast, high-fiber dietary patterns are typically associated with highly diverse GMs, enriched in fibrolytic SCFA-producing bacteria, e.g., Ruminococcus, Faecalibacterium, Eubacterium and Roseburia, along with high fecal levels of SCFAs[49,50], to which multiple beneficial effects are attributed, as detailed below. Moreover, a microbial footprint of this dietary habit is the greater abundance of Prevotella, as also found in hunting-gathering and rural populations who consume a plant-based diet with unprocessed foods[51-53]. It is also worth noting that these dietary habits involve increased intake of polyphenols, which are known to have important GM-mediated health benefits[54], In a recent study in Italian individuals habitually following omnivore, vegetarian or vegan diets[55], we found that high-level adherence to a Mediterranean diet (rich in fruit, legumes and vegetables) was associated with beneficial GM and metabolome profiles, i.e., increased proportions of fiber-degrading bacteria, higher levels of fecal SCFAs and lower urinary levels of trimethylamine N-oxide (TMAO), a risk factor for cardiovascular disease, potentially helping to explain the effectiveness of this diet against obesity, T2D and other inflammatory disorders. Consistently, a 1-year Mediterranean diet intervention was found to positively modulate GM and metabolome (including lower production of secondary BAs and p-cresols) and reduce frailty in elderly subjects, thus paving the way for novel intervention strategies possibly based on Mediterranean diet-responsive taxa and/or metabolites[56]. All this considered, it is not surprising that in recent years, the Paleolithic diet, with a high intake of plant foods while totally excluding industrially processed products and refined sugars, has received a lot attention. Despite some concerns about its long-term adherence, especially due to the consumption of fat and meat[57], it appears to lead to improved metabolic parameters in obese and T2D patients[58] and high levels of GM diversity, similar to those found in traditional rural populations[59].
SCFAs, mainly acetate, propionate and butyrate, are the best-known examples of diet-derived microbial metabolites with several local and systemic functions. Indeed, the human genome encodes only a limited number of carbohydrate-active enzymes (CAZymes), thus requiring complementation by the GM for the degradation of otherwise indigestible dietary fibers (e.g., glycans, xylans, etc.)[60]. SCFAs are the end-products of fermentation of these complex polysaccharides. These metabolites can be variously beneficial to health, as local (butyrate) and peripheral (acetate and pro
On the other hand, branched-chain fatty acids, such as isobutyrate, 2-methylbutyrate and isovalerate, resulting from protein fermentation, have been associated with insulin resistance[63], probably through activation of mammalian target of rapamycin complex 1 (mTORC1)[64]. Protein metabolism by GM may also lead to other po
An admirable but unfortunate example of GM-host co-metabolism of dietary compounds is TMAO. Several gut microbes, e.g., Campylobacter, Shigella and Ruminococcus gnavus, can in fact convert choline and L-carnitine present in seafood, cheese, eggs and red meat, into trimethylamine (TMA) that, once absorbed, circulates to the liver where it is oxidized by host enzymes of the flavin monooxygenase family to TMAO[66-68]. TMAO has recently emerged as a candidate risk factor for cardiovas
The metabolism of BAs is another example of GM-host co-metabolism, through which the GM can modify the composition of the pool of primary and secondary BAs available to the host and therefore modulate their signaling, meaning not only the traditional role in fat absorption but various effects related to glucose, lipid and energy homeostasis, thermogenesis, insulin signaling, immune responses and inflammation[71].
Finally, it is worth mentioning that GM is increasingly recognized as a major, still largely underestimated, player in determining the toxicity of environmental pollutants[72]. With specific regard to diet, for example it can mediate the adverse metabolic effects of non-calorie artificial sweeteners, whose chronic consumption increases the risk of glucose intolerance[73]. In contrast, some GM components have been shown to be involved in the bioremediation of common food processing products, including Maillard reaction products and advanced glycation end-products, which have been implicated in a wide variety of civilization disorders, e.g., atherosclerosis and diabetes[74].
GM is well known to interact with the enteric and central nervous systems via the bidirectional gut-brain axis[75]. On the one hand, GM can be directly influenced by mental health, through the luminal secretion of endocrine mediators capable of interacting with microbial receptors, thus having direct effects on microbial gene expression and signaling mechanisms. At the same time, the microbial community can be indirectly modulated as a result of induced changes in the gut environment. On the other hand, as anticipated above, the GM is capable of producing neuroactive metabolites in a diet-dependent manner, e.g., SCFAs and conjugated fatty acids, which besides exerting peripheral effects can modulate the central nervous system through direct or indirect mechanisms, involving a complex network of neuroendocrine factors and their receptors. Interestingly, these interactions have been shown to affect central appetite, food reward signaling and energy balance[76-79]. GM has also been found to influence eating behaviors through vagal nerve stimulation and immune activation[80]. Perturbations of the gut-microbiome-brain axis could therefore compromise the inhibitory mechanisms normally involved in the regulation of food intake, resulting in unbalanced eating patterns towards cravings, overeating and hedonic-driven eating behavior[79,81]. Regarding neuroactive GM metabolites, it is worth noting that propionate modulates reward pathways by reducing anticipatory reward responses to high-energy foods via striatal pathways[82]. Moreover, tryptophan metabolites have been closely implicated in the modulation of gut-microbiome-brain interactions, and indole propionate has recently been associated with increased food addiction behaviors in obese individuals[83,84]. GM metabolites may also interact with the endocannabinoid system, affecting the homeostatic and hedonic control of appetite and food intake[85], while the dopaminergic mesolimbic system, involved in reward mechanisms and hypothesized to play an important role in the development of obesity, is influenced by GM through the modulation of gut hormone secretion[86]. Not least, it should be remembered that microbes can even produce neurotransmitters, e.g., serotonin and GABA, potentially affecting our mood and feeding[87,88].
In conclusion, particular GM layouts with distinct metabolic activities might have pleiotropic effects on host physiology, including eating behaviors, thus strongly contributing to the development of obesity and eating disorders. In this context, GM modulation or its replacement could be valuable tools to implement and increase the success of current preventive or therapeutic interventions against obesity and related comorbidities, as detailed below.
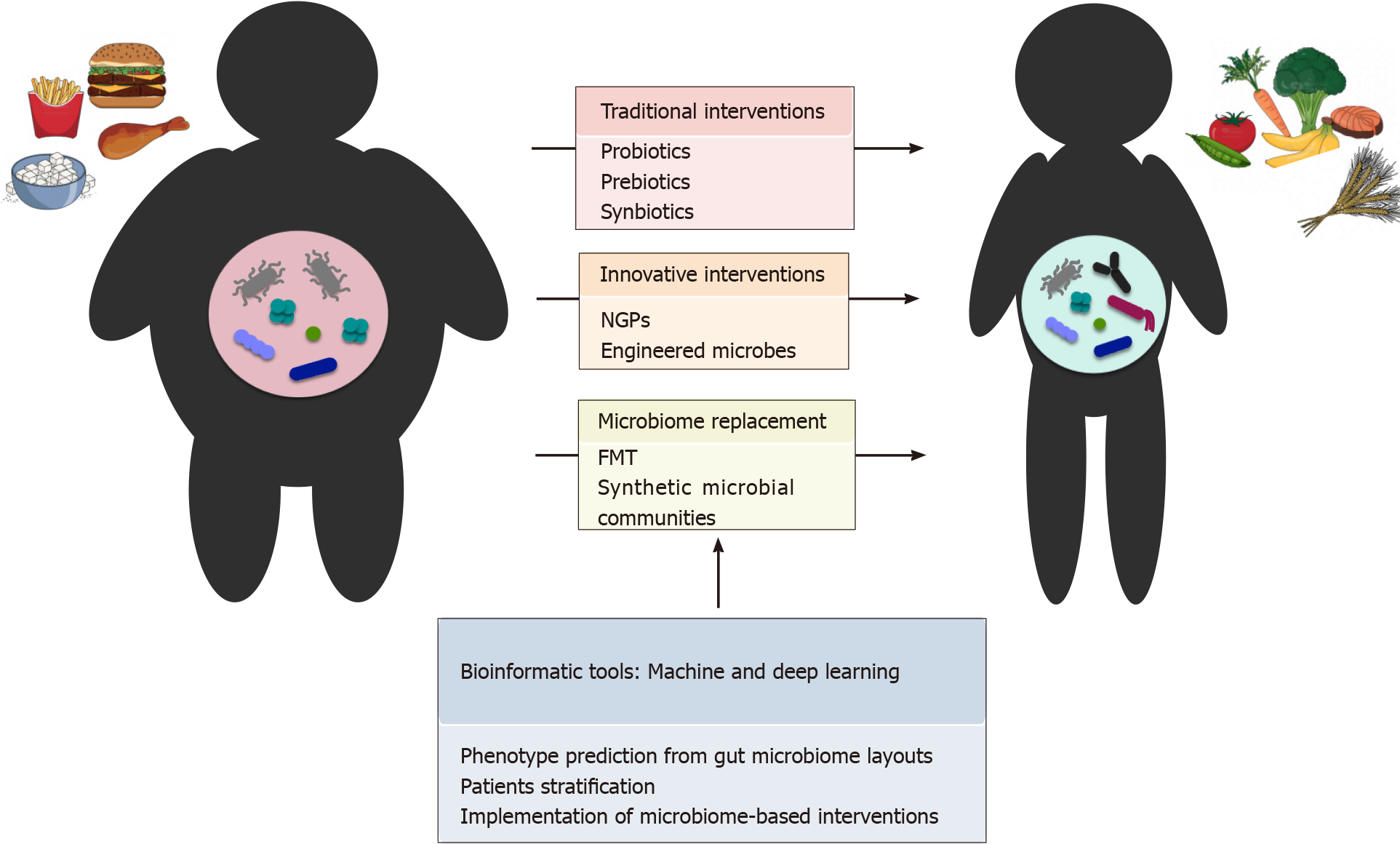
The main microbiome-based strategies that are or could be effective for the prevention and treatment of obesity are discussed below and summarized in Figure 1. In short, traditional (prebiotics, probiotics and synbiotics) and innovative (NGPs and engineered microbes) interventions were considered, along with microbiome replacement strategies, based on FMT and synthetic ecology approaches. Finally, in the next paragraph, the potential of bioinformatics tools, such as machine learning and deep learning, for health risk or outcome prediction is discussed.
Prebiotics are typically referred to as “a substrate that is selectively utilized by host microorganisms conferring a health benefit”[89]. Prebiotic supplementation has been proposed as a means of driving changes in GM while benefitting the host in the context of various disorders, including obesity, for improved glucose homeostasis and enteroendocrine L-cell activity. For instance, dietary supplementation with whole-grain barley and brown rice improved GM diversity, increasing the Firmi
Probiotics, i.e., “live microorganisms that, when administered in adequate amounts, confer a health benefit to the host”[111], represent one of the most widely used GM manipulation tools, for which an increasing number of clinical studies have been carried out in subjects with various pathological conditions, including obesity[112,113]. However, it should be noted that conflicting results have emerged in relation to the ability of probiotics to counterbalance weight loss and obesity-related features. In particular, two meta-analyses of randomized controlled clinical trials[114,115] found almost no efficiency in terms of weight and body mass index (BMI) reduction in obese individuals, especially in short-term interventions. On the other hand, the meta-analysis carried out by Zhang et al[116] on a large number of clinical trials reported substantially different results and a significant reduction in body weight and BMI, with consistent maximum outcomes with multi-strain product supplementation for at least 8 wk. Similar conclusions were drawn by John et al[117] in a similar meta-analysis, showing that probiotic administration was associated with a reduction in all considered parameters (i.e., body weight, BMI and fat mass). As expected, several studies have found strain-specific probiotic effects on body weight and metabolism, with only a few species belonging to Lactobacillus (e.g., L. acidophilus, L. casei, L. rhamnosus, L. reuteri) and Bifidobacterium (e.g., B. bifidum, B. lactis, B. longum) proven effective in overweight/obese individuals[104,118,119]. The greatest decreasing effect on BMI and body weight was reported with high doses of single-strain probiotics[117,120], although John et al[117] observed a considerable reduction in both parameters even at lower doses when interventions continued for more than 12 wk[117]. More recently, a study by our group showed that administering a multi-strain probiotic mixture along with a hypocaloric Mediterranean diet led to weight loss, improvement in oxidative stress markers and an increase in Akkermansia in elderly obese women, even in 15 d[33]. Regardless of the strain, the beneficial effects of probiotics in the treatment of obesity are also attributable to the following general mechanisms of action: (1) Antimicrobial activity, by inhibiting the growth of pathogenic microorganisms and exerting antagonistic effects against colonization of the intestinal mucosa and epithelium adherence; (2) Improvement of the barrier function, reducing intestinal permeability and increasing mucus production; and (3) Immunomodulation, through interaction with innate and adaptive components of the immune system[120]. Taken together, these mechanisms contribute to positively modulate the GM composition towards restoring a health-associated layout, which in turn can help alleviate host metabolic imbalances.
Synbiotics refer to “a mixture comprising live microorganisms and substrates selectively used by host microorganisms that confers a health benefit on the host”[121]. Accumulating evidence has reported the stronger effect of synbiotics in terms of GM modulation than either probiotics or prebiotics alone[122,123], with an overall improvement in lipid metabolism, glycemic status and inflammatory mediators[124,125]. Although the appropriate dose, duration of administration and the composition of a synbiotic product necessary to confer a health benefit are influenced by several factors (e.g., baseline GM layout, medications, habitual diet and lifestyle), the randomized clinical trial conducted by Dao et al[42] on 225 overweight and obese adults resulted in a 4.5% reduction in body fat mass when administering a combination of B. animalis subsp. Lactis 420 and polydextrose[42]. The control of body fat mass in overweight or obese individuals by the aforementioned synbiotic was also confirmed in a second clinical trial, along with a peculiar rearrangement in the GM composition that included an increased abundance of Akkermansia, Christensenellaceae and Methanobrevibacter, all taxa related to improved metabolic health and leanness[126]. GM modulation with increased proportions of potentially beneficial microbial groups (e.g., Lactobacillus) was also observed in a clinical trial in obese individuals following administration of a synbiotic consisting of a multi-strain probiotic formulation (i.e., B. lactis, B. longum, B. bifidum and L. acidophilus) and galactooligosaccharides as a prebiotic component[127].
In conclusion, prebiotics, probiotics and synbiotics are promising dietary agents for the modulation of human GM also in the context of obesity and metabolic disorders. However, given the still conflicting data in the scientific literature on probiotics, further studies are needed to rationally include their prescription as a preventive or therapeutic supplement for obesity.
Moving from the “one-size-fits-all” concept related to traditional probiotics, researchers are striving to thoroughly elucidate the role of each commensal member within the complex ecosystem of the human gastrointestinal tract in influencing host health. Compared to the modest ameliorative effects generally shown by traditional probiotics in obese individuals, emerging NGPs are beginning to reveal their great potential as novel preventive and therapeutic tools[128]. In this perspective, several studies have shown differences in the GM composition between obese and lean individuals, pointing out that an increased abundance of A. muciniphila could lead to an improvement in obesity and metabolic disorders[129,130]. Subsequently, the mechanism underlying the beneficial effect of A. muciniphila was investigated and the involvement of an immunomodulatory membrane protein “Amuc_1100” was proposed, which showed the same beneficial effects as live bacteria[131]. Cani et al[132] demonstrated the ability of this promising NGP to modulate the endocannabinoid system[132], a crucial regulatory system involved in controlling glucose metabolism in obesity, T2D and inflammatory conditions[132]. More recently, as anticipated above, the daily oral administration of live or pasteurized A. muciniphila, for three months to overweight/obese insulin-resistant volunteers, has been shown to be safe and well tolerated, while leading to improved metabolic parameters[44]. However, since some animal studies have reported an increase in A. muciniphila in multiple sclerosis[133] and Parkinson’s disease[134], further studies are needed to fully unravel its effects on host health.
Christensenella minuta also showed potential probiotic effects by ameliorating obesity and associated metabolic disorders through modulation of dysbiotic GM layouts[135], and its abundance was greater in lean individuals with low BMI[136]. However, Yang et al[137] recently highlighted potential pathogenic features (e.g., LPS-mediated triggering of a mild inflammatory response via NF-kB pathway) of C. minuta, suggesting that its application should be limited to therapeutic interventions focused on obesity control[137]. As mentioned above, P. goldsteinii is also a promising anti-obesity and anti-inflammatory NGP candidate. Being selectively enriched in the GM of mice fed a high-fat diet supplemented with oriental medicinal fungi, this commensal bacterium has been associated with increased adipose tissue thermogenesis, reduced levels of inflammation, enhanced intestinal integrity and amelioration of insulin resistance[41]. While promising, these in vivo results have not yet been translated into clinical trials. On the other hand, Faecalibacterium prausnitzii, one of the most promising NGPs due to its well characterized anti-inflammatory activity[138,139], showed a lower clade diversity in intestinal diseases and obese individuals[140]. In light of this finding, caution must be taken in selecting the most suitable strain for the development of therapeutic interventions. Despite the growing amount of NGP candidates so far isolated and characterized, further strain-level functional analyses are required to fully assess the underlying mechanisms by which they could confer health benefits to the host, before pushing them for clinical application.
Synthetic biology approaches have recently been exploited to address disease-specific mechanisms and meet medical needs by designing non-pathogenic and commensal bacteria to deliver therapeutic effectors[141,142]. Regarding the feasibility of using engineered probiotics to alleviate obesity-related characteristics, Long et al[143] demonstrated a decrease in body weight gain in overweight mice given an engineered Bifidobacterium strain secreting oxyntomodulin, an anorexigenic hormone that reduces appetite and food intake[143]. In a similar study, Chen et al[144] developed an engineered probiotic strain of Escherichia coli with increased secretion of N-acyl-phosphatidylethanolamine, the immediate precursor of the anorexigenic metabolite N-acylethanolamide, resulting in reduced weight gain and less accumulation of fat mass in mice fed a high-fat diet[144]. Another potential strategy to alleviate obesity and metabolic disorders has recently been proposed by Bai et al[145]. Administration of Bacillus subtilis SCK6 strain BsS-RS066550 engineered to increase butyric acid production resulted in decreased body weight and food intake in high-fat diet-fed mice, along with beneficial effects on insulin resistance, blood glucose and hepatic biochemistry[145]. In addition, Wang et al[146] showed that administration of genetically engineered Lactococcus lactis expressing GLP-1 significantly reduced body weight and blood glucose of obese mice fed a high-fat diet[146]. Anti-obesity mechanisms included the promotion of fatty acid oxidation and the restoration of GM biodiversity[146]. To date, numerous therapeutic interventions based on engineered live bacteria have entered the early or mid-stage of clinical development[141]. If successfully completed, such studies will be crucial in providing the missing proof of concept required to pave the way for a new class of precision therapeutics. Once their efficacy and risk ratio have been verified and approved for use in humans, engineered bacterial therapeutics will enable specific disease mechanisms and unmet medical needs to be addressed, even in obese patients.
Consisting of the transfer of microbes from healthy individuals to recipients hosting a dysbiotic GM layout with the aim of restoring eubiosis[147,148], FMT has attracted considerable attention in clinical practice, particularly for the treatment of recurrent infections by antibiotic-refractory Clostridioides difficile[149,150]. Recently, the potential of FMT in treating a large number of conditions, including obesity, has been explored. In a pivotal study, Ridaura et al[63] showed that mice receiving GM from obese individuals developed obesity, while those receiving GM from healthy individuals remained lean[63]. Sequencing analysis of post-treatment stools confirmed the successful engraftment of the donor microbiota, along with the transfer of functions associated with either “lean” or “obese” microbial communities. Shortly before, a clinical study on diabetic and obese adult males by Vrieze et al[151] demonstrated improved microbial diversity and insulin sensitivity, along with the expansion of Bacteroidetes and butyrate-producing taxa, following GM transplantation from lean donors[151].
As of April 2021, there were 15 registered clinical trials (ClinicalTrials.gov search terms: “gut microbiota”, “obesity” and “fecal microbiota transplantation”) on obese individuals undergoing FMT for the replacement of the obesogenic microbial community (Table 1). Of these, four have been completed and only two have made the results publicly available. Yu et al[152] performed FMT on 24 obese individuals with the aim of improving metabolic outcomes[152]. Administration of FMT capsules ensured engraftment, persisting for at least 12 wk after treatment, although no clinically significantly metabolic effects were observed. Allegretti et al[153] performed FMT on 22 obese, metabolically uncompromised patients, and although no significant changes in BMI occurred, a sustained shift of obesity-associated microbiomes towards the donor microbiome layout was observed, along with improved BA profiles and decreased fecal levels of taurocholic acid[153]. In a secondary analysis focused on the prevention of metabolic syndrome within the same patient group, the authors found a significant change in glucose and insulin levels after FMT[154]. Similar to other microbiome-based therapeutics, while promising, FMT is still at an early stage for the treatment of obesity and associated comorbidities. Therefore, it should be recognized as a separate pharmacological category, consisting of an entirely novel class of agents and requiring systematic research to fill knowledge gaps, thereby facilitating the development of standardized next-generation microbiota therapeutic interventions with improved safety and efficacy.
| Rank | Title | Status | Results | Condition | Intervention | Location | URL |
| 1 | Fecal Microbiota Transplantation for the Treatment of Obesity | Completed | Available | Obesity | FMT vs placebo | United States | https://ClinicalTrials.gov/show/NCT02741518 |
| 2 | Faecal Microbiota Transplantation in Obesity | Recruiting | Not available | Obesity | FMT vs placebo | Finland | https://ClinicalTrials.gov/show/NCT03391817 |
| 3 | Randomized Controlled Trial of Fecal Microbiota Transplantation in Severe Obesity | Enrolling by invitation | Not available | Obesity | FMT vs placebo | Norway | https://ClinicalTrials.gov/show/NCT03273855 |
| 4 | Fecal Microbiota Transplant (FMT) to Induce Weight Loss in Obese Subjects | Active, not recruiting | Not available | Obesity | FMT and mucosal microbiota assessment | China | https://ClinicalTrials.gov/show/NCT03789461 |
| 5 | FMT and Fiber in Patients With Metabolic Syndrome | Completed | Not available | Obesity, Metabolic Syndrome | FMT and dietary supplement with fiber (cellulose) vs placebo | Canada | https://ClinicalTrials.gov/show/NCT03727321 |
| 6 | Assessment of the Health Improvement of Obese Patients After Fecal Microbiota Transplantation (FMT) | Completed | Not available | Obesity, Type 1 and 2 Diabetes | FMT | Russian Federation | https://ClinicalTrials.gov/show/NCT04579263 |
| 7 | Fecal Microbiota Transplantation for Diabetes Mellitus Type II in Obese Patients | Unknown status | Not available | T2DM, Obesity | FMT and dietary intervention (high-fat low-fiber diet, sham diet or low-fat high-fiber diet) | Israel | https://ClinicalTrials.gov/show/NCT02346669 |
| 8 | Fecal Microbial Transplantation and Fiber Supplementation in Participants With Obesity and Metabolic Syndrome | Active, not recruiting | Not available | Obesity, Metabolic Syndrome | FMT and dietary supplement with fiber (cellulose) or FMT only vs placebo | Canada | https://ClinicalTrials.gov/show/NCT03477916 |
| 9 | Randomised Placebo-controlled Study of FMT to Impact Body Weight and Glycemic Control in Obese Subjects With T2DM | Active, not recruiting | Not available | T2DM, Obesity | FMT and lifestyle modification program or FMT only vs placebo | China | https://ClinicalTrials.gov/show/NCT03127696 |
| 10 | Fecal Microbiota Transplant for Improvement of Metabolism | Completed | Available | Obesity | FMT vs placebo | United States | https://ClinicalTrials.gov/show/NCT02530385 |
| 11 | The Role of Microbiome in Recurrent Obesity | Not yet recruiting | Not available | Obesity | FMT vs placebo | Israel | https://ClinicalTrials.gov/show/NCT04697550 |
| 12 | Effects of Fecal Microbiota Transplantation on Weight in Obese Patients With Non-alcoholic Fatty Liver Disease | Recruiting | Not available | NAFLD | FMT, dietary intervention and physical activity vs placebo | India | https://ClinicalTrials.gov/show/NCT04594954 |
| 13 | Proposal to Examine the Effect of Fecal Transplantation on Obesity | Unknown status | Not available | Obesity | FMT vs placebo | Israel | https://ClinicalTrials.gov/show/NCT02336789 |
| 14 | Safety and Efficacy of Fecal Microbiota Transplantation | Recruiting | Not available | IBD, IBS, Obesity, Metabolic Syndrome, Infections, Others | FMT | China | https://ClinicalTrials.gov/show/NCT04014413 |
| 15 | Transplantation of Microbes for Treatment of Metabolic Syndrome & NAFLD | Completed | Not available | Type 1 and 2 Diabetes, NAFLD, Obesity | FMT | Canada | https://ClinicalTrials.gov/show/NCT02496390 |
It is generally believed that the administration of multi-species microbial consortia is more useful than a single probiotic organism[155], as it probably retains some properties of the community structure, with community members continuously interacting and communicating with each other. Furthermore, a consortium of bacteria possesses a larger gene pool than monocultures, resulting in greater diversity in metabolic pathways. This richness is reflected in a greater ability of the consortium to perform more complex tasks than single organisms, thus exploiting the resources available in the surroundings more efficiently and better adapting to the environment, also through self-organization to form spatial patterns in response to substrates and metabolite gradients[156-160].
Synthetic ecology indicates the rational design of ecosystems, where two or more defined microbial populations are assembled in a well-characterized and controlled environment[161]. This approach requires in vitro controlled environments, biolo
Synthetic microbial communities are systems of known and trimmed-down complexity that can undergo experimental treatments and mathematical modelling, enabling a system-level understanding of the consortium[163,164]. These communities are not only a way to study how the microbial consortium structure emerges and the conditions necessary to generate specific interaction networks among its components, but they can also elucidate the overall function, resistance and resilience of microbial systems[10]. By the so-called microbial resource management[165,166], i.e., the management of consortium parameters such as richness, evenness, predation, abiotic factors, quorum sensing and spatial disposition, the community can be steered to the desired functionality, in order to obtain novel products and processes or potentially improve human health by transplanting the desired synthetic community into a recipient patient.
To date, many studies[167-169] have reported the in vitro assembly of synthetic GM communities with at least two bacterial strains, but clinical applications are still limited. Petrof et al[170] obtained a synthetic consortium consisting of 33 individual microbial species derived from a stool sample from a healthy donor and demonstrated the potential of such synthetic microbiota in the treatment of C. difficile infection. Furthermore, they reported that some of the administered bacteria that were forming the synthetic community stably colonized the recipient’s colon, as opposed to most commercially available probiotics, which only transiently colonize the intestine.
Despite its proven efficacy for the treatment of C. difficile infection, FMT hides some uncertainties that may be resolved by a synthetic stool replacement strategy. Indeed, the synthetic ecology approach has multiple advances over the canonical use of fecal matter from a donor: (1) The exact composition of the administered bacteria is known and can be reproduced; (2) The bacterial composition can be virtually tailored to the specific patient’s needs; (3) The absence of pathogens and viruses can be more reliably guaranteed, improving safety; and (4) The administered microorganisms forming the consortia can be selected based on their sensitivity to antimicrobials, resulting in further improvement of the safety profile.
Applications of synthetic communities have been reported in other fields, such as bioremediation[157] and chemical production[171,172]. Notwithstanding the potential of the synthetic ecology approach in GM replacement therapy, there is still some way to go before synthetic ecology can be translated into the clinic. One major limitation is the lack of a truly representative in vitro system to mimic the in vivo microbiota, but steps in this direction have been achieved by the gut-on-a-chip model[173] and the HuMiX system[173]. In addition, a large and well-documented collection of cultures of human microbiota members will be extremely useful for improving ecology experiments and building mathematical models. In this regard, since 2015, culturomics has led to the discovery of 232 novel human gut species[174] and it is expected that this number will increase in the coming years. The technique is based on the multiplexing of bacterial isolation conditions through serial addition of specific growth promoters and/or inhibitors, coupled with high-throughput identification with MALDI-TOF mass spectrometry, and is capable of overcoming the limitations of conventional single-medium strategies[175-177]. The resulting knowledge could then be used to improve our understanding of the complex gut ecosystem and rationally design efficient and sophisticated synthetic communities, tailored for the treatment of the disease of interest.
Machine learning and, in particular, deep learning have been the subject of intense media hype in recent years. Thanks to the explosion of available data and the rapid growth in the number and size of databases, they have accomplished nothing short of a revolution in the field of modern artificial intelligence, with notable progress in perceptual problems, such as facial and speech recognition[178,179], and have the potential to do the same in medical disciplines[180,181]. However, we are still exploring the full extent of what machine learning and deep learning can do and have just begun to apply them to a wide variety of problems outside of classical algorithms.
In light of the rapid increase in data from microbiome studies induced by the concomitant decrease in sequencing costs (< 10000 times in the past 10 years)[182], there are requirements to apply machine learning and deep learning algorithms to host-microbiome data, exploiting their associations in various diseases. Improved data analytical tools are needed to explore all the information contained in those datasets and identify key features that represent different aspects of the microbiome and that can be linked to host phenotypes. In particular, the possibility of predicting the patient’s phenotype from one’s GM is an integral part of personalized medicine, as it represents not only a way to overcome individual variability, but also a potential therapeutic target for pharmacological interventions. In this field, machine learning might provide new insights, by developing models capable of stratifying individual patients into therapeutic classes, thus paving the way for the fine-tuning of interventions based on the GM structure. An example of machine learning algorithms are Support Vector Machines (SVMs), which were implemented by Cui and Zhang[183] to classify metagenomic samples into inflammatory bowel disease (IBD) and non-IBD classes. More recently, Pasolli et al[184] used SVMs to predict diseases such as liver cirrhosis, colorectal cancer and IBD from fecal metagenomes. To date, deep learning is the most advanced machine learning technique for a variety of applications[185] and has already achieved several results in the microbiome field[186-189], including predicting the microbiome structure in terms of bacterial relative abundance and metabolic layout. Other machine learning methods include Ensemble Methods, which combine multiple classifiers for better performance. The best known ensemble method is Random Forest[190], which has been widely used in microbiome studies for patient stratification[191,192] and biomarker search[193,194].
In a prospective study on obesity in European children, Rampelli et al[31] underscored the importance of the microbiome–host–diet configuration as a possible predictor of obesity. In particular, GM was found to mediate dietary impact on individual metabolic and immunological homeostasis, which, in the context of other individual lifestyle and genetic variables, may be involved in the development of the multifactorial obese phenotype. Such results were based on experimental observations and no machine learning/deep learning algorithms were applied or developed. However, the study suggested that applying a machine learning algorithm to microbiome data could be a feasible method of predicting obesity, when other variables concerning host physiology, diet and lifestyle (e.g., physical activity) are included.
Some attempts have been made in recent years, with poor results, mainly due to a lack of data (often lifestyle and dietary information is not collected or collected in a non-systematic and non-standardized way). Specifically, Pasolli et al[184] developed a Random Forest-based approach that exclusively utilized metagenome data without success in predicting obesity and T2D. A few years later, Fernández-Navarro et al[195] have implemented several machine learning methods, such as decision tree-based methods, ensemble methods and SVMs, to identify predictors of obesity. Starting from serum free fatty acid levels, microbial quantitative polymerase chain reaction information and dietary intake interviews, their model revealed a non-obese profile related to serum eicosapentaenoic acid levels and Bacteroides amount in feces.
Nevertheless, this represents just the tip of the iceberg, as the full deployment of machine learning methods in the human GM sphere for full integration into the field of personalized precision medicine requires additional efforts. Indeed, machine learning often runs like black boxes, which makes it difficult to conduct feature selection. In addition, a large amount of data and computational power are required to train powerful and reliable machine learning-based algorithms. In general, novel technologies have dramatically increased our ability to characterize the human GM, but the way to effectively harness that information is uncertain and presents several key challenges. For example, there is a high need for dimensionality reduction to handle the information of hundreds of thousands of gene markers for just a few hundred samples. In this regard, neural encoder-decoder networks[196] based on a deep learning architecture have proved effective[187], but further efforts are still needed to fully exploit microbiome data. What is certain is that machine learning has already shown its great potential when applied to the microbiome field. In the next years, cutting-edge machine learning-based models might enable a further step towards the microbiome implementation in personalized precision medicine.
As discussed in the respective paragraphs, there are several limitations to the design and application of microbiome-based strategies in clinical routine, with particular reference to the prevention/treatment of obesity and related comorbidities. Although promising data come from the field of NGPs/engineered microbes, synthetic ecology and even FMT, it should be remembered that the evidence is still too little to support the clinical benefit of these novel microbiome modulation tools. Added to this are the sometimes inconsistent results on GM compositional and functional changes in the pathological context, and the lack of a full understanding of the underlying mecha
The emerging role of GM as a contributor to various pathological conditions is fascinating even if not yet easy to untangle. Diseases resulting from multiple factors, such as obesity and metabolic diseases in general, may be difficult to prevent or treat effectively solely relying on currently available therapies. In this scenario, gut microbes and their influence on the host constitute a piece of an intricate puzzle, to be exploited for novel integrated intervention strategies. In a systems biology approach, the advent of –omics technologies and the development of bioinformatics tools are pushing microbiome research to the next level, allowing to extrapolate general principles on community structure and translate the results into rationally designed, personalized microbiome-based interventions aimed at restructuring dysbiotic layouts, thereby contributing to the restoration and maintenance of host health.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cheungpasitporn W S-Editor: Fan JR L-Editor: Webster JR P-Editor: Fan JR
| 1. | Shanahan F, Ghosh TS, O'Toole PW. The healthy microbiome-what is the definition of a healthy gut microbiome? Gastroenterology. 2021;160:483-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 226] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 2. | Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2328] [Cited by in RCA: 2927] [Article Influence: 325.2] [Reference Citation Analysis (0)] |
| 3. | Candela M, Biagi E, Maccaferri S, Turroni S, Brigidi P. Intestinal microbiota is a plastic factor responding to environmental changes. Trends Microbiol. 2012;20:385-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 136] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 4. | Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1428] [Cited by in RCA: 2249] [Article Influence: 281.1] [Reference Citation Analysis (1)] |
| 5. | Fassarella M, Blaak EE, Penders J, Nauta A, Smidt H, Zoetendal EG. Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut. 2021;70:595-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 369] [Article Influence: 92.3] [Reference Citation Analysis (70)] |
| 6. | Robertson RC, Manges AR, Finlay BB, Prendergast AJ. The human microbiome and child growth - first 1000 days and beyond. Trends Microbiol. 2019;27:131-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 481] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 7. | Stanislawski MA, Dabelea D, Wagner BD, Iszatt N, Dahl C, Sontag MK, Knight R, Lozupone CA, Eggesbø M. Gut microbiota in the first 2 years of life and the association with body mass index at age 12 in a Norwegian birth cohort. mBio. 2018;9:e01751-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 8. | Iacob S, Iacob DG, Luminos LM. Intestinal microbiota as a host defense mechanism to infectious threats. Front Microbiol. 2018;9:3328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 109] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 9. | Garcia-Gutierrez E, Mayer MJ, Cotter PD, Narbad A. Gut microbiota as a source of novel antimicrobials. Gut Microbes. 2019;10:1-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 171] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 10. | Turroni S, Brigidi P, Cavalli A, Candela M. Microbiota-host transgenomic metabolism, bioactive molecules from the inside. J Med Chem. 2018;61:47-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 11. | Zimmermann M, Patil KR, Typas A, Maier L. Towards a mechanistic understanding of reciprocal drug-microbiome interactions. Mol Syst Biol. 2021;17:e10116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 12. | Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369-2379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1826] [Cited by in RCA: 2325] [Article Influence: 258.3] [Reference Citation Analysis (0)] |
| 13. | Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20:779-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 565] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 14. | Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, D'Amato M, Bonfiglio F, McDonald D, Gonzalez A, McClure EE, Dunklebarger MF, Knight R, Jansson JK. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 605] [Cited by in RCA: 820] [Article Influence: 102.5] [Reference Citation Analysis (0)] |
| 15. | Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, Hansen TH, Liang S, Feng Q, Zhang C, Pyl PT, Coelho LP, Yang H, Wang J, Typas A, Nielsen MF, Nielsen HB, Bork P, Vilsbøll T, Hansen T, Knop FK, Arumugam M, Pedersen O. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018;3:1255-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 508] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 16. | Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8:1784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 498] [Cited by in RCA: 671] [Article Influence: 83.9] [Reference Citation Analysis (0)] |
| 17. | Lv BM, Quan Y, Zhang HY. Causal inference in microbiome medicine: principles and applications. Trends Microbiol. 2021;29:736-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 18. | NCD Risk Factor Collaboration (NCD-RisC). Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19·2 million participants. Lancet. 2016;387:1377-1396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3952] [Cited by in RCA: 3519] [Article Influence: 391.0] [Reference Citation Analysis (0)] |
| 19. | McAllister EJ, Dhurandhar NV, Keith SW, Aronne LJ, Barger J, Baskin M, Benca RM, Biggio J, Boggiano MM, Eisenmann JC, Elobeid M, Fontaine KR, Gluckman P, Hanlon EC, Katzmarzyk P, Pietrobelli A, Redden DT, Ruden DM, Wang C, Waterland RA, Wright SM, Allison DB. Ten putative contributors to the obesity epidemic. Crit Rev Food Sci Nutr. 2009;49:868-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 455] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 20. | Berrington de Gonzalez A, Hartge P, Cerhan JR, Flint AJ, Hannan L, MacInnis RJ, Moore SC, Tobias GS, Anton-Culver H, Freeman LB, Beeson WL, Clipp SL, English DR, Folsom AR, Freedman DM, Giles G, Hakansson N, Henderson KD, Hoffman-Bolton J, Hoppin JA, Koenig KL, Lee IM, Linet MS, Park Y, Pocobelli G, Schatzkin A, Sesso HD, Weiderpass E, Willcox BJ, Wolk A, Zeleniuch-Jacquotte A, Willett WC, Thun MJ. Body-mass index and mortality among 1.46 million white adults. N Engl J Med. 2010;363:2211-2219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1837] [Cited by in RCA: 1708] [Article Influence: 113.9] [Reference Citation Analysis (0)] |
| 21. | Schwartz MW, Seeley RJ, Zeltser LM, Drewnowski A, Ravussin E, Redman LM, Leibel RL. Obesity pathogenesis: an Endocrine Society Scientific statement. Endocr Rev. 2017;38:267-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 454] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 22. | Blüher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. 2019;15:288-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1741] [Cited by in RCA: 2974] [Article Influence: 495.7] [Reference Citation Analysis (0)] |
| 23. | Wilkins LJ, Monga M, Miller AW. Defining dysbiosis for a cluster of chronic diseases. Sci Rep. 2019;9:12918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 212] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 24. | Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718-15723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4530] [Cited by in RCA: 4417] [Article Influence: 210.3] [Reference Citation Analysis (4)] |
| 25. | Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979-984. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2086] [Cited by in RCA: 1877] [Article Influence: 104.3] [Reference Citation Analysis (0)] |
| 26. | Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9796] [Cited by in RCA: 8762] [Article Influence: 461.2] [Reference Citation Analysis (1)] |
| 27. | Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070-11075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4639] [Cited by in RCA: 4467] [Article Influence: 223.4] [Reference Citation Analysis (1)] |
| 28. | Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 720] [Cited by in RCA: 968] [Article Influence: 107.6] [Reference Citation Analysis (0)] |
| 29. | Dugas LR, Fuller M, Gilbert J, Layden BT. The obese gut microbiome across the epidemiologic transition. Emerg Themes Epidemiol. 2016;13:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 30. | Castaner O, Goday A, Park YM, Lee SH, Magkos F, Shiow STE, Schröder H. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol. 2018;2018:4095789. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 280] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 31. | Rampelli S, Guenther K, Turroni S, Wolters M, Veidebaum T, Kourides Y, Molnár D, Lissner L, Benitez-Paez A, Sanz Y, Fraterman A, Michels N, Brigidi P, Candela M, Ahrens W. Pre-obese children's dysbiotic gut microbiome and unhealthy diets may predict the development of obesity. Commun Biol. 2018;1:222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 32. | Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, de Vos WM, Zoetendal EG. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013;7:707-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 271] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 33. | Cancello R, Turroni S, Rampelli S, Cattaldo S, Candela M, Cattani L, Mai S, Vietti R, Scacchi M, Brigidi P, Invitti C. Effect of short-term dietary intervention and probiotic mix supplementation on the gut microbiota of elderly obese women. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 34. | Candela M, Biagi E, Soverini M, Consolandi C, Quercia S, Severgnini M, Peano C, Turroni S, Rampelli S, Pozzilli P, Pianesi M, Fallucca F, Brigidi P. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br J Nutr. 2016;116:80-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 177] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 35. | Karlsson CL, Onnerfält J, Xu J, Molin G, Ahrné S, Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring). 2012;20:2257-2261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 400] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 36. | Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, Nelson H, Matteson EL, Taneja V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 590] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 37. | Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Dekker Nitert M; SPRING Trial Group. Connections between the gut microbiome and metabolic hormones in early pregnancy in overweight and obese women. Diabetes. 2016;65:2214-2223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 215] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 38. | Frost F, Storck LJ, Kacprowski T, Gärtner S, Rühlemann M, Bang C, Franke A, Völker U, Aghdassi AA, Steveling A, Mayerle J, Weiss FU, Homuth G, Lerch MM. A structured weight loss program increases gut microbiota phylogenetic diversity and reduces levels of Collinsella in obese type 2 diabetics: A pilot study. PLoS One. 2019;14:e0219489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 39. | Gophna U, Konikoff T, Nielsen HB. Oscillospira and related bacteria - from metagenomic species to metabolic features. Environ Microbiol. 2017;19:835-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 297] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 40. | Shen N, Caixàs A, Ahlers M, Patel K, Gao Z, Dutia R, Blaser MJ, Clemente JC, Laferrère B. Longitudinal changes of microbiome composition and microbial metabolomics after surgical weight loss in individuals with obesity. Surg Obes Relat Dis. 2019;15:1367-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 41. | Wu TR, Lin CS, Chang CJ, Lin TL, Martel J, Ko YF, Ojcius DM, Lu CC, Young JD, Lai HC. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut. 2019;68:248-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 42. | Dao MC, Clément K. Gut microbiota and obesity: concepts relevant to clinical care. Eur J Intern Med. 2018;48:18-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 43. | Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Microb Biotechnol. 2019;12:1109-1125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 484] [Article Influence: 80.7] [Reference Citation Analysis (1)] |
| 44. | Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, de Barsy M, Loumaye A, Hermans MP, Thissen JP, de Vos WM, Cani PD. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25:1096-1103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 774] [Cited by in RCA: 1429] [Article Influence: 238.2] [Reference Citation Analysis (0)] |
| 45. | Tavella T, Rampelli S, Guidarelli G, Bazzocchi A, Gasperini C, Pujos-Guillot E, Comte B, Barone M, Biagi E, Candela M, Nicoletti C, Kadi F, Battista G, Salvioli S, O'Toole PW, Franceschi C, Brigidi P, Turroni S, Santoro A. Elevated gut microbiome abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microbes. 2021;13:1-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 176] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 46. | Yadav M, Verma MK, Chauhan NS. A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol. 2018;200:203-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 47. | Lazar V, Ditu LM, Pircalabioru GG, Picu A, Petcu L, Cucu N, Chifiriuc MC. Gut microbiota, host organism, and diet trialogue in diabetes and obesity. Front Nutr. 2019;6:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 126] [Article Influence: 21.0] [Reference Citation Analysis (1)] |
| 48. | Sonnenburg JL, Sonnenburg ED. Vulnerability of the industrialized microbiota. Science. 2019;366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 191] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 49. | Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 998] [Cited by in RCA: 1723] [Article Influence: 191.4] [Reference Citation Analysis (0)] |
| 50. | Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1643] [Cited by in RCA: 1623] [Article Influence: 202.9] [Reference Citation Analysis (0)] |
| 51. | Ayeni FA, Biagi E, Rampelli S, Fiori J, Soverini M, Audu HJ, Cristino S, Caporali L, Schnorr SL, Carelli V, Brigidi P, Candela M, Turroni S. Infant and adult gut microbiome and metabolome in rural Bassa and urban settlers from Nigeria. Cell Rep. 2018;23:3056-3067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 52. | Martínez I, Stegen JC, Maldonado-Gómez MX, Eren AM, Siba PM, Greenhill AR, Walter J. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 2015;11:527-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 403] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 53. | Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, Fiori J, Gotti R, De Bellis G, Luiselli D, Brigidi P, Mabulla A, Marlowe F, Henry AG, Crittenden AN. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 792] [Cited by in RCA: 865] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 54. | Loo YT, Howell K, Chan M, Zhang P, Ng K. Modulation of the human gut microbiota by phenolics and phenolic fiber-rich foods. Compr Rev Food Sci Food Saf. 2020;19:1268-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 55. | De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O'Toole PW, Ercolini D. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812-1821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 844] [Cited by in RCA: 1042] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 56. | Tran TTT, Cousin FJ, Lynch DB, Menon R, Brulc J, Brown JR, O'Herlihy E, Butto LF, Power K, Jeffery IB, O'Connor EM, O'Toole PW. Prebiotic supplementation in frail older people affects specific gut microbiota taxa but not global diversity. Microbiome. 2019;7:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 57. | Genoni A, Christophersen CT, Lo J, Coghlan M, Boyce MC, Bird AR, Lyons-Wall P, Devine A. Long-term Paleolithic diet is associated with lower resistant starch intake, different gut microbiota composition and increased serum TMAO concentrations. Eur J Nutr. 2020;59:1845-1858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 58. | Otten J, Stomby A, Waling M, Isaksson A, Tellström A, Lundin-Olsson L, Brage S, Ryberg M, Svensson M, Olsson T. Benefits of a Paleolithic diet with and without supervised exercise on fat mass, insulin sensitivity, and glycemic control: a randomized controlled trial in individuals with type 2 diabetes. Diabetes Metab Res Rev. 2017;33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 59. | Barone M, Turroni S, Rampelli S, Soverini M, D'Amico F, Biagi E, Brigidi P, Troiani E, Candela M. Gut microbiome response to a modern Paleolithic diet in a Western lifestyle context. PLoS One. 2019;14:e0220619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 60. | El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 892] [Cited by in RCA: 1166] [Article Influence: 97.2] [Reference Citation Analysis (0)] |
| 61. | Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2533] [Cited by in RCA: 4209] [Article Influence: 467.7] [Reference Citation Analysis (0)] |
| 62. | Müller M, Hernández MAG, Goossens GH, Reijnders D, Holst JJ, Jocken JWE, van Eijk H, Canfora EE, Blaak EE. Circulating but not faecal short-chain fatty acids are related to insulin sensitivity, lipolysis and GLP-1 concentrations in humans. Sci Rep. 2019;9:12515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 203] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 63. | Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2415] [Cited by in RCA: 2725] [Article Influence: 227.1] [Reference Citation Analysis (0)] |
| 64. | Yoon MS. The emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients. 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 304] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 65. | Kolodziejczyk AA, Zheng D, Elinav E. Diet-microbiota interactions and personalized nutrition. Nat Rev Microbiol. 2019;17:742-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 555] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 66. | Chen X, Li HY, Hu XM, Zhang Y, Zhang SY. Current understanding of gut microbiota alterations and related therapeutic intervention strategies in heart failure. Chin Med J (Engl). 2019;132:1843-1855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 67. | Yang JJ, Shu XO, Herrington DM, Moore SC, Meyer KA, Ose J, Menni C, Palmer ND, Eliassen H, Harada S, Tzoulaki I, Zhu H, Albanes D, Wang TJ, Zheng W, Cai H, Ulrich CM, Guasch-Ferré M, Karaman I, Fornage M, Cai Q, Matthews CE, Wagenknecht LE, Elliott P, Gerszten RE, Yu D. Circulating trimethylamine N-oxide in association with diet and cardiometabolic biomarkers: an international pooled analysis. Am J Clin Nutr. 2021;113:1145-1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 68. | Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575-1584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2130] [Cited by in RCA: 2410] [Article Influence: 200.8] [Reference Citation Analysis (0)] |
| 69. | Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis Risk. Cell. 2016;165:111-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 947] [Cited by in RCA: 1377] [Article Influence: 153.0] [Reference Citation Analysis (0)] |
| 70. | Wilson A, McLean C, Kim RB. Trimethylamine-N-oxide: a link between the gut microbiome, bile acid metabolism, and atherosclerosis. Curr Opin Lipidol. 2016;27:148-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 71. | Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1317] [Cited by in RCA: 1852] [Article Influence: 205.8] [Reference Citation Analysis (0)] |
| 72. | Claus SP, Guillou H, Ellero-Simatos S. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes. 2016;2:16003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 437] [Cited by in RCA: 427] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 73. | Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, Israeli D, Zmora N, Gilad S, Weinberger A, Kuperman Y, Harmelin A, Kolodkin-Gal I, Shapiro H, Halpern Z, Segal E, Elinav E. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1146] [Cited by in RCA: 1360] [Article Influence: 123.6] [Reference Citation Analysis (0)] |
| 74. | Wolf AR, Wesener DA, Cheng J, Houston-Ludlam AN, Beller ZW, Hibberd MC, Giannone RJ, Peters SL, Hettich RL, Leyn SA, Rodionov DA, Osterman AL, Gordon JI. Bioremediation of a common product of food processing by a human gut bacterium. Cell Host Microbe. 2019;26:463-477.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 75. | Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 969] [Cited by in RCA: 1125] [Article Influence: 86.5] [Reference Citation Analysis (0)] |
| 76. | Fetissov SO. Role of the gut microbiota in host appetite control: bacterial growth to animal feeding behaviour. Nat Rev Endocrinol. 2017;13:11-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 243] [Article Influence: 30.4] [Reference Citation Analysis (1)] |
| 77. | Brown RM, Guerrero-Hreins E, Brown WA, le Roux CW, Sumithran P. Potential gut-brain mechanisms behind adverse mental health outcomes of bariatric surgery. Nat Rev Endocrinol. 2021;17:549-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 78. | Sandhu KV, Sherwin E, Schellekens H, Stanton C, Dinan TG, Cryan JF. Feeding the microbiota-gut-brain axis: diet, microbiome, and neuropsychiatry. Transl Res. 2017;179:223-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 335] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 79. | Torres-Fuentes C, Schellekens H, Dinan TG, Cryan JF. The microbiota-gut-brain axis in obesity. Lancet Gastroenterol Hepatol. 2017;2:747-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 421] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 80. | Dinan TG, Cryan JF. Mood by microbe: towards clinical translation. Genome Med. 2016;8:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 81. | Bliss ES, Whiteside E. The gut-brain axis, the human gut microbiota and their integration in the development of obesity. Front Physiol. 2018;9:900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 82. | Byrne CS, Chambers ES, Alhabeeb H, Chhina N, Morrison DJ, Preston T, Tedford C, Fitzpatrick J, Irani C, Busza A, Garcia-Perez I, Fountana S, Holmes E, Goldstone AP, Frost GS. Increased colonic propionate reduces anticipatory reward responses in the human striatum to high-energy foods. Am J Clin Nutr. 2016;104:5-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 137] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 83. | Osadchiy V, Labus JS, Gupta A, Jacobs J, Ashe-McNalley C, Hsiao EY, Mayer EA. Correlation of tryptophan metabolites with connectivity of extended central reward network in healthy subjects. PLoS One. 2018;13:e0201772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 84. | Dong TS, Mayer EA, Osadchiy V, Chang C, Katzka W, Lagishetty V, Gonzalez K, Kalani A, Stains J, Jacobs JP, Longo VD, Gupta A. A distinct brain-gut-microbiome profile exists for females with obesity and food addiction. Obesity (Silver Spring). 2020;28:1477-1486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 85. | Jager G, Witkamp RF. The endocannabinoid system and appetite: relevance for food reward. Nutr Res Rev. 2014;27:172-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 86. | Schellekens H, Finger BC, Dinan TG, Cryan JF. Ghrelin signalling and obesity: at the interface of stress, mood and food reward. Pharmacol Ther. 2012;135:316-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 155] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 87. | Newman S, Pascal L, Sadeghian K, Baldo BA. Sweetened-fat intake sensitizes gamma-aminobutyric acid-mediated feeding responses elicited from the nucleus accumbens shell. Biol Psychiatry. 2013;73:843-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 88. | Steiger H. Eating disorders and the serotonin connection: state, trait and developmental effects. J Psychiatry Neurosci. 2004;29:20-29. [PubMed] |
| 89. | Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2256] [Cited by in RCA: 3196] [Article Influence: 399.5] [Reference Citation Analysis (0)] |
| 90. | Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, Louk JA, Rose DJ, Kyureghian G, Peterson DA, Haub MD, Walter J. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 410] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 91. | Cani PD, Neyrinck AM, Maton N, Delzenne NM. Oligofructose promotes satiety in rats fed a high-fat diet: involvement of glucagon-like Peptide-1. Obes Res. 2005;13:1000-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 270] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 92. | Christiansen CB, Gabe MBN, Svendsen B, Dragsted LO, Rosenkilde MM, Holst JJ. The impact of short-chain fatty acids on GLP-1 and PYY secretion from the isolated perfused rat colon. Am J Physiol Gastrointest Liver Physiol. 2018;315:G53-G65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 249] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 93. | Delzenne NM, Cani PD, Daubioul C, Neyrinck AM. Impact of inulin and oligofructose on gastrointestinal peptides. Br J Nutr. 2005;93 Suppl 1:S157-S161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 212] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 94. | Flint A, Raben A, Rehfeld JF, Holst JJ, Astrup A. The effect of glucagon-like peptide-1 on energy expenditure and substrate metabolism in humans. Int J Obes Relat Metab Disord. 2000;24:288-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 90] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 95. | Brooks L, Viardot A, Tsakmaki A, Stolarczyk E, Howard JK, Cani PD, Everard A, Sleeth ML, Psichas A, Anastasovskaj J, Bell JD, Bell-Anderson K, Mackay CR, Ghatei MA, Bloom SR, Frost G, Bewick GA. Fermentable carbohydrate stimulates FFAR2-dependent colonic PYY cell expansion to increase satiety. Mol Metab. 2017;6:48-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 187] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 96. | Holz GG 4th, Kühtreiber WM, Habener JF. Pancreatic beta-cells are rendered glucose-competent by the insulinotropic hormone glucagon-like peptide-1(7-37). Nature. 1993;361:362-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 412] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 97. | Zou J, Chassaing B, Singh V, Pellizzon M, Ricci M, Fythe MD, Kumar MV, Gewirtz AT. Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring IL-22-mediated colonic health. Cell Host Microbe. 2018;23:41-53.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 408] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 98. | Bian Y, Wei J, Zhao C, Li G. Natural polyphenols targeting senescence: a novel prevention and therapy strategy for cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 99. | Fang C, Kim H, Yanagisawa L, Bennett W, Sirven MA, Alaniz RC, Talcott ST, Mertens-Talcott SU. Gallotannins and Lactobacillus plantarum WCFS1 mitigate high-fat diet-induced inflammation and induce biomarkers for thermogenesis in adipose tissue in gnotobiotic mice. Mol Nutr Food Res. 2019;63:e1800937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 100. | Noad RL, Rooney C, McCall D, Young IS, McCance D, McKinley MC, Woodside JV, McKeown PP. Beneficial effect of a polyphenol-rich diet on cardiovascular risk: a randomised control trial. Heart. 2016;102:1371-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 101. | Serreli G, Deiana M. In vivo formed metabolites of polyphenols and their biological efficacy. Food Funct. 2019;10:6999-7021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 102. | Nabavi SF, Bilotto S, Russo GL, Orhan IE, Habtemariam S, Daglia M, Devi KP, Loizzo MR, Tundis R, Nabavi SM. Omega-3 polyunsaturated fatty acids and cancer: lessons learned from clinical trials. Cancer Metastasis Rev. 2015;34:359-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 103. | Piazzi G, D'Argenio G, Prossomariti A, Lembo V, Mazzone G, Candela M, Biagi E, Brigidi P, Vitaglione P, Fogliano V, D'Angelo L, Fazio C, Munarini A, Belluzzi A, Ceccarelli C, Chieco P, Balbi T, Loadman PM, Hull MA, Romano M, Bazzoli F, Ricciardiello L. Eicosapentaenoic acid free fatty acid prevents and suppresses colonic neoplasia in colitis-associated colorectal cancer acting on Notch signaling and gut microbiota. Int J Cancer. 2014;135:2004-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 104. | Cao Y, Lu L, Liang J, Liu M, Li X, Sun R, Zheng Y, Zhang P. Omega-3 fatty acids and primary and secondary prevention of cardiovascular disease. Cell Biochem Biophys. 2015;72:77-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 105. | Chiesa G, Busnelli M, Manzini S, Parolini C. Nutraceuticals and bioactive components from fish for dyslipidemia and cardiovascular risk reduction. Mar Drugs. 2016;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 106. | Prossomariti A, Scaioli E, Piazzi G, Fazio C, Bellanova M, Biagi E, Candela M, Brigidi P, Consolandi C, Balbi T, Chieco P, Munarini A, Pariali M, Minguzzi M, Bazzoli F, Belluzzi A, Ricciardiello L. Short-term treatment with eicosapentaenoic acid improves inflammation and affects colonic differentiation markers and microbiota in patients with ulcerative colitis. Sci Rep. 2017;7:7458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 107. | Bellenger J, Bellenger S, Escoula Q, Bidu C, Narce M. N-3 polyunsaturated fatty acids: an innovative strategy against obesity and related metabolic disorders, intestinal alteration and gut microbiota dysbiosis. Biochimie. 2019;159:66-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 108. | White PJ, Marette A. Potential role of omega-3-derived resolution mediators in metabolic inflammation. Immunol Cell Biol. 2014;92:324-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 109. | Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chem Rev. 2011;111:5922-5943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 819] [Cited by in RCA: 760] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 110. | Li J, Li FR, Wei D, Jia W, Kang JX, Stefanovic-Racic M, Dai Y, Zhao AZ. Endogenous ω-3 polyunsaturated fatty acid production confers resistance to obesity, dyslipidemia, and diabetes in mice. Mol Endocrinol. 2014;28:1316-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 111. | Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4055] [Cited by in RCA: 5557] [Article Influence: 505.2] [Reference Citation Analysis (2)] |
| 112. | Rondanelli M, Faliva MA, Perna S, Giacosa A, Peroni G, Castellazzi AM. Using probiotics in clinical practice: where are we now? Gut Microbes. 2017;8:521-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 113. | Schütz F, Figueiredo-Braga M, Barata P, Cruz-Martins N. Obesity and gut microbiome: review of potential role of probiotics. Porto Biomed J. 2021;6:e111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 114. | Borgeraas H, Johnson LK, Skattebu J, Hertel JK, Hjelmesaeth J. Effects of probiotics on body weight, body mass index, fat mass and fat percentage in subjects with overweight or obesity: a systematic review and meta-analysis of randomized controlled trials. Obes Rev. 2018;19:219-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 115. | Park S, Bae JH. Probiotics for weight loss: a systematic review and meta-analysis. Nutr Res. 2015;35:566-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 116. | Zhang Q, Wu Y, Fei X. Effect of probiotics on body weight and body-mass index: a systematic review and meta-analysis of randomized, controlled trials. Int J Food Sci Nutr. 2015;67:571-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 117. | John GK, Wang L, Nanavati J, Twose C, Singh R, Mullin G. Dietary alteration of the gut microbiome and its impact on weight and fat mass: a systematic review and meta-analysis. Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 118. | Kadooka Y, Sato M, Imaizumi K, Ogawa A, Ikuyama K, Akai Y, Okano M, Kagoshima M, Tsuchida T. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur J Clin Nutr. 2010;64:636-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 469] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 119. | Xiao MW, Lin SX, Shen ZH, Luo WW, Wang XY. Systematic review with meta-analysis: the effects of probiotics in nonalcoholic fatty liver disease. Gastroenterol Res Pract. 2019;2019:1484598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 120. | Wang ZB, Xin SS, Ding LN, Ding WY, Hou YL, Liu CQ, Zhang XD. The potential role of probiotics in controlling overweight/obesity and associated metabolic parameters in adults: a systematic review and meta-analysis. Evid Based Complement Alternat Med. 2019;2019:3862971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 121. | Swanson KS, Gibson GR, Hutkins R, Reimer RA, Reid G, Verbeke K, Scott KP, Holscher HD, Azad MB, Delzenne NM, Sanders ME. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol. 2020;17:687-701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 733] [Article Influence: 146.6] [Reference Citation Analysis (0)] |
| 122. | Hadi A, Mohammadi H, Miraghajani M, Ghaedi E. Efficacy of synbiotic supplementation in patients with nonalcoholic fatty liver disease: a systematic review and meta-analysis of clinical trials: synbiotic supplementation and NAFLD. Crit Rev Food Sci Nutr. 2019;59:2494-2505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 123. | Khalesi S, Johnson DW, Campbell K, Williams S, Fenning A, Saluja S, Irwin C. Effect of probiotics and synbiotics consumption on serum concentrations of liver function test enzymes: a systematic review and meta-analysis. Eur J Nutr. 2018;57:2037-2053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 124. | Beserra BT, Fernandes R, do Rosario VA, Mocellin MC, Kuntz MG, Trindade EB. A systematic review and meta-analysis of the prebiotics and synbiotics effects on glycaemia, insulin concentrations and lipid parameters in adult patients with overweight or obesity. Clin Nutr. 2015;34:845-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 125. | McLoughlin RF, Berthon BS, Jensen ME, Baines KJ, Wood LG. Short-chain fatty acids, prebiotics, synbiotics, and systemic inflammation: a systematic review and meta-analysis. Am J Clin Nutr. 2017;106:930-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 145] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 126. | Hibberd AA, Yde CC, Ziegler ML, Honoré AH, Saarinen MT, Lahtinen S, Stahl B, Jensen HM, Stenman LK. Probiotic or synbiotic alters the gut microbiota and metabolism in a randomised controlled trial of weight management in overweight adults. Benef Microbes. 2019;10:121-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 116] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 127. | Sergeev IN, Aljutaily T, Walton G, Huarte E. Effects of synbiotic supplement on human gut microbiota, body composition and weight loss in obesity. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 190] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 128. | Chang CJ, Lin TL, Tsai YL, Wu TR, Lai WF, Lu CC, Lai HC. Next generation probiotics in disease amelioration. J Food Drug Anal. 2019;27:615-622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 210] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 129. | Zhao S, Liu W, Wang J, Shi J, Sun Y, Wang W, Ning G, Liu R, Hong J. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J Mol Endocrinol. 2017;58:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 196] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 130. | Zhai Q, Feng S, Arjan N, Chen W. A next generation probiotic, Akkermansia muciniphila. Crit Rev Food Sci Nutr. 2019;59:3227-3236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 283] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 131. | Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KC, Aalvink S, Martinez LO, Dumas ME, Maiter D, Loumaye A, Hermans MP, Thissen JP, Belzer C, de Vos WM, Cani PD. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 967] [Cited by in RCA: 1461] [Article Influence: 162.3] [Reference Citation Analysis (0)] |
| 132. | Cani PD, Geurts L, Matamoros S, Plovier H, Duparc T. Glucose metabolism: focus on gut microbiota, the endocannabinoid system and beyond. Diabetes Metab. 2014;40:246-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 133. | Cekanaviciute E, Pröbstel AK, Thomann A, Runia TF, Casaccia P, Katz Sand I, Crabtree E, Singh S, Morrissey J, Barba P, Gomez R, Knight R, Mazmanian S, Graves J, Cree BAC, Zamvil SS, Baranzini SE. Multiple Sclerosis-associated changes in the composition and immune functions of spore-forming bacteria. mSystems. 2018;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 134. | Haikal C, Chen QQ, Li JY. Microbiome changes: an indicator of Parkinson's disease? Transl Neurodegener. 2019;8:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 135. | Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. Human genetics shape the gut microbiome. Cell. 2014;159:789-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1911] [Cited by in RCA: 2151] [Article Influence: 215.1] [Reference Citation Analysis (0)] |
| 136. | Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 798] [Cited by in RCA: 717] [Article Influence: 79.7] [Reference Citation Analysis (0)] |
| 137. | Yang Y, Gu H, Sun Q, Wang J. Effects of Christensenella minuta lipopolysaccharide on RAW 264.7 macrophages activation. Microb Pathog. 2018;125:411-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 138. | Martín R, Bermúdez-Humarán LG, Langella P. Searching for the bacterial effector: the example of the multi-skilled commensal bacterium Faecalibacterium prausnitzii. Front Microbiol. 2018;9:346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 139. | Miquel S, Leclerc M, Martin R, Chain F, Lenoir M, Raguideau S, Hudault S, Bridonneau C, Northen T, Bowen B, Bermúdez-Humarán LG, Sokol H, Thomas M, Langella P. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. mBio. 2015;6:e00300-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 206] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 140. | De Filippis F, Pasolli E, Ercolini D. Newly explored Faecalibacterium diversity is connected to age, lifestyle, geography, and disease. Curr Biol. 2020;30:4932-4943.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 141. | Pedrolli DB, Ribeiro NV, Squizato PN, de Jesus VN, Cozetto DA; Team AQA Unesp at iGEM 2017. Engineering microbial living therapeutics: the synthetic biology toolbox. Trends Biotechnol. 2019;37:100-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 142. | Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, Kotula JW, Gerber GK, Way JC, Silver PA. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat Biotechnol. 2017;35:653-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 245] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 143. | Long RT, Zeng WS, Chen LY, Guo J, Lin YZ, Huang QS, Luo SQ. Bifidobacterium as an oral delivery carrier of oxyntomodulin for obesity therapy: inhibitory effects on food intake and body weight in overweight mice. Int J Obes (Lond). 2010;34:712-719. [PubMed] [DOI] [Full Text] |
| 144. | Chen Z, Guo L, Zhang Y, Walzem RL, Pendergast JS, Printz RL, Morris LC, Matafonova E, Stien X, Kang L, Coulon D, McGuinness OP, Niswender KD, Davies SS. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Invest. 2014;124:3391-3406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 191] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 145. | Bai L, Gao M, Cheng X, Kang G, Cao X, Huang H. Engineered butyrate-producing bacteria prevents high fat diet-induced obesity in mice. Microb Cell Fact. 2020;19:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 146. | Wang L, Chen T, Wang H, Wu X, Cao Q, Wen K, Deng KY, Xin H. Engineered Bacteria of MG1363-pMG36e-GLP-1 Attenuated obesity-induced by high fat diet in mice. Front Cell Infect Microbiol. 2021;11:595575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 147. | Khanna S. Microbiota replacement therapies: innovation in gastrointestinal care. Clin Pharmacol Ther. 2018;103:102-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 148. | Vindigni SM, Surawicz CM. Fecal microbiota transplantation. Gastroenterol Clin North Am. 2017;46:171-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 174] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 149. | Gupta A, Saha S, Khanna S. Therapies to modulate gut microbiota: past, present and future. World J Gastroenterol. 2020;26:777-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 150. | Monaghan TM, Seekatz AM, Markham NO, Yau TO, Hatziapostolou M, Jilani T, Christodoulou N, Roach B, Birli E, Pomenya O, Louie T, Lacy DB, Kim P, Lee C, Kao D, Polytarchou C. Fecal microbiota transplantation for recurrent Clostridioides difficile infection associates with functional alterations in circulating microRNAs. Gastroenterology. 2021;161:255-270.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 151. | Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913-6.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1881] [Cited by in RCA: 2016] [Article Influence: 155.1] [Reference Citation Analysis (0)] |
| 152. | Yu EW, Gao L, Stastka P, Cheney MC, Mahabamunuge J, Torres Soto M, Ford CB, Bryant JA, Henn MR, Hohmann EL. Fecal microbiota transplantation for the improvement of metabolism in obesity: the FMT-TRIM double-blind placebo-controlled pilot trial. PLoS Med. 2020;17:e1003051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 207] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 153. | Allegretti JR, Kassam Z, Mullish BH, Chiang A, Carrellas M, Hurtado J, Marchesi JR, McDonald JAK, Pechlivanis A, Barker GF, Miguéns Blanco J, Garcia-Perez I, Wong WF, Gerardin Y, Silverstein M, Kennedy K, Thompson C. Effects of fecal microbiota transplantation with oral capsules in obese patients. Clin Gastroenterol Hepatol. 2020;18:855-863.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 206] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 154. | Allegretti JR, Kassam Z, Hurtado J, Marchesi JR, Mullish BH, Chiang A, Thompson CC, Cummings BP. Impact of fecal microbiota transplantation with capsules on the prevention of metabolic syndrome among patients with obesity. Hormones (Athens). 2021;20:209-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 155. | Timmerman HM, Koning CJ, Mulder L, Rombouts FM, Beynen AC. Monostrain, multistrain and multispecies probiotics--A comparison of functionality and efficacy. Int J Food Microbiol. 2004;96:219-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 355] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 156. | Fu N, Peiris P, Markham J, Bavor J. A novel co-culture process with Zymomonas mobilis and Pichia stipitis for efficient ethanol production on glucose/xylose mixtures. Enzyme Microb Technol. 2009;45:210-217. [RCA] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 70] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 157. | Bernstein HC, Carlson RP. Microbial consortia engineering for cellular factories: in vitro to in silico systems. Comput Struct Biotechnol J. 2012;3:e201210017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 158. | Bader J, Mast-Gerlach E, Popović MK, Bajpai R, Stahl U. Relevance of microbial coculture fermentations in biotechnology. J Appl Microbiol. 2010;109:371-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 153] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 159. | Brenner K, You L, Arnold FH. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 560] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 160. | Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol. 2002;83:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4112] [Cited by in RCA: 2100] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 161. | Leonard E, Nielsen D, Solomon K, Prather KJ. Engineering microbes with synthetic biology frameworks. Trends Biotechnol. 2008;26:674-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 162. | Zomorrodi AR, Segrè D. Synthetic ecology of microbes: mathematical models and applications. J Mol Biol. 2016;428:837-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 127] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 163. | De Roy K, Marzorati M, Van den Abbeele P, Van de Wiele T, Boon N. Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities. Environ Microbiol. 2014;16:1472-1481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 185] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 164. | Johns NI, Blazejewski T, Gomes AL, Wang HH. Principles for designing synthetic microbial communities. Curr Opin Microbiol. 2016;31:146-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 172] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 165. | Verstraete W, Wittebolle L, Heylen K, Vanparys B, de Vos P, van de Wiele T, Boon N. Microbial resource management: the road to go for environmental biotechnology. Eng Life Sci. 2007;7:117-126. [RCA] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 166. | Read S, Marzorati M, Guimarães BC, Boon N. Microbial Resource Management revisited: successful parameters and new concepts. Appl Microbiol Biotechnol. 2011;90:861-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 167. | Gibson GR, Wang X. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J Appl Bacteriol. 1994;77:412-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 394] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 168. | Vázquez-Castellanos JF, Biclot A, Vrancken G, Huys GR, Raes J. Design of synthetic microbial consortia for gut microbiota modulation. Curr Opin Pharmacol. 2019;49:52-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 169. | Touré R, Kheadr E, Lacroix C, Moroni O, Fliss I. Production of antibacterial substances by bifidobacterial isolates from infant stool active against Listeria monocytogenes. J Appl Microbiol. 2003;95:1058-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 170. | Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: 'RePOOPulating' the gut. Microbiome. 2013;1:3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 523] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 171. | Masset J, Calusinska M, Hamilton C, Hiligsmann S, Joris B, Wilmotte A, Thonart P. Fermentative hydrogen production from glucose and starch using pure strains and artificial co-cultures of Clostridium spp. Biotechnol Biofuels. 2012;5:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 172. | Chen Y. Development and application of co-culture for ethanol production by co-fermentation of glucose and xylose: a systematic review. J Ind Microbiol Biotechnol. 2011;38:581-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 173. | Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jäger C, Seguin-Devaux C, Zenhausern F, Wilmes P. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016;7:11535. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 448] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 174. | Bilen M, Dufour JC, Lagier JC, Cadoret F, Daoud Z, Dubourg G, Raoult D. The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome. 2018;6:94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 135] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 175. | Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 741] [Cited by in RCA: 785] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 176. | Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, Dubourg G, Durand G, Mourembou G, Guilhot E, Togo A, Bellali S, Bachar D, Cassir N, Bittar F, Delerce J, Mailhe M, Ricaboni D, Bilen M, Dangui Nieko NP, Dia Badiane NM, Valles C, Mouelhi D, Diop K, Million M, Musso D, Abrahão J, Azhar EI, Bibi F, Yasir M, Diallo A, Sokhna C, Djossou F, Vitton V, Robert C, Rolain JM, La Scola B, Fournier PE, Levasseur A, Raoult D. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 756] [Cited by in RCA: 665] [Article Influence: 73.9] [Reference Citation Analysis (0)] |
| 177. | Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1394] [Cited by in RCA: 1361] [Article Influence: 85.1] [Reference Citation Analysis (0)] |
| 178. | Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. [cited 10 March 2021]. Available from: dl.acm.org/ft_gateway.cfm?id=3065386&type=pdf. |
| 179. | Deng L, Yu D. Deep Convex Net: a scalable architecture for speech pattern classification. 2011. [cited 10 March 2021]. Available from: msr-waypoint.com/pubs/152133/DeepConvexNetwork-Interspeech2011-pub.pdf. |
| 180. | Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017;18:851-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 387] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 181. | Wainberg M, Merico D, Delong A, Frey BJ. Deep learning in biomedicine. Nat Biotechnol. 2018;36:829-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 314] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 182. | Wetterstrand KA. DNA Sequencing Costs: data from the NHGRI Genome Sequencing Program (GSP). [cited 10 March 2021]. Available from: www.genome.gov/sequencingcostsdata. |
| 183. | Cui H, Zhang X. Alignment-free supervised classification of metagenomes by recursive SVM. BMC Genomics. 2013;14:641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 184. | Pasolli E, Truong DT, Malik F, Waldron L, Segata N. Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput Biol. 2016;12:e1004977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 292] [Cited by in RCA: 349] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 185. | Chassagnon G, Vakalopolou M, Paragios N, Revel MP. Deep learning: definition and perspectives for thoracic imaging. Eur Radiol. 2020;30:2021-2030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 186. | Díez López C, Vidaki A, Ralf A, Montiel González D, Radjabzadeh D, Kraaij R, Uitterlinden AG, Haas C, Lao O, Kayser M. Novel taxonomy-independent deep learning microbiome approach allows for accurate classification of different forensically relevant human epithelial materials. Forensic Sci Int Genet. 2019;41:72-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 187. | Le V, Quinn TP, Tran T, Venkatesh S. Deep in the bowel: highly interpretable neural encoder-decoder networks predict gut metabolites from gut microbiome. BMC Genomics. 2020;21:256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 188. | Lee JY, Sadler NC, Egbert RG, Anderton CR, Hofmockel KS, Jansson JK, Song HS. Deep learning predicts microbial interactions from self-organized spatiotemporal patterns. Comput Struct Biotechnol J. 2020;18:1259-1269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 189. | Rampelli S, Fabbrini M, Candela M, Biagi E, Brigidi P, Turroni S. G2S: a new deep learning tool for predicting stool microbiome structure from oral microbiome data. Front Genet. 2021;12:644516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 190. | Breiman L. Random Forests. Mach Learn. 2001;45:5-32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 56052] [Cited by in RCA: 34155] [Article Influence: 2846.3] [Reference Citation Analysis (0)] |
| 191. | Fukui H, Nishida A, Matsuda S, Kira F, Watanabe S, Kuriyama M, Kawakami K, Aikawa Y, Oda N, Arai K, Matsunaga A, Nonaka M, Nakai K, Shinmura W, Matsumoto M, Morishita S, Takeda AK, Miwa H. Usefulness of machine learning-based gut microbiome analysis for identifying patients with irritable bowels syndrome. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 192. | Ai D, Pan H, Han R, Li X, Liu G, Xia LC. Using decision tree aggregation with Random Forest model to identify gut microbes associated with colorectal cancer. Genes. 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 193. | Koohi-Moghadam M, Borad MJ, Tran NL, Swanson KR, Boardman LA, Sun H, Wang J. MetaMarker: a pipeline for de novo discovery of novel metagenomic biomarkers. Bioinformatics. 2019;35:3812-3814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 194. | Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, Brigidi P, Crittenden AN, Henry AG, Candela M. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr Biol. 2015;25:1682-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 259] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 195. | Fernández-Navarro T, Díaz I, Gutiérrez-Díaz I, Rodríguez-Carrio J, Suárez A, de Los Reyes-Gavilán CG, Gueimonde M, Salazar N, González S. Exploring the interactions between serum free fatty acids and fecal microbiota in obesity through a machine learning algorithm. Food Res Int. 2019;121:533-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |