Published online Oct 28, 2021. doi: 10.3748/wjg.v27.i40.6794
Peer-review started: May 11, 2021
First decision: June 12, 2021
Revised: June 15, 2021
Accepted: September 15, 2021
Article in press: September 15, 2021
Published online: October 28, 2021
Processing time: 168 Days and 12.5 Hours
The development of artificial intelligence (AI) has increased dramatically in the last 20 years, with clinical applications progressively being explored for most of the medical specialties. The field of gastroenterology and hepatology, substantially reliant on vast amounts of imaging studies, is not an exception. The clinical applications of AI systems in this field include the identification of premalignant or malignant lesions (e.g., identification of dysplasia or esophageal adenocarcinoma in Barrett’s esophagus, pancreatic malignancies), detection of lesions (e.g., polyp identification and classification, small-bowel bleeding lesion on capsule endoscopy, pancreatic cystic lesions), development of objective scoring systems for risk stratification, predicting disease prognosis or treatment response [e.g., determining survival in patients post-resection of hepatocellular carcinoma), determining which patients with inflammatory bowel disease (IBD) will benefit from biologic therapy], or evaluation of metrics such as bowel preparation score or quality of endoscopic examination. The objective of this comprehensive review is to analyze the available AI-related studies pertaining to the entirety of the gastrointestinal tract, including the upper, middle and lower tracts; IBD; the hepatobiliary system; and the pancreas, discussing the findings and clinical applications, as well as outlining the current limitations and future directions in this field.
Core Tip: Artificial intelligence (AI) clinical applications in gastroenterology and hepatology, which heavily relies on imaging, have dramatically expanded in the last 20 years. These applications include the detection of lesions, identification of pre
- Citation: Kröner PT, Engels MM, Glicksberg BS, Johnson KW, Mzaik O, van Hooft JE, Wallace MB, El-Serag HB, Krittanawong C. Artificial intelligence in gastroenterology: A state-of-the-art review. World J Gastroenterol 2021; 27(40): 6794-6824
- URL: https://www.wjgnet.com/1007-9327/full/v27/i40/6794.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i40.6794
As artificial intelligence (AI) continues to rapidly evolve in medicine, the clinical applications of this technology are becoming increasingly evident[1]. Relying heavily on endoscopic and radiologic imaging, gastroenterology has become an attractive field in which to apply AI. Special interest has already been devoted to several areas, including the detection of gastrointestinal neoplastic lesions to assist with rapid diagnosis, reduction of misdiagnosis, improvement in quality of imaging, reduction of interobserver variability in visual classifications, and radiologic and histopathologic interpretation[2-4].
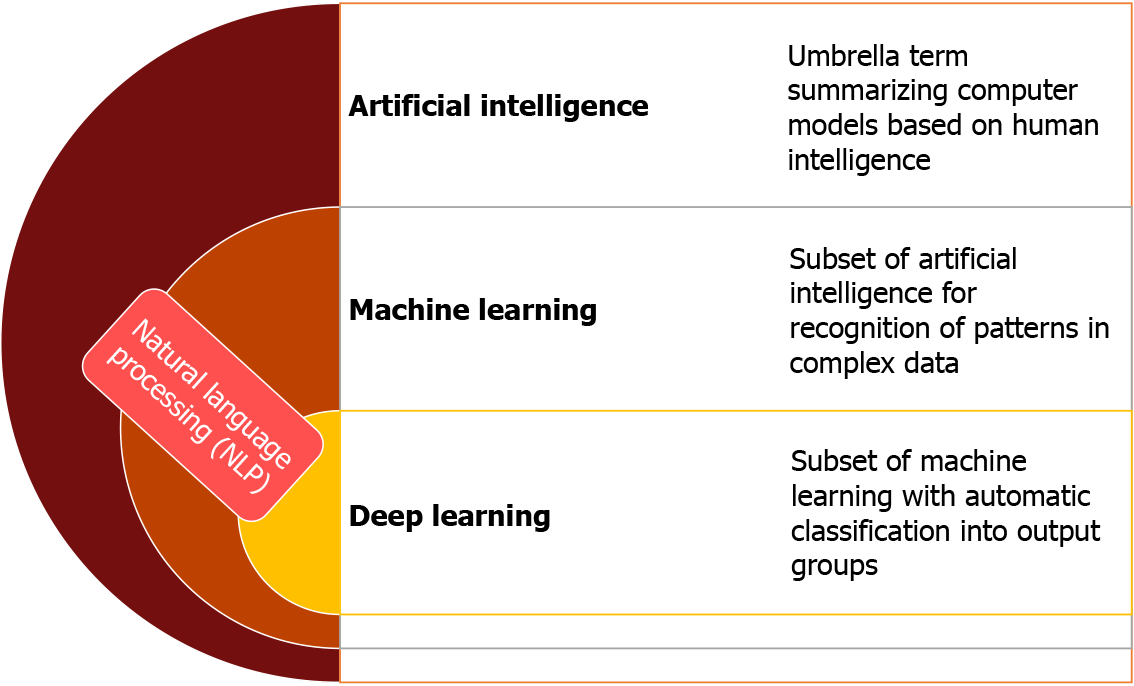
AI is a broad term that encompasses disciplines such as machine learning (ML) and subdisciplines or specific techniques such as deep learning (DL) (Figure 1). The central motivation of ML to use large datasets to recognize patterns of interactions between variables, often ultimately in a way that allows the learned function to be applied to new data[5]. ML is composed of both “supervised” and “unsupervised” learning methods. The goal of supervised learning is to predict a labelled output focusing primarily on classifying data input into specific subgroups, or alternately for prediction of quantitative outcomes[6]. An example of supervised learning is training a system to assist in identifying gastric intestinal metaplasia (GIM) using a large database of lesions that have previously been identified by an operator as corresponding to GIM. In comparison, unsupervised learning does not have an output to predict. It relies on attempting to identify naturally occurring patterns from within the input, often to then group them accordingly (e.g., tissue sample clustering based on similar gene expression values)[6]. DL is a subset of ML, based on artificial neural networks (ANN), which are loosely inspired by the neuronal interplay in the human brain. DL autonomously utilizes the data input to learn, identify, and leverage predictive factors of an outcome, which can use multi-layered systems [i.e., convolutional neural networks (CNN)] to process complex information[3,7]. The realization of the concept of DL has recently become possible with rapid advancements in spe
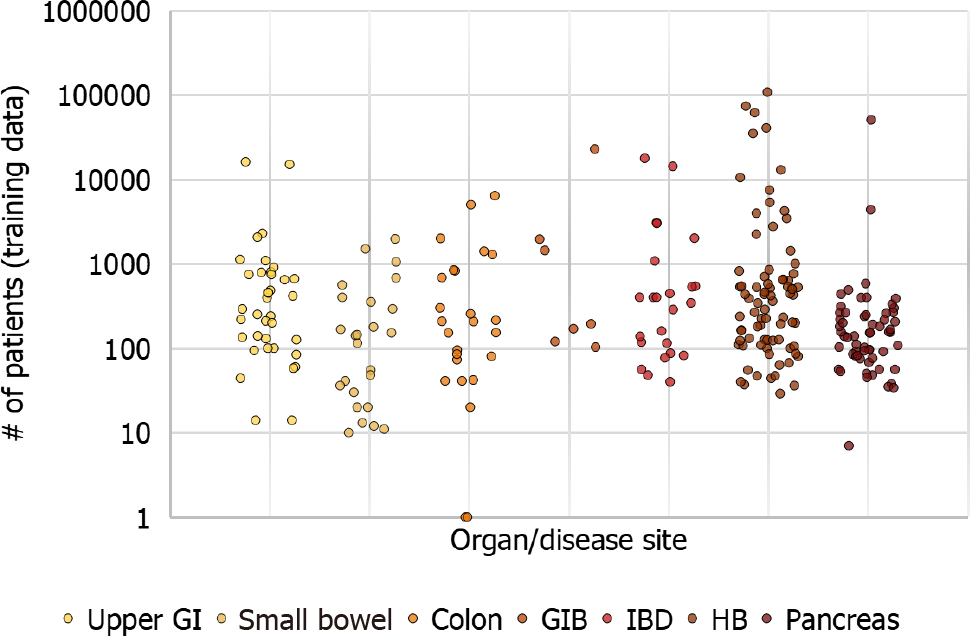
With increased recognition of the importance of AI in gastroenterology, the first global AI in gastroenterology and endoscopy summit was held in Washington D.C. in late 2019, which included multiple experts in the domains of academia, industry and regulatory institutions. The consortium anticipated that in the next 10 years the clinical applications of AI in gastroenterology will positively influence patient care and clinical workflow. The consortium recognized the importance of a close multidisciplinary collaboration between gastroenterologists, industry, and regulatory institutions in the development and application of new technologies in the clinical setting[8]. Therefore, the main objective of this review is to introduce the topic of AI and its clinical applications, outline current limitations and knowledge gaps, and trace the future directions in each field by summarizing the global rapidly expanding pool of ever-changing literature to date (Figures 2 and 3).
The upper gastrointestinal tract has several areas of interest for detection of pre
Despite the fact that histopathologic analysis is the gold standard to establish the diagnosis of BE and determine the presence of dysplasia, it is of paramount importance for endoscopists to obtain targeted biopsies from specific locations that harbor the actual lesion. By identifying areas that may harbor BE with or without dysplasia, AI can orient the clinician in performing directed biopsies instead of relying on random sampling. Importantly, detection of early esophageal neoplasia with conventional white light imaging (WLI) and digital chromoendoscopy [i.e., narrow band imaging (NBI)] represents a challenge, an issue for which AI has been proposed as a possible solution[10,11].
In total, we included 8 studies that examined BE, all of which addressed the detection of dysplasia or early esophageal adenocarcinoma (EAC) based on endo
Similarly, identification of SCC poses a similar clinical challenge, as traditional diagnostic techniques (e.g., chromoendoscopy with lugol or NBI) have relatively low specificity[18]. As complex novel methods that rely heavily on endoscopic imaging such as endocytoscopy or volumetric laser endomicroscopy have been progressively implemented in clinical practice, the interpretation of large volumes of images has been noted to be a challenging and time-consuming issue[19]. Therefore, AI-assisted image interpretation has also found a use in identifying abnormalities in the image inputs[17].
Thirteen studies examining esophageal cancer were examined, of which 11 specifically studied SCC. Nine of the studies targeted to develop DL models for malignancy detection, while two studies aimed to develop models that predict malignancy depth of invasion with DL models. Most studies (8) were based on CNN models, while others used joint diagonalization principal component analysis (JDPCA), VGG16 Net, or GoogLeNet as classifiers. Although the values of accuracy, sensitivity and specificity in esophageal SCC detection varied between the studies, all models performed at least as good as endoscopists in lesion detection and characterization or substantially improved the endoscopists’ lesion detection[20-30]. Fukuda et al[21] developed a DL model aimed at detecting suspicious lesions and characterize SCC using more than 28000 NBI-enhanced images in the construction of the model. The model achieved a higher sensitivity for SCC detection and higher accuracy for SCC characterization from normal tissue than endoscopic experts. Two studies by Nakagawa et al[31] and Shimamoto et al[32] aimed at developing models that pre
GC is the fourth most common cause of cancer-related death in the world. Identification of premalignant lesions or early gastric neoplasia is of paramount importance. Unfortunately, as with esophageal diseases, several studies have shown that conventional endoscopic imaging (e.g., WLI or NBI) or other advanced endoscopic modalities (e.g., magnifying endoscopy, blue laser imaging) have relatively low sensitivity and specificity in identifying premalignant or early neoplastic gastric lesions[33-36].
Twenty-four studies examined gastric malignancy or premalignant conditions. Eleven (46%) studies directly addressed early GC (EGC) detection, of which eight models used DL, two used SVM and one used JDPCA. The sample size in the training and validation datasets varied from less than 100 million to 1.03 million. The accuracy in EGC detection of all models ranged from 86.5%-98.7%, with sensitivity of 80.0%-96.7% and specificity of 89.2%-100%[30-37-51]. Specifically, Wu et al[52] developed a CNN-based model using over 9000 images to train the algorithm, which not only detected EGC lesions with a 92.5% accuracy, 94.0% sensitivity and 91.0% specificity but also performed significantly better than expert endoscopists at this task. Two studies, one based on faster region-based CNNs and another one based on quantum neural networks, used biomarkers, histology and computed tomography (CT) images to predict metastasis to the liver or lymph nodes. These models predicted metastasis to liver and lymph nodes with sensitivity of 66.7%, specificity 97.1% and an area under the curve (AUC) of 0.95[53,54]. Four studies developed models predicting survival in GC or stratification of risk of developing GC[55,56]. While 1 model based on SVM achieved higher accuracy than the tumor, nodes, metastasis cancer staging (TNM) system to predict overall survival and disease-free survival[57], another model based on a preoperative ANN was not superior in predicting survival compared to TNM[58]. Zhu et al[59] developed a ResNet50-based computer-aided detection and diag
Currently, diagnosis of Helicobacter pylori (H. pylori) infection (a known risk factor for the development of peptic ulcer disease and GC) relies on stool or breath testing and histopathology (invasive and expensive). Endoscopic identification of H. pylori has been a target for AI-assisted systems[60,61]. For H. pylori detection and classification, 4 of 4 studies used a CNN model[62-65]. A study by Martin et al[62] using gastric biopsy histopathology images as an input with small number of samples (n = 210 in training dataset and 90-106 in 2 test datasets) had diagnostic accuracy of 98.9%-99.1% for detecting current H. pylori infection. This result was higher than the accuracy achieved in 2 studies using esophagogastroduodenoscopy images as an input with higher number of samples (98.9%-99.1% vs 77.5%-87.7%). The mean accuracy of endoscopists for detecting currently infected H. pylori in 2 studies was around 79.0%-79.4%[62]. A study by Shichijo et al[64] found that the average H. pylori diagnostic time for the AI model was 194 s, while it was 230 ± 65 min for endoscopists. The AI model also had a significantly higher accuracy than the endoscopists by 5.3%, demonstrating that AI algorithms had significantly better accuracy and faster diagnostic time for H. pylori than the endoscopists. More recently, Nakashima et al[63] developed a CAD system based on linked color imaging combined with DL, which achieved 82.5% accuracy, demonstrating comparable diagnostic accuracy of H. pylori to that of experienced endoscopists.
In the area of gastrointestinal bleeding (GIB), risk stratification is of paramount importance not only to identify high-risk patients and guide clinical decision-making but to also identify areas where relatively scarce resources require allocation. In general, risk-stratification tools examine risk factors that are associated with a condition to predict one or several outcomes (e.g., survival, length of hospitalization, rebleeding rates, need for endoscopic therapy, response to treatment)[66-68]. Re
Seven studies examining outcome measures in GIB were included, all of which were developed by reviewing medical record parameters to construct ML algorithms, which were primarily based on ANN[67-73]. Half of the studies had both internal and external validation cohorts, and patient sample sizes varied from 147 to over 22800. Shung et al[69] developed a gradient-boosting ML model that identified patients with upper GIB who met a composite endpoint with superior AUC, sensitivity and specificity to that of the Glasgow-Blatchford (GBS), Rockall and AIMS-65 scores. Seo et al[68] developed 4 different ML algorithms in patients with nonvariceal upper GIB that achieved superior AUC for mortality, rebleeding and hypotension than that of the GBS, particularly with the use of the random forest (RF) analytic model. However, for rebleeding prediction, a gradient-boosting model developed by Ayaru et al[73] showed a higher accuracy than the VC model in the aforementioned study (88% vs 78.5%). Das et al[70] developed a predictive ML model in lower GIB and compared it in internal and external validation cohorts to the validated BLEED score. Both internal and external validation cohorts reached accuracies, sensitivities, and specificities that were superior to the BLEED score.
Direct endoscopic visualization of the colon continues to be the gold standard for detection and resection of colonic premalignant lesions. In addition, patient- and polyp-specific characteristics have also been identified as predictors of missed colorectal cancer (CRC)[74]. Therefore, quality measures such as cecal intubation time, withdrawal time, bowel preparation quality and adenoma detection rates (ADR) have been instituted to attempt and standardize practice and mitigate the rates of missed CRC[74,75]. Since colonoscopy is an operator-dependent procedure, this may lead to variability in polyp detection rates, polyp characterization and estimation of depth of invasion of malignant lesions between endoscopists. CAD has been studied to address these current shortcomings, as well as to improve ADR and reduce CRC risk and colon cancer-related deaths[76] (Supplementary Table 2).
Polyp detection during colonoscopy is the cornerstone of CRC prevention. It is estimated that CRC develops in 2%-6% of cases after colonoscopy but before the next scheduled surveillance and, therefore, could represent missed or new postcolonoscopy CRC[74,77]. A large number of research studies have been devoted to identifying factors that play a role in the detection of premalignant lesions during colonoscopy, as well as factors that are associated with missed CRC[77]. An observation that has gained considerable attention is that the presence of experienced endoscopy nurses, fellows or any trained second observer during the procedure itself improves ADR[78,79]. Therefore, and answering the call for improved polyp detection measures during colonoscopy, AI has become a promising technology serving the role of a more standardized “second observer,” appropriately termed “computer-aided detection” (CADe).
Eighteen studies that evaluated polyp detection were included in this review, thirteen of which employed DL models, while five used other ML models. Nearly all the studies were limited to having internal validation only. The training datasets in these studies utilized still images from colonoscopy videos ranging from 176 to more than 8600 images. Seven studies calculated the ADR of the developed model and compared it to that of endoscopists, most of which found that the AI had significantly higher ADR than endoscopists, with a decreased reaction time, while the remaining studies showed equivalent ADR[80-87]. Repici et al[82] conducted a multicenter study showing that the ADR was 40.4% in the study’s participating endoscopists alone, while it was 54.8% in the group using the CADe-based GI-Genius (®Medtronic, Minneapolis, MN) module. Similarly, Wang et al[85] conducted a study that evaluated over 520 colonoscopies, also examining the use of a CADe model, and found that the ADR increased from 20.34% in endoscopists alone to 29.12% in the CADe-assisted group. Other studies explored the accuracy, sensitivity and specificity of polyp-detection AI models, displaying accuracies of up to 96.4%, sensitivity of up to 99.7% and specificity of up to 93.7%[88-98]. Kominami et al[93] designed an SVM-based model using real-time images with NBI and magnification enhancement to detect colon polyps, achieving an accuracy, sensitivity, and specificity of 93.2%, 93.0%, and 93.3%, respectively. Three studies addressed subjects in the realm of CRC, specifically detecting malignancy on colonoscopy with chromoendoscopy and NBI enhancement, hematoxylin-eosin histopathology slides, or estimating invasion depth on regular WLI colonoscopy images[99-101]. Ito et al[100] developed a DL model based on CNN to detect deeply invasive CRC based on WLI colonoscopy, which utilized over 9900 images from 41 patients to train the model. Furthermore, over 5000 images were used in the testing cohort, achieving an accuracy, sensitivity and specificity in differentiating invasion depth of 81.2%, 67.5%, and 89.0%, respectively. Kudo et al[101] developed a model based on EndoBRAIN (®Cybernet Systems Co., Tokyo, Japan) using colonoscopy with chromoendoscopy and NBI enhancement to detect malignant lesions in the colon. The training sample used over 69000 images and achieved accuracies of 98.0% and 96.0% for chromoendoscopy and NBI enhancement, respec
In an attempt to morphologically classify polyps according to their malignant potential, numerous classification systems have been developed. As an example, the Paris classification classifies polyps according to whether they are pedunculated, sessile, slightly raised, or excavated[102]. These visual characteristics are then used to predict whether the polyp has invasive potential or lymph node involvement and potentially contribute to clinical decision-making, such as determining whether the polyp is resectable endoscopically[103]. Unfortunately, research has shown that interobserver variability with this classification is moderately high, suggesting that this visual classification should not be routinely used in research or practice[104]. In addition, considering that most resected polyps during colonoscopy are diminutive and that histopathologic analysis of resected polyps is costly, optical diagnosis has been proposed in this subset of polyps and termed “optical biopsy”[105]. The concept of optical biopsy has been proposed to support cost-effective strategies in CRC screening such as resecting-and-discarding, as well as diagnosing-and-leaving this subset of polyps[106]. However, the concept of optical biopsy has the same limitations as endoscopic polyp characterization does: relatively high interobserver variability. Hence, computer-aided diagnosis has been proposed as a potential solution to stan
Nine studies pertaining to colonic polyp classification or differentiation were included in this review[107-115]. While some studies did not report on validation methods, others had cross-validation, internal, and external methods. Most studies relied on ML-based algorithms to assess colonoscopy with enhancement measures (e.g., magnification, NBI, endocytoscopy), while deep CNN models were used in studies evaluating real-time polyp differentiation. Sánchez-Montes et al[114] develop
Knowledge on the depth of invasion of early CRC is critical for determining resection modality. Intramucosal and submucosal cancerous lesions can be resected with endoscopic techniques such as endoscopic mucosal resection or endoscopic submucosal dissection[116]. However, lesions that invade the deeper layers have a higher association with lymph node metastasis and, hence, may require a combined surgical and oncologic approach[116,117]. Several features on direct endoscopic visualization have been associated with deep invasion of a lesion, including lesion de
Finally, 6 studies assessed other subjects within the realm of the lower gastro
Video capsule endoscopy: Owing to its ease of use and noninvasiveness, video capsule endoscopy (VCE) is the current diagnostic method of choice to assess the small bowel for conditions such as occult GIB and Crohn’s disease (CD). However, since it lacks self-locomotion, it relies on the motility of the bowel to advance within the gastrointestinal tract. Therefore, it requires a very large number of images (up to 60000 per examination) to be automatically obtained, rendering the process of interpretation lengthy and tedious[124]. VCE has been an area of focus for AI since its early stages, seeing the initial AI classifying methods such as SVM and multilayer perceptron network[125-127]. With faster processors, increased computational power by impro
A total of 31 studies pertaining to VCE were identified. Fifteen studies (48%) developed models to assist in detection of active GIB or angioectasia. Four studies used CNN, five used SVM, and the remaining ones used ML-based analytic models. Five studies reported cross-validation methods. All studies reported accuracy ranging from 94% to 98% in identifying GIB and angioectasias, with sensitivity of 92.0%-100% and specificity of 82.9%-99.9%[130-142]. Specifically, Tsuboi et al[140] developed a DL model based on CNN for angioectasia detection, which achieved an AUC of 0.998, with a sensitivity and specificity of 98.8% and 98.4%, respectively.
Six studies assessed for the presence of small-intestinal ulcers, of which five used DL models based on CNN and one used SVM[128,134,143-146]. Aoki et al[128,143] conducted 2 studies in testing DL models to detect ulcerations, yielding a sensitivity of 88.2%, a specificity of 90.9%, and an AUC of 0.958, respectively. Two more studies evaluated the detection of small-bowel CD by identifying ulcers on VCE[147,148]. Klang et al[147] developed a DL model based on CNN that used over 17000 VCE images to identify CD ulcers, achieving an AUC of 0.99. Two studies examined small intestinal polyp detection using NN-based models, both achieving an accuracy of at least 98%, with sensitivity and specificity of up to 95.5% and 98.5%, respectively[149,150].
Regarding celiac disease (CeD) detection, 4 studies were evaluated, 3 of which used CNN and 1 that used a clinical decision support system[151]. A study by Zhou et al[152] reported 100% sensitivity and specificity in detecting CeD using GoogLeNet as classifier. A study by Wimmer et al[153] using AlexNet, VGGf net, and VGG-16 net as classifiers obtained an optimal accuracy of 92.5% using VGG-16, although the report did not include sample size. More recently, Wang et al[154] developed a DL model using the CNN-based InceptionV3 as well as the SVM-based ResNet50 that was 95.94% accurate, 97.20% sensitive and 95.63% specific in identifying CeD in VCE images.
Hookworm infection represents a significant healthcare issue in the developing world, with an estimated 600 million people harboring this infection[155]. As this helminth typically dwells in the small bowel, it may occasionally be a finding in VCE. Three studies evaluated hookworm detection, of which two used CNN and one used SVM with all using cross-validation[156-158]. Specifically, He et al[157] developed a DL model based on deep CNN, which was trained on a large VCE dataset consisting of more than 440000 images. The model outperformed other handcrafted, feature-based methods, reaching an accuracy of 88.5% and a sensitivity of 84.6% in detecting hookworm in the small bowel.
To assess for quality of bowel preparation during VCE Leenhardt et al[159] and Noorda et al[160] developed a DL models based on CNN. Both models achieved accuracy of over 95%, with sensitivity and specificity of 94.7%-96.18% and 94.0%-94.33%, respectively. Overall, Ding et al[131] developed a DL model based on CNN to identify and categorize all small-bowel ulcers, polyps, bleeding, lymphangiectasia, follicular hyperplasia, protruding lesions, diverticula or inflammation using over 100 million VCE images. The model achieved an impressive 99.90% overall sensitivity and 99.88% specificity, whereas gastroenterologists identified lesions with a 74.57% sensitivity in the per-patient analysis and 76.89% sensitivity in the per-lesion analysis. Furthermore, the reading time per patient was 5.9 min by the CNN model, compared to 96.6 min by conventional reading.
Inflammatory bowel disease: The complex interplay of pathophysiological factors in inflammatory bowel disease (IBD) has led to the realization that differences in patients’ biology may confer differences in disease activity and response to therapy- an example of “precision medicine”[161]. Integration of AI algorithms into the IBD realm brings promise not only in diagnosis or reducing interobserver variability in severity grading but also opens up the possibility of analyzing large databases to identify complex or occult patterns of disease[162-167].
A total of 25 studies pertaining to IBD were included in this review, of which 9 dealt with CD, 6 studied ulcerative colitis (UC,) and 10 investigated both. Five studies aimed to detect CD, UC, or IBD in general from VCE images, endoscopic images, histology, magnetic resonance imaging (MRI) images, and genetics. Most of the studies used cross-validated ML methods, of which SVM was the method of choice; while another study used DL-based CNN. These studies displayed high levels of accuracy in detecting IBD, from 83.3%-90.8%[143,168-171]. Aoki et al[143] developed a DL model based on CNN that used more than 10000 VCE images in the validation dataset to detect CD ulcers in the small bowel, achieving an accuracy, sensitivity and specificity of 90.8%, 88.2% and 90.9%, respectively.
Nine studies that addressed predicting disease severity were primarily based on endoscopic imaging; although other studies also used laboratory studies, demo
To predict therapeutic response in patients with IBD treated with thiopurines or biologics seven studies constructed internally validated ML algorithms, based on RF analytic models. All models achieved an accuracy of 80.0%-89.8% or an AUC of 0.73-0.846 in identifying patients who will respond at 6-8 wk to therapeutic regimens[180-186]. Waljee et al[186] designed a model using only demographic and laboratory data from 401 patients to predict response at 8 wk in patients receiving ustekinumab with an AUC of 0.78, potentially laying the foundation to avoid costly therapeutic drug monitoring. Waljee et al[184] also constructed a model based on laboratory values and demographics with a validation dataset of over 6100 patients to predict IBD-related hospitalization and outpatient steroid use, achieving an AUC of 0.87, suggesting such AI models could be used to identify patients at risk of an IBD flare and enable precision medicine-based therapeutic approaches (Supplementary Table 4).
A further 2 internally validated ML models using SVM and RF were developed to predict the risk of IBD based on a genomic datasets[187,188]. Isakov et al[187] de
Liver diseases are broad and complex, ranging from asymptomatic liver chemistry elevation to life-threatening conditions such as acute liver failure or orthotopic liver transplantation (OLT). Hepatology is can be a fertile ground for applying AI in survival models (e.g., model for end-stage liver disease), disease detection models [e.g., early detection of non-alcoholic fatty liver disease (NAFLD)], disease severity models (e.g., alcoholic hepatitis discriminant function), or disease estimation models (e.g., aspartate aminotransferase-to-platelet ratio index), but also for pattern recognition in radiological images, histopathology and even selection of LT candidates[191,192]. A total of eighty-five studies pertaining to hepatology were examined, out of which twenty assessed prediction of outcome measures, forty-one examined prediction or detection of steatosis, fibrosis or cirrhosis, nine studies examined differentiation of malignant liver neoplasms, six studies explored predictive models for portal hyper
Nine studies developed models that assisted with detection or classification of hepatobiliary neoplastic lesion, six of which involved DL-based CNN[193-202]. Schmauch et al[197] constructed an internally validated CNN-based DL model using ultrasonographic images of the liver to detect and classify focal liver lesions, achieving an overall AUC of 0.891. Six studies constructed models assisting in predicting the presence of portal hypertension complications in patients with all-cause cirrhosis, based on clinical data or radiological images obtained from modalities such as CT scans[203-208]. Dong et al[204] constructed an ML-based model on an RF analytic model that used clinical data and predicted the presence of esophageal varices in patients with cirrhosis with an AUC of 0.82, potentially being useful in performing a better triage of patients who actually require an upper endoscopy for variceal screening. Liu et al[206] developed an ML model that had a higher diagnostic per
Forty-two studies developed models addressing detection of steatosis, fibrosis, or cirrhosis based on clinical data, shear-wave elastography, CT or MRI scans, his
Of 20 studies examining outcome predictors, 8 evaluated predictor models in OLT involving donor-recipient matching, recipient survival at determined time frame, graft survival at different time frames, survival predictors, and morbidity predictors[243-250]. Bertsimas et al[245] constructed an internally-validated decision tree-based ML model using clinical data that predicted 3-mo waitlist mortality or removal with an AUC of 0.895. Five studies designed models for outcome prediction in patients with hepatocellular carcinoma (HCC), including response to trans-arterial chemoembolization (TACE), recurrence or survival after resection, most of which were based on CNN[251-255]. Saillard et al[254] designed an externally-validated model based on a pre-trained CNN based on histology slides from 328 patients that independently predicted survival after HCC resection with a c-index of 0.75. Other studies evaluated outcomes in patients with acetaminophen-related ALF, primary sclerosing cholangitis, predicted the presence of choledocholithiasis, predicted hepatotoxicity of stereotactic body radiation, or mortality in patients with cirrhosis[256-263]. Eaton et al[256] constructed a ML model with XGB that accurately predicted hepatic decompensation in patients with primary sclerosing cholangitis with a C-statistic of 0.90.
Other studies focused on developing models for miscellaneous topics such as estimating liver stiffness from MRI, assessing pretransplant cognitive impairment, detecting spectral differences between normal and hepatitis B virus serum samples, classifying seroconversion to HBeAg, predicting hepatotoxicity in early stages of drug development, predicting fibrosis in hepatitis C virus, or AI-assisted liver tumor seg
Pancreatic diseases contain areas where AI can be effectively applied. Of primary interest is the use of AI in improving existing disease severity scoring systems or prognostic models in complicated acute pancreatitis (AP) or chronic pancreatitis (CP), based on clinical and radiological data, detection and differentiation of pancreas cystic neoplasms (PCN) with prediction of malignant potential, radiologic early detection of pancreatic ductal adenocarcinoma (PDAC), radiologic differentiation between PDAC and benign pancreatic conditions [e.g., autoimmune pancreatitis (AIP)], and histopathologic interpretation of tissue samples[279-282]. A total of 59 studies pertaining to the pancreas were reviewed. Of these, 20 (34%) addressed prediction of outcomes in patients with pancreatic diseases ranging from AP to neoplasia.
Eleven of these studies examined outcome prediction in AP. All of the studies’ ANN models outperformed logistic regression models, Glasgow, and APACHE-II scoring systems in predicting AP severity; while requiring less number of parameters[283-293]. Qiu et al[292] compared the performance of SVM, logistic regression analysis, and ANN models to predict multiorgan failure in AP. All 3 models predicted multiorgan failure, with AUC of 0.840, 0.832, and 0.834, respectively, with ANN requiring a lesser number of parameters.
Seven (12%) studies used AI algorithms to construct clinical registries, segment the pancreas based on imaging, or differentiate between certain pancreatic diseases based on cross-sectional imaging or endoscopic ultrasound (EUS)[294-300]. Zhang et al[298] constructed a DL station classification model and a segmentation model to reduce the difficulty in EUS interpretation for trainees. The trainee station recognition accuracy improved from 67.2% to 78.4% in the crossover study. Interobserver agreement between endoscopists and deep CNN with Cohen’s kappa coefficient was substantial, ranging from 0.826-0.879. The authors conclude that this technology may play a key role in shortening the learning curve of EUS among trainees.
Fifteen studies (25%) addressed prediction of malignancy based on imaging fin
Eleven studies (19%) evaluated differentiation of PCNs by classifying them into their respective subtypes based on their characteristics on imaging[316-326]. Springer et al[324] developed a multimodality ML model that integrated clinical, radiological and genetic/biochemical markers data to determine whether patients with pancreas cyst should undergo surgery, monitoring, or no further surveillance. The model correctly identified serous cystic neoplasms in 65% of the cases with 99% specificity, clearly outperforming the current standard of care of clinical identification in only 18% of cases. The authors conclude that these systems may serve an adjunct role in clinical practice, enabling the clinician to take better-informed clinical decisions[324].
Eight studies addressed PDAC, from developing risk scores for development of PDAC based on urinary biomarkers, predicting clinical performance and response to celiac plexus neurolysis, to prediction of survival time. AI models performed at least as well as the logistic regression models in predicting the selected outcome[327-334].
Six studies directly examined early pancreatic cancer detection in PDAC or PCN by examining the imaging characteristics or identifying high-risk patients on electronic health records based on factors such as family history of pancreatic cancer[335-340]. Roch et al[339] developed NLP-based algorithms based on common terminology used by physicians in describing pancreatic cysts and applied them to automatically conduct searches in electronic health records. The algorithm tracked patients with cysts with a 99.9% sensitivity and 98.8% specificity, demonstrating its utility in capturing patients swiftly and with more ease than manual review. Ozkan et al[338] developed a CAD image-processing system using EUS images to diagnose PDAC, taking patient age into consideration. The accuracy of the model was 87.5, with sensitivity and specificity of 83.3% and 93.3%, respectively (Supplementary Table 6).
Despite the numerous positive advances in AI, there remain several limitations to current studies and obstacles to overcome for future studies. Most current models are based on labeled data and, hence, interpretation is only as good as the observer who labeled the “gold-standard” data. Current algorithms are specifically fitted for a determined dataset. A sizeable proportion of the AI models applied to the clinical setting are only internally validated. Ideally, models should be externally validated on diverse cohorts to ensure that overfitting does not become an issue. Therefore, this issue could be potentially addressed by the creation of a universal, well-annotated, high-quality dataset, and by creating algorithms with more plasticity. However, creating “universal datasets” creates additional challenges, particularly related to data integrity and privacy. A potential solution to this is the decentralized “federated datasets”, which involves combining multiple datasets stored on their respective servers, addressing these challenges[341]. Specific protocols are required for choosing an analytic model and selecting or developing validation techniques (e.g., external-, internal-, cross-validation) for data fine-tuning or augmentation. Algorithms that yield the best accuracy should be promoted. Calibration has translated into improvements in probability prediction, for which they should be instituted in all models. Most current studies were cohort studies, whereas well-designed randomized controlled trials would be needed to better support conclusions. Some studies utilized custom-built models, which are not explained in detail. Therefore, custom-built algorithms should have their background and processes thoroughly declared. The studies presented numerous different newly developed models, which would require vali
ML and DL could assist clinicians in the diagnosis of gastrointestinal and liver neoplasms, bleeding, infection, and inflammatory process, and also predict outcome measures in these conditions.
The initial use of ML or DL models might be used in backing up clinicians in establishing diagnoses or determining a treatment plan.
Given its high predictive value, if AI suggestions match the clinician’s reasoning, clinicians could make a decision more confidently. If the answers are discrepant, careful investigation should be undertaken.
In the future, if ML or DL models find a place to be integrated in standard clinical care, to guide in establishing diagnoses, selecting treatment interventions, predicting outcomes, and influencing clinical decision-making. However, future studies are also necessary to explore avenues of how these measures can be better instituted in clinical practice as a whole.
As demonstrated by this review, AI applications in clinical gastroenterology and hepatology continue to rapidly expand and evolve at many different levels. For general clinical care, the recent proliferation in AI applications is likely to enable “precision medicine” on a broader scale. Clinically, it is predicted that invasive diagnostic interventions will generally fall out of favor for some conditions, as better noninvasive ML-based algorithms pave the way for improved clinical prediction models. Some diagnostic interventions, such as VCE interpretation, may see a considerable decrease in human interpretation, minimizing the human role to that of supervision and attestation of findings of the model. AI-assisted technology will prove important in real-time clinical settings (e.g., polyp detection during colonoscopy). Integration of monitoring devices (e.g., smartphones, smart watches) with ML in the management of selected diseases is also predicted to significantly receive more attention the coming years. The creation of a universal, large, high-quality, well-labelled dataset is a necessity, from which algorithms could be developed to better define the epidemiology and risk factors of diseases. Well-harnessed AI assistance should decrease physician workload or at least maximize their productivity by allowing them to shift from menial tasks to faster, more accurate clinical decision-making. ML algorithms based on these datasets can also be used for other quality measures, such as improvement of process efficiency or identifying cost-effective interventions. In terms of data analysis, traditional analytic models (e.g., logistic regression and clinical scoring systems) may be substituted or augmented by ML algorithms to achieve greater capability and accuracy. Developing and maintaining multidisciplinary teams of data scientists, physicians, content subject experts and industry is of paramount importance in the advancement of AI in gastroenterology and hepatology. Finally, educating clinicians and patients in the future paths of AI applications is critical to increase understanding of future value and decrease reluc
The latest advances in AI in gastroenterology and hepatology are promising for aspect many fields of clinical care, from detection of neoplastic lesions on endoscopic assessment and improving current survival models to predicting treatment response. The application of AI to large and complex datasets may assist in the identification of new associations between variables, potentially leading to changes in clinical practice. Furthermore, the use of AI-assisted technologies has the potential to dramatically improve the quality of care. Finally, the time for assisted precision medicine is at hand, with the AI being able to tailor a treatment regimen or potentially predict the response to treatment in a specific patient based on extensive amounts of clinical data from large patient datasets. It is important to realize that, while AI currently does not substitute human clinical reasoning, it has a bright future in the betterment of patient care.
The authors are grateful to for Wang Z, PhD, Zhang HJ, PhD, Sun T, MD, PhD, Hassan Virk H, MD, and Aiumtrakul N, MD assistance with the additional literature search.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shafqat S S-Editor: Gao CC L-Editor: A P-Editor: Yuan YY
| 1. | Noorbakhsh-Sabet N, Zand R, Zhang Y, Abedi V. Artificial Intelligence Transforms the Future of Health Care. Am J Med. 2019;132:795-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 229] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 2. | Calderaro J, Kather JN. Artificial intelligence-based pathology for gastrointestinal and hepatobiliary cancers. Gut. 2021;70:1183-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 3. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 324] [Article Influence: 64.8] [Reference Citation Analysis (1)] |
| 4. | Kather JN, Calderaro J. Development of AI-based pathology biomarkers in gastrointestinal and liver cancer. Nat Rev Gastroenterol Hepatol. 2020;17:591-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 5. | Hoogenboom SA, Bagci U, Wallace MB. Artificial intelligence in gastroenterology. The current state of play and the potential. How will it affect our practice and when? Tech Gastrointest Endosc. 2019;22:42-47. [RCA] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 6. | Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1155] [Cited by in RCA: 1960] [Article Influence: 217.8] [Reference Citation Analysis (6)] |
| 7. | Wu J, Chen J, Cai J. Application of Artificial Intelligence in Gastrointestinal Endoscopy. J Clin Gastroenterol. 2021;55:110-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 8. | Parasa S, Wallace M, Bagci U, Antonino M, Berzin T, Byrne M, Celik H, Farahani K, Golding M, Gross S, Jamali V, Mendonca P, Mori Y, Ninh A, Repici A, Rex D, Skrinak K, Thakkar SJ, van Hooft JE, Vargo J, Yu H, Xu Z, Sharma P. Proceedings from the First Global Artificial Intelligence in Gastroenterology and Endoscopy Summit. Gastrointest Endosc. 2020;92:938-945.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 9. | Mori Y, Kudo SE, Mohmed HEN, Misawa M, Ogata N, Itoh H, Oda M, Mori K. Artificial intelligence and upper gastrointestinal endoscopy: Current status and future perspective. Dig Endosc. 2019;31:378-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 10. | Hashimoto R, Requa J, Dao T, Ninh A, Tran E, Mai D, Lugo M, El-Hage Chehade N, Chang KJ, Karnes WE, Samarasena JB. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett's esophagus (with video). Gastrointest Endosc. 2020;91:1264-1271.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 11. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (2)] |
| 12. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 222] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 13. | de Groof J, van der Sommen F, van der Putten J, Struyvenberg MR, Zinger S, Curvers WL, Pech O, Meining A, Neuhaus H, Bisschops R, Schoon EJ, de With PH, Bergman JJ. The Argos project: The development of a computer-aided detection system to improve detection of Barrett's neoplasia on white light endoscopy. United European Gastroenterol J. 2019;7:538-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 14. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Prinz F, de Souza LA Jr, Papa J, Palm C, Messmann H. Real-time use of artificial intelligence in the evaluation of cancer in Barrett's oesophagus. Gut. 2020;69:615-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 124] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 15. | Riaz F, Ribeiro MD, Pimentel-Nunes P, Coimbra MT. Integral scale histogram local binary patterns for classification of narrow-band gastroenterology images. Annu Int Conf IEEE Eng Med Biol Soc. 2013;2013:3714-3717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Swager AF, van der Sommen F, Klomp SR, Zinger S, Meijer SL, Schoon EJ, Bergman JJGHM, de With PH, Curvers WL. Computer-aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 17. | Trindade AJ, McKinley MJ, Fan C, Leggett CL, Kahn A, Pleskow DK. Endoscopic Surveillance of Barrett's Esophagus Using Volumetric Laser Endomicroscopy With Artificial Intelligence Image Enhancement. Gastroenterology. 2019;157:303-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Morita FH, Bernardo WM, Ide E, Rocha RS, Aquino JC, Minata MK, Yamazaki K, Marques SB, Sakai P, de Moura EG. Narrow band imaging vs lugol chromoendoscopy to diagnose squamous cell carcinoma of the esophagus: a systematic review and meta-analysis. BMC Cancer. 2017;17:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 19. | Pannala R, Krishnan K, Melson J, Parsi MA, Schulman AR, Sullivan S, Trikudanathan G, Trindade AJ, Watson RR, Maple JT, Lichtenstein DR. Artificial intelligence in gastrointestinal endoscopy. VideoGIE. 2020;5:598-613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 20. | Cai SL, Li B, Tan WM, Niu XJ, Yu HH, Yao LQ, Zhou PH, Yan B, Zhong YS. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2019;90:745-753.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 21. | Fukuda H, Ishihara R, Kato Y, Matsunaga T, Nishida T, Yamada T, Ogiyama H, Horie M, Kinoshita K, Tada T. Comparison of performances of artificial intelligence vs expert endoscopists for real-time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2020;92:848-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 22. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 23. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 272] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 24. | Kumagai Y, Takubo K, Kawada K, Aoyama K, Endo Y, Ozawa T, Hirasawa T, Yoshio T, Ishihara S, Fujishiro M, Tamaru JI, Mochiki E, Ishida H, Tada T. Diagnosis using deep-learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus. 2019;16:180-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 25. | Li B, Cai SL, Tan WM, Li JC, Yalikong A, Feng XS, Yu HH, Lu PX, Feng Z, Yao LQ, Zhou PH, Yan B, Zhong YS. Comparative study on artificial intelligence systems for detecting early esophageal squamous cell carcinoma between narrow-band and white-light imaging. World J Gastroenterol. 2021;27:281-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 26. | Ohmori M, Ishihara R, Aoyama K, Nakagawa K, Iwagami H, Matsuura N, Shichijo S, Yamamoto K, Nagaike K, Nakahara M, Inoue T, Aoi K, Okada H, Tada T. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2020;91:301-309.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 27. | Tan MC, Bhushan S, Quang T, Schwarz R, Patel KH, Yu X, Li Z, Wang G, Zhang F, Wang X, Xu H, Richards-Kortum RR, Anandasabapathy S. Automated software-assisted diagnosis of esophageal squamous cell neoplasia using high-resolution microendoscopy. Gastrointest Endosc. 2021;93:831-838.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 29. | Liu G, Hua J, Wu Z, Meng T, Sun M, Huang P, He X, Sun W, Li X, Chen Y. Automatic classification of esophageal lesions in endoscopic images using a convolutional neural network. Ann Transl Med. 2020;8:486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 30. | Liu DY, Gan T, Rao NN, Xing YW, Zheng J, Li S, Luo CS, Zhou ZJ, Wan YL. Identification of lesion images from gastrointestinal endoscope based on feature extraction of combinational methods with and without learning process. Med Image Anal. 2016;32:281-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 31. | Nakagawa K, Ishihara R, Aoyama K, Ohmori M, Nakahira H, Matsuura N, Shichijo S, Nishida T, Yamada T, Yamaguchi S, Ogiyama H, Egawa S, Kishida O, Tada T. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc. 2019;90:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 32. | Shimamoto Y, Ishihara R, Kato Y, Shoji A, Inoue T, Matsueda K, Miyake M, Waki K, Kono M, Fukuda H, Matsuura N, Nagaike K, Aoi K, Yamamoto K, Nakahara M, Nishihara A, Tada T. Real-time assessment of video images for esophageal squamous cell carcinoma invasion depth using artificial intelligence. J Gastroenterol. 2020;55:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 33. | Pimentel-Nunes P, Libânio D, Lage J, Abrantes D, Coimbra M, Esposito G, Hormozdi D, Pepper M, Drasovean S, White JR, Dobru D, Buxbaum J, Ragunath K, Annibale B, Dinis-Ribeiro M. A multicenter prospective study of the real-time use of narrow-band imaging in the diagnosis of premalignant gastric conditions and lesions. Endoscopy. 2016;48:723-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 186] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 34. | White JR, Sami SS, Reddiar D, Mannath J, Ortiz-Fernández-Sordo J, Beg S, Scott R, Thiagarajan P, Ahmad S, Parra-Blanco A, Kasi M, Telakis E, Sultan AA, Davis J, Figgins A, Kaye P, Robinson K, Atherton JC, Ragunath K. Narrow band imaging and serology in the assessment of premalignant gastric pathology. Scand J Gastroenterol. 2018;53:1611-1618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Kimura-Tsuchiya R, Dohi O, Fujita Y, Yagi N, Majima A, Horii Y, Kitaichi T, Onozawa Y, Suzuki K, Tomie A, Okayama T, Yoshida N, Kamada K, Katada K, Uchiyama K, Ishikawa T, Takagi T, Handa O, Konishi H, Kishimoto M, Naito Y, Yanagisawa A, Itoh Y. Magnifying Endoscopy with Blue Laser Imaging Improves the Microstructure Visualization in Early Gastric Cancer: Comparison of Magnifying Endoscopy with Narrow-Band Imaging. Gastroenterol Res Pract. 2017;2017:8303046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Mouri R, Yoshida S, Tanaka S, Oka S, Yoshihara M, Chayama K. Evaluation and validation of computed virtual chromoendoscopy in early gastric cancer. Gastrointest Endosc. 2009;69:1052-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 37. | Guimarães P, Keller A, Fehlmann T, Lammert F, Casper M. Deep-learning based detection of gastric precancerous conditions. Gut. 2020;69:4-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 38. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 427] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 39. | Ishioka M, Hirasawa T, Tada T. Detecting gastric cancer from video images using convolutional neural networks. Dig Endosc. 2019;31:e34-e35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 40. | Korhani Kangi A, Bahrampour A. Predicting the Survival of Gastric Cancer Patients Using Artificial and Bayesian Neural Networks. Asian Pac J Cancer Prev. 2018;19:487-490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 41. | Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, Yang X, Li J, Chen M, Jin C, Chen C, Yu C. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. 2020;23:126-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 42. | Miyaki R, Yoshida S, Tanaka S, Kominami Y, Sanomura Y, Matsuo T, Oka S, Raytchev B, Tamaki T, Koide T, Kaneda K, Yoshihara M, Chayama K. A computer system to be used with laser-based endoscopy for quantitative diagnosis of early gastric cancer. J Clin Gastroenterol. 2015;49:108-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 61] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 43. | Togo R, Yamamichi N, Mabe K, Takahashi Y, Takeuchi C, Kato M, Sakamoto N, Ishihara K, Ogawa T, Haseyama M. Detection of gastritis by a deep convolutional neural network from double-contrast upper gastrointestinal barium X-ray radiography. J Gastroenterol. 2019;54:321-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 44. | Wang S, Zhu Y, Yu L, Chen H, Lin H, Wan X, Fan X, Heng PA. RMDL: Recalibrated multi-instance deep learning for whole slide gastric image classification. Med Image Anal. 2019;58:101549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 45. | Luo H, Xu G, Li C, He L, Luo L, Wang Z, Jing B, Deng Y, Jin Y, Li Y, Li B, Tan W, He C, Seeruttun SR, Wu Q, Huang J, Huang DW, Chen B, Lin SB, Chen QM, Yuan CM, Chen HX, Pu HY, Zhou F, He Y, Xu RH. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645-1654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 46. | Namikawa K, Hirasawa T, Nakano K, Ikenoyama Y, Ishioka M, Shiroma S, Tokai Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Artificial intelligence-based diagnostic system classifying gastric cancers and ulcers: comparison between the original and newly developed systems. Endoscopy. 2020;52:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 47. | Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Hojo M, Yao T, Nagahara A, Tada T. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol Hepatol. 2021;36:482-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 48. | Zhou C, Hu J, Wang Y, Ji MH, Tong J, Yang JJ, Xia H. A machine learning-based predictor for the identification of the recurrence of patients with gastric cancer after operation. Sci Rep. 2021;11:1571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 49. | Zhang Y, Li F, Yuan F, Zhang K, Huo L, Dong Z, Lang Y, Zhang Y, Wang M, Gao Z, Qin Z, Shen L. Diagnosing chronic atrophic gastritis by gastroscopy using artificial intelligence. Dig Liver Dis. 2020;52:566-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 50. | Kanesaka T, Lee TC, Uedo N, Lin KP, Chen HZ, Lee JY, Wang HP, Chang HT. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc. 2018;87:1339-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 51. | Sakai Y, Takemoto S, Hori K, Nishimura M, Ikematsu H, Yano T, Yokota H. Automatic detection of early gastric cancer in endoscopic images using a transferring convolutional neural network. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:4138-4141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 52. | Wu L, Zhou W, Wan X, Zhang J, Shen L, Hu S, Ding Q, Mu G, Yin A, Huang X, Liu J, Jiang X, Wang Z, Deng Y, Liu M, Lin R, Ling T, Li P, Wu Q, Jin P, Chen J, Yu H. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy. 2019;51:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 53. | Gao Y, Zhang ZD, Li S, Guo YT, Wu QY, Liu SH, Yang SJ, Ding L, Zhao BC, Lu Y. Deep neural network-assisted computed tomography diagnosis of metastatic lymph nodes from gastric cancer. Chin Med J (Engl). 2019;132:2804-2811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 54. | Jagric T, Potrc S, Jagric T. Prediction of liver metastases after gastric cancer resection with the use of learning vector quantization neural networks. Dig Dis Sci. 2010;55:3252-3261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 55. | Chen T, Zhang C, Liu Y, Zhao Y, Lin D, Hu Y, Yu J, Li G. A gastric cancer LncRNAs model for MSI and survival prediction based on support vector machine. BMC Genomics. 2019;20:846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Nakahira H, Ishihara R, Aoyama K, Kono M, Fukuda H, Shimamoto Y, Nakagawa K, Ohmori M, Iwatsubo T, Iwagami H, Matsuno K, Inoue S, Matsuura N, Shichijo S, Maekawa A, Kanesaka T, Yamamoto S, Takeuchi Y, Higashino K, Uedo N, Matsunaga T, Tada T. Stratification of gastric cancer risk using a deep neural network. JGH Open. 2020;4:466-471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 57. | Jiang Y, Xie J, Han Z, Liu W, Xi S, Huang L, Huang W, Lin T, Zhao L, Hu Y, Yu J, Zhang Q, Li T, Cai S, Li G. Immunomarker Support Vector Machine Classifier for Prediction of Gastric Cancer Survival and Adjuvant Chemotherapeutic Benefit. Clin Cancer Res. 2018;24:5574-5584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 58. | Que SJ, Chen QY, Qing-Zhong, Liu ZY, Wang JB, Lin JX, Lu J, Cao LL, Lin M, Tu RH, Huang ZN, Lin JL, Zheng HL, Li P, Zheng CH, Huang CM, Xie JW. Application of preoperative artificial neural network based on blood biomarkers and clinicopathological parameters for predicting long-term survival of patients with gastric cancer. World J Gastroenterol. 2019;25:6451-6464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 59. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc. 2019;89:806-815.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 60. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging-bright and linked color imaging: a single-center prospective study. Ann Gastroenterol. 2018;31:462-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (1)] |
| 61. | Shichijo S, Endo Y, Aoyama K, Takeuchi Y, Ozawa T, Takiyama H, Matsuo K, Fujishiro M, Ishihara S, Ishihara R, Tada T. Application of convolutional neural networks for evaluating Helicobacter pylori infection status on the basis of endoscopic images. Scand J Gastroenterol. 2019;54:158-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 62. | Martin DR, Hanson JA, Gullapalli RR, Schultz FA, Sethi A, Clark DP. A Deep Learning Convolutional Neural Network Can Recognize Common Patterns of Injury in Gastric Pathology. Arch Pathol Lab Med. 2020;144:370-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 63. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Endoscopic three-categorical diagnosis of Helicobacter pylori infection using linked color imaging and deep learning: a single-center prospective study (with video). Gastric Cancer. 2020;23:1033-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 64. | Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K, Tada T. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. EBioMedicine. 2017;25:106-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 65. | Zheng W, Zhang X, Kim JJ, Zhu X, Ye G, Ye B, Wang J, Luo S, Li J, Yu T, Liu J, Hu W, Si J. High Accuracy of Convolutional Neural Network for Evaluation of Helicobacter pylori Infection Based on Endoscopic Images: Preliminary Experience. Clin Transl Gastroenterol. 2019;10:e00109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 66. | Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol. 2019;25:1666-1683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 211] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (5)] |
| 67. | Wong GL, Ma AJ, Deng H, Ching JY, Wong VW, Tse YK, Yip TC, Lau LH, Liu HH, Leung CM, Tsang SW, Chan CW, Lau JY, Yuen PC, Chan FK. Machine learning model to predict recurrent ulcer bleeding in patients with history of idiopathic gastroduodenal ulcer bleeding. Aliment Pharmacol Ther. 2019;49:912-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (1)] |
| 68. | Seo DW, Yi H, Park B, Kim YJ, Jung DH, Woo I, Sohn CH, Ko BS, Kim N, Kim WY. Prediction of Adverse Events in Stable Non-Variceal Gastrointestinal Bleeding Using Machine Learning. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 69. | Shung DL, Au B, Taylor RA, Tay JK, Laursen SB, Stanley AJ, Dalton HR, Ngu J, Schultz M, Laine L. Validation of a Machine Learning Model That Outperforms Clinical Risk Scoring Systems for Upper Gastrointestinal Bleeding. Gastroenterology. 2020;158:160-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 70. | Das A, Ben-Menachem T, Cooper GS, Chak A, Sivak MV Jr, Gonet JA, Wong RC. Prediction of outcome in acute lower-gastrointestinal haemorrhage based on an artificial neural network: internal and external validation of a predictive model. Lancet. 2003;362:1261-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 150] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 71. | Das A, Ben-Menachem T, Farooq FT, Cooper GS, Chak A, Sivak MV Jr, Wong RC. Artificial neural network as a predictive instrument in patients with acute nonvariceal upper gastrointestinal hemorrhage. Gastroenterology. 2008;134:65-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 72. | Loftus TJ, Brakenridge SC, Croft CA, Smith RS, Efron PA, Moore FA, Mohr AM, Jordan JR. Neural network prediction of severe lower intestinal bleeding and the need for surgical intervention. J Surg Res. 2017;212:42-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 73. | Ayaru L, Ypsilantis PP, Nanapragasam A, Choi RC, Thillanathan A, Min-Ho L, Montana G. Prediction of Outcome in Acute Lower Gastrointestinal Bleeding Using Gradient Boosting. PLoS One. 2015;10:e0132485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 74. | Bressler B, Paszat LF, Chen Z, Rothwell DM, Vinden C, Rabeneck L. Rates of new or missed colorectal cancers after colonoscopy and their risk factors: a population-based analysis. Gastroenterology. 2007;132:96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 449] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 75. | Jover R, Zapater P, Polanía E, Bujanda L, Lanas A, Hermo JA, Cubiella J, Ono A, González-Méndez Y, Peris A, Pellisé M, Seoane A, Herreros-de-Tejada A, Ponce M, Marín-Gabriel JC, Chaparro M, Cacho G, Fernández-Díez S, Arenas J, Sopeña F, de-Castro L, Vega-Villaamil P, Rodríguez-Soler M, Carballo F, Salas D, Morillas JD, Andreu M, Quintero E, Castells A; COLONPREV study investigators. Modifiable endoscopic factors that influence the adenoma detection rate in colorectal cancer screening colonoscopies. Gastrointest Endosc. 2013;77:381-389.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 76. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. 2021;93:77-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 308] [Article Influence: 77.0] [Reference Citation Analysis (1)] |
| 77. | Samadder NJ, Curtin K, Tuohy TM, Pappas L, Boucher K, Provenzale D, Rowe KG, Mineau GP, Smith K, Pimentel R, Kirchhoff AC, Burt RW. Characteristics of missed or interval colorectal cancer and patient survival: a population-based study. Gastroenterology. 2014;146:950-960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 233] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 78. | Lee CK, Park DI, Lee SH, Hwangbo Y, Eun CS, Han DS, Cha JM, Lee BI, Shin JE. Participation by experienced endoscopy nurses increases the detection rate of colon polyps during a screening colonoscopy: a multicenter, prospective, randomized study. Gastrointest Endosc. 2011;74:1094-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 79. | Aslanian HR, Shieh FK, Chan FW, Ciarleglio MM, Deng Y, Rogart JN, Jamidar PA, Siddiqui UD. Nurse observation during colonoscopy increases polyp detection: a randomized prospective study. Am J Gastroenterol. 2013;108:166-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 80. | Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, Wang Z, Zhou W, An P, Huang X, Jiang X, Li Y, Wan X, Hu S, Chen Y, Hu X, Xu Y, Zhu X, Li S, Yao L, He X, Chen D, Huang L, Wei X, Wang X, Yu H. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 265] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 81. | Klare P, Sander C, Prinzen M, Haller B, Nowack S, Abdelhafez M, Poszler A, Brown H, Wilhelm D, Schmid RM, von Delius S, Wittenberg T. Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc. 2019;89:576-582.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 82. | Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, Ferrara E, Spadaccini M, Alkandari A, Fugazza A, Anderloni A, Galtieri PA, Pellegatta G, Carrara S, Di Leo M, Craviotto V, Lamonaca L, Lorenzetti R, Andrealli A, Antonelli G, Wallace M, Sharma P, Rosch T, Hassan C. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology. 2020;159:512-520.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 393] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 83. | Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, Li GC, Liu GQ, He YS, Zuo XL, Li YQ. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc. 2020;91:415-424.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 208] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 84. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 433] [Article Influence: 61.9] [Reference Citation Analysis (1)] |
| 85. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 86. | Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, Xiong F, Wang H, Song Y, Pan Y, Zhou G. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. 2020;5:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 294] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 87. | Liu WN, Zhang YY, Bian XQ, Wang LJ, Yang Q, Zhang XD, Huang J. Study on detection rate of polyps and adenomas in artificial-intelligence-aided colonoscopy. Saudi J Gastroenterol. 2020;26:13-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 126] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 88. | Bernal J, Tajkbaksh N, Sanchez FJ, Matuszewski BJ, Hao Chen, Lequan Yu, Angermann Q, Romain O, Rustad B, Balasingham I, Pogorelov K, Sungbin Choi, Debard Q, Maier-Hein L, Speidel S, Stoyanov D, Brandao P, Cordova H, Sanchez-Montes C, Gurudu SR, Fernandez-Esparrach G, Dray X, Jianming Liang, Histace A. Comparative Validation of Polyp Detection Methods in Video Colonoscopy: Results From the MICCAI 2015 Endoscopic Vision Challenge. IEEE Trans Med Imaging. 2017;36:1231-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 183] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 89. | Blanes-Vidal V, Baatrup G, Nadimi ES. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 2019;58:S29-S36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 90. | Fernández-Esparrach G, Bernal J, López-Cerón M, Córdova H, Sánchez-Montes C, Rodríguez de Miguel C, Sánchez FJ. Exploring the clinical potential of an automatic colonic polyp detection method based on the creation of energy maps. Endoscopy. 2016;48:837-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 91. | Figueiredo PN, Figueiredo IN, Pinto L, Kumar S, Tsai YR, Mamonov AV. Polyp detection with computer-aided diagnosis in white light colonoscopy: comparison of three different methods. Endosc Int Open. 2019;7:E209-E215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 92. | Hassan C, Wallace MB, Sharma P, Maselli R, Craviotto V, Spadaccini M, Repici A. New artificial intelligence system: first validation study vs experienced endoscopists for colorectal polyp detection. Gut. 2020;69:799-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 93. | Kominami Y, Yoshida S, Tanaka S, Sanomura Y, Hirakawa T, Raytchev B, Tamaki T, Koide T, Kaneda K, Chayama K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 94. | Lequan Yu, Hao Chen, Qi Dou, Jing Qin, Pheng Ann Heng. Integrating Online and Offline Three-Dimensional Deep Learning for Automated Polyp Detection in Colonoscopy Videos. IEEE J Biomed Health Inform. 2017;21:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 106] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 95. | Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology. 2018;154:2027-2029.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 269] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 96. | Misawa M, Kudo SE, Mori Y, Hotta K, Ohtsuka K, Matsuda T, Saito S, Kudo T, Baba T, Ishida F, Itoh H, Oda M, Mori K. Development of a computer-aided detection system for colonoscopy and a publicly accessible large colonoscopy video database (with video). Gastrointest Endosc. 2021;93:960-967.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 105] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 97. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 355] [Article Influence: 50.7] [Reference Citation Analysis (1)] |
| 98. | Wang P, Xiao X, Glissen Brown JR, Berzin TM, Tu M, Xiong F, Hu X, Liu P, Song Y, Zhang D, Yang X, Li L, He J, Yi X, Liu J, Liu X. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng. 2018;2:741-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 270] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 99. | Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, Heij LR, Tan X, Richman SD, Krause J, Alwers E, Jenniskens J, Offermans K, Gray R, Brenner H, Chang-Claude J, Trautwein C, Pearson AT, Boor P, Luedde T, Gaisa NT, Hoffmeister M, Kather JN. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology. 2020;159:1406-1416.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 220] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 100. | Ito N, Kawahira H, Nakashima H, Uesato M, Miyauchi H, Matsubara H. Endoscopic Diagnostic Support System for cT1b Colorectal Cancer Using Deep Learning. Oncology. 2019;96:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 101. | Kudo SE, Misawa M, Mori Y, Hotta K, Ohtsuka K, Ikematsu H, Saito Y, Takeda K, Nakamura H, Ichimasa K, Ishigaki T, Toyoshima N, Kudo T, Hayashi T, Wakamura K, Baba T, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin Gastroenterol Hepatol. 2020;18:1874-1881.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 102. | . The Paris endoscopic classification of superficial neoplastic lesions: esophagus, stomach, and colon: November 30 to December 1, 2002. Gastrointest Endosc. 2003;58:S3-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1117] [Cited by in RCA: 1325] [Article Influence: 60.2] [Reference Citation Analysis (4)] |
| 103. | Endoscopic Classification Review Group. Update on the paris classification of superficial neoplastic lesions in the digestive tract. Endoscopy. 2005;37:570-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 559] [Cited by in RCA: 646] [Article Influence: 32.3] [Reference Citation Analysis (1)] |
| 104. | van Doorn SC, Hazewinkel Y, East JE, van Leerdam ME, Rastogi A, Pellisé M, Sanduleanu-Dascalescu S, Bastiaansen BA, Fockens P, Dekker E. Polyp morphology: an interobserver evaluation for the Paris classification among international experts. Am J Gastroenterol. 2015;110:180-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (1)] |
| 105. | Wang TD, Van Dam J. Optical biopsy: a new frontier in endoscopic detection and diagnosis. Clin Gastroenterol Hepatol. 2004;2:744-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 93] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 106. | Hassan C, Pickhardt PJ, Rex DK. A resect and discard strategy would improve cost-effectiveness of colorectal cancer screening. Clin Gastroenterol Hepatol. 2010;8:865-869, 869.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 221] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 107. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 412] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 108. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 276] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 109. | Gross S, Trautwein C, Behrens A, Winograd R, Palm S, Lutz HH, Schirin-Sokhan R, Hecker H, Aach T, Tischendorf JJ. Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc. 2011;74:1354-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 110. | Horiuchi H, Tamai N, Kamba S, Inomata H, Ohya TR, Sumiyama K. Real-time computer-aided diagnosis of diminutive rectosigmoid polyps using an auto-fluorescence imaging system and novel color intensity analysis software. Scand J Gastroenterol. 2019;54:800-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 111. | Misawa M, Kudo SE, Mori Y, Nakamura H, Kataoka S, Maeda Y, Kudo T, Hayashi T, Wakamura K, Miyachi H, Katagiri A, Baba T, Ishida F, Inoue H, Nimura Y, Mori K. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology. 2016;150:1531-1532.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 125] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 112. | Mori Y, Kudo SE, Chiu PW, Singh R, Misawa M, Wakamura K, Kudo T, Hayashi T, Katagiri A, Miyachi H, Ishida F, Maeda Y, Inoue H, Nimura Y, Oda M, Mori K. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy. 2016;48:1110-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 113. | Mori Y, Kudo SE, Wakamura K, Misawa M, Ogawa Y, Kutsukawa M, Kudo T, Hayashi T, Miyachi H, Ishida F, Inoue H. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest Endosc. 2015;81:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 114. | Sánchez-Montes C, Sánchez FJ, Bernal J, Córdova H, López-Cerón M, Cuatrecasas M, Rodríguez de Miguel C, García-Rodríguez A, Garcés-Durán R, Pellisé M, Llach J, Fernández-Esparrach G. Computer-aided prediction of polyp histology on white light colonoscopy using surface pattern analysis. Endoscopy. 2019;51:261-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 115. | Tischendorf JJ, Gross S, Winograd R, Hecker H, Auer R, Behrens A, Trautwein C, Aach T, Stehle T. Computer-aided classification of colorectal polyps based on vascular patterns: a pilot study. Endoscopy. 2010;42:203-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 116. | Ikehara H, Saito Y, Matsuda T, Uraoka T, Murakami Y. Diagnosis of depth of invasion for early colorectal cancer using magnifying colonoscopy. J Gastroenterol Hepatol. 2010;25:905-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 117. | Kitajima K, Fujimori T, Fujii S, Takeda J, Ohkura Y, Kawamata H, Kumamoto T, Ishiguro S, Kato Y, Shimoda T, Iwashita A, Ajioka Y, Watanabe H, Watanabe T, Muto T, Nagasako K. Correlations between lymph node metastasis and depth of submucosal invasion in submucosal invasive colorectal carcinoma: a Japanese collaborative study. J Gastroenterol. 2004;39:534-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 468] [Cited by in RCA: 486] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 118. | Matsuda T, Saito Y, Nakajima T, Sakamoto T, Ikematsu H, Sano Y, Fu KI, Fujii T. Macroscopic estimation of submucosal invasion in the colon. Tech Gastrointest Endosc. 2011;13:24-32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 119. | Mori Y, Kudo SE, East JE, Rastogi A, Bretthauer M, Misawa M, Sekiguchi M, Matsuda T, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Kudo T, Mori K. Cost savings in colonoscopy with artificial intelligence-aided polyp diagnosis: an add-on analysis of a clinical trial (with video). Gastrointest Endosc. 2020;92:905-911.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 120. | Thakkar S, Carleton NM, Rao B, Syed A. Use of Artificial Intelligence-Based Analytics From Live Colonoscopies to Optimize the Quality of the Colonoscopy Examination in Real Time: Proof of Concept. Gastroenterology. 2020;158:1219-1221.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 121. | Reichling C, Taieb J, Derangere V, Klopfenstein Q, Le Malicot K, Gornet JM, Becheur H, Fein F, Cojocarasu O, Kaminsky MC, Lagasse JP, Luet D, Nguyen S, Etienne PL, Gasmi M, Vanoli A, Perrier H, Puig PL, Emile JF, Lepage C, Ghiringhelli F. Artificial intelligence-guided tissue analysis combined with immune infiltrate assessment predicts stage III colon cancer outcomes in PETACC08 study. Gut. 2020;69:681-690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 122. | Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA, Gaiser T, Marx A, Valous NA, Ferber D, Jansen L, Reyes-Aldasoro CC, Zörnig I, Jäger D, Brenner H, Chang-Claude J, Hoffmeister M, Halama N. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019;16:e1002730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 492] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 123. | Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, Farstad IN, Domingo E, Church DN, Nesbakken A, Shepherd NA, Tomlinson I, Kerr R, Novelli M, Kerr DJ, Danielsen HE. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 363] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 124. | Yang YJ. The Future of Capsule Endoscopy: The Role of Artificial Intelligence and Other Technical Advancements. Clin Endosc. 2020;53:387-394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 125. | Liu G, Yan G, Kuang S, Wang Y. Detection of small bowel tumor based on multi-scale curvelet analysis and fractal technology in capsule endoscopy. Comput Biol Med. 2016;70:131-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 126. | Li B, Meng MQ. Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Trans Inf Technol Biomed. 2012;16:323-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 127. | Li B, Meng MQ. Computer-based detection of bleeding and ulcer in wireless capsule endoscopy images by chromaticity moments. Comput Biol Med. 2009;39:141-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 128. | Aoki T, Yamada A, Aoyama K, Saito H, Fujisawa G, Odawara N, Kondo R, Tsuboi A, Ishibashi R, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Clinical usefulness of a deep learning-based system as the first screening on small-bowel capsule endoscopy reading. Dig Endosc. 2020;32:585-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 129. | Soffer S, Klang E, Shimon O, Nachmias N, Eliakim R, Ben-Horin S, Kopylov U, Barash Y. Deep learning for wireless capsule endoscopy: a systematic review and meta-analysis. Gastrointest Endosc. 2020;92:831-839.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 115] [Article Influence: 23.0] [Reference Citation Analysis (3)] |
| 130. | Aoki T, Yamada A, Kato Y, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J Gastroenterol Hepatol. 2020;35:1196-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 131. | Ding Z, Shi H, Zhang H, Meng L, Fan M, Han C, Zhang K, Ming F, Xie X, Liu H, Liu J, Lin R, Hou X. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology. 2019;157:1044-1054.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 209] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 132. | Fu Y, Zhang W, Mandal M, Meng MQ. Computer-aided bleeding detection in WCE video. IEEE J Biomed Health Inform. 2014;18:636-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 133. | Hassan AR, Haque MA. Computer-aided gastrointestinal hemorrhage detection in wireless capsule endoscopy videos. Comput Methods Programs Biomed. 2015;122:341-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 134. | Iakovidis DK, Koulaouzidis A. Automatic lesion detection in capsule endoscopy based on color saliency: closer to an essential adjunct for reviewing software. Gastrointest Endosc. 2014;80:877-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 135. | Leenhardt R, Vasseur P, Li C, Saurin JC, Rahmi G, Cholet F, Becq A, Marteau P, Histace A, Dray X; CAD-CAP Database Working Group. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc. 2019;89:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 136. | Lv G, Yan G, Wang Z. Bleeding detection in wireless capsule endoscopy images based on color invariants and spatial pyramids using support vector machines. Annu Int Conf IEEE Eng Med Biol Soc. 2011;2011:6643-6646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 137. | Noya F, Alvarez-Gonzalez MA, Benitez R. Automated angiodysplasia detection from wireless capsule endoscopy. Annu Int Conf IEEE Eng Med Biol Soc. 2017;2017:3158-3161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 138. | Pan G, Yan G, Song X, Qiu X. BP neural network classification for bleeding detection in wireless capsule endoscopy. J Med Eng Technol. 2009;33:575-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 139. | Sainju S, Bui FM, Wahid KA. Automated bleeding detection in capsule endoscopy videos using statistical features and region growing. J Med Syst. 2014;38:25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 140. | Tsuboi A, Oka S, Aoyama K, Saito H, Aoki T, Yamada A, Matsuda T, Fujishiro M, Ishihara S, Nakahori M, Koike K, Tanaka S, Tada T. Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images. Dig Endosc. 2020;32:382-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 141. | Xing X, Jia X, Meng MQ. Bleeding Detection in Wireless Capsule Endoscopy Image Video Using Superpixel-Color Histogram and a Subspace KNN Classifier. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 142. | Yuan Y, Li B, Meng MQ. Bleeding Frame and Region Detection in the Wireless Capsule Endoscopy Video. IEEE J Biomed Health Inform. 2016;20:624-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 143. | Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Tanaka S, Koike K, Tada T. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2019;89:357-363.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 175] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 144. | Otani K, Nakada A, Kurose Y, Niikura R, Yamada A, Aoki T, Nakanishi H, Doyama H, Hasatani K, Sumiyoshi T, Kitsuregawa M, Harada T, Koike K. Automatic detection of different types of small-bowel lesions on capsule endoscopy images using a newly developed deep convolutional neural network. Endoscopy. 2020;52:786-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 145. | Fan S, Xu L, Fan Y, Wei K, Li L. Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys Med Biol. 2018;63:165001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 146. | Wang S, Xing Y, Zhang L, Gao H, Zhang H. A systematic evaluation and optimization of automatic detection of ulcers in wireless capsule endoscopy on a large dataset using deep convolutional neural networks. Phys Med Biol. 2019;64:235014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 147. | Klang E, Barash Y, Margalit RY, Soffer S, Shimon O, Albshesh A, Ben-Horin S, Amitai MM, Eliakim R, Kopylov U. Deep learning algorithms for automated detection of Crohn's disease ulcers by video capsule endoscopy. Gastrointest Endosc. 2020;91:606-613.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 148. | Charisis VS, Hadjileontiadis LJ. Potential of hybrid adaptive filtering in inflammatory lesion detection from capsule endoscopy images. World J Gastroenterol. 2016;22:8641-8657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 149. | Saito H, Aoki T, Aoyama K, Kato Y, Tsuboi A, Yamada A, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2020;92:144-151.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (1)] |
| 150. | Yuan Y, Meng MQ. Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys. 2017;44:1379-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 151. | Tenório JM, Hummel AD, Cohrs FM, Sdepanian VL, Pisa IT, de Fátima Marin H. Artificial intelligence techniques applied to the development of a decision-support system for diagnosing celiac disease. Int J Med Inform. 2011;80:793-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 152. | Zhou T, Han G, Li BN, Lin Z, Ciaccio EJ, Green PH, Qin J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput Biol Med. 2017;85:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 153. | Wimmer G, Vécsei A, Uhl A. CNN transfer learning for the automated diagnosis of celiac disease. IEEE. 2016;1-6. [DOI] [Full Text] |
| 154. | Wang X, Qian H, Ciaccio EJ, Lewis SK, Bhagat G, Green PH, Xu S, Huang L, Gao R, Liu Y. Celiac disease diagnosis from videocapsule endoscopy images with residual learning and deep feature extraction. Comput Methods Programs Biomed. 2020;187:105236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 155. | Fenwick A. The global burden of neglected tropical diseases. Public Health. 2012;126:233-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 156. | Chen H, Chen J, Peng Q, Sun G, Gan T. Automatic hookworm image detection for wireless capsule endoscopy using hybrid color gradient and contourlet transform. Int Conf Biomed Eng Inf. 2013;116-120. [DOI] [Full Text] |
| 157. | He JY, Wu X, Jiang YG, Peng Q, Jain R. Hookworm Detection in Wireless Capsule Endoscopy Images With Deep Learning. IEEE Trans Image Process. 2018;27:2379-2392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 158. | Wu X, Chen H, Gan T, Chen J, Ngo CW, Peng Q. Automatic Hookworm Detection in Wireless Capsule Endoscopy Images. IEEE Trans Med Imaging. 2016;35:1741-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 159. | Leenhardt R, Souchaud M, Houist G, Le Mouel JP, Saurin JC, Cholet F, Rahmi G, Leandri C, Histace A, Dray X. A neural network-based algorithm for assessing the cleanliness of small bowel during capsule endoscopy. Endoscopy. 2021;53:932-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 160. | Noorda R, Nevárez A, Colomer A, Pons Beltrán V, Naranjo V. Automatic evaluation of degree of cleanliness in capsule endoscopy based on a novel CNN architecture. Sci Rep. 2020;10:17706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 161. | Borg-Bartolo SP, Boyapati RK, Satsangi J, Kalla R. Precision medicine in inflammatory bowel disease: concept, progress and challenges. F1000Res. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 162. | Seyed Tabib NS, Madgwick M, Sudhakar P, Verstockt B, Korcsmaros T, Vermeire S. Big data in IBD: big progress for clinical practice. Gut. 2020;69:1520-1532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 143] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 163. | Olivera P, Danese S, Jay N, Natoli G, Peyrin-Biroulet L. Big data in IBD: a look into the future. Nat Rev Gastroenterol Hepatol. 2019;16:312-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 164. | Bossuyt P, Vermeire S, Bisschops R. Scoring endoscopic disease activity in IBD: artificial intelligence sees more and better than we do. Gut. 2020;69:788-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 165. | Iacucci M, Furfaro F, Matsumoto T, Uraoka T, Smith S, Ghosh S, Kiesslich R. Advanced endoscopic techniques in the assessment of inflammatory bowel disease: new technology, new era. Gut. 2019;68:562-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 166. | Ozawa T, Ishihara S, Fujishiro M, Saito H, Kumagai Y, Shichijo S, Aoyama K, Tada T. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest Endosc. 2019;89:416-421.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 167. | Zezos P, Borowski K, Bajaj G, Boland K, Sheasgreen C, Tessolini JM, Silverberg MS. 439 - Toward Computer-Based Automated Mayo Score Classification in Ulcerative Colitis through Classical and Deep Machine Learning. Gastroenterology. 2018;154:S-99. [DOI] [Full Text] |
| 168. | Khorasani HM, Usefi H, Peña-Castillo L. Detecting ulcerative colitis from colon samples using efficient feature selection and machine learning. Sci Rep. 2020;10:13744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (1)] |
| 169. | Kumar R, Zhao Q, Seshamani S, Mullin G, Hager G, Dassopoulos T. Assessment of Crohn's disease lesions in wireless capsule endoscopy images. IEEE Trans Biomed Eng. 2012;59:355-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 170. | Mahapatra D, Vos FM, Buhmann JM. Active learning based segmentation of Crohns disease from abdominal MRI. Comput Methods Programs Biomed. 2016;128:75-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 171. | Mossotto E, Ashton JJ, Coelho T, Beattie RM, MacArthur BD, Ennis S. Classification of Paediatric Inflammatory Bowel Disease using Machine Learning. Sci Rep. 2017;7:2427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 172. | Maeda Y, Kudo SE, Mori Y, Misawa M, Ogata N, Sasanuma S, Wakamura K, Oda M, Mori K, Ohtsuka K. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. 2019;89:408-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 165] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 173. | Matalka II, Al-Omari FA, Salama RM, Mohtaseb AH. A novel approach for quantitative assessment of mucosal damage in inflammatory bowel disease. Diagn Pathol. 2013;8:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 174. | Niehaus KE, Uhlig HH, Clifton DA. Phenotypic characterisation of Crohn's disease severity. Annu Int Conf IEEE Eng Med Biol Soc. 2015;2015:7023-7026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 175. | Reddy BK, Delen D, Agrawal RK. Predicting and explaining inflammation in Crohn's disease patients using predictive analytics methods and electronic medical record data. Health Informatics J. 2019;25:1201-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 176. | Stidham RW, Enchakalody B, Waljee AK, Higgins PDR, Wang SC, Su GL, Wasnik AP, Al-Hawary M. Assessing Small Bowel Stricturing and Morphology in Crohn's Disease Using Semi-automated Image Analysis. Inflamm Bowel Dis. 2020;26:734-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 177. | Stidham RW, Liu W, Bishu S, Rice MD, Higgins PDR, Zhu J, Nallamothu BK, Waljee AK. Performance of a Deep Learning Model vs Human Reviewers in Grading Endoscopic Disease Severity of Patients With Ulcerative Colitis. JAMA Netw Open. 2019;2:e193963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 175] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 178. | Takenaka K, Ohtsuka K, Fujii T, Negi M, Suzuki K, Shimizu H, Oshima S, Akiyama S, Motobayashi M, Nagahori M, Saito E, Matsuoka K, Watanabe M. Development and Validation of a Deep Neural Network for Accurate Evaluation of Endoscopic Images From Patients With Ulcerative Colitis. Gastroenterology. 2020;158:2150-2157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 181] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 179. | Yao H, Najarian K, Gryak J, Bishu S, Rice MD, Waljee AK, Wilkins HJ, Stidham RW. Fully automated endoscopic disease activity assessment in ulcerative colitis. Gastrointest Endosc. 2021;93:728-736.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 83] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 180. | Doherty MK, Ding T, Koumpouras C, Telesco SE, Monast C, Das A, Brodmerkel C, Schloss PD. Fecal Microbiota Signatures Are Associated with Response to Ustekinumab Therapy among Crohn's Disease Patients. mBio. 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 181. | Douglas GM, Hansen R, Jones CMA, Dunn KA, Comeau AM, Bielawski JP, Tayler R, El-Omar EM, Russell RK, Hold GL, Langille MGI, Van Limbergen J. Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn's disease. Microbiome. 2018;6:13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 182. | Waljee AK, Joyce JC, Wang S, Saxena A, Hart M, Zhu J, Higgins PD. Algorithms outperform metabolite tests in predicting response of patients with inflammatory bowel disease to thiopurines. Clin Gastroenterol Hepatol. 2010;8:143-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 183. | Waljee AK, Lipson R, Wiitala WL, Zhang Y, Liu B, Zhu J, Wallace B, Govani SM, Stidham RW, Hayward R, Higgins PDR. Predicting Hospitalization and Outpatient Corticosteroid Use in Inflammatory Bowel Disease Patients Using Machine Learning. Inflamm Bowel Dis. 2017;24:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 184. | Waljee AK, Liu B, Sauder K, Zhu J, Govani SM, Stidham RW, Higgins PDR. Predicting corticosteroid-free endoscopic remission with vedolizumab in ulcerative colitis. Aliment Pharmacol Ther. 2018;47:763-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 185. | Waljee AK, Sauder K, Patel A, Segar S, Liu B, Zhang Y, Zhu J, Stidham RW, Balis U, Higgins PDR. Machine Learning Algorithms for Objective Remission and Clinical Outcomes with Thiopurines. J Crohns Colitis. 2017;11:801-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 186. | Waljee AK, Wallace BI, Cohen-Mekelburg S, Liu Y, Liu B, Sauder K, Stidham RW, Zhu J, Higgins PDR. Development and Validation of Machine Learning Models in Prediction of Remission in Patients With Moderate to Severe Crohn Disease. JAMA Netw Open. 2019;2:e193721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 187. | Isakov O, Dotan I, Ben-Shachar S. Machine Learning-Based Gene Prioritization Identifies Novel Candidate Risk Genes for Inflammatory Bowel Disease. Inflamm Bowel Dis. 2017;23:1516-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 188. | Wei Z, Wang W, Bradfield J, Li J, Cardinale C, Frackelton E, Kim C, Mentch F, Van Steen K, Visscher PM, Baldassano RN, Hakonarson H; International IBD Genetics Consortium. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet. 2013;92:1008-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 189. | Hou JK, Chang M, Nguyen T, Kramer JR, Richardson P, Sansgiry S, D'Avolio LW, El-Serag HB. Automated identification of surveillance colonoscopy in inflammatory bowel disease using natural language processing. Dig Dis Sci. 2013;58:936-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 190. | Firouzi F, Rashidi M, Hashemi S, Kangavari M, Bahari A, Daryani NE, Emam MM, Naderi N, Shalmani HM, Farnood A, Zali M. A decision tree-based approach for determining low bone mineral density in inflammatory bowel disease using WEKA software. Eur J Gastroenterol Hepatol. 2007;19:1075-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 191. | Ahn JC, Connell A, Simonetto DA, Hughes C, Shah VH. Application of Artificial Intelligence for the Diagnosis and Treatment of Liver Diseases. Hepatology. 2021;73:2546-2563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 101] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 192. | Spann A, Yasodhara A, Kang J, Watt K, Wang B, Goldenberg A, Bhat M. Applying Machine Learning in Liver Disease and Transplantation: A Comprehensive Review. Hepatology. 2020;71:1093-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 193. | Chen T, Tu S, Wang H, Liu X, Li F, Jin W, Liang X, Zhang X, Wang J. Computer-aided diagnosis of gallbladder polyps based on high resolution ultrasonography. Comput Methods Programs Biomed. 2020;185:105118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 194. | Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Duncan JS, Weinreb JC, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29:3338-3347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 207] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 195. | Kim DW, Lee G, Kim SY, Ahn G, Lee JG, Lee SS, Kim KW, Park SH, Lee YJ, Kim N. Deep learning-based algorithm to detect primary hepatic malignancy in multiphase CT of patients at high risk for HCC. Eur Radiol. 2021;31:7047-7057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 196. | Kim JW, Ye Q, Forgues M, Chen Y, Budhu A, Sime J, Hofseth LJ, Kaul R, Wang XW. Cancer-associated molecular signature in the tissue samples of patients with cirrhosis. Hepatology. 2004;39:518-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 111] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 197. | Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, Luciani A, Lassau N, Jégou S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100:227-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 198. | Singal AG, Mukherjee A, Elmunzer BJ, Higgins PD, Lok AS, Zhu J, Marrero JA, Waljee AK. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol. 2013;108:1723-1730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 203] [Article Influence: 16.9] [Reference Citation Analysis (1)] |
| 199. | Sun C, Xu A, Liu D, Xiong Z, Zhao F, Ding W. Deep Learning-Based Classification of Liver Cancer Histopathology Images Using Only Global Labels. IEEE J Biomed Health Inform. 2020;24:1643-1651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 200. | Wang CJ, Hamm CA, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Weinreb JC, Duncan JS, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol. 2019;29:3348-3357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 201. | Yasaka K, Akai H, Abe O, Kiryu S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology. 2018;286:887-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 394] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 202. | Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S. Liver Fibrosis: Deep Convolutional Neural Network for Staging by Using Gadoxetic Acid-enhanced Hepatobiliary Phase MR Images. Radiology. 2018;287:146-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (1)] |
| 203. | Abd El-Salam SM, Ezz MM, Hashem S, Elakel W, Salama R, ElMakhzangy H, ElHefnawi M. Performance of machine learning approaches on prediction of esophageal varices for Egyptian chronic hepatitis C patients. Inform Med Unlocked. 2019;17:100267. [RCA] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 204. | Dong TS, Kalani A, Aby ES, Le L, Luu K, Hauer M, Kamath R, Lindor KD, Tabibian JH. Machine Learning-based Development and Validation of a Scoring System for Screening High-Risk Esophageal Varices. Clin Gastroenterol Hepatol. 2019;17:1894-1901.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 205. | Hong WD, Ji YF, Wang D, Chen TZ, Zhu QH. Use of artificial neural network to predict esophageal varices in patients with HBV related cirrhosis. Hepat Mon. 2011;11:544-547. [PubMed] |
| 206. | Liu Y, Ning Z, Örmeci N, An W, Yu Q, Han K, Huang Y, Liu D, Liu F, Li Z, Ding H, Luo H, Zuo C, Liu C, Wang J, Zhang C, Ji J, Wang W, Wang Z, Yuan M, Li L, Zhao Z, Wang G, Li M, Liu Q, Lei J, Tang T, Akçalar S, Çelebioğlu E, Üstüner E, Bilgiç S, Ellik Z, Asiller ÖÖ, Liu Z, Teng G, Chen Y, Hou J, Li X, He X, Dong J, Tian J, Liang P, Ju S, Zhang Y, Qi X. Deep Convolutional Neural Network-Aided Detection of Portal Hypertension in Patients With Cirrhosis. Clin Gastroenterol Hepatol. 2020;18:2998-3007.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 207. | Marozas M, Zykus R, Sakalauskas A, Kupčinskas L, Lukoševičius A. Noninvasive Evaluation of Portal Hypertension Using a Supervised Learning Technique. J Healthc Eng. 2017;2017:6183714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 208. | Qi X, An W, Liu F, Qi R, Wang L, Liu Y, Liu C, Xiang Y, Hui J, Liu Z, Qi X, Peng B, Ding H, Yang Y, He X, Hou J, Tian J, Li Z. Virtual Hepatic Venous Pressure Gradient with CT Angiography (CHESS 1601): A Prospective Multicenter Study for the Noninvasive Diagnosis of Portal Hypertension. Radiology. 2019;290:370-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 209. | Ahmed Y, Hussein RS, Basha TA, Khalifa AM, Ibrahim AS, Abdelmoaty AS, Abdella HM, Fahmy AS. Detecting liver fibrosis using a machine learning-based approach to the quantification of the heart-induced deformation in tagged MR images. NMR Biomed. 2020;33:e4215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 210. | Byra M, Styczynski G, Szmigielski C, Kalinowski P, Michałowski Ł, Paluszkiewicz R, Ziarkiewicz-Wróblewska B, Zieniewicz K, Sobieraj P, Nowicki A. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg. 2018;13:1895-1903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 211. | Chang EK, Yu CY, Clarke R, Hackbarth A, Sanders T, Esrailian E, Hommes DW, Runyon BA. Defining a Patient Population With Cirrhosis: An Automated Algorithm With Natural Language Processing. J Clin Gastroenterol. 2016;50:889-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 212. | Chen Y, Luo Y, Huang W, Hu D, Zheng RQ, Cong SZ, Meng FK, Yang H, Lin HJ, Sun Y, Wang XY, Wu T, Ren J, Pei SF, Zheng Y, He Y, Hu Y, Yang N, Yan H. Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis B. Comput Biol Med. 2017;89:18-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 213. | Choi KJ, Jang JK, Lee SS, Sung YS, Shim WH, Kim HS, Yun J, Choi JY, Lee Y, Kang BK, Kim JH, Kim SY, Yu ES. Development and Validation of a Deep Learning System for Staging Liver Fibrosis by Using Contrast Agent-enhanced CT Images in the Liver. Radiology. 2018;289:688-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 151] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 214. | Cui E, Long W, Wu J, Li Q, Ma C, Lei Y, Lin F. Predicting the stages of liver fibrosis with multiphase CT radiomics based on volumetric features. Abdom Radiol (NY). 2021;46:3866-3876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 215. | Forlano R, Mullish BH, Giannakeas N, Maurice JB, Angkathunyakul N, Lloyd J, Tzallas AT, Tsipouras M, Yee M, Thursz MR, Goldin RD, Manousou P. High-Throughput, Machine Learning-Based Quantification of Steatosis, Inflammation, Ballooning, and Fibrosis in Biopsies From Patients With Nonalcoholic Fatty Liver Disease. Clin Gastroenterol Hepatol. 2020;18:2081-2090.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 216. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. A new computer aided diagnosis system for evaluation of chronic liver disease with ultrasound shear wave elastography imaging. Med Phys. 2016;43:1428-1436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 217. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. A Machine-Learning Algorithm Toward Color Analysis for Chronic Liver Disease Classification, Employing Ultrasound Shear Wave Elastography. Ultrasound Med Biol. 2017;43:1797-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 218. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. Temporal stability assessment in shear wave elasticity images validated by deep learning neural network for chronic liver disease fibrosis stage assessment. Med Phys. 2019;46:2298-2309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 219. | Heinemann F, Birk G, Stierstorfer B. Deep learning enables pathologist-like scoring of NASH models. Sci Rep. 2019;9:18454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 220. | Huang H, Shiffman ML, Friedman S, Venkatesh R, Bzowej N, Abar OT, Rowland CM, Catanese JJ, Leong DU, Sninsky JJ, Layden TJ, Wright TL, White T, Cheung RC. A 7 gene signature identifies the risk of developing cirrhosis in patients with chronic hepatitis C. Hepatology. 2007;46:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 228] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 221. | Konerman MA, Beste LA, Van T, Liu B, Zhang X, Zhu J, Saini SD, Su GL, Nallamothu BK, Ioannou GN, Waljee AK. Machine learning models to predict disease progression among veterans with hepatitis C virus. PLoS One. 2019;14:e0208141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 222. | Konerman MA, Zhang Y, Zhu J, Higgins PD, Lok AS, Waljee AK. Improvement of predictive models of risk of disease progression in chronic hepatitis C by incorporating longitudinal data. Hepatology. 2015;61:1832-1841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 223. | Kuppili V, Biswas M, Sreekumar A, Suri HS, Saba L, Edla DR, Marinho RT, Sanches JM, Suri JS. Extreme Learning Machine Framework for Risk Stratification of Fatty Liver Disease Using Ultrasound Tissue Characterization. J Med Syst. 2017;41:152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 224. | Lara J, López-Labrador F, González-Candelas F, Berenguer M, Khudyakov YE. Computational models of liver fibrosis progression for hepatitis C virus chronic infection. BMC Bioinformatics. 2014;15 Suppl 8:S5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 225. | Li W, Huang Y, Zhuang BW, Liu GJ, Hu HT, Li X, Liang JY, Wang Z, Huang XW, Zhang CQ, Ruan SM, Xie XY, Kuang M, Lu MD, Chen LD, Wang W. Multiparametric ultrasomics of significant liver fibrosis: A machine learning-based analysis. Eur Radiol. 2019;29:1496-1506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (1)] |
| 226. | Meffert PJ, Baumeister SE, Lerch MM, Mayerle J, Kratzer W, Völzke H. Development, external validation, and comparative assessment of a new diagnostic score for hepatic steatosis. Am J Gastroenterol. 2014;109:1404-1414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 227. | Perakakis N, Polyzos SA, Yazdani A, Sala-Vila A, Kountouras J, Anastasilakis AD, Mantzoros CS. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: A proof of concept study. Metabolism. 2019;101:154005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 228. | Piscaglia F, Cucchetti A, Benlloch S, Vivarelli M, Berenguer J, Bolondi L, Pinna AD, Berenguer M. Prediction of significant fibrosis in hepatitis C virus infected liver transplant recipients by artificial neural network analysis of clinical factors. Eur J Gastroenterol Hepatol. 2006;18:1255-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 229. | Raoufy MR, Vahdani P, Alavian SM, Fekri S, Eftekhari P, Gharibzadeh S. A novel method for diagnosing cirrhosis in patients with chronic hepatitis B: artificial neural network approach. J Med Syst. 2011;35:121-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 230. | Redman JS, Natarajan Y, Hou JK, Wang J, Hanif M, Feng H, Kramer JR, Desiderio R, Xu H, El-Serag HB, Kanwal F. Accurate Identification of Fatty Liver Disease in Data Warehouse Utilizing Natural Language Processing. Dig Dis Sci. 2017;62:2713-2718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 231. | Sowa JP, Heider D, Bechmann LP, Gerken G, Hoffmann D, Canbay A. Novel algorithm for non-invasive assessment of fibrosis in NAFLD. PLoS One. 2013;8:e62439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 232. | Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, Wu C, Liu C, Huang L, Jiang T, Meng F, Lu Y, Ai H, Xie XY, Yin LP, Liang P, Tian J, Zheng R. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68:729-741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 354] [Article Influence: 59.0] [Reference Citation Analysis (1)] |
| 233. | Wei R, Wang J, Wang X, Xie G, Wang Y, Zhang H, Peng CY, Rajani C, Kwee S, Liu P, Jia W. Clinical prediction of HBV and HCV related hepatic fibrosis using machine learning. EBioMedicine. 2018;35:124-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 234. | Lee JH, Joo I, Kang TW, Paik YH, Sinn DH, Ha SY, Kim K, Choi C, Lee G, Yi J, Bang WC. Deep learning with ultrasonography: automated classification of liver fibrosis using a deep convolutional neural network. Eur Radiol. 2020;30:1264-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 235. | Canbay A, Kälsch J, Neumann U, Rau M, Hohenester S, Baba HA, Rust C, Geier A, Heider D, Sowa JP. Non-invasive assessment of NAFLD as systemic disease-A machine learning perspective. PLoS One. 2019;14:e0214436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 236. | Docherty M, Regnier SA, Capkun G, Balp MM, Ye Q, Janssens N, Tietz A, Löffler J, Cai J, Pedrosa MC, Schattenberg JM. Development of a novel machine learning model to predict presence of nonalcoholic steatohepatitis. J Am Med Inform Assoc. 2021;28:1235-1241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 237. | Fialoke S, Malarstig A, Miller MR, Dumitriu A. Application of Machine Learning Methods to Predict Non-Alcoholic Steatohepatitis (NASH) in Non-Alcoholic Fatty Liver (NAFL) Patients. AMIA Annu Symp Proc. 2018;2018:430-439. [PubMed] |
| 238. | Sowa JP, Atmaca Ö, Kahraman A, Schlattjan M, Lindner M, Sydor S, Scherbaum N, Lackner K, Gerken G, Heider D, Arteel GE, Erim Y, Canbay A. Non-invasive separation of alcoholic and non-alcoholic liver disease with predictive modeling. PLoS One. 2014;9:e101444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 239. | Taylor-Weiner A, Pokkalla H, Han L, Jia C, Huss R, Chung C, Elliott H, Glass B, Pethia K, Carrasco-Zevallos O, Shukla C, Khettry U, Najarian R, Taliano R, Subramanian GM, Myers RP, Wapinski I, Khosla A, Resnick M, Montalto MC, Anstee QM, Wong VW, Trauner M, Lawitz EJ, Harrison SA, Okanoue T, Romero-Gomez M, Goodman Z, Loomba R, Beck AH, Younossi ZM. A Machine Learning Approach Enables Quantitative Measurement of Liver Histology and Disease Monitoring in NASH. Hepatology. 2021;74:133-147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 240. | Van Vleck TT, Chan L, Coca SG, Craven CK, Do R, Ellis SB, Kannry JL, Loos RJF, Bonis PA, Cho J, Nadkarni GN. Augmented intelligence with natural language processing applied to electronic health records for identifying patients with non-alcoholic fatty liver disease at risk for disease progression. Int J Med Inform. 2019;129:334-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 241. | Vanderbeck S, Bockhorst J, Kleiner D, Komorowski R, Chalasani N, Gawrieh S. Automatic quantification of lobular inflammation and hepatocyte ballooning in nonalcoholic fatty liver disease liver biopsies. Hum Pathol. 2015;46:767-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 36] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 242. | Vanderbeck S, Bockhorst J, Komorowski R, Kleiner DE, Gawrieh S. Automatic classification of white regions in liver biopsies by supervised machine learning. Hum Pathol. 2014;45:785-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 243. | Andres A, Montano-Loza A, Greiner R, Uhlich M, Jin P, Hoehn B, Bigam D, Shapiro JAM, Kneteman NM. A novel learning algorithm to predict individual survival after liver transplantation for primary sclerosing cholangitis. PLoS One. 2018;13:e0193523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 244. | Ayllón MD, Ciria R, Cruz-Ramírez M, Pérez-Ortiz M, Gómez I, Valente R, O'Grady J, de la Mata M, Hervás-Martínez C, Heaton ND, Briceño J. Validation of artificial neural networks as a methodology for donor-recipient matching for liver transplantation. Liver Transpl. 2018;24:192-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 245. | Bertsimas D, Kung J, Trichakis N, Wang Y, Hirose R, Vagefi PA. Development and validation of an optimized prediction of mortality for candidates awaiting liver transplantation. Am J Transplant. 2019;19:1109-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 246. | Bhat V, Tazari M, Watt KD, Bhat M. New-Onset Diabetes and Preexisting Diabetes Are Associated With Comparable Reduction in Long-Term Survival After Liver Transplant: A Machine Learning Approach. Mayo Clin Proc. 2018;93:1794-1802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 247. | Briceño J, Cruz-Ramírez M, Prieto M, Navasa M, Ortiz de Urbina J, Orti R, Gómez-Bravo MÁ, Otero A, Varo E, Tomé S, Clemente G, Bañares R, Bárcena R, Cuervas-Mons V, Solórzano G, Vinaixa C, Rubín A, Colmenero J, Valdivieso A, Ciria R, Hervás-Martínez C, de la Mata M. Use of artificial intelligence as an innovative donor-recipient matching model for liver transplantation: results from a multicenter Spanish study. J Hepatol. 2014;61:1020-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 248. | Kazemi A, Kazemi K, Sami A, Sharifian R. Identifying Factors That Affect Patient Survival After Orthotopic Liver Transplant Using Machine-Learning Techniques. Exp Clin Transplant. 2019;17:775-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 249. | Lau L, Kankanige Y, Rubinstein B, Jones R, Christophi C, Muralidharan V, Bailey J. Machine-Learning Algorithms Predict Graft Failure After Liver Transplantation. Transplantation. 2017;101:e125-e132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 250. | Lee HC, Yoon SB, Yang SM, Kim WH, Ryu HG, Jung CW, Suh KS, Lee KH. Prediction of Acute Kidney Injury after Liver Transplantation: Machine Learning Approaches vs. Logistic Regression Model. J Clin Med. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 251. | Abajian A, Murali N, Savic LJ, Laage-Gaupp FM, Nezami N, Duncan JS, Schlachter T, Lin M, Geschwind JF, Chapiro J. Predicting Treatment Response to Intra-arterial Therapies for Hepatocellular Carcinoma with the Use of Supervised Machine Learning-An Artificial Intelligence Concept. J Vasc Interv Radiol. 2018;29:850-857.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 252. | Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 588] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 253. | Morshid A, Elsayes KM, Khalaf AM, Elmohr MM, Yu J, Kaseb AO, Hassan M, Mahvash A, Wang Z, Hazle JD, Fuentes D. A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization. Radiol Artif Intell. 2019;1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 254. | Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky JM, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology. 2020;72:2000-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 189] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 255. | Shan QY, Hu HT, Feng ST, Peng ZP, Chen SL, Zhou Q, Li X, Xie XY, Lu MD, Wang W, Kuang M. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging. 2019;19:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 133] [Article Influence: 22.2] [Reference Citation Analysis (1)] |
| 256. | Eaton JE, Vesterhus M, McCauley BM, Atkinson EJ, Schlicht EM, Juran BD, Gossard AA, LaRusso NF, Gores GJ, Karlsen TH, Lazaridis KN. Primary Sclerosing Cholangitis Risk Estimate Tool (PREsTo) Predicts Outcomes of the Disease: A Derivation and Validation Study Using Machine Learning. Hepatology. 2020;71:214-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 257. | Garcia MS, Agarwal B, Mookerjee RP, Jalan R, Doyle G, Ranco G, Arroyo V, Pavesi M, Garcia E, Saliba F, Banares R, Fernandez J. An Accurate Data Preparation Approach for the Prediction of Mortality in ACLF Patients using the CANONIC Dataset. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:1371-1377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 258. | Ibragimov B, Toesca D, Chang D, Yuan Y, Koong A, Xing L. Development of deep neural network for individualized hepatobiliary toxicity prediction after liver SBRT. Med Phys. 2018;45:4763-4774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 259. | Jovanovic P, Salkic NN, Zerem E. Artificial neural network predicts the need for therapeutic ERCP in patients with suspected choledocholithiasis. Gastrointest Endosc. 2014;80:260-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 260. | Kanwal F, Taylor TJ, Kramer JR, Cao Y, Smith D, Gifford AL, El-Serag HB, Naik AD, Asch SM. Development, Validation, and Evaluation of a Simple Machine Learning Model to Predict Cirrhosis Mortality. JAMA Netw Open. 2020;3:e2023780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 261. | Speiser JL, Karvellas CJ, Wolf BJ, Chung D, Koch DG, Durkalski VL. Predicting daily outcomes in acetaminophen-induced acute liver failure patients with machine learning techniques. Comput Methods Programs Biomed. 2019;175:111-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 262. | Speiser JL, Lee WM, Karvellas CJ; US Acute Liver Failure Study Group. Predicting outcome on admission and post-admission for acetaminophen-induced acute liver failure using classification and regression tree models. PLoS One. 2015;10:e0122929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 263. | Konerman MA, Lu D, Zhang Y, Thomson M, Zhu J, Verma A, Liu B, Talaat N, Balis U, Higgins PDR, Lok ASF, Waljee AK. Assessing risk of fibrosis progression and liver-related clinical outcomes among patients with both early stage and advanced chronic hepatitis C. PLoS One. 2017;12:e0187344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 264. | Ai H, Chen W, Zhang L, Huang L, Yin Z, Hu H, Zhao Q, Zhao J, Liu H. Predicting Drug-Induced Liver Injury Using Ensemble Learning Methods and Molecular Fingerprints. Toxicol Sci. 2018;165:100-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 265. | Banerjee I, Choi HH, Desser T, Rubin DL. A Scalable Machine Learning Approach for Inferring Probabilistic US-LI-RADS Categorization. AMIA Annu Symp Proc. 2018;2018:215-224. [PubMed] |
| 266. | Dickerson LK, Rouhizadeh M, Korotkaya Y, Bowring MG, Massie AB, McAdams-Demarco MA, Segev DL, Cannon A, Guerrerio AL, Chen PH, Philosophe BN, Mogul DB. Language impairment in adults with end-stage liver disease: application of natural language processing towards patient-generated health records. NPJ Digit Med. 2019;2:106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 267. | He L, Li H, Dudley JA, Maloney TC, Brady SL, Somasundaram E, Trout AT, Dillman JR. Machine Learning Prediction of Liver Stiffness Using Clinical and T2-Weighted MRI Radiomic Data. AJR Am J Roentgenol. 2019;213:592-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 268. | Khan S, Ullah R, Khan A, Ashraf R, Ali H, Bilal M, Saleem M. Analysis of hepatitis B virus infection in blood sera using Raman spectroscopy and machine learning. Photodiagnosis Photodyn Ther. 2018;23:89-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 269. | Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans Med Imaging. 2018;37:2663-2674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 873] [Article Influence: 124.7] [Reference Citation Analysis (0)] |
| 270. | Mueller-Breckenridge AJ, Garcia-Alcalde F, Wildum S, Smits SL, de Man RA, van Campenhout MJH, Brouwer WP, Niu J, Young JAT, Najera I, Zhu L, Wu D, Racek T, Hundie GB, Lin Y, Boucher CA, van de Vijver D, Haagmans BL. Machine-learning based patient classification using Hepatitis B virus full-length genome quasispecies from Asian and European cohorts. Sci Rep. 2019;9:18892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 271. | Williams DP, Lazic SE, Foster AJ, Semenova E, Morgan P. Predicting Drug-Induced Liver Injury with Bayesian Machine Learning. Chem Res Toxicol. 2020;33:239-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 272. | Shousha HI, Awad AH, Omran DA, Elnegouly MM, Mabrouk M. Data Mining and Machine Learning Algorithms Using IL28B Genotype and Biochemical Markers Best Predicted Advanced Liver Fibrosis in Chronic Hepatitis C. Jpn J Infect Dis. 2018;71:51-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 273. | Garcia-Carretero R, Vigil-Medina L, Barquero-Perez O, Ramos-Lopez J. Relevant Features in Nonalcoholic Steatohepatitis Determined Using Machine Learning for Feature Selection. Metab Syndr Relat Disord. 2019;17:444-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 274. | Ma H, Xu CF, Shen Z, Yu CH, Li YM. Application of Machine Learning Techniques for Clinical Predictive Modeling: A Cross-Sectional Study on Nonalcoholic Fatty Liver Disease in China. Biomed Res Int. 2018;2018:4304376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 275. | Moccia S, Mattos LS, Patrini I, Ruperti M, Poté N, Dondero F, Cauchy F, Sepulveda A, Soubrane O, De Momi E, Diaspro A, Cesaretti M. Computer-assisted liver graft steatosis assessment via learning-based texture analysis. Int J Comput Assist Radiol Surg. 2018;13:1357-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 276. | Perveen S, Shahbaz M, Keshavjee K, Guergachi A. A Systematic Machine Learning Based Approach for the Diagnosis of Non-Alcoholic Fatty Liver Disease Risk and Progression. Sci Rep. 2018;8:2112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 277. | Wu CC, Yeh WC, Hsu WD, Islam MM, Nguyen PAA, Poly TN, Wang YC, Yang HC, Jack Li YC. Prediction of fatty liver disease using machine learning algorithms. Comput Methods Programs Biomed. 2019;170:23-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 121] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 278. | Yip TC, Ma AJ, Wong VW, Tse YK, Chan HL, Yuen PC, Wong GL. Laboratory parameter-based machine learning model for excluding non-alcoholic fatty liver disease (NAFLD) in the general population. Aliment Pharmacol Ther. 2017;46:447-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 279. | Gorris M, Hoogenboom SA, Wallace MB, van Hooft JE. Artificial intelligence for the management of pancreatic diseases. Dig Endosc. 2021;33:231-241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 280. | Marya NB, Powers PD, Chari ST, Gleeson FC, Leggett CL, Abu Dayyeh BK, Chandrasekhara V, Iyer PG, Majumder S, Pearson RK, Petersen BT, Rajan E, Sawas T, Storm AC, Vege SS, Chen S, Long Z, Hough DM, Mara K, Levy MJ. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut. 2021;70:1335-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (1)] |
| 281. | Tonozuka R, Mukai S, Itoi T. The Role of Artificial Intelligence in Endoscopic Ultrasound for Pancreatic Disorders. Diagnostics (Basel). 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 282. | van den Heever M, Mittal A, Haydock M, Windsor J. The use of intelligent database systems in acute pancreatitis--a systematic review. Pancreatology. 2014;14:9-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 283. | Andersson B, Andersson R, Ohlsson M, Nilsson J. Prediction of severe acute pancreatitis at admission to hospital using artificial neural networks. Pancreatology. 2011;11:328-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 284. | Fei Y, Gao K, Li WQ. Artificial neural network algorithm model as powerful tool to predict acute lung injury following to severe acute pancreatitis. Pancreatology. 2018;18:892-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 285. | Fei Y, Gao K, Li WQ. Prediction and evaluation of the severity of acute respiratory distress syndrome following severe acute pancreatitis using an artificial neural network algorithm model. HPB (Oxford). 2019;21:891-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 286. | Fei Y, Hu J, Li WQ, Wang W, Zong GQ. Artificial neural networks predict the incidence of portosplenomesenteric venous thrombosis in patients with acute pancreatitis. J Thromb Haemost. 2017;15:439-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 287. | Hong WD, Chen XR, Jin SQ, Huang QK, Zhu QH, Pan JY. Use of an artificial neural network to predict persistent organ failure in patients with acute pancreatitis. Clinics (Sao Paulo). 2013;68:27-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 288. | Keogan MT, Lo JY, Freed KS, Raptopoulos V, Blake S, Kamel IR, Weisinger K, Rosen MP, Nelson RC. Outcome analysis of patients with acute pancreatitis by using an artificial neural network. Acad Radiol. 2002;9:410-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 289. | Mofidi R, Duff MD, Madhavan KK, Garden OJ, Parks RW. Identification of severe acute pancreatitis using an artificial neural network. Surgery. 2007;141:59-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 290. | Pearce CB, Gunn SR, Ahmed A, Johnson CD. Machine learning can improve prediction of severity in acute pancreatitis using admission values of APACHE II score and C-reactive protein. Pancreatology. 2006;6:123-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 291. | Pofahl WE, Walczak SM, Rhone E, Izenberg SD. Use of an artificial neural network to predict length of stay in acute pancreatitis. Am Surg. 1998;64:868-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 292. | Qiu Q, Nian YJ, Guo Y, Tang L, Lu N, Wen LZ, Wang B, Chen DF, Liu KJ. Development and validation of three machine-learning models for predicting multiple organ failure in moderately severe and severe acute pancreatitis. BMC Gastroenterol. 2019;19:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 293. | Qiu Q, Nian YJ, Tang L, Guo Y, Wen LZ, Wang B, Chen DF, Liu KJ. Artificial neural networks accurately predict intra-abdominal infection in moderately severe and severe acute pancreatitis. J Dig Dis. 2019;20:486-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 294. | Al-Haddad MA, Friedlin J, Kesterson J, Waters JA, Aguilar-Saavedra JR, Schmidt CM. Natural language processing for the development of a clinical registry: a validation study in intraductal papillary mucinous neoplasms. HPB (Oxford). 2010;12:688-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 295. | Gao X, Wang X. Performance of deep learning for differentiating pancreatic diseases on contrast-enhanced magnetic resonance imaging: A preliminary study. Diagn Interv Imaging. 2020;101:91-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 296. | Mashayekhi R, Parekh VS, Faghih M, Singh VK, Jacobs MA, Zaheer A. Radiomic features of the pancreas on CT imaging accurately differentiate functional abdominal pain, recurrent acute pancreatitis, and chronic pancreatitis. Eur J Radiol. 2020;123:108778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 297. | Roth HR, Lu L, Lay N, Harrison AP, Farag A, Sohn A, Summers RM. Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med Image Anal. 2018;45:94-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 144] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 298. | Zhang J, Zhu L, Yao L, Ding X, Chen D, Wu H, Lu Z, Zhou W, Zhang L, An P, Xu B, Tan W, Hu S, Cheng F, Yu H. Deep learning-based pancreas segmentation and station recognition system in EUS: development and validation of a useful training tool (with video). Gastrointest Endosc. 2020;92:874-885.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 299. | Zheng H, Chen Y, Yue X, Ma C, Liu X, Yang P, Lu J. Deep pancreas segmentation with uncertain regions of shadowed sets. Magn Reson Imaging. 2020;68:45-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 300. | Zhu J, Wang L, Chu Y, Hou X, Xing L, Kong F, Zhou Y, Wang Y, Jin Z, Li Z. A new descriptor for computer-aided diagnosis of EUS imaging to distinguish autoimmune pancreatitis from chronic pancreatitis. Gastrointest Endosc. 2015;82:831-836.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 301. | Chu LC, Park S, Kawamoto S, Fouladi DF, Shayesteh S, Zinreich ES, Graves JS, Horton KM, Hruban RH, Yuille AL, Kinzler KW, Vogelstein B, Fishman EK. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma From Normal Pancreatic Tissue. AJR Am J Roentgenol. 2019;213:349-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 302. | Das A, Nguyen CC, Li F, Li B. Digital image analysis of EUS images accurately differentiates pancreatic cancer from chronic pancreatitis and normal tissue. Gastrointest Endosc. 2008;67:861-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 303. | Gao X, Wang X. Deep learning for World Health Organization grades of pancreatic neuroendocrine tumors on contrast-enhanced magnetic resonance images: a preliminary study. Int J Comput Assist Radiol Surg. 2019;14:1981-1991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 304. | Kaissis G, Ziegelmayer S, Lohöfer F, Steiger K, Algül H, Muckenhuber A, Yen HY, Rummeny E, Friess H, Schmid R, Weichert W, Siveke JT, Braren R. A machine learning algorithm predicts molecular subtypes in pancreatic ductal adenocarcinoma with differential response to gemcitabine-based vs FOLFIRINOX chemotherapy. PLoS One. 2019;14:e0218642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 305. | Kaissis GA, Ziegelmayer S, Lohöfer FK, Harder FN, Jungmann F, Sasse D, Muckenhuber A, Yen HY, Steiger K, Siveke J, Friess H, Schmid R, Weichert W, Makowski MR, Braren RF. Image-Based Molecular Phenotyping of Pancreatic Ductal Adenocarcinoma. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 306. | E L, Xu Y, Wu Z, Li L, Zhang N, Yang H, Schwartz LH, Lu L, Zhao B. Differentiation of Focal-Type Autoimmune Pancreatitis From Pancreatic Ductal Adenocarcinoma Using Radiomics Based on Multiphasic Computed Tomography. J Comput Assist Tomogr. 2020;44:511-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 307. | Momeni-Boroujeni A, Yousefi E, Somma J. Computer-assisted cytologic diagnosis in pancreatic FNA: An application of neural networks to image analysis. Cancer Cytopathol. 2017;125:926-933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 308. | Norton ID, Zheng Y, Wiersema MS, Greenleaf J, Clain JE, Dimagno EP. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest Endosc. 2001;54:625-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 309. | Qiu W, Duan N, Chen X, Ren S, Zhang Y, Wang Z, Chen R. Pancreatic Ductal Adenocarcinoma: Machine Learning-Based Quantitative Computed Tomography Texture Analysis For Prediction Of Histopathological Grade. Cancer Manag Res. 2019;11:9253-9264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 310. | Săftoiu A, Vilmann P, Dietrich CF, Iglesias-Garcia J, Hocke M, Seicean A, Ignee A, Hassan H, Streba CT, Ioncică AM, Gheonea DI, Ciurea T. Quantitative contrast-enhanced harmonic EUS in differential diagnosis of focal pancreatic masses (with videos). Gastrointest Endosc. 2015;82:59-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 311. | Săftoiu A, Vilmann P, Gorunescu F, Gheonea DI, Gorunescu M, Ciurea T, Popescu GL, Iordache A, Hassan H, Iordache S. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest Endosc. 2008;68:1086-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 174] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 312. | Săftoiu A, Vilmann P, Gorunescu F, Janssen J, Hocke M, Larsen M, Iglesias-Garcia J, Arcidiacono P, Will U, Giovannini M, Dietrich CF, Havre R, Gheorghe C, McKay C, Gheonea DI, Ciurea T; European EUS Elastography Multicentric Study Group. Efficacy of an artificial neural network-based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin Gastroenterol Hepatol. 2012;10:84-90.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 313. | Yeaton P, Sears RJ, Ledent T, Salmon I, Kiss R, Decaestecker C. Discrimination between chronic pancreatitis and pancreatic adenocarcinoma using artificial intelligence-related algorithms based on image cytometry-generated variables. Cytometry. 1998;32:309-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 314. | Zhu M, Xu C, Yu J, Wu Y, Li C, Zhang M, Jin Z, Li Z. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: a diagnostic test. PLoS One. 2013;8:e63820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (2)] |
| 315. | Luo Y, Chen X, Chen J, Song C, Shen J, Xiao H, Chen M, Li ZP, Huang B, Feng ST. Preoperative Prediction of Pancreatic Neuroendocrine Neoplasms Grading Based on Enhanced Computed Tomography Imaging: Validation of Deep Learning with a Convolutional Neural Network. Neuroendocrinology. 2020;110:338-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 316. | Chakraborty J, Midya A, Gazit L, Attiyeh M, Langdon-Embry L, Allen PJ, Do RKG, Simpson AL. CT radiomics to predict high-risk intraductal papillary mucinous neoplasms of the pancreas. Med Phys. 2018;45:5019-5029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 317. | Corral JE, Hussein S, Kandel P, Bolan CW, Bagci U, Wallace MB. Deep Learning to Classify Intraductal Papillary Mucinous Neoplasms Using Magnetic Resonance Imaging. Pancreas. 2019;48:805-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 318. | Dmitriev K, Kaufman AE, Javed AA, Hruban RH, Fishman EK, Lennon AM, Saltz JH. Classification of Pancreatic Cysts in Computed Tomography Images Using a Random Forest and Convolutional Neural Network Ensemble. Med Image Comput Comput Assist Interv. 2017;10435:150-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 319. | Kurita Y, Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Koda H, Tajika M, Shimizu Y, Nakajima A, Kubota K, Niwa Y. Diagnostic ability of artificial intelligence using deep learning analysis of cyst fluid in differentiating malignant from benign pancreatic cystic lesions. Sci Rep. 2019;9:6893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 320. | Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Kurita Y, Koda H, Toriyama K, Onishi S, Ishihara M, Tanaka T, Tajika M, Niwa Y. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin Transl Gastroenterol. 2019;10:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 128] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 321. | Li H, Shi K, Reichert M, Lin K, Tselousov N, Braren R, Fu D, Schmid R, Li J, Menze B. Differential Diagnosis for Pancreatic Cysts in CT Scans Using Densely-Connected Convolutional Networks. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:2095-2098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 322. | Okoń K, Tomaszewska R, Nowak K, Stachura J. Application of neural networks to the classification of pancreatic intraductal proliferative lesions. Anal Cell Pathol. 2001;23:129-136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 323. | Song JW, Lee JH, Choi JH, Chun SJ. Automatic differential diagnosis of pancreatic serous and mucinous cystadenomas based on morphological features. Comput Biol Med. 2013;43:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 324. | Springer S, Masica DL, Dal Molin M, Douville C, Thoburn CJ, Afsari B, Li L, Cohen JD, Thompson E, Allen PJ, Klimstra DS, Schattner MA, Schmidt CM, Yip-Schneider M, Simpson RE, Fernandez-Del Castillo C, Mino-Kenudson M, Brugge W, Brand RE, Singhi AD, Scarpa A, Lawlor R, Salvia R, Zamboni G, Hong SM, Hwang DW, Jang JY, Kwon W, Swan N, Geoghegan J, Falconi M, Crippa S, Doglioni C, Paulino J, Schulick RD, Edil BH, Park W, Yachida S, Hijioka S, van Hooft J, He J, Weiss MJ, Burkhart R, Makary M, Canto MI, Goggins MG, Ptak J, Dobbyn L, Schaefer J, Sillman N, Popoli M, Klein AP, Tomasetti C, Karchin R, Papadopoulos N, Kinzler KW, Vogelstein B, Wolfgang CL, Hruban RH, Lennon AM. A multimodality test to guide the management of patients with a pancreatic cyst. Sci Transl Med. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 134] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 325. | Wei R, Lin K, Yan W, Guo Y, Wang Y, Li J, Zhu J. Computer-Aided Diagnosis of Pancreas Serous Cystic Neoplasms: A Radiomics Method on Preoperative MDCT Images. Technol Cancer Res Treat. 2019;18:1533033818824339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 326. | Yang J, Guo X, Ou X, Zhang W, Ma X. Discrimination of Pancreatic Serous Cystadenomas From Mucinous Cystadenomas With CT Textural Features: Based on Machine Learning. Front Oncol. 2019;9:494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 327. | Blyuss O, Zaikin A, Cherepanova V, Munblit D, Kiseleva EM, Prytomanova OM, Duffy SW, Crnogorac-Jurcevic T. Development of PancRISK, a urine biomarker-based risk score for stratified screening of pancreatic cancer patients. Br J Cancer. 2020;122:692-696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 328. | Facciorusso A, Del Prete V, Antonino M, Buccino VR, Muscatiello N. Response to repeat echoendoscopic celiac plexus neurolysis in pancreatic cancer patients: A machine learning approach. Pancreatology. 2019;19:866-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 329. | Hayward J, Alvarez SA, Ruiz C, Sullivan M, Tseng J, Whalen G. Machine learning of clinical performance in a pancreatic cancer database. Artif Intell Med. 2010;49:187-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 330. | Kaissis G, Ziegelmayer S, Lohöfer F, Algül H, Eiber M, Weichert W, Schmid R, Friess H, Rummeny E, Ankerst D, Siveke J, Braren R. A machine learning model for the prediction of survival and tumor subtype in pancreatic ductal adenocarcinoma from preoperative diffusion-weighted imaging. Eur Radiol Exp. 2019;3:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 331. | Li K, Xiao J, Yang J, Li M, Xiong X, Nian Y, Qiao L, Wang H, Eresen A, Zhang Z, Hu X, Wang J, Chen W. Association of radiomic imaging features and gene expression profile as prognostic factors in pancreatic ductal adenocarcinoma. Am J Transl Res. 2019;11:4491-4499. [PubMed] |
| 332. | Walczak S, Velanovich V. An Evaluation of Artificial Neural Networks in Predicting Pancreatic Cancer Survival. J Gastrointest Surg. 2017;21:1606-1612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 333. | Xu W, Liu Y, Lu Z, Jin ZD, Hu YH, Yu JG, Li ZS. A new endoscopic ultrasonography image processing method to evaluate the prognosis for pancreatic cancer treated with interstitial brachytherapy. World J Gastroenterol. 2013;19:6479-6484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 334. | Zhang Y, Lobo-Mueller EM, Karanicolas P, Gallinger S, Haider MA, Khalvati F. CNN-based survival model for pancreatic ductal adenocarcinoma in medical imaging. BMC Med Imaging. 2020;20:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 335. | Li S, Jiang H, Wang Z, Zhang G, Yao YD. An effective computer aided diagnosis model for pancreas cancer on PET/CT images. Comput Methods Programs Biomed. 2018;165:205-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 336. | Liu SL, Li S, Guo YT, Zhou YP, Zhang ZD, Lu Y. Establishment and application of an artificial intelligence diagnosis system for pancreatic cancer with a faster region-based convolutional neural network. Chin Med J (Engl). 2019;132:2795-2803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 337. | Mehrabi S, Krishnan A, Roch AM, Schmidt H, Li D, Kesterson J, Beesley C, Dexter P, Schmidt M, Palakal M, Liu H. Identification of Patients with Family History of Pancreatic Cancer--Investigation of an NLP System Portability. Stud Health Technol Inform. 2015;216:604-608. [PubMed] |
| 338. | Ozkan M, Cakiroglu M, Kocaman O, Kurt M, Yilmaz B, Can G, Korkmaz U, Dandil E, Eksi Z. Age-based computer-aided diagnosis approach for pancreatic cancer on endoscopic ultrasound images. Endosc Ultrasound. 2016;5:101-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 339. | Roch AM, Mehrabi S, Krishnan A, Schmidt HE, Kesterson J, Beesley C, Dexter PR, Palakal M, Schmidt CM. Automated pancreatic cyst screening using natural language processing: a new tool in the early detection of pancreatic cancer. HPB (Oxford). 2015;17:447-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 340. | Zhang MM, Yang H, Jin ZD, Yu JG, Cai ZY, Li ZS. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointest Endosc. 2010;72:978-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 341. | Liu X, Rivera SC, Moher D, Calvert MJ, Denniston AK; SPIRIT-AI and CONSORT-AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI Extension. BMJ. 2020;370:m3164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 209] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 342. | Krittanawong C, Johnson KW, Rosenson RS, Wang Z, Aydar M, Baber U, Min JK, Tang WHW, Halperin JL, Narayan SM. Deep learning for cardiovascular medicine: a practical primer. Eur Heart J. 2019;40:2058-2073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 203] [Article Influence: 33.8] [Reference Citation Analysis (0)] |