Published online Jul 7, 2021. doi: 10.3748/wjg.v27.i25.3863
Peer-review started: February 8, 2021
First decision: March 28, 2021
Revised: April 8, 2021
Accepted: April 20, 2021
Article in press: April 20, 2021
Published online: July 7, 2021
Processing time: 147 Days and 13.3 Hours
The association between PPARGC1A rs8192678 and nonalcoholic fatty liver disease (NAFLD) requires further confirmation. In addition, it is still unknown whether PPARGC1A rs8192678 is associated with hepatic histological features in NAFLD in the Chinese population.
To investigate the interaction between PPARGC1A rs8192678 and nonalcoholic steatohepatitis (NASH), and whether this polymorphism is associated with hepatic histological features.
Fifty-nine patients with liver biopsy-proven NAFLD and 93 healthy controls were recruited to a cohort representing the Chinese Han population. The SAF (steatosis, activity, and fibrosis) scoring system was used for hepatic histopathological evaluation. The polymorphisms of PPARGC1A rs8192678 and patatin-like phospholipase domain-containing protein 3 (PNPLA3) rs738409 were genotyped. The intrahepatic mRNA expression of PPARGC1A was evaluated by real-time polymerase chain reaction.
Thirty-seven patients with NAFLD had NASH, of which 12 were nonobese. The PPARGC1A rs8192678 risk A allele (carrying GA and AA genotypes) had the lowest P value in the dominant model; the odds ratio (OR) for NAFLD was 2.321 [95% confidence interval (CI): 1.121-4.806]. After adjusting for age, sex, and the PNPLA3 rs738409 risk G allele, the PPARGC1A rs8192678 A allele was a risk factor for NAFLD (OR 2.202, 95%CI: 1.030-4.705, P = 0.042). The genetic analysis showed that patients with NAFLD, moderate-to-severe steatosis (S2-3), and Activity 2-4 (A ≥ 2) were more likely to carry A in PPARGC1A rs8192678 (OR 5.000, 95%CI: 1.343-18.620, P = 0.012; and OR 4.071, 95%CI: 1.076-15.402, P = 0.031). The multivariate logistic regression analysis showed that PPARGC1A rs8192678 risk A allele was also independently associated with S2-3, A ≥ 2, and NASH (OR 6.190, 95%CI: 1.508-25.410, P = 0.011; OR 4.506, 95%CI 1.070-18.978, P = 0.040; and OR 6.337, 95%CI: 1.135-35.392, P = 0.035, respectively) after adjusting for age, sex, body mass index, and PNPLA3 rs738409 risk G allele. The results also showed that this polymorphism was associated with nonobese NASH (OR 22.000, 95%CI: 1.540-314.292, P = 0.021). The intrahepatic expression of PPARGC1A mRNA was significantly lower in the group of patients who carried the risk A allele (P = 0.014).
The PPARGC1A rs8192678 risk A allele is associated with NAFLD, and with S2-3, A ≥ 2 and NASH in NAFLD patients, independent of PNPLA3 rs738409, and may be associated with nonobese NASH.
Core Tip: The PPARGC1A rs8192678 A allele was found to be a risk factor for liver biopsy-proven nonalcoholic fatty liver disease (NAFLD) after adjusting for age, sex, and patatin-like phospholipase domain-containing protein 3 rs738409 polymorphism. The multivariate logistic regression analysis showed that the PPARGC1A rs8192678 risk A allele was also independently associated with the severity of hepatic histological features (S2-3 and A ≥ 2) and nonalcoholic steatohepatitis (NASH), and might also be associated with nonobese NASH, indicating that the PPARGC1A rs8192678 risk A allele was associated with NAFLD in the Chinese Han adult population. Also, the intrahepatic expression of PPARGC1A mRNA was significantly lower in the patients who carried the risk A allele, implying that it might be a genetic contribution to the pathogenesis of NAFLD.
- Citation: Zhang RN, Shen F, Pan Q, Cao HX, Chen GY, Fan JG. PPARGC1A rs8192678 G>A polymorphism affects the severity of hepatic histological features and nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease . World J Gastroenterol 2021; 27(25): 3863-3876
- URL: https://www.wjgnet.com/1007-9327/full/v27/i25/3863.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i25.3863
Nonalcoholic fatty liver disease (NAFLD) has become one of the most common forms of chronic liver diseases worldwide. The overall global prevalence of NAFLD is about 25%. The clinical spectrum of NAFLD ranges from nonalcoholic fatty liver (NAFL), nonalcoholic steatohepatitis (NASH), liver fibrosis and cirrhosis, and finally hepatocellular carcinoma. NAFLD also has high risk and is associated with multisystem diseases, such as obesity, type 2 diabetes mellitus (T2DM), cardiovascular diseases, chronic kidney disease, and malignancy[1,2].
The pathogenesis of NAFLD is multifactorial and strongly associated with obesity, insulin resistance, T2DM, and metabolic syndrome. Although NAFLD has a high prevalence in obese patients, nonobese patients and even lean patients, were recently found to have NAFLD as well. The anthropometric data on body mass index (BMI) was used to define nonobese or lean. The BMI values of 25 and 30 kg/m2 are the thresholds to define overweight and obese participants, respectively, as recommended by the World Health Organization. In Asia, BMI < 25 kg/m2 is defined as nonobese, whereas BMI < 23 kg/m2 is defined as lean[3]. A meta-analysis was carried out to estimate the overall prevalence of NAFLD in nonobese and lean populations (15.7% and 10.2%, respectively)[4].
The differences in prevalence, clinical profile, hepatic histology severity, and outcomes of NAFLD among different ethnic groups suggested that both environmental and genetic factors influenced susceptibility to NAFLD in individuals[5]. Genetic factors have been implicated in the occurrence and development of NAFLD; the gene of peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), encoded by PPARGC1A, is also a susceptibility candidate gene for NAFLD[6-8]. PGC-1α is a transcriptional coactivator with a crucial impact on multiple aspects of cellular energy metabolism including mitochondrial biogenesis and cellular respiration, regulation of adaptive thermogenesis, adipocyte cell development, and lipid and glucose metabolism. A number of genetic variants have been identified in PPARGC1A, and the Gly482Ser polymorphism (a G-to-A transition that predicted a glycine (G) to serine (S) substitution at amino acid position 482 in exon 8, rs8192678) is the most common variant, which was associated with T2DM, obesity, and hypertension in several studies[9-13]. However, the results were controversial in the Chinese Han adult population, which require further confirmation[14,15]. Whether this polymo
The present study aimed to determine the relationship between polymorphism rs8192678 of PPARGC1A gene and the risk of NAFLD, especially in NASH, in a Chinese Han population, and to investigate the association between PPARGC1A rs8192678 G>A polymorphism and the severity of hepatic histological features.
A total of 152 unrelated adult participants (18-70 years old) were recruited between March 2012 and March 2013. Patients with NAFLD were enrolled from Xinhua Hospital, Shanghai, China. All participants were Han Chinese in origin. Each patient had undergone an ultrasound-guided percutaneous liver biopsy and met the diagnostic criteria for NAFLD. The liver stiffness measurement (LSM) and controlled attenuation parameter (CAP) values were tested using FibroScan 502 (Echosens, Paris, France) by previously described methods.
The exclusion criteria were as follows: (1) excessive alcohol consumption (> 30 g/d for men and > 20 g/d for women); other diseases that led to fatty liver, such as chronic hepatitis C, drug-induced liver injury, Wilson’s disease, total parenteral nutrition, and autoimmune hepatitis; (2) previous liver transplantation; (3) other end-stage diseases or malignancies; and (4) contraindications to FibroScan examination (e.g., ascites, implanted pacemakers, nonhealing wounds in the upper-right quadrant of the abdomen, and pregnancy) or unreliable measurement of LSM and CAP values.
All control participants were confirmed to be free of liver diseases by both B-mode ultrasound and FibroScan 502 examination (CAP ≤ 240 dB/m and LSM values < 7.0 kPa)[16]; they all had normal liver function test results and lacked evidence of etiologies of liver injury. The study protocol was approved by the Ethics Committees of Xinhua Hospital, and the study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants.
Demographic information, such as age and sex, was collected. Venous blood samples were obtained from the participants after overnight fasting (12 h) to measure the levels of alanine transaminase (ALT), alkaline phosphatase (ALP), γ-glutamyl transpeptidase (GGT), total bilirubin, directed bilirubin, fasting blood glucose (FBG), total cholesterol (TC), triglycerides (TG), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and uric acid (UA). All laboratory biochemical parameters were measured in a conventional automated analyzer (Hitachi 7600, Tokyo, Japan).
Liver biopsy specimens were fixed with 10% buffered formalin, embedded in paraffin, sliced, and stained with hematoxylin & eosin (HE), reticulin, and Masson trichrome. A minimum 15-mm biopsy specimen or the presence of at least 10 complete portal tracts was required. For each liver biopsy, the steatosis, activity, and fibrosis (SAF) score summarizing the main histological lesions was defined. The SAF score was designed by the fatty liver inhibition of progression Pathology Consortium. The steatosis (0-3) was categorized as follows: S0: < 5%; S1: 5%-33%; S2: 34%-66 %; and S3: ≥ 67%. The grade of hepatic inflammatory activity (A, from 0 to 4) was calculated by adding together the grades for hepatocyte ballooning (0-2) and lobular inflammation (0-2). Liver fibrosis was staged as follows: F0, none; F1, perisinusoidal or portal fibrosis; F2, perisinusoidal and periportal fibrosis without bridging; F3, bridging fibrosis; and F4, cirrhosis. A case presenting with at least grade 1 of each of the three features (steatosis, ballooning, and lobular inflammation) was classified as NASH; other cases were diagnosed as NAFL.
Polymorphism genotyping of genomic DNA was prepared from each blood sample using the QiAamp DNA Mini Kit (Qiagen, Hilden, Germany). The custom Ion AmpliSeq panel (Life Technologies, MA, United States) of PPARGC1A and patatin-like phospholipase domain-containing protein 3 (PNPLA3) was designed. The polymerase chain reaction (PCR) of the template was performed using the Ion OneTouch 2 System (Life Technologies, MA, United States) following the manu
Total RNA was isolated from samples of liver biopsy specimens using the TRIzol reagent (Takara, Dalian, China) and reverse transcribed using the PrimeScript RT Reagent Kit (Takara, Dalian, China). The mRNA expression of PPARGC1A and β-actin was measured by real-time PCR (RT-PCR) using the SYBR Premix Ex Taq Kit (Takara, Dalian, China) and an ABI 7500 RT-PCR System (Applied Biosystems, United States). Target mRNA levels were normalized to β-actin expression levels. The following primers were used: PPARGC1A (F: 5'-GAGTGACATCGAGTGTGCTG-3'; R: 5'-GGGCAATCCGTCTTCATCCA-3'); β-actin (F: 5'-TCCTTCCTGGGCATGGAGT-3'; R: 5'-CAGGAGGAGCAATGATCTTGAT-3'). A duplicate of this experiment was performed for the same reaction. The 2-ΔΔCt method was used to calculate the relative expression levels of each gene. Thermal cycling was performed as follows: Initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 34 s.
Continuous variables were expressed as mean ± SD, median and interquartile range for those with a skewed distribution and categorical variables as the frequency or percentage. The t test and chi-squared test were used where appropriate. Multiple logistic regression models were used to assess the factors independently associated with the severity of histological parameters. In the dominant model, the frequencies of the participants with the allele 1 (allele 1 homozygote and heterozygote) were compared with the participants homozygous for the allele 2 using a 2 × 2 contingency table. The odds ratio (OR) and the 95% confidence interval (CI) were calculated using Woolf’s method. In all analyses, GA and AA for PPARGC1A rs8192678 was considered when choosing a dominant model of inheritance. Analyses were performed using SPSS 23.0 (SPSS Inc., IL, United States). A two-sided P < 0.05 indicated a statistically significant difference.
The clinical characteristics of 152 participants (59 patients with NAFLD and 93 controls) are listed in Table 1. Age, BMI, ALT, GGT, TC, TG, LDL-C, UA, CAP, and LSM values were significantly higher in the NAFLD group. No differences in sex, ALP, HDL-C, and FBG were observed between the two groups. Among the patients with NAFLD, 37 (62.71%) had NASH, and 22 (37.29%) had NAFL. Furthermore, 69.49% (41/59) were obese, and 30.51% (18/59) were nonobese. Patients with NAFLD also showed a significantly higher frequency of hypertension and T2DM. The ALT, GGT, CAP, and LSM levels were significantly higher in patients with NASH compared to patients with NAFL, with no significant difference in the frequency of obesity, hypertension, and T2DM (Tables 1 and 2).
| Variables | NAFLD (n = 59) | Controls (n = 93) | P value |
| Sex (M/F) | 43/16 | 59/34 | 0.227 |
| Age (year) | 38.20 ± 13.78 | 42.20 ± 11.05 | 0.039 |
| BMI (kg/m2) | 27.31 ± 3.31 | 23.06 ± 2.74 | < 0.0001 |
| ALT (U/L) | 70.56 ± 47.6 | 24.98 ± 16.07 | < 0.0001 |
| GGT (U/L) | 72.75 ± 55.22 | 25.94 ± 18.17 | < 0.0001 |
| ALP (U/L) | 97.07 ± 44.73 | 96.50 ± 32.54 | 0.347 |
| TC (mmol/L) | 4.86 ± 0.86 | 4.39 ± 0.89 | 0.001 |
| TG (mmol/L) | 1.99 ± 1.25 | 1.14 ± 0.74 | < 0.0001 |
| HDL-C (mmol/L) | 1.20 ± 0.28 | 1.23 ± 0.28 | 0.177 |
| LDL-C (mmol/L) | 2.83 ± 0.7 | 2.24 ± 0.50 | < 0.0001 |
| FBG (mmol/L) | 5.93 ± 2.62 | 5.31 ± 0.79 | 0.941 |
| UA (μmol/L) | 361.32 ± 105.78 | 274.25 ± 76.53 | < 0.0001 |
| CAP (dB/m) | 313 (277-351) | 212 (172-232) | < 0.0001 |
| LSM (kPa) | 7.7 (5.6-12) | 5.25 (4.1-7.0) | < 0.0001 |
| Hypertension, n (%) | 22 (37.29%) | 15 (16.13%) | 0.003 |
| T2DM, n (%) | 7 (11.86%) | 1 (1.08%) | 0.005 |
| Obesity, n (%) | 41 (69.49%) | 19 (20.43%) | < 0.0001 |
| Steatosis (S, n) | |||
| S1/S2/S3 | 25/25/9 | / | / |
| Activity (A, n) | |||
| A0/A1/A2/A3/A4 | 1/12/16/22/8 | / | / |
| Fibrosis (F, n) | |||
| F0/F1/F2/F3/F4 | 11/29/8/7/4 | / | / |
| Variable | NAFL (n = 22) | NASH (n = 37) | P value |
| Sex (M/F) | 17/5 | 26/11 | 0.559 |
| Age (year) | 37.23 ± 13.64 | 38.78 ± 14.01 | 0.605 |
| BMI (kg/m2) | 27.07 ± 3.69 | 27.38 ± 3.17 | 0.675 |
| ALP (U/L) | 102.96 ± 52.34 | 93.56 ± 39.91 | 0.982 |
| GGT (U/L) | 38.00 (23.80-69.03) | 69.70 (39.00-85.50) | 0.023 |
| ALT (U/L) | 46.00 (30.33-64.95) | 71.40 (46.9-111.70) | 0.006 |
| TC (mmol/L) | 5.03 ± 1.04 | 4.76 ± 0.72 | 0.801 |
| TG (mmol/L) | 1.91 ± 0.74 | 2.04 ± 1.48 | 0.532 |
| HDL-C (mmol/L) | 1.21 ± 0.28 | 1.19 ± 0.28 | 0.903 |
| LDL-C (mmol/L) | 2.97 ± 0.91 | 2.72 ± 0.49 | 0.444 |
| FBG (mmol/L) | 6.43 ± 3.59 | 5.62 ± 1.77 | 0.318 |
| UA (μmol/L) | 360.91 ± 107.80 | 361.62 ± 106.22 | 0.482 |
| CAP (dB/m) | 295.50 (249.25-345.00) | 333.00 (296.25-356.25) | 0.038 |
| LSM (kPa) | 6.20 (4.55-9.95) | 9.60 (6.50-12.85) | 0.008 |
| Obesity, n (%) | 16 (72.73) | 25 (60.97) | 0.677 |
| Hypertension, n (%) | 8 (36.36) | 14 (37.84) | 0.910 |
| T2DM, n (%) | 3 (13.64) | 4 (10.81) | 0.746 |
The percentage of PPARGC1A rs8192678 GA genotype was significantly higher in the NAFLD group than in the control group (62.71% vs 39.78 %), suggesting that carriers of this genotype had a significantly increased risk of NAFLD (OR 2.786, 95%CI: 1.300-5.962, P = 0.008) (Table 3). The carriers of the homozygous AA genotype had no significantly increased risk of NAFLD compared to participants with GG homozygous alleles (P = 0.609). The close association was further revealed in the dominant model (combined GA and AA genotypes, GA + AA vs GG, OR 2.321, 95%CI: 1.121-4.806, P = 0.022) (Tables 3 and 4).
| Genotype | NAFLD (n = 59) | Controls (n = 93) | χ2 | P value | OR (95%CI) |
| PPARGC1A rs8192678 | |||||
| GG | 14 (23.73%) | 39 (41.94%) | |||
| GA | 37 (62.71%) | 37 (39.78%) | 7.148 | 0.008 | 2.786 (1.300-5.962) |
| AA | 8 (13.56%) | 17 (18.28%) | 0.262 | 0.609 | 1.311 (0.464-3.704) |
| GA+AA | 45 (76.27%) | 54 (58.06%) | 5.269 | 0.022 | 2.321 (1.121-4.806) |
| PNPLA3 rs738409 | |||||
| CC | 12 (20.34%) | 35 (37.63%) | |||
| CG | 27 (45.76%) | 43 (46.24%) | 2.152 | 0.142 | 1.831 (0.812-4.131) |
| GG | 20 (33.90%) | 15 (16.13%) | 8.423 | 0.004 | 3.889 (1.524-9.926) |
| CG + GG | 47 (79.56%) | 58 (62.37%) | 5.055 | 0.025 | 2.364 (1.105-5.055) |
| PPARGC1A rs8192678 | Case | Control | Dominant model | ||||||
| GG | GA | AA | GG | GA | AA | χ2 | P value | OR (95%CI) | |
| NAFLD vs Control | 14 | 37 | 8 | 39 | 37 | 17 | 5.269 | 0.022 | 2.321 (1.121-4.806) |
| NASH vs Control | 5 | 28 | 4 | 39 | 37 | 17 | 9.550 | 0.002 | 4.622 (1.653-12.929) |
| NASH vs NAFL | 5 | 28 | 4 | 9 | 9 | 4 | 5.721 | 0.017 | 4.431 (1.245-15.763) |
| Obese vs nonobese participants | 17 | 33 | 10 | 36 | 41 | 15 | 1.864 | 0.172 | 1.626 (0.807-3.276) |
| Obese NAFLD vs nonobese NAFLD | 9 | 27 | 5 | 5 | 10 | 3 | 0.235 | 0.628 | 1.368 (0.384-4.865) |
| Obese NASH vs nonobese NASH | 4 | 19 | 2 | 1 | 9 | 2 | 0.408 | 0.348 | 0.477 (0.047-4.806) |
| Obese NAFL vs nonobese NAFL | 5 | 8 | 3 | 4 | 1 | 1 | 2.264 | 0.132 | 4.400 (0.596-32.501) |
| Obese NASH vs obese NAFL | 4 | 19 | 2 | 5 | 8 | 3 | 1.324 | 0.250 | 2.386 (0.531-10.734) |
| Nonobese NASH vs nonobese NAFL | 1 | 9 | 2 | 4 | 1 | 1 | 6.785 | 0.021 | 22.000 (1.540-314.292) |
The percentage of the PNPLA3 rs738409 GG genotype was also significantly higher in patients with NAFLD than in controls (33.90% vs 16.13%, P = 0.004). The OR of NAFLD was 3.889-fold (95%CI: 1.534-9.926, P = 0.004) higher in participants with the GG genotype than in those with the CC genotype; and the participants carrying G allele also had a high risk of NAFLD (GG + GC vs CC, OR 2.364, 95%CI: 1.105-5.055, P = 0.025) (Table 3). The multiple logistic regression analysis revealed that the PPARGC1A rs8192678 A allele was a risk factor for NAFLD (OR 2.202, 95%CI: 1.030-4.705, P = 0.042) independent of age, sex, and PNPLA3 rs738409 polymorphism (Table 5).
| Variables | B | S.E. | Wald | OR (95%CI) | P value |
| Age (year) | -0.031 | 0.015 | 4.236 | 0.970 (0.942-0.999) | 0.040 |
| Sex (Male) | -0.233 | 0.385 | 0.365 | 0.793 (0.373-1.686) | 0.546 |
| PPARGC1A rs8192678 A allele | 0.789 | 0.388 | 4.148 | 2.202 (1.030-4.705) | 0.042 |
| PNPLA3 rs738409 G allele | 0.857 | 0.403 | 4.524 | 2.356 (1.070-5.191) | 0.033 |
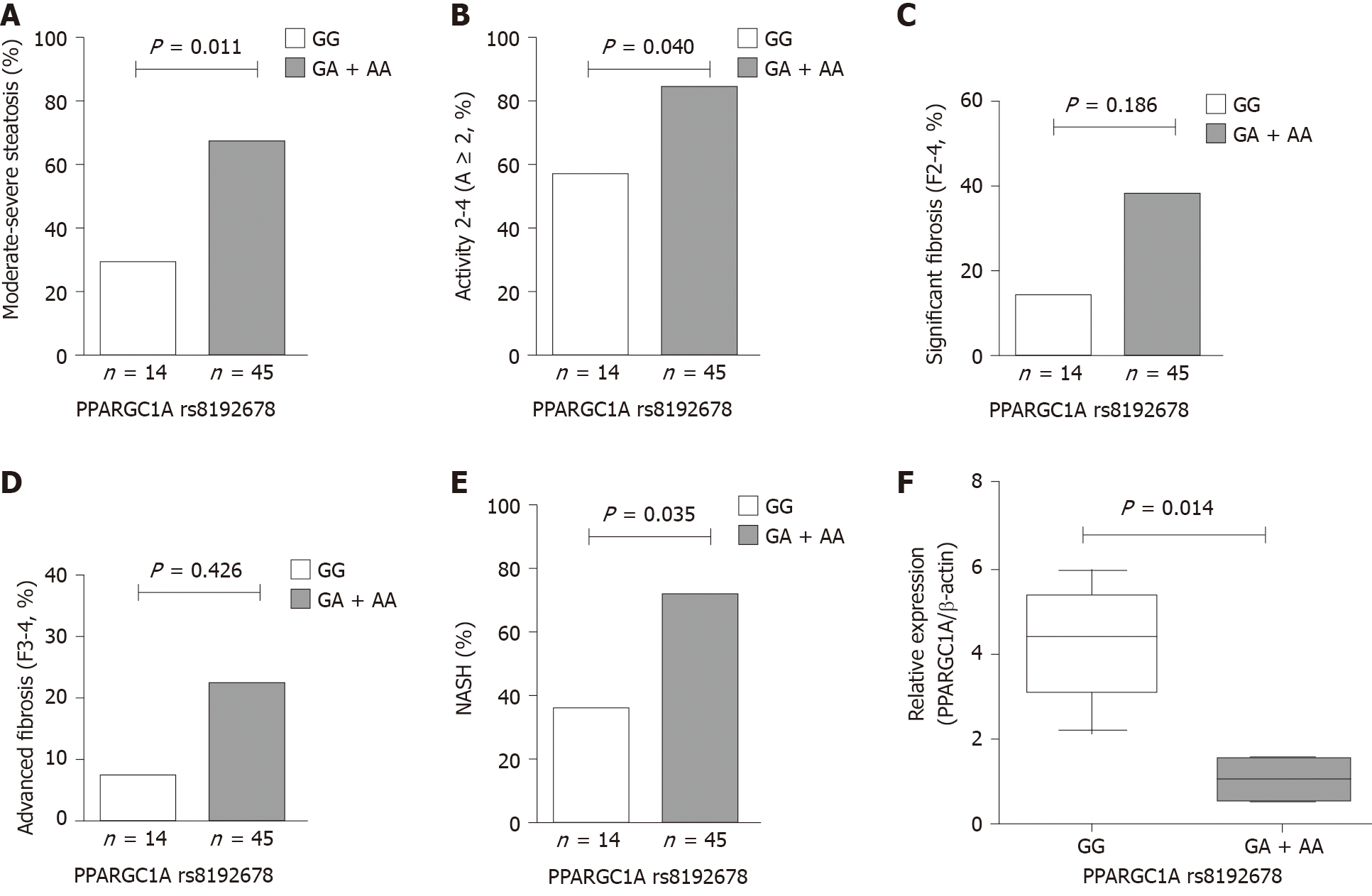
The hepatic histological features, including steatosis, activity (lobular inflammation and ballooning), and fibrosis, were assessed in patients with NAFLD. The frequencies of PPARGC1A rs8192678 GA + AA genotypes were remarkably higher in patients with moderate-to-severe steatosis (S2-3) and activity 2-4 (A ≥ 2) compared with those with mild steatosis (S1) and activity 0-1 (A < 2). Also, the carriers with risk A allele (carrying GA and AA genotypes) had a significantly increased risk in NAFLD patients with S2-3 and A ≥ 2 (OR 5.000, 95%CI: 1.343-18.620, P = 0.012 and OR 4.071, 95%CI: 1.076-15.402, P = 0.031). The multivariate analysis showed that PPARGC1A rs8192678 A allele was also significantly associated with S2-3 and A ≥ 2 (OR 6.190, 95%CI: 1.508-25.410, P = 0.011 and OR 4.506, 95%CI: 1.070-18.978, P = 0.040) after adjusting for age, sex, BMI, and PNPLA3 rs738409 risk G allele. However, no significant differences were found in significant fibrosis 2-4 (F2-4) and advanced fibrosis 3-4 (F3-4) compared with F0-1 and F0-2 (P = 0.186 and P = 0.426) (Figure 1A-E and Table 6).
| Histological features | Genotypes | Dominant model | Multivariate analysis1 | ||||
| GG (n = 14) | GA + AA (n = 45) | P value | OR (95%CI) | P value | OR (95%CI) | ||
| Steatosis (S) | S1 | 10 (71.43%) | 15 (33.33%) | ||||
| S2-3 | 4 (28.57%) | 30 (66.67%) | 0.012 | 5.000 (1.343-18.620) | 0.011 | 6.190 (1.508-25.410) | |
| Activity (A) | A < 2 | 6 (42.86%) | 7 (15.56%) | ||||
| A ≥ 2 | 8 (57.14%) | 38 (84.44%) | 0.031 | 4.071 (1.076-15.402) | 0.040 | 4.506 (1.070-18.978) | |
| Significant fibrosis | F0-1 | 12 (85.71%) | 28 (62.22%) | ||||
| F2-4 | 2 (14.29%) | 17 (37.78%) | 0.100 | 3.643 (0.726-18.292) | 0.186 | 3.449 (0.551-21.598) | |
| Advanced fibrosis | F0-2 | 13 (92.86%) | 35 (77.78%) | ||||
| F3-4 | 1 (7.14%) | 10 (22.22%) | 0.206 | 3.714 (0.432-31.949) | 0.426 | 2.549 (0.255-25.490) | |
| NASH | NAFL | 9 (64.28%) | 13 (28.89%) | ||||
| NASH | 5 (35.71%) | 32 (71.11%) | 0.017 | 4.431 (1.245-15.763) | 0.035 | 6.337 (1.135-35.392) | |
The patients with NASH carrying PPARGC1A rs8192678 GA + AA genotypes had an OR of 4.431 (95%CI: 1.245-15.763, P = 0.017) when compared with NAFL (Table 4). The multivariate analysis also showed that PPARGC1A rs8192678 risk A allele was independently associated with NASH after adjusting for age, sex, BMI, and PNPLA3 rs738409 risk G allele in patients with NAFLD (OR 6.337, 95%CI: 1.135-35.392, P = 0.035) (Table 6).
The GA + AA genotypes were not associated with obese NAFLD, obese NASH, and obese NAFL in the dominant model compared with nonobese NAFLD, nonobese NASH, and nonobese NAFL, respectively. Genetic analysis also showed no significant difference in the patients carrying A allele in PPARGC1A rs8192678 between obese NASH and obese NAFL groups (Table 4).
The remaining anthropometric values, biochemical test values, and PNPLA3 polymorphism did not show any significant differences between the two groups, apart from a trend in a higher level of GGT (P = 0.068) and CAP (P = 0.056) in nonobese NASH. However, genetic analysis showed that the proportion of PPARGC1A rs8192678 GA + AA genotypes was significantly higher in the patients with nonobese NASH compared with patients with nonobese NAFL, which was significantly associated with nonobese NASH (OR 22.000, 95%CI: 1.540-341.292, P = 0.021) (Tables 4 and 7).
| Variables | NAFL (n = 6) | NASH (n = 12) | P value |
| Sex (M/F) | 4/2 | 10/2 | 0.569 |
| Age (year) | 35.33 ± 18.44 | 39.67 ± 4.80 | 0.452 |
| BMI (kg/m2) | 22.83 ± 1.61 | 23.69 ± 0.90 | 0.122 |
| ALP (U/L) | 123.32 ± 77.62 | 84.25 ± 23.28 | 0.426 |
| GGT (U/L) | 28.50 (21.18-85.00) | 73.25 (54.75-99.25) | 0.068 |
| ALT (U/L) | 42.00 (21.78-78.50) | 71.15 (41.10-115.25) | 0.325 |
| TC (mmol/L) | 5.11 ± 0.55 | 4.96 ± 1.02 | 0.606 |
| TG (mmol/L) | 2.00 ± 0.43 | 1.77 ± 0.95 | 0.371 |
| HDL-C (mmol/L) | 1.10 ± 0.16 | 1.24 ± 0.44 | 0.943 |
| LDL-C (mmol/L) | 3.16 ± 0.46 | 2.82 ± 0.80 | 0.524 |
| FBG (mmol/L) | 5.03 ± 0.94 | 5.24 ± 0.85 | 0.587 |
| UA (μmol/L) | 360.86 ± 167.34 | 354.17 ± 67.43 | 0.684 |
| CAP (dB/m) | 257.00 ± 57.03 | 316.73 ± 43.48 | 0.056 |
| LSM (kPa) | 6.95 ± 4.07 | 6.79 ± 2.06 | 0.421 |
| PPARGC1A rs8162678 | |||
| GG, n (%) | 4 (66.67) | 1 (8.33) | |
| GA + AA, n (%) | 2 (33.33) | 11 (91.67) | 0.021 |
| PNPLA3 rs738409 | |||
| CC, n (%) | 2 (33.33) | 1 (8.33) | |
| CG + GG, n (%) | 4 (66.67) | 11 (91.67) | 0.245 |
The expression of PPARGC1A mRNA in the liver biopsy specimens of patients with NAFLD was evaluated by RT-PCR. The results obtained by RT-PCR in the GA or AA group (n = 5) were compared with the results in the GG group (n = 5) at PPARGC1A rs8192678. As shown in Figure 1F, the expression of PPARGC1A mRNA transcripts in the liver was significantly lower in the GA or AA group than in the GG group (P = 0.014).
Genetic factors may be important in the development of NAFLD, in either obese or nonobese patients. The present study further indicated that PPARGC1A rs8192678 risk A allele was associated with liver biopsy-proven NAFLD susceptibility in a Chinese Han adult population, and it was especially associated with NASH compared with NAFL. The PPARGC1A rs8192678 risk A allele was also associated with nonobese NASH in patients with nonobese NAFLD. In addition, this polymorphism was associated with moderate-to-severe steatosis and higher histological activity, but not with significant and advanced fibrosis.
PPARGC1A/PGC-1α is a coactivator for a number of transcription factors, including the peroxisome proliferator-activated receptor. PGC-1α is a critical regulator of adaptive thermogenesis, cellular respiration, and energy metabolism[17]. Recent studies suggested that PGC-1α also regulates lipid metabolism, PGC-1α stimulates the expression of farnesoid X receptor (FXR) target genes in hepatic cells and reduces TG secretion by enhancing FXR activity[18]. In addition, PGC-1α can transactivate the hepatocyte nuclear factor 4α-dependent human hepatic lipase gene promoter to increase the production of hepatic lipase. The PGC-1α is also highly expressed in the liver and coordinates the induction of fatty acid oxidation when fasting. Multiple studies showed that decreased PGC-1α expression might be the mechanism underlying metabolic diseases. Tissue-specific reduction of PGC-1α expression in adipose, liver, and muscle reduced glucose tolerance and insulin sensitivity in mice[19,20], supporting the hypothesis that decreased PGC-1α expression in insulin-sensitive tissues might contribute to a higher risk of T2DM. PPARGC1A mRNA expression decreased in muscle and adipose tissue of human participants with T2DM[21], and a correlation existed between adipose PGC-1α protein levels and decreased insulin sensitivity[22]. T2DM was also a risk factor for NAFLD. Another reported PPARGC1A polymorphism rs2290602 also decreased the expression of PPARGC1A in patients with NAFLD[6]. Therefore, genetic factors that alter the expression of PGC-1α might participate in the pathogenesis of NAFLD. The present study also indicated that the expression of PPARGC1A was significantly lower in patients who carried the A allele, which supported the hypothesis that decreased PGC-1α expression in steatosis liver tissues might contribute to a higher risk of NAFLD.
Recently, the association between different single nucleotide polymorphisms (SNPs) and metabolic diseases, such as NAFLD, was reported[23]. Therefore, different polymorphisms of PPARGC1A associated with different metabolic diseases were investigated. Iglseder et al[24] showed that the -3974T/C (rs2970865) polymorphism of the PPARGC1A gene was associated with the severity of carotid atherosclerosis. The variations in rs2290602 in the PPARGC1A gene was expected to affect lipid and glucose metabolism and result in the development of NAFLD and NASH[6]. Another common PPARGC1A rs8192678 polymorphism might contribute to the risk of coronary artery disease in the Chinese population[9,24], and was also associated with obesity, hypertension, and T2DM. This polymorphism in PPARGC1A was also reported to be related to the susceptibility of NAFLD in different populations and ethnicities. The present study showed that the variation of PPARGC1A rs8192678 GA genotype was significantly higher in patients with NAFLD, but PPARGC1A rs8192678 AA genotype showed no difference between NAFLD and control groups, and the percentage of AA genotype was only 13.56% (8/59) in NAFLD patients, which was consistent with the findings of Lin et al[7] in obese children in Taiwan. The explanation of this finding in Lin et al[7] concluded two probable reasons. First, the number with PPARGC1A rs8192678 AA genotype was relatively small, because of the sample variation and inadequate statistical power, the role of AA genotype in NAFLD was not presented. Second, the cellular effects of PGC-1α were complicated, and the mechanism by which PGC-1α influenced the pathogenesis of NAFLD was still unclear[7]. However, a comparison between GA + AA and GG genotypes revealed that the participants who carried the PPARGC1A rs8192678 A allele were found to be significantly associated with not only liver biopsy-proven NAFLD but also NASH, independent of age, sex, BMI, and PNPLA3 rs738409 risk G allele in the dominant model. No study analyzed the association between the PPARGC1A rs8192678 A allele and hepatic histological features in NAFLD. Significant associations of PPARGC1A rs8192678 risk A allele with moderate-to-severe steatosis and higher histological activity in patients with NAFLD were found in this study. However, significant fibrosis and advanced fibrosis were not associated with PPARGC1A rs8192678 risk A allele. This suggested that PPARGC1A rs8192678 risk A allele was associated with steatosis and liver damage, contributing to the occurrence of NASH.
Several studies reported that PPARGC1A rs8192678 was significantly associated with obesity; this polymorphism conferred a higher risk of obesity in certain populations[25,26]. The patients who carried PPARGC1A rs8192678 AA genotype benefited more from interventions aimed at weight loss, including caloric restriction, bariatric surgery, and acarbose treatment[27,28]. The prevalence of lean NAFLD and nonobese NAFLD has increased in eastern and western countries in recent years. The genetic, metabolic, microbiome, metabolomics, and inflammatory factors may also be the causes of NAFLD in lean or nonobese individuals. Genetic factors may also be important in the occurrence and development of nonobese NAFLD. IFNL3 rs12979860 polymorphism has been shown to be associated with hepatic inflammation and fibrosis in patients with nonobese NAFLD[29]. Two SNPs rs12447924 and rs12597002 of the cholesteryl ester transfer protein gene were reported to be associated with a risk of steatosis in nonobese individuals[30]. Wang et al[31] found a significantly higher prevalence of advanced fibrosis (F ≥ 3) and LSM values in patients with nonobese NASH compared with obese NASH; also, a higher prevalence of TM6SF2 T allele was observed in patients with nonobese NASH, indicating that patients with nonobese NASH had more severe hepatic histological changes and genetic susceptibility in the Chinese population[31]. The present study found that the PPARGC1A rs8192678 A allele was not significantly closely associated with obese participants, patients with obese NAFLD, and patients with obese NASH. However, the PPARGC1A rs8192678 A allele was relatively more common in patients with nonobese NASH compared with patients with obese NASH. The results also showed that the PPARGC1A rs8192678 GA + AA genotypes were remarkably associated with nonobese NASH, indicating that the PPARGC1A rs8192678 A allele was associated with a high risk of NASH. This, especially, increased the risk of nonobese NASH, and the PPARGC1A rs8192678 polymorphism might be the pathogenesis of nonobese NASH. Despite no significant difference in TC, TG, ALT, GGT, FBG, UA, and CAP values and PNAPL3 rs738409 between the nonobese NAFL and NASH groups, the levels of GGT (P = 0.068) and CAP (P = 0.056) followed an increasing trend in nonobese NASH. This suggested that genetic background might also be a risk factor for nonobese NASH. The influence of metabolic factors, such as lipid and glucose, needs further exploration using a large sample.
The association between PPARGC1A rs8192678 and NAFLD in the Asian population is controversial. Hui et al[15] and Zhou et al[14] reported no association between the PPARGC1A rs8192678 variant and ultrasound-defined NAFLD in Chinese adults[14,15]. However, Tai et al[8] reported that the PPARGC1A variant rs8192678 was associated with liver biopsy-proven NASH in severely obese Taiwanese patients[8]. Saremi et al[32] also indicated that the AA genotype and A allele of PPARGC1A increased in Iranian patients with liver biopsy-proven NAFLD[32]. The frequency of the PPARGC1A rs8192678 A allele was 0.453 in Han Chinese, 0.350 in Europeans, and 0.040 in Africans according to the National Center for Biotechnology Information human SNP database, which inferred that the PPARGC1A rs8192678 risk A allele conferred a higher genetic susceptibility to NAFLD in Asians than in Europeans and Africans[7]. Also, the PPARGC1A rs8192678 was associated with NAFLD, especially NASH, in the Chinese Han adult population with liver biopsy-proven NAFLD. This contradictory conclusion might be due to the different criteria used for the definition of NAFLD; ultrasonography or liver biopsy was carried out in different studies. In the present study, the gold diagnostic standard for NAFLD, liver histopathology, was used to avoid the potential image bias in Chinese Han patients with NAFLD.
However, this study also had limitations. First, obtaining liver biopsies was difficult, and hence patients with NAFLD were relatively few, especially those with nonobese NAFLD. Consequently, no statistically significant differences were identified. Second, the risk factors such as hypertension and diabetes, which might influence obese/nonobese NAFLD, were not mentioned in this study. Therefore, a large number of patients with liver biopsy-proven NAFLD should be included in further studies, and well-designed case-control studies should be performed to confirm and support these findings.
In conclusion, the present study showed that PPARGC1A rs8192678 risk A allele was associated with liver biopsy-proven NAFLD/NASH, which also had an effect on the severity of hepatic histological features (S2-3 and A ≥ 2), and might also be associated with nonobese NASH in the Chinese Han adult population. These findings also suggested that PPARGC1A polymorphism rs8192678 risk A allele and lower expression of PPARGC1A mRNA might contribute to the etiology of NAFLD. Further studies with large samples are required to confirm these findings.
Nonalcoholic fatty liver disease (NAFLD) has become the most common chronic liver disease and is a significant global burden worldwide. It is also common in obese and nonobese populations. Although the mechanism of NAFLD is unclear, genetic susceptibility plays a vital role in NAFLD. The association between PPARGC1A rs8192678 polymorphism and NAFLD has been reported in several studies, but it is controversial in the Asian population. Whether PPARGC1A rs8192678 is associated with hepatic histological features in NAFLD in the Chinese population is unknown.
The association between PPARGC1A rs8192678 polymorphism and NAFLD requires further investigation, and the association with hepatic histological features and nonobese NAFLD in the Chinese population is unknown.
The aim was to investigate the association between PPARGC1A rs8192678 polymorphism and nonalcoholic steatohepatitis (NASH) and to determine whether PPARGC1A rs8192678 is associated with the hepatic histological features of NASH.
Patients with NAFLD and healthy controls were recruited to a cohort representing the Chinese Han population. Patients with NAFLD were proven by liver biopsy. The SAF (Steatosis, activity, and fibrosis) scoring system was used for histopathological evaluation. The polymorphisms of PPARGC1A rs8192678 and patatin-like phospho
In NAFLD patients, 37 patients had NASH, of which 12 were nonobese NASH. The PPARGC1A rs8192678 risk A allele (carrying GA and AA genotypes) increased the risk of NAFLD in the dominant model. The PPARGC1A rs8192678 A allele was also found to be a risk factor for NAFLD after adjusting for age, sex, and PNPLA3 rs738409 polymorphism. In the hepatic histological features of NAFLD patients, moderate-to-severe steatosis (S2-3), and Activity 2-4 (A ≥ 2) were more likely to carry PPARGC1A rs8192678 risk A allele. After adjusting for age, sex, body mass index, and PNPLA3 rs738409 risk G allele, the PPARGC1A rs8192678 risk A allele was also independently associated with S2-3, A ≥ 2, and NASH. The results also showed that this polymorphism was associated with nonobese NASH. In the group of patients who carried A allele (GA or AA genotypes), the intrahepatic expression of PPARGC1A mRNA was significantly lower than that in patients with GG genotype.
The PPARGC1A rs8192678 A allele is a risk factor for NAFLD, and is associated with the severity of hepatic histological features (S2-3 and A ≥ 2) and NASH in NAFLD patients, independent of PNPLA3 rs738409, and might be associated with nonobese NASH.
The result that PPARGC1A rs8192678 was associated with liver biopsy-proven NASH and had an additive effect on the severity of hepatic histological features was further confirmed in the Chinese Han adult population. PPARGC1A rs8192678 might contribute to the etiology of NAFLD. PPARGC1A rs8192678 might be a useful tool for diagnosing NASH and predicting the severity of the hepatic histological features of NAFLD.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Victor DW S-Editor: Fan JR L-Editor: Webster JR P-Editor: Ma YJ
| 1. | Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263-2273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1508] [Cited by in RCA: 1755] [Article Influence: 175.5] [Reference Citation Analysis (0)] |
| 2. | Fan JG, Jia JD, Li YM, Wang BY, Lu LG, Shi JP, Chan LY; Chinese Association for the Study of Liver Disease. Guidelines for the diagnosis and management of nonalcoholic fatty liver disease: update 2010: (published in Chinese on Chinese Journal of Hepatology 2010; 18:163-166). J Dig Dis. 2011;12:38-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 3. | Kim D, Kim WR. Nonobese Fatty Liver Disease. Clin Gastroenterol Hepatol. 2017;15:474-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 259] [Article Influence: 32.4] [Reference Citation Analysis (1)] |
| 4. | Shi Y, Wang Q, Sun Y, Zhao X, Kong Y, Ou X, Jia J, Wu S, You H. The Prevalence of Lean/Nonobese Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. J Clin Gastroenterol. 2020;54:378-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 5. | Adams LA, Waters OR, Knuiman MW, Elliott RR, Olynyk JK. NAFLD as a risk factor for the development of diabetes and the metabolic syndrome: an eleven-year follow-up study. Am J Gastroenterol. 2009;104:861-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 320] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 6. | Yoneda M, Hotta K, Nozaki Y, Endo H, Uchiyama T, Mawatari H, Iida H, Kato S, Hosono K, Fujita K, Yoneda K, Takahashi H, Kirikoshi H, Kobayashi N, Inamori M, Abe Y, Kubota K, Saito S, Maeyama S, Wada K, Nakajima A. Association between PPARGC1A polymorphisms and the occurrence of nonalcoholic fatty liver disease (NAFLD). BMC Gastroenterol. 2008;8:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Lin YC, Chang PF, Chang MH, Ni YH. A common variant in the peroxisome proliferator-activated receptor-γ coactivator-1α gene is associated with nonalcoholic fatty liver disease in obese children. Am J Clin Nutr. 2013;97:326-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | Tai CM, Huang CK, Tu HP, Hwang JC, Yeh ML, Huang CF, Huang JF, Dai CY, Chuang WL, Yu ML. Interactions of a PPARGC1A Variant and a PNPLA3 Variant Affect Nonalcoholic Steatohepatitis in Severely Obese Taiwanese Patients. Medicine (Baltimore). 2016;95:e3120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 9. | Zhang Y, Xu W, Li X, Tang Y, Xie P, Ji Y, Fan L, Chen Q. Association between PPARGC1A gene polymorphisms and coronary artery disease in a Chinese population. Clin Exp Pharmacol Physiol. 2008;35:1172-1177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Vimaleswaran KS, Radha V, Ghosh S, Majumder PP, Deepa R, Babu HN, Rao MR, Mohan V. Peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1alpha) gene polymorphisms and their relationship to Type 2 diabetes in Asian Indians. Diabet Med. 2005;22:1516-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 75] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Muller YL, Bogardus C, Pedersen O, Baier L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes. 2003;52:895-898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Zhang KH, Huang Q, Dai XP, Yin JY, Zhang W, Zhou G, Zhou HH, Liu ZQ. Effects of the peroxisome proliferator activated receptor-γ coactivator-1α (PGC-1α) Thr394Thr and Gly482Ser polymorphisms on rosiglitazone response in Chinese patients with type 2 diabetes mellitus. J Clin Pharmacol. 2010;50:1022-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Villegas R, Williams SM, Gao YT, Long J, Shi J, Cai H, Li H, Chen CC, Tai ES; AGEN-T2D Consortium; Hu F, Cai Q, Zheng W, Shu XO. Genetic variation in the peroxisome proliferator-activated receptor (PPAR) and peroxisome proliferator-activated receptor gamma co-activator 1 (PGC1) gene families and type 2 diabetes. Ann Hum Genet. 2014;78:23-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 14. | Zhou YJ, Li YY, Nie YQ, Yang H, Zhan Q, Huang J, Shi SL, Lai XB, Huang HL. Influence of polygenetic polymorphisms on the susceptibility to non-alcoholic fatty liver disease of Chinese people. J Gastroenterol Hepatol. 2010;25:772-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Hui Y, Yu-Yuan L, Yu-Qiang N, Wei-Hong S, Yan-Lei D, Xiao-Bo L, Yong-Jian Z. Effect of peroxisome proliferator-activated receptors-gamma and co-activator-1alpha genetic polymorphisms on plasma adiponectin levels and susceptibility of non-alcoholic fatty liver disease in Chinese people. Liver Int. 2008;28:385-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Wong VW, Vergniol J, Wong GL, Foucher J, Chan HL, Le Bail B, Choi PC, Kowo M, Chan AW, Merrouche W, Sung JJ, de Lédinghen V. Diagnosis of fibrosis and cirrhosis using liver stiffness measurement in nonalcoholic fatty liver disease. Hepatology. 2010;51:454-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 876] [Cited by in RCA: 970] [Article Influence: 64.7] [Reference Citation Analysis (1)] |
| 17. | Liu C, Lin JD. PGC-1 coactivators in the control of energy metabolism. Acta Biochim Biophys Sin (Shanghai). 2011;43:248-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 18. | Buler M, Aatsinki SM, Skoumal R, Hakkola J. Energy sensing factors PGC-1α and SIRT1 modulate PXR expression and function. Biochem Pharmacol. 2011;82:2008-2015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Besse-Patin A, Léveillé M, Oropeza D, Nguyen BN, Prat A, Estall JL. Estrogen Signals Through Peroxisome Proliferator-Activated Receptor-γ Coactivator 1α to Reduce Oxidative Damage Associated With Diet-Induced Fatty Liver Disease. Gastroenterology. 2017;152:243-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 20. | Sczelecki S, Besse-Patin A, Abboud A, Kleiner S, Laznik-Bogoslavski D, Wrann CD, Ruas JL, Haibe-Kains B, Estall JL. Loss of Pgc-1α expression in aging mouse muscle potentiates glucose intolerance and systemic inflammation. Am J Physiol Endocrinol Metab. 2014;306:E157-E167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 21. | Ghosh S, Lertwattanarak R, Lefort N, Molina-Carrion M, Joya-Galeana J, Bowen BP, Garduno-Garcia Jde J, Abdul-Ghani M, Richardson A, DeFronzo RA, Mandarino L, Van Remmen H, Musi N. Reduction in reactive oxygen species production by mitochondria from elderly subjects with normal and impaired glucose tolerance. Diabetes. 2011;60:2051-2060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 22. | Hammarstedt A, Jansson PA, Wesslau C, Yang X, Smith U. Reduced expression of PGC-1 and insulin-signaling molecules in adipose tissue is associated with insulin resistance. Biochem Biophys Res Commun. 2003;301:578-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 122] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 23. | Woo HJ, Reifman J. Genetic interaction effects reveal lipid-metabolic and inflammatory pathways underlying common metabolic disease risks. BMC Med Genomics. 2018;11:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Iglseder B, Oberkofler H, Felder TK, Klein K, Paulweber B, Krempler F, Tregouet DA, Patsch W. Associations of PPARGC1A haplotypes with plaque score but not with intima-media thickness of carotid arteries in middle-aged subjects. Stroke. 2006;37:2260-2265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 25. | Weng SW, Lin TK, Wang PW, Chen IY, Lee HC, Chen SD, Chuang YC, Liou CW. Gly482Ser polymorphism in the peroxisome proliferator-activated receptor gamma coactivator-1alpha gene is associated with oxidative stress and abdominal obesity. Metabolism. 2010;59:581-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Vázquez-Del Mercado M, Guzmán-Ornelas MO, Corona Meraz FI, Ríos-Ibarra CP, Reyes-Serratos EA, Castro-Albarran J, Ruíz-Quezada SL, Navarro-Hernández RE. The 482Ser of PPARGC1A and 12Pro of PPARG2 Alleles Are Associated with Reduction of Metabolic Risk Factors Even Obesity in a Mexican-Mestizo Population. Biomed Res Int. 2015;2015:285491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Goyenechea E, Crujeiras AB, Abete I, Parra D, Martínez JA. Enhanced short-term improvement of insulin response to a low-caloric diet in obese carriers the Gly482Ser variant of the PGC-1alpha gene. Diabetes Res Clin Pract. 2008;82:190-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Geloneze SR, Geloneze B, Morari J, Matos-Souza JR, Lima MM, Chaim EA, Pareja JC, Velloso LA. PGC1α gene Gly482Ser polymorphism predicts improved metabolic, inflammatory and vascular outcomes following bariatric surgery. Int J Obes (Lond). 2012;36:363-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Petta S, Valenti L, Tuttolomondo A, Dongiovanni P, Pipitone RM, Cammà C, Cabibi D, Di Marco V, Fracanzani AL, Badiali S, Nobili V, Fargion S, Grimaudo S, Craxì A. Interferon lambda 4 rs368234815 TT>δG variant is associated with liver damage in patients with nonalcoholic fatty liver disease. Hepatology. 2017;66:1885-1893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 30. | Adams LA, Marsh JA, Ayonrinde OT, Olynyk JK, Ang WQ, Beilin LJ, Mori T, Palmer LJ, Oddy WW, Lye SJ, Pennell CE. Cholesteryl ester transfer protein gene polymorphisms increase the risk of fatty liver in females independent of adiposity. J Gastroenterol Hepatol. 2012;27:1520-1527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Wang Q, You H, Ou X, Zhao X, Sun Y, Wang M, Wang P, Wang Y, Duan W, Wang X, Wu S, Kong Y, Saxena R, Gouw ASH, Jia J. Non-obese histologically confirmed NASH patients with abnormal liver biochemistry have more advanced fibrosis. Hepatol Int. 2019;13:766-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 32. |
Saremi L, Lotfıpanah S, Mohammadi M, Hosseinzadeh H, Hosseini-Khah Z, Johari B, Saltanatpour Z.
Association between PPARGC1A single nucleotide polymorphisms and increased risk of nonalcoholic fatty liver disease among Iranian patients with type 2 diabetes mellitus |