Published online Jun 14, 2021. doi: 10.3748/wjg.v27.i22.2979
Peer-review started: February 3, 2021
First decision: February 27, 2021
Revised: March 10, 2021
Accepted: April 22, 2021
Article in press: April 22, 2021
Published online: June 14, 2021
Processing time: 125 Days and 4.3 Hours
The landscape of gastrointestinal endoscopy continues to evolve as new technologies and techniques become available. The advent of image-enhanced and magnifying endoscopies has highlighted the step toward perfecting endoscopic screening and diagnosis of gastric lesions. Simultaneously, with the development of convolutional neural network, artificial intelligence (AI) has made unprecedented breakthroughs in medical imaging, including the ongoing trials of computer-aided detection of colorectal polyps and gastrointestinal bleeding. In the past demi-decade, applications of AI systems in gastric cancer have also emerged. With AI’s efficient computational power and learning capacities, endoscopists can improve their diagnostic accuracies and avoid the missing or mischaracterization of gastric neoplastic changes. So far, several AI systems that incorporated both traditional and novel endoscopy technologies have been developed for various purposes, with most systems achieving an accuracy of more than 80%. However, their feasibility, effectiveness, and safety in clinical practice remain to be seen as there have been no clinical trials yet. Nonetheless, AI-assisted endoscopies shed light on more accurate and sensitive ways for early detection, treatment guidance and prognosis prediction of gastric lesions. This review summarizes the current status of various AI applications in gastric cancer and pinpoints directions for future research and clinical practice implementation from a clinical perspective.
Core Tip: Artificial intelligence-assisted endoscopy can assist physicians in the screening and diagnosis of gastric cancer. Most of the systems developed so far, applied in images and videos and using white light imaging and narrow-band imaging endoscopies, have achieved accuracies and sensitivities of at least 80%. However, the efficacy of artificial intelligence applications in gastric cancer depends on its intended role in clinical practice, and there have not been any attempts of clinical trials yet. This review summarizes the existing artificial intelligence applications in gastric cancer and pinpoints future research directions for their clinical practice implementation.
- Citation: Hsiao YJ, Wen YC, Lai WY, Lin YY, Yang YP, Chien Y, Yarmishyn AA, Hwang DK, Lin TC, Chang YC, Lin TY, Chang KJ, Chiou SH, Jheng YC. Application of artificial intelligence-driven endoscopic screening and diagnosis of gastric cancer. World J Gastroenterol 2021; 27(22): 2979-2993
- URL: https://www.wjgnet.com/1007-9327/full/v27/i22/2979.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i22.2979
Diagnostic and therapeutic endoscopies play a major role in the management of gastric cancer (GC). Endoscopy is the mainstay for the diagnosis and treatment of early adenocarcinoma and lesions and the palliation of advanced cancer[1-5]. GC, being the fifth most common cancer and the third leading cause of cancer-related deaths worldwide, affects more than one million people and causes approximately 780000 deaths annually[6-10]. Continued development in endoscopy aims to strengthen its quality indicators. These developments include using higher resolution and magnification endoscopies, chromoendoscopy and optical techniques based on the modulation of the light source, such as narrow-band imaging (NBI), fluorescence endoscopy and elastic scattering spectroscopy[11-13]. New tissue sampling methods to identify the stages of a patient’s risk for cancer are also being developed to decrease the burden on patients and clinicians during endoscopy.
Statistically, the relative 5-year survival rate of GC is less than 40%[7,9,14,15], often attributed to the late onset of symptoms and delayed diagnosis[10]. Although early diagnosis is difficult as most patients are asymptomatic in the early stage, the diagnosis point largely determines the patient’s prognosis[16,17]. In other words, endoscopic detection of GC at an earlier stage is the only and most effective way to reduce its recurrence and to prolong patient survival. This early diagnosis of GC provides the opportunity for minimally invasive therapy methods such as endoscopic mucosal resection or submucosal dissection[18-20]. The 5-year survival rate was reported to be more than 90% among patients with GC detected at an early stage[21-23]. Yet, the false negative rate of GC detected by esophagogastroduodenoscopy, the current standard diagnostic procedure, was reported to be between 4.6% and 25.8%[24-29]. In terms of the common diagnostic methods, esophagogastroduodenoscopy is the preferred diagnostic modality for patients with suspected GC; the combination of lymph node dissection, endoscopic ultrasonography and computed tomographic scanning is involved in staging the tumor[30,31]. From the differential diagnosis between GC and gastritis, prediction of the horizontal extent of GC to characterizing the depth of invasion of GC, the early abnormal symptoms of GC and its advanced aggressive malignancy as well as the heavy workload of image analysis present ample inevitable challenges for endoscopists[32-34]. With large variations in the diagnostic ability of endoscopists, long-term training and experience may not guarantee their consistency and accuracy of diagnosis[35-37].
In recent years, artificial intelligence (AI) has caught considerable attention in various medical fields, including skin cancer classification[38-41], diagnosis in radiation oncology[42-45] and analysis of brain magnetic resonance imaging[46-49]. Although its applications have shown impressive accuracy and sensitivity identifying and characterizing imaging abnormalities, its improved sensitivity also meant the detection of subtle and indeterminately significant changes[50,51]. For example, in the analysis of brain magnetic resonance imaging, despite the promise of early diagnosis with machine learning, the relationship between subtle parenchymal brain alterations detected by AI and its neurological outcomes is unknown in the absence of a well-defined abnormality[52]. In other words, the use of AI in diagnostic imaging in various medical fields is continuously undergoing extensive evaluation.
In the field of gastroenterology, AI applications in capsule endoscopy[53-56] and in the detection, localization and segmentation of colonic polyps have been reported as well[57-59]. In particular, in the late 2010s, there was an explosion of interest in GC. The use of AI has proven to provide better diagnostic capabilities, although further validation and extensions are necessary to augment their quality and interpretability. An AI system’s quality is often described with statistical measures of sensitivity, specificity, positive predictive value and accuracy.
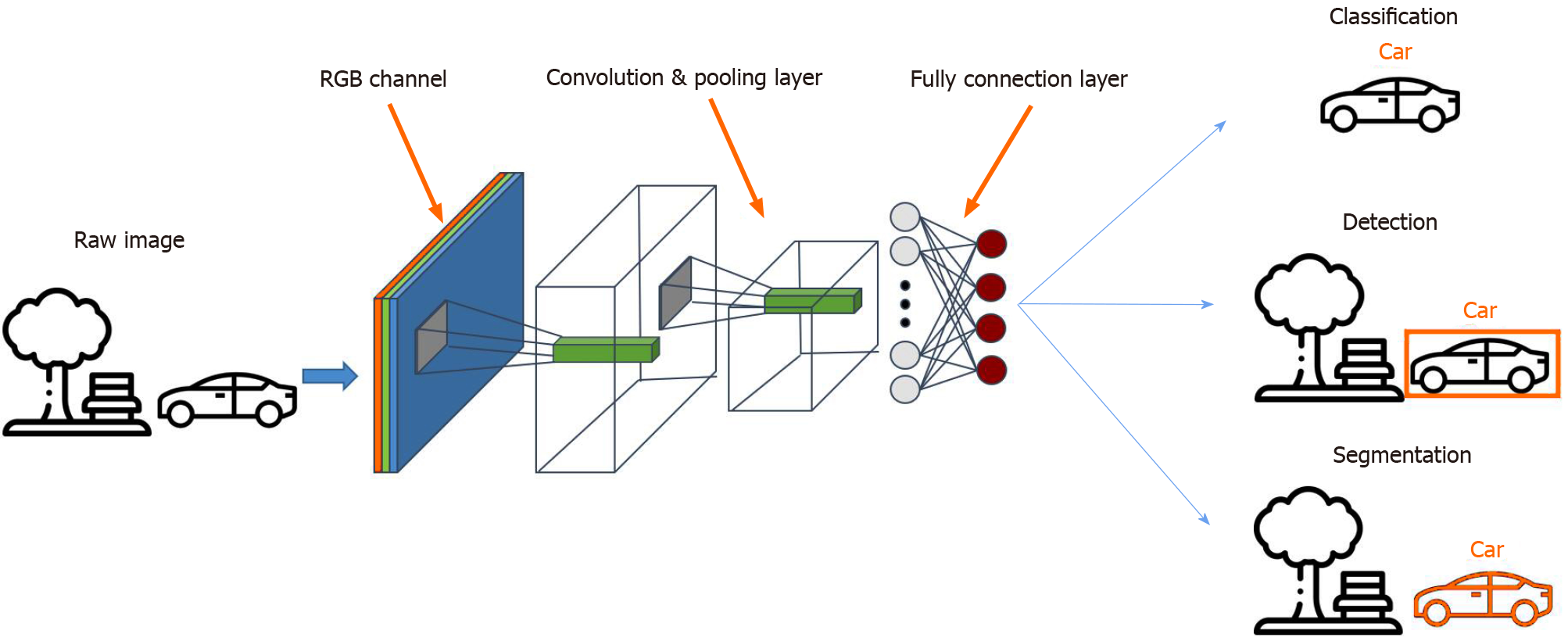
Among the different AI models, the convolutional neural network (CNN) is a method most commonly used in medical imaging[60,61] as it allows the detection, segmentation and classification of image patterns[62] (Figure 1). CNN uses the mathematical operation of convolution to classify the images after recognizing patterns from the raw image pixel. Because the 7-layer Le-Net-5 program was first pioneered by LeCun et al in 1998, CNN architectures have been rapidly developing. Today, other widely-used CNNs include AlexNet (2012) with about 15.3% error rate, 22-layer GoogLeNet (2014) with a 6.67% error rate but only 4 million parameters, 19-layer visual geometry group (VGG) Net (2014) with 7.3% error rate and 138 million parameters, and Microsoft’s ResNet (2015) with an error rate of 3.6% that can be trained with as many as 152 layers[63-65]. While scholars have lauded AI for the potential and performance it has displayed, some have cast doubts on its generalizability and role in the holistic assessment of gastric abnormalities.
In the beginning of an AI-assisted diagnostic imaging revolution, we have to anticipate and meticulously assess the potential perils, in the context of its capabilities, to ensure effective and safe incorporation into clinical practice[66]. In this paper, we thereby review the current status of AI applications in screening and diagnosing GC. We explore with emphasis on two broad categories: namely, the identification of pathogenic infection and the qualitative diagnosis of GC. Finally, we considered some directions for further research and the future of its introduction into clinical practice.
AI applications in identifying pathogenic infections have been widely explored[67] (Table 1). Gastric epithelium Helicobacter pylori (H. pylori) infection is associated with functional dyspepsia, peptic ulcers, mucosal atrophy, intestinal metaplasia, atrophic gastritis and GC[68,69]. Because most gastric malignancies correlate with H. pylori infection, identifying H. pylori infection at its early stage is essential in preventing H. pylori-aggravated comorbidities[70-74]. Although physicians usually use the C13 urea breath test to diagnose H. pylori, most subclinical H. pylori infection cases still rely on the time-consuming and invasive biopsy examination to avoid the risk of a false negative diagnosis. Moreover, the Kyoto Classification, as the current gold standard of H. pylori severity classification, requires examiners to measure lesions by their bare eyes. Such a method is a subjective judgment that usually comes with interoperator bias[75-77]. Compelled by such ambiguity, researchers have turned to devising a next-generation semi-automatic standard examination protocol, that is AI.
| Ref. | Endoscopic modality | Training dataset | Validation dataset | Accuracy | Sensitivity | Specificity | PPV |
| Huang et al[78], 2004 | WLI | 30 patients | 74 patients | 85.1 (avg)1 | 78.8 (avg) | 90.2 (avg) | - |
| Shichijo et al[79], 2017 | WLI | 32208 images, 1768 patients | 11481 images, 397 patients | 87.7 | 88.9 | 87.4 | - |
| Itoh et al[81], 2018 | WLI | 149 images, 139 patients | 30 images, 30 patients | - | 86.7 | 86.7 | - |
| Nakashima et al[84], 2018 | WLI, BLI and LCI | 162 patients | 60 patients | - | 96.7 | - | - |
| Shichijo et al[80], 2019 | WLI | 98564 images, 4494 patients | 23699 images, 847 patients | Infected: 66.0; post-eradication: 86.0 | - | - | - |
| Zheng et al[82], 2019 | WLI | 11729 images, 1507 patients | 3755 images, 452 patients | 84.5 | 81.4 | 90.1 | - |
| Zhu et al[100], 2019 | WLI | 790 images | 203 images | 89.2 | 76.5 | 95.6 | 89.7 |
| Nakashima et al[85], 2020 | WLI, BLI and LCI | 12887 images, 395 patients | 120 patients | 80.0 (avg)2 | 61.3 (avg) | 89.4 (avg) | 74.7 (avg) |
As early as 2004, before CNN took the lead in machine-assisted image diagnosis, Huang et al[78] deployed the refined feature selection neural network to process endoscopic images and return the results of H. pylori infection probability and severity. By training AI with 30 patients’ endoscopic images including crops of antrum, body and cardia locations of the stomach, they established an algorithm that achieved an average of 78.8% sensitivity, 90.2% specificity and 85.1% accuracy in an independent cohort of 74 patient images. The overall prediction accuracy was better than the one demonstrated by young physicians and fellow doctors, who scored 68.4% and 78.4%, respectively. It was the first model demonstrating the potential of computer-aided diagnosis of H. pylori infection by endoscope images.
However, since the introduction of the 7-layer Le-Net-5 program by LeCun et al in 1998, the CNNs have gradually taken over in the field of medical image processing. To name a few examples, Shichijo et al[79] used 32208 images of 735 H. pylori-positive and 1015 H. pylori-negative cases to develop an H. pylori identifying AI system based on the architecture of 22-layer GoogLeNet. The sensitivity, specificity, accuracy and time consumption were 81.9%, 83.4%, 83.1% and 198 s for the first CNN and 88.9%, 87.4%, 87.7% and 194 s for the secondary CNN developed, respectively, compared with that of 79.0%, 83.2%, 82.4% and 230 min by the endoscopists. Later, still using GoogLeNet, Shichijo et al[80] developed another system that further classified the current infection, post-eradication and current noninfection statuses of H. pylori, obtaining an accuracy of 48%, 84%, and 80%, respectively. In this system, the CNN was trained with 98564 images from 4494 patients and tested with 23699 images from 847 independent cases. Itoh et al[81] also developed a CNN based on GoogLeNet, trained with 149 endoscopic images obtained from 139 patients and tested with 30 images from 30 patients, which could detect and diagnose H. pylori with sensitivity and specificity of 86.7%. Additionally, the use of ResNet CNN architecture was reported by Zheng et al[82] in 2019, achieving a sensitivity, specificity and accuracy of 81.4%, 90.1% and 84.5%, respectively. In this study, the system was trained with 11729 images from 1507 patients and tested with 3755 images from 452 patients using a 50-layer ResNet-50 (Microsoft) CNN system and PyTorch (Facebook) deep learning framework.
Recently, AI has also been applied to linked color imaging (LCI) and blue laser imaging, two novel image-enhanced endoscopy technologies[83]. It helped diagnose[84] and classify[85] the H. pylori infection and has shown greater effectiveness. In 2018, Nakashima et al[84] developed a system on a training set of 162 patients and a test set of 60 patients that could diagnose H. pylori infection with an area under the curve of 0.96 and 0.95 and sensitivity of 96.7% and 96.7% for blue laser imaging-bright and LCI, respectively. Such performance is superior to systems that use conventional white light imaging (WLI) (with 0.66 area under the curve and sensitivities as mentioned earlier in other studies) as well as that of experienced endoscopists. Another 2020 study by the same team also showed that classifying the H. pylori infection status (uninfected, infected and post-eradication) by incorporating deep learning and image-enhanced endoscopies yields more accurate results. The system was trained with 6639 WLI and 6248 LCI images from 395 patients and tested with images from 120 patients[85].
However, there are some limitations of AI in identifying H. pylori that remain to be overcome amongst the developed systems and findings. First, the histological time frame, especially for the eradicated infection, was not considered in the AI systems[86]. Second, both the training data sets and test data sets were obtained from a single center for all existing systems. A continued and even more rigorous external validation, which uses more diverse sources of images and endoscopies, is necessary to evaluate each system’s generalizability[87,88]. Additionally, the application of CNN algorithms is also still confined to the existing models of CNN algorithms (mostly GoogLeNet and a few ResNet). Further technical refinements may overcome current limitations faced by endoscopists. They also shed light on the possibility of a system that distinguishes between H. pylori-infected and H. pylori-eradicated patients, determines different parts of the stomach (cardia, body, angle and pylorus) and provides real-time evaluations of H. pylori. These will be considerations vital for its implementation in clinical practice in the future.
Besides H. pylori infection, computer-aided pattern recognition with endoscopic images has also been applied to diagnose wall invasion depth (Table 2). An accurate diagnosis of invasion depth and subsequent staging is the basis for determining the appropriate treatment modality, especially for suspected early GCs (EGC)[89-91]. Classified based on the 7th TNM classification of tumors[92,93], EGC is categorized as tumor invasion of the mucosa (T1a) or invasion of the submucosa (T1b) stages. While endoscopic ultrasonography is useful for T-staging of GC by delineating each gastric wall layer[94,95], conventional endoscopy is still arguably superior to endoscopic ultrasonography for T-staging of EGC[96,97]. However, there remains room for improvement, such as by utilizing AI, to increase its accuracy. In 2012, Kubota et al[98] first explored the system with a relatively high sensitivity of 68.9% and 63.6% in T1a and T1b GCs, achieving high accuracy, especially in early tumors. The accuracy for T1 tumors was 77.2% compared to that of 49.1%, 51.0%, and 55.3% for T2, T3 and T4 tumors, respectively. Another system developed by Hirasawa et al[99] achieved a high sensitivity of 92.2% of CNN, though at the expense of a low positive predictive value (30.6%). Zhu et al[100] later demonstrated a CNN-computer assisted diagnosis system that achieved much higher accuracy (by 17.25%; 95% confidence interval: 11.63-22.59) and specificity (by 32.21%; 95% confidence interval: 26.78-37.44) compared to human endoscopists. These preliminary findings showed that AI is a potentially helpful diagnostic procedure in EGC detection and pointed towards developing an AI system that can differentiate between malignant and benign lesions.
| Ref. | Application | Endoscopic modality | Training dataset | Validation dataset | Accuracy | Sensitivity | Specificity | PPV | NPV |
| Kubota et al[98], 2012 | Prediction of invasion depth | WLI | 344 patients, 902 images | - | 77.2 (T1) | - | - | 80.1 (T1) | |
| Miyaki et al[137], 2013 | Differentiation of cancerous areas from noncancerous areas | WLI and magnified FICE | 493 images | 46 images | 85.9 | 84.8 | 87.0 | 86.7 | 85.1 |
| Hirasawa et al[99], 2018 | Differentiation of cancerous areas from noncancerous areas | WLI | 13584 images | 2296 images, 69 patients | 92.2 | 92.2 | - | 30.6 | - |
| Kanesaka et al[138], 2018 | Detection of EGC | Magnified NBI | 126 images | 81 images | 96.3 | 96.7 | 95.0 | 98.3 | - |
| Horiuchi et al[103], 2020 | Differentiation of EGC from gastritis | Magnified NBI | 2570 images | 258 images | 85.3 | 95.4 | 71.0 | 82.3 | 91.7 |
| Yoon et al[101], 2019 | Detection of EGC and prediction of EGC invasion depth | WLI | 11686 images, 800 patients | - | 79.2 | 77.8 | 79.3 | 77.7 | |
| Horiuchi et al[105], 2020 | Detection of EGC | Magnified NBI | 2570 images | 174 videos, 82 patients | 85.1 | 87.4 | 82.8 | 83.5 | 86.7 |
| Li et al[139], 2020 | Differentiation of EGC from noncancerous lesions | Magnified NBI | 2088 images | 342 images | 90.9 | 91.2 | 90.6 | 90.6 | 91.2 |
| Nagao et al[102], 2020 | Prediction of invasion depth | WLI, nonmagnifying NBI and indigo-carmine dye contrast imaging (Indigo) | 16557 images, 1084 patients | - | 94.4 | 89.2 | 98.7 | 98.3 | 91.7 |
| Namikawa et al[104], 2020 | Differentiation of cancerous areas from noncancerous areas | WLI, nonmagnifying NBI and indigo-carmine dye contrast imaging (Indigo) | 18410 images | 1459 images | 95.9 | 99.0 | 93.3 | 92.5 | - |
Given that the difference in EGC depth in endoscopic images is subtler and more difficult to discern, Yoon et al[101] identified that more sophisticated image classification methods but not merely conventional CNN models are required. The team developed a system that classifies endoscopic images into EGC (T1a or T1b) or non-EGC. This system used the combination of the CNN-based visual geometry group-16 network pretrained on ImageNet and a novel method of the weighted sum of gradient-weighted class activation mapping. This system focused on learning the visual features of EGC regions rather than those of other gastric textures, achieving both high accuracy of 91.0% and high area under the curve of 0.981. In another study in 2020, Nagao et al[102] used the state-of-the-art ResNet50 CNN architecture to develop a system via training the images from different angles and distances. This system predicted the invasion depth of GC with an image-based accuracy as high as 94.5%.
However, when using these AI systems for invasion depth diagnosis, distinguishing superficially depressed and differentiated-type intramucosal cancers from gastritis remains a challenge. The diagnostic accuracy of determining invasion depth is largely affected by its histological characteristics. For instance, the system developed by Yoon et al[101] achieved an accuracy of 77.1% for differentiated-type tumors in contrast to that of 65.5% for undifferentiated type. Horiuchi et al[103] made a substantial effort and developed another system that could differentiate EGC from gastritis using magnifying endoscopy with NBI (M-NBI). The system achieved an accuracy of 85.3% and sensitivity and negative predictive value of 95.4% and 91.7%. Another attempt was made by Namikawa et al[104], who developed a system that was trained by gastritis images and tested to classify GC and gastric ulcers. A continued development of AI systems that consider the differentiated type histology will shed light on the future of AI-assisted differentiation of T1a from T1b GC and that of T1a and T1b cancers from the later stages of GC.
To bring AI-assisted systems one step closer to real-time clinical applications, video-based systems have also been explored. In 2020, Horiuchi et al[105] used the video-based systems and achieved a comparable accuracy of 85.1% in distinguishing EGC and noncancerous lesions. Based on the CNN-CAD system, their system was trained with 2570 images (1492 cancerous and 1078 noncancerous images) and tested with 174 videos. This preliminary success in the video-based CNN-CAD system pointed out the potential of real-time AI-assisted diagnosis, which could be a promising technique for detecting EGC for clinicians in the future. Early detection of GC means an early treatment of endoscopic dissection in accordance with the works promoted by the Japanese Gastric Cancer Association since 2014[106].
Over the past decade, AI has displayed its potential diagnosing GC to amplify human endoscopist capacities. Although the diagnosis of GC requires a holistic set of assessments, AI is applicable and helpful in some parts. A system that detects GC with high sensitivity regardless of its accuracy in determining invasion depth could provide great clinical assistance for physicians to decide if biopsy and endoscopic submucosal dissection are necessary. In the near future, there should be some other diagnostic procedures that can be explored with AI. For example, macroscopic characteristics, namely the “nonextension signs” commonly used to distinguish between SM1 and SM2 invasion depths of GCs[107] have yet to be explored with AI.
Clinically, there are also some distinct markers that endoscopists use to evaluate gastric surface and color changes. Distinguishing the markers such as changes in light reflection and spontaneous bleeding are clinical skills[108,109] that AI could potentially learn and interpret. In clinical practice, antiperistaltic agents are suggested for polyethersulfone preparation, and indigo carmine chromoendoscopy could help diagnose elevated superficial lesions with an irregular surface pattern[110] with which their efficacy could be evaluated by real-time AI endoscopy in the near future.
Although several studies have attempted to apply AI in different types of endoscopies, ranging from WLI to LCI to blue laser imaging, these studies can also continue to extend AI to NBI and other nonconventional endoscopies. For instance, endocytoscopy with NBI has shown higher diagnostic accuracy compared to M-NBI [78.8% (76.4%-83.0%) vs 72.2% (69.3%-73.6%), P < 0.0001][111]. An AI system that is trained with WLI images and tested with NBI images instead will also have clinical significance[112,113]. Proposed in 2016 was the Magnifying Endoscopy Simple Diagnostic Algorithms for EGC that suggested a systematic approach to WLI magnifying endoscopy. It is recommended that if a suspicious lesion is detected, M-NBI should be performed to distinguish if the lesion is cancer or noncancer[114]. According to this algorithm, changing from WLI to M-NBI endoscopy is therefore critical for diagnosis, and the future development of AI systems can consider accounting for such changes.
In the AI systems developed over the past decade, we summarize the following common limitations faced. First, there seems to be a common lack of high-quality datasets for machine learning development, a problem faced in clinical practice even without AI[115]. Simultaneously, some studies reported that low-quality images result in higher chances of misdiagnosis by the AI system[116], and most studies excluded large numbers of poor-quality images[99-101]. The call for cross-validation with multicenter observational studies has also been discussed in several studies in hopes of picking out any potential overfitting and spectrum bias that is foreseeable in deep image classification models[117-119]. Some authors have argued that the AI system they have developed is institution-specific and that the validation with the dataset from external sources is necessary[103,105]. In this regard, multicenter studies have been widely used in other medical fields to evaluate deep learning systems[120-122], though there have been no such studies in the field of GC.
Another challenge that remains is seen in the imbalanced class distributions, a common classification problem in which the distribution of samples across the known classes is biased or skewed. For example, in the study reported by Hirasawa et al[99] in 2018, there are few samples for the later stages (only 32.5% of samples were T2-T4 cancers) than for early cancer (67.5% of samples). Such imbalanced classifications pose a challenge as machine learning models are primarily designed on the assumption of an equal number of samples for each class[123]. Without sufficient samples for certain classes of the training dataset, their existence might be misperceived as other classes as the AI model becomes more sensitive to classification errors. It may result in poor predictive performance, especially for the minority class and subsequently an overall increased misdiagnosis rate. For example, in the cases of the AI model for GC staging, a misdiagnosis of late-stage cancer for gastritis or nonmalignancy has dangerous implications[124-126] if the AI system was used for its diagnosis alone. However, in most cases, advanced-stage GC might have already metastasized to other parts of the body[127-129], and its diagnosis based only on the AI system alone is unlikely. Nonetheless, in the development of AI systems for such medical applications, these technical problems of imbalanced classification should not be overlooked. It has been discussed by other reviews how modifications can be made to AI models to recognize targets, no matter how frequent or rare they are, to minimize the possibility of misdiagnosis[130,131].
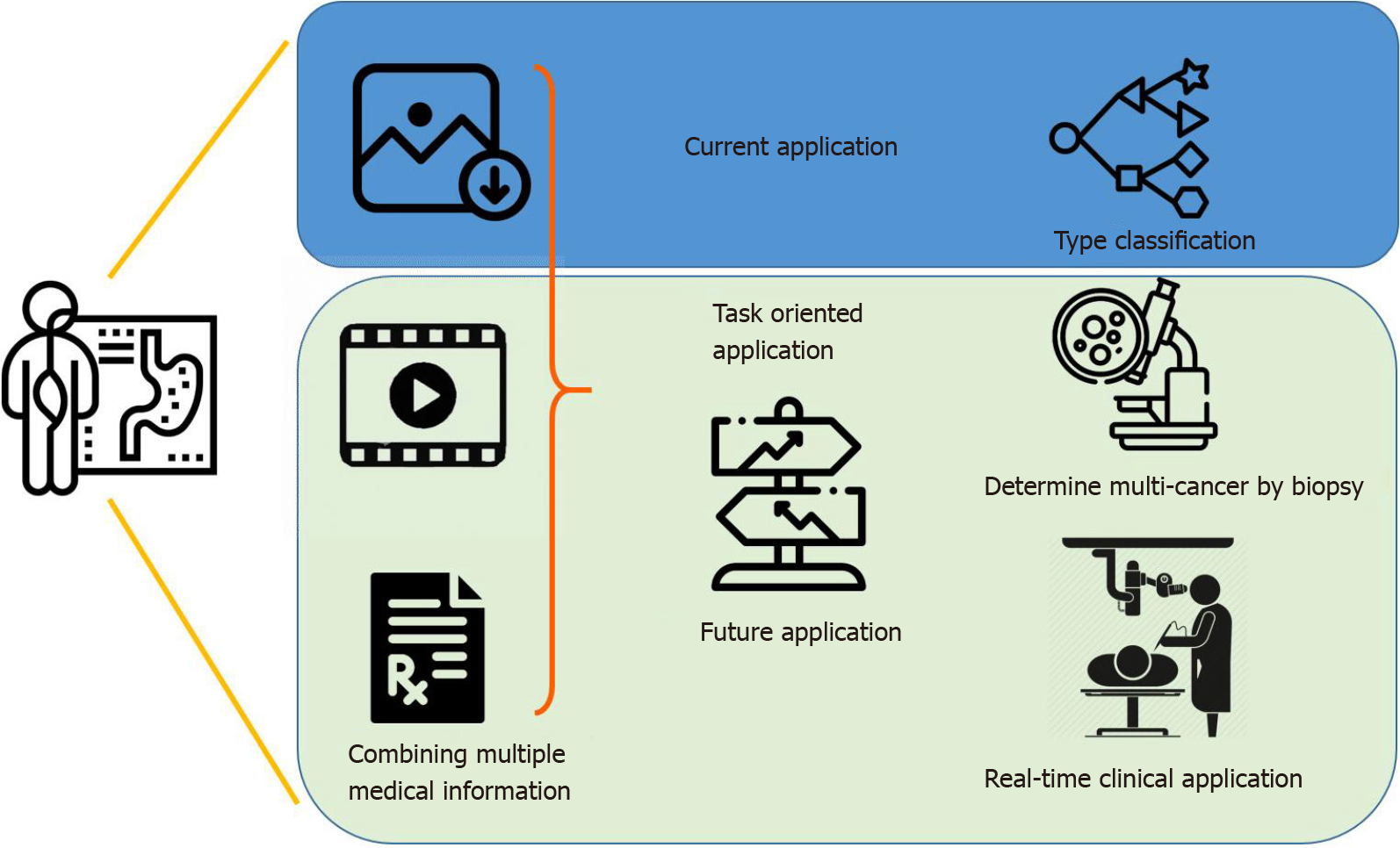
Overall, the potential for AI applications in GC is extensive yet highly specific. In an upcoming era of AI-assisted diagnosis, by combining image information, medical history and laboratory data, endoscopists can look forward to the continued development of new systems for varying purposes (Figure 2). AI systems are specific and unlikely to be generalized[132,133], and it is fallacious to compare a single statistical performance measure across different AI systems. The efficacy of an AI system depends on the intended role it plays in clinical practice. For example, an AI system with a high positive predictive value is desirable in determining which multicancer to send for biopsy, while high sensitivity suffices for a system that helps differentiate cancerous from noncancerous clinical signs, especially for amateur endoscopists. In the foreseeable future, AI can be incorporated in the differential diagnosis of the malignancy and stages of gastric lesions, using various endoscopic technologies and techniques.
Overall, the application of AI in gastroenterology is in its infancy. At present, there exist several retrospective models applied in both images and videos and using both WLI and NBI endoscopies that have proven to have better performance for the same tasks carried out by experienced endoscopists. However, there have not been any attempts of clinical trials. In contrast to the ongoing trials for detecting colorectal polyps[134-136], AI applications in GC and its corresponding diagnostic methods are still preliminary. The limitations of existing efforts point towards the importance of continued research in the field that can go a long way in making quicker, more accurate and precise evaluations of GC risk. While we witnessed its rapid and steep growth in the past decade, future studies are needed to streamline the machine learning process and define its role in the computer-aided diagnosis of H. pylori infections and GC in real-life clinical scenarios.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Taiwan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Santos-García G S-Editor: Gao CC L-Editor: Filipodia P-Editor: Liu JH
| 1. | Drewes AM, Olesen AE, Farmer AD, Szigethy E, Rebours V, Olesen SS. Gastrointestinal pain. Nat Rev Dis Primers. 2020;6:1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 178] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 2. | Hoffman A, Manner H, Rey JW, Kiesslich R. A guide to multimodal endoscopy imaging for gastrointestinal malignancy - an early indicator. Nat Rev Gastroenterol Hepatol. 2017;14:421-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 3. | Mannath J, Ragunath K. Role of endoscopy in early oesophageal cancer. Nat Rev Gastroenterol Hepatol. 2016;13:720-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 4. | Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, Bleotu C, Diaconu CC, Chivu-Economescu M. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029-2044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 307] [Cited by in RCA: 299] [Article Influence: 49.8] [Reference Citation Analysis (3)] |
| 5. | Yang D, Wagh MS, Draganov PV. The status of training in new technologies in advanced endoscopy: from defining competence to credentialing and privileging. Gastrointest Endosc. 2020;92:1016-1025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55853] [Article Influence: 7979.0] [Reference Citation Analysis (132)] |
| 7. | Dicken BJ, Bigam DL, Cass C, Mackey JR, Joy AA, Hamilton SM. Gastric adenocarcinoma: review and considerations for future directions. Ann Surg. 2005;241:27-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 505] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 8. | Lyons K, Le LC, Pham YT, Borron C, Park JY, Tran CTD, Tran TV, Tran HT, Vu KT, Do CD, Pelucchi C, La Vecchia C, Zgibor J, Boffetta P, Luu HN. Gastric cancer: epidemiology, biology, and prevention: a mini review. Eur J Cancer Prev. 2019;28:397-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 9. | Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 726] [Article Influence: 103.7] [Reference Citation Analysis (1)] |
| 10. | Vingeliene S, Chan DSM, Vieira AR, Polemiti E, Stevens C, Abar L, Navarro Rosenblatt D, Greenwood DC, Norat T. An update of the WCRF/AICR systematic literature review and meta-analysis on dietary and anthropometric factors and esophageal cancer risk. Ann Oncol. 2017;28:2409-2419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 11. | Cummins G, Cox BF, Ciuti G, Anbarasan T, Desmulliez MPY, Cochran S, Steele R, Plevris JN, Koulaouzidis A. Gastrointestinal diagnosis using non-white light imaging capsule endoscopy. Nat Rev Gastroenterol Hepatol. 2019;16:429-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Kiesslich R, Goetz M, Hoffman A, Galle PR. New imaging techniques and opportunities in endoscopy. Nat Rev Gastroenterol Hepatol. 2011;8:547-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Kuipers EJ, Haringsma J. Diagnostic and therapeutic endoscopy. J Surg Oncol. 2005;92:203-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Cenitagoya GF, Bergh CK, Klinger-Roitman J. A prospective study of gastric cancer. 'Real' 5-year survival rates and mortality rates in a country with high incidence. Dig Surg. 1998;15:317-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Tarver T. Cancer Facts & Figures 2012. American Cancer Society (ACS). J Consum Health Internet. 2012;16:366-367. [RCA] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 16. | Becker KF, Keller G, Hoefler H. The use of molecular biology in diagnosis and prognosis of gastric cancer. Surg Oncol. 2000;9:5-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 17. | Nakamura K, Ueyama T, Yao T, Xuan ZX, Ambe K, Adachi Y, Yakeishi Y, Matsukuma A, Enjoji M. Pathology and prognosis of gastric carcinoma. Findings in 10,000 patients who underwent primary gastrectomy. Cancer. 1992;70:1030-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 18. | Gotoda T, Jung HY. Endoscopic resection (endoscopic mucosal resection/ endoscopic submucosal dissection) for early gastric cancer. Dig Endosc. 2013;25 Suppl 1:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Nishizawa T, Yahagi N. Endoscopic mucosal resection and endoscopic submucosal dissection: technique and new directions. Curr Opin Gastroenterol. 2017;33:315-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 20. | Uraoka T, Saito Y, Yamamoto K, Fujii T. Submucosal injection solution for gastrointestinal tract endoscopic mucosal resection and endoscopic submucosal dissection. Drug Des Devel Ther. 2009;2:131-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Asaka M, Mabe K. Strategies for eliminating death from gastric cancer in Japan. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90:251-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Katai H, Ishikawa T, Akazawa K, Isobe Y, Miyashiro I, Oda I, Tsujitani S, Ono H, Tanabe S, Fukagawa T, Nunobe S, Kakeji Y, Nashimoto A; Registration Committee of the Japanese Gastric Cancer Association. Five-year survival analysis of surgically resected gastric cancer cases in Japan: a retrospective analysis of more than 100,000 patients from the nationwide registry of the Japanese Gastric Cancer Association (2001-2007). Gastric Cancer. 2018;21:144-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 359] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 23. | Wu MS, Lin JT, Lee WJ, Yu SC, Wang TH. [Gastric cancer in Taiwan]. J Formos Med Assoc. 1994;93 Suppl 2:S77-S89. [PubMed] |
| 24. | Amin A, Gilmour H, Graham L, Paterson-Brown S, Terrace J, Crofts TJ. Gastric adenocarcinoma missed at endoscopy. J R Coll Surg Edinb. 2002;47:681-684. [PubMed] |
| 25. | Hosokawa O, Hattori M, Douden K, Hayashi H, Ohta K, Kaizaki Y. Difference in accuracy between gastroscopy and colonoscopy for detection of cancer. Hepatogastroenterology. 2007;54:442-444. [PubMed] |
| 26. | Hosokawa O, Tsuda S, Kidani E, Watanabe K, Tanigawa Y, Shirasaki S, Hayashi H, Hinoshita T. Diagnosis of gastric cancer up to three years after negative upper gastrointestinal endoscopy. Endoscopy. 1998;30:669-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 114] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 27. | Menon S, Trudgill N. How commonly is upper gastrointestinal cancer missed at endoscopy? Endosc Int Open. 2014;2:E46-E50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 230] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 28. | Voutilainen ME, Juhola MT. Evaluation of the diagnostic accuracy of gastroscopy to detect gastric tumours: clinicopathological features and prognosis of patients with gastric cancer missed on endoscopy. Eur J Gastroenterol Hepatol. 2005;17:1345-1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Yalamarthi S, Witherspoon P, McCole D, Auld CD. Missed diagnoses in patients with upper gastrointestinal cancers. Endoscopy. 2004;36:874-879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 139] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 30. | Park CH, Kim B, Chung H, Lee H, Park JC, Shin SK, Lee SK, Lee YC. Endoscopic quality indicators for esophagogastroduodenoscopy in gastric cancer screening. Dig Dis Sci. 2015;60:38-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Willis S, Truong S, Gribnitz S, Fass J, Schumpelick V. Endoscopic ultrasonography in the preoperative staging of gastric cancer. Surg Endosc. 2000;14:951-954. [RCA] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 93] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 32. | Dixon BJ, Chan H, Daly MJ, Vescan AD, Witterick IJ, Irish JC. The effect of augmented real-time image guidance on task workload during endoscopic sinus surgery. Int Forum Allergy Rhinol. 2012;2:405-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 33. | Hudler P. Challenges of deciphering gastric cancer heterogeneity. World J Gastroenterol. 2015;21:10510-10527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 59] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 34. | Reinhart K, Bannert C, Dunkler D, Salzl P, Trauner M, Renner F, Knoflach P, Ferlitsch A, Weiss W, Ferlitsch M. Prevalence of flat lesions in a large screening population and their role in colonoscopy quality improvement. Endoscopy. 2013;45:350-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Anderson ML, Heigh RI, McCoy GA, Parent K, Muhm JR, McKee GS, Eversman WG, Collins JM. Accuracy of assessment of the extent of examination by experienced colonoscopists. Gastrointest Endosc. 1992;38:560-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 36. | Hawkins SC, Osborne A, Schofield SJ, Pournaras DJ, Chester JF. Improving the accuracy of self-assessment of practical clinical skills using video feedback--the importance of including benchmarks. Med Teach. 2012;34:279-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 37. | Higashi R, Uraoka T, Kato J, Kuwaki K, Ishikawa S, Saito Y, Matsuda T, Ikematsu H, Sano Y, Suzuki S, Murakami Y, Yamamoto K. Diagnostic accuracy of narrow-band imaging and pit pattern analysis significantly improved for less-experienced endoscopists after an expanded training program. Gastrointest Endosc. 2010;72:127-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 38. | Jinnai S, Yamazaki N, Hirano Y, Sugawara Y, Ohe Y, Hamamoto R. The Development of a Skin Cancer Classification System for Pigmented Skin Lesions Using Deep Learning. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (1)] |
| 39. | Nelson CA, Pérez-Chada LM, Creadore A, Li SJ, Lo K, Manjaly P, Pournamdari AB, Tkachenko E, Barbieri JS, Ko JM, Menon AV, Hartman RI, Mostaghimi A. Patient Perspectives on the Use of Artificial Intelligence for Skin Cancer Screening: A Qualitative Study. JAMA Dermatol. 2020;156:501-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 141] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 40. | Tschandl P, Rinner C, Apalla Z, Argenziano G, Codella N, Halpern A, Janda M, Lallas A, Longo C, Malvehy J, Paoli J, Puig S, Rosendahl C, Soyer HP, Zalaudek I, Kittler H. Human-computer collaboration for skin cancer recognition. Nat Med. 2020;26:1229-1234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 361] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 41. | Zakhem GA, Fakhoury JW, Motosko CC, Ho RS. Characterizing the role of dermatologists in developing artificial intelligence for assessment of skin cancer: A systematic review. J Am Acad Dermatol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 42. | Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1552] [Cited by in RCA: 1853] [Article Influence: 264.7] [Reference Citation Analysis (2)] |
| 43. | Huynh E, Hosny A, Guthier C, Bitterman DS, Petit SF, Haas-Kogan DA, Kann B, Aerts HJWL, Mak RH. Artificial intelligence in radiation oncology. Nat Rev Clin Oncol. 2020;17:771-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 211] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 44. | Kocher M. Artificial intelligence and radiomics for radiation oncology. Strahlenther Onkol. 2020;196:847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Syed K, Sleeman IV WC, Nalluri JJ, Kapoor R, Hagan M, Palta J, Ghosh P. Artificial intelligence methods in computer-aided diagnostic tools and decision support analytics for clinical informatics. Artificial Intelligence in Precision Health. Elsevier, 2020: 31-59. |
| 46. | Bhanumurthy M, Anne K. An automated detection and segmentation of tumor in brain MRI using artificial intelligence. Proceedings of the 2014 IEEE International Conference on Computational Intelligence and Computing Research; IEEE, 2014: 1-6. |
| 47. | Ghafoorian M, Mehrtash A, Kapur T, Karssemeijer N, Marchiori E, Pesteie M, Guttmann CR, de Leeuw FE, Tempany CM, Van Ginneken B. Transfer learning for domain adaptation in MRI: Application in brain lesion segmentation. Proceedings of the International conference on medical image computing and computer-assisted intervention; New York: Springer: 2017: 516-524. |
| 48. | Rauschecker AM, Rudie JD, Xie L, Wang J, Duong MT, Botzolakis EJ, Kovalovich AM, Egan J, Cook TC, Bryan RN, Nasrallah IM, Mohan S, Gee JC. Artificial Intelligence System Approaching Neuroradiologist-level Differential Diagnosis Accuracy at Brain MRI. Radiology. 2020;295:626-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 49. | Shahamat H, Saniee Abadeh M. Brain MRI analysis using a deep learning based evolutionary approach. Neural Netw. 2020;126:218-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 50. | Cho J, Park KS, Karki M, Lee E, Ko S, Kim JK, Lee D, Choe J, Son J, Kim M, Lee S, Lee J, Yoon C, Park S. Improving Sensitivity on Identification and Delineation of Intracranial Hemorrhage Lesion Using Cascaded Deep Learning Models. J Digit Imaging. 2019;32:450-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 51. | Simon R. Sensitivity, Specificity, PPV, and NPV for Predictive Biomarkers. J Natl Cancer Inst. 2015;107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 52. | Arbabshirani MR, Fornwalt BK, Mongelluzzo GJ, Suever JD, Geise BD, Patel AA, Moore GJ. Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digit Med. 2018;1:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 242] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 53. | Leenhardt R, Vasseur P, Li C, Saurin JC, Rahmi G, Cholet F, Becq A, Marteau P, Histace A, Dray X; CAD-CAP Database Working Group. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc. 2019;89:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 54. | Soffer S, Klang E, Shimon O, Nachmias N, Eliakim R, Ben-Horin S, Kopylov U, Barash Y. Deep learning for wireless capsule endoscopy: a systematic review and meta-analysis. Gastrointest Endosc 2020; 92: 831-839. e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 115] [Article Influence: 23.0] [Reference Citation Analysis (3)] |
| 55. | Wang S, Xing Y, Zhang L, Gao H, Zhang H. A systematic evaluation and optimization of automatic detection of ulcers in wireless capsule endoscopy on a large dataset using deep convolutional neural networks. Phys Med Biol. 2019;64:235014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 56. | Xia J, Xia T, Pan J, Gao F, Wang S, Qian YY, Wang H, Zhao J, Jiang X, Zou WB, Wang YC, Zhou W, Li ZS, Liao Z. Use of artificial intelligence for detection of gastric lesions by magnetically controlled capsule endoscopy. Gastrointest Endosc 2021; 93: 133-139. e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 57. | Sánchez-Peralta LF, Bote-Curiel L, Picón A, Sánchez-Margallo FM, Pagador JB. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif Intell Med. 2020;108:101923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 58. | Summers RM, Jerebko AK, Franaszek M, Malley JD, Johnson CD. Colonic polyps: complementary role of computer-aided detection in CT colonography. Radiology. 2002;225:391-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 93] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 59. | Wang KW, Dong M. Potential applications of artificial intelligence in colorectal polyps and cancer: Recent advances and prospects. World J Gastroenterol. 2020;26:5090-5100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 60. | Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5573] [Cited by in RCA: 4942] [Article Influence: 617.8] [Reference Citation Analysis (0)] |
| 61. | Wieslander H, Forslid G, Bengtsson E, Wahlby C, Hirsch J-M, Runow Stark C, Kecheril Sadanandan S. Deep convolutional neural networks for detecting cellular changes due to malignancy. Proceedings of the Proceedings of the IEEE International Conference on Computer Vision Workshops; 2017 Oct 22-29; Venice, Italy. IEEE, 2017: 82-89. |
| 62. | Ebigbo A, Palm C, Probst A, Mendel R, Manzeneder J, Prinz F, de Souza LA, Papa JP, Siersema P, Messmann H. A technical review of artificial intelligence as applied to gastrointestinal endoscopy: clarifying the terminology. Endosc Int Open. 2019;7:E1616-E1623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 63. | Mortazi A, Bagci U. Automatically designing CNN architectures for medical image segmentation. 2018 Preprint. Available from: arXiv:1807.07663. |
| 64. | Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29:102-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 780] [Cited by in RCA: 804] [Article Influence: 114.9] [Reference Citation Analysis (0)] |
| 65. | Serte S, Serener A, Al-Turjman F. Deep learning in medical imaging: A brief review. Transactions on Emerging Telecommunications Technologies. Hoboken: John Wiley & Sons, 2020: e4080. |
| 66. | Abe S, Oda I. How can endoscopists adapt and collaborate with artificial intelligence for early gastric cancer detection? Dig Endosc. 2021;33:98-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 67. | Malfertheiner P. Diagnostic methods for H. pylori infection: Choices, opportunities and pitfalls. United European Gastroenterol J. 2015;3:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 68. | Correa P, Piazuelo MB. The gastric precancerous cascade. J Dig Dis. 2012;13:2-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 525] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 69. | Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2805] [Cited by in RCA: 2739] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 70. | Bornschein J, Malfertheiner P. Gastric carcinogenesis. Langenbecks Arch Surg. 2011;396:729-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 71. | Kato T, Yagi N, Kamada T, Shimbo T, Watanabe H, Ida K; Study Group for Establishing Endoscopic Diagnosis of Chronic Gastritis. Diagnosis of Helicobacter pylori infection in gastric mucosa by endoscopic features: a multicenter prospective study. Dig Endosc. 2013;25:508-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 72. | Laine L, Cohen H, Sloane R, Marin-Sorensen M, Weinstein WM. Interobserver agreement and predictive value of endoscopic findings for H. pylori and gastritis in normal volunteers. Gastrointest Endosc. 1995;42:420-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 73. | Goodwin CS. Helicobacter pylori gastritis, peptic ulcer, and gastric cancer: clinical and molecular aspects. Clin Infect Dis. 1997;25:1017-1019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 74. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3126] [Cited by in RCA: 3187] [Article Influence: 132.8] [Reference Citation Analysis (0)] |
| 75. | Christensen AH, Gjørup T, Hilden J, Fenger C, Henriksen B, Vyberg M, Ostergaard K, Hansen BF. Observer homogeneity in the histologic diagnosis of Helicobacter pylori. Latent class analysis, kappa coefficient, and repeat frequency. Scand J Gastroenterol. 1992;27:933-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 50] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 76. | de Martel C, Plummer M, van Doorn LJ, Vivas J, Lopez G, Carillo E, Peraza S, Muñoz N, Franceschi S. Comparison of polymerase chain reaction and histopathology for the detection of Helicobacter pylori in gastric biopsies. Int J Cancer. 2010;126:1992-1996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 77. | el-Zimaity HM, Graham DY, al-Assi MT, Malaty H, Karttunen TJ, Graham DP, Huberman RM, Genta RM. Interobserver variation in the histopathological assessment of Helicobacter pylori gastritis. Hum Pathol. 1996;27:35-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 166] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 78. | Huang CR, Sheu BS, Chung PC, Yang HB. Computerized diagnosis of Helicobacter pylori infection and associated gastric inflammation from endoscopic images by refined feature selection using a neural network. Endoscopy. 2004;36:601-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 79. | Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K, Tada T. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. EBioMedicine. 2017;25:106-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 80. | Shichijo S, Endo Y, Aoyama K, Takeuchi Y, Ozawa T, Takiyama H, Matsuo K, Fujishiro M, Ishihara S, Ishihara R, Tada T. Application of convolutional neural networks for evaluating Helicobacter pylori infection status on the basis of endoscopic images. Scand J Gastroenterol. 2019;54:158-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 81. | Itoh T, Kawahira H, Nakashima H, Yata N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open. 2018;6:E139-E144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 82. | Zheng W, Zhang X, Kim JJ, Zhu X, Ye G, Ye B, Wang J, Luo S, Li J, Yu T, Liu J, Hu W, Si J. High Accuracy of Convolutional Neural Network for Evaluation of Helicobacter pylori Infection Based on Endoscopic Images: Preliminary Experience. Clin Transl Gastroenterol. 2019;10:e00109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 83. | Kanzaki H, Takenaka R, Kawahara Y, Kawai D, Obayashi Y, Baba Y, Sakae H, Gotoda T, Kono Y, Miura K, Iwamuro M, Kawano S, Tanaka T, Okada H. Linked color imaging (LCI), a novel image-enhanced endoscopy technology, emphasizes the color of early gastric cancer. Endosc Int Open. 2017;5:E1005-E1013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 84. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging-bright and linked color imaging: a single-center prospective study. Ann Gastroenterol. 2018;31:462-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (1)] |
| 85. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Endoscopic three-categorical diagnosis of Helicobacter pylori infection using linked color imaging and deep learning: a single-center prospective study (with video). Gastric Cancer. 2020;23:1033-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 86. | Kodama M, Murakami K, Okimoto T, Sato R, Uchida M, Abe T, Shiota S, Nakagawa Y, Mizukami K, Fujioka T. Ten-year prospective follow-up of histological changes at five points on the gastric mucosa as recommended by the updated Sydney system after Helicobacter pylori eradication. J Gastroenterol. 2012;47:394-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 87. | Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17:195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1023] [Cited by in RCA: 952] [Article Influence: 158.7] [Reference Citation Analysis (0)] |
| 88. | Luo H, Xu G, Li C, He L, Luo L, Wang Z, Jing B, Deng Y, Jin Y, Li Y, Li B, Tan W, He C, Seeruttun SR, Wu Q, Huang J, Huang DW, Chen B, Lin SB, Chen QM, Yuan CM, Chen HX, Pu HY, Zhou F, He Y, Xu RH. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645-1654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 89. | Goto O, Fujishiro M, Kodashima S, Ono S, Omata M. Outcomes of endoscopic submucosal dissection for early gastric cancer with special reference to validation for curability criteria. Endoscopy. 2009;41:118-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 145] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 90. | Maruyama K. The most important factors for gastric cancer patients. A studyusing univariate and multivariate analyses. Scand J Gastroenterol. 1987;22:63-68. [RCA] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 151] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 91. | Wang J, Yu JC, Kang WM, Ma ZQ. Treatment strategy for early gastric cancer. Surg Oncol. 2012;21:119-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 92. | Marrelli D, Morgagni P, de Manzoni G, Coniglio A, Marchet A, Saragoni L, Tiberio G, Roviello F; Italian Research Group for Gastric Cancer (IRGGC). Prognostic value of the 7th AJCC/UICC TNM classification of noncardia gastric cancer: analysis of a large series from specialized Western centers. Ann Surg. 2012;255:486-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 93. | Wittekind C. The development of the TNM classification of gastric cancer. Pathol Int. 2015;65:399-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 94. | Mocellin S, Marchet A, Nitti D. EUS for the staging of gastric cancer: a meta-analysis. Gastrointest Endosc. 2011;73:1122-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 95. | Yanai H, Matsumoto Y, Harada T, Nishiaki M, Tokiyama H, Shigemitsu T, Tada M, Okita K. Endoscopic ultrasonography and endoscopy for staging depth of invasion in early gastric cancer: a pilot study. Gastrointest Endosc. 1997;46:212-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 111] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 96. | Choi J, Kim SG, Im JP, Kim JS, Jung HC, Song IS. Comparison of endoscopic ultrasonography and conventional endoscopy for prediction of depth of tumor invasion in early gastric cancer. Endoscopy. 2010;42:705-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 169] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 97. | Pei Q, Wang L, Pan J, Ling T, Lv Y, Zou X. Endoscopic ultrasonography for staging depth of invasion in early gastric cancer: A meta-analysis. J Gastroenterol Hepatol. 2015;30:1566-1573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 98. | Kubota K, Kuroda J, Yoshida M, Ohta K, Kitajima M. Medical image analysis: computer-aided diagnosis of gastric cancer invasion on endoscopic images. Surg Endosc. 2012;26:1485-1489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (1)] |
| 99. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 428] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 100. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc 2019; 89: 806-815. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 101. | Yoon HJ, Kim S, Kim JH, Keum JS, Oh SI, Jo J, Chun J, Youn YH, Park H, Kwon IG, Choi SH, Noh SH. A Lesion-Based Convolutional Neural Network Improves Endoscopic Detection and Depth Prediction of Early Gastric Cancer. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 102. | Nagao S, Tsuji Y, Sakaguchi Y, Takahashi Y, Minatsuki C, Niimi K, Yamashita H, Yamamichi N, Seto Y, Tada T, Koike K. Highly accurate artificial intelligence systems to predict the invasion depth of gastric cancer: efficacy of conventional white-light imaging, nonmagnifying narrow-band imaging, and indigo-carmine dye contrast imaging. Gastrointest Endosc 2020; 92: 866-873. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 103. | Horiuchi Y, Aoyama K, Tokai Y, Hirasawa T, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Convolutional Neural Network for Differentiating Gastric Cancer from Gastritis Using Magnified Endoscopy with Narrow Band Imaging. Dig Dis Sci. 2020;65:1355-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (1)] |
| 104. | Namikawa K, Hirasawa T, Nakano K, Ikenoyama Y, Ishioka M, Shiroma S, Tokai Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Artificial intelligence-based diagnostic system classifying gastric cancers and ulcers: comparison between the original and newly developed systems. Endoscopy. 2020;52:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 105. | Horiuchi Y, Hirasawa T, Ishizuka N, Tokai Y, Namikawa K, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Performance of a computer-aided diagnosis system in diagnosing early gastric cancer using magnifying endoscopy videos with narrow-band imaging (with videos). Gastrointest Endosc 2020; 92: 856-865. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 106. | Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2010 (ver. 3). Gastric Cancer. 2011;14:113-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1723] [Cited by in RCA: 1897] [Article Influence: 135.5] [Reference Citation Analysis (0)] |
| 107. | Nagahama T, Yao K, Imamura K, Kojima T, Ohtsu K, Chuman K, Tanabe H, Yamaoka R, Iwashita A. Diagnostic performance of conventional endoscopy in the identification of submucosal invasion by early gastric cancer: the "non-extension sign" as a simple diagnostic marker. Gastric Cancer. 2017;20:304-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 108. | Cappellani A, Zanghi A, Di Vita M, Zanet E, Veroux P, Cacopardo B, Cavallaro A, Piccolo G, Lo Menzo E, Murabito P, Berretta M. Clinical and biological markers in gastric cancer: update and perspectives. Front Biosci (Schol Ed). 2010;2:403-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 109. | Li T, Huang A, Zhang M, Lan F, Zhou D, Wei H, Liu Z, Qin X. Increased Red Blood Cell Volume Distribution Width: Important Clinical Implications in Predicting Gastric Diseases. Clin Lab. 2017;63:1199-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 110. | Yao K. The endoscopic diagnosis of early gastric cancer. Ann Gastroenterol. 2013;26:11-22. [PubMed] |
| 111. | Horiuchi Y, Hirasawa T, Ishizuka N, Hatamori H, Ikenoyama Y, Tokura J, Ishioka M, Tokai Y, Namikawa K, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J. Diagnostic performance in gastric cancer is higher using endocytoscopy with narrow-band imaging than using magnifying endoscopy with narrow-band imaging. Gastric Cancer. 2021;24:417-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 112. | Ang TL, Pittayanon R, Lau JY, Rerknimitr R, Ho SH, Singh R, Kwek AB, Ang DS, Chiu PW, Luk S, Goh KL, Ong JP, Tan JY, Teo EK, Fock KM. A multicenter randomized comparison between high-definition white light endoscopy and narrow band imaging for detection of gastric lesions. Eur J Gastroenterol Hepatol. 2015;27:1473-1478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 113. | Hazewinkel Y, López-Cerón M, East JE, Rastogi A, Pellisé M, Nakajima T, van Eeden S, Tytgat KM, Fockens P, Dekker E. Endoscopic features of sessile serrated adenomas: validation by international experts using high-resolution white-light endoscopy and narrow-band imaging. Gastrointest Endosc. 2013;77:916-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 114. | Miyaoka M, Yao K, Tanabe H, Kanemitsu T, Otsu K, Imamura K, Ono Y, Ishikawa S, Yasaka T, Ueki T, Ota A, Haraoka S, Iwashita A. Diagnosis of early gastric cancer using image enhanced endoscopy: a systematic approach. Transl Gastroenterol Hepatol. 2020;5:50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 115. | Yamamoto S, Nishida T, Kato M, Inoue T, Hayashi Y, Kondo J, Akasaka T, Yamada T, Shinzaki S, Iijima H, Tsujii M, Takehara T. Evaluation of endoscopic ultrasound image quality is necessary in endosonographic assessment of early gastric cancer invasion depth. Gastroenterol Res Pract. 2012;2012:194530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 116. | Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Hojo M, Yao T, Nagahara A, Tada T. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol Hepatol. 2021;36:482-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 117. | Hawkins DM. The problem of overfitting. J Chem Inf Comput Sci. 2004;44:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1358] [Cited by in RCA: 925] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 118. | Qian L, Hu L, Zhao L, Wang T, Jiang R. Sequence-Dropout Block for Reducing Overfitting Problem in Image Classification. IEEE Access. 2020;8:62830-62840. [DOI] [Full Text] |
| 119. | Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014 Preprint. Available from: arXiv:14091556. |
| 120. | Betancur J, Commandeur F, Motlagh M, Sharir T, Einstein AJ, Bokhari S, Fish MB, Ruddy TD, Kaufmann P, Sinusas AJ, Miller EJ, Bateman TM, Dorbala S, Di Carli M, Germano G, Otaki Y, Tamarappoo BK, Dey D, Berman DS, Slomka PJ. Deep Learning for Prediction of Obstructive Disease From Fast Myocardial Perfusion SPECT: A Multicenter Study. JACC Cardiovasc Imaging. 2018;11:1654-1663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 226] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 121. | Commandeur F, Goeller M, Razipour A, Cadet S, Hell MM, Kwiecinski J, Chen X, Chang HJ, Marwan M, Achenbach S, Berman DS, Slomka PJ, Tamarappoo BK, Dey D. Fully Automated CT Quantification of Epicardial Adipose Tissue by Deep Learning: A Multicenter Study. Radiol Artif Intell. 2019;1:e190045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 122. | Nakajima K, Kudo T, Nakata T, Kiso K, Kasai T, Taniguchi Y, Matsuo S, Momose M, Nakagawa M, Sarai M, Hida S, Tanaka H, Yokoyama K, Okuda K, Edenbrandt L. Diagnostic accuracy of an artificial neural network compared with statistical quantitation of myocardial perfusion images: a Japanese multicenter study. Eur J Nucl Med Mol Imaging. 2017;44:2280-2289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 123. |
Liu H, Motoda H.
Data reduction |
| 124. | Horiuchi Y, Fujisaki J, Yamamoto N, Ida S, Yoshimizu S, Ishiyama A, Yoshio T, Hirasawa T, Yamamoto Y, Nagahama M, Takahashi H, Tsuchida T. Pretreatment diagnosis factors associated with overtreatment with surgery in patients with differentiated-type early gastric cancer. Sci Rep. 2019;9:15356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 125. | Kuroki K, Oka S, Tanaka S, Yorita N, Hata K, Kotachi T, Boda T, Arihiro K, Chayama K. Clinical significance of endoscopic ultrasonography in diagnosing invasion depth of early gastric cancer prior to endoscopic submucosal dissection. Gastric Cancer. 2021;24:145-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 126. | Yamada S, Shimada M, Utsunomiya T, Morine Y, Imura S, Ikemoto T, Mori H, Hanaoka J, Iwahashi S, Saitoh Y, Asanoma M. Hepatic screlosed hemangioma which was misdiagnosed as metastasis of gastric cancer: report of a case. J Med Invest. 2012;59:270-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 127. | Ahn JB, Ha TK, Kwon SJ. Bone metastasis in gastric cancer patients. J Gastric Cancer. 2011;11:38-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 128. | Akagi T, Shiraishi N, Kitano S. Lymph node metastasis of gastric cancer. Cancers (Basel). 2011;3:2141-2159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 129. | Esaki Y, Hirayama R, Hirokawa K. A comparison of patterns of metastasis in gastric cancer by histologic type and age. Cancer. 1990;65:2086-2090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 130. | He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21:1263-1284. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4687] [Cited by in RCA: 2112] [Article Influence: 132.0] [Reference Citation Analysis (0)] |
| 131. | Melillo P, De Luca N, Bracale M, Pecchia L. Classification tree for risk assessment in patients suffering from congestive heart failure via long-term heart rate variability. IEEE J Biomed Health Inform. 2013;17:727-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 132. | Harari YN. 21 Lessons for the 21st century. London: Jonathan Cape, 2018. |
| 133. | Fjelland R. Why general artificial intelligence will not be realized. Humanit Soc Sci Commun. 2020;7:1-9. [DOI] [Full Text] |
| 134. | King's College Hospital. Computer Aided Diagnosis of Colorectal Polyps. [accessed August 25, 2020]. In: ClinicalTrials.gov [Internet]. London: National Library of Medicine. Available from: https://ClinicalTrials.gov/show/NCT04510545 ClinicalTrials.gov Identifier: NCT04510545. |
| 135. | Neumann H. AI for Colorectal Polyp Detection in Endoscopy. [accessed April 28, 2020]. In: ClinicalTrials.gov [Internet]. Mainz: National Library of Medicine. Available from: https://ClinicalTrials.gov/show/NCT04339855 ClinicalTrials.gov Identifier: NCT04339855. |
| 136. | Petz Aladar County Teaching Hospital. Polyp Artificial Intelligence Study. [accessed June 29, 2020]. In: ClinicalTrials.gov [Internet]. Gyor: National Library of Medicine. Available from: https://ClinicalTrials.gov/show/NCT04425941 ClinicalTrials.gov Identifier: NCT04425941. |
| 137. | Miyaki R, Yoshida S, Tanaka S, Kominami Y, Sanomura Y, Matsuo T, Oka S, Raytchev B, Tamaki T, Koide T, Kaneda K, Yoshihara M, Chayama K. Quantitative identification of mucosal gastric cancer under magnifying endoscopy with flexible spectral imaging color enhancement. J Gastroenterol Hepatol. 2013;28:841-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 138. | Kanesaka T, Lee TC, Uedo N, Lin KP, Chen HZ, Lee JY, Wang HP, Chang HT. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc. 2018;87:1339-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 139. | Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, Yang X, Li J, Chen M, Jin C, Chen C, Yu C. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. 2020;23:126-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |