Published online Jun 14, 2021. doi: 10.3748/wjg.v27.i22.2944
Peer-review started: January 29, 2021
First decision: February 24, 2021
Revised: March 6, 2021
Accepted: April 25, 2021
Article in press: April 25, 2021
Published online: June 14, 2021
Processing time: 131 Days and 2.1 Hours
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 is a global pandemic and poses a major threat to human health worldwide. In addition to respiratory symptoms, COVID-19 is usually accompanied by systemic inflammation and liver damage in moderate and severe cases. Nuclear factor erythroid 2-related factor 2 (NRF2) is a transcription factor that regulates the expression of antioxidant proteins, participating in COVID-19-mediated inflammation and liver injury. Here, we show the novel reciprocal regulation between NRF2 and inflammatory mediators associated with COVID-19-related liver injury. Additionally, we describe some mechanisms and treatment strategies.
Core Tip: Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 is a global pandemic and poses a major threat to human health worldwide. In this review, we provide the most comprehensive guide to nuclear factor erythroid 2-related factor 2 and inflammatory mediators in COVID-19-related liver injury, emphasizing their potential as targets for COVID-19 therapy.
- Citation: Zhu DD, Tan XM, Lu LQ, Yu SJ, Jian RL, Liang XF, Liao YX, Fan W, Barbier-Torres L, Yang A, Yang HP, Liu T. Interplay between nuclear factor erythroid 2-related factor 2 and inflammatory mediators in COVID-19-related liver injury. World J Gastroenterol 2021; 27(22): 2944-2962
- URL: https://www.wjgnet.com/1007-9327/full/v27/i22/2944.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i22.2944
Coronavirus disease 2019 (COVID-19), a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared to be a global pandemic by the World Health Organization in 2020. COVID-19 is caused by infection with SARS-CoV-2, which has a single-stranded RNA approximately 26-32 kb in length and belongs to the same coronavirus family as Middle East respiratory syndrome coronavirus and SARS-CoV-1[1,2].
Viral toxicity, body immunity, and the induced inflammatory response all affect the occurrence and progression of COVID-19[3]. The commonly accepted method of transmission is that SARS-CoV-2 binds to the receptor angiotensin-converting enzyme 2 (ACE2) on cells through its spike (S) glycoprotein protein, which has two domains, S1 and S2, with the former binding the peptidase domain of ACE2, called the receptor-binding domain, and the latter catalyzing membrane fusion to release genetic material into the cell[4,5]. After entry, SARS-CoV-2 interferes with the host’s immune defenses, evades host immune surveillance, and rapidly replicates. The replication of the virus activates monocytes, macrophages and granulocytes, accompanied by the release of many reactive oxygen species (ROS) and inflammatory cytokine storms, which eventually lead to tissue inflammatory cell infiltration, necrosis, and fibrosis[3,6].
The pathological mechanism of SARS-CoV-2 infection is very complex and unclear. It is now widely believed to be associated with the host’s immune response, the inflammatory response, and oxidative stress[3]. It manifests with moderate to severe respiratory symptoms and is usually accompanied by multiple organ dysfunction syndrome (MODS)[7]. Based on the pathogenesis of COVID-19, it has been hypothesized that SARS-CoV-2 might directly bind to the receptor ACE2, which is widely distributed among body tissues, such as liver tissue, and is considered a target for SARS-CoV-2 entry, which causes inflammation, oxidative stress, and proapoptotic reactions, ultimately leading to liver injury[5]. SARS-CoV-2 can directly cause bile duct epithelial cell dysfunction and affect hepatocyte regeneration and the immune response[8]. Liver dysfunction is a common complication and mainly manifests as elevated transaminase and bilirubin levels[9-12]. According to statistics, 5.1%-69.8% of patients with COVID-19 suffer from liver dysfunction, occurring more commonly in patients with other severe symptoms[13,14], and 2.1%-4.1% of patients with COVID-19 undergo pre-existing liver disease[14]. Interestingly, liver dysfunction appears earlier in COVID-19-related MODS, most likely because of the number of Kupffer cells, natural killer (NK) cells and NK T cells. The expression of endothelial adhesion molecules is higher in the liver than in other organs[15], which demonstrates that the inflammatory response, oxidative stress, and immune response may play important roles in COVID-19-related liver injury. Moreover, SARS-CoV-2-induced cytotoxicity, ischemia, drug toxicity, and triggering of pre-existing liver diseases are also possible contributors to the pathogenesis of COVID-19-related liver injury[16,17]. However, the exact mechanism of COVID-19-related liver injury remains unclear.
Nuclear factor erythroid 2-related factor 2 (NRF2) is a nuclear transcription factor that is activated by oxidative stress and belongs to the family of Cap'n'Collar family of basic leucine zippers[18,19]. Kelch-like ECH-associated protein-1 (KEAP1), a redox-regulated substrate adaptor protein, which interacts with the Cul3-dependent ubiquitin ligase complex[20]. The antioxidant response element (ARE), an electrophilic response element, is a cis-regulatory element[21]. NRF2 is recognized as a regulator of oxidative stress, and KEAP1-NRF2-ARE is one of the most important pathways of antioxidant and anti-inflammatory element signaling[22]. Under homeostasis, NRF2 binds to KEAP1 and is anchored to the cytoplasm[23]. When cells are attacked by ROS or other stimuli, NRF2 dissociates from KEAP1 and is quickly translocated to the nucleus, forming a heterodimer with the transcriptional regulator and binding to an ARE[24]. The complex ultimately activates the expression of antioxidant enzymes and thus leads to the removal of excessive ROS in the body, thereby exerting antioxidant effects[25]. Then, NRF2 is degraded in the nucleus through the β-transducin repeat-containing protein-glycogen synthase kinase 3 (β-TrCP-GSK3) axis or by translocating to the cytoplasm and becoming rapidly degraded by KEAP1[26]. Since the outbreak of COVID-19, continuous studies have shown that enhancing immunity, establishing resistance to the inflammatory response, and reducing oxidative stress are keys to COVID-19 treatment, while NRF2 may be an important target for COVID-19 treatment and plays a protective role because of its dual anti-viral and anti-inflammatory properties [27]. Currently, NRF2 activators, such as 4-octyl-itaconate (4-OI) and dimethyl fumarate (DMF), can be used for the treatment of COVID-19[28]. As most liver injury is associated with increased oxidative stress and an overloaded antioxidant defense system, NRF2 may also be an ideal therapeutic target for treating chronic liver disease. Indeed, NRF2 has previously been shown to protect liver cells in viral hepatitis[29,30], drug-induced liver injury[31-33], cholestasis liver injury[34-36], fatty liver[37,38], and other liver diseases by reversing insulin resistance, inhibiting gluconeogenesis and lipid accumulation, and enhancing the anti-inflammatory and antioxidative effects[39]. We speculate that NRF2 may also play a regulatory role in COVID-19-related liver injury. In this review, we systematically describe the reciprocal regulation between NRF2 and inflammatory mediators in COVID-19-related liver injury.
Cytokines are small-molecule messengers secreted by cells, and they can exert effects on the same type of cell in the organ periphery and cells in other organs[40]. Most cytokines are in a lytic state, and some exist in membrane form. Cytokines can be secreted by most cells and possess a wide range of functions. Interleukins (ILs) are a group of cytokines that were first seen to be expressed by white blood cells. Chemically attractive cytokines are called chemokines. Cytokines that interfere with viral replication are called interferons (IFNs).
When the body is stimulated by endogenous or exogenous stimuli such as viruses, it usually triggers the immune defense system and oxidative stress response to maintain homeostasis. This process is often accompanied by the production of many cytokines[41], which interfere with mitochondrial function and lead to the production of a large amount of ROS, thereby inducing the expression of IL-1, IL-6, tumor necrosis factor alpha (TNF-α), IL-8 receptor-β (CXCR2), monocyte chemotactic protein-1, and cell adhesion molecule 1 and promoting the aggregation of macrophages and chemotactic neutrophils[42-44]. Then neutrophils produce myeloperoxidase and catalyze the production of large amounts of hypochlorous acid, which kills cells through strong oxidation reactions[45]. Cell injury induces the expression of high mobility group box 1, osteopontin, and Toll-like receptor 4, recruiting more neutrophils, amplifying the inflammatory response, and further aggravating tissue injury[46].
It is thought that COVID-19 patients suffer from cytokine storm symptoms and an uncontrolled release of proinflammatory cytokines. The levels of proinflammatory factors such as IL-1β, IL-7, IL-8, IL-9, IL-10, TNF-α, and other inflammatory factors are higher in COVID-19 patients than in healthy adults[47,48]. Serum inflammatory cytokine levels are positively correlated with indicators of liver dysfunction in patients, suggesting potential mechanisms between liver injury and the inflammatory response[49]. NRF2 negatively regulates the expression of these inflammatory cytokines. For example, in human macrophages, NRF2 can inhibit the expression of inflammatory cytokines, such as IL-1β, IL-6, and TNF-α, by blocking the recruitment of RNA polymerase II, thus reducing the inflammatory response. Moreover, an agonist of NRF2 represses the replication of SARS-CoV-2 and the inflammatory response[50].
The NRF2-activating compound resveratrol is a growth suppressor and apoptosis promoter in liver cancer cells[51]. Resveratrol can reduce the expression of IL-1β. Another NRF2-activating compound, PB125, which contains the active ingredients carnosol, withaferin A, and luteolin, significantly downregulates IL-1β, IL6, TNF-α, intercellular adhesion molecule 1, vascular cell adhesion molecule 1 (VCAM1), E-selectin, and IFN-γ-induced gene expression[52]. The inflammatory response and oxidative stress are the main pathological features of COVID-19, and disease progression has been related to redox imbalance. The NRF2 antioxidant pathway is suppressed in COVID-19 patients and cells infected with SARS-CoV-2 show activated GSK-3, which is a conserved serine/threonine kinase that degrades NRF2[53]. The NRF2 agonists 4-OI and DMF can inhibit the replication of SARS-CoV-2[28]. All of these findings support the idea that NRF2 has dual antiviral and anti-inflammatory properties and may be a potential therapeutic target for the treatment of COVID-19.
Pre-existing liver disease may play an important role in COVID-19-related liver injury, since an investigation found that a small proportion of patients with COVID-19 also have pre-existing liver diseases[54-56]. This effect may be due to the significant decrease in the number of lymphocytes, CD4+ T cells, CD8+ T cells, B cells, and NK cells in patients with COVID-19, which prevents a proper immune response[57], ultimately leading to the recurrence of pre-existing liver disease. Glucocorticoids effectively reduce mortality in patients with severe COVID-19[58]. In addition to its anti-inflammatory function, glucocorticoids also have immunosuppressive effects, which can also participate in the reactivation of pre-existing liver diseases.
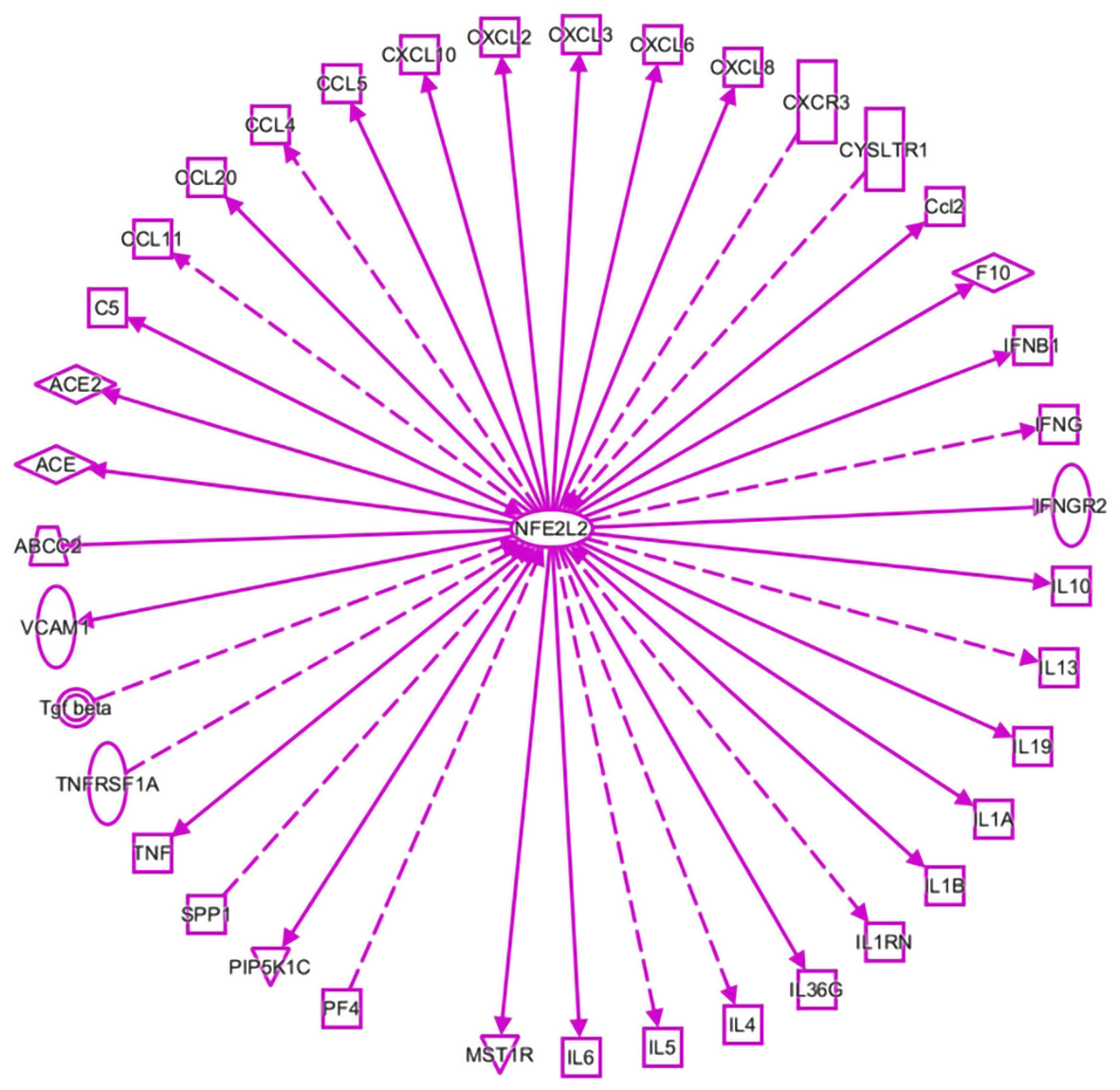
As shown in Figure 1, inflammatory molecules are involved in the interactions with NRF2 in viral infection, as listed in a dataset generated by ingenuity pathway analysis (IPA). They include IL1A, IL1B, IL4, IL5, IL6, IL10, IL13, IL19, IL36G, IL1RN, IFNGR2, IFNB1, CCL2, CYSLTR1, EXCR3, CXCL8, CXCL6, CXCL3, CXCL2, CXCL10, CCL5, CCL4, CCL20 CCL11, TGF-β, TNFRSFA, TNF, and MST1R. Current literature shows that these molecules are related to the COVID-19-mediated inflammatory response [41-48].
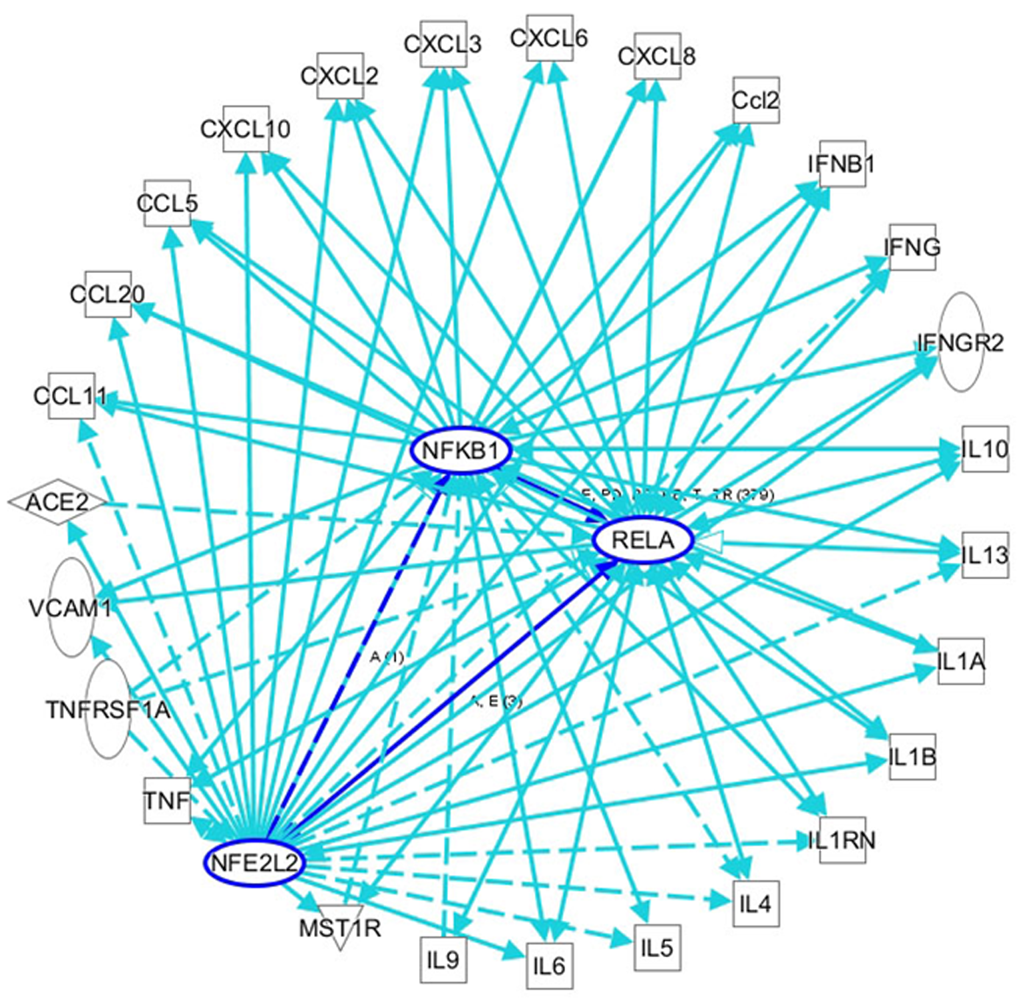
Activated nuclear factor kappa-B (NF-κB) is a key transcription factor in the inflammatory response and oxidative stress. The NF-κB signal transduction pathway is a typical proinflammatory pathway[59]. There are five members in the NF-κB family: p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), RelB, and c-Rel[60]. Under physiological conditions, NF-κB and inhibitor kappa B (IκB) combine and are maintained in the cytoplasm in an inactive state. Upon viral infection or other stimuli, IκB is phosphory
KEAP1 is the intermediate link between NRF2 and NF-κB. ARE is a cis-acting element, and the NRF2-ARE pathway is the most important endogenous antioxidant excitation pathway[73]. On one hand, p65 can increase the nuclear level of KEAP1 to inhibit the NRF2-ARE pathway, thus reducing the expression of cytoprotective enzymes. On the other hand, KEAP1 can initiate the autophagic degradation of inhibitor of NF-κB kinase subunit β (IKKβ) by preventing the binding of heat shock protein 90 (HSP90) to IKKβ. Additionally, KEAP1 can specifically bind to IKKβ and then connect with the ubiquitin connexin CUL3-RBX1 complex, which promotes IKKβ degradation and inhibits the expression of NF-kB[74].
COVID-19 is always accompanied by an increase in inflammatory cells and proinflammatory factors. Oxidative stress is an important mechanism for the occurrence and development of COVID-19, and is closely related to the prognosis of the disease[12]. ROS are the key agents in oxidative stress; some researchers propose monitoring ROS levels as diagnostic criteria for COVID-19[75]. As a key regulator of oxidative stress and inflammation, NF-κB plays an important role in the progression of COVID-19. In patients with COVID-19, a severe systemic inflammatory response is always followed by acute respiratory distress syndrome (ARDS) and sepsis, which leads to tissue ischemia and hypoxia and the generation of ROS, thus activating the NF-κB pathway[76]. In addition, Hirano et al[77] proposed that ACE2 is the key factor in the early stage of COVID-19, and the IL-6-signal transducer and activator of transcription 3 (STAT3) axis plays an important role in the inflammatory cytokine storm in the late stage of the disease. ACE2 is necessary for SARS-CoV-2 entry, and it can also inactivate angiotensin 2 (AngII), which can play a proinflammatory role via the angiotensin 2 receptor (AT1R). SARS-CoV, has high homology with SARS-CoV-2 and binds to ACE2 on the cell membrane, which results in an increase in AngII in serum[78]. In animal models, AT1R inhibitors can prevent ARDS caused by SARS-CoV infection[78]. The AngII-AT1R axis also activates NF-κB and ADAM metallopeptidase domain 17 (ADAM17), which can promote the production of an epidermal growth factor ligand (EGFR) ligand and TNF-α, and this EGFR ligand and TNF-α can also stimulate NF-κB[79]. ADAM17 can also activate STAT3 via IL-6, thus activating NF-κB[80]. The NF-κB-triggered positive feedback loop for IL-6 signaling in type 1 collagen+ nonimmune cells is termed an IL-6 amplifier. Indeed, it has been proven that both the NF-κB pathway and STAT3 pathway are activated in COVID-19, promoting an increase in proinflammatory factors and inflammatory cells by activating the IL-6 amplifier, thus forming a vicious cycle and eventually leading to a systemic inflammatory response and MODS[80]. Among the various organs affected by MODS, COVID-19-related liver injury has a high incidence, ranging from 5.1% to 69.8% according to recent reports, and manifests mainly as elevated transaminase and bilirubin levels (Table 1). Liver injury greatly affects the progression and prognosis of COVID-19. However, the exact mechanisms of liver injury are unclear due to limited data. In addition to the application of drugs that cause liver injury, direct viral cytotoxicity and reactivity of existing liver disease, systemic inflammatory response, ischemia, and hypoxia may play crucial roles, as increased oxidative stress and an overloaded antioxidant defense system have always been acknowledged as key factors in the occurrence and development of various liver injuries[81]. Oxidative stress can stimulate Kupffer cells to produce cytokines that promote the apoptosis of hepatocytes and can also induce stellate cell activation/proliferation and collagen synthesis to promote fibrosis and cirrhosis[82]. To maintain redox homeostasis, the liver fights oxidative stress through a complex antioxidant system. NRF2 is associated with enhancement of the endogenous antioxidant system and has been identified as a target to prevent liver injury, inhibit liver fibrosis, and promote liver regeneration[74,83-86]. As shown through IPA analyses of the interactions among NRF2, NF-κB, and inflammatory cytokines in viral infections, the interaction and balance between NRF2 and NF-kB may play important roles in COVID-19-related liver injury, and targeting their interactions may be an important therapeutic strategy for COVID-19 treatment.
| Ref. | Total number of patients | Proportion of liver injury, (%) | Proportions of pre-existing liver diseases, n (%) | Others |
| Guan et al[87] | 1099 | Elevated AST 168/757 (22.2); Elevated ALT 158/741 (21.3); Elevated TB 76/722 (10.5) | 23 (2.1) | |
| Lian et al[56] | 465 | 61 (13.1) | 19 (4.1) | Liver injury was the most common complication among these patients |
| Chen et al[88] | 503 | 301 (69.8) | NA | Liver damage is an independent prognostic factor of COVID-19 |
| Jin et al[55] | 651 | 64 (9.8) | 25 (3.8) | |
| Xie et al[89] | 79 | Elevated ALT (31.6); Elevated AST (35.4); Elevated TB (5.1) | NA | COVID-19 with liver injury in non-ICU hospitalized patients was common |
| Wang et al[90] | 293 | 19 (6.5) | 12 (4.1) | Compared with the survival group, the non-survival group was more prone to complications of acute liver injury |
| Chen et al[91] | 274 | Elevated AST 84 (31) | 11 (4) | The frequency of liver injury in the deceased patients was significantly higher than that in the recovered patients |
| Yu et al[54] | 1633 | Elevated ALT 298/1445 (20.6); Elevated AST 303/1445 (21.0) | 38 (2.3) | Severe patients are more likely to have liver injury than mild patients |
Glutathione (GSH) is an important molecule involved in redox homeostasis, and NRF2 promotes its synthesis by regulating glutamate-cysteine ligase catalytic subunit (GCLC), glutamate-cysteine ligase regulatory subunit (GCLM), glutamine synthetase (GS or GSr1), GSH reductase (GR), GSH peroxidase (GPx), and GSH S-transferases (GSTs)[92]. The tricarboxylic acid cycle (TCA cycle) is the central link and cross hub of glucose metabolism, lipid metabolism, and protein metabolism. NRF2 participates in the TCA cycle by regulating the expression of glucose 6-phosphate dehydrogenase (G6PD), malic enzyme 1, and isocitrate dehydrogenase (IDH), which are involved in the production of NADPH, a key cofactor in promoting antioxidant reactions[93-95]. NRF2 can also affect intermediate metabolism, increases the availability of substrates for the mitochondrial respiratory chain, and increases nicotinamide adenine dinucleotide phosphate (NAPDH) production through crosstalk between the glycoly
Many researchers have proposed the hypothesis that COVID-19-related liver damage may be caused by systemic inflammatory response to viral infection. After SARS-CoV-2 infection and the increased production of ROS[98], the inflammation response of the body is activated and the expression of proinflammatory cytokines including IL-1β, IL-6, IL-8, and TNF-α is greatly increased, and immune cells are continuously activated and are accumulating[99,100]. The combined effects of many immune cells and proinflammatory factors lead to a cytokine storm[4,99-102], which may further result in multiple organ function damage, including liver dysfunction, and even the death of the patient[103,104]. Moreover, there seems to be a positive correlation between the severity of COVID-19 and the level of inflammation [57,100,105-107], and the incidence of liver damage is related to the severity of COVID-19[13], which also reveals the close relationship between COVID-19-related liver injury and the inflammatory response. Since many cases of limb[108], gastrointestinal[109], heart[110], and cerebral[111] ischemia have been reported, it is reasonable to suspect that tissue ischemia is among the causes of COVID-19-related liver injury, in which the inflammatory response may cause local blood stasis, vascular endothelial destruction and excessive activation of the coagulation system, leading to local thrombosis and tissue ischemia, ultimately resulting in liver injury. Moreover, severe inflammatory reactions, such as sepsis[76], which cause hemodynamic instability and lead to ischemia, may also be among the causes of liver injury. Strong evidence points to the major causes for liver injury involve the systemic inflammatory response and oxidative stress following SARS-CoV-2 infection.
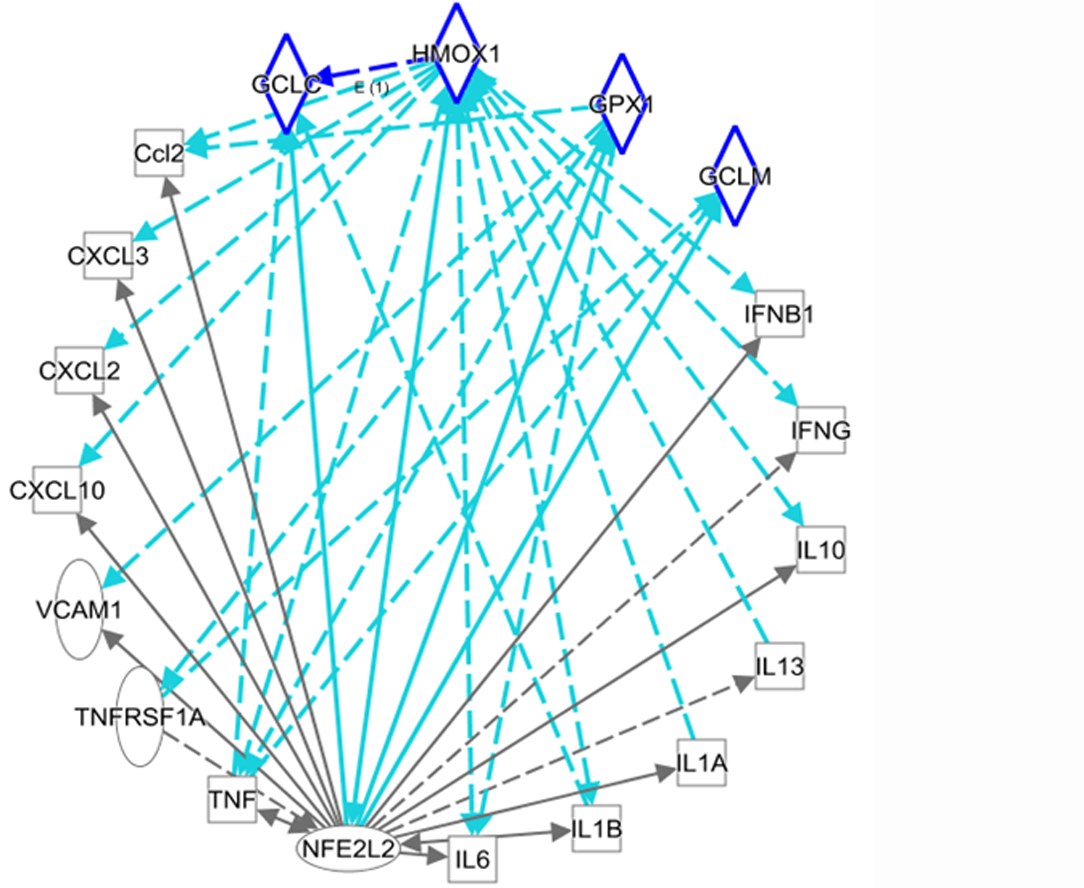
Regarding the mechanisms of direct damage and inflammation-induced damage after SARS-CoV-2 infection, heme oxygenase 1 (HO-1) seems to be involved and could represent a therapeutic strategy. HO-1 can inhibit the replication of influenza A virus, human respiratory syncytial virus, and hepatitis B virus, among others. In all cases, viral replication blockade is achieved through the type I IFN response mediated by HO-1[112]. Therefore, it is reasonable to assume that HO-1 inhibits SARS-CoV-2 replication by the same mechanism. HO-1 and its metabolites have significant anti-inflammatory effects via NRF2. HO-1 metabolizes free heme into carbon monoxide (CO), biliverdin and iron, exhibiting cell protection and anti-apoptosis and immune regulation properties[113]. In animal models of sepsis and renal ischemia-reperfusion injury with significant systemic inflammatory responses, HO-1 showed great anti-inflammatory effects and its overexpression provided cell protection[112]. Hence, it can be speculated that HO-1 counteracts the cytokine storm produced by COVID-19. Additionally, the CO product of HO-1 can stimulate the release of hydrogen sulfide (H2S)[114]. H2S is a natural defense against enveloped RNA virus infections that can activate regulatory T cells, maintain the body’s immune homeostasis, and exert anti-viral effects[114]. H2S can also oversulfate the KATP channel on the white blood cell membrane to keep it open, preventing white blood cells adherence to the endothelium and triggering the inflammatory cascade[114]. On the one hand, H2S regulates endothelial cell function and attenuates ischemia. On the other hand, H2S controls the body’s inflammatory response and prevents liver injury. Therefore, HO-1 activated by NRF2 can inhibit viral replication to control viral infection and can also balance host defenses through the HO-1/CO/H2S axis, protecting endothelial structures, improving ischemia, and protecting the liver from damage induced by SARS-CoV-2 infection.
Figure 3 shows reciprocal regulation between NRF2/redox-related genes and inflammatory mediators. In particular, upregulation of NRF2/HO-1 (HMOX1) may be an important strategy for the treatment of the liver damage caused by COVID-19.
NRF2 consists of seven highly conserved domains, NRF2-ECH homology 1 (Neh1) to Neh7. Each of the Neh domains executes distinct functions[18,19]. Neh1 is a CNC-bZIP domain that allows NRF2 to heterodimerize with small musculoaponeurotic fibrosarcoma (MAF) proteins (MAFF, MAFG, and MAFK)[115]. Under normal physiological conditions, NRF2 is degraded via the ubiquitin-proteasome pathway in a KEAP1-dependent manner and maintained at a basal level[36]. Under stress conditions, the cysteine residues in KEAP1 are modified when they are oxidized, causing the loss of its adaptor activity[20]. Then NRF2 is disengaged from the regulatory complex, is phosphorylated, and accumulates in the nucleus, where it heterodimerizes with small MAF proteins and regulates ARE-driven genes[115]. These proteins regulate transcription when they dimerize among themselves[116,117]; that is, they seem to serve as transcriptional activators by dimerizing with other basic zipper proteins such as NFE2L2[115]. The interaction of MAFG and NFE2L2 results in activation of the ARE, which is present at the promoter-proximal region of many genes involved in the antioxidant defense and triggers the expression of certain genes[115]. MAFG expression is induced by oxidative stressors, such as hydrogen peroxide and electrophilic compounds, increasing NRF2 and GSH expression and activating the NRF2-ARE pathway[118]. NRF2 exerts protective effects on various liver diseases [119,120]. Therefore, NRF2 can also be a therapeutic target and NRF2 activators can also be used for the treatment of COVID-19-related liver injury. MAFG, as a molecular chaperone is required for the function of NRF2, may also be a target for the treatment of COVID-19-related liver injury.
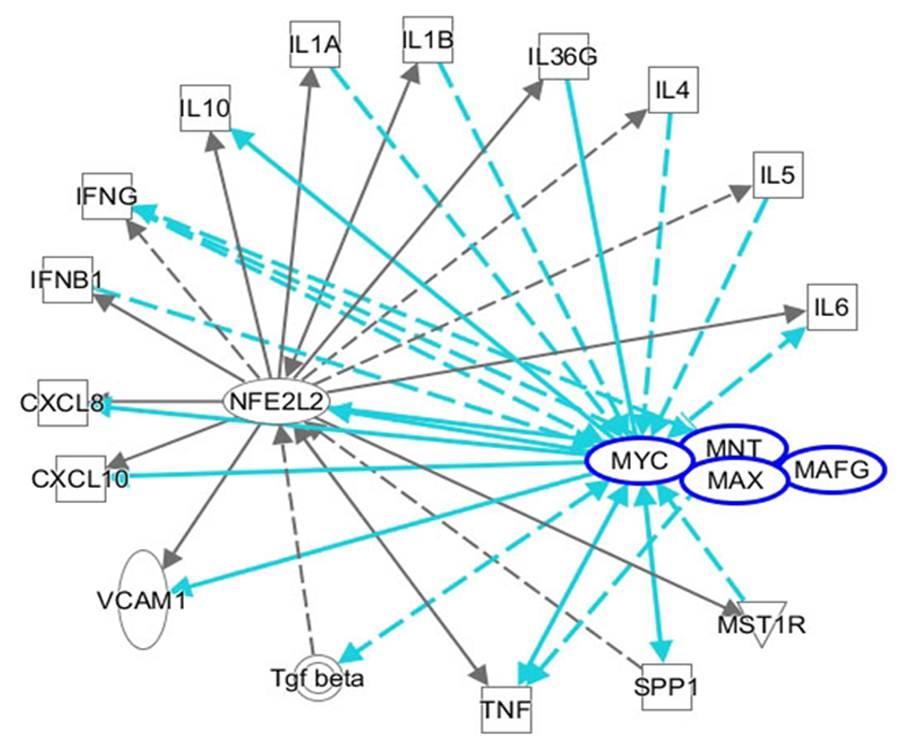
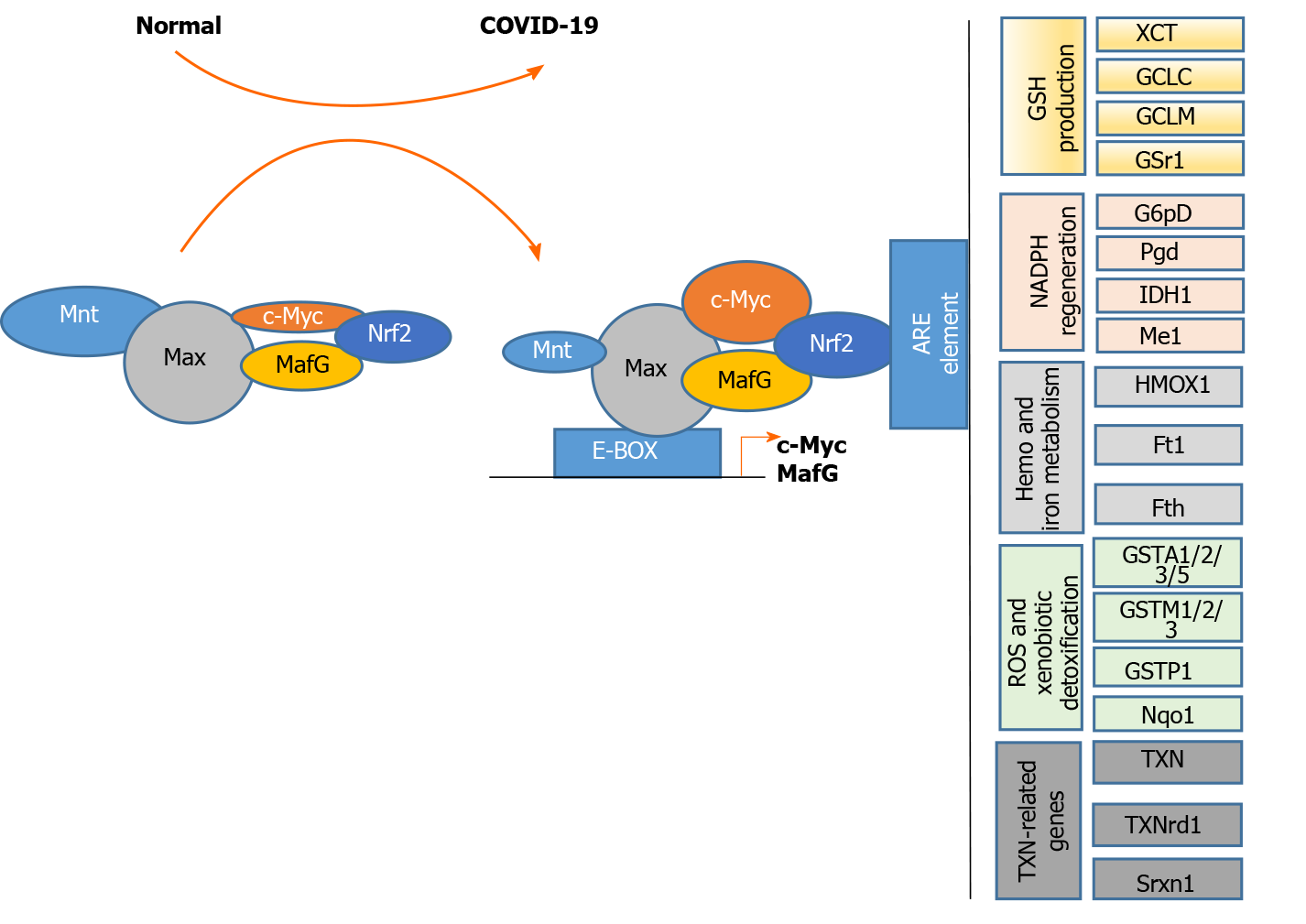
C-MYC is a family of proto-oncogenes and encodes a nuclear phosphoprotein that plays roles in cell cycle progression, apoptosis, and cell transformation[121]. MNT is a MAX network transcriptional repressor[122]. C-MYC and MNT each have a basic helix-loop-helix-zipper domain (bHLHzip) with which they bind the canonical DNA sequence CACGTG, known as an E-box, following heterodimerization with MAX proteins. The protein complexes bind to the E-box DNA sequence and regulate the transcription of specific target genes[122]. The MYC-MAX and MNT-MAX networks comprise transcription factors that regulate gene-specific transcription[122]. The heterodimer MNT-MAX is a transcriptional repressor, and the heterodimer MYC-MAX is a transcriptional activator[123-125]. We also reported that c-MYC can interact with NRF2 to repress NRF2-mediated transactivation of the ARE[126]. Their interaction may regulate the oxidative stress pathway. An IPA showed the interactions among NFE2L2/MAF/MYC/MNT and inflammatory mediators. NRF2/MYC/MNT/ MAFG are involved with inflammatory mediators IL10, IL1A, IL1B, IL36G, IL4, IL5, IL6, IFNG, IFNG, CXCL8, VCAM1, TNF, TGF-β, SPP1 and MST1R (Figure 4).
Upon analyses of a protein-protein interaction network with 65 intersecting targets, c-MYC was found to be one of the core targets for COVID-19 treatment[127]. MAFG and c-MYC are upregulated and MNT is downregulated in liver injury[125,128]. MAX directly interacts with the c-MYC, MAFG, MNT, and NRF2 proteins, whereas c-MYC and MAFG interact with NRF2. As shown in Figure 5, these complexes bind to the E-box region in the promoters of the c-MYC and MAFG genes, leading to gene activation. The complexes can also bind to AREs, leading to the dysregulation of the following genes: TXN-related genes (TXN, TXNrd1, and Srxn1); ROS and xenobiotic detoxification-related genes (GSTA1/2 and GSTM1/2/3), GSTP1, and Nqo1); heme and ion metabolism-related genes (HMOX1 or HO-1, Ft1, and Fth); NADPH regeneration-related genes (G6pD, PgD, IDH1 and Me1); and GSH production-related genes (XCT, GCLC, GCLM, and GSR1). This gene dysregulation may lead to COVID-19-related cytokine storm and liver injury.
Alanine aminotransferase (ALT) was selected to represent liver injury rather than aspartate aminotransferase (AST) because the sources of extrahepatic AST are very common, including the heart and skeletal muscle during related muscle breakdown. Therefore, in patients infected with SAR-CoV-2, we classified liver damage into normal/mild (< 2 fold the upper limit of normal, upper limit of normal [ULN]), moderate (2- to 5-fold ULN), and severe (> 5 fold ULN) according to the degree of ALT elevation (Table 2)[129].
| Ref. | Date | Cohort | The severity of liver injury | How the ULN was defined | ||
| Normal/mild (< 2 times ULN) | moderate (2-5 times ULN) | Severe (> 5 times ULN) | ||||
| Phipps et al[129] | March 8, 2020-April 14, 2020 | 2273 | 1784 (78.5) | 344 (15.1) | 145 (6.4) | ALT = 50 U/L |
| Mendizabal et al[130] | April 15, 2020-July 31, 2020 | 1611 | 322 (84.7) | 185 (11.5) | 61 (3.8) | ALT (the reference value of different institutions) |
| Hundt et al[131] | March 14, 2020-April, 2020 | 1753 | 1123 (64.1) | 408 (23.3) | 222 (12.7) | ALT = 34 U / L |
| Yip et al[132] | January 23, 2020-May 1, 2020 | 816 | 635 (77.8) | 141 (17.3) | 40 (4.9) | ALT = 40 U / L |
Normal and mild liver injury account for the largest proportion (60%-90%) of SAR-CoV-2-infected patients. Severe liver injury accounts for less than 20% of these patients. In addition, moderate to severe liver injury is more common in patients requiring intensive care unit (ICU) care. However, in general, the most common liver injury is mild liver injury, and the degree of liver injury is independently related to the occurrence of end events (ICU admission, death, mechanical injury)[129-131]. Only 5% of COVID-19 patients acquire severe liver damage[132]. The Cai et al[133] study showed that the incidence of liver injury in hospitals was higher than that at the time of admission. For these patients, few other factors affected liver test abnormalities, such as potential liver disease or drug use. Therefore, it can be speculated that the occurrence of liver damage is caused by COVID-19. Zhao et al[134] compared patients with mild COVID-19 systems with non-infected patients. The absolute value of lymphocyte level differences in the two groups was low, and there was no difference in C-reactive protein or IL-6 level. ALT was not elevated in patients with pneumonia unrelated to COVID-19 when they were admitted to the hospital, but it was elevated in 28% of COVID-19 patients. Compared with the general inflammation caused by other pathogens, the specific inflammation caused by COVID-19 is more likely to cause abnormal liver function. Additionally, severe liver damage caused by SARS-CoV-2 infection is associated with serum inflammatory markers (white blood cell count and neutrophil-to-lymphocyte ratio are significantly higher). Age, hypertension, and the presence of diabetes are negatively correlated with severe liver damage in COVID-19 patients[129]. It can be speculated that young patients may have a stronger immune and inflammatory response to infection, which may lead to more severe liver damage. In other words, the inflammatory response can cause liver damage and determine the degree of liver damage. Therefore, inflammation plays an important role in the development of liver injury induced by COVID-19. Because of the individual response to the virus, different patients may exhibit different degrees of liver injury; therefore, it is necessary to grade the degree of liver injury. We described the anti-inflammatory effect of NRF2 above, which is of great benefit to the prevention or treatment of COVID-19-mediated liver injury, and is also crucial for improving the prognosis of patients.
There are many reasons for COVID-19-related liver injury including the inflammatory response and oxidative stress, ischemia, drug toxicity, SARS-CoV-2 cytotoxicity, and pre-existing liver disease. Regardless of the etiology, inflammatory cell infiltration, cell necrosis, and liver tissue fibrosis may be the final pathological result. As an important transcription factor of antioxidant stress and anti-inflammation, NRF2 can exert protective effects on COVID-19-related liver injury. The KEAP1-NRF2-ARE, NRF2-HO-1, NRF2-MYC/MNT/MAFG, and NRF2-NF-κB pathways are potential preventive and therapeutic targets in COVID-19-related liver injury.
We thank the authors of the primary studies for their responses to our information requests.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Boscá L S-Editor: Gao CC L-Editor: Filipodia P-Editor: Ma YJ
| 1. | Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front Cell Infect Microbiol. 2020;10:587269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 474] [Cited by in RCA: 527] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 2. | Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020; 181: 914-921. e10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1550] [Cited by in RCA: 1550] [Article Influence: 310.0] [Reference Citation Analysis (0)] |
| 3. | Mrityunjaya M, Pavithra V, Neelam R, Janhavi P, Halami PM, Ravindra PV. Immune-Boosting, Antioxidant and Anti-inflammatory Food Supplements Targeting Pathogenesis of COVID-19. Front Immunol. 2020;11:570122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 185] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 4. | Cuadrado A, Pajares M, Benito C, Jiménez-Villegas J, Escoll M, Fernández-Ginés R, Garcia Yagüe AJ, Lastra D, Manda G, Rojo AI, Dinkova-Kostova AT. Can Activation of NRF2 Be a Strategy against COVID-19? Trends Pharmacol Sci. 2020;41:598-610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 163] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 5. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020; 181: 271-280. e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14274] [Article Influence: 2854.8] [Reference Citation Analysis (0)] |
| 6. | Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529-539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1895] [Cited by in RCA: 1791] [Article Influence: 223.9] [Reference Citation Analysis (0)] |
| 7. | Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2713] [Cited by in RCA: 2765] [Article Influence: 553.0] [Reference Citation Analysis (0)] |
| 8. | Wang Y, Liu S, Liu H, Li W, Lin F, Jiang L, Li X, Xu P, Zhang L, Zhao L, Cao Y, Kang J, Yang J, Li L, Liu X, Li Y, Nie R, Mu J, Lu F, Zhao S, Lu J, Zhao J. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J Hepatol. 2020;73:807-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 457] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 9. | Jorgensen SCJ, Tse CLY, Burry L, Dresser LD. Baricitinib: A Review of Pharmacology, Safety, and Emerging Clinical Experience in COVID-19. Pharmacotherapy. 2020;40:843-856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 10. | Cha MH, Regueiro M, Sandhu DS. Gastrointestinal and hepatic manifestations of COVID-19: A comprehensive review. World J Gastroenterol. 2020;26:2323-2332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 124] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (2)] |
| 11. | Zhang C, Shi L, Wang FS. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5:428-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1348] [Cited by in RCA: 1295] [Article Influence: 259.0] [Reference Citation Analysis (4)] |
| 12. | Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5228] [Cited by in RCA: 5786] [Article Influence: 1157.2] [Reference Citation Analysis (2)] |
| 13. | Wang Q, Zhao H, Liu LG, Wang YB, Zhang T, Li MH, Xu YL, Gao GJ, Xiong HF, Fan Y, Cao Y, Ding R, Wang JJ, Cheng C, Xie W. Pattern of liver injury in adult patients with COVID-19: a retrospective analysis of 105 patients. Mil Med Res. 2020;7:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 14. | Jothimani D, Venugopal R, Abedin MF, Kaliamoorthy I, Rela M. COVID-19 and the liver. J Hepatol. 2020;73:1231-1240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 358] [Article Influence: 71.6] [Reference Citation Analysis (1)] |
| 15. | Caraballo C, Jaimes F. Organ Dysfunction in Sepsis: An Ominous Trajectory From Infection To Death. Yale J Biol Med. 2019;92:629-640. [PubMed] |
| 16. | Sun J, Aghemo A, Forner A, Valenti L. COVID-19 and liver disease. Liver Int. 2020;40:1278-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 219] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 17. | Karakike E, Giamarellos-Bourboulis EJ. Macrophage Activation-Like Syndrome: A Distinct Entity Leading to Early Death in Sepsis. Front Immunol. 2019;10:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 215] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 18. | Tonelli C, Chio IIC, Tuveson DA. Transcriptional Regulation by Nrf2. Antioxid Redox Signal. 2018;29:1727-1745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 923] [Cited by in RCA: 1543] [Article Influence: 220.4] [Reference Citation Analysis (0)] |
| 19. | Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1217] [Cited by in RCA: 1637] [Article Influence: 148.8] [Reference Citation Analysis (0)] |
| 20. | Kopacz A, Kloska D, Forman HJ, Jozkowicz A, Grochot-Przeczek A. Beyond repression of Nrf2: An update on Keap1. Free Radic Biol Med. 2020;157:63-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 189] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 21. | Lu MC, Ji JA, Jiang ZY, You QD. The Keap1-Nrf2-ARE Pathway As a Potential Preventive and Therapeutic Target: An Update. Med Res Rev. 2016;36:924-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 595] [Article Influence: 66.1] [Reference Citation Analysis (0)] |
| 22. | Sajadimajd S, Khazaei M. Oxidative Stress and Cancer: The Role of Nrf2. Curr Cancer Drug Targets. 2018;18:538-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 295] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 23. | David JA, Rifkin WJ, Rabbani PS, Ceradini DJ. The Nrf2/Keap1/ARE Pathway and Oxidative Stress as a Therapeutic Target in Type II Diabetes Mellitus. J Diabetes Res. 2017;2017:4826724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 193] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 24. | Tang W, Jiang YF, Ponnusamy M, Diallo M. Role of Nrf2 in chronic liver disease. World J Gastroenterol. 2014;20:13079-13087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 156] [Cited by in RCA: 178] [Article Influence: 16.2] [Reference Citation Analysis (1)] |
| 25. | Cui Y, Xin H, Tao Y, Mei L, Wang Z. Arenaria kansuensis attenuates pulmonary fibrosis in mice via the activation of Nrf2 pathway and the inhibition of NF-kB/TGF-beta1/Smad2/3 pathway. Phytother Res. 2021;35:974-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 26. | Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121-1133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 650] [Cited by in RCA: 640] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 27. | Lin CY, Yao CA. Potential Role of Nrf2 Activators with Dual Antiviral and Anti-Inflammatory Properties in the Management of Viral Pneumonia. Infect Drug Resist. 2020;13:1735-1741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 28. | Olagnier D, Farahani E, Thyrsted J, Blay-Cadanet J, Herengt A, Idorn M, Hait A, Hernaez B, Knudsen A, Iversen MB, Schilling M, Jørgensen SE, Thomsen M, Reinert LS, Lappe M, Hoang HD, Gilchrist VH, Hansen AL, Ottosen R, Nielsen CG, Møller C, van der Horst D, Peri S, Balachandran S, Huang J, Jakobsen M, Svenningsen EB, Poulsen TB, Bartsch L, Thielke AL, Luo Y, Alain T, Rehwinkel J, Alcamí A, Hiscott J, Mogensen TH, Paludan SR, Holm CK. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat Commun. 2020;11:4938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 298] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 29. | Espinoza JA, González PA, Kalergis AM. Modulation of Antiviral Immunity by Heme Oxygenase-1. Am J Pathol. 2017;187:487-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 30. | Ruan Y, Jiang Y, Wang W, Ma J, He B, Chen M. HBV down-regulates PTEN expression via Nrf2/GSK3β signaling pathway. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2020;45:1009-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 31. | Goldring CE, Kitteringham NR, Elsby R, Randle LE, Clement YN, Williams DP, McMahon M, Hayes JD, Itoh K, Yamamoto M, Park BK. Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology. 2004;39:1267-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 164] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 32. | Jadeja RN, Urrunaga NH, Dash S, Khurana S, Saxena NK. Withaferin-A Reduces Acetaminophen-Induced Liver Injury in Mice. Biochem Pharmacol. 2015;97:122-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 33. | Liu J, Wu KC, Lu YF, Ekuase E, Klaassen CD. Nrf2 protection against liver injury produced by various hepatotoxicants. Oxid Med Cell Longev. 2013;2013:305861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 34. | Tan KP, Wood GA, Yang M, Ito S. Participation of nuclear factor (erythroid 2-related), factor 2 in ameliorating lithocholic acid-induced cholestatic liver injury in mice. Br J Pharmacol. 2010;161:1111-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Okada K, Shoda J, Taguchi K, Maher JM, Ishizaki K, Inoue Y, Ohtsuki M, Goto N, Sugimoto H, Utsunomiya H, Oda K, Warabi E, Ishii T, Yamamoto M. Nrf2 counteracts cholestatic liver injury via stimulation of hepatic defense systems. Biochem Biophys Res Commun. 2009;389:431-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 72] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 36. | Aleksunes LM, Slitt AL, Maher JM, Dieter MZ, Knight TR, Goedken M, Cherrington NJ, Chan JY, Klaassen CD, Manautou JE. Nuclear factor-E2-related factor 2 expression in liver is critical for induction of NAD(P)H:quinone oxidoreductase 1 during cholestasis. Cell Stress Chaperones. 2006;11:356-363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 37. | Sugimoto H, Okada K, Shoda J, Warabi E, Ishige K, Ueda T, Taguchi K, Yanagawa T, Nakahara A, Hyodo I, Ishii T, Yamamoto M. Deletion of nuclear factor-E2-related factor-2 Leads to rapid onset and progression of nutritional steatohepatitis in mice. Am J Physiol Gastrointest Liver Physiol. 2010;298:G283-G294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 131] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 38. | Lamlé J, Marhenke S, Borlak J, von Wasielewski R, Eriksson CJ, Geffers R, Manns MP, Yamamoto M, Vogel A. Nuclear factor-eythroid 2-related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology. 2008;134:1159-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 166] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 39. | Cuadrado A, Rojo AI, Wells G, Hayes JD, Cousin SP, Rumsey WL, Attucks OC, Franklin S, Levonen AL, Kensler TW, Dinkova-Kostova AT. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat Rev Drug Discov. 2019;18:295-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 801] [Cited by in RCA: 963] [Article Influence: 160.5] [Reference Citation Analysis (0)] |
| 40. | Oppenheim JJ. Cytokines: past, present, and future. Int J Hematol. 2001;74:3-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 143] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 41. | Dinarello CA. Historical insights into cytokines. Eur J Immunol. 2007;37 Suppl 1:S34-S45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 674] [Cited by in RCA: 581] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 42. | Konishi T, Schuster RM, Goetzman HS, Caldwell CC, Lentsch AB. Cell-specific regulatory effects of CXCR2 on cholestatic liver injury. Am J Physiol Gastrointest Liver Physiol. 2019;317:G773-G783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 43. | Saito JM, Maher JJ. Bile duct ligation in rats induces biliary expression of cytokine-induced neutrophil chemoattractant. Gastroenterology. 2000;118:1157-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 94] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Jones H, Alpini G, Francis H. Bile acid signaling and biliary functions. Acta Pharm Sin B. 2015;5:123-128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 45. | Li M, Cai SY, Boyer JL. Mechanisms of bile acid mediated inflammation in the liver. Mol Aspects Med. 2017;56:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 188] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 46. | Wenzel P, Kossmann S, Münzel T, Daiber A. Redox regulation of cardiovascular inflammation - Immunomodulatory function of mitochondrial and Nox-derived reactive oxygen and nitrogen species. Free Radic Biol Med. 2017;109:48-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 121] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 47. | Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2604] [Cited by in RCA: 3195] [Article Influence: 639.0] [Reference Citation Analysis (0)] |
| 48. | Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4960] [Cited by in RCA: 5518] [Article Influence: 1103.6] [Reference Citation Analysis (1)] |
| 49. | Yang RX, Zheng RD, Fan JG. Etiology and management of liver injury in patients with COVID-19. World J Gastroenterol. 2020;26:4753-4762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 58] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (2)] |
| 50. | Robledinos-Antón N, Fernández-Ginés R, Manda G, Cuadrado A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxid Med Cell Longev. 2019;2019:9372182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 394] [Article Influence: 65.7] [Reference Citation Analysis (0)] |
| 51. | Yang H, Zheng Y, Li TW, Peng H, Fernandez-Ramos D, Martínez-Chantar ML, Rojas AL, Mato JM, Lu SC. Methionine adenosyltransferase 2B, HuR, and sirtuin 1 protein cross-talk impacts on the effect of resveratrol on apoptosis and growth in liver cancer cells. J Biol Chem. 2013;288:23161-23170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 52. | McCord JM, Hybertson BM, Cota-Gomez A, Geraci KP, Gao B. Nrf2 Activator PB125® as a Potential Therapeutic Agent against COVID-19. Antioxidants (Basel). 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 53. | Rana AK, Rahmatkar SN, Kumar A, Singh D. Glycogen synthase kinase-3: A putative target to combat severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. Cytokine Growth Factor Rev. 2021;58:92-101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 54. | Yu C, Lei Q, Li W, Wang X, Liu W. Epidemiological and clinical characteristics of 1663 hospitalized patients infected with COVID-19 in Wuhan, China: a single-center experience. J Infect Public Health. 2020;13:1202-1209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 55. | Jin X, Lian JS, Hu JH, Gao J, Zheng L, Zhang YM, Hao SR, Jia HY, Cai H, Zhang XL, Yu GD, Xu KJ, Wang XY, Gu JQ, Zhang SY, Ye CY, Jin CL, Lu YF, Yu X, Yu XP, Huang JR, Xu KL, Ni Q, Yu CB, Zhu B, Li YT, Liu J, Zhao H, Zhang X, Yu L, Guo YZ, Su JW, Tao JJ, Lang GJ, Wu XX, Wu WR, Qv TT, Xiang DR, Yi P, Shi D, Chen Y, Ren Y, Qiu YQ, Li LJ, Sheng J, Yang Y. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002-1009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 944] [Cited by in RCA: 871] [Article Influence: 174.2] [Reference Citation Analysis (0)] |
| 56. | Lian J, Jin X, Hao S, Jia H, Cai H, Zhang X, Hu J, Zheng L, Wang X, Zhang S, Ye C, Jin C, Yu G, Gu J, Lu Y, Yu X, Xiang D, Li L, Liang T, Sheng J, Yang Y. Epidemiological, clinical, and virological characteristics of 465 hospitalized cases of coronavirus disease 2019 (COVID-19) from Zhejiang province in China. Influenza Other Respir Viruses. 2020;14:564-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 57. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30123] [Article Influence: 6024.6] [Reference Citation Analysis (3)] |
| 58. | Angus DC, Derde L, Al-Beidh F, Annane D, Arabi Y, Beane A, van Bentum-Puijk W, Berry L, Bhimani Z, Bonten M, Bradbury C, Brunkhorst F, Buxton M, Buzgau A, Cheng AC, de Jong M, Detry M, Estcourt L, Fitzgerald M, Goossens H, Green C, Haniffa R, Higgins AM, Horvat C, Hullegie SJ, Kruger P, Lamontagne F, Lawler PR, Linstrum K, Litton E, Lorenzi E, Marshall J, McAuley D, McGlothin A, McGuinness S, McVerry B, Montgomery S, Mouncey P, Murthy S, Nichol A, Parke R, Parker J, Rowan K, Sanil A, Santos M, Saunders C, Seymour C, Turner A, van de Veerdonk F, Venkatesh B, Zarychanski R, Berry S, Lewis RJ, McArthur C, Webb SA, Gordon AC; Writing Committee for the REMAP-CAP Investigators; Angus D, Cheng A, De Jong M, Gordon A, Lawler P, Webb S, Campbell L, Forbes A, Gattas D, Heritier S, Higgins L, Peake S, Presneill J, Seppelt I, Trapani T, Young P, Bagshaw S, Daneman N, Ferguson N, Misak C, Hullegie S, Pletz M, Rohde G, Alexander B, Basile K, Girard T, Huang D, Vates J, Beasley R, Fowler R, McGloughlin S, Morpeth S, Paterson D, Uyeki T, Baillie K, Duffy E, Hills T, Orr K, Patanwala A, Tong S, Netea M, Bihari S, Carrier M, Fergusson D, Goligher E, Haidar G, Hunt B, Kumar A, Laffan M, Lawless P, Lother S, McCallum P, Middeldopr S, McQuilten Z, Neal M, Pasi J, Schutgens R, Stanworth S, Turgeon A, Weissman A, Adhikari N, Anstey M, Brant E, de Man A, Lamonagne F, Masse MH, Udy A, Arnold D, Begin P, Charlewood R, Chasse M, Coyne M, Cooper J, Daly J, Gosbell I, Harvala-Simmonds H, MacLennan S, Menon D, McDyer J, Pridee N, Roberts D, Shankar-Hari M, Thomas H, Tinmouth A, Triulzi D, Walsh T, Wood E, Calfee C, O’Kane C, Shyamsundar M, Sinha P, Thompson T, Young I, Hodgson C, Laffey J, Orford N, Neto A, Lewis R, McGlothlin A, Miller E, Singh V, Zammit C, van Bentum Puijk W, Bouwman W, Mangindaan Y, Parker L, Peters S, Rietveld I, Raymakers K, Ganpat R, Brillinger N, Markgraf R, Ainscough K, Brickell K, Anjum A, Lane JB, Richards-Belle A, Saull M, Wiley D, Bion J, Connor J, Gates S, Manax V, van der Poll T, Reynolds J, van Beurden M, Effelaar E, Schotsman J, Boyd C, Harland C, Shearer A, Wren J, Clermont G, Garrard W, Kalchthaler K, King A, Ricketts D, Malakoutis S, Marroquin O, Music E, Quinn K, Cate H, Pearson K, Collins J, Hanson J, Williams P, Jackson S, Asghar A, Dyas S, Sutu M, Murphy S, Williamson D, Mguni N, Potter A, Porter D, Goodwin J, Rook C, Harrison S, Williams H, Campbell H, Lomme K, Williamson J, Sheffield J, van’t Hoff W, McCracken P, Young M, Board J, Mart E, Knott C, Smith J, Boschert C, Affleck J, Ramanan M, D’Souza R, Pateman K, Shakih A, Cheung W, Kol M, Wong H, Shah A, Wagh A, Simpson J, Duke G, Chan P, Cartner B, Hunter S, Laver R, Shrestha T, Regli A, Pellicano A, McCullough J, Tallott M, Kumar N, Panwar R, Brinkerhoff G, Koppen C, Cazzola F, Brain M, Mineall S, Fischer R, Biradar V, Soar N, White H, Estensen K, Morrison L, Cooper M, Health M, Shehabi Y, Al-Bassam W, Hulley A, Whitehead C, Lowrey J, Gresha R, Walsham J, Meyer J, Harward M, Venz E, Kurenda C, Smith K, Smith M, Garcia R, Barge D, Byrne D, Byrne K, Driscoll A, Fortune L, Janin P, Yarad E, Hammond N, Bass F, Ashelford A, Waterson S, Wedd S, McNamara R, Buhr H, Coles J, Schweikert S, Wibrow B, Rauniyar R, Myers E, Fysh E, Dawda A, Mevavala B, Ferrier J, Nair P, Buscher H, Reynolds C, Santamaria J, Barbazza L, Homes J, Smith R, Murray L, Brailsford J, Forbes L, Maguire T, Mariappa V, Simpson S, Maiden M, Bone A, Horton M, Salerno T, Sterba M, Geng W, Depuydt P, De Waele J, De Bus L, Fierens J, Bracke S, Reeve B, Dechert W, Chassé M, Carrier FM, Boumahni D, Benettaib F, Ghamraoui A, Bellemare D, Cloutier È, Francoeur C, D’Aragon F, Carbonneau E, Leblond J, Vazquez-Grande G, Marten N, Wilson M, Albert M, Serri K, Cavayas A, Duplaix M, Williams V, Rochwerg B, Karachi T, Oczkowski S, Centofanti J, Millen T, Duan E, Tsang J, Patterson L, English S, Watpool I, Porteous R, Miezitis S, McIntyre L, Brochard L, Burns K, Sandhu G, Khalid I, Binnie A, Powell E, McMillan A, Luk T, Aref N, Andric Z, Cviljevic S, Đimoti R, Zapalac M, Mirković G, Baršić B, Kutleša M, Kotarski V, Vujaklija Brajković A, Babel J, Sever H, Dragija L, Kušan I, Vaara S, Pettilä L, Heinonen J, Kuitunen A, Karlsson S, Vahtera A, Kiiski H, Ristimäki S, Azaiz A, Charron C, Godement M, Geri G, Vieillard-Baron A, Pourcine F, Monchi M, Luis D, Mercier R, Sagnier A, Verrier N, Caplin C, Siami S, Aparicio C, Vautier S, Jeblaoui A, Fartoukh M, Courtin L, Labbe V, Leparco C, Muller G, Nay MA, Kamel T, Benzekri D, Jacquier S, Mercier E, Chartier D, Salmon C, Dequin P, Schneider F, Morel G, L’Hotellier S, Badie J, Berdaguer FD, Malfroy S, Mezher C, Bourgoin C, Megarbane B, Voicu S, Deye N, Malissin I, Sutterlin L, Guitton C, Darreau C, Landais M, Chudeau N, Robert A, Moine P, Heming N, Maxime V, Bossard I, Nicholier TB, Colin G, Zinzoni V, Maquigneau N, Finn A, Kreß G, Hoff U, Friedrich Hinrichs C, Nee J, Hagel S, Ankert J, Kolanos S, Bloos F, Petros S, Pasieka B, Kunz K, Appelt P, Schütze B, Kluge S, Nierhaus A, Jarczak D, Roedl K, Weismann D, Frey A, Klinikum Neukölln V, Reill L, Distler M, Maselli A, Bélteczki J, Magyar I, Fazekas Á, Kovács S, Szőke V, Szigligeti G, Leszkoven J, Collins D, Breen P, Frohlich S, Whelan R, McNicholas B, Scully M, Casey S, Kernan M, Doran P, O’Dywer M, Smyth M, Hayes L, Hoiting O, Peters M, Rengers E, Evers M, Prinssen A, Bosch Ziekenhuis J, Simons K, Rozendaal W, Polderman F, de Jager P, Moviat M, Paling A, Salet A, Rademaker E, Peters AL, de Jonge E, Wigbers J, Guilder E, Butler M, Cowdrey KA, Newby L, Chen Y, Simmonds C, McConnochie R, Ritzema Carter J, Henderson S, Van Der Heyden K, Mehrtens J, Williams T, Kazemi A, Song R, Lai V, Girijadevi D, Everitt R, Russell R, Hacking D, Buehner U, Williams E, Browne T, Grimwade K, Goodson J, Keet O, Callender O, Martynoga R, Trask K, Butler A, Schischka L, Young C, Lesona E, Olatunji S, Robertson Y, José N, Amaro dos Santos Catorze T, de Lima Pereira TNA, Neves Pessoa LM, Castro Ferreira RM, Pereira Sousa Bastos JM, Aysel Florescu S, Stanciu D, Zaharia MF, Kosa AG, Codreanu D, Marabi Y, Al Qasim E, Moneer Hagazy M, Al Swaidan L, Arishi H, Muñoz-Bermúdez R, Marin-Corral J, Salazar Degracia A, Parrilla Gómez F, Mateo López MI, Rodriguez Fernandez J, Cárcel Fernández S, Carmona Flores R, León López R, de la Fuente Martos C, Allan A, Polgarova P, Farahi N, McWilliam S, Hawcutt D, Rad L, O’Malley L, Whitbread J, Kelsall O, Wild L, Thrush J, Wood H, Austin K, Donnelly A, Kelly M, O’Kane S, McClintock D, Warnock M, Johnston P, Gallagher LJ, Mc Goldrick C, Mc Master M, Strzelecka A, Jha R, Kalogirou M, Ellis C, Krishnamurthy V, Deelchand V, Silversides J, McGuigan P, Ward K, O’Neill A, Finn S, Phillips B, Mullan D, Oritz-Ruiz de Gordoa L, Thomas M, Sweet K, Grimmer L, Johnson R, Pinnell J, Robinson M, Gledhill L, Wood T, Morgan M, Cole J, Hill H, Davies M, Antcliffe D, Templeton M, Rojo R, Coghlan P, Smee J, Mackay E, Cort J, Whileman A, Spencer T, Spittle N, Kasipandian V, Patel A, Allibone S, Genetu RM, Ramali M, Ghosh A, Bamford P, London E, Cawley K, Faulkner M, Jeffrey H, Smith T, Brewer C, Gregory J, Limb J, Cowton A, O’Brien J, Nikitas N, Wells C, Lankester L, Pulletz M, Birch J, Wiseman S, Horton S, Alegria A, Turki S, Elsefi T, Crisp N, Allen L, McCullagh I, Robinson P, Hays C, Babio-Galan M, Stevenson H, Khare D, Pinder M, Selvamoni S, Gopinath A, Pugh R, Menzies D, Mackay C, Allan E, Davies G, Puxty K, McCue C, Cathcart S, Hickey N, Ireland J, Yusuff H, Isgro G, Brightling C, Bourne M, Craner M, Watters M, Prout R, Davies L, Pegler S, Kyeremeh L, Arbane G, Wilson K, Gomm L, Francia F, Brett S, Sousa Arias S, Elin Hall R, Budd J, Small C, Collins E, Henning J, Bonner S, Hugill K, Cirstea E, Wilkinson D, Karlikowski M, Sutherland H, Wilhelmsen E, Woods J, North J, Sundaran D, Hollos L, Coburn S, Walsh J, Turns M, Hopkins P, Noble H, Depante MT, Clarey E, Laha S, Verlander M, Williams A, Huckle A, Hall A, Cooke J, Gardiner-Hill C, Maloney C, Qureshi H, Flint N, Nicholson S, Southin S, Nicholson A, Borgatta B, Turner-Bone I, Reddy A, Wilding L, Chamara Warnapura L, Agno Sathianathan R, Golden D, Hart C, Jones J, Bannard-Smith J, Henry J, Birchall K, Pomeroy F, Quayle R, Makowski A, Misztal B, Ahmed I, KyereDiabour T, Naiker K, Stewart R, Mwaura E, Mew L, Wren L, Willams F, Innes R, Doble P, Hutter J, Shovelton C, Plumb B, Szakmany T, Hamlyn V, Hawkins N, Lewis S, Dell A, Gopal S, Ganguly S, Smallwood A, Harris N, Metherell S, Lazaro JM, Newman T, Fletcher S, Nortje J, Fottrell-Gould D, Randell G, Zaman M, Elmahi E, Jones A, Hall K, Mills G, Ryalls K, Bowler H, Sall J, Bourne R, Borrill Z, Duncan T, Lamb T, Shaw J, Fox C, Moreno Cuesta J, Xavier K, Purohit D, Elhassan M, Bakthavatsalam D, Rowland M, Hutton P, Bashyal A, Davidson N, Hird C, Chhablani M, Phalod G, Kirkby A, Archer S, Netherton K, Reschreiter H, Camsooksai J, Patch S, Jenkins S, Pogson D, Rose S, Daly Z, Brimfield L, Claridge H, Parekh D, Bergin C, Bates M, Dasgin J, McGhee C, Sim M, Hay SK, Phull MK, Zaidi A, Pogreban T, Rosaroso LP, Harvey D, Lowe B, Meredith M, Ryan L, Hormis A, Walker R, Collier D, Kimpton S, Oakley S, Rooney K, Rodden N, Hughes E, Thomson N, McGlynn D, Walden A, Jacques N, Coles H, Tilney E, Vowell E, Schuster-Bruce M, Pitts S, Miln R, Purandare L, Vamplew L, Spivey M, Bean S, Burt K, Moore L, Day C, Gibson C, Gordon E, Zitter L, Keenan S, Baker E, Cherian S, Cutler S, Roynon-Reed A, Harrington K, Raithatha A, Bauchmuller K, Ahmad N, Grecu I, Trodd D, Martin J, Wrey Brown C, Arias AM, Craven T, Hope D, Singleton J, Clark S, Rae N, Welters I, Hamilton DO, Williams K, Waugh V, Shaw D, Puthucheary Z, Martin T, Santos F, Uddin R, Somerville A, Tatham KC, Jhanji S, Black E, Dela Rosa A, Howle R, Tully R, Drummond A, Dearden J, Philbin J, Munt S, Vuylsteke A, Chan C, Victor S, Matsa R, Gellamucho M, Creagh-Brown B, Tooley J, Montague L, De Beaux F, Bullman L, Kersiake I, Demetriou C, Mitchard S, Ramos L, White K, Donnison P, Johns M, Casey R, Mattocks L, Salisbury S, Dark P, Claxton A, McLachlan D, Slevin K, Lee S, Hulme J, Joseph S, Kinney F, Senya HJ, Oborska A, Kayani A, Hadebe B, Orath Prabakaran R, Nichols L, Worner R, Faulkner B, Gendall E, Hayes K, Hamilton-Davies C, Mfuko C, Abbass H, Mandadapu V, Leaver S, Forton D, Patel K, Paramasivam E, Powell M, Gould R, Wilby E, Howcroft C, Banach D, Fernández de Pinedo Artaraz Z, Cabreros L, White I, Croft M, Holland N, Pereira R, Zaki A, Johnson D, Jackson M, Garrard H, Juhaz V, Roy A, Rostron A, Woods L, Cornell S, Pillai S, Harford R, Rees T, Ivatt H, Sundara Raman A, Davey M, Lee K, Barber R, Chablani M, Brohi F, Jagannathan V, Clark M, Purvis S, Wetherill B, Dushianthan A, Cusack R, de Courcy-Golder K, Smith S, Attwood B, Parsons P, Page V, Zhao XB, Oza D, Rhodes J, Anderson T, Morris S, Xia Le Tai C, Thomas A, Keen A, Digby S, Cowley N, Southern D, Reddy H, Campbell A, Watkins C, Smuts S, Touma O, Barnes N, Alexander P, Felton T, Ferguson S, Sellers K, Bradley-Potts J, Yates D, Birkinshaw I, Kell K, Marshall N, Carr-Knott L, Summers C. Effect of Hydrocortisone on Mortality and Organ Support in Patients With Severe COVID-19: The REMAP-CAP COVID-19 Corticosteroid Domain Randomized Clinical Trial. JAMA. 2020;324:1317-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 636] [Cited by in RCA: 621] [Article Influence: 124.2] [Reference Citation Analysis (0)] |
| 59. | Tóbon-Velasco JC, Cuevas E, Torres-Ramos MA. Receptor for AGEs (RAGE) as mediator of NF-kB pathway activation in neuroinflammation and oxidative stress. CNS Neurol Disord Drug Targets. 2014;13:1615-1626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 235] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 60. | Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-kappa B. Annu Rev Cell Biol. 1994;10:405-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1634] [Cited by in RCA: 1655] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 61. | Hariharan A, Hakeem AR, Radhakrishnan S, Reddy MS, Rela M. The Role and Therapeutic Potential of NF-kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology. 2021;29:91-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 211] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 62. | Sivandzade F, Prasad S, Bhalerao A, Cucullo L. NRF2 and NF-қB interplay in cerebrovascular and neurodegenerative disorders: Molecular mechanisms and possible therapeutic approaches. Redox Biol. 2019;21:101059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 474] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 63. | Bellezza I, Giambanco I, Minelli A, Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim Biophys Acta Mol Cell Res. 2018;1865:721-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 1268] [Article Influence: 181.1] [Reference Citation Analysis (0)] |
| 64. | Sies H, Berndt C, Jones DP. Oxidative Stress. Annu Rev Biochem. 2017;86:715-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1456] [Cited by in RCA: 2270] [Article Influence: 283.8] [Reference Citation Analysis (0)] |
| 65. | Wardyn JD, Ponsford AH, Sanderson CM. Dissecting molecular cross-talk between Nrf2 and NF-κB response pathways. Biochem Soc Trans. 2015;43:621-626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 880] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 66. | Zelová H, Hošek J. TNF-α signalling and inflammation: interactions between old acquaintances. Inflamm Res. 2013;62:641-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 598] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 67. | Leist M, Gantner F, Bohlinger I, Tiegs G, Germann PG, Wendel A. Tumor necrosis factor-induced hepatocyte apoptosis precedes liver failure in experimental murine shock models. Am J Pathol. 1995;146:1220-1234. [PubMed] |
| 68. | Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, Itoh N, Suda T, Nagata S. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1429] [Cited by in RCA: 1429] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 69. | Ren J, Su D, Li L, Cai H, Zhang M, Zhai J, Li M, Wu X, Hu K. Anti-inflammatory effects of Aureusidin in LPS-stimulated RAW264.7 macrophages via suppressing NF-κB and activating ROS- and MAPKs-dependent Nrf2/HO-1 signaling pathways. Toxicol Appl Pharmacol. 2020;387:114846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 70. | Liu GH, Qu J, Shen X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim Biophys Acta. 2008;1783:713-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 548] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 71. | Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol. 2009;29:2658-2672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 335] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 72. | Wang B, Zhu X, Kim Y, Li J, Huang S, Saleem S, Li RC, Xu Y, Dore S, Cao W. Histone deacetylase inhibition activates transcription factor Nrf2 and protects against cerebral ischemic damage. Free Radic Biol Med. 2012;52:928-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 176] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 73. | Yu M, Li H, Liu Q, Liu F, Tang L, Li C, Yuan Y, Zhan Y, Xu W, Li W, Chen H, Ge C, Wang J, Yang X. Nuclear factor p65 interacts with Keap1 to repress the Nrf2-ARE pathway. Cell Signal. 2011;23:883-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 252] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 74. | Probst BL, McCauley L, Trevino I, Wigley WC, Ferguson DA. Cancer Cell Growth Is Differentially Affected by Constitutive Activation of NRF2 by KEAP1 Deletion and Pharmacological Activation of NRF2 by the Synthetic Triterpenoid, RTA 405. PLoS One. 2015;10:e0135257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 75. | Miripour ZS, Sarrami-Forooshani R, Sanati H, Makarem J, Taheri MS, Shojaeian F, Eskafi AH, Abbasvandi F, Namdar N, Ghafari H, Aghaee P, Zandi A, Faramarzpour M, Hoseinyazdi M, Tayebi M, Abdolahad M. Real-time diagnosis of reactive oxygen species (ROS) in fresh sputum by electrochemical tracing; correlation between COVID-19 and viral-induced ROS in lung/respiratory epithelium during this pandemic. Biosens Bioelectron. 2020;165:112435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 76. | Cecchini R, Cecchini AL. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med Hypotheses. 2020;143:110102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 258] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 77. | Hirano T, Murakami M. COVID-19: A New Virus, but a Familiar Receptor and Cytokine Release Syndrome. Immunity. 2020;52:731-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 436] [Cited by in RCA: 584] [Article Influence: 116.8] [Reference Citation Analysis (0)] |
| 78. | Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875-879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2710] [Cited by in RCA: 2645] [Article Influence: 132.3] [Reference Citation Analysis (0)] |
| 79. | Eguchi S, Kawai T, Scalia R, Rizzo V. Understanding Angiotensin II Type 1 Receptor Signaling in Vascular Pathophysiology. Hypertension. 2018;71:804-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 80. | Murakami M, Kamimura D, Hirano T. Pleiotropy and Specificity: Insights from the Interleukin 6 Family of Cytokines. Immunity. 2019;50:812-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 358] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 81. | Cichoż-Lach H, Michalak A. Oxidative stress as a crucial factor in liver diseases. World J Gastroenterol. 2014;20:8082-8091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 910] [Cited by in RCA: 794] [Article Influence: 72.2] [Reference Citation Analysis (8)] |
| 82. | Yu HH, Qiu YX, Li B, Peng CY, Zeng R, Wang W. Kadsura heteroclita stem ethanol extract protects against carbon tetrachloride-induced liver injury in mice via suppression of oxidative stress, inflammation, and apoptosis. J Ethnopharmacol. 2021;267:113496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 83. | Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, Misra V, Biswal S, Yamamoto M, Kensler TW. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal. 2010;3:ra52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 180] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 84. | Duarte TL, Caldas C, Santos AG, Silva-Gomes S, Santos-Gonçalves A, Martins MJ, Porto G, Lopes JM. Genetic disruption of NRF2 promotes the development of necroinflammation and liver fibrosis in a mouse model of HFE-hereditary hemochromatosis. Redox Biol. 2017;11:157-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 85. | Wu KC, Liu JJ, Klaassen CD. Nrf2 activation prevents cadmium-induced acute liver injury. Toxicol Appl Pharmacol. 2012;263:14-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 86. | Luedde T, Schwabe RF. NF-κB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1094] [Cited by in RCA: 1089] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 87. | Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS; China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708-1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19202] [Cited by in RCA: 18878] [Article Influence: 3775.6] [Reference Citation Analysis (7)] |
| 88. | Chen LY, Chu HK, Bai T, Tu SJ, Wei Y, Li ZL, Hu LL, Zhu R, Zhang L, Han CQ, Xiao L, He Q, Song J, Liu WH, Zhu QJ, Chen H, Yang L, Hou XH. Liver damage at admission is an independent prognostic factor for COVID-19. J Dig Dis. 2020;21:512-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 89. | Xie H, Zhao J, Lian N, Lin S, Xie Q, Zhuo H. Clinical characteristics of non-ICU hospitalized patients with coronavirus disease 2019 and liver injury: A retrospective study. Liver Int. 2020;40:1321-1326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 213] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 90. | Wang Z, Ye D, Wang M, Zhao M, Li D, Ye J, Liu J, Xu Y, Zhang J, Pan W, Liu M, Luo Z, Wan J. Clinical Features of COVID-19 Patients with Different Outcomes in Wuhan: A Retrospective Observational Study. Biomed Res Int. 2020;2020:2138387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 91. | Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14869] [Cited by in RCA: 12977] [Article Influence: 2595.4] [Reference Citation Analysis (1)] |
| 92. | Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. 2009;30:42-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1643] [Cited by in RCA: 1504] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 93. | Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196-5203. [PubMed] |
| 94. | Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 947] [Cited by in RCA: 1108] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 95. | Wu KC, Cui JY, Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci. 2011;123:590-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 299] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 96. | Holmström KM, Kostov RV, Dinkova-Kostova AT. The multifaceted role of Nrf2 in mitochondrial function. Curr Opin Toxicol. 2016;1:80-91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 261] [Cited by in RCA: 288] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 97. | Hu Q, Ren J, Li G, Wu J, Wu X, Wang G, Gu G, Ren H, Hong Z, Li J. The mitochondrially targeted antioxidant MitoQ protects the intestinal barrier by ameliorating mitochondrial DNA damage via the Nrf2/ARE signaling pathway. Cell Death Dis. 2018;9:403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 142] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 98. | Codo AC, Davanzo GG, Monteiro LB, de Souza GF, Muraro SP, Virgilio-da-Silva JV, Prodonoff JS, Carregari VC, de Biagi Junior CAO, Crunfli F, Jimenez Restrepo JL, Vendramini PH, Reis-de-Oliveira G, Bispo Dos Santos K, Toledo-Teixeira DA, Parise PL, Martini MC, Marques RE, Carmo HR, Borin A, Coimbra LD, Boldrini VO, Brunetti NS, Vieira AS, Mansour E, Ulaf RG, Bernardes AF, Nunes TA, Ribeiro LC, Palma AC, Agrela MV, Moretti ML, Sposito AC, Pereira FB, Velloso LA, Vinolo MAR, Damasio A, Proença-Módena JL, Carvalho RF, Mori MA, Martins-de-Souza D, Nakaya HI, Farias AS, Moraes-Vieira PM. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab 2020; 32: 437-446. e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 365] [Cited by in RCA: 536] [Article Influence: 107.2] [Reference Citation Analysis (0)] |
| 99. | Martínez-Sánchez G, Schwartz A, Donna VD. Potential Cytoprotective Activity of Ozone Therapy in SARS-CoV-2/COVID-19. Antioxidants (Basel). 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 100. | Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620-2629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2835] [Cited by in RCA: 3419] [Article Influence: 683.8] [Reference Citation Analysis (0)] |
| 101. | Iddir M, Brito A, Dingeo G, Fernandez Del Campo SS, Samouda H, La Frano MR, Bohn T. Strengthening the Immune System and Reducing Inflammation and Oxidative Stress through Diet and Nutrition: Considerations during the COVID-19 Crisis. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 485] [Cited by in RCA: 457] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 102. | Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453-R462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3416] [Cited by in RCA: 4598] [Article Influence: 459.8] [Reference Citation Analysis (0)] |
| 103. | Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol Sin. 2020;35:266-271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 512] [Cited by in RCA: 504] [Article Influence: 100.8] [Reference Citation Analysis (0)] |
| 104. | Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 655] [Article Influence: 131.0] [Reference Citation Analysis (0)] |
| 105. | Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, Tian DS. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762-768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2495] [Cited by in RCA: 3341] [Article Influence: 668.2] [Reference Citation Analysis (0)] |
| 106. | Zhang W, Zhao Y, Zhang F, Wang Q, Li T, Liu Z, Wang J, Qin Y, Zhang X, Yan X, Zeng X, Zhang S. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin Immunol. 2020;214:108393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 812] [Cited by in RCA: 861] [Article Influence: 172.2] [Reference Citation Analysis (0)] |
| 107. | Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1000] [Cited by in RCA: 938] [Article Influence: 187.6] [Reference Citation Analysis (0)] |
| 108. | Bellosta R, Luzzani L, Natalini G, Pegorer MA, Attisani L, Cossu LG, Ferrandina C, Fossati A, Conti E, Bush RL, Piffaretti G. Acute limb ischemia in patients with COVID-19 pneumonia. J Vasc Surg. 2020;72:1864-1872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 297] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 109. | Low SW, Swanson KL, McCain JD, Sen A, Kawashima A, Pasha SF. Gastric ischemia and portal vein thrombosis in a COVID-19-infected patient. Endoscopy. 2020;52:E465-E466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 110. | Shafi AMA, Shaikh SA, Shirke MM, Iddawela S, Harky A. Cardiac manifestations in COVID-19 patients-A systematic review. J Card Surg. 2020;35:1988-2008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 111. | Garg RK. Spectrum of Neurological Manifestations in Covid-19: A Review. Neurol India. 2020;68:560-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 112. | Rossi M, Piagnerelli M, Van Meerhaeghe A, Zouaoui Boudjeltia K. Heme oxygenase-1 (HO-1) cytoprotective pathway: A potential treatment strategy against coronavirus disease 2019 (COVID-19)-induced cytokine storm syndrome. Med Hypotheses. 2020;144:110242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 113. | Calay D, Mason JC. The multifunctional role and therapeutic potential of HO-1 in the vascular endothelium. Antioxid Redox Signal. 2014;20:1789-1809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 114. | Dattilo M. The role of host defences in Covid 19 and treatments thereof. Mol Med. 2020;26:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 115. | Raghunath A, Sundarraj K, Nagarajan R, Arfuso F, Bian J, Kumar AP, Sethi G, Perumal E. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018;17:297-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 342] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 116. | Motohashi H, Katsuoka F, Miyoshi C, Uchimura Y, Saitoh H, Francastel C, Engel JD, Yamamoto M. MafG sumoylation is required for active transcriptional repression. Mol Cell Biol. 2006;26:4652-4663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 117. | Shavit JA, Motohashi H, Onodera K, Akasaka J, Yamamoto M, Engel JD. Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev. 1998;12:2164-2174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 87] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 118. | Katsuoka F, Otsuki A, Takahashi M, Ito S, Yamamoto M. Direct and Specific Functional Evaluation of the Nrf2 and MafG Heterodimer by Introducing a Tethered Dimer into Small Maf-Deficient Cells. Mol Cell Biol. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 119. | Xu D, Xu M, Jeong S, Qian Y, Wu H, Xia Q, Kong X. The Role of Nrf2 in Liver Disease: Novel Molecular Mechanisms and Therapeutic Approaches. Front Pharmacol. 2018;9:1428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 194] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 120. | Sun J, Fu J, Li L, Chen C, Wang H, Hou Y, Xu Y, Pi J. Nrf2 in alcoholic liver disease. Toxicol Appl Pharmacol. 2018;357:62-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 121. | Qin SJ, Wang BY, Li BR, Zheng K, Cai Y, Li RB, Zeng M, Xiao F, Xu XY. [Effect of c-myc gene silence on the expression of oncogenes and apoptotic genes in hepatocytes treated with PM(2).5]. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2020;38:657-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 122. | Wahlström T, Henriksson M. Mnt takes control as key regulator of the myc/max/mxd network. Adv Cancer Res. 2007;97:61-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 123. | Yang H, Li TW, Peng J, Tang X, Ko KS, Xia M, Aller MA. A mouse model of cholestasis-associated cholangiocarcinoma and transcription factors involved in progression. Gastroenterology 2011; 141: 378-388, 388.e1-388. e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 124. | Yang H, Li TW, Ko KS, Xia M, Lu SC. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mouse and human hepatocytes. Hepatology. 2009;49:860-870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 125. | Yang H, Liu T, Wang J, Li TW, Fan W, Peng H, Krishnan A, Gores GJ, Mato JM, Lu SC. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans(‡). Hepatology. 2016;64:439-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 126. | Yang H, Li TW, Zhou Y, Peng H, Liu T, Zandi E, Martínez-Chantar ML, Mato JM, Lu SC. Activation of a novel c-Myc-miR27-prohibitin 1 circuitry in cholestatic liver injury inhibits glutathione synthesis in mice. Antioxid Redox Signal. 2015;22:259-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 127. | Zeng Y, Lou G, Ren Y, Li T, Zhang X, Wang J, Huang Q. Network pharmacology-based analysis of Zukamu granules for the treatment of COVID-19. Eur J Integr Med. 2021;42:101282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 128. | Li Y, Lu L, Tu J, Zhang J, Xiong T, Fan W, Wang J, Li M, Chen Y, Steggerda J, Peng H, Li TWH, Zhou ZG, Mato JM, Seki E, Liu T, Yang H, Lu SC. Reciprocal Regulation Between Forkhead Box M1/NF-κB and Methionine Adenosyltransferase 1A Drives Liver Cancer. Hepatology. 2020;72:1682-1700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 129. | Phipps MM, Barraza LH, LaSota ED, Sobieszczyk ME, Pereira MR, Zheng EX, Fox AN, Zucker J, Verna EC. Acute Liver Injury in COVID-19: Prevalence and Association with Clinical Outcomes in a Large U.S. Cohort. Hepatology. 2020;72:807-817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 272] [Article Influence: 54.4] [Reference Citation Analysis (2)] |
| 130. | Mendizabal M, Piñero F, Ridruejo E, Anders M, Silveyra MD, Torre A, Montes P, Urzúa A, Pages J, Toro LG, Díaz J, Gonzalez Ballerga E, Miranda-Zazueta G, Peralta M, Gutiérrez I, Michelato D, Venturelli MG, Varón A, Vera-Pozo E, Tagle M, García M, Tassara A, Brutti J, Ruiz García S, Bustios C, Escajadillo N, Macias Y, Higuera-de la Tijera F, Gómez AJ, Dominguez A, Castillo-Barradas M, Contreras F, Scarpin A, Schinoni MI, Toledo C, Girala M, Mainardi V, Sanchez A, Bessone F, Rubinstein F, Silva MO. Prospective Latin American cohort evaluating outcomes of patients with COVID-19 and abnormal liver tests on admission. Ann Hepatol. 2021;21:100298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 131. | Hundt MA, Deng Y, Ciarleglio MM, Nathanson MH, Lim JK. Abnormal Liver Tests in COVID-19: A Retrospective Observational Cohort Study of 1,827 Patients in a Major U.S. Hospital Network. Hepatology. 2020;72:1169-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 197] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 132. | Yip TC, Lui GC, Wong VW, Chow VC, Ho TH, Li TC, Tse YK, Hui DS, Chan HL, Wong GL. Liver injury is independently associated with adverse clinical outcomes in patients with COVID-19. Gut. 2021;70:733-742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 133. | Cai Q, Huang D, Yu H, Zhu Z, Xia Z, Su Y, Li Z, Zhou G, Gou J, Qu J, Sun Y, Liu Y, He Q, Chen J, Liu L, Xu L. COVID-19: Abnormal liver function tests. J Hepatol. 2020;73:566-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 623] [Cited by in RCA: 661] [Article Influence: 132.2] [Reference Citation Analysis (0)] |
| 134. | Zhao D, Yao F, Wang L, Zheng L, Gao Y, Ye J, Guo F, Zhao H, Gao R. A Comparative Study on the Clinical Features of Coronavirus 2019 (COVID-19) Pneumonia With Other Pneumonias. Clin Infect Dis. 2020;71:756-761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 294] [Article Influence: 58.8] [Reference Citation Analysis (0)] |