Published online May 28, 2021. doi: 10.3748/wjg.v27.i20.2545
Peer-review started: January 28, 2021
First decision: February 24, 2021
Revised: March 27, 2021
Accepted: April 29, 2021
Article in press: April 29, 2021
Published online: May 28, 2021
Processing time: 111 Days and 12.8 Hours
Machine learning (ML)- and deep learning (DL)-based imaging modalities have exhibited the capacity to handle extremely high dimensional data for a number of computer vision tasks. While these approaches have been applied to numerous data types, this capacity can be especially leveraged by application on histopathological images, which capture cellular and structural features with their high-resolution, microscopic perspectives. Already, these methodologies have demonstrated promising performance in a variety of applications like disease classification, cancer grading, structure and cellular localizations, and prognostic predictions. A wide range of pathologies requiring histopathological evaluation exist in gastroenterology and hepatology, indicating these as disciplines highly targetable for integration of these technologies. Gastroenterologists have also already been primed to consider the impact of these algorithms, as development of real-time endoscopic video analysis software has been an active and popular field of research. This heightened clinical awareness will likely be important for future integration of these methods and to drive interdisciplinary collaborations on emerging studies. To provide an overview on the application of these methodologies for gastrointestinal and hepatological histopathological slides, this review will discuss general ML and DL concepts, introduce recent and emerging literature using these methods, and cover challenges moving forward to further advance the field.
Core Tip: Machine learning- and deep learning-based imaging approaches have been increasingly applied to histopathological slides and hold much potential in areas spanning diagnosis, disease grading and characterizations, academic research, and clinical decision support mechanisms. As these studies have entered into translational applications, tracking the current state of these methodologies and the clinical areas in which impact is most likely is of high importance. This review will thus provide a background of major concepts and terminologies while highlighting emerging literature regarding histopathological applications of these techniques and challenges and opportunities moving forward.
- Citation: Kobayashi S, Saltz JH, Yang VW. State of machine and deep learning in histopathological applications in digestive diseases. World J Gastroenterol 2021; 27(20): 2545-2575
- URL: https://www.wjgnet.com/1007-9327/full/v27/i20/2545.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i20.2545
The past decade has seen the growing popularity of machine learning (ML) and deep learning (DL) applications across numerous domains, and the medical field has been no exception. A search for DL publications in the domains of Medical Informatics, Sensing, Bioinformatics, Imaging, and Public Health shows a 5-fold to 6-fold increase in publication counts from 2010-2015[1], and this trend continues today. Applications of DL in healthcare have been particularly wide ranged, covering proteomics, genomics and expression data, electronic health records for patient characterizations, as well as image analysis for histopathology, magnetic resonance images (MRI) scans, positron emission topography scans, computerized topography (CT) scans, and endoscopy videos. DL image analysis methodologies have the potential to automate and speed up pathologists’ tasks with high accuracy and precision. Recent applications have also illustrated the capacity for DL methodologies to extract information from histopathological images unseen to the human eye, such as expression data. Importantly, these ML and DL image analysis applications have the benefit of requiring no additional sample collection from patients, as inputs are typically biomedical images already collected within the clinical workflow.
The majority of ML and DL focus in the gastroenterology and hepatology communities has been in endoscopy, and this is highlighted by the recent Breakthrough Device Designation granted by the United States Food and Drug Administration (FDA) for a DL-based endoscopic, real-time diagnostic software for gastric cancer[2]. However, the application of ML and DL methodologies on histopathological images is a blossoming field with significant potential for clinical impact. Imaging modalities like hematoxylin and eosin (HE)- or immunohistochemistry (IHC)-stained slides, unlike others such as CT, MRIs, or endoscopies, provide microscopic perspectives into tissue sections, allowing for the algorithms to utilize cellular and nuclear features like shape, size, color, and texture. Hence, the goal of this review is: (1) To cover major terminology and trainable tasks by ML and DL; (2) To briefly review the history of digital pathology; (3) To provide an overview of the current ML and DL histopathological imaging-based approaches in gastroenterology and hepatology; and (4) To discuss challenges and opportunities moving forward.
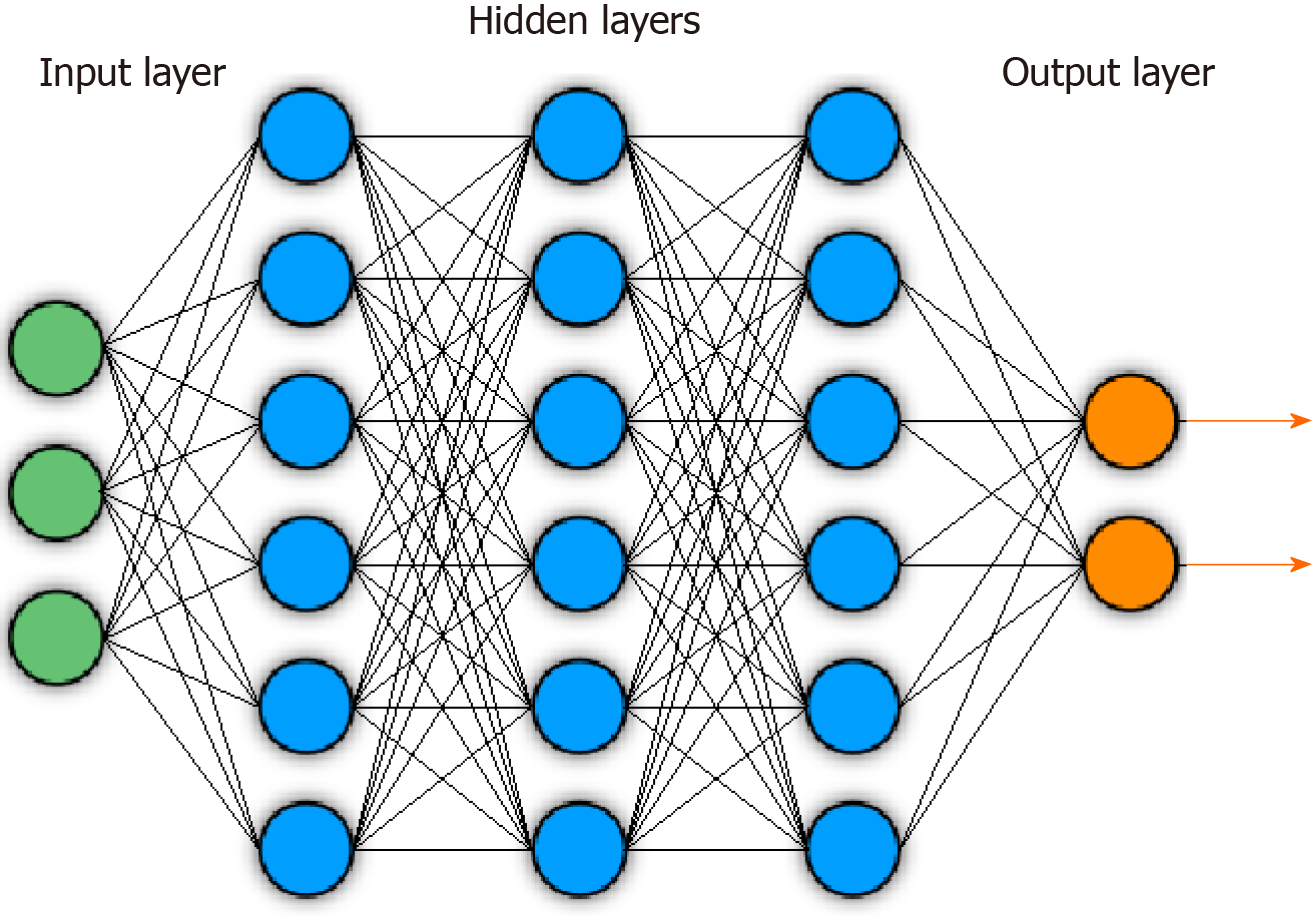
The FDA defines ML as “an artificial intelligence technique that can be used to design and train software algorithms to learn from and act on data”[3], where artificial intelligence is the development of computer systems capable of tasks deemed to require human intelligence. ML involves representation of samples or inputs by a fixed, user-determined set of features, then the application of a classifier that can distinguish and separate classes or types within the set of samples based off those selected features. There have historically been a range of popular ML techniques. Some examples include logic-based approaches, such as decision tree-based methods like Random Forest (RF) classifiers, statistic-based approaches, such as Bayesian networks or nearest neighbor algorithms, and support vector machines (SVMs), which aim to find optimal hyperplanes to separate classes on high dimensional data feature spaces[4]. DL represents a modern, specific subset of ML that uses deep neural network architectures for feature extraction and predictions. A schematic of a deep neural network is shown in Figure 1.
The general goal of a DL algorithm is to connect an input, such as an image, to a desired output. The hidden layers in the network act as feature extractors, and a final layer aggregates and utilizes these extracted features to generate the desired output. Specifically, deep neural networks have an input layer that is followed by successive hidden layers, each containing nodes. Starting at the input layer containing data, nodes in each hidden layer compute weighted sums from outputs in the previous layer. Within each node, these weighted sums are then passed into activation functions, which are critical for neural networks as they introduce non-linear transformations onto data. Each hidden layer thus introduces additional mathematical complexity in an effort to transform the input into new, informative representations within a new feature space. This process of defining representations for inputs in this new feature space is called embedding, and the representations are deemed informative when they can be effectively utilized by the final output layer in the network to carry out desired predictions. Some popular activation functions include the sigmoid, tanh, and Rectified Linear Unit functions.
To train the model, gradient descent, a popular optimization method, is utilized to minimize the “loss function”, which quantifies model performance. Specifically, gradient descent minimizes the “loss function” by adjusting algorithmic weights at layer nodes, which directly affect the weighted sum calculations. As a result, the embedding process is iteratively improved to gradually tune and train the model for the task at hand. More detail is provided in the Model Training and Gradient Descent subsection below.
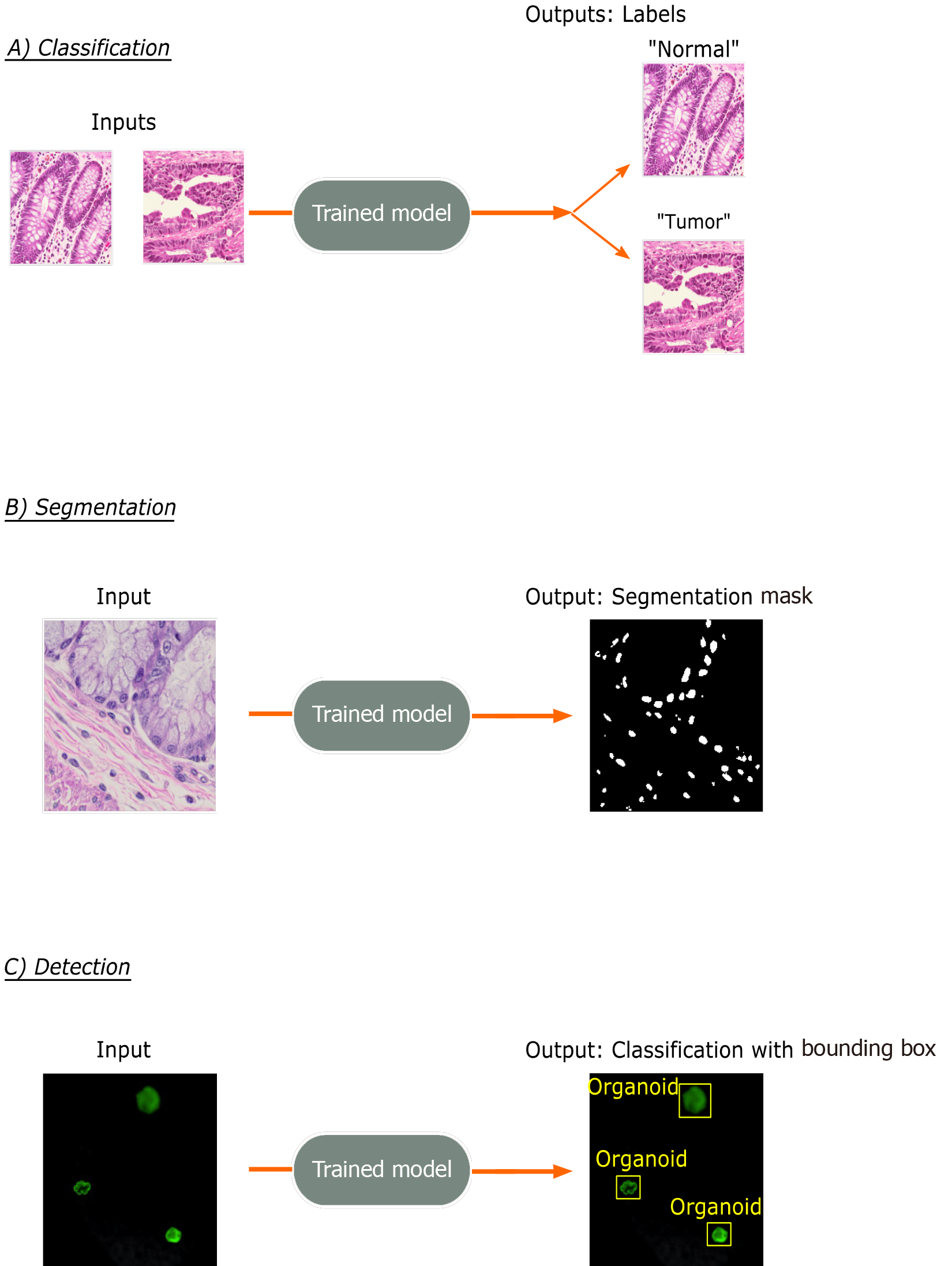
The three most common tasks for DL approaches in imaging applications is in classification, segmentation, and detection (Figure 2). Classification involves the prediction of a label for an input image, such as “Normal” vs “Cancer”. Segmentation involves the identification and localization of objects within a single image and outputs pixel-level designation of classes. Therefore, output segmentation maps will commonly have objects in the image colored or shaded based on their predicted class type. Lastly, detection, which is not a focus in this review, involves the identification of object classes in an image with a bounding box placed around it, such as in facial detection.
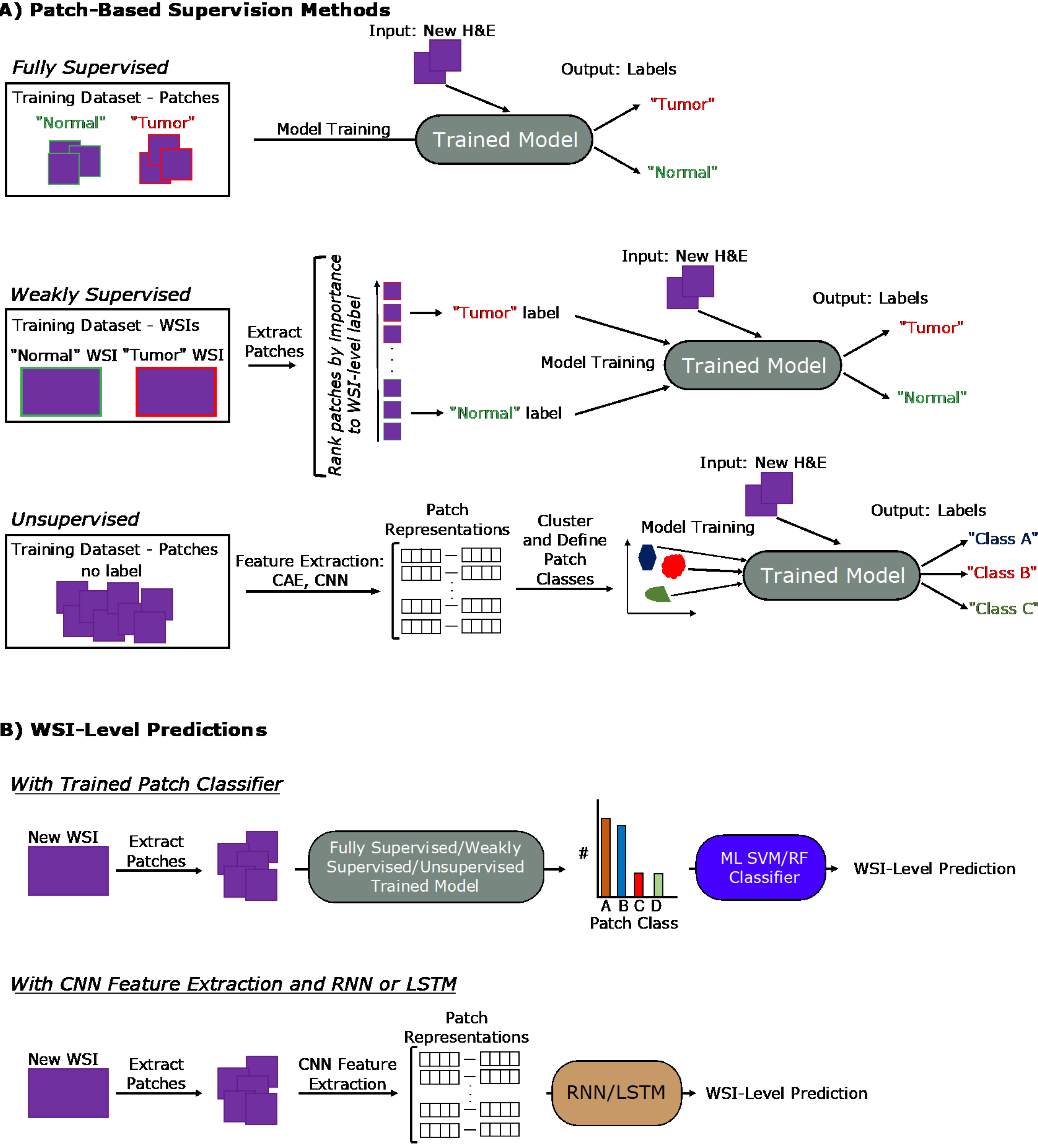
An important aspect of these studies is image annotation of correct class labels. Due to the tremendous file size of these high-resolution images upon digitization of histological slides into whole-slide images (WSIs), analysis over an entire WSI at once is computationally infeasible. As such, WSIs are typically broken up into equally sized patches and require training patch-level models. Labeling therefore can occur at the level of the WSI and at the level of the patches.
When labelling the classes of individual patches from a WSI, this can be done in a fully supervised, weakly supervised, or unsupervised manner. This section will cover these levels of supervision in the context of classification tasks. An overview of these approaches is covered in Figure 3A.
The fully supervised approach involves dataset-wide annotation at the patch-level. For example, this may involve a dataset of patches extracted from WSIs with pathologist-annotated labels for each patch as “cancer” or “normal”. Thus, cancer positive WSIs will likely contain both types of patch classes. Though training iterations, the model will eventually learn to correctly predict the patch labels from just input patch images.
Weakly supervised methods concern annotations provided only at the WSI-level. Extracted patches from these WSIs are then run through algorithms that determine which patches were most important for the WSI-level label. Some possible approaches involve expectation-maximization methods[5] or multiple instance learning (MIL)[6-8]. In the context of the cancer positive WSIs, this would mean that the model eventually learns that the cancerous patches were most responsible for the WSI label, while healthy patches were not.
Unsupervised methods require no annotations to generate patch classes. While WSI-level labels may be provided, they are not utilized in patch class definition. These methods typically involve feature extraction from patches across the training dataset, followed by clustering approaches to define patch classes. For example, analysis of a dataset of cancer positive and negative WSIs may reveal dataset-wide patch-level classes for tumor, healthy, and fibrosis, although not all types may be present in each WSI. These approaches identify implicit patterns in the data to define these classes.
Fully supervised approaches require a tedious annotation process to provide correct output labels. As such, weakly supervised and unsupervised approaches hold the additional benefit of circumventing this labeling process and may be important in increasing throughput by decreasing annotation-related load. Another possible solution that is an active field of research is the generation of synthetic data that is indistinguishable from real world data. An example of this is general adversarial networks (GANs). GANs create synthetic data then have a ‘discriminator’ module that attempts to determine whether generated data is synthetic or real. The worse this discriminator performs, the better the GAN is at generating synthetic data. As such, computational approaches that effectively generate synthetic data across different classes may help develop labeled training datasets at high throughput.
Although the above methods discuss patch-level classifications, many biomedical imaging studies require a prediction at the WSI-level, such as a diagnosis. Often, this patch level information is aggregated by an additional classifier, and an overview of general approaches is provided in Figure 3B. This can be a ML classifier, such as an SVM or RF classifier that takes as input the relative counts of the different patch types per WSI to output a WSI-level prediction. This classifier can also be in the form of neural networks like recurrent neural networks (RNNs) and long short-term memory (LSTM) networks, covered later in this section, that take in variable length sequences of patches or patch representations as inputs to generate a WSI-level prediction. As the WSI-level label is typically clinically or biologically-informed, such as a diagnosis, prognosis, or grading, this part of the process typically receives supervision.
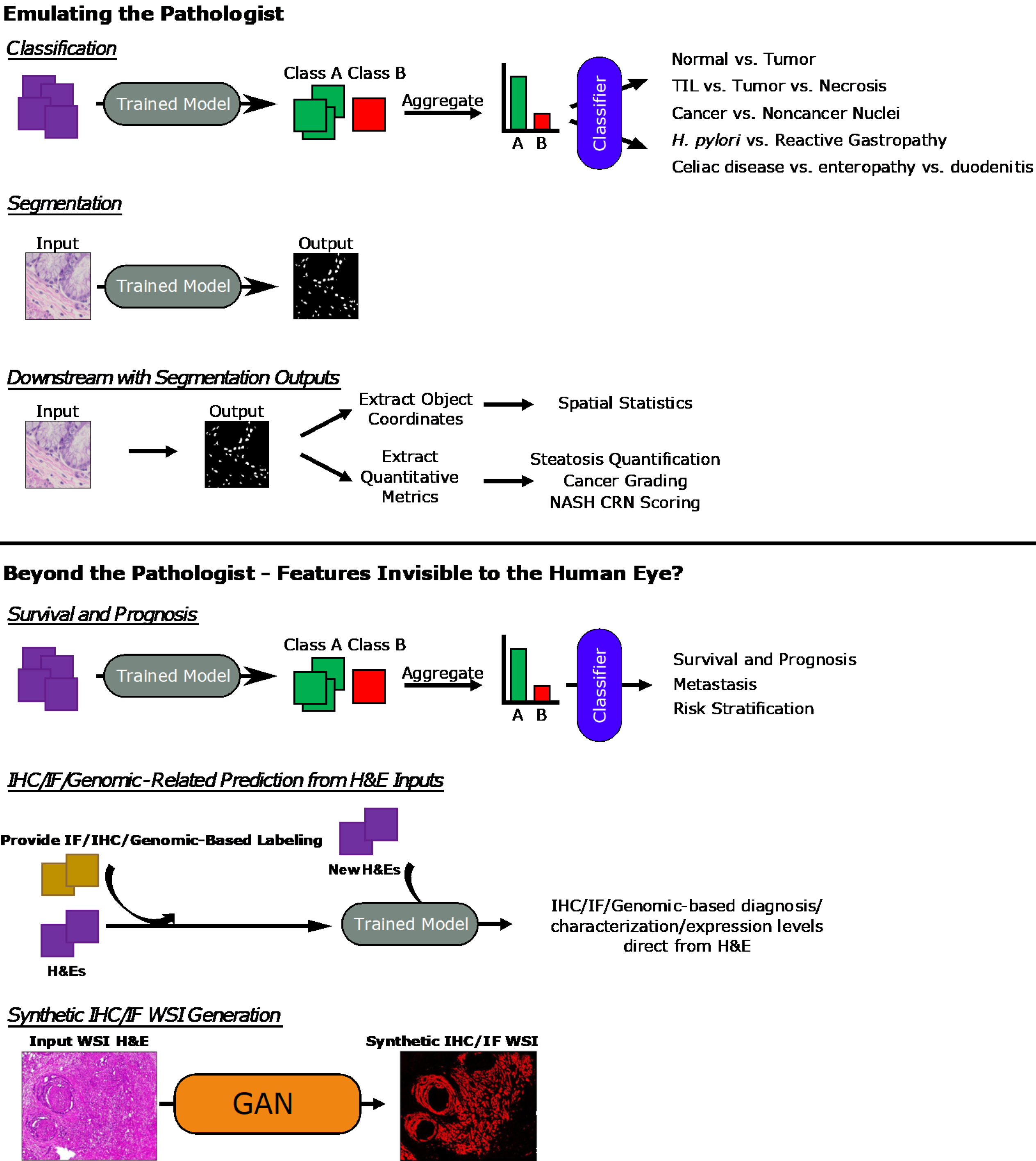
A variety of studies encompassing these approaches will be covered in this review in the two sections “Emulating the Pathologist” and “Beyond the Pathologist – Features Invisible to the Human Eye?”. A general diagrammatic overview of the approaches used in provided in Figure 4.
In practice, DL is performed in response to the quantification by a “loss function” of how well the neural network performed across the training dataset. As loss functions quantify model performance, they require knowledge of the correct output for each sample and are easiest to introduce with supervised learning concepts. For classification, the output involves patch-level labels, and, for segmentation, the output will be images of the same dimensions as the inputs, where object classes in the image are distinguished by pixel-level color designation of classes (e.g., shading cancerous areas with one color and shading healthy areas with another).
For classification, on each epoch, or iteration, of training, the algorithm attempts to predict the label of every image, then calculates, from the loss function, a scalar loss value that captures the degree to which the model-predicted output labels were different from the correct, user-designated output labels across the dataset. A lower loss value would therefore indicate better performance of the model in predicting the correct image labels. As segmentation involves correct, human-designated outputs at the pixel-level, the loss function quantifies correct class predictions across every pixel in a segmentation map output.
Gradient descent is an optimization method that iteratively moves in the direction of the steepest slope to approach minimums and is utilized in DL to minimize the loss functions. Gradient descent starts from the loss function and propagates through previous layers to the first, identifying the gradients for each algorithmic weight at every network layer node, then incrementally adjusting these weights according to the gradients. This process occurs every epoch with the overall goal of improving model performance by minimizing the loss function. These weights affect the non-linear mathematical operations performed at each hidden layer node and thus serves as a way for the network to tweak these operations to eventually determine a feature space and sample representations most effective for the task at hand. As opposed to ML techniques that depend on human-designated features for classification, the DL-based sample representations can be interpreted as an extraction of features deemed best and driven by the neural network’s gradient descent optimization with respect to the loss function. This therefore introduces the common “black box” issue, where the meanings of these final representations, or extracted features, cannot be defined due to the high amount of mathematical complexity introduced onto the input image tensor by each of the layers in the network.
Although gradient descent and loss functions are covered here, these are basic descriptions. For instance, the introduction of more complex loss functions that incorporate different learning constraints, study of approaches to take in multimodal inputs, and the development of novel network layers are all examples of highly active fields of research that add complexity to these concepts. Additionally, subcategories of gradient descent exist based on batch size, the number of images inputted before updating weights, such as stochastic gradient descent and mini-batch gradient descent. Other optimization algorithms like Adam optimization exist as well. Finally, various hyperparameters that affect model learning typically need to be tested over a range of values and each can affect different portions of training. Some major hyperparameters include learning rate, momentum, batch size, and number of epochs. Many of the common alternatives and hyperparameters are covered in this review by Shrestha and Mahmood[9].
For imaging data, a specific type of neural network, called convolutional neural networks (CNNs), need to be utilized as inputs are in the form of 3-dimensional matrices, or tensors. A key concept is that any image can be represented by its numerical, pixel intensity values. For example, a 224 × 224 pixels grayscale image will be a 224 × 224 tensor with all values on a range from 0-1. A 224 × 224 RGB image, however, will be a 224 × 224 × 3 tensor with all values on a range from 0-255, though values are typically normalized to a range from 0-1.
To work with these tensors, convolutions need to be implemented in the form of convolutional network layers. Convolutions can be interpreted as the sliding of another tensor, or filter, typically of much smaller size than the input, over the input tensor. The filter slides from left to right of the input tensor, then moves down and repeats the process from left to right again. The mathematical operations can be considered as an expansion of the weighted sum and activation function approaches described earlier in this section. As the filter slides over the input, a weighted sum is calculated incorporating every cell overlap between the two tensors and generates a new output tensor that is then passed to an activation function. Like the layer nodes, convolution filters have weights that are trainable by gradient descent. Each hidden layer will perform similar operations on outputs from the previous layer but may have varying filter sizes or activation functions. Since the non-linearities introduced by the various layers sequentially add complexity, the earlier layers are believed to encode simpler features like edges, while the latter layers capture even more abstract features.
Analysis of images requires consideration of relationships between adjacent regions to capture spatial information. Though the weighted sum calculation in convolutions does account for neighboring pixels, the receptive fields are still quite small. Pooling layers are also carried out by filters and further aggregate local information from previous layers. The two major types are max and average pooling. For a 4 × 4 pooling filter, this would mean selecting the maximum value within the 4 × 4 receptive field in max pooling or averaging the 16 values for average pooling, as opposed to performing the weighted sum calculations that would occur in convolutions. Importantly, pooling layers do not have any trainable weights and represent fixed operations.
Convolutional layers typically reduce tensor height or width while increasing number of channel dimensions. Pooling layers do not affect channel dimensions but reduce tensor height and width. Thus, a series of convolutional and pooling layers will serve to reduce tensor height and width and increase channel dimensions relative to the original input.
The outputs of convolutional and pooling layers are often 3-dimensional and need to be flattened into a 1-dimensional vector towards the end of the network. The flattened 1-dimensional vectors then feed into a full-connected layer, a feed forward layer where nodes calculate weighted sums from the flattened vector input and pass values to an activation function. Finally, these outputs are utilized for the final classification layer, which typically uses a softmax activation function in classification tasks. The softmax layer will have the same number of nodes as the number of possible classes to predict. The outputs of this layer will sum up to 1 and can be interpreted as the relative probabilities for each class prediction with each class corresponding to one softmax node.
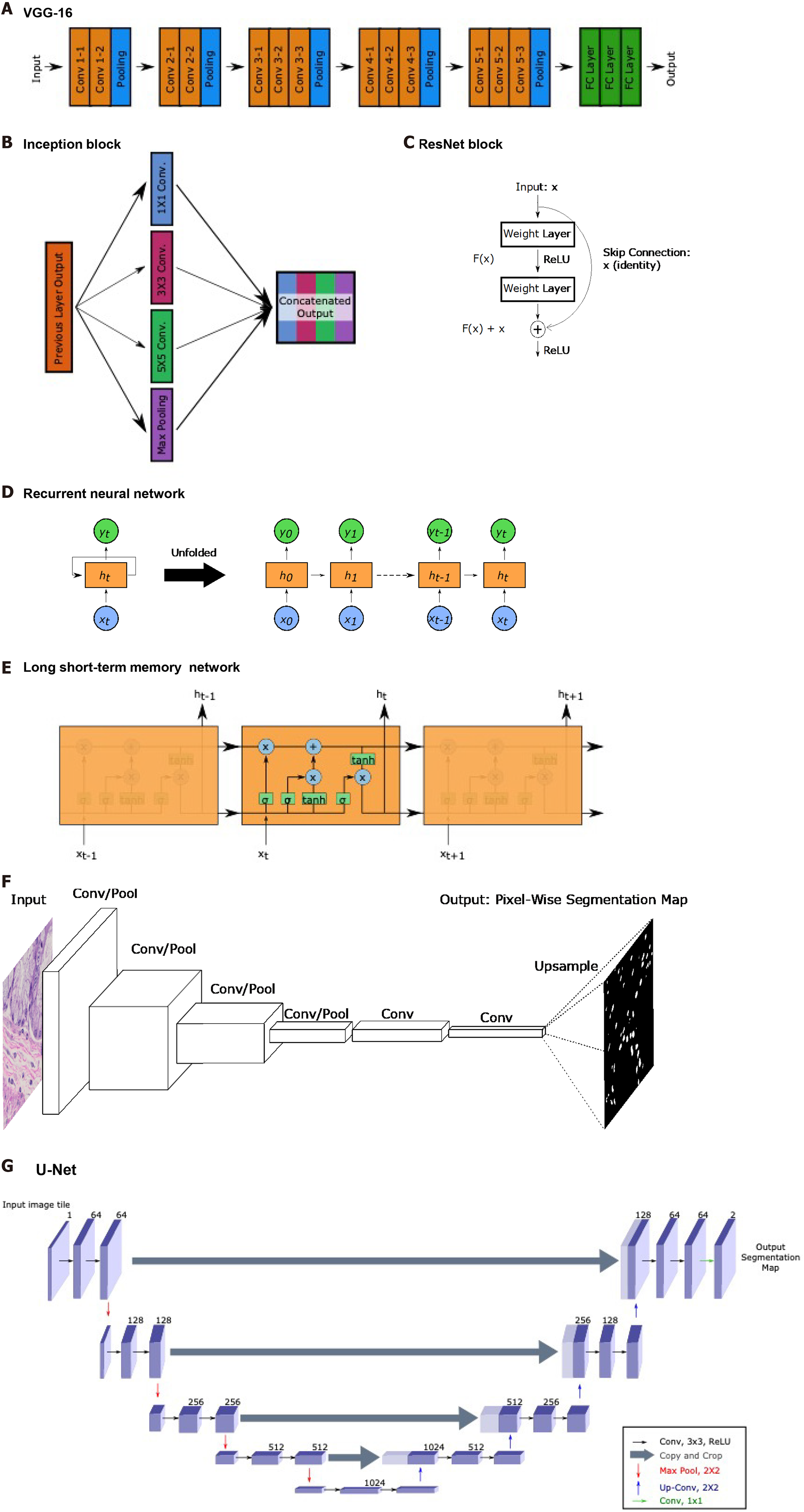
Although the focus of this review is not to delve deeply into the different types of neural network architectures, those that appear will be covered briefly here to provide background. An overview of the network structures is shown in Figure 5.
The visual geometry group (VGG)-16 and VGG-19 networks published by Simonyan and Zisserman[10] consist of sequences of convolutions and pooling operations followed by fully connected layers for a total of 16 or 19 layers, respectively. The authors incorporated very small 3 × 3 convolutional filters and demonstrated the capacity to create a network that had a lot of layers relative to other networks at time of publication.
The Inception network was initially published in 2015 by Szegedy et al[11], though several improved versions, such as the Inception-v3 used by some studies in this review, have since been developed. The major contribution of these networks is the introduction of the inception module that performs 1 × 1 convolutions, 3 × 3 convolutions, 5 × 5 convolutions, and max pooling at the same layer. An n × n convolution refers to a convolutional layer with an n × n dimension filter. The general concept is that predicting the optimal convolution filter size may depend on the image at hand. Instead of selecting a single filter size, more may be learned by incorporating information from convolutions with different receptive fields along with max pooling.
He et al[12] introduced ResNets which contain a the residual block with a skip connection. Deep neural networks with many layers often experience the issue of vanishing or exploding gradients. With the high amount of mathematical complexity introduced by many layers, backprogating these gradients can approach local minima and maxima and impede training. Since calculated gradient values are used to update layer node weights, a value of zero means the weight barely shifts, while infinity causes too significant of a change. Without getting into much detail, these residual blocks allow for skipping of portions of the network where this occurs to allow for continued training. This has allowed for networks like the ResNet-34 and ResNet-50, which have 34 and 50 layers, respectively, and for extraction of even higher dimensional features.
Though typically for sequential or temporal data, RNNs and LSTM networks are utilized by a few studies covered in this review. RNNs were developed earlier and process sequences of data. Each layer performs the same task; however, the decisions made at each layer is dependent on previous outputs. This capacity has been important for speech data as words typically have a relationship with the previous word in a sentence. LSTMs have similar use cases but with a superior ability to identify longer term dependencies and relationships than RNNs. In the context of this review, RNNs and LSTMs are useful in being able to take in a variable length sequences of inputs to provide one output. As WSIs are composed of varying numbers of patches, these networks are implemented to aggregate patch information into a WSI-level output. In these studies, patch sequences are typically shuffled to ensure input patch ordering has no effect on the output and focus on leveraging the capacity to take in variable length inputs as opposed to the temporal component. While not utilized in studies covered in this review, bidirectional encoder representations from transformers is a more modern technique that, instead of only reading sequences left-to-right or right-to-left, considers bidirectional contexts when making predictions[13].
Segmentation tasks, which generate a segmentation map output of the same dimensions as the input, require specific types of networks. Long et al[14] introduced the fully convolutional network (FCN), which replaces the fully connected layers described earlier in the CNN description, with additional convolutional layers. Forgoing the flattening operation and fully connected layers maintains the spatial relationships in the 3-dimensional tensors. The FCN then performs deconvolutions, also known as transpose convolutions, which in practice perform the opposite function of convolutions. Deconvolutions increase the height and width dimensions of inputs, allowing for eventual generation of an output with the same dimensions as input. FCNs are able to generate probability heatmaps of possible segmentation classes.
Ronneberg et al[15] built upon the FCN by developing the U-Net, named due to its U-shaped network structure. U-Net has 4 convolutional layers to generate a bottleneck tensor, then 4 deconvolutional layers to up-sample the bottleneck tensor back to the original input dimensions. Additionally, each deconvolutional layer receives input feature maps from the corresponding convolutional layer. As the convolutional layers occur earlier and encode spatial relationships more, concatenating these feature maps to the deconvolutional layer outputs helps the network with localization, which is important for segmentation tasks.
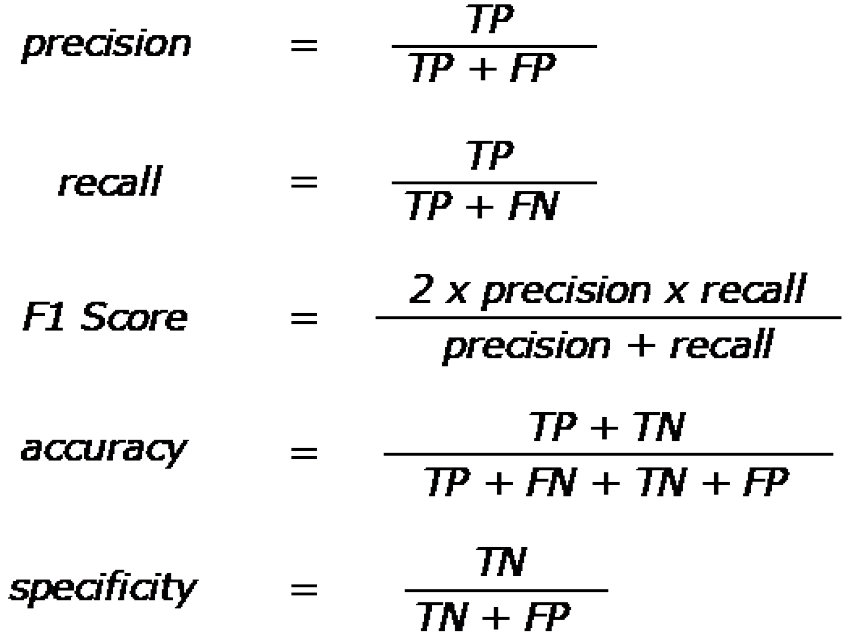
Lastly, it is important to understand the quantitative metrics used to assess model performance. The most popular are accuracy, precision, recall, and F1 score. These metrics are all calculated based off the total number of true positives, true negatives, false positives, and false negatives, and their formulas are shown in Figure 6. Area under the curves (AUCs) are also a common metric and involve calculating this metric from a plot of sensitivity vs (1-specificity).
While ML and DL approaches have been applied to many input data types, the fields of computer vision and image analysis owe much of its popularity to the emergence of digital pathology. An early milestone for digital pathology was the development of software to view histology images, such as the Virtual Microscope developed from 1996 to 1998 that had to take advantage of existing methods for handling satellite and earth science data[16]. The Virtual Microscope was further refined to allow for capabilities like data caching, support for simultaneous queries from multiple users, and precomputed image pyramids. Modern viewers have continued to grow capabilities and allow for collaborative, multi-user work on the same images, annotations, the ability to zoom and inspect WSIs, and construction of imaging datasets and cohorts.
At the time the Virtual Microscope was being developed, WSI scanners were not yet available, so histology sections had to be digitally tiled up before uploading into viewing systems. Today, many commercially available WSI scanners that can scan entire slides exist, and this issue can be avoided. Furthermore, there are now two FDA-approved digital pathology platforms: the Phillips IntelliSite Pathology Solution[17] and the Sectra Digital Pathology Module[18].
Digital pathology comes with some clear benefits, including the ease of sample storage and access through software and the capacity to perform image analysis directly on digitized WSIs. However, the utilization of WSIs comes with its own set of quality concerns, which are covered nicely in the review by Kothari et al[19]. In brief, these methods can introduce image artifacts and batch effects. Image artifacts can occur both from scanning or preparation of samples. Some examples include blurring of tissue regions due to microscope autofocusing mechanisms, shadows in the image, pen marks from pathologists, or folding of tissue. In these cases, care needs to be taken to remove the artifacts, such as in the case of pen marks, or to filter out image areas with issues like blurring. Batch effects can occur due to the individual preparing the sample, the specific reagents used, the site of acquisition, or the microscope type. To address some of these concerns, studies frequently apply methods to normalize the color or pixel intensity values across their images. However, certain factors into batch effect exist that cannot be addressed computationally, such as varying patient population demographics based on location. As such, there is a strong need to incorporate multicenter data sources to develop more generalizable models, or a realization that certain models may only be used within specific demographics.
So long as these quality control concerns are recognized, however, digitized WSIs have the potential to be further adopted within practices. They are high-resolution gigapixel scale images that can be stored digitally and distributed for research. Furthermore, Al-Janabi et al[20] examined the feasibility of utilizing WSIs instead of classic light microscopy for diagnosis of gastrointestinal tract pathologies. For 100 cases of biopsies and resections along the entire gastrointestinal tract that had been diagnosed by light microscopy a year earlier, the authors recruited the same pathologists to re-diagnose their own cases using digitized WSIs. The study showed a 95% concordance between light microscopy- and WSI-based diagnoses with the discordant 5% of cases showing no clinical implications, highlighting the potential for the adoption of WSIs into the diagnostic workflow.
Finally, the growing popularity of DL imaging approaches owes itself to the development of computational hardware and software[21]. Graphical Processing Units (GPUs) were primarily used in the setting of video games, but their high capacities for parallel computation were found to be ideal for DL methodologies. GPUs significantly increased DL model training speeds relative to Central Processing Units and played a strong part in the popularity growth of these methodologies. In addition, the DL community has been aided by the presence of open-source libraries for efficient DL GPU implementation. Some examples include PyTorch, Caffe, and Tensorflow. These libraries can easily load up and train major, landmark network architectures, simplify design of new networks relative to manual coding, and encourage cohesion amongst researchers by having standardized coding styles and pre-defined functions for common operations within each library. The emergence of digital pathology viewers, WSI scanners, GPUs, and open-source DL libraries has led to Pathomics, defined as the generation of quantitative imaging features to describe the diverse phenotypes found in tissue sample WSIs[22]. The foundations for DL-based imaging fields have thus been set and have welcomed a new influx of applications and studies within the biomedical disciplines.
One clear application of these ML- and DL-based methodologies is to replicate the tasks of pathologists. A well-trained model has the benefits of eliminating interobserver variability amongst pathologists and of achieving a level of throughput impossible to humans. This section will cover research in classification for both cancer and non-cancer pathologies and in segmentation tasks aimed at the identification of structures or cell types within images.
The most popular histopathological application of these methods in gastroenterology and hepatology occurs in the classification of cancers. The general concept is that if these classifications can be made with the human eye, then the models should be able to learn to make such distinctions themselves.
Colorectal: Thakur et al[23] recently published a comprehensive review of artificial intelligence applications in colorectal cancer pathology image analysis, but several papers will still be highlighted in this review.
In an earlier study, Yoon et al[24] trained a customized VGG-based network architecture on 28 normal and 29 colorectal cancer HE-stained slides that were tiled up into 256 × 256 pixel patches. After testing several, custom VGG-based networks, the best model had an accuracy of 93.5% with a sensitivity and specificity of about 95% and 93%, respectively, in determining if an image patch was cancer vs healthy. This study showed promise in the relatively simpler binary classification task of tumor vs normal.
In a study published since, Sena et al[25] took the classification task another step further to train a model to classify between normal mucosa, early neoplastic lesion, adenoma, and cancer in HE-stained samples. The authors used a custom network architecture similar to VGG with four sequential convolutional and pooling layers followed by dense layers. Even with this relatively simple network architecture, the model achieved about a 95% accuracy in predicting the exact label for its larger 864x648 pixel patches across the four classes.
While the above patch-level performances are encouraging, clinical diagnoses are typically at the slide level. To address this, researchers often train additional classifiers, in addition to the patch-level ones, that can make a prediction at the WSI-level by aggregating patch-level information. An example of this is the study by Iizuka et al[26], which initially trained the Inception-v3 network to classify between non-neoplastic, adenoma, and adenocarcinoma for 512 × 512 pixel patches from HE-stained colorectal and gastric biopsy WSIs. The authors utilized the trained Inception-v3 classifier as a feature extractor to generate 715-length feature vectors, the representations, for each patch. The sequence of feature vectors of every patch in a WSI are used as the input and the WSI-label as the output in training a subsequent RNN. Though RNNs are typically used in temporal data, they have the advantage of being able to take in variable length, sequential inputs in generating final output labels. This is an important feature considering that every WSI has a varying number of total patches sampled. The trained RNN can thus predict the WSI diagnosis by aggregating extracted feature vectors of all patches in that sample. The study achieved WSI-level prediction AUCs of up to 0.97 and 0.99 for gastric adenocarcinoma and adenoma, respectively, and 0.96 and 0.99 for colonic adenocarcinoma and adenoma, respectively. Of note, the gastric classifier model outperformed pathologists in classification accuracy when pathologists were given a 30 s time limit, which is the average amount of time the model takes per WSI. The gastric model achieved an accuracy of 95.6% compared to the 85.89% ± 1.401% (n = 23) for the pathologists.
Russakovsky et al[27] utilized the AlexNet architecture pretrained on ImageNet, a large collection of non-biomedical, natural images with 1000 classes, as a feature extractor for classification and patch-based segmentation tasks on brain and colorectal HE datasets[28]. To address the common lack of annotated training datasets for these DL methodologies, the authors took this approach to demonstrate the potential of CNNs pretrained on non-biomedical images as feature extractors in biomedical applications. For the classification task, CNN-extracted patch representations for each WSI were pooled and condensed by feature selection methods before input into a SVM classifier to generate a WSI prediction. In colorectal cancer classification, the network was trained for a binary classification task to recognize tumor vs normal, and a multiclass classification task to recognize between adenocarcinoma, mucinous carcinoma, serrated carcinoma, papillary carcinoma, cribriform comedo-type adenocarcinoma, and normal. SVM with these CNN features as inputs outperformed SVM with a set of manually extracted feature as inputs in both classification tasks, achieving a 98.0% accuracy in binary and 87.2% in multiclass classification compared to 90.1% and 75.75%, respectively. The segmentation task involved no feature pooling as a SVM was trained to utilize patch-level CNN features to generate a patch classification prediction. By utilizing overlapping patches, pixel-level class predictions can be designated based off an ensemble method aggregating overlapping patch predictions. Again, the SVM with CNN prediction inputs outperformed the SVM with manually extracted feature inputs, showing an overall accuracy of 93.2% compared to 77.0%.
Although the above studies demonstrate the value of patch-level classifications in determining a WSI-level prediction, the annotations required for such a training dataset are highly time-consuming. Additionally, clinically archived tissue specimens are typically accompanied only by the WSI or patient-level diagnosis. MIL encompasses approaches to obtain insight into patches or patch-level features most critical for designation of the WSI-level label. MIL thus represents a possible way to generate effective patch classifier models utilizing only WSI-level annotations.
In MIL, each WSI is considered a bag in which multiple instances, or patches, are contained. If any one of these patches are positive for cancer presence, then the WSI can be determined to be cancer positive. While the instance-level patches have their own classes, these are unprovided or unknown. As such, the goal of MIL is to train an instance-level classifier based on the WSI-level labels to determine these unknown patch labels.
Xu et al[7] applied the MIL-Boost algorithm for HE colorectal slides for binary cancer vs non-cancer classification. In brief, the MIL-Boost algorithm trains the instance-level classifier by “boosting”. “Boosting” refers to the successive training of weak classifiers, where each classifier improves by adding weights to incorrect predictions made by the previous classifier. Here, weak classifier weights are iteratively updated by gradient descent on the bag-level classifier loss function. Backpropagation occurs along patch instances that most negatively affected the predicted bag-level cancer positivity relative to the true WSI-level label and adjusts algorithmic weights to reduce these errors on the next iteration of the weak, instance level classifiers. This process with weak classifiers is repeated until the loss function is minimized and an effective instance-level classifier is developed. The authors demonstrated superior performance of this approach (96.30% accuracy) as opposed to a fully supervised, patch-annotated approach (95.40%) in the binary cancer vs normal classification task.
As patch instances make up a WSI bag, a MIL-type bag representation can be considered to be a collection of patch instance representations where positive instances are provided a higher weight. To this end, Ilse et al[6] utilized a CNN to extract patch feature representations, then incorporated an attention mechanism to output a weighted average of all instances in a bag. Notably, the attention mechanism weights are determined by a two-layer neural network, meaning they are trainable unlike conventional MIL pooling operators that calculate maxes and means. These weighted bag representations can also be used to identify the most important instances for the bag prediction. The authors utilized a published HE colorectal cancer dataset[29] with annotated nuclear patches for epithelial, inflammatory, fibroblast, and miscellaneous and formed the MIL problem so that a bag is considered positive if at least one epithelium-positive patch exists in a WSI. This MIL approach trained an epithelial patch classifier with an accuracy and F1 score of approximately 90% and AUC of 96.8%. Furthermore, the authors could threshold for only instances with high weights, leading to the visualization of epithelial regions in the original HE WSI. Although the focus of this paper was on superior performance of the neural network tunable attention-mechanism relative to fixed alternatives, the final performance metrics lend support to the capacity of MIL approaches in training patch-level classifiers from WSI-level annotations.
Another major part of the colorectal cancer field is in the histopathological evaluation of HE-stained polyps to determine cancerous potential. In 2017, Korbar et al[30,31] trained a ResNet-based network to detect between hyperplastic polyps, sessile serrated polyps, traditional serrated adenoma, tubular adenoma, and tubulovillous/villous adenoma. The authors trained a patch-based classifier, then designated WSI-level predictions according to the patch-level class prediction that was most prevalent in the sample, given that at least 5 patches outputted that prediction[31]. This model achieved a 93.0% overall accuracy [95% confidence interval (CI): 89.0-95.9]. In another study, the authors utilized the same network architecture to identify the 5 classes but focused on implementing Gradient-weighted Class Activation Mapping (Grad-CAM) approaches to address model interpretability[30]. Grad-CAM can backpropagate from a patch’s predicted class label to identify the regions in the input image that contributed most to the prediction. Though this was an early approach, the study showed promising potential for these Grad-CAM approaches to help identify regions of interests (ROIs) that were most influential in the patch-level polyp classification.
Esophageal: While different from Grad-CAM, Tomita et al[32] utilized a related concept in implementing attention-based mechanisms for weakly supervised training to detect 4 classes—normal, Barrett’s esophagus without dysplasia, Barrett’s esophagus with dysplasia, and esophageal adenocarcinoma—from HE-stained esophageal and gastroesophageal junction biopsies. The approach involved breaking up a WSI into patches, from which a CNN would extract features. Each WSI could then be represented as a feature map that is an aggregated patch grid of extracted feature vectors. These feature maps serve as inputs to the attention-based model, the goal of which is to identify the regions of the input feature maps most important for the output label classifications. Therefore, a concept is shared with Grad-CAM in identifying input image regions most influential to the class predictions. Unlike Grad-CAM, the attention-based model will learn to add weights to influential areas in the feature map to aid in final model classification performance. Of note, this process is considered weakly supervised because image output labels are only provided at the WSI-level, as opposed to the patch-level, yet the most influential patch types can be distinguished. The model manages to learn on its own the most salient image features and regions that were most important for the WSI label. The approach here achieved an overall accuracy of 83.0% (95%CI: 80-86) in identifying the 4 classes, outperforming the supervised baseline with an overall accuracy of 76% (95%CI: 73-80) that depends upon extraction of patches from ROI tediously annotated by pathologists. It should be noted, however, that the model achieves an F1 score of 0.59 (95%CI: 0.52-0.66) and the supervised baseline an F1 score of 0.50 (95%CI: 0.43-0.56) possibly indicating a high rate of false positives and negatives.
Moving even further away from supervised learning, Sali et al[33] demonstrated superior performance of unsupervised approaches in classifying HE-stained WSIs to be dysplastic Barrett’s esophagus, non-dysplastic Barrett’s esophagus, and squamous tissue relative to supervised methods. The supervised approach was analogous to Iizuka et al[26]. Training patches labeled by pathologists were used to train the model, then an SVM or RF classifier aggregated the patch-level information for the WSI-level prediction.
The unsupervised feature extraction approach involved a deep convolutional autoencoder (CAE). Deep CAEs are broken up into an encoder and decoder branch. The encoder branch typically applies a series of convolution and pooling operations to act as a feature extractor that outputs a bottleneck feature vector. The decoder branch upsamples back from the bottleneck feature vector and reproduces the original image. Here, the loss function minimizes the differences between the input image and reproduced version, thereby enforcing that the bottleneck feature vector is an effective representation of the input. A helpful analogy is when one zips files on the computer. The process compresses the original file to a smaller memory size (encoding), but then still allows one to re-generate the full-size, original file (decoding). As one knows the zipping mechanism works, he or she can confidently share zipped file versions to others, instead of the larger, original file.
Once the deep CAE is trained, it can be utilized as a feature extractor for all patches in one’s training dataset. Then, by performing clustering approaches, such as k-nearest neighbors (k-NN) or Gaussian mixture models (GMM), on all feature vector-transformed patches, patch types or classes across the dataset can be defined. A SVM or RF classifier can be trained to predict the WSI class by using the relative proportions of the different patch class types in the sample. For WSI-level inference, the deep CAE extracts feature vectors from all patches in the WSI, bins and counts the number of patches per clustering-defined patch type, then utilizes the trained SVM or RF classifier to generate the WSI prediction. This process is called unsupervised because the different types of patches in the WSI are determined by the algorithm independent of any labelling. This is in contrast to the supervised approach, where a CNN was trained to classify between human-defined Barrett’s esophagus, non-dysplastic Barrett’s esophagus, and squamous tissue patch types. The unsupervised GMM method showed good performance with weighted averages for accuracy, AUC, F1, precision, and recall all above 90%. In contrast, the metrics for the supervised approaches ranged from 50%-80%.
Gastric: Though gastric pathologies and cancers will be covered further in other sections of this review, not a tremendous amount of literature exists regarding just classification of gastric cancers. Leon et al[34] demonstrated that, in gastric cancer classification, inputting image patches as a whole into a custom, Keras sequential model shows superior performance than utilizing nuclei extracted from these image patches as input. This may be explained by the fact that the whole image patch contains morphological features that might be important for classification, while the cell input approach sacrifices those portions of the image. The other major study to note is the one by Iizuka et al[26] mentioned earlier, which showed impressive performance in classifying gastric and colorectal adenomas and adenocarcinomas.
Liver: As in the mentioned studies by Iizuka et al[26] and Ilse et al[6], CNNs can be used to extract patch feature representations. These representations are 1-dimensional vectors comprised of numerical, float values, and higher values can be interpreted as features most important, or highly activated nodes, for the prediction at hand, while lower values may be interpreted as important for the other non-predicted class.
Since these feature values can be reflective of their relative importance in the predicted class, Sun et al[8] used a CNN to extract patch representations from HE-stained WSIs, performed a pooling operation to aggregate patch features at the image level, then sorted the representations to organize activation values from high to low importance in terms of liver cancer prediction. The authors selected a range of top-k and bottom-k features from this sorted list to use in patch representations, driven by the idea that high activations should indicate features important for cancer classifications, while the lower activations should correspond to normal. The variable length representations dependent on k were tested to generate condensed patch representations in training a binary cancer vs normal classifier. A value of 100 for k was deemed optimal, and the authors used the patch classifications to predict WSI cancer vs normal status. The approach achieved an accuracy of 98%, a recall of 1.0, and an F1 score of 0.99.
In addition to the design of effective image classification algorithms, the incorporation of these methodologies into clinical workflow is important to consider. Kiani et al[35] trained a DenseNet CNN to classify between hepatocellular carcinoma and cholangiocarcinoma from HE image patches and developed a diagnostic support tool that outputs predicted classes with probabilities and class activation maps (CAMs) to highlight areas of the input patch important for the prediction. The effects of the diagnostic support tool were analyzed and revealed that, while correct classifier predictions significantly improved accuracy, incorrect classifier predictions significantly decreased accuracy of diagnosing pathologists. Thus, this study highlights the important notion that the damaging effects of incorrect and misleading classifiers need to be strongly considered before clinical implementations.
Pancreatic neuroendocrine: IHC stains are another common technique applied to histopathological samples. The ability to detect specific antigens can be important for the characterization of certain cancer types. In pancreatic neoplasms, for example, the Ki67 stain is used to define proliferative rate and assign grades to pancreatic neuroendocrine tumors (NETs). However, this process is complicated by Ki67 stain positivity in both tumor and non-tumor regions. To address this issue, Niazi et al[36] trained an Inception-v3 network pretrained on ImageNet in a transfer learning setting to detect tumor and non-tumor regions on Ki67-stained pancreatic NET WSIs. As with Xu et al[28], the concept is that learned features from training on ImageNet should be beneficial within the biomedical setting. By freezing weights on all layers except for the final classification layer, the authors ensured that the feature extraction portion of the network remains unchanged. Training thus affects only the manner in which the classification layer utilizes patch representations instead of affecting the feature extraction itself. The trained model was used to create probability maps for tumor and non-tumor predictions for every pixel in the WSI, then thresholded by 0.5 to generate masks for each class. As each pixel in the image was then assigned to its most probable class, the output generated a segmentation map-type output that is shaded by predicted classes. In identifying tumor and non-tumor regions on a Ki67-stained IHC slide, the model showed about 96%-99% overall accuracy with 97.8% sensitivity and 88.8% specificity.
Cancer lymphocyte interactions: In addition to the cancer itself, other cell types exist within the microenvironment. To address this, Saltz et al[37] trained a VGG-16 network to identify tumor-infiltrating lymphocyte (TIL) containing patches across 13 The Cancer Genome Atlas (TCGA) HE-stained tumor types. The study identified four types of TIL infiltration patterns: Brisk Diffuse, Brisk Band-like, Non-Brisk Multifocal, and Non-Brisk Focal. The study also found associations between TIL infiltration patterns, cancer type, inflammatory response subtype, and molecular cancer subtypes and supports the notion that spatial phenotypes have the exciting potential to correlate with molecular findings.
Cancer nuclei classification: Another avenue of classification tasks in cancer applications has been in the study of nuclei. Pathologists are able to utilize visual, nuclear information, such as aberrant chromatin structures, to identify cancerous cells. Thus, groups have worked on replication this task of nuclei classification.
Chang et al[38] extracted HE-stained nuclei, used immunofluorescence (IF) pan-cytokeratin (panCK) stains aligned to the HE slide by image registration methods to label the HE-extracted nuclei as cancerous or non-cancerous, then trained a CNN to make these distinctions from just an HE input. The panCK-defined cancer positivity approach eliminated the need for tedious, pathologist annotations on the HE images and achieved a 91.3% accuracy with 89.9% sensitivity, 92.8% specificity, and 92.6% precision in classifying cancerous vs non-cancerous nuclei on the independent test set.
Sirinukunwattana et al[29] implemented a spatially constrained CNN to identify pixels most likely to represent the center of nuclei, then trained a subsequent CNN classifier to predict whether the nuclei came from an epithelial, inflammatory, fibroblast, or miscellaneous cell in colon cancer. The authors also implement a Neighboring Ensemble Predictor in the nuclei classifications, which, when predicting the class of a nuclei, incorporated the predictions from all neighboring patches. This approach achieved a weighted average F1 score of 0.784 and AUC of 0.917 in the nucleus classification tasks and a weighted average F1 score of 0.692 in the combined nucleus detection and classification tasks. In a follow up study since, Shapcott et al[39] utilized this nuclei classification algorithm to quantify the four cell types to correlate cellular proportions with different clinical variables in TCGA colorectal cancer patients. This led to findings such as samples with metastasis having more fibroblasts with fewer epithelial and inflammatory cells, samples with residual tumor having more fibroblasts and fewer epithelial and inflammatory cells, and that both venous and vascular invasion were associated with more fibroblasts.
Though much focus in image classification has been in cancers, other image classification applications exist and are highlighted here.
Celiac disease, environmental enteropathy, and nonspecific duodenitis: Wei et al[40] trained a ResNet-based model to classify between celiac disease, normal tissue, and nonspecific duodenitis on HE-stained WSIs with accuracies of 95.3%, 91.0%, and 89.2%, respectively. This was a supervised, patch-based approach for training, and WSIs were predicted to be nonspecific duodenitis if more than 5 patches were classified as such or predicted to be the dominant patch class otherwise.
In a similar supervised fashion, Srivastava et al[41] trained a ResNet model on duodenal HE biopsies to classify between celiac disease, environmental enteropathy, and normal tissue. Patch classifications were aggregated for the WSI prediction and returned an overall 97.6% accuracy.
Sali et al[42] also trained a ResNet model, but for the task of Marsh Score-based grading of celiac disease severity using HE-stained duodenal biopsies. The authors utilized a CAE to generate patch representations, then performed a 2-class k-NN clustering to filter out useless, non-tissue containing patches. The tissue-containing patches were then used for supervised training of the ResNet model to recognize between Marsh scores of I, IIIa, IIIb, and IIIC. Again, patch predictions were aggregated for a WSI-level prediction. The model showed an accuracy and F1 score of around 80-90% for all classes and also implemented CAM approaches to localize certain cell subsets contributing to some of these Marsh Score categories.
In another study, Sali et al[43] took a novel, hierarchical approach towards training a VGG classifier to detect 7 classes: Duodenum-celiac disease, Duodenum-Environmental enteropathy, Duodenum-normal, Ileum-Crohn’s, Ileum-normal, Esophagus-eosinophilic esophagitis, and Esophagus-normal. In addition to having the classifier predict the disease type with the final classification layer, the approach incorporated another output branch in the VGG network to predict anatomic location. The loss function combined outputs of the two branches and enforced the network to learn both anatomic origin and specific disease type. Additionally, the anatomic origin branch occurs before the final classification layer, meaning that the network needs to correctly determine the anatomic origin first, before homing in on the specific diagnosis. Across all 7 classes, the model exhibited F1 scores ranging from 0.714 for Duodenum-normal to 0.950 for Duodenum- Environmental enteropathy.
Helicobacter pylori gastritis and reactive gastropathy: Similar to the other examples, these represent diagnoses that can be made from HE-stained specimens. Martin et al[44] trained the commercially available HALO-AI CNN to classify between Helicobacter pylori, reactive gastropathy, and normal in gastric biopsies. The model achieved sensitivity/specificity pairings of 73.7%/79.6%, 95.7%/100%, 100%/62.5% for normal, Helicobacter pylori, and reactive gastropathy, respectively.
Klein et al[45] developed a model that combines image processing techniques with DL. The authors utilized image processing techniques on both Giemsa- and HE-stained slides to identify potential Helicobacter pylori regions, then had experts review these as being positive or negative for Helicobacter pylori presence. These could then be utilized as input-output pairs to train a VGG-style network. The main goal of this paper, however, was to create a clinical decision support system that utilized the trained model and directs pathologists to Helicobacter pylori hotspots using Grad-CAM-style methodologies. Although this clinical decision support approach showed higher sensitivity than just microscopic diagnosis (100% vs 68.4%), specificity was lower than with just microscopic diagnosis (66.2% vs 92.6%).
Segmentation generally refers to operations that localize and detect cells and structures within a WSI. As pathologists can detect these objects within a sample, the goal is to train models to replicate these tasks.
The gland segmentation in colon histology images challenge contest challenge: A key contributor to the progression of computer vision disciplines has been the presence of challenges that provide a dataset and rank submitted models based off of performance-related quantitative metrics such as F1 scores or AUC values. One example of this is the gland segmentation in colon histology images challenge contest (GlaS) that was held in 2015[46]. These challenges help to stimulate computational disciplines. For one, the announcement of the challenge itself encourages researchers worldwide to address and tackle the problem. Compared to standalone papers, these challenges also have the advantage of pitting the best models against each other to generate a clear benchmark for state-of-the-art performance.
Furthermore, even after completion of the challenge, groups will continue to optimize their algorithms and will have the ability to compare performance to previous high rankers in the challenge. Even since the GlaS challenge, numerous groups have continued to work on gland segmentation models by incorporating novel mechanisms. In 2016, Xu et al[47] added multichannel feature extractions for region and edge probability maps that were then fed into the final CNN for instance segmentation. Also in 2016, BenTaieb et al[48] applied topological and geometric loss functions into their FCN-based model. In 2019, Graham et al[49] introduced a new network component, the minimal information loss unit, that re-introduces resized versions of the original input image to combat the loss of information that accompanies downsampling from the successive convolution and max-pooling operations that occur in neural networks. Most recently in 2020, Zhao et al[50] incorporated spatial attention to weight important spatial locations and channel attention to weight important features to improve gland segmentation performance.
Non-colon gland segmentations: In general, segmentation methodologies require an additional step of development compared to classification tasks. For example, identifying glands in colonic mucosa is an important task but needs additional interpretation to be useful in the clinic. Some possibilities include quantifying the total number of glands or extracting shape-based glandular information to feed into a colorectal cancer classifier. Classification tasks like “Tumor” vs “Healthy”, on the other hand, often already have a clear path towards clinical integration within the pathologist diagnostic workflow.
Reflective of this, many histopathological segmentation studies in gastroenterology and hepatology tend to be focused on optimizing segmentation results themselves, as opposed to continuing onto the translational application. However, high performance segmentations are critical in developing the downstream, clinically impactful algorithms. While some studies have continued onto the next step, the next few years will likely see some more of these segmentation studies bridging into more translational studies.
To highlight some examples, Xiao et al[51] segment out liver portal area components for eventual hepatitis grading. Extraction of features from these segmented structures to train a classifier to grade hepatitis will likely be the next step of this process. Xu et al[52] used a patch-based segmentation approach to identify epithelial and stromal regions in HE-stained breast and epithelial growth factor receptor-stained colon cancer slides as tumor-stroma ratios are recognized to have prognostic value. Here, the next step would be to assess the impact of algorithm-derived epithelium and stroma ratios in patient prognosis or cancer classification. Similarly, to address the eventual use case of segmenting tumors to assess pre-surgical tumor burden, Wang et al[53] used multitask and ensemble learning techniques for pixel-wise HE hepatocellular carcinoma segmentation. For eventual use in computer-assisted diagnosis systems, Qaiser et al[54] develop a fast HE colorectal segmentation algorithm that defines persistent homology profiles to capture morphological differences between normal and cancer nuclei. The emergence of more directly translational follow up studies and validations should be exciting and will be important to monitor.
Moving downstream with segmentation outputs: Some studies have entered this second phase and will be highlighted in this section. Awan et al[55] utilized a modified version of U-Net to perform colon gland segmentation on HE-stained colorectal adenocarcinoma patches, then extracted quantitative measures of glandular aberrance to train a SVM classifier for normal vs tumor classification and for normal vs low grade vs high grade classification. Glandular aberrance correlated with tumor grade, and this method achieved an accuracy of 97% and 91% for the two-class and three-class classifications, respectively. Thus, application of segmentation outputs in this manner can allow for the definition and extraction of novel quantitative features to aid in classification tasks and may provide a look into how these segmentation algorithms will be clinically implemented in the future.
Multiplex IHC (mIHC) involves concurrent histological staining of 6 cell markers or more, and Abousamra et al[56] developed an autoencoder-based color deconvolution algorithm to segment these different stains within a WSI. In a follow-up study, Fassler et al[57] utilized this algorithm on mIHC-stained pancreatic ductal adenocarcinoma (PDAC) WSIs to detect and perform spatial analyses on the cell types. Results indicated that CD16+ myeloid cells dominated the immune microenvironment and on average were of closer distance to tumor cells than CD3+, CD4+, CD8+, or CD20+ lymphocyte populations. In contrast to the study by Awan et al[55] that used segmentation outputs to inform a clinical task, Fassler et al[57] targeted a research application. A pipeline to detect all cell types from mIHC-stained WSIs, quantify, and perform special statistics would serve a wide audience of basic and translational researchers, and, in elevating analytical capacities, may stimulate research output.
A popular translational application of segmentation outputs has been in the field of hepatic steatosis quantification, which is important in the assessment of patients with fatty liver disease or to assess donor liver-quality for transplantation. In an earlier study, Lee et al[58] demonstrated correlation of steatosis quantification by image processing methods on WSIs with MRI measurements, pathologist visual scoring, and several clinical parameters, serving to validate the potential of image feature extraction from WSIs for these applications.
Forlano et al[59] took a ML-based approach to quantify the four histological features used in the Nonalcoholic Steatohepatitis Clinical Research Network (NASH CRN) Scoring System, in an effort to automate the process and assess how their computationally extracted, quantitative histological metrics correlate with the semi-quantitative, categorial metrics of the NASH CRN Scoring System. The authors used image processing techniques to segment out and calculate percentages of fat, inflammation, ballooning, and collagen proportionate area, then fed the values into a binary logistic regression classifier to predict the presence of NASH. The authors argued that the traditional, semiquantitative approaches are outdated, due to their categorical nature and unavoidable interobserver variability, and demonstrated an AUC of 0.802 for their pipeline’s capacity to predict NASH.
Sun et al[60] took a modified VGG-16 patch-based segmentation approach to quantify macrovesicular steatosis in HE-stained frozen, donor liver biopsies. The network was trained on patches extracted from WSIs with steatosis regions annotated by pathologists. As such, the final portion of their network could be trained against the pathologist-annotated steatosis maps to output pixel-wise steatosis prediction maps from HE patch inputs. Steatosis percent could then be calculated by summing steatosis probabilities from the predictions maps and dividing by total tissue area. Overall, the model had a sensitivity of 71.4% and specificity of 97.3% in predicting samples with over 30% steatosis, which is the threshold used by some centers for donor rejection.
Roy et al[61] trained a network to segment foreground steatosis droplet pixels from background, a network to recognize steatosis droplet boundaries, and a third neural network that took both of those outputs as input to generate the final segmentation map. Their segmentation results allowed for the calculation of steatosis pixel percentage (DSP%) and steatosis droplet count percentage (DSC%). DSC% most strongly correlated with histologically determined macrovesicular steatosis percentage (rho = 0.90, P < 0.001) and total steatosis percentage (rho = 0.90, P < 0.001). DSP% showed the best correlation with MRI fat quantification (rho = 0.85, P < 0.001).
Lastly, Salvi et al[62] gained the capacity to quantify both micro- and macrosteatosis on HE-stained liver WSIs. The algorithm achieved an overall accuracy of 97.27% on the test set for steatosis segmentation and showed the lowest average error of 1.07% when comparing automated steatosis quantification with manual quantification methods.
While the emulation and automation of pathologist tasks is a clear and exciting application of these methodologies, recent studies have shown that extraction of information that typically requires other sources of data or that are not obvious to the human eye are possible. This section will cover emerging research that utilizes histopathological specimens to extract such information.
Although pathologists and physicians can estimate cancer patient prognosis, these determinations often require more than microscopically examining a histological slide. For example, the tumor-node-metastasis (TNM) staging system, though informative, can require information like tumor size, typically gathered from CT scans, or nodal and distal metastases status, which is not evident from a single histopathological slide. In other cases, pathologists may perform genetic testing or IHC-staining for further molecular characterization and subtyping of cancers. Recent work shows that these ML- and DL-based methodologies may be able to learn to predict such information from just histopathological samples.
Bychkov et al[63] developed an approach to predict 5-year survival from HE-stained tumor microarrays from colorectal cancer patients. As with Xu et al[28] and Niazi et al[36], the authors took the VGG-16 network pretrained on the ImageNet dataset[27] as a feature extractor. Each WSI’s collection of extracted patch features were then used to train a three-layer 1D LSTM network, since, similar to the RNN approach used by Iizuka et al[26], LSTM networks can take in a sequence of patch feature inputs. The LSTM model in this study was trained to generate a WSI-level 5-year prognosis probability. While the model’s capacity to predict disease-specific survival was not extremely high (AUC = 0.69), it outperformed histological grade (AUC = 0.57) and Visual Risk Score (AUC = 0.58).
Yue et al[64] incorporated an unsupervised patch clustering method to define patch types, trained a VGG-16 network to recognize the patch types, then implemented an SVM classifier to predict 5-year disease-specific survival. For the unsupervised patch clustering, patch features were extracted by a CNN and pooled, dimensionality reduction was performed by principal component analysis, and the k-means clustering was performed in this lower-dimensional feature space to define patch clusters. While the best performing model generated an accuracy and F1 score of 100%, the approach needs to be validated given the small dataset of 75 WSIs. However, this study is another example of how these unsupervised patch clustering methodologies might be effective in determining patch classes.
To generate a more interpretable model for colorectal cancer survival, Kather et al[65] developed a prognostically predictive “deep stromal score” that utilizes outputs from a CNN trained to recognize adipose, background (glass slide), colorectal adenocarcinoma epithelium, debris, lymphocyte, mucus, smooth muscle, normal colon mucosa, and cancer-associated stroma. The authors used their NCT-CRC-HE-100K dataset that contains 100000 image patches covering these nine tissue classes to train and compare several models in classification performance. The top performing VGG-19 model was then applied to a held-out portion of their dataset. When fitting univariate Cox proportional hazard models to each of these 9 classes across the held-out dataset, the authors found that higher activation of five of the nine classes correlated with poor survival, though three were not significant (NS): Adipose [hazard ratio (HR) = 1.150 (NS)]; debris [HR = 5.967 (P = 0.004)]; lymphocytes [HR = 1.226 (NS)]; muscle [HR = 3.761 (P= 0.025)]; stroma [R = 1.154 (NS)] These five class activations were combined to form the prognostic deep stromal score and validated independently on colorectal adenocarcinoma cases from TCGA program. Multivariate analysis should that the deep stromal score was significant as a prognostic metric for overall survival [HR = 1.63 (P = 0.008)], disease-specific survival [HR = 2.29 (P = 0.0004)], and relapse-free survival [HR = 1.92 (P = 0.0004)].
Focusing on Stage III colon cancer patients, Jiang et al[66] used the NCT-CRC-HE-100K dataset generated by Kather et al[65] to train a classifier to determine the proportion of these tissue types in WSIs, then predict prognosis. Like Kather et al[65], the authors tested several networks on the classification task. They identified InceptionResNetV2 as their top performing model, which was utilized to extract proportions of the nine different tissue types from their own colorectal Stage III cancer dataset. The tissue proportions were fed into several ML classifiers, and the Gradient Boosting Decision Tree was identified as the top performer for prognostic predictions. On Stage III colorectal adenocarcinoma cases from TCGA, this top-performing approach correctly allocated patients into high- and low-risk recurrence groups to predict disease-free survival risk by both univariate and multivariate Cox regression analysis [univariate: HR = 4.324 (P = 0.004); multivariate: HR = 10.273 (P = 0.003)]. In addition, this approach also showed the capacity to predict overall survival risk on the TCGA dataset [univariate: HR = 5.766 (P = 0.000); multivariate: HR = 5.033 (P = 0.002)]. These results highlight a potential avenue for more interpretable ML- and DL-based algorithms and also are evidence of the importance of groups like Kather et al[65] making datasets publicly available to help advance the field as a whole.
For prediction of survival after hepatocellular carcinoma resection, Saillard et al[67] compared an weakly approach with and without an additional, supervised attention mechanism. In both approaches, a pre-trained CNN first extracts features from all patches in the WSI. For the weakly supervised approach (CHOWDER), these patch features are fed into the network along with WSI-level survival data to eventually determine the patches most influential to the survival outcome in an iterative learning process. For the approach with additional supervision (SCHOWDER), the weakly supervised mechanism in CHOWDER is further coupled by an attention mechanism that localizes to tumoral slide regions annotated by pathologists to identify the most influential patches. The SCHOWDER and CHOWDER surprisingly generated highly similar c-indices for survival prediction on the discovery set with 0.78 and 0.75, respectively, further supporting the potential of these weakly supervised methodologies to rival more supervised ones. As CHOWDER assigns risk scores to tiles, the authors could also re-extract then visually inspect types of tiles indicated to be most high and low risk. This involved tumor presence, macrotrabecular architectural tumoral pattern, and vascular spaces in the tumor as high-risk, and tumoral and non-tumoral fibrosis and non-tumoral immune cells as low-risk.
Also in a weakly supervised fashion, Wulczyn et al[68] developed a DL system (DLS) to predict disease specific survival across 10 cancer types, including colon and stomach adenocarcinoma, from TCGA using only WSI-level survival data. Assuming the frequency of informative patches for cancer diagnosis on a slide is P, the probability of a randomly sampled patch being uninformative is 1-P. As such, as one samples n patches, the probability of not sampling any informative patches exponentially approaches zero as (1-P)n. The authors leverage this property by randomly sampling multiple patches per slide, extracting features from each with CNNs that share weights, and performing average pooling of the extracted features. The outputs can then be fed into a fully-connected layer before the final logistic regression layer. Here, logistic regression is used as the authors found that discretizing time-to-event periods into specific intervals improved performance. In a combined cohort of all 10 cancers, the DLS was significantly associated with disease specific survival [HR = 1.58 (P < 0.0001)] after adjusting for cancer type, stage, age, and sex in multivariable Cox regression analysis. The DLS also outputs a risk score, allowing for stratification of stage II (P = 0025) and stage III (P < 0.001) patients. Finally, as with Saillard et al[67], high- and low-risk patches can be extracted and visualized for qualitative evaluation.
Although more of an intermediate endpoint, cancer metastasis is a specific event that correlates with reduced survival. Takamatsu et al[69] utilized image processing techniques to extract features from cytokeratin IHC-stained endoscopic resection samples and trained a RF ML classifier to predict lymph node metastasis. Their method demonstrated comparable performance to the predictive capacity of conventional histological features extracted from HE-stained slides. On the cross-validation approach, the ML achieved an average AUC of 0.822, compared to 0.855 for the conventional method. Although ML performance was not superior, comparable results by these algorithms are accompanied with the additional benefit of reduced interobserver variability. Furthermore, given the fact that the conventionally extracted HE features showed decent predictive power for lymph node metastasis, it will be interesting to see if a predictive classifier can be built off the HE images directly.
IF and IHC methods are often applied for further characterization of samples. However, these methodologies are time-consuming, can be costly, and require an unstained portion of the tissue. This section will thus focus on recent literature that trains using HE-stained images as input with output information that typically requires these additional staining techniques.
With the introduction of targeted molecular therapies for human epidermal growth factor receptor 2 (HER2) in gastric cancer, determining patient HER2 status has become increasingly important[70]. As this process requires IHC staining for HER2, automated extraction of such information from routinely collected HE samples has advantages in time, cost, and consistency. Sharma et al[71] utilized HER2 IHC-stained sections to define HER2 positive and negative regions on HE-stained sections from the same patient. HER2-stained IHC WSIs and HE-stained WSIs were aligned via semi-atuomatic image-registration approaches to define HER2+ and HER2- tumor regions on the HE based off of corresponding IHC positivity. Training on just the HE patches, the authors trained a custom CNN with three sequential convolution and pooling layers to classify between HER2+ tumor (69.6% accuracy), HER2- tumor (58.1% accuracy), and non-tumor patches (82.0%). Although the performance was modest in predicting IHC-defined output labels from HE, this was one of the earliest studies exploring this type of multimodal approach and used a relatively simple network architecture.
IHC can also be applied to metastatic NET samples to identify primary sites of origin. Redemann et al[72] trained the commercially available HALO-AI CNN on HE-stained samples of metastatic NET with known sites of origin to compare the algorithm’s capacity to make such predictions against IHC-based approaches. While the algorithm achieved a worse overall accuracy of 72% compared to 82% for IHC-based diagnosis, the results are promising given the relatively comparable performance to the gold standard IHC approach and the author’s training of an off-the-shelf, commercial algorithm. A comprehensive comparison of classification performance across multiple models may identify one with superior performance.
Govind et al[73] developed a DL-based pipeline to automate gastrointestinal NET grading, which classically involves IHC detection of a Ki-67-positive tumor hotspot region, then manual counting to obtain the percentage of Ki-67-positive tumor cells. The authors trained one model to detect Ki-67 hotpots and calculate a Ki-67 index from those hotspots similar to pathologists’ workflow, and another model that generates Ki-67 index-based heat maps to classify hot-spot-sized tiles in the WSI as background, non-tumor, G1 tumor, or G2 tumor. Importantly, both models used synaptophysin- and Ki-67-double-stained (DS) WSIs as input. As DS WSIs are not common in clinical practice, the authors computationally merged synaptophysin- and Ki-67-single-stained IHC WSIs to generate DS WSIs to be used for this study.
The first model, SKIE, detects synaptophysin-positive, Ki-67-dense hotspots from these DS WSIs and emulates current pathologist workflow by calculating Ki-67 indices that capture proportional Ki-67 tumor positivity within these hotspots. The second model, Deep-SKIE, was trained by extracting hot-spot sized patches, then assigning correct, output labels according to SKIE outputs on those patches. Specifically, the four classes were background (class 0: if the tile has > 70% background pixels), non-tumor (class 1: < 20% synaptophysin stain), tumor grade 1 (class 2: Ki-67 index < 3%) and tumor grade 2 (class 3: 3% < Ki-67 index < 20%). In predicting these hot-spot patch labels, Deep-SKIE trained on DS WSIs exhibited an overall accuracy of 90.98% compared to 84.84% when trained on SS WSIs, indicating that the additional information from multiple markers aids in classification performance.
The most interesting contribution of this paper with respect to this section, however, is the authors’ decision to train a cycle GAN to generate DS WSIs from SS WSI inputs. In brief, GANs attempt to generate synthetic imaging data that is indistinguishable from the real samples. There is typically a ‘discriminator’ module that attempts to correctly distinguish between the GAN-generated synthetic data and the real-world data. The worse your discriminator performs, the better your GAN is at generating synthetic data.
Here, the authors used a cycle GAN to generate synthetic DS WSIs from SS WSIs that are highly similar to the real DS WSIs already in the dataset. This approach has the ability to generate highly informative DS WSIs in the clinic without the required time and costs associated with additional stains. The authors showed that Deep-SKIE when trained on the GAN-generated DS WSIs showed an accuracy of 87.08% in predicting the four patch classes that was still significantly higher than 84.84% when trained on the SS WSIs.
IF staining is the other major staining technique. Unlike IHC, IF comes with an additional disadvantage regarding sample stability. As signal is carried by fluorophores, signals are often lost within a week, and samples are thus typically imaged immediately after staining. As such, DL approaches outputting IF-related data is not only informative and cost-efficient but may simplify acquisition of such data.
Burlingame et al[74] developed an experimental protocol allowing for HE and panCK IF staining in the same section of tissue, then trained a conditional GAN to output virtual panCK IF WSIs from HE PDAC WSI inputs. Similar to the cycle GAN used by Govind et al[73], the conditional GAN here depends upon a discriminator attempting to distinguish between real and virtual IF WSIs. As the protocol allows for HE and panCK IF staining on the same tissue, the authors have HE-IF WSI input-output pairs to train the conditional GAN. The virtual IF WSIs showed high similarity to the real IF WSIs in terms of structural similarity metrics, and the authors also present preliminary data on virtual IF generation for alpha-smooth muscle actin, a stromal marker.
This section has covered two categories of methodologies that circumvent the need for staining. The first category involves, directly from HE-stained inputs, the extraction of information that typically necessitates additional staining. These approaches may one day assist pathologists in quicker, cheaper molecular characterizations of patients. The second category involves the generation of synthetic staining outputs directly from HE inputs. These have the potential to complement the first category of these methods in outputting a virtual staining for pathologists to reference and may improve model interpretability. The other exciting avenue for this second category is research. Currently, many HE-stained imaging databases exist. A reliable methodology to generate high-quality, synthetic IF or IHC WSIs can augment these datasets for researchers worldwide. The addition of these new types of WSI data to existing datasets allows for the application of methods not previously utilized on these HE-only datasets. For example, synthetic IHC WSIs allow for segmentation approaches to identify cell types by marker positivity and allow for types of additional characterizations previously impossible with just the original HE WSIs.
A string of recent studies has begun to explore the capacity of these approaches in extracting genomic or expression data from histopathological samples. As molecular subtypes affect underlying biology, the concept is that morphological shifts occur in the image and can be detected by algorithms.
Mutational panels are commonly used to further subtype cancers in the clinic. A method to detect these mutations directly from HE will allow for rapid subtyping without the need for additional genetic testing. Chen et al[75] developed an Inception-v3 model to detect hepatocellular carcinoma and predict mutational status for CTNNB1, FMN2, TP53, and ZFX4 from frozen sections stained by HE. The mutational status prediction model was trained on the ten most significantly mutated genes in liver cancers. To validate the model, patients were split into mutated and wild-type groups for all of the ten genes, then the mutation prediction probabilities were examined across these cohorts. The four mentioned genes showed significant differences between the mutated and wild-type cohorts, indicating the model’s ability to predict these mutations. As with the cancer classification tasks, mutational subtyping from HE has a clear path to clinical integration, and these results are encouraging in supporting the value of imaging-based molecular phenotypes.
The consensus molecular subtypes (CMS) transcriptionally distinguish four groups of colorectal cancer with different clinical behaviors and biology, so Sirinukunwattana et al[76] trained a model to designate image-based CMS (imCMS) classes to HE-stained slides. The authors used two resection cohorts (TCGA, FOCUS) and one biopsy cohort (GRAMPIAN), and patches were extracted from WSI regions annotated by pathologists to be tumor. The FOCUS dataset was used for training, as their associated transcriptional data could be utilized to provide CMS labels to the extracted tumoral patches. ImCMS predictions were thus CMS predictions made on a patch-level from histology, and WSI-level subtypes were assigned according to the most prevalent patch subtype prediction. On external validation, the trained model achieved a macro-average classification AUC ranging from 0.80 to 0.83 across the TCGA and GRAMPIAN datasets.
Interestingly, to improve model generalizability, the authors implemented domain-adversarial training. Similar to the GANs mentioned earlier, the goal is to reduce the discriminative ability of this domain-adversarial module in determining whether the input data came from the TCGA, FOCUS, or GRAMPIAN datasets. In practice, the network learns to identify input features important to determine imCMS, while lessening the importance of features that simply vary based off of dataset origin. This improved macro average AUCs in TCGA to 0.84 and in GRAMPIAN to 0.85.
This study by Sirinukunwattana et al[76] offered two additional novelties in CMS classification. First, patches with high prediction confidence for imCMS subtypes could be extracted to examine histological patterns. imCMS1 was associated with mucinous differentiation and lymphocytic infiltration, imCMS2 with cribriform growth patterns and comedo-like necrosis, imCMS3 with ectatic, mucin-filled glandular structures, and imCMS4 with prominent desmoplasia. Secondly, samples with tumoral heterogeneity are currently considered unclassifiable by CMS. The authors here compared agreement between the second most prominent CMS, determined transcriptionally, and the second most prominent imCMS, predicted through this pipeline. The authors noted a high degree of significant, cosine similarity between all four CMS-imCMS pairs. This ability to identify imCMS tumor heterogeneity may improve colorectal cancer classifications and is a nice example of how current molecular subtyping approaches may be augmented by improved spatial granularity.
Microsatellite instability (MSI) is another prognostic indicator in colorectal cancers that can be diagnosed via genetic analyses. Kather et al[77] thus trained a classifier to recognize MSI and microsatellite stability (MSS) from HE-stained TCGA slides. The approach involved training an initial ResNet-18 model to recognize tumor vs normal to eventually extract only tumor-containing patches from the WSIs. Patches were then assigned MSI or MSS labels based on the patient’s TCGA-recorded MSI status or as MSI-positive if patients have an unknown status but a mutation count over 1000. This labeled data was then used to train another ResNet-18 model to predict MSI and MSS. In external validation on colorectal cases, the model exhibited a patient-level AUC of 0.84 (95%CI: 0.72-0.92). Interestingly, the gastric MSI-detection model achieved a lower AUC of 0.69 (95%CI: 0.52-0.82) on a Japanese cohort, likely reflective of the TCGA stomach adenocarcinoma training cohort being composed of 80% non-Asians and indicates the necessity of multi-center training data for more generalizable models. Finally, patient MSI-levels could be correlated with transcriptomic data. Higher MSI-levels correlated with lymphocyte gene expression in gastric cancer and with PD-L1 expression and interferon- γ signal in colorectal cancer.
This model was further refined by Kather et al[78] in a follow-up study. Specifically, the authors trained the ShuffleNet network, a more lightweight architecture that performed comparably to the more complex ones. After validating on the capacity to classify MSI in colorectal cancer, the model was trained on other tasks and found to be able detect the mutation of at least one clinically actionable mutation in 13 of 14 cancers from TCGA.
To take the next step of validation, Echle et al[79] applied this refined model to a multicenter dataset across Europe. This required the authors to form the MSIDETECT consortium and led to the generation of an 8000-patient dataset with molecular alterations. The algorithm was trained using data from multiple sources, including the TCGA, a German, a United Kingdom, and a Netherlands cohort. The study achieved an impressive AUC of 0.96 for detecting MSI in a large, international validation cohort and exemplifies the importance of multi-source training data. Compared to the study from[77], where authors showed an inability of the model trained on an 80% non-Asian TCGA dataset to perform well on a Japanese dataset, this study improved model generalizability by incorporating a more global patient distribution during training.
While the above studies targeted a specific molecular phenotype in MSI, Schmauch et al[80] set to explore whether general RNA-sequencing expression could be inferred from HE-stained tumor WSIs. The authors extracted up to 8000 tissue-containing patches per WSI from all TCGA cancer types and utilized an ImageNet-pre-trained ResNet-50 to extract 2048-length feature vectors from each. By utilizing k-means clustering, the authors generated 100 supertiles per WSI from these patches on the basis of location. The supertile representation consisted of averaged values for all contained patches over the 2048 ResNet-50 features. For each slide, the feature-extracted supertiles could be fed into another network where the output classification layer contains nodes corresponding to every gene. Thus, the inputs represent supertile feature vectors for every supertile in a WSI, and the network deconvolutes TCGA patient-level transcriptional levels across the WSI supertiles. The network is thus able to detect relationships between supertile features and WSI expression levels to identify the supertile-features most important for certain gene expressions. This is then translated into gene expression outputs at the supertile-level, which could be aggregated to generate gene expression heatmaps at the WSI-level.
Results were validated by comparing predicted expression of CD3 and CD20 with actual IHC-stained sections. For both markers, expression predicted by the model highly correlated with the percentage of cells positively stained in the IHC sections (P < 0.0001 for both). Furthermore, this predictive ability for expression data allowed the authors to implement lists of genes involved in major cancer pathways, including angiogenesis, hypoxia, deregulation of DNA repair, cell-cycling, B-cell responses, and T-cell responses for Gene Set Enrichment Analysis. The authors were able to assess the activation of these pathways across a wide range of cancers.
Finally, as MSI should be defined in part by some changes in expression levels, the authors tested whether the transcriptomic representations learned in their approach can improve the MSI detection performance relative to Kather et al[77]. When applied to HE-stained TCGA colorectal cancer slides, the model achieved a superior AUC of 0.81 compared to 0.68 for the method by Kather et al[77] in predicting MSI. The takeaway message is that, since their model can generate transcriptomic representations to the level of predicting supertile-level, gene-specific expression data, using this model as a feature extractor will generate sets of feature inputs superior in the subsequent MSI vs MSS classification compared to simply inputting the patches themselves. A method to detect expression levels with a patch-level resolution can impact not only these sorts of expression-based molecular characterizations, but also serves to provide the research community with a powerful tool to further leverage existing HE WSI datasets.
The above studies all occurred within the last few years and represent a growing application of these approaches. Historically in public datasets like TCGA, WSIs have been underutilized relative to the genomic data. These recent advances demonstrate promise in proving direct connection of imaging features with underlying genomic and expression data. Furthermore, even with the gradual decrease in sequencing costs, finances still inhibit widespread adoption and availability of these technologies. Thus, these sorts of algorithms may eventually alter clinical landscapes by providing access to genomic and expression characterizations worldwide at a fraction of the cost.
Although the field is in an exciting, fast-moving period, several challenges exist moving forward. In terms of clinical integration, the highly discussed “black box” problem still persists. Deep neural networks and the representations they learn in making their predictions lack in interpretability. This issue is magnified in healthcare where discussion of clinical decisions between patient and physician is critical and is therefore a field of highly active research. For imaging-based studies, CAM visualizations like the one utilized by Kiani et al[35] can direct pathologists’ attention the image locations utilized for generating final predictions. In addition, the decision of Kiani et al[35] to output prediction probabilities and confidences for every possible class provided pathologists with more transparency. This allowed for a clinical decision support system that operated as a tool to help inform pathologists in their decisions and may be more easily integrable into the clinic than algorithms that simply generate a prediction. The study importantly identified, however, the detrimental effects on pathologist performance when the classifier outputs an incorrect prediction. Future care must be taken to fully consider the ramifications of incorrect predictions on patient care and to manage liability of pathologists in these situations.
Another general feature of ML- and DL-based algorithms as a whole is their highly specialized nature. As might be evident from this review, models are typically focused on specific pathologies within a specific organ site. For example, there is no current study that attempts to tackle classification of gastric carcinoma, nonspecific duodenitis, and Helicobacter pylori gastritis at once. As algorithms for specific pathologies get adopted into clinical care, the feasibility of this approach will likely be tested. Special care will likely need to be taken for cataloguing a wide range of models per healthcare system, keeping up with library updates and decisions to keep or update code with deprecated support of certain functions, and maintaining constant quality control mechanisms to ensure high model performance. Each of these will be amplified by the addition of more models into a healthcare system.
As many of these studies rely upon sample annotation for certain use cases, models often become highly specialized. However, more generalizable approaches may become necessary. Hosseini et al[81] addressed this issue by establishing an “Atlas of Digital Pathology” that contains around 18000 annotated images of different tissue types across the human body. The dataset contains images across three levels, with the top level addressing the general tissue types and subsequent levels addressing subtypes. An example from top to lowest level would be Epithelium - Simple Epithelium - Simple Squamous Epithelium. Using this Atlas of Digital Pathology, Chan et al[82] then trained a model that can segment out 31 of the tissue types in the database across over more than 10 organ types. The generalizability of the model may be attributed to the non-organ-specific nature of the tissue types in the Atlas of Digital Pathology.
Binder et al[83] developed a gland segmentation algorithm for colon adenocarcinoma and breast invasive cancer by utilizing stromal masks. Here, this may be due to stroma appearing more similar across breast and colon than the glands themselves. Analogous approaches leveraging shared features across organ sites may thus help for future multi-organ models.
These research fields are highly interdisciplinary, requiring collaboration between the more quantitative computer scientists and the more biological physicians and academics. While the strength of these fields derives from the complementary nature of the two sides’ highly specialized skillsets, efforts should be made to further increase their cohesion. To illustrate a difference between the two sides, physicians and academics may be surprised to find that a large number of high impact computational studies are in the forms of conference papers or open-access online publications. This is in contrast to biomedical conferences typically being restricted to abstracts, posters, and oral presentations, and the emphasis on peer-reviewed journals.
The computer vision challenges, such as the GlaS challenge[46], may represent an avenue to introduce more cohesion. As mentioned earlier, these challenges serve an important role of generating excitement for an application and leads to submissions of high performing models from a multitude of groups. At present, however, endpoints tend to be metrics like F1 score that focus on the computational performance for the task at hand. Introduction of challenges with more biomedical endpoints may invite design and competition of pipelines that incorporate these methodologies for more directly translational tasks. An example of this would be hepatic steatosis quantification. As opposed to having a challenge to only improve lipid droplet segmentation, a challenge that provides an HE dataset and evaluates submissions by ability to translate segmentation outputs into highly accurate steatosis quantification might fast track the development of methods for specific clinical tasks. Furthermore, these sorts of challenges would require collaboration of computer scientists and physicians during submission, standardizing the concept of interdisciplinary workflows at a more preclinical point.
In a similar vein, Louis et al[84] argue the field of computational pathology faces an important need to “create a culture that considers the computer and computation as being as central to pathology as the microscope”. Integral to this, the authors posit, is the early exposure to computational concepts ideally during medical school. A certain level of computational awareness and literacy on the physicians’ side is integral to perpetuate excitement of these methodologies and for clinical integration. A similar need, however, exists on the computational side. Emerging computational scientists should be provided the opportunity and made aware of the various biomedical applications of their methodologies. This exposure benefits both sides by instilling experience of collaboration at an early stage, recognition of constraints on the other side, and a cultural adoption of the notion that the two sides should be integrated.
Overall, computational pathology is in an exciting time with rapid advancements. The imaging applications have advanced to the point of defining imaging phenotypes and correlating with some clinical variables. As covered in the NCI workshop report by Colen et al[85], the integration of imaging approaches with -omics information will be a powerful strategy to further characterize and direct clinical care but necessitates the definition of imaging standards. Though the workshop focused on radiological phenotypes, the ideas translate similarly to histopathology. Standardization of methods for data analysis, feature extraction, data integration, and data acquisition will likely be important for robust comparison of methodologies for clinical evaluation. These steps will allow for decreased uncertainties when comparing across different clinical sites and provide a confidence in the imaging phenotypes that will be necessary when beginning to correlate with other -omic phenotypes. While some studies in this review excitingly extracted -omic information directly from histopathological slides, future clinical decision support systems will likely still need to aggregate histopathological information with -omic data to a degree to inform users.
Lastly, these ML- and DL-based technologies are unique in their capacity to continuously learn in response to new data. The FDA has recognized this and published a discussion paper soliciting feedback for a potential premarket review approach for these technologies[86]. Specifically, the FDA has proposed a “Total Product Lifecycle Regulatory Approach” that covers not only premarket review and methods for transparent real-world monitoring upon rollout, but also required proposals for any anticipated changes and steps to be taken for model alterations. These changes include retraining based on new data, incorporation of new target demographics based on new data, increasing capability to different input types but with same intended use (being able to take in MRI in addition to CT for a particular diagnosis), or changing intended use (now able to diagnose an additional type of cancer). As indicated by the steps the FDA has already taken, these algorithms will need to be regulated in a unique way that maximizes the capacity for continued improvement.
In summary, these ML- and DL-based imaging methodologies are rapidly expanding and being increasingly applied in the biomedical domains. Even at this point in time, we are seeing studies that are focused on optimizing the computational tasks, on bridging into translational applications, and on integrating these technologies into clinical decision support tools. In addition to the exciting performance and potential over a wide range of topics, this field also represents an opportunity to further bring together computational scientists with their physician and academic counterparts. Future adoption of these technologies into the clinic will likely be accompanied by increased dependence of healthcare systems on computer scientists who can understand and manage the software and will hopefully encourage cultural standardization of these interdisciplinary workflows.
The application ML- and DL-based methodologies on histopathological slides in the context of gastroenterology and hepatology continues to rise. Though still largely preclinical, recent studies have exhibited the exciting performance of these models in classification and segmentation tasks and in the extraction of features unseen to the human eye, such as prognosis, information that typically necessitates additional stains, or genomic and expression data. The field is in a time rife with studies demonstrating these potential applications, and the FDA has already taken steps to begin considering the adoption of these technologies into healthcare systems. As such, it will be of importance and interest to monitor not only the methodologies themselves, but the considerations necessary in developing clinical tools.
We would like to thank Dr. Williams J for allowing us to use colorectal cancer and adjacent normal hematoxylin and eosin-stained slides, for which she helped organized IRB-approved collection, in our illustrative Figure 2A. IRB information is as follows: IRB Net #: 245765, CORIHS#: 2014-2821, Title: The Mechanisms of Intestinal Tumorigenesis, PI Name: Yang VW. We would like to thank Dr. Orzechowska E as well for providing the mouse organoid figures used in our illustrative Figure 2C.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: American Gastroenterological Association.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cabezuelo AS S-Editor: Gao CC L-Editor: A P-Editor: Wang LL
| 1. | Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, Yang GZ. Deep Learning for Health Informatics. IEEE J Biomed Health Inform. 2017;21:4-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 634] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 2. | Cision PR Newswire. AI Medical Service Inc. announces FDA Breakthrough Device Designation for endoscopic AI system. [cited 22 January 2021]. In: Cision PR Newswire [Internet]. Available from: https://www.prnewswire.com/news-releases/ai-medical-service-inc-announces-fda-breakthrough-device-designation-for-endoscopic-ai-system-300953301.html. |
| 3. | U.S. Food and Drug Administration. Artificial Intelligence and Machine Learning in Software as a Medical Device. [cited 20 December 2020]. In: U.S. Food and Drug Administration [Internet]. Available from: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device. |
| 4. | Kotsiantis SB, Zaharakis ID, Pintelas PE. Machine learning: a review of classification and combining techniques. Artif Intell Rev. 2006;26:159-190. [RCA] [DOI] [Full Text] [Cited by in Crossref: 707] [Cited by in RCA: 345] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 5. | Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH. Patch-based Convolutional Neural Network for Whole Slide Tissue Image Classification. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2016;2016:2424-2433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 304] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 6. | Ilse M, Tomczak J, Welling M. Attention-based deep multiple instance learning. International conference on machine learning. Proceedings of Machine Learning Research; 2018. |
| 7. | Xu Y, Mo T, Feng Q, Zhong P, Lai M, Eric I, Chang C. Deep learning of feature representation with multiple instance learning for medical image analysis. IEEE. 2014;1626-1630. [DOI] [Full Text] |
| 8. | Sun C, Xu A, Liu D, Xiong Z, Zhao F, Ding W. Deep Learning-Based Classification of Liver Cancer Histopathology Images Using Only Global Labels. IEEE J Biomed Health Inform. 2020;24:1643-1651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 9. | Shrestha A, Mahmood A. Review of deep learning algorithms and architectures. IEEE Access. 2019;7:53040-53065. [DOI] [Full Text] |
| 10. | Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014 Preprint. Available from: arXiv:14091556. |
| 11. | Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions. IEEE CVPR. 2015;1-9. [DOI] [Full Text] |
| 12. | He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2015 Preprint. Available from: arXiv:1512.03385. |
| 13. | Devlin J, Chang MW, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. 2018 Preprint. Available from: arXiv:1810.04805. |
| 14. | Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. 2015 Preprint. Available from: arXiv:1411.4038. |
| 15. | Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science: Springer, 2015: 234-241. |
| 16. | Pantanowitz L, Sharma A, Carter AB, Kurc T, Sussman A, Saltz J. Twenty Years of Digital Pathology: An Overview of the Road Travelled, What is on the Horizon, and the Emergence of Vendor-Neutral Archives. J Pathol Inform. 2018;9:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 135] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 17. | U.S. Food and Drug Administration. 510(k) Substantial Equivalence Determination Summary for Phillips Intellisite Pathology Solution (PIPS). [cited 22 January 2021]. In: U.S. Food and Drug Administration [Internet]. Available from: https://www.accessdata.fda.gov/cdrh_docs/reviews/K172174.pdf. . |
| 18. | U.S. Food and Drug Administration. 510(k) Substantial Equivalence Determination Summary for Sectra Digital Pathology Module. [cited 22 January 2021]. In: U.S. Food and Drug Administration [Internet]. Available from: https://www.accessdata.fda.gov/cdrh_docs/reviews/K193054.pdf. . |
| 19. | Kothari S, Phan JH, Stokes TH, Wang MD. Pathology imaging informatics for quantitative analysis of whole-slide images. J Am Med Inform Assoc. 2013;20:1099-1108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 153] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 20. | Al-Janabi S, Huisman A, Vink A, Leguit RJ, Offerhaus GJ, ten Kate FJ, van Diest PJ. Whole slide images for primary diagnostics of gastrointestinal tract pathology: a feasibility study. Hum Pathol. 2012;43:702-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5573] [Cited by in RCA: 4932] [Article Influence: 616.5] [Reference Citation Analysis (0)] |
| 22. | Gupta R, Kurc T, Sharma A, Almeida JS, Saltz J. The emergence of pathomics. Curr Pathobiol Rep. 2019;7:73-84. |
| 23. | Thakur N, Yoon H, Chong Y. Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 24. | Yoon H, Lee J, Oh JE, Kim HR, Lee S, Chang HJ, Sohn DK. Tumor Identification in Colorectal Histology Images Using a Convolutional Neural Network. J Digit Imaging. 2019;32:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 25. | Sena P, Fioresi R, Faglioni F, Losi L, Faglioni G, Roncucci L. Deep learning techniques for detecting preneoplastic and neoplastic lesions in human colorectal histological images. Oncol Lett. 2019;18:6101-6107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Iizuka O, Kanavati F, Kato K, Rambeau M, Arihiro K, Tsuneki M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci Rep. 2020;10:1504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 215] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 27. | Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211-252. |
| 28. | Xu Y, Jia Z, Wang LB, Ai Y, Zhang F, Lai M, Chang EI. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics. 2017;18:281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 194] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 29. | Sirinukunwattana K, Ahmed Raza SE, Tsang YW, Snead DR, Cree IA, Rajpoot NM. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans Med Imaging. 2016;35:1196-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 784] [Cited by in RCA: 510] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 30. | Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, Suriawinata AA, Hassanpour S. Looking under the hood: Deep neural network visualization to interpret whole-slide image analysis outcomes for colorectal polyps. IEEE CVPRW. 2017;821-827. [DOI] [Full Text] |
| 31. | Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, Suriawinata AA, Hassanpour S. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J Pathol Inform. 2017;8:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 168] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 32. | Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw Open. 2019;2:e1914645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 33. | Sali R, Moradinasab N, Guleria S, Ehsan L, Fernandes P, Shah TU, Syed S, Brown DE. Deep Learning for Whole-Slide Tissue Histopathology Classification: A Comparative Study in the Identification of Dysplastic and Non-Dysplastic Barrett's Esophagus. J Pers Med. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 34. | Leon F, Gelvez M, Jaimes Z, Gelvez T, Arguello H. Supervised classification of histopathological images using convolutional neuronal networks for gastric cancer detection. IEEE STSIVA. 2019;1-5. [DOI] [Full Text] |
| 35. | Kiani A, Uyumazturk B, Rajpurkar P, Wang A, Gao R, Jones E, Yu Y, Langlotz CP, Ball RL, Montine TJ, Martin BA, Berry GJ, Ozawa MG, Hazard FK, Brown RA, Chen SB, Wood M, Allard LS, Ylagan L, Ng AY, Shen J. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med. 2020;3:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 36. | Niazi MKK, Tavolara TE, Arole V, Hartman DJ, Pantanowitz L, Gurcan MN. Identifying tumor in pancreatic neuroendocrine neoplasms from Ki67 images using transfer learning. PLoS One. 2018;13:e0195621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 37. | Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, Samaras D, Shroyer KR, Zhao T, Batiste R, Van Arnam J; Cancer Genome Atlas Research Network; Shmulevich I, Rao AUK, Lazar AJ, Sharma A, Thorsson V. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep 2018; 23: 181-193. e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 611] [Article Influence: 87.3] [Reference Citation Analysis (0)] |
| 38. | Chang YH, Thibault G, Madin O, Azimi V, Meyers C, Johnson B, Link J, Margolin A, Gray JW. Deep learning based Nucleus Classification in pancreas histological images. Annu Int Conf IEEE Eng Med Biol Soc. 2017;2017:672-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 39. | Shapcott M, Hewitt KJ, Rajpoot N. Deep Learning With Sampling in Colon Cancer Histology. Front Bioeng Biotechnol. 2019;7:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 40. | Wei JW, Wei JW, Jackson CR, Ren B, Suriawinata AA, Hassanpour S. Automated Detection of Celiac Disease on Duodenal Biopsy Slides: A Deep Learning Approach. J Pathol Inform. 2019;10:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 41. | Srivastava A, Sengupta S, Kang SJ, Kant K, Khan M, Ali SA, Moore SR, Amadi BC, Kelly P, Syed S. Deep learning for detecting diseases in gastrointestinal biopsy images. IEEE SIEDS. 2019;1-4. [DOI] [Full Text] |
| 42. | Sali R, Ehsan L, Kowsari K, Khan M, Moskaluk CA, Syed S, Brown DE. Celiacnet: Celiac disease severity diagnosis on duodenal histopathological images using deep residual networks. IEEE BIBM. 2019;962-967. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 43. | Sali R, Adewole S, Ehsan L, Denson LA, Kelly P, Amadi BC, Holtz L, Ali SA, Moore SR, Syed S. Hierarchical Deep Convolutional Neural Networks for Multi-category Diagnosis of Gastrointestinal Disorders on Histopathological Images. 2020 Preprint. Available from: arXiv:2005.03868. |
| 44. | Martin DR, Hanson JA, Gullapalli RR, Schultz FA, Sethi A, Clark DP. A Deep Learning Convolutional Neural Network Can Recognize Common Patterns of Injury in Gastric Pathology. Arch Pathol Lab Med. 2020;144:370-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 45. | Klein S, Gildenblat J, Ihle MA, Merkelbach-Bruse S, Noh KW, Peifer M, Quaas A, Büttner R. Deep learning for sensitive detection of Helicobacter Pylori in gastric biopsies. BMC Gastroenterol. 2020;20:417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 46. | Sirinukunwattana K, Pluim JPW, Chen H, Qi X, Heng PA, Guo YB, Wang LY, Matuszewski BJ, Bruni E, Sanchez U, Böhm A, Ronneberger O, Cheikh BB, Racoceanu D, Kainz P, Pfeiffer M, Urschler M, Snead DRJ, Rajpoot NM. Gland segmentation in colon histology images: The glas challenge contest. Med Image Anal. 2017;35:489-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 287] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 47. | Xu Y, Li Y, Liu M, Wang Y, Lai M, Eric I, Chang C. Gland instance segmentation by deep multichannel side supervision. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. Lecture Notes in Computer Science: Springer, 2016: 496-504. |
| 48. | BenTaieb A, Hamarneh G. Topology aware fully convolutional networks for histology gland segmentation. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. Lecture Notes in Computer Science: Springer, 2016: 460-468. |
| 49. | Graham S, Chen H, Gamper J, Dou Q, Heng PA, Snead D, Tsang YW, Rajpoot N. MILD-Net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Med Image Anal. 2019;52:199-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 129] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 50. | Zhao P, Zhang J, Fang W, Deng S. SCAU-Net: Spatial-Channel Attention U-Net for Gland Segmentation. Front Bioeng Biotechnol. 2020;8:670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 51. | Xiao QE, Chung PC, Tsai HW, Cheng KS, Chow NH, Juang YZ, Tsai HH, Wang CH, Hsieh TA. Hematoxylin and Eosin (H&E) Stained Liver Portal Area Segmentation Using Multi-Scale Receptive Field Convolutional Neural Network. IEEE J Emerg Sel Top Circuits Syst. 2019;9:623-634. |
| 52. | Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing. 2016;191:214-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 232] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 53. | Wang X, Fang Y, Yang S, Zhu D, Wang M, Zhang J, Tong KY, Han X. A hybrid network for automatic hepatocellular carcinoma segmentation in H&E-stained whole slide images. Med Image Anal. 2021;68:101914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 54. | Qaiser T, Tsang YW, Taniyama D, Sakamoto N, Nakane K, Epstein D, Rajpoot N. Fast and accurate tumor segmentation of histology images using persistent homology and deep convolutional features. Med Image Anal. 2019;55:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 55. | Awan R, Sirinukunwattana K, Epstein D, Jefferyes S, Qidwai U, Aftab Z, Mujeeb I, Snead D, Rajpoot N. Glandular Morphometrics for Objective Grading of Colorectal Adenocarcinoma Histology Images. Sci Rep. 2017;7:16852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 56. | Abousamra S, Fassler D, Hou L, Zhang Y, Gupta R, Kurc T, Escobar-Hoyos LF, Samaras D, Knudson B, Shroyer K. Weakly-Supervised Deep Stain Decomposition for Multiplex IHC Images. IEEE ISBI. 2020;481-485. [DOI] [Full Text] |
| 57. | Fassler DJ, Abousamra S, Gupta R, Chen C, Zhao M, Paredes D, Batool SA, Knudsen BS, Escobar-Hoyos L, Shroyer KR, Samaras D, Kurc T, Saltz J. Deep learning-based image analysis methods for brightfield-acquired multiplex immunohistochemistry images. Diagn Pathol. 2020;15:100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 58. | Lee MJ, Bagci P, Kong J, Vos MB, Sharma P, Kalb B, Saltz JH, Martin DR, Adsay NV, Farris AB. Liver steatosis assessment: correlations among pathology, radiology, clinical data and automated image analysis software. Pathol Res Pract. 2013;209:371-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 59. | Forlano R, Mullish BH, Giannakeas N, Maurice JB, Angkathunyakul N, Lloyd J, Tzallas AT, Tsipouras M, Yee M, Thursz MR, Goldin RD, Manousou P. High-Throughput, Machine Learning-Based Quantification of Steatosis, Inflammation, Ballooning, and Fibrosis in Biopsies From Patients With Nonalcoholic Fatty Liver Disease. Clin Gastroenterol Hepatol 2020; 18: 2081-2090. e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 60. | Sun L, Marsh JN, Matlock MK, Chen L, Gaut JP, Brunt EM, Swamidass SJ, Liu TC. Deep learning quantification of percent steatosis in donor liver biopsy frozen sections. EBioMedicine. 2020;60:103029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 61. | Roy M, Wang F, Vo H, Teng D, Teodoro G, Farris AB, Castillo-Leon E, Vos MB, Kong J. Deep-learning-based accurate hepatic steatosis quantification for histological assessment of liver biopsies. Lab Invest. 2020;100:1367-1383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 62. | Salvi M, Molinaro L, Metovic J, Patrono D, Romagnoli R, Papotti M, Molinari F. Fully automated quantitative assessment of hepatic steatosis in liver transplants. Comput Biol Med. 2020;123:103836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 63. | Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, Walliander M, Lundin M, Haglund C, Lundin J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8:3395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 427] [Cited by in RCA: 364] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 64. | Yue X, Dimitriou N, Arandjelovic O. Colorectal cancer outcome prediction from H&E whole slide images using machine learning and automatically inferred phenotype profiles. 2019 Preprint. Available from: arXiv:1902.03582. |
| 65. | Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA, Gaiser T, Marx A, Valous NA, Ferber D, Jansen L, Reyes-Aldasoro CC, Zörnig I, Jäger D, Brenner H, Chang-Claude J, Hoffmeister M, Halama N. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019;16:e1002730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 491] [Article Influence: 81.8] [Reference Citation Analysis (0)] |
| 66. | Jiang D, Liao J, Duan H, Wu Q, Owen G, Shu C, Chen L, He Y, Wu Z, He D, Zhang W, Wang Z. A machine learning-based prognostic predictor for stage III colon cancer. Sci Rep. 2020;10:10333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 67. | Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky JM, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology. 2020;72:2000-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 187] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 68. | Wulczyn E, Steiner DF, Xu Z, Sadhwani A, Wang H, Flament-Auvigne I, Mermel CH, Chen PC, Liu Y, Stumpe MC. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS One. 2020;15:e0233678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 69. | Takamatsu M, Yamamoto N, Kawachi H, Chino A, Saito S, Ueno M, Ishikawa Y, Takazawa Y, Takeuchi K. Prediction of early colorectal cancer metastasis by machine learning using digital slide images. Comput Methods Programs Biomed. 2019;178:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 70. | Abrahao-Machado LF, Scapulatempo-Neto C. HER2 testing in gastric cancer: An update. World J Gastroenterol. 2016;22:4619-4625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 155] [Cited by in RCA: 207] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 71. | Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 171] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 72. | Redemann J, Schultz FA, Martinez C, Harrell M, Clark DP, Martin DR, Hanson JA. Comparing Deep Learning and Immunohistochemistry in Determining the Site of Origin for Well-Differentiated Neuroendocrine Tumors. J Pathol Inform. 2020;11:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 73. | Govind D, Jen KY, Matsukuma K, Gao G, Olson KA, Gui D, Wilding GE, Border SP, Sarder P. Improving the accuracy of gastrointestinal neuroendocrine tumor grading with deep learning. Sci Rep. 2020;10:11064. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 74. | Burlingame EA, McDonnell M, Schau GF, Thibault G, Lanciault C, Morgan T, Johnson BE, Corless C, Gray JW, Chang YH. SHIFT: speedy histological-to-immunofluorescent translation of a tumor signature enabled by deep learning. Sci Rep. 2020;10:17507. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 75. | Chen M, Zhang B, Topatana W, Cao J, Zhu H, Juengpanich S, Mao Q, Yu H, Cai X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol. 2020;4:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 76. | Sirinukunwattana K, Domingo E, Richman SD, Redmond KL, Blake A, Verrill C, Leedham SJ, Chatzipli A, Hardy C, Whalley CM, Wu CH, Beggs AD, McDermott U, Dunne PD, Meade A, Walker SM, Murray GI, Samuel L, Seymour M, Tomlinson I, Quirke P, Maughan T, Rittscher J, Koelzer VH; S:CORT consortium. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut. 2021;70:544-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 148] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 77. | Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 992] [Cited by in RCA: 755] [Article Influence: 125.8] [Reference Citation Analysis (0)] |
| 78. | Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, Krause J, Niehues JM, Sommer KAJ, Bankhead P, Kooreman LFS, Schulte JJ, Cipriani NA, Buelow RD, Boor P, Ortiz-Brüchle NN, Hanby AM, Speirs V, Kochanny S, Patnaik A, Srisuwananukorn A, Brenner H, Hoffmeister M, van den Brandt PA, Jäger D, Trautwein C, Pearson AT, Luedde T. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer. 2020;1:789-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 342] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 79. | Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, Heij LR, Tan X, Richman SD, Krause J, Alwers E, Jenniskens J, Offermans K, Gray R, Brenner H, Chang-Claude J, Trautwein C, Pearson AT, Boor P, Luedde T, Gaisa NT, Hoffmeister M, Kather JN. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 2020; 159: 1406-1416. e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 219] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 80. | Schmauch B, Romagnoni A, Pronier E, Saillard C, Maillé P, Calderaro J, Kamoun A, Sefta M, Toldo S, Zaslavskiy M, Clozel T, Moarii M, Courtiol P, Wainrib G. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat Commun. 2020;11:3877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 261] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 81. | Hosseini MS, Chan L, Tse G, Tang M, Deng J, Norouzi S, Rowsell C, Plataniotis KN, Damaskinos S. Atlas of digital pathology: A generalized hierarchical histological tissue type-annotated database for deep learning. IEEE CVPR. 2019;11747-11756. |
| 82. | Chan L, Hosseini MS, Rowsell C, Plataniotis KN, Damaskinos S. Histosegnet: Semantic segmentation of histological tissue type in whole slide images. IEEE ICCV. 2019;10661-10670. [DOI] [Full Text] |
| 83. | Binder T, Tantaoui EM, Pati P, Catena R, Set-Aghayan A, Gabrani M. Multi-Organ Gland Segmentation Using Deep Learning. Front Med (Lausanne). 2019;6:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 84. | Louis DN, Feldman M, Carter AB, Dighe AS, Pfeifer JD, Bry L, Almeida JS, Saltz J, Braun J, Tomaszewski JE, Gilbertson JR, Sinard JH, Gerber GK, Galli SJ, Golden JA, Becich MJ. Computational Pathology: A Path Ahead. Arch Pathol Lab Med. 2016;140:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 85. | Colen R, Foster I, Gatenby R, Giger ME, Gillies R, Gutman D, Heller M, Jain R, Madabhushi A, Madhavan S, Napel S, Rao A, Saltz J, Tatum J, Verhaak R, Whitman G. NCI Workshop Report: Clinical and Computational Requirements for Correlating Imaging Phenotypes with Genomics Signatures. Transl Oncol. 2014;7:556-569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 86. | U.S. Food and Drug Administration. Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD). [cited 22 January 2021]. In: U.S. Food and Drug Administration [Internet]. Available from: https://www.fda.gov/media/122535/download. |