Published online Oct 21, 2020. doi: 10.3748/wjg.v26.i39.5911
Peer-review started: July 27, 2020
First decision: August 8, 2020
Revised: August 18, 2020
Accepted: September 23, 2020
Article in press: September 23, 2020
Published online: October 21, 2020
Processing time: 86 Days and 1 Hours
Colonoscopy remains the standard strategy for screening for colorectal cancer around the world due to its efficacy in both detecting adenomatous or pre-cancerous lesions and the capacity to remove them intra-procedurally. Computer-aided detection and diagnosis (CAD), thanks to the brand new developed innovations of artificial intelligence, and especially deep-learning techniques, leads to a promising solution to human biases in performance by guarantying decision support during colonoscopy. The application of CAD on real-time colonoscopy helps increasing the adenoma detection rate, and therefore contributes to reduce the incidence of interval cancers improving the effectiveness of colonoscopy screening on critical outcome such as colorectal cancer related mortality. Furthermore, a significant reduction in costs is also expected. In addition, the assistance of the machine will lead to a reduction of the examination time and therefore an optimization of the endoscopic schedule. The aim of this opinion review is to analyze the clinical applications of CAD and artificial intelligence in colonoscopy, as it is reported in literature, addressing evidence, limitations, and future prospects.
Core Tip: Artificial intelligence is an emerging technology which application is rapidly increasingin numerous medical fields. The several applications of artificial intelligence in gastroenterology are showing promising results, especially in the setting of gastrointestinal oncology. Among these, the techniques able to increase the Adenoma detection rate will play a key role in reducing the colorectal cancer incidence and its related mortality caused by undetected or missclassified interval cancers. Furthermore, a significant reduction in costs is also expected. In addition the assistance of the Computer-aided detection and diagnosis systems will lead to a reduction of the examination time and therefore an optimization of the endoscopic schedule.
- Citation: Sinagra E, Badalamenti M, Maida M, Spadaccini M, Maselli R, Rossi F, Conoscenti G, Raimondo D, Pallio S, Repici A, Anderloni A. Use of artificial intelligence in improving adenoma detection rate during colonoscopy: Might both endoscopists and pathologists be further helped. World J Gastroenterol 2020; 26(39): 5911-5918
- URL: https://www.wjgnet.com/1007-9327/full/v26/i39/5911.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i39.5911
Colorectal cancer (CRC) is a major healthcare issue all over the world. It is the third most common cancer in males and second in females, and the fourth cause for cancer death worldwide[1-3]. As of now, almost 60%-70% of recognized cases in symptomatic patients are diagnosed at an advanced stage[4]. Early stage detection would offer better outcome in terms of decreasing the disease burden[5].
Globally, colonoscopy remains the standard strategy for CRC screening due to its efficacy in both detecting adenomatous or pre-cancerous lesions and the possibility to remove them intra-procedurally[5,6]. However, the identification as well as the classification of lesions are strictly operator-dependent, human errors in this setting increases the chances of progression to CRC[6]. Although uncertain, indirect evidence highlights that endoscopists might fail to detect lesions even when within the endoscopic field of view[7-11], therefore increasing the occurrence of interval colorectal cancer, defined as a CRC diagnosed within 60 mo after a negative colonoscopy.
Computer-aided detection and diagnosis (CAD), thanks to the brand new developed innovations of artificial intelligence (AI), and especially deep-learning techniques, leads to a promising solution to human biases in performance by guarantying decision support during colonoscopy[11]. The aim of this opinion review is to analyze the clinical applications of CAD and AI in colonoscopy, as it is reported in literature, addressing evidence, limitations, and future prospects.
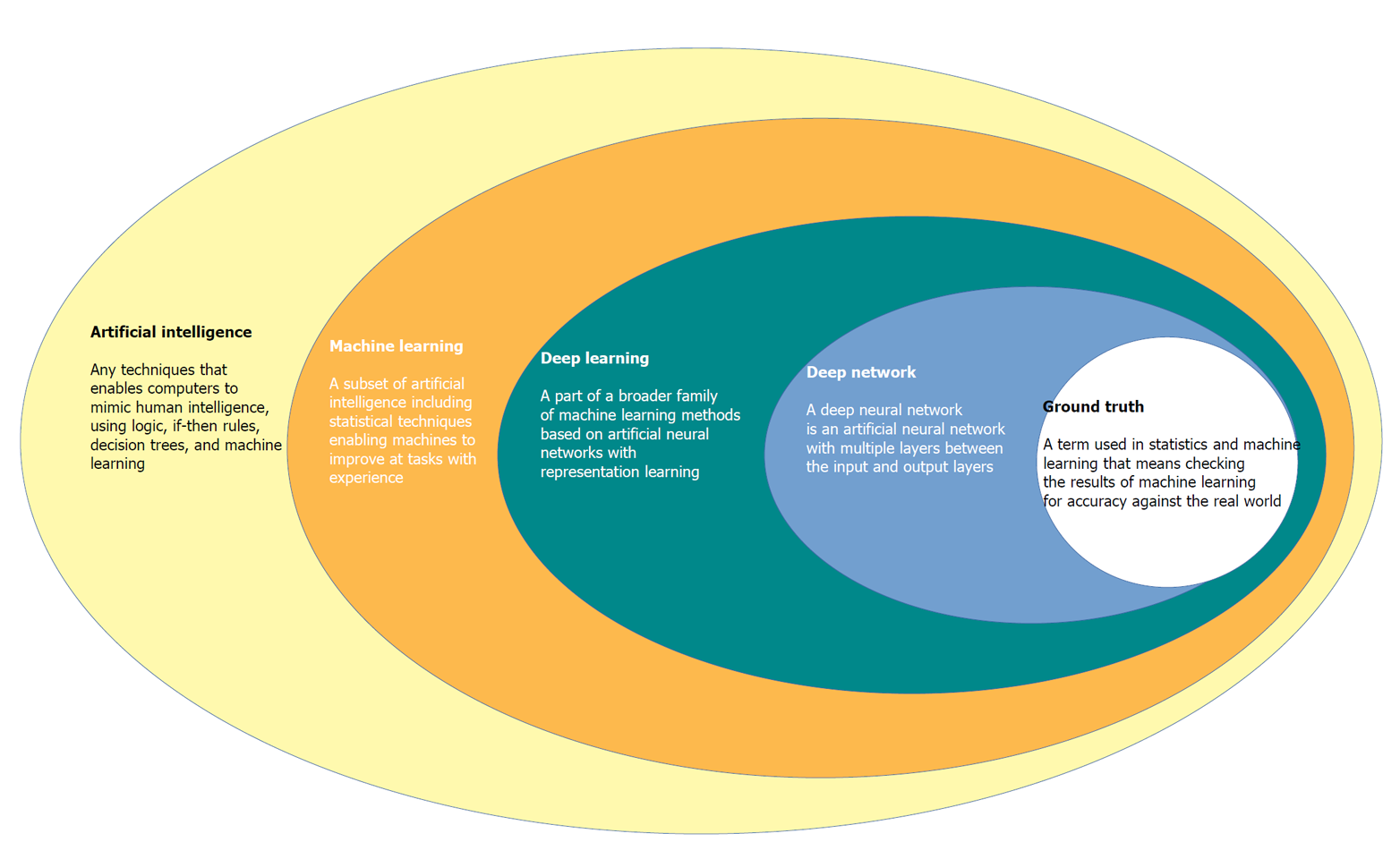
CAD systems by the use of advanced AI techniques represent an innovative technology that will likely lead to a paradigm move in the field of diagnostic colonoscopy[12,13]. Since endoscopy is generally related to computer vision technology, this technique allows computers to “see” and decipher visual content[14]. Through processes of machine learning, and more recently deep learning, AI systems can be trained to recognise “normal” characteristics by connecting a gold standard to suitable images[14] (Figure 1).
Deep learning is a recent machine learning method that utilizes a deep neural network automatically extracting specific features from data without human power if a very high numbers of learning samples are available[15,16].
Initial AI development is based on creating an algorithm that uses highly accurate datasets, coded in an independently organized manner by a group of specialists[16]. This process is called the “ground-truth” setting, where the computer is prepared to distinguish between ‘abnormal’ and ‘normal’ tissue[16]. Once an algorithm is made, deep convoluted neural networks use vast datasets to enable the algorithm to become more skilled[16]. This intelligence can extend to the algorithm becoming agnostic to manual data input and it can learn by developing its own rules and classifiers[16,17]. AI is expected to have at least 2 major roles in colonoscopy practice: Polyp detection (CADe) and polyp characterization (CADx). CADe has the potential to decrease the polyp miss rate, contributing to improve adenoma detection, whereas CADx can improve the accuracy of colorectal polyp optical diagnosis, leading to reduction of unnecessary removal of non-neoplastic lesions, potential implementation of a resect-and-discard strategy, and proper application of advanced resection techniques[17-20].
Adenoma detection rate (ADR) is defined as the proportion of patients with at least one colorectal adenoma detected at first-time screening colonoscopy, among all the patients examined by an endoscopist[21]. The benchmark for ADRs is 25% overall (30% in males, 20% in females)[22] and it is reported that a 1% increase in the ADR is associated with a 3% decrease in interval CRC incidence[20]. A recent systematic review with Meta-Analysis reported that about 25% of colorectal neoplasm are missed at screening colonoscopy[18], resulting in an unacceptable variability in the key quality indicator ADR, among endoscopists[19,20].
The rationale on the working process and therefore on the aid of AI CADe system is the Deep Learning properties. CADe systems are fed with data derived from video recordings of colonoscopies that are post-hoc revised by experts. Among them, a Convolutional Neural Network (GI-genius, Medtronic) for instance was trained and validated (99.7% sensitivity, 0.9% of activation noise due to false-positive frames)[23] using a video library of 2684 Lesions that were confirmed at histology, in a high-quality clinical trial[24]. This CAD system collects input from the digital frames of the standard endoscopic processor and outputs the coordinates of a signal box only when instance of the target lesions are recognized in the image. Real-time assessment is conceived thanks to the fact that this input-output process, leading to the CADe signal on the endoscopy display, is not perceivable as a matter of time by the endoscopist, because it appears immediately (1.52 ± 0.08 μs i.e., 1.52 millionth of a second)[25].
The average video recording of a 30 min colonoscopy is made of approximately 50000 frames, corresponding to 25-30 frames per second, and one polyp may be recognizable even only in few frames, explaining how failure in polyp recognition is likely to occur, irrespectively of the endoscopy setting. In this way this cutting-edge new technology could provide the essential help in decreasing the CRC incidence and related mortality. In fact, CADe systems have been recently introduced in the real-time endoscopic setting to overcome the colossal miss lesions rate. The outcomes reported by differentmono- and multi-centric randomized clinical trials (RCTs) seem to be stronglypromising: The overall ADR of these studies was significantly higher while aided by CADe systems as shown in Table 1[25-29]. Repici et al[25] conducted in 2019 a multicentered study in which 700 consecutive 40 to 80 years old patients were randomized 1: 1 in CADe arm and Control. ADR was significantly higher in the CADe arm 54.8% vs 40.4%. They also reported the Adenoma Per Colonoscopy, that still was higher in the CADe group (1.05 ± 1.55 vs 0.7 ± 1.19)[25]. Wang et al[26] in 2019 performed an open, non-blinded trial, in which 1058 consecutive patients were prospectively randomized to undergo diagnostic colonoscopy with (n = 522) or without (n = 536) assistance of a real-time automatic polyp detection system. Thanks to the aid of AI, ADR increased from 20% to 29%[26]. Wang et al[26] in 2020 performed a double blind monocenter RCT enrolling 1010 patients that were randomized in a CAD aided arm (n = 508) and a Control one (n = 502), reporting an higher ADR in the CAD assisted arm: 34.1% vs 28%. Gong et al[28] in 2020 performed and RCT in which 704 patients were randomly allocated with the ENDOANGEL CADe system (n = 355) or unassisted (control) colonoscopy (n = 349). ADR was significantly higher in the CADe group: 16.7% vs 8.2%. Liu et al[29] enrolled 1026 patients in a RCT and randomized them in a Control arm (n = 518) and a CADe assisted one (n = 508), also in this case ADR was reported to be significantly higher on the CADe assisted group: 39.2% vs 24%. This type of AI could also help in standardizing colonoscopy regardless the operator and the endoscopic setting by eliminating the subjective biases.
| Ref. | Study type | CADe system | ARM | ADR |
| Repici et al[25], 2020 | Multicenter RCT | GI Genius | WL | 40, 40% |
| CADe | 54, 80% | |||
| Wang et al[26], 2019 | Monocenter RCT | Endoscreener | WL | 20% |
| CADe | 29% | |||
| Wang et al[27], 2020 | Monocenter RCT | Endoscreener | WL | 28% |
| CADe | 34, 10% | |||
| Gong et al[28], 2020 | Monocenter RCT | ENDOANGEL | WL | 8, 20% |
| CADe | 16, 70% | |||
| Liu et al[29], 2020 | Monocenter RCT | HenanTongyu | WL | 24% |
| CADe | 39, 20% |
In order to reduce the incidence and related mortality of CRC, there is also a major factor other than missing-lesions that need to be demolished: The accuracy of characterizing and classifying small (< 5 mm) polyps by trained endoscopists is reported to be < 80%[30]. Furthermore, the resection of non-neoplastic/hyperplastic polyps, that are almost never evolving in a malignant form, represent a financial burden, an unnecessary intervention performed on an healthy tissue, and both the resection or/and the observation of these irrelevant polyps represent an unnecessary waste of time[31]. Recently, CADx system AI for polyp characterization and diagnosis, optical biopsy and histology prediction classificationhas been developed by multiple groups that used computational analysis to predict polyp histology[32-34]. CADx are applications of AI where the system helps the physician in interpreting the content of a medical image by assigning a label to the image itself. Consequently, AI systems that support the real time optical biopsy by providing the classification of a colon polyp into a category (e.g., adenoma, hyperplastic, etc.) are CADx. The Convolutional Neural Networks that are able to perform such task are called image classifiers, and the research in this field has been extremely active in the past five years. Both accuracy, sensitivity, specificity, positive predictive value and negative predictive value (NPV) as well as feasibility of these systems have been recently described: Aihara et al[35] initially published a prospective study of 32 patients with 102 colorectal lesions, with the use of CAD to differentiate neoplastic from non-neoplastic lesions using a CADx system that enables “real-time” color analysis of colorectal lesions when applied to autofluorescence endoscopy to perform color-tone sampling aiming to the reduction of unnecessary treatments for non-neoplastic lesions; all lesions greater than 5 mm were endoscopically removed and lesions less than 5 mm were biopsied. They concluded that lesions with a green/red ratio of less than 1.01 were neoplastic while those above that cut-off were considered non-neoplastic, with sensitivity, specificity and NPV of 94.2%, 88.9% and 85.2% respectively[35]. Kuiper et al[36] used a system called WavSTAT for real-time optical diagnosis based on laser-induced autofluorescence spectroscopy on 87 patients with 207 small (< 10 mm) colorectal lesions. During colonoscopy, the endoscopists tried to differenciate real-time adenomas vs non-adenomas as a low or high confidence call. Then, all lesions were analyzed using the system. Histopathology was used as the reference standard. The accuracy and NPV of WavSTAT were 74.4% and 73.5% respectively for WavSTAT alone, while they were 79.2% and 73.9% respectively combining WavSTAT with high resolution endoscopy[36]. The study concluded that it did not fulfill the American Society of Gastrointestinal Endoscopy (ASGE) performance thresholds for the assessment of diminutive and small lesions. Rath et al[37] used the WavSTAT4 optical biopsy forceps system designed by Spectrascience Inc, San Diego, California, United States. For prediction of histology using laser-induced autofluorescence spectroscopy on 27 patients with 137 diminutive (≤ 5 mm) polyps[37]. The accuracy was 84.7% along with sensitivity of 81.8%, specificity of 85.2% and NPV of 96.1%. They concluded that this new WavSTAT4 systemhad the potential to meet the ASGE thresholds for the “resect and discard” strategy. Kominami et al[38] used a narrow band imaging (NBI) magnification colonoscopy system for CAD real-time image recognition system with a support vector machine outputof colorectal polyps histology on 41 patients with 118 colorectal lesions[38]. Concordance between the endoscopists and CAD was 97.5%. The accuracy, sensitivity, specificity, positive predictive value and NPV were 93.2%, 93%, 93.3%, 93% and 93.3% respectively, concluding that this CAD system may satisfy the Preservation and Incorporation of Valuable Endoscopic Innovations committee recommendations of ASGE for the resect and discard strategy. Mori et al[39] used × 520 ultramagnifying colonoscopes providing microvascular and cellular visualization of colorectal polyps after application of NBI and methylene blue staining modes to assess the performance of real-time CAD. To assist with CAD and characterization in 325 patients with 466 diminutive polyps. The accuracy was 98.1% with a NPV of 96.4% and 96.5% for methylene blue staining modes and NBI modes respectively. They concluded that their CAD system can achieve the thresholds of preservation and incorporation of valuable endoscopic innovations for diminutive, non-neoplastic rectosigmoid polyps[39]. Study data are reported in Table 2. Given the novelty of this type of AI, there is still a lack of Randomized Clinical Trials that would be needed to define the exact efficacy of CADx system.
| Ref. | Target | System | Performance | Year | Country |
| Aihara et al[35] | All polyps (n = 102) | AFE | Sensitivity: 94.2 % Specificity: 88.9% NPV: 85.2% | 2013 | Japan |
| Kuiper et al[36] | < 10 mm polyps (n = 207) | WAVSTAT, WAVSTAT + HRE | Accuracy: 74.4% Accuracy + HRE: 79.2% NPV: 73.5% NPV + HRE: 73.9 % | 2015 | Netherlands |
| Rath et al[37] | ≤ 5 mm polyps (n = 137) | WAVSTAT4 + LIFS | Accuracy: 84.7% Sensitivity: 81.8% Specificity: 85.2% NPV: 96.1% | 2016 | Germany |
| Kominami et al[38] | All polyps (n = 118) | NBI | Concordance: 97.5% Accuracy: 93.2 % Sensitivity: 93% Specificity: 93.3% PPV: 93% NPV: 93.3% | 2016 | Japan |
| Mori et al[39] | ≤ 5 mm polyps (n = 466) | EC-NBI-CAD EC-MB-CAD | Accuracy: 98.1% NPV EC-NBI-CAD:96.5% NPV EC-MB-CAD: 96.4% | 2018 | Japan |
Anyhow, these technologies are rapidly evolving andare expected also to lead to a reduction of the costs related to unnecessary polypectomy, “resect and discard” and perhaps, in the future, to substitute completely the pathological examination of the resected tissue, therefore defining the specific histological characteristic and related malignancy potential as well as potential consequent follow ups immediately, during colonoscopy itself. In addition, most of the proposed AI are aiming to create a system which is able to write automatic endoscopic report. The main advantages of developing this aspect would increase the accuracy of the report itself that nowadays can stillbe compromised by the subjectivity bias of different endoscopists both in describing location and characteristics of the polyps, such as dimensions or pit pattern etc. In this way, we would expect that this technology would help standardizing characterization real-time of the lesions, and the endoscopic reports as well. The automatic report would also help the endoscopist to avoid time wasting in the writing of the report and could therefore also increase the rate of endoscopic exams and interventions performed by the digestive endoscopic unit.
AI is an emerging technology which application is rapidly increasing in numerous medical fields. The several applications of AI in gastroenterology are showing promising results, especially in the setting of gastrointestinal oncology. Among these, the techniques able to increase the ADR will play a key role in reducing the CRC incidence and its related mortality caused by undetected or misclassified interval cancers. Furthermore, a significant reduction in costs is also expected. In addition, the assistance of the machine will lead to a reduction of the examination time and therefore an optimization of the endoscopic schedule.
Moreover, the resect and discharge policy, supported by good sensitivity and specificity, will lead to decrease the direct costs of unnecessary polypectomies. Furthermore, greater diagnostic accuracy in identifying pre-neoplastic and neoplastic lesions will lead to a reduction in the secondary costs of the prevented neoplasms.
Despite the promising results, AI techniques for detection and identification of colorectal lesions, need to be furtherly investigated in the near future. In particular, since most of results come from trials performed in highly specialized centers, representing a limitation for the generalizability of results, they must be also confirmed in clinical practice.
Finally, the integration of AI in a human-base medicine setting has to be taken into consideration: AI is not conceived, nor now, not ever, to substitute the endoscopist, on the contrary, it seems to be an extremely helpful tool to be used from the endoscopist himself that, given his ability and skills, is the only one able to process and interpret all the AI information to make decisions on the patient management.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: Società Italiana Di Gastroenterologia Ed Endoscopia Digestiva.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cheng X, Coghlan E S-Editor: Zhang L L-Editor: A P-Editor: Ma YJ
| 1. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, Hamadeh RR, Moore A, Werdecker A, Gessner BD, Te Ao B, McMahon B, Karimkhani C, Yu C, Cooke GS, Schwebel DC, Carpenter DO, Pereira DM, Nash D, Kazi DS, De Leo D, Plass D, Ukwaja KN, Thurston GD, Yun Jin K, Simard EP, Mills E, Park EK, Catalá-López F, deVeber G, Gotay C, Khan G, Hosgood HD 3rd, Santos IS, Leasher JL, Singh J, Leigh J, Jonas JB, Sanabria J, Beardsley J, Jacobsen KH, Takahashi K, Franklin RC, Ronfani L, Montico M, Naldi L, Tonelli M, Geleijnse J, Petzold M, Shrime MG, Younis M, Yonemoto N, Breitborde N, Yip P, Pourmalek F, Lotufo PA, Esteghamati A, Hankey GJ, Ali R, Lunevicius R, Malekzadeh R, Dellavalle R, Weintraub R, Lucas R, Hay R, Rojas-Rueda D, Westerman R, Sepanlou SG, Nolte S, Patten S, Weichenthal S, Abera SF, Fereshtehnejad SM, Shiue I, Driscoll T, Vasankari T, Alsharif U, Rahimi-Movaghar V, Vlassov VV, Marcenes WS, Mekonnen W, Melaku YA, Yano Y, Artaman A, Campos I, MacLachlan J, Mueller U, Kim D, Trillini M, Eshrati B, Williams HC, Shibuya K, Dandona R, Murthy K, Cowie B, Amare AT, Antonio CA, Castañeda-Orjuela C, van Gool CH, Violante F, Oh IH, Deribe K, Soreide K, Knibbs L, Kereselidze M, Green M, Cardenas R, Roy N, Tillmann T, Li Y, Krueger H, Monasta L, Dey S, Sheikhbahaei S, Hafezi-Nejad N, Kumar GA, Sreeramareddy CT, Dandona L, Wang H, Vollset SE, Mokdad A, Salomon JA, Lozano R, Vos T, Forouzanfar M, Lopez A, Murray C, Naghavi M. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1945] [Cited by in RCA: 2055] [Article Influence: 205.5] [Reference Citation Analysis (0)] |
| 2. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21366] [Article Influence: 2136.6] [Reference Citation Analysis (3)] |
| 3. | Maida M, Morreale G, Sinagra E, Ianiro G, Margherita V, Cirrone Cipolla A, Camilleri S. Quality measures improving endoscopic screening of colorectal cancer: a review of the literature. Expert Rev Anticancer Ther. 2019;19:223-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 4. | Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, Ederer F. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328:1365-1371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2174] [Article Influence: 67.9] [Reference Citation Analysis (1)] |
| 5. | Maida M, Macaluso FS, Ianiro G, Mangiola F, Sinagra E, Hold G, Maida C, Cammarota G, Gasbarrini A, Scarpulla G. Screening of colorectal cancer: present and future. Expert Rev Anticancer Ther. 2017;17:1131-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 6. | Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, Zauber AG, de Boer J, Fireman BH, Schottinger JE, Quinn VP, Ghai NR, Levin TR, Quesenberry CP. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:1298-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1251] [Cited by in RCA: 1559] [Article Influence: 141.7] [Reference Citation Analysis (0)] |
| 7. | Thakkar S, Carleton NM, Rao B, Syed A. Use of Artificial Intelligence-Based Analytics From Live Colonoscopies to Optimize the Quality of the Colonoscopy Examination in Real Time: Proof of Concept. Gastroenterology 2020; 158: 1219-1221. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 8. | Crockett SD, Gourevitch RA, Morris M, Carrell DS, Rose S, Shi Z, Greer JB, Schoen RE, Mehrotra A. Endoscopist factors that influence serrated polyp detection: a multicenter study. Endoscopy. 2018;50:984-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 9. | McGill SK, Kaltenbach T, Friedland S, Soetikno R. The learning curve for detection of non-polypoid (flat and depressed) colorectal neoplasms. Gut. 2015;64:184-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Coe SG, Crook JE, Diehl NN, Wallace MB. An endoscopic quality improvement program improves detection of colorectal adenomas. Am J Gastroenterol. 2013;108:219-26; quiz 227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 11. | Ahmad OF, Soares AS, Mazomenos E, Brandao P, Vega R, Seward E, Stoyanov D, Chand M, Lovat LB. Artificial intelligence and computer-aided diagnosis in colonoscopy: current evidence and future directions. Lancet Gastroenterol Hepatol. 2019;4:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 12. | Byrne MF, Shahidi N, Rex DK. Will Computer-Aided Detection and Diagnosis Revolutionize Colonoscopy? Gastroenterology. 2017;153:1460-1464.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 13. | Ahmad OF, Stoyanov D, Lovat LB. Human-machine collaboration: bringing artificial intelligence into colonoscopy. Frontline Gastroenterol. 2019;10:198-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology 2018; 154: 2027-2029. e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 269] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 15. | Tran D, Bourdev L, Fergus R, Torresani L, Paluri M. Learning spatiotemporal features with 3d convolutional networks. Proceedings of the IEEE International Conference on Computer Vision. 2015: 4489-4497. [DOI] [Full Text] |
| 16. | Alagappan M, Brown JRG, Mori Y, Berzin TM. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J Gastrointest Endosc. 2018;10:239-249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 106] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 17. | Vinsard DG, Mori Y, Misawa M, Kudo SE, Rastogi A, Bagci U, Rex DK, Wallace MB. Quality assurance of computer-aided detection and diagnosis in colonoscopy. Gastrointest Endosc. 2019;90:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 18. | Wieszczy P, Regula J, Kaminski MF. Adenoma detection rate and risk of colorectal cancer. Best Pract Res Clin Gastroenterol. 2017;31:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 19. | Rex DK, Schoenfeld PS, Cohen J, Pike IM, Adler DG, Fennerty MB, Lieb JG 2nd, Park WG, Rizk MK, Sawhney MS, Shaheen NJ, Wani S, Weinberg DS. Quality indicators for colonoscopy. Gastrointest Endosc. 2015;81:31-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 836] [Article Influence: 83.6] [Reference Citation Analysis (0)] |
| 20. | Corley DA, Levin TR, Doubeni CA. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:2541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 21. | Zhao S, Wang S, Pan P, Xia T, Chang X, Yang X, Guo L, Meng Q, Yang F, Qian W, Xu Z, Wang Y, Wang Z, Gu L, Wang R, Jia F, Yao J, Li Z, Bai Y. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology 2019; 156: 1661-1674. e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 374] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 22. | Rex DK, Cutler CS, Lemmel GT, Rahmani EY, Clark DW, Helper DJ, Lehman GA, Mark DG. Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies. Gastroenterology. 1997;112:24-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1089] [Cited by in RCA: 1048] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 23. | Hassan C, Wallace MB, Sharma P, Maselli R, Craviotto V, Spadaccini M, Repici A. New artificial intelligence system: first validation study versus experienced endoscopists for colorectal polyp detection. Gut. 2020;69:799-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 24. | Repici A, Wallace MB, East JE, Sharma P, Ramirez FC, Bruining DH, Young M, Gatof D, Irene Mimi Canto M, Marcon N, Cannizzaro R, Kiesslich R, Rutter M, Dekker E, Siersema PD, Spaander M, Kupcinskas L, Jonaitis L, Bisschops R, Radaelli F, Bhandari P, Wilson A, Early D, Gupta N, Vieth M, Lauwers GY, Rossini M, Hassan C. Efficacy of Per-oral Methylene Blue Formulation for Screening Colonoscopy. Gastroenterology 2019; 156: 2198-2207. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 25. | Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, Ferrara E, Spadaccini M, Alkandari A, Fugazza A, Anderloni A, Galtieri PA, Pellegatta G, Carrara S, Di Leo M, Craviotto V, Lamonaca L, Lorenzetti R, Andrealli A, Antonelli G, Wallace M, Sharma P, Rosch T, Hassan C. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology 2020; 159: 512-520. e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 393] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 26. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 27. | Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, Xiong F, Wang H, Song Y, Pan Y, Zhou G. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. 2020;5:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 294] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 28. | Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, Wang Z, Zhou W, An P, Huang X, Jiang X, Li Y, Wan X, Hu S, Chen Y, Hu X, Xu Y, Zhu X, Li S, Yao L, He X, Chen D, Huang L, Wei X, Wang X, Yu H. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 265] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 29. | Liu WN, Zhang YY, Bian XQ, Wang LJ, Yang Q, Zhang XD, Huang J. Study on detection rate of polyps and adenomas in artificial-intelligence-aided colonoscopy. Saudi J Gastroenterol. 2020;26:13-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 126] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 30. | Rees CJ, Rajasekhar PT, Wilson A, Close H, Rutter MD, Saunders BP, East JE, Maier R, Moorghen M, Muhammad U, Hancock H, Jayaprakash A, MacDonald C, Ramadas A, Dhar A, Mason JM. Narrow band imaging optical diagnosis of small colorectal polyps in routine clinical practice: the Detect Inspect Characterise Resect and Discard 2 (DISCARD 2) study. Gut. 2017;66:887-895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 31. | El Hajjar A, Rey JF. Artificial intelligence in gastrointestinal endoscopy: general overview. Chin Med J (Engl). 2020;133:326-334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 32. | Puig I, Kaltenbach T. Optical Diagnosis for Colorectal Polyps: A Useful Technique Now or in the Future? Gut Liver. 2018;12:385-392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Mori Y, Kudo SE, Chiu PW, Singh R, Misawa M, Wakamura K, Kudo T, Hayashi T, Katagiri A, Miyachi H, Ishida F, Maeda Y, Inoue H, Nimura Y, Oda M, Mori K. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy. 2016;48:1110-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 34. | Lee BI, Matsuda T. Estimation of Invasion Depth: The First Key to Successful Colorectal ESD. Clin Endosc. 2019;52:100-106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Aihara H, Saito S, Inomata H, Ide D, Tamai N, Ohya TR, Kato T, Amitani S, Tajiri H. Computer-aided diagnosis of neoplastic colorectal lesions using 'real-time' numerical color analysis during autofluorescence endoscopy. Eur J Gastroenterol Hepatol. 2013;25:488-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Kuiper T, Alderlieste YA, Tytgat KM, Vlug MS, Nabuurs JA, Bastiaansen BA, Löwenberg M, Fockens P, Dekker E. Automatic optical diagnosis of small colorectal lesions by laser-induced autofluorescence. Endoscopy. 2015;47:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 37. | Rath T, Tontini GE, Vieth M, Nägel A, Neurath MF, Neumann H. In vivo real-time assessment of colorectal polyp histology using an optical biopsy forceps system based on laser-induced fluorescence spectroscopy. Endoscopy. 2016;48:557-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 38. | Kominami Y, Yoshida S, Tanaka S, Sanomura Y, Hirakawa T, Raytchev B, Tamaki T, Koide T, Kaneda K, Chayama K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 39. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 355] [Article Influence: 50.7] [Reference Citation Analysis (1)] |