Published online Sep 14, 2020. doi: 10.3748/wjg.v26.i34.5090
Peer-review started: May 22, 2020
First decision: June 20, 2020
Revised: July 1, 2020
Accepted: August 12, 2020
Article in press: August 12, 2020
Published online: September 14, 2020
Processing time: 109 Days and 23.5 Hours
Since the advent of artificial intelligence (AI) technology, it has been constantly studied and has achieved rapid development. The AI assistant system is expected to improve the quality of automatic polyp detection and classification. It could also help prevent endoscopists from missing polyps and make an accurate optical diagnosis. These functions provided by AI could result in a higher adenoma detection rate and decrease the cost of polypectomy for hyperplastic polyps. In addition, AI has good performance in the staging, diagnosis, and segmentation of colorectal cancer. This article provides an overview of recent research focusing on the application of AI in colorectal polyps and cancer and highlights the advances achieved.
Core Tip: In recent years, the application of artificial intelligence in the diagnosis and treatment of colorectal polyps and cancer has increased. These applications include automated polyp detection and classification as well as qualitative and staging diagnosis of colorectal cancer. This article provides an overview of recent research focusing on the application of artificial intelligence in colorectal polyps and cancer and highlights the advances achieved.
- Citation: Wang KW, Dong M. Potential applications of artificial intelligence in colorectal polyps and cancer: Recent advances and prospects. World J Gastroenterol 2020; 26(34): 5090-5100
- URL: https://www.wjgnet.com/1007-9327/full/v26/i34/5090.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i34.5090
Artificial intelligence (AI) is a wide-ranging branch of computer science concerned with building smart machines capable of performing tasks that typically require human intelligence. AI technology has made great progress, mainly owing to the development of analytical methods such as support vector machines and deep learning. Through continuous learning from data and experience accumulation, the task processing ability of the machine is greatly enhanced. AI has been improved through algorithm learning and knowledge management. It has gradually been applied in imaging and pathological diagnosis, disease management, drug research and development[1], and promoting the development of genetics and molecular medicine. The research results of Theofilatos et al[2] confirm this view. In their research, AI was used to find new treatment methods through a protein interaction algorithm, which may become a new direction in the development of molecular medicine. AI systems that use deep learning have been utilized in images of lesions such as esophageal cancer, glaucoma, and skin cancer with good performance[3-5].
In recent years, the application of AI in the diagnosis and treatment of colorectal polyps and cancer has also increased[6-8]. In the field of gastroenterology, there has been considerable interest in utilizing AI as an adjunctive detection technique in endoscopy. AI provides the promise of increasing polyp detection and even optical polyp diagnosis, all requiring minimal training of the endoscopist. For instance, a fast detection algorithm named ResYOLO was pretrained with a large database of nonmedical images and then refined with images extracted from colonoscopic videos. Evaluated on 17574 frames from 18 endoscopic videos, the proposed method could find frames with polyps with an accuracy of 88.6%, recall of 71.6%, and a processing speed of 0.15 s per frame[6]. With the advent of deep learning algorithms and significant advances in computer capabilities, more and more AI assistance, some of which may be used in real time during colonoscopy, is now being implemented.
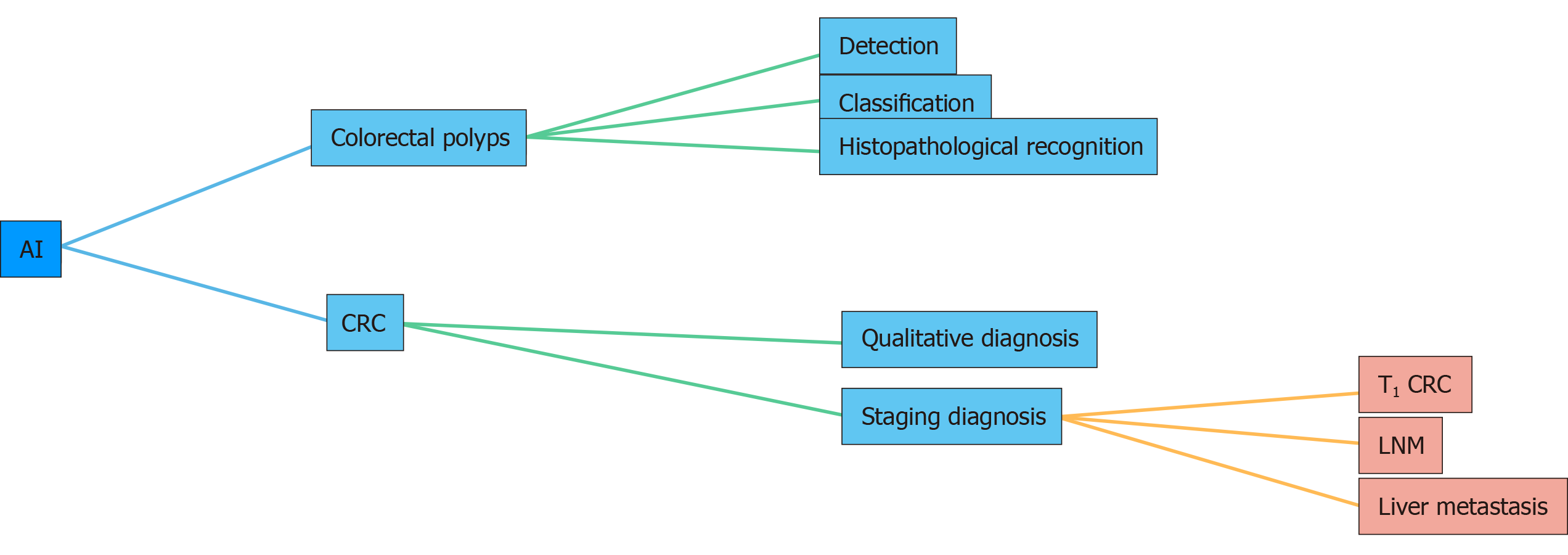
We searched for relevant literature in the MEDLINE and PubMed databases (2015–2020) using the following keywords: “deep learning,” “computer-assisted diagnosis,” “artificial intelligence,” “colorectal polyps,” and “colorectal cancer.” We only reviewed full journal articles published in English. The inclusion criteria were as follows: (1) Studies that associated AI with the detection and classification of colorectal polyps; and (2) Studies that associated AI with the diagnosis of colorectal cancer (CRC). This review highlights recent advances in the application of AI in colorectal polyps and cancer in the past 5 years (Figure 1).
At present, colonoscopy is still the most important diagnostic method for colorectal polyps. It is estimated that the prevalence of precancerous polyps in the 50+ years old screening population will be more than 50%[9]. Adenoma is the most common precancerous polyp. The adenoma detection rate (ADR) is an indicator of the colonoscopist’s ability to detect adenomas. However, the ADR by colonoscopists varies from 7% to 53%[10]. Many studies have shown that endoscopists with a higher ADR in screening colonoscopy can more effectively protect patients from the subsequent risk of colon cancer[10,11]. Corley et al[10] evaluated 314872 colonoscopies performed by 136 colonoscopists. The results showed that for every 1.0% increase in ADR, the risk of CRC was reduced by 3.0%. However, the rate of missed adenoma during colonoscopy is still high and estimated to be between 6% and 27%[12]. Thus, new techniques are required to increase the ADR during colonoscopy. In recent years, more and more scholars have investigated the application of AI in the diagnosis of colonic polyps[13-26]. All these studies/applications with detailed data are summarized in Tables 1 and 2.
| Ref. | Study type | Algorithm | Imaging modality | Image type | Training set | Testing set | Processing time |
| Mori et al[13] | Pilot study | - | EC | Real-time | - | - | 0.3 s/image |
| Misawa et al[14] | Ex vivo | Machine learning: SVM | EC, NBI | Still | 979 images (381 non-neoplasms, 598 neoplasms) | 100 images (50 non-neoplasms, 50 neoplasms) | 0.3 s/image |
| Kominami et al[15] | - | Machine learning: SVM | Colonoscopy, NBI | Real-time | 2247 cutout training images from 1262 colorectal lesions | 118 images | 20 frame/s |
| Mori et al[16] | International web-based trial | Machine learning: SVM | EC | Still | 6051 endocytoscopic images | 205 small polyps (147 neoplastic and 58 non-neoplastic) | 0.2 s/image |
| Misawa et al[17] | Pilot study | Machine learning: SVM | EC, NBI | Still | 1661 EC-NBI images (1213 neoplasm images, 448 non-neoplastic images) | 124 (19 neoplastic and 105 non-neoplastic) | - |
| Chen et al[18] | Pilot study | Deep neural network | Colonoscopy, magnifying NBI | Still | 2157 (1476 neoplastic polyps vs 681 hyperplastic polyps) | 284 (96 hyperplastic and 188 neoplastic polyps) | 0.45 s/image |
| Misawa et al[19] | Ex vivo | Machine learning | Colonoscopy, WL | Video | 411 (105 positive and 306 negative) | 135 (50 positive and 85 negative) | - |
| Shin et al[20] | Pilot study | Machine learning | Colonoscopy, WL | Video | 1525 (561 polyp patches and 964 normal patches) | 366 (196 polyp patches and 170 normal patches) | 95 ms/frame |
| Wang et al[21] | Ex vivo | Deep learning | Colonoscopy, WL | Still | 5545 (3634 images contained polyps and 1911 images did not contain polyps) | 27 113 (5541 images contained polyps and 21572 images did not contain polyps) | - |
| Kudo et al[22] | Pilot study | Texture analysis | EC stained or NBI image | Still | 69 142 EC images (43197 stained images and 25945 NBI images) | 100 polyps | 0.4 s/image |
| Min et al[23] | Pilot study | Gaussian mixture model | Colonoscopy, linked color imaging | Still | 139 images of adenomatous polyps and 69 images of non-adenomatous polyps | 115 images of adenomatous polyps and 66 images of non-adenomatous polyps | - |
| Sánchez-Montes et al[24] | Pilot study | SVM | Colonoscopy, WL | Still | - | - | - |
| Horiuchi et al[25] | Pilot study | - | Colonoscopy, autofluorescence imaging | Real-time | - | - | - |
| Byrne et al[26] | Ex vivo | Convolutional neural network | EC, NBI | Video | 223 polyp videos | 125 polyp videos | 50 ms/frame |
| Ref. | Patients, n | Samples, n | Sensitivity, % | Specificity, % | Accuracy, % | NPV, % | PPV, % |
| Mori et al[13] | 152 | 176 | 92.0 | 79.5 | 89.2 | - | |
| Misawa et al[14] | - | 100 | 84.5 | 97.6 | 90.0 | 82.0 | 98.0 |
| Kominami et al[15] | 41 | 118 | 95.9 | 93.3 | 94.9 | 93.3 | 95.9 |
| Mori et al[16] | 123 | 205 | 89.0 | 88.0 | 89.0 | 76.0 | 95.0 |
| Misawa et al[17] | 58 | 64 | 94.3 | 71.4 | 87.8 | 83.3 | 89.2 |
| Chen et al[18] | 193 | 284 | 96.3 | 78.1 | 90.1 | 91.5 | 89.6 |
| Misawa et al[19] | 73 | 155 | 90.0 | 63.3 | 76.5 | - | - |
| Shin et al[20] | - | 366 | 95.9 | 95.9 | 95.9 | - | 96.4 |
| Wang et al[21] | 1138 | 27113 | 94.4 | 95.9 | - | - | - |
| Kudo et al[22] | 89 | 100 | 96.9 (stained) | 100.0 | 98.0 | 94.6 | 100.0 |
| 96.9 (NBI) | 94.3 | 96.0 | 94.3 | 96.9 | |||
| Min et al[23] | 91 | 181 | 83.3 | 70.1 | 78.4 | 71.2 | 82.6 |
| Sánchez-Montes et al[24] | - | 225 | 92.3 | 89.2 | 91.1 | 87.1 | 93.6 |
| Horiuchi et al[25] | 77 | 258 | 80.0 | 95.3 | 91.5 | 93.4 | 85.2 |
| Byrne et al[26] | - | 106 | 98.0 | 83.0 | 94.0 | 97.0 | 90.0 |
AI is increasingly applied in gastrointestinal endoscopy, especially in the detection of colorectal polyps[27,28]. The ideal automatic detection tools for polyps should have a high sensitivity for polyp detection, a low rate of false positives, and a low latency so that polyps can be tracked and identified during real-time colonoscopy. Bowel preparation quality is an important factor affecting the accuracy of routine colonoscopy. Becq et al[29] evaluated the performance of a deep learning method for polyp detection during routine colonoscopy with variable bowel preparation quality and found that the deep learning method could effectively identify polyps by colonoscopy, even in the setting of variable bowel preparation quality.
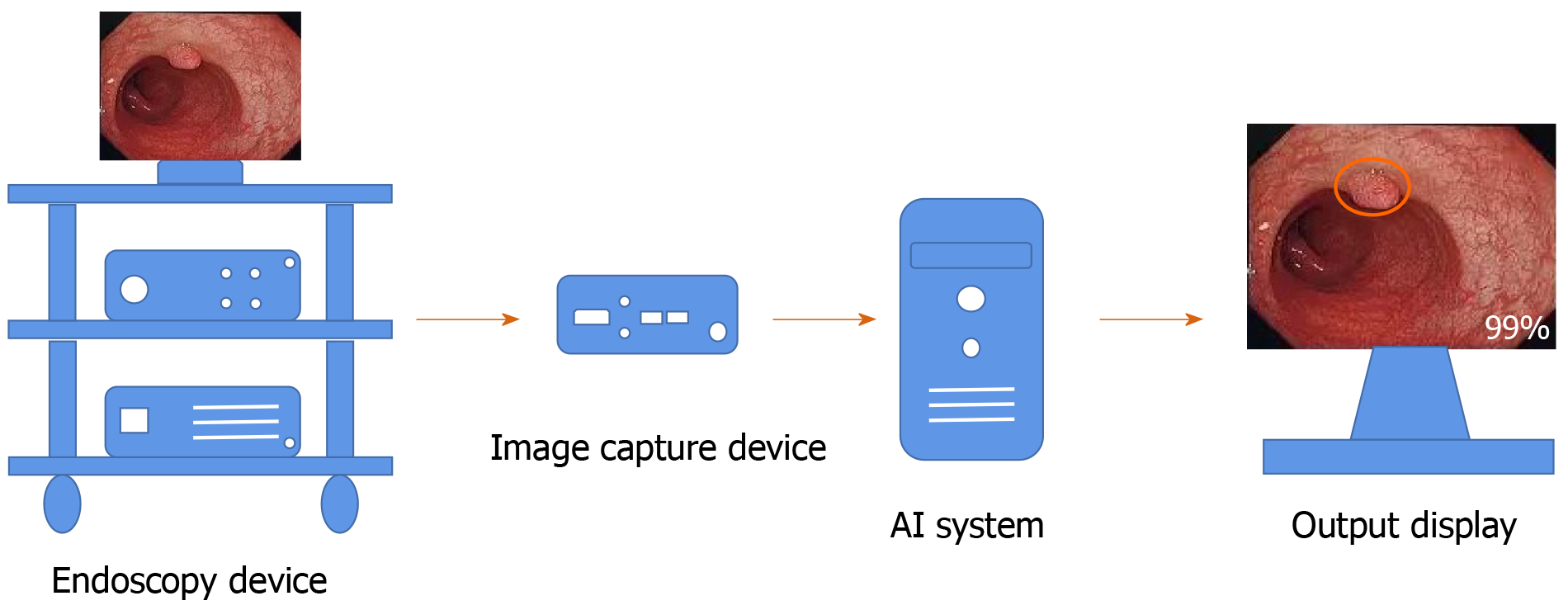
In recent years, many studies have found that an AI system can remind the endoscopist in real time to avoid the omission of nonpolypoid lesions and other abnormalities during colonoscopy, which increases the ADR[30,31] (Figure 2). However, this requires validation in large multicenter trials. In addition to conventional computer-assisted diagnosis (CAD), a convolutional neural network (CNN) system using AI has rapidly developed over the past 5 years[32]. A novel online and offline three-dimensional deep learning integration framework based on a three-dimensional fully convolutional network was proposed by Yu et al[33]. This framework can learn more representative spatiotemporal features from colonoscopy videos and has stronger recognition ability compared with previous methods such as two-dimensional CNN or hand-crafted features[33]. Recently, a novel AI system (GI-Genius, Medtronic) was reported to have a sensitivity of 99.7% in the detection of colorectal polyps. The proportion of false positive frames found from colonoscopy was less than 1% of the total frames. Furthermore, the reaction time was shorter using this novel AI system compared with visual inspection by endoscopists in 82% of the cases[34]. A meta-analysis including six studies of AI on polyp detection showed a pooled area under the receiver operating characteristic curve (AUC) of 0.90. The pooled sensitivity and specificity of AI for polyp detection were 95.0% and 88.0%, respectively[35].
Wireless capsule endoscopy (WCE) is a noninvasive alternative to conventional endoscopes and is an essential tool for diagnostic inspection of the gastrointestinal tract. Due to large amounts of data captured by WCE, it takes a few hours for the doctor to make a diagnostic decision as the images need to be checked frame by frame. Therefore, an automatic CAD system is essential to assist physicians in analyzing and separating polyp images from whole data. For this purpose, Yuan et al[36] proposed a novel deep feature learning algorithm, named stacked sparse autoencoder with image manifold constraint, to identify polyps in the WCE images. The average accuracy of this algorithm for WCE images was 98.0%. Although this accuracy is high, it is far from perfect. The proposed algorithm did not perform well if inhomogeneous illuminations existed in the WCE images. Thus, there is still a lot of work to do to improve this method.
In the preservation and incorporation of valuable endoscopic innovations of the American Society for Gastrointestinal Endoscopy recommendation, endoscopists are required to receive intensive training on image-enhanced endoscopy to achieve a negative predictive value of > 90% in predicting the absence of adenomatous histology[37]. At present, many AI systems have reached the above standards. A total of 7680 colonic polyp images from 18 studies were included in a meta-analysis of polyp histology prediction utilizing an AI system. The pooled sensitivity in polyp histology prediction was 92.3%, and pooled specificity was 89.8%. The AUC of the AI in polyp histology prediction was 0.96[35]. When compared with visual inspection by endoscopists, the results of one study show that AI had similar precision (87.3% vs 86.4%) but a higher recall rate (87.6% vs 77.0%) and higher accuracy (85.9% vs 74.3%)[38]. Sánchez-Montes et al[24] developed a CAD system that can help the identification of dysplastic lesions. This system includes three stages: (1) Image preprocessing; (2) Extraction of textons. They used three texton feature images (branching, tubularity, and contrast) generated from textons extracted from the input image; and (3) Characterization. The sensitivity, specificity, accuracy, negative predictive value, and positive predictive value were 92.3%, 89.2%, 91.1%, 87.1%, and 93.6%, respectively[24].
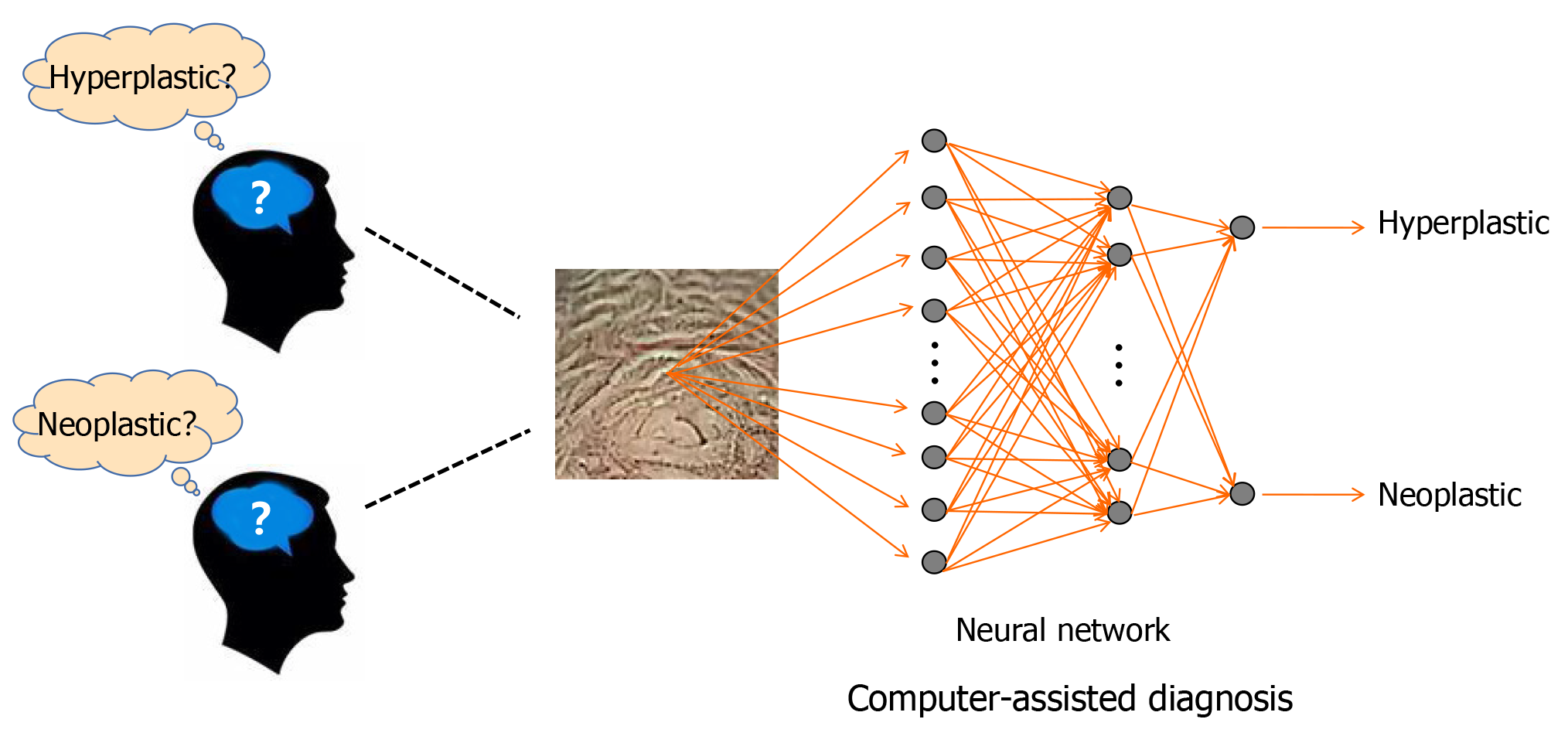
More and more studies have revealed that deep learning using CNNs is a good option for colonic polyp classification[39] (Figure 3). Song et al[40] reported that the overall diagnostic accuracy of CAD using a deep learning model was 81.3%-82.4%, which was significantly higher than that of the trainees (63.8%-71.8%, P < 0.01) and comparable with that of experts (82.4%-87.3%)[40]. This result suggests that CAD using deep learning is helpful for trainees in diagnosing colorectal polyps. Similar results were also seen in the evaluation of diminutive (< 5 mm) colorectal polyps by a CNN model, which also significantly reduced the time of diagnosis by endoscopists (from 3.92 s to 3.37 s/polyp, P = 0.042)[41]. An optical diagnosis model based on CNN was specifically designed to identify hyperplastic/serrated and adenomatous polyps, and the performance of this model exceeded the threshold of preservation and incorporation of valuable endoscopic innovations for both “diagnose and leave” and “resect and discard” strategies independent of narrow-band imaging utilization[42]. With the development of AI systems, endoscopists may accurately predict the pathology of polyps less than 3 mm in diameter[43].
The above studies were all retrospective. In order to further verify the effectiveness of AI in recent years, more and more scholars have begun to carry out prospective research to examine the application of AI in the diagnosis of colorectal polyps[44-48]. Wang et al[46] conducted a nonblinded, prospective randomized controlled study from September 2017 to February 2018, which included the largest sample size to date. This prospective study enrolled 1058 patients including 522 randomized to colonoscopy with CAD and 536 randomized to standard colonoscopy. The results showed that the AI system significantly increased the ADR (29.1% vs 20.3%, P < 0.001) and the mean number of adenomas per patient (0.53 vs 0.31, P < 0.001)[46]. In order to eliminate the operational bias in their nonblinded study and evaluate the effectiveness of the CAD system more rigorously, the authors performed a randomized, double-blind trial from September 2018 in a single center. The ADR was significantly higher in the CAD group than in the sham group (34% vs 28%, P = 0.03)[47].
Histopathological characterization of colorectal polyps is still the gold standard for diagnosis of polyps. It is critical for determining future endoscopic resection or regular follow-up in patients. However, this characterization is a challenging task and suffers significant intra- and interobserver variability. Thus, an automatic image analysis AI system that can help pathologists to identify different types of colorectal polyps accurately is necessary. In recent years, many scholars have begun to probe into this area[49-51]. Korbar et al[50] proposed an AI system based on a deep neural network model to identify the types of colorectal polyps on whole slide, hematoxylin and eosin-stained images. The results of this system showed a precision of 89.7%, F1 score of 88.8%, recall of 88.3%, and accuracy of 93.0%[50]. In another study, a deep learning model was proposed to recognize four different stages of cancerous tissue development, including normal mucosa, early preneoplastic lesion, adenoma, and cancer. An overall accuracy of > 95% was achieved[51].
CRC usually begins with a benign tumor, initially in the form of polyps, which will develop into cancer over time. It is the third most common cancer and second most common cause of cancer-related mortality worldwide[52]. The number of patients with new onset CRC is approximately 12 million a year with 600000 deaths[53]. The high mortality and poor prognosis of CRC make this disease a huge threat to the social economy and people’s health. Early diagnosis and treatment patients with CRC have always been the focus of clinical work. The systematic research in application of AI in the diagnosis of CRC is still lacking. However, with the continuous development of AI and more applications in the field of medicine, it has now emerged in the diagnosis of CRC.
The diagnosis of colorectal tumors can be divided into qualitative diagnosis and staging diagnosis. Qualitative diagnosis refers to colonoscopy and pathological biopsy to determine the presence of colorectal tumors. Colonoscopy has been an effective tool in the early detection of neoplastic lesions. Although magnifying endoscopy[54], narrow-band imaging[55], endocytoscopy[56], and confocal laser endomicroscopy[57] have a higher accuracy, the results are operator dependent. It is difficult to train all endoscopists to perform all methods well. Thus, a CAD system for endocytoscopy was developed to solve this problem. Takeda et al[58] carried out a study to evaluate the diagnostic ability of a CAD system for endocytoscopy for invasive CRC. In this study, a CAD system for endocytoscopy analyzed endocytoscopy images that are based on the information from texture analysis and nuclei. All 296 features (288 from texture analysis, 8 from nuclei) are used as the data for evaluating endocytoscopy images. A support vector machine analyzed these features and classified the images into three histological groups: Invasive cancer, adenoma, and non-neoplasm. The sensitivity, specificity, accuracy, negative predictive value, and positive predictive value were 89.4%, 98.9%, 94.1%, 90.1%, and 98.8%, respectively[58]. This technology is expected to bridge the gap in diagnosis quality for endoscopists at different levels. Histopathological diagnosis can be made by pathologists based on images of tissues obtained from a colonoscopic biopsy. Recently, many scholars have begun to explore the application of AI in identifying histopathological images of CRC[59,60]. Yoon et al[60] evaluated the performance of the CNN model in histologic diagnosis. The results for sensitivity, specificity, and accuracy were 95.10%, 92.76%, and 93.48%, respectively. The CNN model correctly classified 294 of 309 normal images and 667 of 719 tumor images[60].
AI is also used in the staging diagnosis of CRC. Computed tomography, magnetic resonance imaging (MRI), and other imaging techniques are commonly used to stage CRC. Ito et al[60] used CNN to assist in the diagnosis of cT1b CRC. With CNN learning, the sensitivity, specificity, and accuracy were 67.5%, 89.0%, and 81.2%, respectively, and the AUC was 0.871[61]. Whether additional surgery is required after endoscopic resection of T1 CRC is currently based on international guidelines. A recent study reported that an AI model predicted positivity or negativity for lymph node metastasis by analyzing 45 clinicopathological factors of T1 CRC. The sensitivity, specificity, and accuracy were 100%, 66%, and 69%, respectively, which were higher compared to the current guidelines[62]. These results suggested that AI may help to reduce unnecessary additional surgery after endoscopic resection of T1 CRC. MRI is the best method for confirming the diagnosis of pelvic lymph node metastasis before surgery. Radiologists make diagnostic decisions usually based on their subjective experience. Thus, this diagnosis lacks accuracy and objectivity. To address this problem, a faster region-based CNN was trained to read pelvic MRI images and to make diagnoses with an AUC of 0.912. The diagnosis time in one case by faster region-based CNN was 20 s, which was much shorter than that (600 s) for the radiologists’ diagnoses[63,64]. These results suggest that the faster region-based CNN enables an accurate and rapid diagnosis of CRC lymph node metastasis.
Liver is another common metastatic site of CRC. Therefore, screening of CRC patients with a high risk of liver metastasis is very important for individualized surveillance. One hundred and fifty-two tumor features extracted from computed tomography imaging and six clinical factors were used to develop a new noninvasive AI model for this task. The hybrid model, which combined relevant imaging features and clinical variables, improved accuracy of both training (90.63%) and validation (85.50%) sets with an AUC of (0.96; 0.87)[65].
Colorectal tumor segmentation is an important step in the analysis and diagnosis of CRC. However, the manual delineation of tumors is a time-consuming procedure and requires a high level of expertise. Thus, many deep learning models for automatic localization and segmentation of rectal cancers on MRI images have been proposed with high accuracy in recent years[66,67]. These AI systems can save radiologists a lot of time, but more randomized controlled studies are required to verify the stability of the results.
Although the results from previous studies appear to be promising, supporting evidence of AI systems applied in colonoscopy is still lacking as most studies were designed retrospectively. Due to the retrospective nature of most studies and the potential selection bias involved, further prospective double-blinded clinical trials are required to confirm the role of AI-assisted colonoscopy in clinical practice. We suggest the following for further research: (1) A prospective evaluation with real-time use of AI is required; (2) In order for the proposed method to have practical value in clinical trials, further testing with a large number of pathologically proved data sets is very important to verify the effectiveness and stability of the proposed classification method; (3) A study in an international, multicenter setting should be conducted to ensure the reproducibility and stability of the results; and (4) The efficacy of AI should be evaluated in all types of colorectal lesions. Other important types of lesions such as sessile serrated lesions, ulcerative colitis, or colitis-associated cancer should be also investigated as targets of an AI system. In addition, the establishment of a clinical AI system requires the use of a large amount of clinical data from patients. Compared with research in other directions, the application of medical data also involves protection of patient privacy and ethical issues. Once the information is leaked, it may cause unpredictable consequences. Therefore, the safe management of medical data should also be a key issue. When these problems are appropriately addressed, AI can be used clinically for colorectal diseases.
AI is an exciting new field in colorectal diseases. AI technologies such as deep learning can speed up the processing of large amounts of imaging or clinical data, allowing machines to assist physicians in many important tasks such as colorectal polyp detection and classification as well as qualitative and staging diagnosis of CRC. In order to utilize AI wisely, clinicians should strive to understand the feasibility of AI and mitigate its drawbacks.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bramhall SR, Caboclo JF, Tomizawa M S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Wang LL
| 1. | de Grey AD. Artificial Intelligence and Medical Research: Time to Aim Higher? Rejuvenation Res. 2016;19:105-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | Theofilatos K, Pavlopoulou N, Papasavvas C, Likothanassis S, Dimitrakopoulos C, Georgopoulos E, Moschopoulos C, Mavroudi S. Predicting protein complexes from weighted protein-protein interaction graphs with a novel unsupervised methodology: Evolutionary enhanced Markov clustering. Artif Intell Med. 2015;63:181-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 3. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 271] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 4. | Li Z, He Y, Keel S, Meng W, Chang RT, He M. Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. Ophthalmology. 2018;125:1199-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 443] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 5. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Corrigendum: Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;546:686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 107] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 6. | Zhang R, Zheng Y, Poon CCY, Shen D, Lau JYW. Polyp detection during colonoscopy using a regression-based convolutional neural network with a tracker. Pattern Recognit. 2018;83:209-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 7. | Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, Li GC, Liu GQ, He YS, Zuo XL, Li YQ. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc. 2020;91:415-424.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 206] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 8. | Zhang S, Han F, Liang Z, Tan J, Cao W, Gao Y, Pomeroy M, Ng K, Hou W. An investigation of CNN models for differentiating malignant from benign lesions using small pathologically proven datasets. Comput Med Imaging Graph. 2019;77:101645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Leufkens AM, van Oijen MG, Vleggaar FP, Siersema PD. Factors influencing the miss rate of polyps in a back-to-back colonoscopy study. Endoscopy. 2012;44:470-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 200] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 10. | Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, Zauber AG, de Boer J, Fireman BH, Schottinger JE, Quinn VP, Ghai NR, Levin TR, Quesenberry CP. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:1298-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1251] [Cited by in RCA: 1552] [Article Influence: 141.1] [Reference Citation Analysis (0)] |
| 11. | Coe SG, Wallace MB. Assessment of adenoma detection rate benchmarks in women versus men. Gastrointest Endosc. 2013;77:631-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Ahn SB, Han DS, Bae JH, Byun TJ, Kim JP, Eun CS. The Miss Rate for Colorectal Adenoma Determined by Quality-Adjusted, Back-to-Back Colonoscopies. Gut Liver. 2012;6:64-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 13. | Mori Y, Kudo SE, Wakamura K, Misawa M, Ogawa Y, Kutsukawa M, Kudo T, Hayashi T, Miyachi H, Ishida F, Inoue H. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest Endosc. 2015;81:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 14. | Misawa M, Kudo SE, Mori Y, Nakamura H, Kataoka S, Maeda Y, Kudo T, Hayashi T, Wakamura K, Miyachi H, Katagiri A, Baba T, Ishida F, Inoue H, Nimura Y, Mori K. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology. 2016;150:1531-1532.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 125] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 15. | Kominami Y, Yoshida S, Tanaka S, Sanomura Y, Hirakawa T, Raytchev B, Tamaki T, Koide T, Kaneda K, Chayama K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 16. | Mori Y, Kudo SE, Chiu PW, Singh R, Misawa M, Wakamura K, Kudo T, Hayashi T, Katagiri A, Miyachi H, Ishida F, Maeda Y, Inoue H, Nimura Y, Oda M, Mori K. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy. 2016;48:1110-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 17. | Misawa M, Kudo SE, Mori Y, Takeda K, Maeda Y, Kataoka S, Nakamura H, Kudo T, Wakamura K, Hayashi T, Katagiri A, Baba T, Ishida F, Inoue H, Nimura Y, Oda M, Mori K. Accuracy of computer-aided diagnosis based on narrow-band imaging endocytoscopy for diagnosing colorectal lesions: comparison with experts. Int J Comput Assist Radiol Surg. 2017;12:757-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 18. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 275] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 19. | Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology. 2018;154:2027-2029.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 267] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 20. | Shin Y, Balasingham I. Automatic polyp frame screening using patch based combined feature and dictionary learning. Comput Med Imaging Graph. 2018;69:33-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Wang P, Xiao X, Glissen Brown JR, Berzin TM, Tu M, Xiong F, Hu X, Liu P, Song Y, Zhang D, Yang X, Li L, He J, Yi X, Liu J, Liu X. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng. 2018;2:741-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 269] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 22. | Kudo SE, Misawa M, Mori Y, Hotta K, Ohtsuka K, Ikematsu H, Saito Y, Takeda K, Nakamura H, Ichimasa K, Ishigaki T, Toyoshima N, Kudo T, Hayashi T, Wakamura K, Baba T, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin Gastroenterol Hepatol. 2020;18:1874-1881.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 151] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 23. | Min M, Su S, He W, Bi Y, Ma Z, Liu Y. Computer-aided diagnosis of colorectal polyps using linked color imaging colonoscopy to predict histology. Sci Rep. 2019;9:2881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 24. | Sánchez-Montes C, Sánchez FJ, Bernal J, Córdova H, López-Cerón M, Cuatrecasas M, Rodríguez de Miguel C, García-Rodríguez A, Garcés-Durán R, Pellisé M, Llach J, Fernández-Esparrach G. Computer-aided prediction of polyp histology on white light colonoscopy using surface pattern analysis. Endoscopy. 2019;51:261-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 25. | Horiuchi H, Tamai N, Kamba S, Inomata H, Ohya TR, Sumiyama K. Real-time computer-aided diagnosis of diminutive rectosigmoid polyps using an auto-fluorescence imaging system and novel color intensity analysis software. Scand J Gastroenterol. 2019;54:800-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 410] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 27. | Figueiredo PN, Figueiredo IN, Pinto L, Kumar S, Tsai YR, Mamonov AV. Polyp detection with computer-aided diagnosis in white light colonoscopy: comparison of three different methods. Endosc Int Open. 2019;7:E209-E215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Tajbakhsh N, Gurudu SR, Liang J. Automated Polyp Detection in Colonoscopy Videos Using Shape and Context Information. IEEE Trans Med Imaging. 2016;35:630-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 273] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 29. | Becq A, Chandnani M, Bharadwaj S, Baran B, Ernest-Suarez K, Gabr M, Glissen-Brown J, Sawhney M, Pleskow DK, Berzin TM. Effectiveness of a Deep-learning Polyp Detection System in Prospectively Collected Colonoscopy Videos With Variable Bowel Preparation Quality. J Clin Gastroenterol. 2020;54:554-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Yamada M, Saito Y, Imaoka H, Saiko M, Yamada S, Kondo H, Takamaru H, Sakamoto T, Sese J, Kuchiba A, Shibata T, Hamamoto R. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci Rep. 2019;9:14465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 31. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 428] [Article Influence: 61.1] [Reference Citation Analysis (1)] |
| 32. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 33. | Lequan Yu, Hao Chen, Qi Dou, Jing Qin, Pheng Ann Heng. Integrating Online and Offline Three-Dimensional Deep Learning for Automated Polyp Detection in Colonoscopy Videos. IEEE J Biomed Health Inform. 2017;21:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 34. | Hassan C, Wallace MB, Sharma P, Maselli R, Craviotto V, Spadaccini M, Repici A. New artificial intelligence system: first validation study versus experienced endoscopists for colorectal polyp detection. Gut. 2020;69:799-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 35. | Lui TKL, Guo CG, Leung WK. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: a systematic review and meta-analysis. Gastrointest Endosc. 2020;92:11-22.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 36. | Yuan Y, Meng MQ. Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys. 2017;44:1379-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 37. | Gupta N, Brill JV, Canto M, DeMarco D, Fennerty BM, Laine L, Lieberman D, Lightdale C, Montgomery E, Odze R, Rex D, Sharma P, Tokar JL, Kochman ML. AGA White Paper: Training and Implementation of Endoscopic Image Enhancement Technologies. Clin Gastroenterol Hepatol. 2017;15:820-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Zhang R, Zheng Y, Mak TW, Yu R, Wong SH, Lau JY, Poon CC. Automatic Detection and Classification of Colorectal Polyps by Transferring Low-Level CNN Features From Nonmedical Domain. IEEE J Biomed Health Inform. 2017;21:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 166] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 39. | Ribeiro E, Uhl A, Wimmer G, Häfner M. Exploring Deep Learning and Transfer Learning for Colonic Polyp Classification. Comput Math Methods Med. 2016;2016:6584725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 40. | Song EM, Park B, Ha CA, Hwang SW, Park SH, Yang DH, Ye BD, Myung SJ, Yang SK, Kim N, Byeon JS. Endoscopic diagnosis and treatment planning for colorectal polyps using a deep-learning model. Sci Rep. 2020;10:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 41. | Jin EH, Lee D, Bae JH, Kang HY, Kwak MS, Seo JY, Yang JI, Yang SY, Lim SH, Yim JY, Lim JH, Chung GE, Chung SJ, Choi JM, Han YM, Kang SJ, Lee J, Chan Kim H, Kim JS. Improved Accuracy in Optical Diagnosis of Colorectal Polyps Using Convolutional Neural Networks with Visual Explanations. Gastroenterology. 2020;158:2169-2179.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 42. | Zachariah R, Samarasena J, Luba D, Duh E, Dao T, Requa J, Ninh A, Karnes W. Prediction of Polyp Pathology Using Convolutional Neural Networks Achieves "Resect and Discard" Thresholds. Am J Gastroenterol. 2020;115:138-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 43. | Shahidi N, Rex DK, Kaltenbach T, Rastogi A, Ghalehjegh SH, Byrne MF. Use of Endoscopic Impression, Artificial Intelligence, and Pathologist Interpretation to Resolve Discrepancies Between Endoscopy and Pathology Analyses of Diminutive Colorectal Polyps. Gastroenterology. 2020;158:783-785.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 44. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 353] [Article Influence: 50.4] [Reference Citation Analysis (1)] |
| 45. | Klare P, Sander C, Prinzen M, Haller B, Nowack S, Abdelhafez M, Poszler A, Brown H, Wilhelm D, Schmid RM, von Delius S, Wittenberg T. Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc. 2019;89:576-582.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 46. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 47. | Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, Xiong F, Wang H, Song Y, Pan Y, Zhou G. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. 2020;5:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 48. | Liu WN, Zhang YY, Bian XQ, Wang LJ, Yang Q, Zhang XD, Huang J. Study on detection rate of polyps and adenomas in artificial-intelligence-aided colonoscopy. Saudi J Gastroenterol. 2020;26:13-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 49. | Kainz P, Pfeiffer M, Urschler M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ. 2017;5:e3874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 50. | Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, Suriawinata AA, Hassanpour S. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J Pathol Inform. 2017;8:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 168] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 51. | Sena P, Fioresi R, Faglioni F, Losi L, Faglioni G, Roncucci L. Deep learning techniques for detecting preneoplastic and neoplastic lesions in human colorectal histological images. Oncol Lett. 2019;18:6101-6107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 52. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55686] [Article Influence: 7955.1] [Reference Citation Analysis (132)] |
| 53. | Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1965] [Cited by in RCA: 2282] [Article Influence: 207.5] [Reference Citation Analysis (1)] |
| 54. | Kudo S, Tamura S, Nakajima T, Yamano H, Kusaka H, Watanabe H. Diagnosis of colorectal tumorous lesions by magnifying endoscopy. Gastrointest Endosc. 1996;44:8-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 721] [Cited by in RCA: 707] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 55. | Atkinson NSS, Ket S, Bassett P, Aponte D, De Aguiar S, Gupta N, Horimatsu T, Ikematsu H, Inoue T, Kaltenbach T, Leung WK, Matsuda T, Paggi S, Radaelli F, Rastogi A, Rex DK, Sabbagh LC, Saito Y, Sano Y, Saracco GM, Saunders BP, Senore C, Soetikno R, Vemulapalli KC, Jairath V, East JE. Narrow-Band Imaging for Detection of Neoplasia at Colonoscopy: A Meta-analysis of Data From Individual Patients in Randomized Controlled Trials. Gastroenterology. 2019;157:462-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 113] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 56. | Neumann H, Kudo SE, Kiesslich R, Neurath MF. Advanced colonoscopic imaging using endocytoscopy. Dig Endosc. 2015;27:232-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 57. | De Palma GD, Maione F, Esposito D, Luglio G, Giglio MC, Siciliano S, Gennarelli N, Cassese G, Campione S, D'Armiento FP, Bucci L. In vivo assessment of tumour angiogenesis in colorectal cancer: the role of confocal laser endomicroscopy. Colorectal Dis. 2016;18:O66-O73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 58. | Takeda K, Kudo SE, Mori Y, Misawa M, Kudo T, Wakamura K, Katagiri A, Baba T, Hidaka E, Ishida F, Inoue H, Oda M, Mori K. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy. 2017;49:798-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 59. | Rączkowski Å, Możejko M, Zambonelli J, Szczurek E. ARA: accurate, reliable and active histopathological image classification framework with Bayesian deep learning. Sci Rep. 2019;9:14347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 60. | Yoon H, Lee J, Oh JE, Kim HR, Lee S, Chang HJ, Sohn DK. Tumor Identification in Colorectal Histology Images Using a Convolutional Neural Network. J Digit Imaging. 2019;32:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 61. | Ito N, Kawahira H, Nakashima H, Uesato M, Miyauchi H, Matsubara H. Endoscopic Diagnostic Support System for cT1b Colorectal Cancer Using Deep Learning. Oncology. 2019;96:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 62. | Ichimasa K, Kudo SE, Mori Y, Misawa M, Matsudaira S, Kouyama Y, Baba T, Hidaka E, Wakamura K, Hayashi T, Kudo T, Ishigaki T, Yagawa Y, Nakamura H, Takeda K, Haji A, Hamatani S, Mori K, Ishida F, Miyachi H. Artificial intelligence may help in predicting the need for additional surgery after endoscopic resection of T1 colorectal cancer. Endoscopy. 2018;50:230-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 63. | Lu Y, Yu Q, Gao Y, Zhou Y, Liu G, Dong Q, Ma J, Ding L, Yao H, Zhang Z, Xiao G, An Q, Wang G, Xi J, Yuan W, Lian Y, Zhang D, Zhao C, Yao Q, Liu W, Zhou X, Liu S, Wu Q, Xu W, Zhang J, Wang D, Sun Z, Gao Y, Zhang X, Hu J, Zhang M, Wang G, Zheng X, Wang L, Zhao J, Yang S. Identification of Metastatic Lymph Nodes in MR Imaging with Faster Region-Based Convolutional Neural Networks. Cancer Res. 2018;78:5135-5143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 64. | Ding L, Liu GW, Zhao BC, Zhou YP, Li S, Zhang ZD, Guo YT, Li AQ, Lu Y, Yao HW, Yuan WT, Wang GY, Zhang DL, Wang L. Artificial intelligence system of faster region-based convolutional neural network surpassing senior radiologists in evaluation of metastatic lymph nodes of rectal cancer. Chin Med J (Engl). 2019;132:379-387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 65. | Li Y, Eresen A, Shangguan J, Yang J, Lu Y, Chen D, Wang J, Velichko Y, Yaghmai V, Zhang Z. Establishment of a new non-invasive imaging prediction model for liver metastasis in colon cancer. Am J Cancer Res. 2019;9:2482-2492. [PubMed] |
| 66. | Trebeschi S, van Griethuysen JJM, Lambregts DMJ, Lahaye MJ, Parmar C, Bakers FCH, Peters NHGM, Beets-Tan RGH, Aerts HJWL. Deep Learning for Fully-Automated Localization and Segmentation of Rectal Cancer on Multiparametric MR. Sci Rep. 2017;7:5301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 195] [Cited by in RCA: 178] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 67. | Wang J, Lu J, Qin G, Shen L, Sun Y, Ying H, Zhang Z, Hu W. Technical Note: A deep learning-based autosegmentation of rectal tumors in MR images. Med Phys. 2018;45:2560-2564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |