Published online Aug 21, 2020. doi: 10.3748/wjg.v26.i31.4607
Peer-review started: March 19, 2020
First decision: April 18, 2020
Revised: May 27, 2020
Accepted: July 22, 2020
Article in press: July 22, 2020
Published online: August 21, 2020
Processing time: 155 Days and 4.4 Hours
Early diagnosis of hepatocellular carcinoma may help to ensure that patients have a chance for long-term survival; however, currently available biomarkers lack sensitivity and specificity.
To characterize the serum metabolome of hepatocellular carcinoma in order to develop a new metabolomics diagnostic model and identifying novel biomarkers for screening hepatocellular carcinoma based on the pattern recognition method.
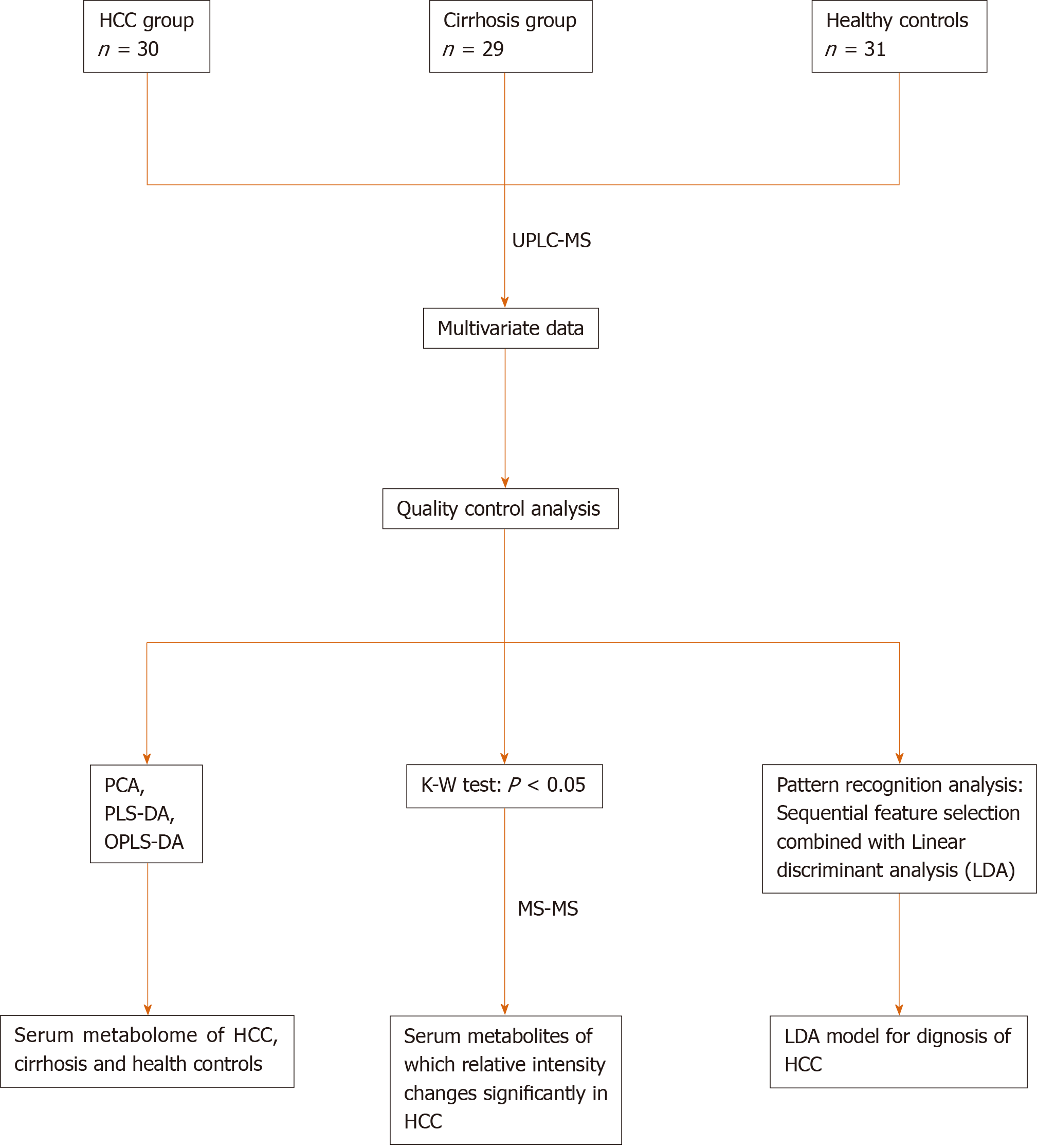
Ultra-performance liquid chromatography-mass spectroscopy was used to characterize the serum metabolome of hepatocellular carcinoma (n = 30) and cirrhosis (n = 29) patients, followed by sequential feature selection combined with linear discriminant analysis to process the multivariate data.
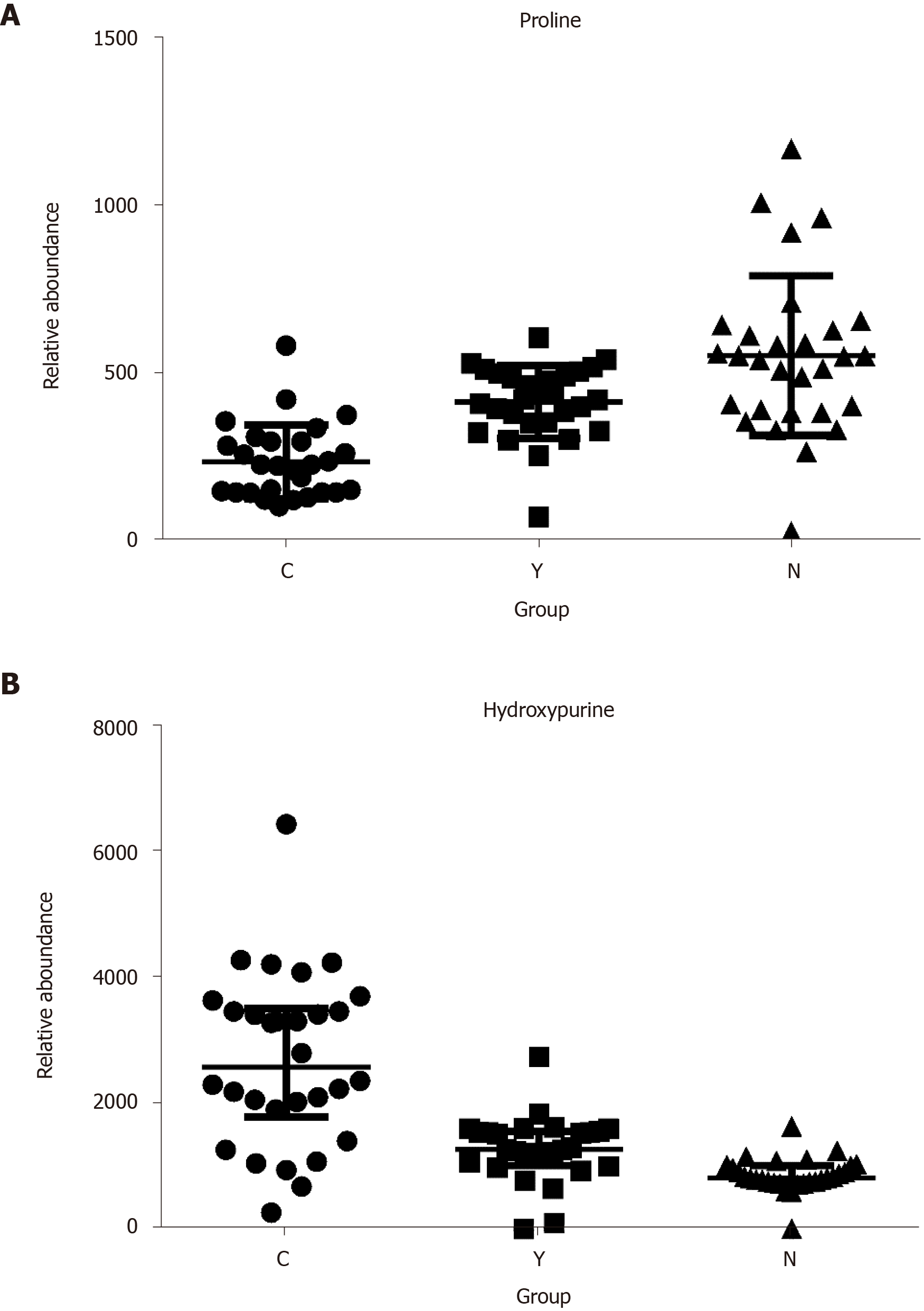
The concentrations of most metabolites, including proline, were lower in patients with hepatocellular carcinoma, whereas the hydroxypurine levels were higher in these patients. As ordinary analysis models failed to discriminate hepatocellular carcinoma from cirrhosis, pattern recognition analysis was used to establish a pattern recognition model that included hydroxypurine and proline. The leave-one-out cross-validation accuracy and area under the receiver operating characteristic curve analysis were 95.00% and 0.90 [95% Confidence Interval (CI): 0.81-0.99] for the training set, respectively, and 78.95% and 0.84 (95%CI: 0.67-1.00) for the validation set, respectively. In contrast, for α-fetoprotein, the accuracy and area under the receiver operating characteristic curve were 65.00% and 0.69 (95%CI: 0.52-0.86) for the training set, respectively, and 68.42% and 0.68 (95%CI: 0.41-0.94) for the validation set, respectively. The Z test revealed that the area under the curve of the linear discriminant analysis model was significantly higher than the area under the curve of α-fetoprotein (P < 0.05) in both the training and validation sets.
Hydroxypurine and proline might be novel biomarkers for hepatocellular carcinoma, and this disease could be diagnosed by the metabolomics model based on pattern recognition.
Core tip: We used ultra-performance liquid chromatography-mass spectroscopy to characterize the metabolome of serum samples from patients with hepatocellular carcinoma. We processed multivariate data using pattern recognition analysis and established a diagnostic model that included hydroxypurine and proline. The accuracy and area under the curve were 95.00% and 0.90 for the training set, respectively, and 78.95% and 0.84 for the validation set, respectively. The Z test revealed that the area under the curve of the model was significantly higher than that of α-fetoprotein. The results suggest that hydroxypurine and proline might be novel biomarkers for hepatocellular carcinoma, and the pattern recognition metabolomics model could be used to diagnose hepatocellular carcinoma.
- Citation: Zhou PC, Sun LQ, Shao L, Yi LZ, Li N, Fan XG. Establishment of a pattern recognition metabolomics model for the diagnosis of hepatocellular carcinoma. World J Gastroenterol 2020; 26(31): 4607-4623
- URL: https://www.wjgnet.com/1007-9327/full/v26/i31/4607.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i31.4607
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of death due to cancer worldwide[1]. In particular, approximately 50% of the total patients with HCC in the world are from China, owing to the highest carrier prevalence of hepatitis B[2-4]. Early diagnosis of HCC offers patients a better chance for long-term survival[5]. Although imaging technologies such as magnetic resonance imaging and ultrasonography, and serum biomarkers [notably α-fetoprotein (AFP)] are widely used to diagnose HCC in the clinic[6], they are far from satisfactory because they lack sensitivity and specificity[7]. Therefore, there is an urgent and unmet desire for novel screening methods and new biomarkers.
The emergence of metabolomics has provided a powerful tool for discovering novel biomarkers and revealing metabolic pathways of cancer and liver diseases[8,9]. A metabolomics approach to screen individual metabolites or their combinations for the diagnosis of HCC[10] identified a series of potential biomarkers including phenylalanyl-tryptophan, glycocholate, concanavanine succinic acid, bile acid, long chain fatty acid, and so on for future clinical application[5,7,11]. However, none of these markers have thus far been validated for clinical applications. Metabolomics datasets commonly contain hundreds to thousands of variables; however, biomarkers are identified using conventional data processing methods such as principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA), orthogonal partial least squares discriminant analysis (OPLS-DA), and binary logistic regression[11,12]. With the advent of data processing technology to handle big data, it is incumbent upon researchers in this area to adopt advanced methods such as pattern recognition to seek new biomarkers and to establish mathematical models that facilitate screening for HCC.
In previous studies, we established a pattern recognition metabolomics method based on sequential feature selection combined with linear discriminant analysis (LDA) to evaluate the severity of fulminant hepatic failure and for the differential diagnosis of Clostridium difficile infection[13,14]. In the current study, ultra-performance liquid chromatography-mass spectroscopy (UPLC-MS) was used to characterize the serum metabolomes of patients with HCC, patients with cirrhosis, and healthy controls. Furthermore, the pattern recognition method developed herein was used to process multivariate data with the aim of developing a novel metabolomics diagnostic model and identifying novel biomarkers for HCC screening purposes.
Between March and August 2016, samples from patients who met the inclusion criteria of HCC diagnosis set by the Ministry of Health were collected[15]. HCC confirmation required histological evidence or two different imaging techniques, or the combination of one imaging technique and an AFP level of > 400 ng/mL. Patients with cirrhosis meeting the criteria described elsewhere[16] based on clinical manifestations, laboratory examinations, and imaging results were included. HCC patients (C group, n = 30) all had cirrhosis, and cirrhosis patients without HCC were included in Y group (n = 29). The Child-Pugh Score in patients in the C group and Y group patients was A or B. Healthy controls (N group, n = 31) were chosen from the general population. The exclusion criteria were Child-Pugh Score C patients, malignant neoplasm (except HCC for C group), metabolic diseases, autoimmune disease, excess alcohol consumption, and known history of toxic exposure. Whole blood samples (3-5 mL) were collected on an empty stomach in the morning in BD Vacutainer® blood specimen collection tubes (Weigao Group, Weihai, China). Whole blood samples were stored at 4°C immediately after collection and were transported to the laboratory in < 30 min. After centrifugation at 3000 × g for 10 min at 4°C, a portion of the serum from the samples was used for biochemical assays and the remaining serum was aliquoted into fresh Eppendorf® tubes and stored at -80°C for metabolomic analysis. Fresh surgical tumor tissue samples were obtained from patients following informed consent.
Hepatitis B virus (HBV) and HCV antigens and a biochemical panel including alanine aminotransferase, aspartate aminotransferase, glutamic-oxaloacetic transaminase, total bilirubin, direct bilirubin, total protein, and albumin were assayed in the clinical laboratory. Histopathological samples were prepared as described previously[13].
Acetonitrile and methanol (HPLC grade) were purchased from Merck (Darmstadt, Germany). Distilled water was purified using a Milli-Q system (Darmstadt, Germany). Fatty acids, amino acids, bile acid, and nucleotide standards were purchased from Sigma-Aldrich (St. Louis, MO, United States). Citric acid, pantothenic acid, and malonic acid were purchased from Supelco (Bellefonte, PA, United States). Lysophosphatidyl cholines (LysoPCs) and lysophosphatidyl ethanolamine were purchased from Avanti Polar Lipids, Inc. (AL, United States).
Prior to the assay, all samples were thawed on ice. Pooled aliquots (1 μL) of each sample formed the quality control (QC) sample. Metabolites in serum were extracted by methanol (serum/methanol (V/V) = 1:3). The mixture (100 μL) was vortexed for 60 s, and then centrifuged at 14000 × g for 10 min at 4°C. Supernatants were dried by nitrogen flow and then re-dissolved in 100 μL methanol. The mixture was again centrifuged at 14000 ×g for 5 min at 4°C. The resulting clear supernatant was transferred into UPLC vials and stored at 4°C.
An aliquot (2 μL) of the clear supernatant obtained above was chromatographed on a Thermo Fisher Scientific UltiMate 3000 UPLC system using an ACQUITY UPLC BEH C18 analytical column (i.d. 2.1 mm × 100 mm, particle size 1.7 mm, pore size 130 A˚). Mobile phase A and mobile phase B were water/formic acid (99.9: 0.1, V/V) and acetonitrile/formic acid (99.9: 0.1, V/V), respectively, and the flow rate was 200 μL/min. A linear gradient was optimized as follows: the initial composition of the mobile phase was 95% A and 5% B; 0-2 min, 95% A; 2-9 min, 95%-62% A; 9-14 min, 62%–32% A; 14-22 min, 32%-0% A; 22-30 min, 0-95% A. The column eluent was directed to the mass spectrometer for analyses.
Mass spectrometry was performed on a Thermo Fisher Scientific Q-Exactive Focus Mass Spectrometer operating in positive ion electrospray mode. The instrument parameters were set as follows: Mass range scanned from 50 to 1000, spray voltage was 4000 V, atomization temperature was 300°C, nebulizer pressure was 45 bar, capillary temperature was 350°C, and the capillary voltage was set to 4.00 kV; the sampling cone voltage was set to 35.0 V. The instrument parameters for MS/MS analysis were set at different collision energies according to the stability of metabolites (collision energy was set from 15 to 35 eV).
Five injections of QC samples were performed to equilibrate the UPLC-MS systems prior to testing individual patient samples. QC samples were injected after every six patient samples at regular intervals throughout the analytical run. Patient samples were tested in a random manner.
The raw UPLC-MS data of the samples were extracted using MZmine2.3 software and Xcalibur software (Thermo Fisher Scientific), which enabled detection, integration and normalization of the intensities of the peaks to the sum of peaks within the sample and to create a multivariate dataset containing the retention time, m/z, and relative abundances. The parameters were set as follows: Retention time ranging from 0 to 30 min, mass range m/z from 50 to 1000, and mass tolerance at 0.05 Da. For peak integration, peak width at 5% of the height was 1 s, peak-to-peak baseline noise was 0, peak intensity threshold was 100, and retention time window was 0.20 s.
The statistical analysis is shown in Figure 1. In brief, we used SIMCA-P + 12.0 software (Umetrics, AB, Sweden) to perform PCA, PLS-DA, and OPLS-DA. Pattern recognition analysis based on sequential feature selection combined with LDA for diagnosis of HCC, and the Z test [for comparison of area under curve (AUC)] were performed using Matlab Version 8.1 (R2013a) software (MathWorks Inc., Natick, MA, United States). One-way ANOVA, the Chi-square test, and Kruskal–Wallis test were conducted using SPSS v16.0 software (SPSS Inc. Chicago, IL, United States). Differences were considered statistically significant at P < 0.05.
The compounds were identified by searching the Human Metabolome Database (http://hmdb.ca/), PubChem compound database (http://www.ncbi.nlm.nih.gov), and our own compound database that includes metabolites previously identified by us. Finally, the compound was verified by comparing the mass spectra and retention time of potential biomarkers with authentic standards (Supplementary Figures 1-5).
Demographic data and clinical characteristics of the subjects are shown in Table 1. Thirty patients with HCC (all with cirrhosis, C group), 29 patients with cirrhosis (all without HCC, Y group), and 31 healthy controls (N group) were enrolled. There were no significant differences in age and sex among the three groups, and no significant differences in the causes of liver injury and Child-Pugh Score between C group and Y group. The levels of AFP, glutamic-oxaloacetic transaminase, and alanine aminotransferase were relatively higher and the level of albumin was relatively lower in patients with HCC than in patients with cirrhosis and healthy controls. The histopathology results of patients with HCC are shown in Supplementary Figure 6. We used the Chinese staging system to stage HCC[15], and 11 cases were stage IIIa, 12 cases were stageIIb, one case was stageIIa, 5 cases were stageIb, and one case was stageIa.
| Characteristics | C (n = 30) | Y (n = 29) | N (n = 31) | P value | |
| Sex (Male/Female) | 25/5 | 21/8 | 25/6 | 0.565 | |
| Age (yr) | 52.93 ± 11.01 | 56.63 ± 9.15 | 51.23 ± 11.79 | 0.148 | |
| Pathogens | HBV | 25 | 24 | / | 0.720 |
| HCV | 1 | 2 | |||
| HBV + HCV | 1 | 0 | |||
| None | 3 | 3 | |||
| AFP (ng/mL) | > 200 | 11 | 0 | / | 0.000 |
| 50-199 | 4 | 1 | |||
| < 50 | 15 | 28 | |||
| ALT (U/L) | 162.32 ± 201.06 | 91.02 ± 156.39 | 20.34 ± 8.43 | 0.000 | |
| AST (U/L) | 146.35 ± 112.70 | 114.49 ± 191.67 | 21.59 ± 4.51 | 0.012 | |
| TBIL (μmol/L) | 39.21 ± 68.38 | 40.87 ± 42.41 | 9.66 ± 2.66 | 0.015 | |
| DBIL (μmol/L) | 17.91 ± 34.43 | 17.90 ± 23.03 | 4.49 ± 1.38 | 0.044 | |
| TP (g/L) | 62.42 ± 10.95 | 74.14 ± 8.05 | 72.31 ± 3.96 | 0.000 | |
| ALB (g/L) | 33.51 ± 6.30 | 37.65 ± 7.64 | 45.36 ± 2.62 | 0.000 | |
| Child-Pugh score (A/B) | 18/12 | 15/14 | / | 0.353 | |
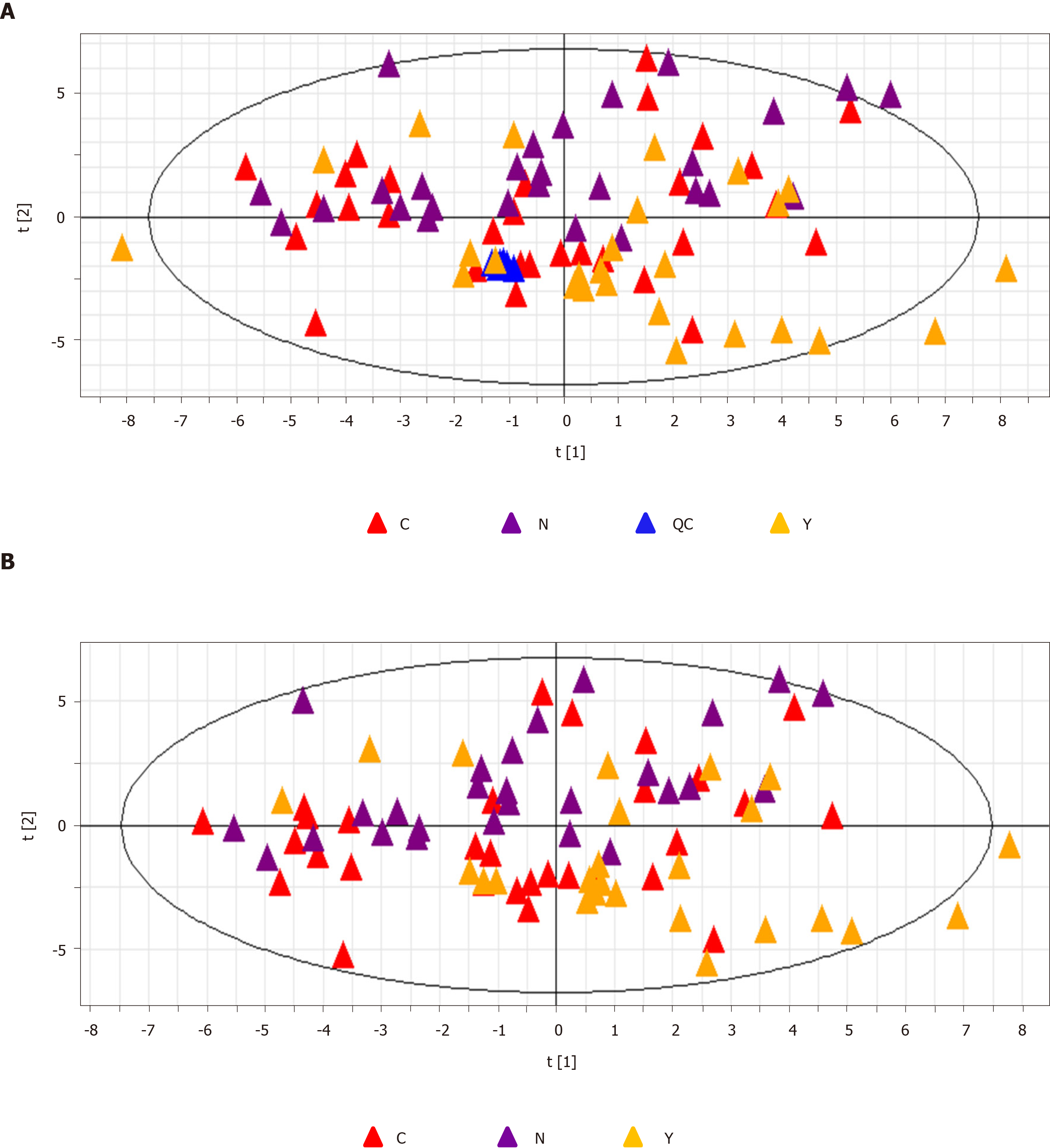
QC samples clustered compactly in the middle of the PCA score plot (Figure 2A). The coefficient of variation (CV) of identified metabolites in QC samples ranged from 2.09% to 16.27% with a median CV of 7.83% (Table 2).
| Retention time | m/z | Metabolites | Adduction | Adduct mass | Delta ppm | Coefficient of variation (%) | Comparison | ||
| 3.52 | C vs N | Y vs N | C vs Y | ||||||
| 9.57 | 166.0862 | Phenylalanine | M + H | 166.0863 | 1.00 | 8.36 | D | U | NS |
| 3.49 | 118.0864 | Valine | M + H | 118.0863 | 1.00 | 6.76 | D | NS | NS |
| 6.63 | 132.1019 | Leucine | M + H | 132.1019 | 0.00 | 3.34 | NS | U | NS |
| 3.58 | 116.0708 | Proline | M + H | 116.0706 | 1.00 | 14.23 | D | D | D |
| 5.42 | 182.0811 | Tyrosine | M + H | 182.0812 | 0.00 | 6.12 | NS | U | NS |
| 6.05 | 132.1019 | Isoleucine | M + H | 132.1019 | 0.00 | 3.37 | NS | U | D |
| 4.89 | 150.0583 | Methionine | M + H | 150.0583 | 0.00 | 9.28 | NS | U | NS |
| 3.16 | 156.0766 | Histidine | M + H | 156.0768 | 1.00 | 10.83 | D | U | D |
| 3.44 | 148.0602 | Glutamic acid | M + H | 148.0604 | 2.00 | 6.38 | U | D | U |
| 3.32 | 106.0502 | Serine | M + H | 106.0499 | 3.00 | 2.59 | D | U | D |
| 3.38 | 147.0762 | Glutamine | M + H | 147.0764 | 1.00 | 11.41 | NS | D | D |
| 3.44 | 90.0554 | Alanine | M + H | 90.0550 | 5.00 | 12.28 | D | D | NS |
| 5.43 | 165.0546 | Hydroxycinnamic acid | M + H | 165.0546 | 0.00 | 16.17 | D | NS | D |
| 5.43 | 123.0442 | Benzoic acid | M + H | 123.0441 | 1.00 | 10.11 | D | U | NS |
| 9.57 | 149.0596 | Cinnamic acid | M + H | 149.0597 | 1.00 | 12.29 | D | U | NS |
| 24.40 | 190.0497 | Kynurenic acid | M + H | 190.0499 | 1.00 | 6.51 | D | D | U |
| 26.39 | 169.0495 | Vanillic acid | M + H | 169.0495 | 0.00 | 3.41 | D | D | U |
| 13.83 | 239.0912 | Trimethoxycinnamic acid | M + H | 239.0914 | 1.00 | 5.08 | D | U | NS |
| 18.85 | 279.2318 | Linolenic acid | M + H | 279.2319 | 0.00 | 11.26 | D | NS | D |
| 3.10 | 130.0862 | Pipecolinic acid | M + H | 130.0863 | 0.00 | 10.58 | D | U | NS |
| 29.42 | 494.3235 | LysoPC 16:1 | M + H | 494.3241 | 1.00 | 4.32 | NS | D | D |
| 22.87 | 542.3234 | LysoPC 20:5 | M + H | 542.3241 | 1.00 | 3.09 | NS | D | NS |
| 17.33 | 548.3705 | LysoPC 20:2 | M + H | 548.3711 | 1.00 | 2.31 | D | D | NS |
| 21.65 | 550.3857 | LysoPC 20:1 | M + H | 550.3867 | 2.00 | 5.58 | D | D | NS |
| 23.13 | 468.3078 | LysoPC 14:0 | M + H | 468.3085 | 1.00 | 6.27 | D | D | D |
| 19.25 | 478.2926 | LysoPE 20:1 | M + H | 478.2928 | 0.00 | 8.72 | D | NS | NS |
| 17.58 | 181.0857 | Propylparaben | M + H | 181.0859 | 1.00 | 6.39 | D | NS | NS |
| 5.42 | 136.0756 | Acetylarylamine | M + H | 136.0757 | 0.00 | 2.59 | D | U | D |
| 18.20 | 127.0390 | Trihydroxybenzene | M + H | 127.0390 | 0.00 | 13.83 | D | U | D |
| 22.22 | 191.1428 | Damascenone | M + H | 191.1430 | 1.00 | 10.02 | U | D | NS |
| 10.70 | 181.0718 | Myoinositol | M + H | 181.0707 | 6.00 | 8.55 | D | NS | NS |
| 4.88 | 137.0457 | Hydroxypurine | M + H | 137.0458 | 1.00 | 9.74 | U | D | U |
| 3.48 | 114.0664 | Creatinine | M + H | 114.0662 | 2.00 | 7.83 | D | NS | NS |
| 3.82 | 72.0815 | Pyrrolidine | M + H | 72.0808 | 10.00 | 2.09 | U | U | NS |
| 11.71 | 195.0875 | Methyl lucopyranoside | M + H | 195.0863 | 6.00 | 12.43 | D | NS | NS |
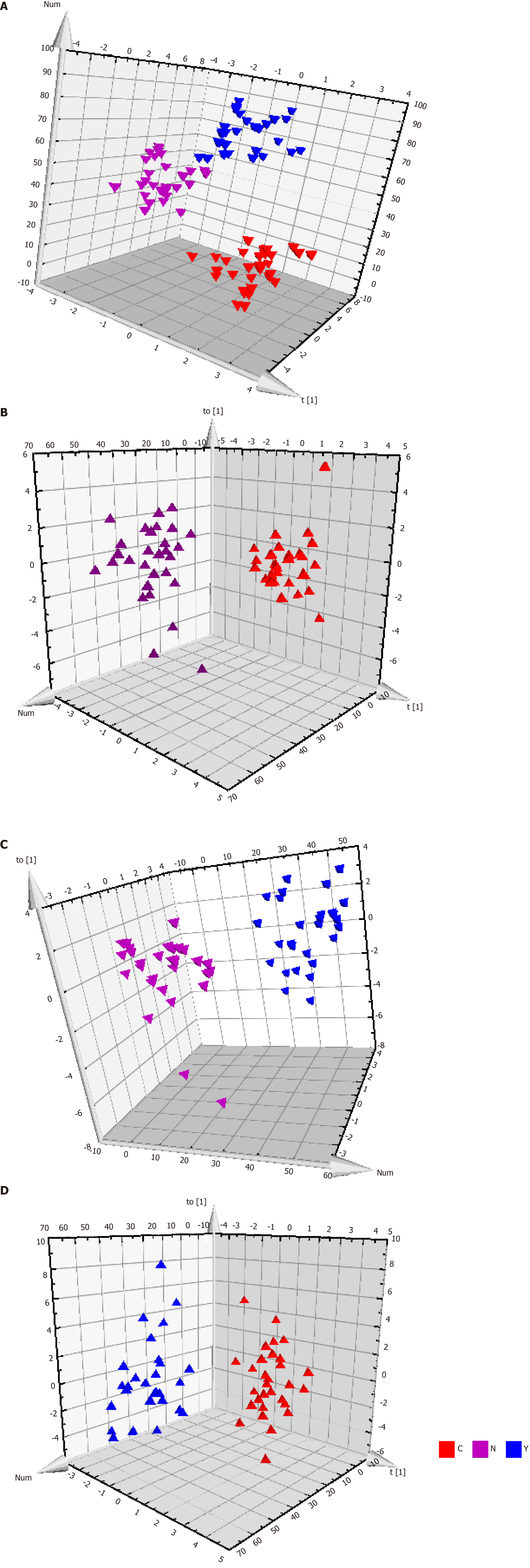
Patients with HCC, patients with cirrhosis, and healthy controls showed no significant differences in the base peak intensity chromatogram (Supplementary Figure 7). The three groups intermixed with each other in the PCA score plot, although there was a tendency to separate along PC1 (Figure 2B). Characterization of metabolic differences among the three groups using PLS-DA and OPLS-DA showed that the three groups also intermixed with each other in the PLS-DA score plot (Supplementary Figure 8). The PLS-DA score plot of the HCC group vs the cirrhosis group also intermixed with each other (Supplementary Figure 9). Validation plots of the PLS-DA models acquired through 20 permutation tests were used for cross-validation purposes (Supplementary Figures 10 and 11). Analysis of the PLS-DA score plot for all three groups revealed that R2 = (0.0, 0.401) and Q2 = (0.0, -0.35); cross-validation of the PLS-DA score plot of C group and Y group revealed that R2 = (0.0, 0.645) and Q2 = (0.0, -0.507). Although the PLS-DA model showed intermixing of the three groups, they could be separated in the OPLS-DA model (Figure 3A). OPLS-DA score plots of the HCC group vs healthy controls (Figure 3B), the cirrhosis group vs healthy controls (Figure 3C), and the HCC group vs the cirrhosis group (Figure 3D) demonstrated very clear separation. However, the R2 and Q2 values were not high enough in the three OPLS-DA models.
Potential biomarkers were characterized by variable importance in the projection values retrieved from the PLS-DA model combined with the Kruskal–Wallis test (P < 0.05). Potential biomarkers were identified by a preliminary search of the HMDB and PubChem compound databases and verified by comparing the mass spectra and retention time of potential biomarkers with authentic standards. As shown in Table 2 and Supplementary Figure 12, the levels of most metabolites, including proline, were lower in patients with HCC than in healthy controls and patients with cirrhosis (Figure 4A). However, the levels of glutamic acid, pyrrolidine, and damascenone were higher in patients with HCC than in healthy controls; glutamic acid, kynurenic acid, vanillic acid, and hydroxypurine (Figure 4B) were higher in patients with HCC than in patients with cirrhosis.
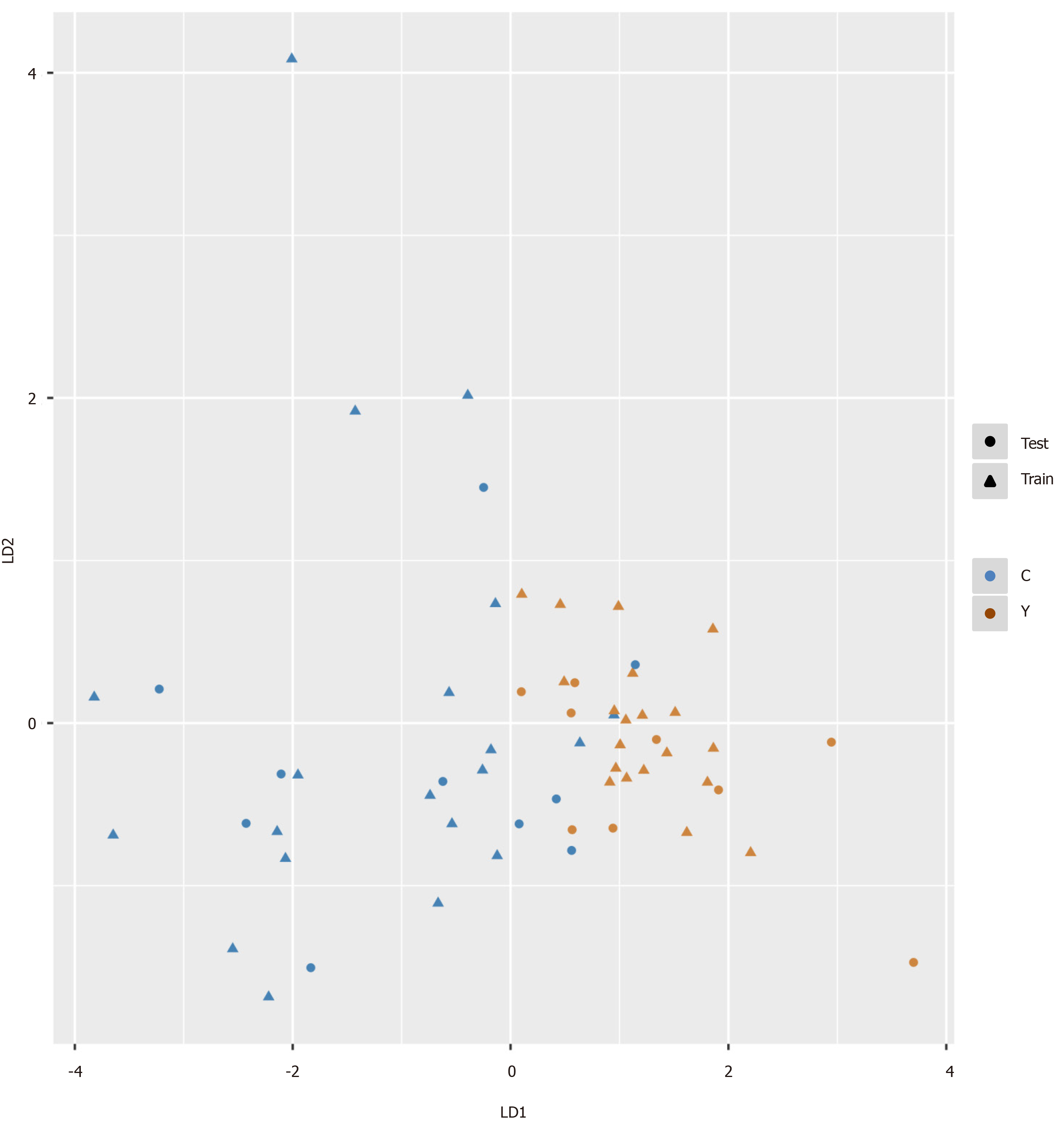
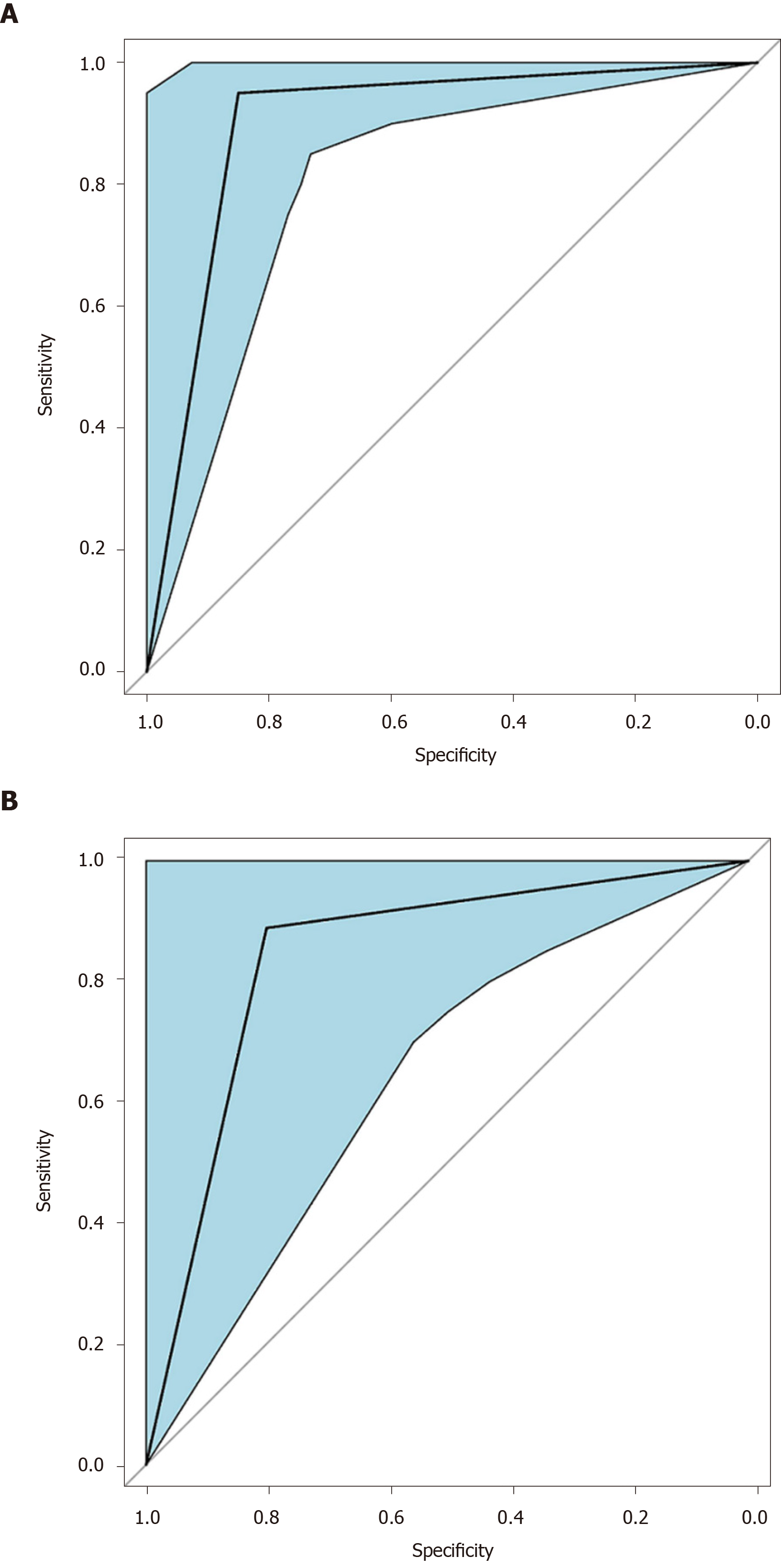
We intended to establish a PLS-DA model or OPLS-DA model with the aim of distinguishing patients with HCC from patients with cirrhosis. However, as the metabolomes of HCC and cirrhosis are not very different, the efficiency of the models was not robust enough to discriminate the two groups using ordinary PLS-DA or OPLS-DA models. Therefore, we used pattern recognition, an advance data processing method, to achieve our aim. To enable this, the dataset was randomly split into a training set and a validation set. The training set comprised 20 HCC samples and 20 cirrhosis samples, and the validation set comprised 10 HCC samples and nine cirrhosis samples. We used sequential feature selection to select the most suitable metabolites for constructing the best performing LDA model based on the training set. The validation set was used to confirm the reliability of the model for discriminating patients with HCC from patients with cirrhosis. When the metabolites hydroxypurine and proline were included in the LDA model, a differential distribution pattern between HCC and cirrhosis began to emerge in the LDA plot (Figure 5). The leave-one-out cross-validation analysis provided accuracy, sensitivity, specificity, a positive predictive value, and a negative predictive value of 95.00%, 100.00%, 90.00%, 0.91, and 1.00, respectively, for the training set, and 78.95%, 100.00%, 60.00%, 0.69, and 1.00, respectively, for the external validation set (Table 3). Validation of AFP as a biomarker to discriminate HCC and cirrhosis provided accuracy, sensitivity, specificity, a positive predictive value, and a negative predictive value of 65.00%, 30.00%, 100.00%, 1.00 and 0.59, respectively, for training samples, and 68.42%, 40.00%, 100.00%, 1.00 and 0.60, respectively, for test samples. For the training samples, the AUC in the LDA model (AUCLDA) was 0.90 (95%CI: 0.81–0.99, P < 0.05, Figure 6A), and AUCAFP was 0.69 (95%CI: 0.52–0.86, P < 0.05, Supplementary Figure 13); AUCLDA was significantly more than AUCAFP (P < 0.05, Z test). For validation samples, AUCLDA was 0.84 (95%CI: 0.67–1.00, P < 0.05, Figure 6B), and AUCAFP was 0.68 (95%CI: 0.41–0.94, P = 0.191, Supplementary Figure 14); AUCLDA was significantly larger than AUCAFP (P < 0.05, Z test).
| Model | Accuracy (%) | Sensitivity (%) | Specificity (%) | Positive predictive value | Negative predictive value | ROC-AUC (95%CI) | P value | |
| Training set | LDA | 95.00 | 100.00 | 90.00 | 0.91 | 1.00 | 0.90 (0.81-0.99) | < 0.05 |
| AFP | 65.00 | 30.00 | 100.00 | 1.00 | 0.59 | 0.69 (0.52-0.86) | ||
| Validation set | LDA | 78.95 | 100.00 | 60.00 | 0.69 | 1.00 | 0.84 (0.67-1.00) | < 0.05 |
| AFP | 68.42 | 40.00 | 100.00 | 1.00 | 0.60 | 0.68 (0.41-0.94) | ||
In this study, the serum metabolomes of patients with HCC, patients with cirrhosis, and healthy controls were profiled by UPLC-MS to establish a metabolomics model for the diagnosis of HCC. This approach not only enabled elucidation of HCC pathogenesis but also provided a mathematical model based on possible biomarkers for screening HCC.
The stability of metabolomics data and the comparability of demographic data are the two crucial issues that should be considered prior to statistical analysis[17]. In this study, the reproducibility and stability of metabolomics data are reflected in the compact clustering of QC samples in the PCA score plot, as well as in the low CV of specific metabolites of the QC samples. There were no statistical differences in age and sex among the patients with HCC, patients with cirrhosis, and healthy controls. Also, the constituent ratio of etiology of liver injury (pathogenesis) was comparable between the HCC and cirrhosis groups, all of which confirm the reliability of the UPLC-MS assay and optimal homogeneity of baseline characteristics[9].
The liver is the principal organ for metabolism of carbohydrates, lipids, amino acids etc[18]. Particularly in HCC, liver disease always results in apparent metabolic dysregulation[19], as in the case of glutamine addiction, a hallmark feature of HCC[20]. The decrease in serum metabolites in patients with HCC is largely due to uptake and utilization of metabolites by the tumor to feed its malignant behavior, as in the case of glutamine addiction[20]. This is evident in HCC tissue that has 20 times higher glutaminase 1 concentration than normal liver tissue[21], leading to 10 times faster consumption of glutamine resulting in diminished glutamine levels in the serum of patients with HCC. On the contrary, an increase in the concentration of serum metabolites in HCC may reflect tumor necrosis. The best illustration of this process is the increase in hydroxypurine in the serum of patients with HCC, likely due to the release of nucleic acids from tumor tissues, which then metabolizes into hydroxypurine under necrotic conditions[22].
Our findings are in line with previous studies that demonstrated diminished levels of serum phospholipid metabolites in patients with liver diseases (including HCC, liver cirrhosis, hepatitis, and liver failure)[7,9]. Indeed, through an untargeted metabolomics approach, we found significantly reduced amounts of phospholipid metabolites in patients with HCC. Reduced serum LysoPC, a molecule associated with malignancies, autoimmune disease, inflammation, and cell signaling[23], is an indicator of liver injury; LysoPC correlates with model for end-stage liver disease score, independently of age, sex, and diet. As the patients with HCC in our cohort also had concurrent liver cirrhosis, the serum LysoPC of C group was lower than that of healthy controls. However, since the severity of liver injury was similar between C and Y groups, the serum LysoPC concentration was not significantly different between these groups. Low levels of LysoPC may be attributed to the inhibition of phospholipase A2 or LCAT activity or perturbed LysoPC acyltransferase activity[7]. More recently, based on studies from our group and others, it was postulated that excessive consumption of LysoPC results in an anti-inflammatory response, leading to low levels of serum and severe immunosuppression in patients with liver diseases[9,23].
The reduced levels of serum creatinine found in patients with HCC in this study may be attributed to the diminished hepatic conversion of creatine to creatinine in patients with hepatic disease[5]. Another reason may be the decrease in levels of serine and alanine, involved in the synthesis of creatine, in HCC[5]. Down regulation of fatty acids was also found in patients with HCC compared with cirrhotic patients and heathy controls. Fatty acids can be transported into the mitochondria for beta-oxidation to generate adenosine triphosphate (ATP) energy, and its metabolism could be perturbed in patients with chronic liver disease[24]. Thus, we hypothesized that differential levels of metabolites in HCC may enable biomarker identification for the diagnosis of HCC.
As the PCA and PLS-DA models suffered from relatively poor efficiencies in our study and were overfit for the dataset, they were therefore unable to discriminate patients with HCC from patients with cirrhosis. Hence, a pattern recognition approach, based on sequential feature selection combined with LDA, was adopted to find the most suitable combination of biomarkers. This resulted in the generation of an LDA model for the diagnosis of HCC, which included two novel biomarkers, hydroxypurine and proline, highlighting the rapid growth and necrotic characteristics of HCC. As the accuracy, sensitivity, negative predictive value, and AUCLDA were higher in the LDA model compared to those in the AFP diagnostic model, the relatively better efficiency of the LDA model could ensure proper discrimination of patients with HCC. However, the specificity and positive predictive value of the LDA model were lower than those in the AFP diagnostic model, suggesting that AFP remains a useful biomarker for discriminating patients with HCC from those with cirrhosis. If AFP levels reach the threshold of ≥ 400 ng/mL[15], patients are very likely to be diagnosed with HCC. Our results suggest that the two methods are complementary to each other, and the combination of the two approaches may offer better validation of diagnostic results. Further more, our findings indicated that pattern recognition analysis was better than conventional multivariate statistical analysis for data processing.
In conclusion, competitive access to nutrition and necrosis can be identified in HCC using a metabolomics model based on sequential feature selection combined with LDA, which may be an ideal method for novel biomarker discovery.
Early diagnosis of hepatocellular carcinoma (HCC) offers patients a better chance for long-term survival. The current biomarkers are far from satisfactory as they lack sensitivity and specificity. The emergence of metabolomics has provided a powerful tool for discovering novel biomarkers. In previous studies, we established a pattern recognition metabolomics method based on sequential feature selection combined with linear discriminant analysis for differential diagnosis.
There is an urgent and unmet desire for novel screening methods and new biomarkers for the diagnosis of HCC. Whether the pattern recognition method mentioned above could be used to establish a metabolomics model for the diagnosis of HCC is still unknown.
We aimed to use the pattern recognition method to develop a metabolomics diagnostic model and identify new biomarkers for HCC screening.
We used ultra-performance liquid chromatography-mass spectroscopy to characterize the serum metabolome of HCC and cirrhosis patients. We then processed the multivariate data using sequential feature selection combined with linear discriminant analysis.
The concentrations of most metabolites, including proline, were lower in patients with HCC, whereas hydroxypurine levels were higher in these patients. As ordinary analysis models failed to discriminate hepatocellular carcinoma from cirrhosis, pattern recognition analysis was used to establish a pattern recognition model that included hydroxypurine and proline. The leave-one-out cross-validation accuracy and area under curve (AUC) were 95.00% and 0.90 (95% confidence interval (CI): 0.81–0.99) for the training set, respectively, and 78.95% and 0.84 (95%CI: 0.67–1.00) for the validation set, respectively. The Z test revealed that the AUC of the model was significantly higher than the AUC (P < 0.05) in both the training and validation sets.
Hydroxypurine and proline might be novel biomarkers for HCC, and the disease could be diagnosed by the metabolomics model based on pattern recognition.
This study determined the applicability of the pattern recognition metabolomics model for the diagnosis of HCC. Two novel biomarkers for HCC were also found. Future studies should verify the validity of the model and the applicability of the biomarkers in the early diagnosis of patients with HCC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lopez-Guerrero J, Sallustio F S-Editor: Zhang L L-Editor: Webster JR P-Editor: Wang LL
| 1. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, Hamadeh RR, Moore A, Werdecker A, Gessner BD, Te Ao B, McMahon B, Karimkhani C, Yu C, Cooke GS, Schwebel DC, Carpenter DO, Pereira DM, Nash D, Kazi DS, De Leo D, Plass D, Ukwaja KN, Thurston GD, Yun Jin K, Simard EP, Mills E, Park EK, Catalá-López F, deVeber G, Gotay C, Khan G, Hosgood HD 3rd, Santos IS, Leasher JL, Singh J, Leigh J, Jonas JB, Sanabria J, Beardsley J, Jacobsen KH, Takahashi K, Franklin RC, Ronfani L, Montico M, Naldi L, Tonelli M, Geleijnse J, Petzold M, Shrime MG, Younis M, Yonemoto N, Breitborde N, Yip P, Pourmalek F, Lotufo PA, Esteghamati A, Hankey GJ, Ali R, Lunevicius R, Malekzadeh R, Dellavalle R, Weintraub R, Lucas R, Hay R, Rojas-Rueda D, Westerman R, Sepanlou SG, Nolte S, Patten S, Weichenthal S, Abera SF, Fereshtehnejad SM, Shiue I, Driscoll T, Vasankari T, Alsharif U, Rahimi-Movaghar V, Vlassov VV, Marcenes WS, Mekonnen W, Melaku YA, Yano Y, Artaman A, Campos I, MacLachlan J, Mueller U, Kim D, Trillini M, Eshrati B, Williams HC, Shibuya K, Dandona R, Murthy K, Cowie B, Amare AT, Antonio CA, Castañeda-Orjuela C, van Gool CH, Violante F, Oh IH, Deribe K, Soreide K, Knibbs L, Kereselidze M, Green M, Cardenas R, Roy N, Tillmann T, Li Y, Krueger H, Monasta L, Dey S, Sheikhbahaei S, Hafezi-Nejad N, Kumar GA, Sreeramareddy CT, Dandona L, Wang H, Vollset SE, Mokdad A, Salomon JA, Lozano R, Vos T, Forouzanfar M, Lopez A, Murray C, Naghavi M. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1945] [Cited by in RCA: 2055] [Article Influence: 205.5] [Reference Citation Analysis (0)] |
| 2. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13214] [Article Influence: 1468.2] [Reference Citation Analysis (3)] |
| 3. | Fu S, Li N, Zhou PC, Huang Y, Zhou RR, Fan XG. Detection of HBV DNA and antigens in HBsAg-positive patients with primary hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41:415-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 4. | Xiao Y, Sun L, Fu Y, Huang Y, Zhou R, Hu X, Zhou P, Quan J, Li N, Fan XG. High mobility group box 1 promotes sorafenib resistance in HepG2 cells and in vivo. BMC Cancer. 2017;17:857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, Qi X, Cao Y, Su M, Wang X, Xu LX, Yen Y, Liu P, Jia W. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10:M110.004945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 246] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 6. | Ren B, Zou G, Xu F, Huang Y, Xu G, He J, Li Y, Zhu H, Yu P. Serum levels of anti-sperm-associated antigen 9 antibody are elevated in patients with hepatocellular carcinoma. Oncol Lett. 2017;14:7608-7614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Wang B, Chen D, Chen Y, Hu Z, Cao M, Xie Q, Chen Y, Xu J, Zheng S, Li L. Metabonomic profiles discriminate hepatocellular carcinoma from liver cirrhosis by ultraperformance liquid chromatography-mass spectrometry. J Proteome Res. 2012;11:1217-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 113] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 8. | Peng F, Liu Y, He C, Kong Y, Ouyang Q, Xie X, Liu T, Liu Z, Peng J. Prediction of platinum-based chemotherapy efficacy in lung cancer based on LC-MS metabolomics approach. J Pharm Biomed Anal. 2018;154:95-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Zhou P, Shao L, Zhao L, Lv G, Pan X, Zhang A, Li J, Zhou N, Chen D, Li L. Efficacy of Fluidized Bed Bioartificial Liver in Treating Fulminant Hepatic Failure in Pigs: A Metabolomics Study. Sci Rep. 2016;6:26070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57:2072-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 11. | Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, Yin Z, Xie X, Wang X, Chen W, Zhou L, Wang X, Li Y, Chen H, Gao L, Lu X, Wu T, Wang H, Niu J, Xu G. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67:662-675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 284] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 12. | Huang Q, Tan Y, Yin P, Ye G, Gao P, Lu X, Wang H, Xu G. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013;73:4992-5002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 338] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 13. | Zhou P, Li J, Shao L, Lv G, Zhao L, Huang H, Zhang A, Pan X, Liu W, Xie Q, Chen D, Guo Y, Hao S, Xu W, Li L. Dynamic Patterns of serum metabolites in fulminant hepatic failure pigs. Metabolomics. 2012;8:869-879. [DOI] [Full Text] |
| 14. | Zhou P, Zhou N, Shao L, Li J, Liu S, Meng X, Duan J, Xiong X, Huang X, Chen Y, Fan X, Zheng Y, Ma S, Li C, Wu A. Diagnosis of Clostridium difficile infection using an UPLC-MS based metabolomics method. Metabolomics. 2018;14:102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Zhou J, Sun HC, Wang Z, Cong WM, Wang JH, Zeng MS, Yang JM, Bie P, Liu LX, Wen TF, Han GH, Wang MQ, Liu RB, Lu LG, Ren ZG, Chen MS, Zeng ZC, Liang P, Liang CH, Chen M, Yan FH, Wang WP, Ji Y, Cheng WW, Dai CL, Jia WD, Li YM, Li YX, Liang J, Liu TS, Lv GY, Mao YL, Ren WX, Shi HC, Wang WT, Wang XY, Xing BC, Xu JM, Yang JY, Yang YF, Ye SL, Yin ZY, Zhang BH, Zhang SJ, Zhou WP, Zhu JY, Liu R, Shi YH, Xiao YS, Dai Z, Teng GJ, Cai JQ, Wang WL, Dong JH, Li Q, Shen F, Qin SK, Fan J. Guidelines for Diagnosis and Treatment of Primary Liver Cancer in China (2017 Edition). Liver Cancer. 2018;7:235-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 444] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 16. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1538] [Article Influence: 139.8] [Reference Citation Analysis (40)] |
| 17. | Chen E, Lu J, Chen D, Zhu D, Wang Y, Zhang Y, Zhou N, Wang J, Li J, Li L. Dynamic changes of plasma metabolites in pigs with GalN-induced acute liver failure using GC-MS and UPLC-MS. Biomed Pharmacother. 2017;93:480-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Chen R, Zhu S, Fan XG, Wang H, Lotze MT, Zeh HJ, Billiar TR, Kang R, Tang D. High mobility group protein B1 controls liver cancer initiation through yes-associated protein -dependent aerobic glycolysis. Hepatology. 2018;67:1823-1841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 19. | Fitian AI, Cabrera R. Disease monitoring of hepatocellular carcinoma through metabolomics. World J Hepatol. 2017;9:1-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
| 20. | Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1408] [Cited by in RCA: 1343] [Article Influence: 89.5] [Reference Citation Analysis (0)] |
| 21. | Tremosini S, Forner A, Boix L, Vilana R, Bianchi L, Reig M, Rimola J, Rodríguez-Lope C, Ayuso C, Solé M, Bruix J. Prospective validation of an immunohistochemical panel (glypican 3, heat shock protein 70 and glutamine synthetase) in liver biopsies for diagnosis of very early hepatocellular carcinoma. Gut. 2012;61:1481-1487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 22. | Howard SC, Jones DP, Pui CH. The tumor lysis syndrome. N Engl J Med. 2011;364:1844-1854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 705] [Cited by in RCA: 595] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 23. | McPhail MJW, Shawcross DL, Lewis MR, Coltart I, Want EJ, Antoniades CG, Veselkov K, Triantafyllou E, Patel V, Pop O, Gomez-Romero M, Kyriakides M, Zia R, Abeles RD, Crossey MME, Jassem W, O'Grady J, Heaton N, Auzinger G, Bernal W, Quaglia A, Coen M, Nicholson JK, Wendon JA, Holmes E, Taylor-Robinson SD. Multivariate metabotyping of plasma predicts survival in patients with decompensated cirrhosis. J Hepatol. 2016;64:1058-1067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 24. | Xiao JF, Varghese RS, Zhou B, Nezami Ranjbar MR, Zhao Y, Tsai TH, Di Poto C, Wang J, Goerlitz D, Luo Y, Cheema AK, Sarhan N, Soliman H, Tadesse MG, Ziada DH, Ressom HW. LC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. J Proteome Res. 2012;11:5914-5923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 104] [Article Influence: 8.0] [Reference Citation Analysis (0)] |