Published online Jan 14, 2020. doi: 10.3748/wjg.v26.i2.168
Peer-review started: October 8, 2019
First decision: December 5, 2019
Revised: December 11, 2019
Accepted: December 22, 2019
Article in press: December 22, 2019
Published online: January 14, 2020
Processing time: 96 Days and 13.6 Hours
Hepatitis C virus (HCV) infection and its consequent complications are undeniably a public health burden worldwide, particularly in Egypt. Emerging evidence suggests that many lncRNAs have relevant roles in viral infections and antiviral responses.
To investigate the expression profiles of circulating lncRNAGAS5, lncRNAHEIH, lncRNABISPR and mRNABST2 in naïve, treated and relapsed HCV Egyptian patients, to elucidate relation to HCV infection and their efficacy as innovative biomarkers for the diagnosis and prognosis of HCV GT4.
One hundred and thirty HCV-infected Egyptian patients and 20 healthy controls were included in this study. Serum lncRNAs and mRNABST2 were measured using quantitative real-time polymerase chain reaction (qRT-PCR).
Our results indicated that serum lncRNAGAS5 and LncRNABISPR were upregulated, whereas mRNA BST2 and LncRNA HEIH were downregulated in naïve patients. In contrast, HCV patients treated with sofosbuvir and simeprevir; with sofosbuvir and daclatasvir; or with sofosbuvir, daclatasvir and ribavirin exhibited lower levels of lncRNAGAS5 and lncRNABISPR with higher mRNABST2 compared to naïve patients. Notably, patients relapsed from sofosbuvir and simeprevir showed higher levels of these lncRNAs with lower mRNABST2 compared to treated patients. LncRNAGAS5 and lncRNABISPR were positively correlated with viral load and ALT at P < 0.001, whereas mRNABST2 was negatively correlated with viral load at P < 0.001 and ALT at P < 0.05. Interestingly, a significant positive correlation between lncRNA HEIH and AFP was observed at P < 0.001.
Differential expression of these RNAs suggests their involvement in HCV pathogenesis or antiviral response and highlights their promising roles in diagnosis and prognosis of HCV.
Core tip: The expression profiles of the studied RNAs in naïve, treated and relapsed hepatitis C virus (HCV) Egyptian patients suggest their involvement in HCV-pathogenesis or antiviral response. Additionally, lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and mRNA BST2 could serve as potential diagnostic biomarkers in HCV GT4 Egyptian patients while, lncRNA GAS5, lncRNA BISPR and mRNA BST2 could also be considered novel prognostic biomarkers for treatment in HCV patients. Importantly, lncRNA HEIH might represent a powerful prognostic marker for differentiating relapsed patients from SOF + SIM treatment. Finally, these biomarkers can be used in combination to complete the whole picture of diagnosis, prognosis and follow-up of HCV.
- Citation: El Samaloty NM, Shabayek MI, Ghait RS, El-Maraghy SA, Rizk SM, El-Sawalhi MM. Assessment of lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and its mRNA BST2 as serum innovative non-invasive biomarkers: Recent insights into Egyptian patients with hepatitis C virus type 4. World J Gastroenterol 2020; 26(2): 168-183
- URL: https://www.wjgnet.com/1007-9327/full/v26/i2/168.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i2.168
Viral hepatitis is the seventh leading cause of mortality worldwide. Hepatitis C virus (HCV) comprises almost half of this mortality, ultimately leading to cirrhosis and hepatocellular carcinoma (HCC)[1]. In 2008, the predominance of HCV infection in Egypt was the most noteworthy in the world, near 15% of the populace were seropositive, and approximately 90% of patients were infected with genotype 4 (GT4). In 2015, the National Demographic Health Survey proved a noticeable drop in HCV burden to 6.3% among the population[2]. Actually, Egypt launched one of the largest treatment programmes for controlling viral hepatitis with direct-acting antiviral (DAAs)-based regimens that aim to treat above 250000 chronically infected individuals per year[3].
The era of DAAs was successful for HCV elimination, the sustained viral response (SVR) outcomes improved significantly, and many studies reported decreased risk of liver-related morality with antiviral treatment of HCV[4,5]. Sofosbuvir combination-based regimens were designed as a promising regimen for the treatment of HCV GT4 patients[6].
A substantial effort has been directed to elucidate the molecular mechanisms of HCV, identifying new diagnostic and prognostic markers and therapeutic targets to improve the clinical outcome of HCV patients. LncRNAs are transcripts greater than 200 nucleotides (nt) with poor coding potential[7], that play important roles in regulating gene expression[8]. Emerging evidence suggests that lncRNAs play relevant roles in viral infection and in antiviral responses[9]. A recent study described deregulation of the lncRNA transcriptome in HCV-infected cells[10].
Growth arrest-specific 5 (GAS5) is a 650 nt tumour suppressor lncRNA that has been reported to suppress migration and invasion of some HCC cell lines[11]. It also inhibited the pathogenesis of liver fibrosis via RNAs crosstalk[12]. In HCV-infected Huh7 cells, lncRNA GAS5 has been shown to be upregulated and exhibit a potential antiviral activity. It did not affect virus entry but suppressed HCV replication[13].
Another potential lncRNA is bone marrow stromal cell antigen 2 lncBST2, which has been renamed lncRNA BISPR from BST2 IFN-stimulated positive regulator. LncRNA BISPR was found to be upregulated after treatment with different doses of interferon (IFN), type I IFNα2, in different cells. Moreover, lncRNA BISPR was reported to be upregulated in HCV-infected livers and cultured cells, probably, because of the induction of the IFN signaling pathway in these cells[10,14].
A third interesting lncRNA, called high expression in hepatocellular carcinoma (HEIH), is an oncogenic lncRNA that promotes tumor progression and has been proposed as key regulatory hubs in HCC progression. The expression level of lncRNA-HEIH in HBV-related HCC was significantly associated with recurrence and was identified as an independent prognostic factor for survival[15]. In addition, lncRNA-HEIH in serum and exosomes was identified as a potential biomarker in the HCV-related HCC[16].
mRNA bone marrow stromal cell antigen 2 (BST-2 or Tetherin), which is expressed in most human tissues, has been revealed to inhibit the release of several enveloped viruses[17]. In HCV Huh 7.5 cells, it was demonstrated that BST2 moderately restricted HCV release and production, while the virus lacks mechanisms to respond to this restriction[18].
To date, the expression profiles of the above-mentioned RNAs in HCV GT4 patients and their clinical relevance as biomarkers for HCV have not been studied yet. Therefore, we aimed to assess the serum levels of lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and mRNA BST2 in naïve, treated and relapsed Egyptian HCV patients to examine their relation to HCV infection and their potential usefulness as new diagnostic and prognostic biomarkers for HCV GT4. Additionally, the possible correlations between these RNAs and the clinical data were also analysed.
One hundred and thirty HCV-infected Egyptian patients and 20 healthy controls were recruited in this study. The HCV patient samples were collected from the Department of Internal Medicine, Gastroenterology and Hepatology Clinic- Ain Shams Hospital from March 2017- March 2018. The study included six groups; group I healthy controls (n = 20). Group II naïve HCV patients without treatment (n = 30). Groups from III to V comprised HCV patients treated daily with three different 12-week oral treatment regimens as follows: Group III (SOF + SIM) (n = 30) received combination of sofosbuvir (SOF 400 mg) and simeprevir (SIM 150 mg). Group IV (SOF + DAC) (n = 20) received combination of SOF (400 mg) and daclatasvir (DAC 60 mg). Group V (SOF + DAC + RBV) (n = 20) received fixed dose combination of SOF (400 mg) and DAC (60 mg) with ribavirin (RBV) at weight-based doses of 600, 800 and 1000 mg for patients with body weight less than 60 kg, between 60-80 kg, and more than 80 kg respectively[19]. Group VI included HCV patients who relapsed after 12-wk treatment with SOF + SIM (n = 30). Patients on therapy showed SVR (undetectable HCV RNA at the end of 12-wk treatment and remained free from HCV RNA for further 12 wk). In contrast, relapsed patients showed undetectable HCV RNA after completion of 12-week treatment however, after further 12 wk the HCV RNA was detected and was nearly high as those of naïve patients. All enrolled HCV patients presented positive outcomes when tested for serum anti-HCV antibodies with detectable serum HCV RNA GT4, and they had abnormal serum aminotransferases for 6 months. Naïve patients had not previously received any HCV treatment or antiviral therapy. Patients with cirrhotic liver, HCC, alcohol-induced liver injury, HBV antigen or antibody, thyroid dysfunction, hypertension, renal insufficiency, and other major diseases were excluded. All participants were age and gender matched. The study protocol was approved by the Research Ethics Committee for Experimental and Clinical studies at the Faculty of Pharmacy, Cairo University, Cairo, Egypt (approval number: BC 1955) and was conducted in accordance with the ethical guidelines of the Declaration of Helsinki. All participants received the required information regarding the study, and their written informed consents were obtained.
Venous blood samples were collected from all participants using serum collection tubes. The separated sera were aliquoted and stored at -80 °C for the analysis of lncRNAs and mRNA expressions. An aliquot of the serum was used to assess the routine workup; serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), prothrombin time (PT), international normalized ratio (INR), albumin, total bilirubin; which were analysed spectrophotometrically (Spectrum Diagnostic, Cairo, Egypt). Alpha-fetoprotein (AFP), hepatitis B surface antigen, anti-HCV titres, anti-schistosomal antibodies, and hepatitis B core antibodies were assessed by enzyme-linked immunosorbent assay (Aviva System Biology, CA, United States).
Serum HCV RNA was extracted by a viral RNA extraction kit (Qiagen, CA, United States) according to the manufacturer’s protocol, and it was quantified by quantitative Real Time-PCR (qRT-PCR) (TaqMan assay reagents and Ambion, the RNA Company-one step, CA, United States). Genotyping was done based on the core region sequence using the Ohno method. This method used genotype-specific primers and depends on the PCR amplification of the HCV core gene[20].
RNA extraction: Total RNA was extracted from 200 μL serum by the miRNeasy Mini Kit (Qiagen, Hilden, German) using QIAZOL lysis reagent according to the manufacturer's instructions. The extracted RNA was dissolved in 50 μL RNase-free water and stored at −80 °C until analysis. The quality of RNA was determined using nanodrop (Thermo Scientific, United States).
Reverse transcription: Reverse transcription was done using RT2 first strand Kit (Qiagen, Hilden, Germany), 8 μL total RNA template were reverse transcribed in a final reaction mix volume of 20 μL. For synthesis of cDNA, the RT reaction was incubated for 60 min at 37 °C, and for 5 min at 95 °C. The cDNA produced were stored at −20 °C till analysis.
qRT-PCR: Relative expression levels of the LncRNAs and mRNA were evaluated using the RT2 SYBR Green Master Mix kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The housekeeping gene, Hs_GAPDH_1_SG was selected as the internal control. Briefly, for analysis of the LncRNAs, 2 μL cDNA product was used as a template in a 25 μL total reaction volume containing 12.5 μL RT2 SYBR Green PCR Master Mix, 9.5 μL nuclease-free water, and 1 μL RT2 LncRNA PCR primer assay. Readily made primers by Qiagen were used for amplification. The primer sequences used are as follows: lncRNA GAS5 (sense: CACACA GGCATTAGACAGA, antisense: GCTCCACACAGTGTAGTCA), lncRNA BISPR (sense: GCCAACAAACAATGTCGGTCT, antisense: CAGAGACACAGATG CTGCCTAA) and lncRNA HEIH (sense: CCTCTTGTGCCCCTTTCTT, antisense: ATGGCTTCTCGCATCCTAT). qRT-PCR was performed with a Qiagen Rotor Gene Q6 Plex Real-Time PCR system (Qiagen, Hilden, Germany), with a PCR initial activation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Data were analysed with Rotor Gene Q software with the automatic threshold cycle (Ct) setting. The relative expression for each LncRNA after normalization to Hs_GAPDH_1_SG was calculated using the 2-ΔΔct method.
For the assessment of mRNA BST2 relative expression, qRT-PCR was performed in 25 µL reaction mixture prepared by mixing 12.5 µL master mix, 2.5 µL primer assay, 5 µL cDNA, and 5 µL RNAase-free water. The primer sequence for the mRNA BST2 analysis is (sense: TCAGGAGTCCCTGGAGAAGA, antisense: ATGGAG CTGCCAGAGTTCAC). The reaction was performed with a PCR initial activation at 95 °C for 15 min followed by 40 cycles at 95 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s. The data were examined with Rotor Gene Q software with the automatic threshold cycle (Ct) setting. The relative expression of mRNA BST2 after normalization to Hs_GAPDH_1_SG was calculated using the 2-ΔΔct method.
The results were reported as the mean ± SD. Ordinary one-way analysis of variance (ANOVA) was used for normally distributed variables followed by the Tukey HSD multiple comparisons test. Viral load levels were log transformed to enable parametric statistical tests. The normality of the variable’s distribution was assessed using the Shapiro Wilks test. To test the association between lncRNA Gas 5, lncRNA BISPR, lncRNA HEIH and mRNA BST2 and different biochemical parameters, Pearson’s linear correlation and multiple regression analysis were performed. P values < 0.05 were considered significant, with a 95%CI. The diagnostic precision of lncRNAs and mRNAs was assessed by receiver-operating-characteristic (ROC) analysis. Univariate and multiple binary logistic regression analyses were completed to detect predictors associated with the risk of HCV and of treatment or relapsed outcome. The significant data in the univariate analysis were further analysed by multiple analysis to determine the independent variables that affected the response. All statistical analyses were performed using SPSS Statistics v. 20 for Windows (SPSS, Chicago, IL). The graphs were plotted using GraphPad Prism 7.
Naïve HCV patients had significantly higher levels of viral load, AST, ALT, ALP, AFP, PT, bilirubin and INR along with lower albumin levels compared to healthy controls. Meanwhile, HCV-infected patients treated with various treatment regimen showed undetected viral load together with significantly lower serum levels of hepatic enzyme markers compared to naïve HCV patients. On the other hand, relapsed patients exhibited marked increase in viral load levels together with significantly higher serum ALP and AFP compared to SOF + SIM treated group as illustrated in Table 1.
| Parameters | Healthy control | Naïve | SOF + SIM | SOF + DAC | SOF + DAC + RBV | Relapsed |
| (n = 20) | (n = 30) | (n = 30) | (n = 20) | (n = 20) | (n = 30) | |
| Age (yr) | 40.2 ± 5 | 44.3 ± 6.9 | 42.2 ± 6.7 | 42.8 ± 7.2 | 39.7 ± 5.1 | 44.1 ± 6.5 |
| Gender (M/F) | 16/4 | 26/4 | 27/3 | 19/1 | 19/1 | 25/5 |
| ALT (IU/L) | 9.2 ± 6.2 | 37.2 ± 25b | 18.7 ± 8.2ad | 17.6 ± 5.6ad | 26.7 ± 2.4adeg | 23.7 ± 6.9d |
| AST (IU/L) | 10.3 ± 4.2 | 34.4 ± 16.4b | 27.4 ± 13.7bc | 26.9 ± 6.9bc | 32.7 ± 5.2b | 23.3 ± 6.3d |
| Albumin (g/dL) | 3.8 ± 0.27 | 2.7 ± 0.43b | 3.2 ± 0.5bd | 3.9 ± 0.3dh | 4 ± 0.4adh | 3 ± 0.4d |
| ALP (IU/L) | 53.7 ± 6.8 | 130.6 ± 21b | 86.1 ± 16bd | 78.4 ± 16bd | 81.2 ± 12.4bd | 99.4 ± 11.2bf |
| Bilirubin (mg/dL) | 0.9 ± 0.1 | 1.1 ± 0.4a | 1.2 ± 0.5b | 1.1 ± 0.4 | 0.9 ± 0.3ce | 1.2 ± 0.4 |
| INR | 1.06 ± 0.07 | 1.4 ± 0.25b | 1.5 ± 0.25b | 1.3 ± 0.19b | 1.2 ± 0.22adfh | 1.4 ± 0.27 |
| AFP (ng/mL) | 7.6 ± 1.3 | 15 ± 8.6b | 7.3 ± 0.8d | 7 ± 1d | 6.9 ± 1d | 32 ± 9.6bf |
| PT | 11.7 ± 0.5 | 14.8 ± 2b | 13.7 ± 1.4bd | 13.1 ± 1.2bd | 12.6 ± 1.5de | 14.1 ± 1.1 |
| Viral load (log copies/mL) | 0 | 6.47 ± 0.7c | 0d | 0d | 0d | 6.38 ± 0.39f |
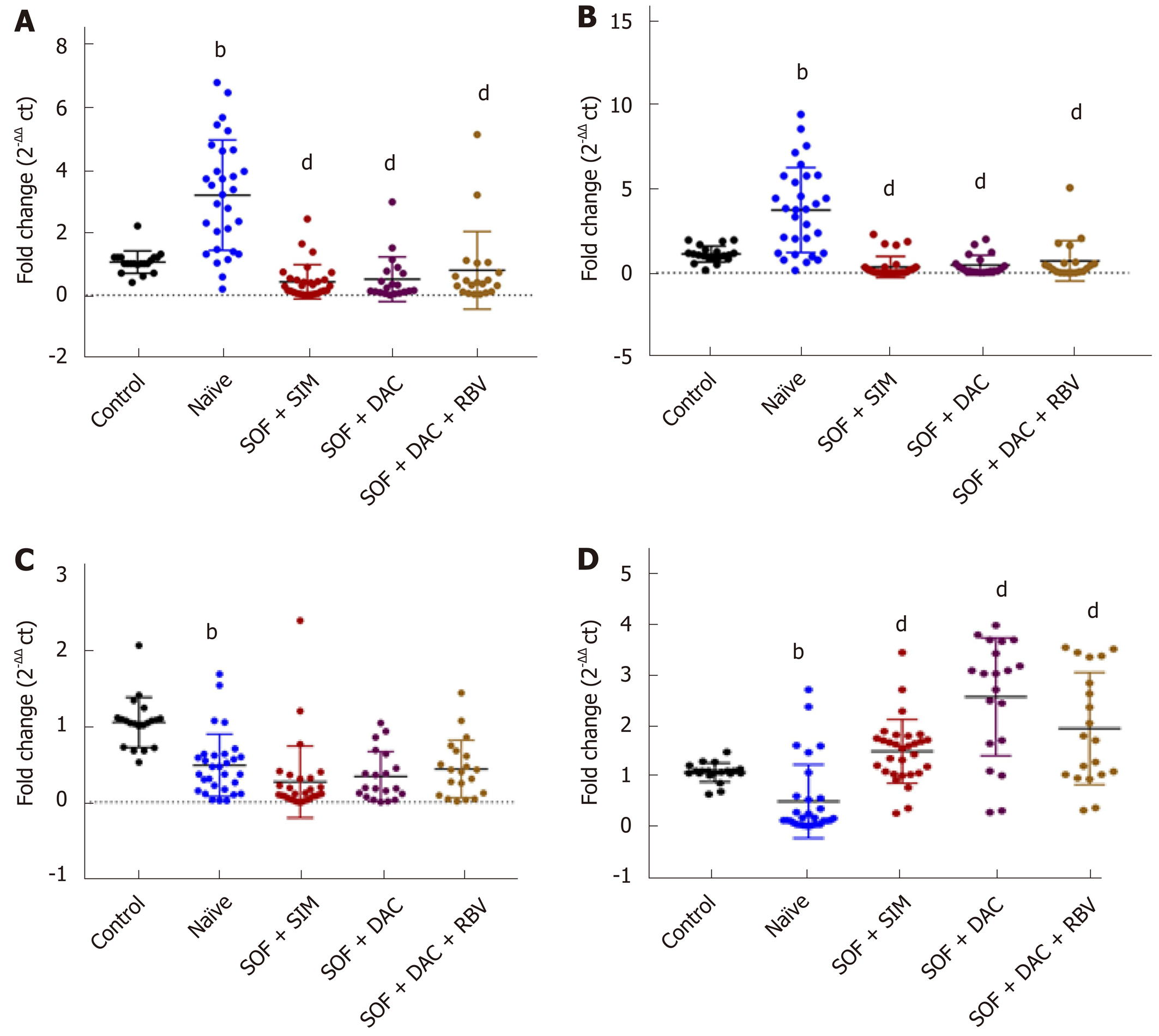
Compared to healthy controls, serum expression levels of LncRNA GAS5 and LncRNA BISPR were upregulated, while, LncRNA HEIH and mRNA BST2 were downregulated in the naïve HCV group (Figure 1).
The levels of lncRNA GAS5 and lncRNA BISPR were significantly lower in the three treated groups, whereas, the serum levels of mRNA BST2 were significantly higher compared with that of the naïve group.
However, no significant difference was observed between the relative expression of lncRNA HEIH in the three treated groups versus that in the naïve HCV patients (Figure 1).
Pearson’s correlation analyses of lncRNA GAS5, lncRNA BISPR, lncRNA HEIH and mRNA BST2 expression with all the studied parameters are presented in Table 2. Expression of lncRNA GAS5 revealed significant positive correlations with ALT, AST, ALP, AFP, PT, viral load, lncRNA BISPR and lncRNA HEIH. LncRNA GAS5 levels were negatively correlated with albumin and mRNA BST2. Meanwhile, lncRNA BISPR was positively correlated with ALT, ALP, INR, AFP, PT and viral load, whereas lncRNA BISPR was negatively correlated with albumin and mRNA BST2. Additionally, serum lncRNA HEIH levels were significantly positively correlated with AFP and negatively correlated with AST and mRNA BST2. On the other hand, serum expression of mRNA BST2 was significantly negatively correlated with ALT, ALP, AFP, PT and viral load and was positively correlated with albumin.
| Variables | LncRNA Gas5 | LncRNA BISPR | LncRNA HEIH | MRNA BST2 | ||||
| r | P value | r | P value | r | P value | r | P value | |
| Age (yr) | 0.051 | 0.53 | 0.138 | 0.09 | 0.095 | 0.24 | -0.043 | 0.60 |
| Sex (M/F) | 0.070 | 0.39 | -0.043 | 0.60 | 0.037 | 0.65 | -0.062 | 0.44 |
| ALT (IU/L) | 0.332 | 0.001c | 0.287 | 0.001c | -0.072 | 0.37 | -0.194 | 0.01a |
| AST (IU/L) | 0.016 | 0.04a | 0.141 | 0.08 | -0.222 | 0.006b | 0.008 | 0.92 |
| Albumin (g/dL) | -0.454 | 0.001c | -0.419 | 0.001c | -0.0157 | 0.05 | 0.434 | 0.001c |
| Bilirubin (mg/dL) | 0.088 | 0.28 | 0.049 | 0.54 | 0.034 | 0.67 | -0.083 | 0.31 |
| ALP (IU/L) | 0.589 | 0.001c | 0.517 | 0.001c | 0.044 | 0.59 | -0.346 | 0.001c |
| INR | 0.136 | 0.09 | 0.172 | 0.03a | -0.062 | 0.45 | 0.021 | 0.79 |
| AFP | 0.476 | 0.001c | 0.176 | 0.03a | 0.640 | 0.001c | -0.347 | 0.001c |
| PT | 0.405 | 0.001c | 0.300 | 0.001c | 0.081 | 0.32 | -0.206 | 0.01a |
| Viral load | 0.276 | 0.001c | 0.374 | 0.001c | 0.056 | 0.49 | -0.284 | 0.001c |
| (log copies/mL) | ||||||||
| LncRNA Gas5 | 0.607 | 0.001c | 0.318 | 0.001c | -0.465 | 0.001c | ||
| LncRNA BST2 | 0.607 | 0.001c | 0.040 | 0.62 | -0.371 | 0.001c | ||
| LncRNA HEIH | 0.318 | 0.001c | 0.040 | 0.62 | -0.235 | 0.004b | ||
| mRNA BST2 | -0.465 | 0.001c | -0.371 | 0.001c | -0.235 | 0.004b | ||
On performing multiple linear regression calculations, using lncRNA GAS5 as the dependent variable and AST, ALT, bilirubin, albumin, INR, AFP, viral load, PT, ALP, lncRNA BISPR, lncRNA HEIH and mRNA BST2 as independent variables, only AFP, ALP, lncRNA BISPR, lncRNA HEIH and mRNA BST2 remained significant (β = 0.16, P < 0.05; β = 0.28, P < 0.001; β = 0.37, P < 0.001, β = 0.15, P < 0.05; β = -0.13, P < 0.05, respectively). However, in using lncRNA BISPR as the dependent variable and AST, ALT, bilirubin, albumin, INR, AFP, viral load, PT, ALP, lncRNA GAS5, lncRNA HEIH and mRNA BST2 as independent variables, only viral load and lncRNA GAS5 were significant (β = 0.14, P < 0.05; β = 0.57, P < 0.001, respectively).
Via multiple regression analysis using lncRNA HEIH as the dependent variable and AST, ALT, bilirubin, albumin, INR, AFP, viral load, PT, ALP, lncRNA GAS5, lncRNA BISPR and mRNA BST2 as independent variables, only AFP and lncRNA GAS5, were significant (β = 0.66, P < 0.001, β = 0.19, P < 0.05, respectively). However, in multiple regression analysis using mRNA BST2 as the dependent variable and AST, ALT, bilirubin, albumin, INR, AFP, viral load, PT, ALP, lncRNA GAS5, lncRNA BISPR and lncRNA HEIH as independent variables, only albumin and lncRNA GAS5 were still significant (β = 0.24, P < 0.01; β = -0.22, P < 0.05, respectively).
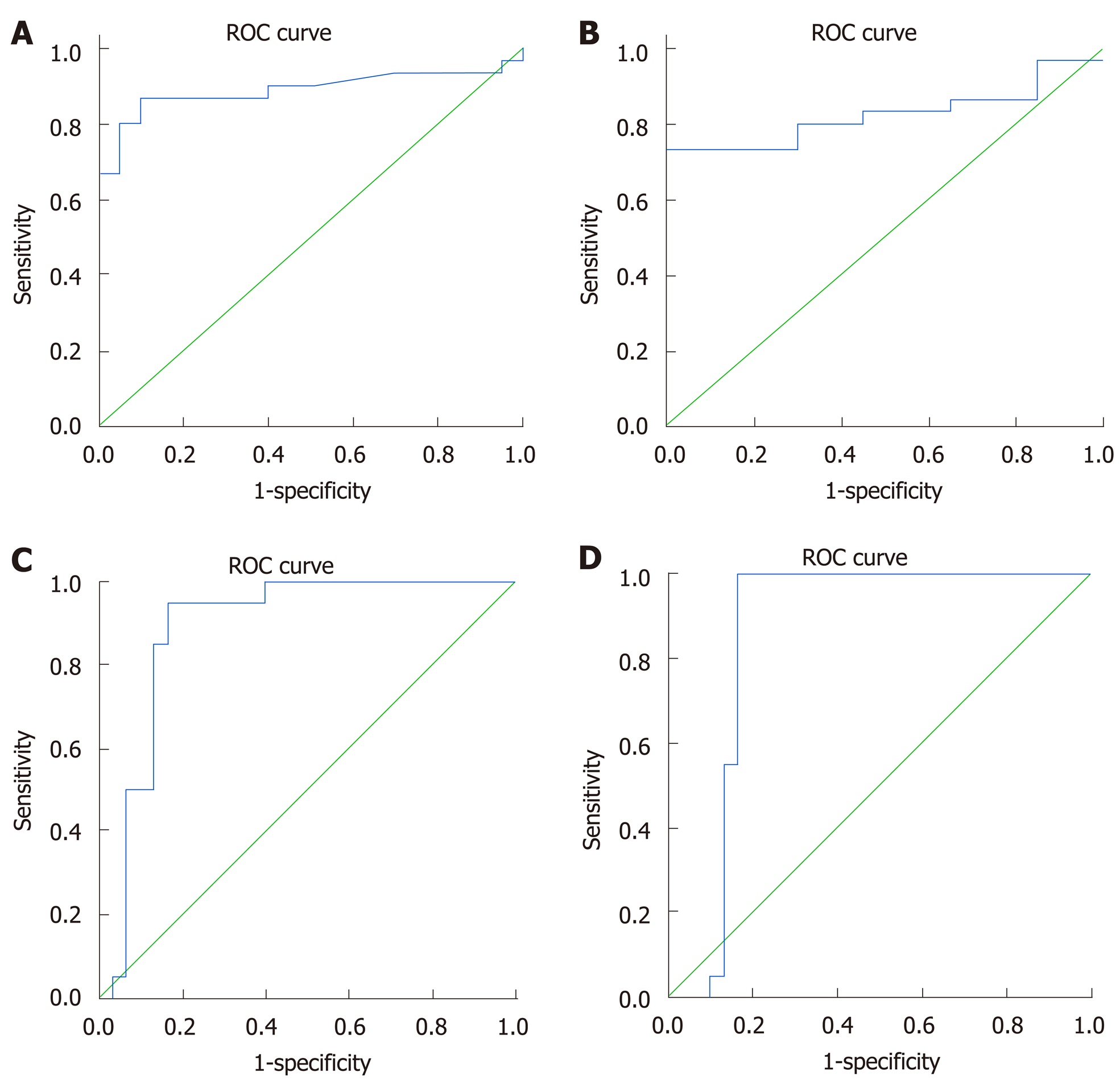
ROC analysis demonstrated that the studied lncRNAs and mRNA could differentiate between the naïve group and the healthy controls (Figure 2) with an AUC = 0.88 for LncRNA GAS5, sensitivity = 86%, specificity = 90% at a cut-off = 1.2-fold. For LncRNA BISPR, the AUC = 0.82, the sensitivity = 80%, and the specificity = 70% at a cut-off = 1.1-fold. For lncRNA HEIH and mRNA BST2, the AUC = 0.88, the sensitivity = 90%, and the specificity = 80% at a cut-off = 0.76-fold, and AUC = 0.85 sensitivity = 95%, specificity = 84% at a cut-off = 0.65-fold, respectively.
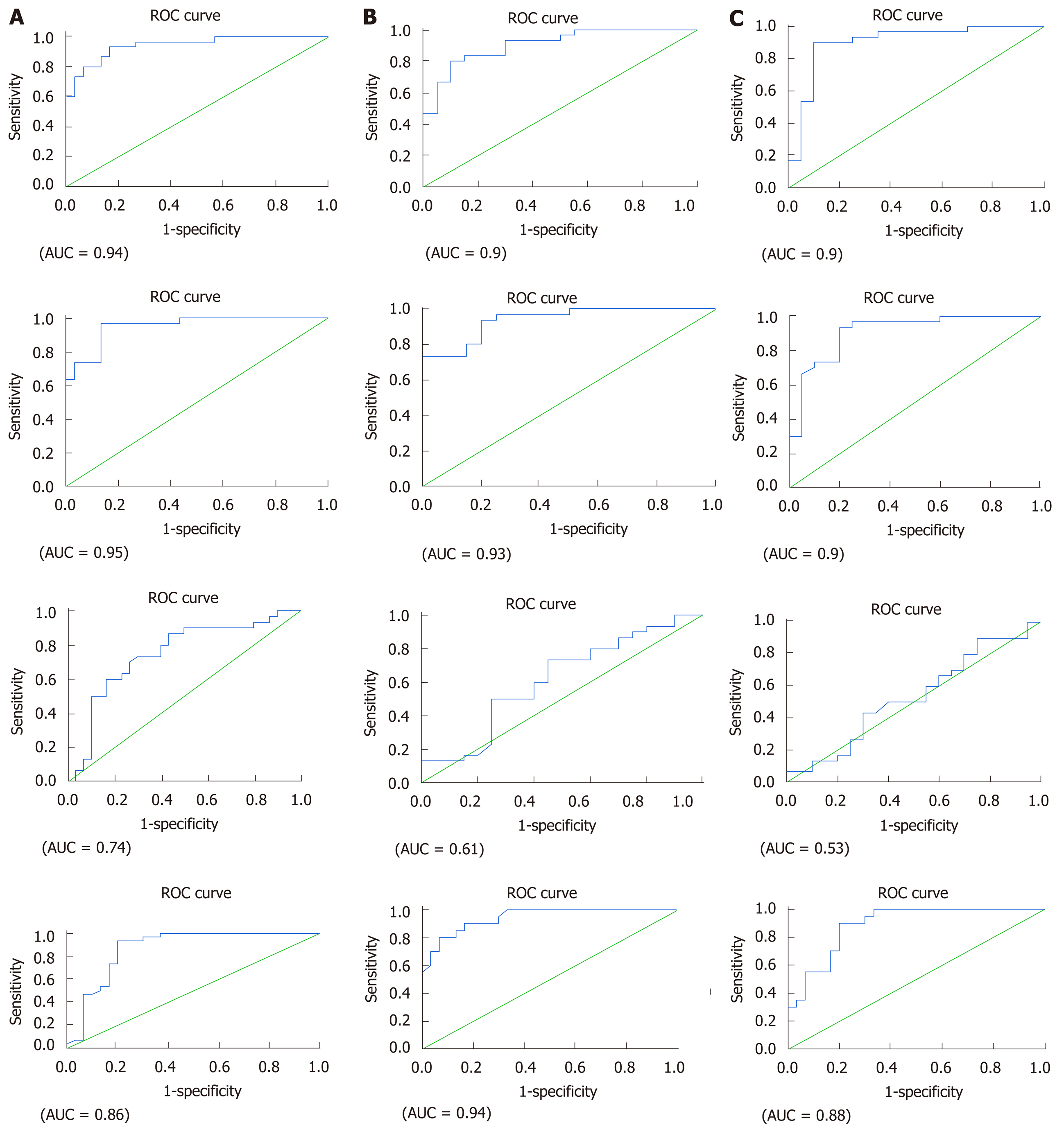
ROC analysis revealed that all the studied lncRNAs and mRNA could also discriminate between the naïve group and the HCV patients treated with different treatment regimens (Figure 3). The prognostic accuracy of lncRNA GAS5 in SOF + SIM treatment had an AUC = 0.94, sensitivity = 93%, specificity = 84% at a cut-off = 0.95 fold, while in SOF + DAC with an AUC = 0.90, sensitivity = 83%, specificity = 85% at a cut-off = 0.94-fold, and SOF + DAC + RBV with an AUC = 0.90, sensitivity = 90%, specificity = 90% at a cut-off = 1.1-fold. The prognostic relevance of lncRNA BISPR in SOF + SIM treatment showed AUC = 0.95, sensitivity = 93%, specificity = 87% at a cut-off = 0.7-fold, in SOF + DAC with an AUC = 0.93, sensitivity = 93%, specificity = 80% at a cut-off = 0.77-fold and in SOF+DAC+RBV with an AUC = 0.90, sensitivity = 93%, specificity = 80% at a cut-off = 0.77-fold. In addition, the significance of lncRNA HEIH in the SOF + SIM, SOF + DAC and SOF + DAC + RBV treatment groups had AUC = 0.74, 0.61, and 0.53, respectively, with sensitivity and specificity values of 73% and 70%, 60% and 60%, and 66% and 40% at cut-off values of 0.2-fold, 0.36-fold, 0.28-fold, respectively. Additionally, the prognostic significance of mRNA BST2 in the SOF + SIM, SOF + DAC and SOF + DAC + RBV treatment groups demonstrated AUC = 0.86, 0.94, and 0.88, respectively, with sensitivity and specificity values of 90% and 80%, 85% and 87%, and 90% and 80% at cut-off values of at 0.83-fold, at 1.1-fold, at 0.76-fold, respectively.
Analysis of univariate and multiple logistic regression were employed to discriminate between naïve patients and healthy controls (Table 3). In the univariate analysis, ALT, AST, ALP, AFP, albumin, and expression levels of lncRNA GAS5, lncRNA BISPR, lncRNA HEIH and mRNA BST2 were designated as significant predictors associated with the diagnosis of naïve HCV patients. On conducting multiple analysis, LncRNA GAS5 and mRNA BST2 were designated as significant independent variables that could be used in the diagnosis of HCV.
| Parameter | B | SE | P value | Odds ratio | Odds ratio (95%CI) |
| Univariate analysis | |||||
| LncRNA-Gas5 | 1.95 | 0.675 | 0.004b | 7.08 | 1.88-26.59 |
| LncRNA-BISPR | 1.2 | 0.421 | 0.004b | 3.32 | 1.45-7.57 |
| LncRNA-HEIH | -3.8 | 1.079 | 0.005b | 0.02 | 0.003-0.184 |
| mRNA BST2 | -1.71 | 0.606 | 0.003b | 0.17 | 0.05-0.587 |
| ALT (IU/L) | 0.2 | 0.06 | 0.001b | 1.22 | 1.09-1.38 |
| AST (IU/L) | 0.42 | 0.135 | 0.002b | 1.52 | 1.16-1.98 |
| AFP (ng/ml) | 0.358 | 0.165 | 0.03a | 1.43 | 1.03-1.97 |
| ALP (IU/L) | 0.209 | 0.067 | 0.002b | 1.23 | 1.080-1.4 |
| Albumin (mg/dl) | -20.26 | 9.95 | 0.04a | 0.01 | 0.00-0.471 |
| Multiple analysis | |||||
| LncRNA-Gas5 | 2.4 | 0.766 | 0.002b | 11.12 | 2.47-49.88 |
| mRNA-BST2 | -2.71 | 1.013 | 0.007b | 0.066 | 0.009-0.484 |
Concerning the result variables in response to SOF + SIM, SOF + DAC or SOF + DAC + RBV therapies, univariate and multiple logistic regression analyses were performed (Table 4). Expression levels of lncRNA GAS5, lncRNA BISPR and mRNA BST2 and serum levels of ALT, AFP, ALP and albumin were nominated as significant predictor variables in the univariate analysis of SOF + SIM treatment. In multiple analysis, lncRNA GAS5, lncRNA BISPR and mRNA BST2 were chosen as significant independent variables that might predict the response to SOF + SIM therapy. Serum levels of ALT, AFP, and ALP and expression levels of lncRNA GAS5, lncRNA BISPR and mRNA BST2 were selected as significant predictor variables in the univariate analysis of SOF + DAC treatment (Table 4). When multiple analysis was employed, lncRNA GAS5 and lncRNA BISPR showed the highest odds ratios upon holding all variables constant. Expression levels of lncRNA GAS5, lncRNA BISPR, mRNA BST2 and AFP were selected as significant predictors in the univariate analysis of SOF + DAC + RBV treatment (Table 4). Upon multiple analysis, lncRNA GAS5 and lncRNA BISPR were designated as significant independent variables that might predict the response to SOF + DAC + RBV therapy.
| Paramete | B | SE | P value | Odds ratio | Odds ratio (95%CI) |
| Univariate analysis (SOF + SIM) | |||||
| LncRNA-Gas5 | 2.135 | 0.583 | < 0.001c | 8.45 | 2.69-26.50 |
| LncRNA-BISPR | 1.733 | 0.482 | < 0.001c | 5.65 | 2.19-14.56 |
| mRNA BST2 | -2.029 | 0.501 | < 0.001c | 0.13 | 0.04-0.351 |
| ALT (IU/L) | 0.084 | 0.03 | 0.005b | 1.08 | 1.025-1.153 |
| AFP (ng/mL) | 0.715 | 0.299 | 0.01a | 2.044 | 1.137-3.675 |
| ALP (IU/L) | 0.209 | 0.067 | 0.002b | 1.232 | 1.080-1.405 |
| Albumin (mg/dL) | -1.947 | 0.635 | 0.002b | 0.143 | 0.041-0.495 |
| Multivariate analysis | |||||
| LncRNA-Gas5 | 1.397 | 0.704 | 0.04a | 4.04 | 1.01-16.073 |
| LncRNA-BISPR | 1.613 | 820 | 0.04a | 5.01 | 1.007-25.02 |
| mRNA BST2 | -3.149 | 1.307 | 0.01a | 0.043 | 0.003-0.556 |
| Univariate analysis (SOF + DAC) | |||||
| LncRNA-Gas5 | 1.54 | 0.48 | 0.002b | 4.69 | 1.80-12.26 |
| LncRNA-BISPR | 1.86 | 0.61 | 0.003b | 6.43 | 1.91-21.59 |
| mRNA BST2 | -1.677 | 0.415 | < 0.001c | 0.187 | 0.083-0.422 |
| ALT (IU/L) | 0.125 | 0.048 | 0.01a | 1.13 | 1.031-1.245 |
| AFP (ng/mL) | 0.715 | 0.299 | 0.01a | 2.044 | 1.137-3.675 |
| ALP (IU/L) | 0.729 | 0.296 | 0.01a | 2.073 | 1.161-3.699 |
| Multivariate analysis | |||||
| LncRNA-Gas5 | 1.17 | 0.54 | 0.03a | 3.05 | 1.06-8.79 |
| LncRNA-BISPR | 1.42 | 0.69 | 0.03a | 4.17 | 1.07-16.1 |
| Univariate analysis (SOF + DAC + RBV) | |||||
| LncRNA-Gas5 | 1.12 | 0.33 | 0.001b | 3.07 | 1.6-5.88 |
Remarkably, lncRNA GAS5 and lncRNA BISPR levels were positive predictors, whereas the level of mRNA BST2 was an independent negative predictor of treatment responses in all treatment groups.
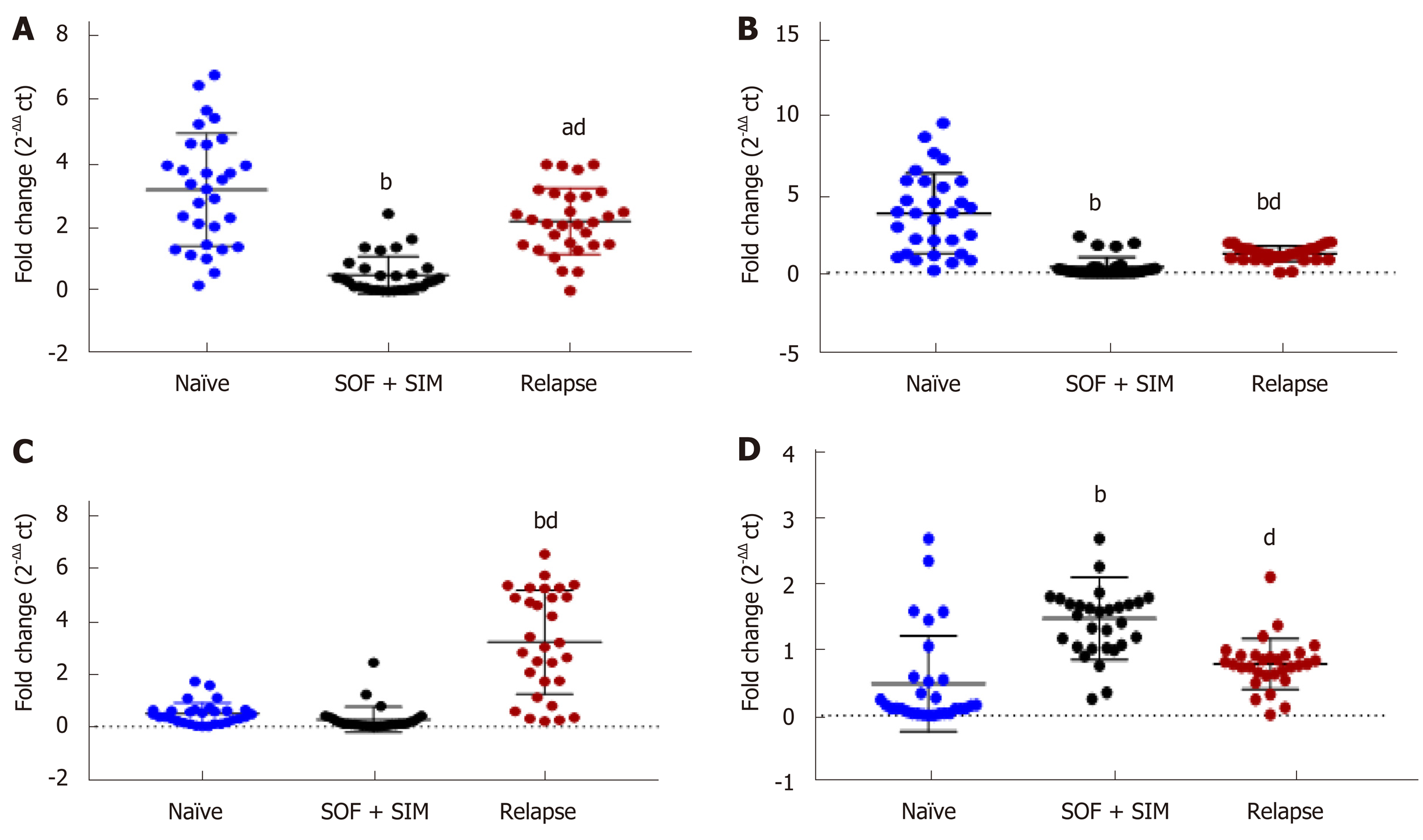
In relapsed patients, expression of lncRNA GAS5, lncRNA BISPR and lncRNA HEIH revealed significantly higher levels than those of the SOF + SIM treated group. In contrast, mRNA expression levels were significantly lower than in the SOF + SIM-treated group (Figure 4). Moreover, relapsed patients showed significantly lower lncRNA GAS5and lncRNA BISPR values compared to those of the naïve group, whereas lncRNA HEIH demonstrated a significantly higher value than that of naïve group (Figure 4).
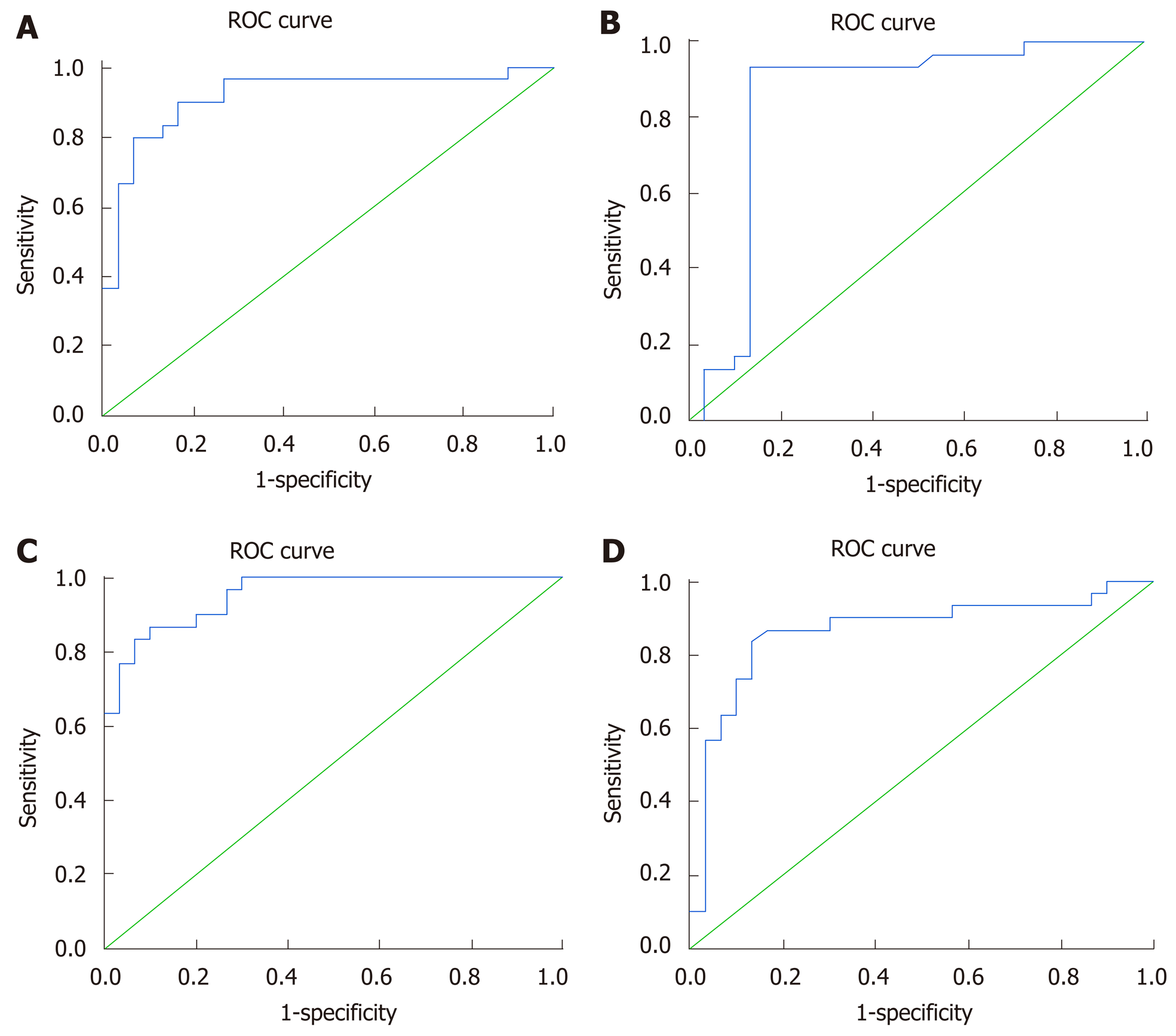
ROC curve analysis showed that serum lncRNA GAS5, lncRNA BISPR, lncRNA HEIH and mRNA BST2 could discriminate the relapsed group from the SOF + SIM-treated group with AUC = 0.91, 0.84, 0.95, and 0.86 respectively, with sensitivities and specificities at cut-off values of 90% and 84% at 0.96-fold, 90% and 87% at 0.77-fold, 90% and 80% at 0.32-fold, and 86% and 84% at 0.98-fold, respectively (Figure 5).
Univariate and multiple logistic regression analyses were presented to choose the predictor variables related with relapsed HCV patients from SOF + SIM therapy (Table 5). Based on univariate analysis, serum ALT, ALP, lncRNA GAS5, lncRNA BISPR and lncRNA HEIH were designated as positive significant predictor variables, while mRNA BST2 was designated as a significant negative predictor variable. In multiple analysis, only serum lncRNA GAS5 and lncRNA HEIH could be significant independent predictors of relapsed HCV patients from SOF + SIM therapy.
| Parameter | B | SE | P value | Odds ratio | Odds ratio (95%CI) |
| Univariate analysis | |||||
| LncRNA-Gas5 | 2.35 | 0.564 | < 0.001b | 10.48 | 3.47-31.67 |
| LncRNA-BISPR | 2.27 | 0.555 | < 0.001b | 9.69 | 3.26-28.73 |
| LncRNA-HEIH | 1.99 | 0.599 | 0.001b | 7.31 | 2.26-23.63 |
| mRNA BST2 | -3.15 | 0.858 | < 0.001b | 0.042 | 0.08-0.228 |
| ALT (IU/L) | 0.87 | 0.037 | 0.01a | 1.09 | 1.01-1.17 |
| ALP (IU/L) | 0.069 | 0.022 | 0.002b | 1.07 | 1.02-1.11 |
| Multivariate analysis | |||||
| LncRNA-Gas5 | 1.46 | 0.69 | 0.03a | 4.322 | 1.12-16.70 |
| LncRNA-HEIH | 1.4 | 0.714 | 0.04a | 8.885 | 1.00-16.51 |
HCV infection is a complex multifactorial process that involves multiple viral and host interactions. Besides, the high mutation rate of HCV that enables the virus to generate escape mutants resistant to treatment[21], emphasizing the need for a proper understanding of the pathogenesis of HCV infection, and various research efforts should be oriented to develop novel diagnostic and prognostic molecular tools.
The present study demonstrated for the first-time differential expression of lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and mRNA BST2 in the sera of naïve, treated and relapsed HCV GT4 Egyptian patients.
We found that naïve patients exhibited elevated serum levels of lncRNA GAS5 and lncRNA BISPR compared to healthy controls. A recent study in Huh 7 cells reported upregulation of lncRNA GAS5 expression during the progression of HCV infection[13]. The authors showed that GAS5 overexpression inhibited HCV infection, while GAS5 knockdown promoted HCV infection. Furthermore, GAS5 was found to directly bound HCV NS3 protein with its 5′ end 200 sequences and acted as a decoy to inhibit the function of this key viral protein that is essential for viral replication. Here, the observed increase in serum lncRNA GAS5 in naïve patients supports these in vitro findings and implicates GAS5 in the antiviral response that occurs along with HCV infection.
Meanwhile, the observed upregulation of lncRNA BISPR in the sera of our naïve HCV patients confirms the findings of Barriocanal et al[14], in which significant increases in lncRNA BISPR levels were demonstrated in HCV-infected Huh 7 cells and in liver samples from HCV-infected donors compared to HCV-negative controls. These findings also showed that lncRNA BISPR expression was highly upregulated in HuH7 cells in response to IFN, suggesting that lncRNA BISPR could be induced by IFN to control the potency of the antiviral IFN response.
Unlike lncRNA GAS5 and lncRNA BISPR, the current study revealed significant downregulation of lncRNA HEIH and mRNA BST2 levels in the sera of naïve HCV patients versus healthy controls. To our knowledge, no previous studies had assessed the expression levels of lncRNA HEIH in the serum of naïve HCV patients in relation to healthy controls. Nevertheless, a recent study stated that serum exosomal lncRNA-HEIH levels in HCV-related HCC patients were remarkably higher than its levels in patients with HCV-induced cirrhosis and both were higher than lncRNA-HEIH levels in HCV patients[16]. Additionally, Zhefeng et al[22] found an association between the expression of LncRNA-HEIH in plasma and HCC risk. Accordingly, in the current study, it can be suggested that naïve HCV patients with low levels of lncRNA HEIH might be less susceptible to HCC risk at this stage of the disease; however, additional prospective studies are needed to confirm this assumption.
According to the present findings, mRNA BST2 was significantly downregulated in naïve HCV patients versus healthy controls. Additionally, there was a significant negative correlation between lncRNA BISPR and its mRNA, indicating that upregulation of lncRNA BISPR expression might lead to downregulation of its mRNA. In contrast, a previous study using isolated HuH7 cells showed that inhibition of lncBISPR by RNA interference led to decreased levels of mRNA BST2[14]. However, our study is the first to assess the expression of both lncRNA BISPR and its mRNA in HCV G4 Egyptian patients; further studies are required to explore the exact mechanistic relevance of lncRNA BISPR and mRNA BST2 in those patients.
Of note, the study’s results indicated that lncRNA Gas5, lncRNA BISPR and lncRNA HEIH and mRNA BST2 could discriminate naïve HCV patients from controls by ROC curve analysis; thus, they could be promising diagnostic biomarkers in HCV infection.
One of the main novelties in present study was the significant decline in circulating lncRNA GAS5 and lncRNA BISPR in HCV patients treated with SOF + SIM, SOF + DAC and SOF + DAC + RBV compared to naïve HCV patients. Recently, it has been reported that treatment of the cells with IFN and ruxolitinib, an inhibitor of the JAK/STAT pathway, or transfection of siRNAs that target STAT2 in IFN-treated cells, significantly reduced BISPR levels compared to those of control cells[10]. It should be noted that in this study, the expression levels of lncRNA GAS5and lncRNA BISPR were positively correlated with viral load, ALT, ALP and AFP, implicating both lncRNA GAS5 and lncRNA BISPR as key players in HCV disease progression and/or host viral response against HCV. Herein, both lncRNA GAS5 and lncRNA BISPR were excellent in discriminating the HCV groups treated with SOF + SIM, SOF + DAC or SOF + DAC + RBV from naïve HCV patients. Interestingly, lncRNA GAS5 and lncRNA BISPR had the highest prognostic accuracy in the SOF + SIM treatment group.
In the current investigations the serum levels of mRNA BST2 were significantly higher in all treated HCV patients compared to the levels in naïve HCV patients. In fact, it has been reported that BISPR protein may regulate host response to viral infection either by inhibiting the release of nascent viral particles or by restricting HCV production[23]. Furthermore, mRNA BST2 was negatively correlated with viral load, ALT, ALP, and AFP, supporting the pivotal role of BST2 protein in eradicating HCV in those treated patients. As far as we are aware, there are no reports on lncRNA GAS5, lncRNA BISPR and mRNA BST2 expression in treated HCV patients, and no data were available to oppose or correspond with our results.
To the best of the author’s knowledge, this study is the first to address the expression level of lncRNAs and mRNA in the sera of Egyptian HCV G4 patients who experienced a viral relapse or disease reactivation after 12-wk treatment with SOF + SIM. It is well known that HCV attains alternative strategies to efficiently replicate in hepatocytes, thus HCV can become less susceptible to anti-viral drugs, and patients who have been treated can relapse[24]. Indeed, the most interesting finding was the remarkable upregulation of lncRNA HEIH in relapsed HCV patients when compared to SOF + SIM-treated patients. Moreover, significantly higher expression of lncRNA GAS5 and lncRNA BISPR along with a significant reduction in mRNA BST2 expression were observed in relapsed HCV patients when compared to SOF + SIM-treated patients. lncRNA HEIH was the best prognostic marker to distinguish relapsed HCV patients from SOF + SIM-treated HCV patients. In fact, lncRNA HEIH has been suggested to be a potential biomarker in HCV-related HCC[16]. Our results elaborated a significant positive correlation between lncRNA HEIH and AFP and confirms that lncRNA HEIH could be a potential biomarker for the prognosis of/and the monitoring of the progression of relapsed HCV patients.
This study is limited by the relatively small number of patients and the fact that all the patients were enrolled from a single centre (Gastroenterology and Hepatology Clinic- Ain Shams Hospital in Egypt). Further multicenter studies with large number of participants infected with either HCV or other infectious agents unlike HCV should be conducted to justify the specificity of these markers. Moreover, a prospective longitudinal study is needed to follow the same group of patients before and after treatment to confirm the potential of these biomarkers as prognostic tools for HCV and occurrence of relapse after therapy.
In conclusion, differential expression of lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and mRNA BST2 in naïve, treated and relapsed HCV Egyptian patients suggests their involvement in HCV-pathogenesis or antiviral response. In addition, lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and mRNA BST2 could serve as potential diagnostic biomarkers in HCV GT4 Egyptian patients while, lncRNA GAS5, lncRNA BISPR and mRNA BST2 could also be considered novel prognostic biomarkers for treatment in HCV patients. Importantly, lncRNA HEIH might represent a powerful prognostic marker for differentiating relapsed patients from SOF + SIM treatment. Finally, these biomarkers can be used in combination to complete the whole picture of diagnosis, prognosis and follow-up of HCV.
Hepatitis C virus (HCV) infection is a complex multifactorial process that involves multiple viral and host interactions. Besides, the high mutation rate of HCV that enables the virus to generate escape mutants resistant to treatment, emphasizing the need for a proper understanding of the pathogenesis of HCV infection, and various research efforts should be oriented to develop novel diagnostic and prognostic molecular tools.
lncRNAs are transcripts greater than 200 nucleotides with poor coding potential, that play important roles in regulating gene expression. Emerging evidence suggests that lncRNAs play relevant roles in viral infection and in antiviral responses. To date, the expression profiles of lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and its mRNA BST2 in HCV GT4 patients and their clinical relevance as biomarkers for HCV infection have not been studied yet.
To assess the serum levels of lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and mRNA BST2 in naïve, treated and relapsed Egyptian HCV patients to examine their relation to HCV infection and their potential usefulness as new diagnostic and prognostic biomarkers for HCV GT4.
Serum lncRNAs and mRNABST2 were measured using quantitative real-time polymerase chain reaction. The study included six groups; group I healthy controls and group II naïve HCV patients without treatment. Groups from III to V comprised HCV patients treated daily with three different 12-wk oral treatment regimens as follows: Group III received combination of sofosbuvir and simeprevir. Group IV received combination of sofosbuvir and daclatasvir. Group V received sofosbuvir and daclatasvir with ribavirin. Group VI included HCV patients who relapsed after 12-week treatment with sofosbuvir and simeprevir.
We found that serum levels of lncRNAGAS5 and LncRNABISPR were upregulated, whereas mRNA BST2 and LncRNA HEIH levels were downregulated in naïve patients compared to healthy controls. In contrast, HCV patients treated with sofosbuvir and simeprevir; with sofosbuvir and daclatasvir; or with sofosbuvir, daclatasvir and ribavirin exhibited lower levels of lncRNAGAS5 and lncRNABISPR with higher mRNABST2 compared to naïve patients. Notably, patients relapsed from sofosbuvir and simeprevir showed higher levels of these lncRNAs with lower mRNABST2 compared to treated patients. Moreover, lncRNA GAS5, lncRNA BISPR and mRNA BST2 could differentiate naïve patients from controls and treated patients, whereas lncRNA HEIH best differentiated relapsed from treated patients.
Differential expression of lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and mRNA BST2 in naïve, treated and relapsed HCV Egyptian patients suggests their involvement in HCV-pathogenesis or antiviral response. In addition, lncRNA GAS5, lncRNA HEIH, lncRNA BISPR and mRNA BST2 could serve as potential diagnostic biomarkers in HCV GT4 Egyptian patients while, lncRNA GAS5, lncRNA BISPR and mRNA BST2 could also be considered novel prognostic biomarkers for treatment in HCV patients. Importantly, lncRNA HEIH might represent a powerful prognostic marker for differentiating relapsed patients from sofosbuvir and simeprevir treatment.
Further multicentre studies with large number of participants infected with either HCV or other infectious agents unlike HCV should be conducted to justify the specificity of these markers. Moreover, a prospective longitudinal study is needed to follow the same group of patients before and after treatment to confirm the potential of these biomarkers as prognostic tools for HCV and occurrence of relapse after therapy.
Manuscript source: Unsolicited Manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Egypt
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cui J S-Editor: Ma YJ L-Editor: A E-Editor: Zhang YL
| 1. | Jefferies M, Rauff B, Rashid H, Lam T, Rafiq S. Update on global epidemiology of viral hepatitis and preventive strategies. World J Clin Cases. 2018;6:589-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 171] [Cited by in RCA: 195] [Article Influence: 27.9] [Reference Citation Analysis (20)] |
| 2. | Kouyoumjian SP, Chemaitelly H, Abu-Raddad LJ. Characterizing hepatitis C virus epidemiology in Egypt: systematic reviews, meta-analyses, and meta-regressions. Sci Rep. 2018;8:1661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 3. | Ayoub HH, Abu-Raddad LJ. Impact of treatment on hepatitis C virus transmission and incidence in Egypt: A case for treatment as prevention. J Viral Hepat. 2017;24:486-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 4. | van der Meer AJ, Veldt BJ, Feld JJ, Wedemeyer H, Dufour JF, Lammert F, Duarte-Rojo A, Heathcote EJ, Manns MP, Kuske L, Zeuzem S, Hofmann WP, de Knegt RJ, Hansen BE, Janssen HL. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA. 2012;308:2584-2593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1165] [Cited by in RCA: 1167] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 5. | Lee MH, Yang HI, Lu SN, Jen CL, You SL, Wang LY, Wang CH, Chen WJ, Chen CJ; R. E.V.E.A.L.-HCV Study Group. Chronic hepatitis C virus infection increases mortality from hepatic and extrahepatic diseases: a community-based long-term prospective study. J Infect Dis. 2012;206:469-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 426] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 6. | Abdel-Moneim A, Abood A, Abdel-Gabaar M, Zanaty MI, Ramadan M. Effectiveness of sofosbuvir/pegylated-interferon plus ribavirin in treatment of hepatitis C virus genotype 4 patients. Clin Exp Hepatol. 2018;4:191-196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 7. | Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1788] [Cited by in RCA: 2147] [Article Influence: 214.7] [Reference Citation Analysis (0)] |
| 8. | Wu R, Su Y, Wu H, Dai Y, Zhao M, Lu Q. Characters, functions and clinical perspectives of long non-coding RNAs. Mol Genet Genomics. 2016;291:1013-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 9. | Fortes P, Morris KV. Long noncoding RNAs in viral infections. Virus Res. 2016;212:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | Barriocanal M, Fortes P. Long Non-coding RNAs in Hepatitis C Virus-Infected Cells. Front Microbiol. 2017;8:1833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y, Yang X, Shen J, Liu Q, Zhang J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691-2702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 12. | Littlejohn GO. Fibrositis/fibromyalgia syndrome in the workplace. Rheum Dis Clin North Am. 1989;15:45-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 131] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Qian X, Xu C, Zhao P, Qi Z. Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology. 2016;492:155-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Barriocanal M, Carnero E, Segura V, Fortes P. Long Non-Coding RNA BST2/BISPR is Induced by IFN and Regulates the Expression of the Antiviral Factor Tetherin. Front Immunol. 2014;5:655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 15. | Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 555] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 16. | Zhang C, Yang X, Qi Q, Gao Y, Wei Q, Han S. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018;21:651-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 17. | Erikson E, Adam T, Schmidt S, Lehmann-Koch J, Over B, Goffinet C, Harter C, Bekeredjian-Ding I, Sertel S, Lasitschka F, Keppler OT. In vivo expression profile of the antiviral restriction factor and tumor-targeting antigen CD317/BST-2/HM1.24/tetherin in humans. Proc Natl Acad Sci USA. 2011;108:13688-13693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 18. | Dafa-Berger A, Kuzmina A, Fassler M, Yitzhak-Asraf H, Shemer-Avni Y, Taube R. Modulation of hepatitis C virus release by the interferon-induced protein BST-2/tetherin. Virology. 2012;428:98-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | Dahari H, Canini L, Graw F, Uprichard SL, Araújo ES, Penaranda G, Coquet E, Chiche L, Riso A, Renou C, Bourliere M, Cotler SJ, Halfon P. HCV kinetic and modeling analyses indicate similar time to cure among sofosbuvir combination regimens with daclatasvir, simeprevir or ledipasvir. J Hepatol. 2016;64:1232-1239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 20. | Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, Mukaide M, Williams R, Lau JY. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201-207. [PubMed] |
| 21. | Wyles D, Mangia A, Cheng W, Shafran S, Schwabe C, Ouyang W, Hedskog C, McNally J, Brainard DM, Doehle BP, Svarovskaia E, Miller MD, Mo H, Dvory-Sobol H. Long-term persistence of HCV NS5A resistance-associated substitutions after treatment with the HCV NS5A inhibitor, ledipasvir, without sofosbuvir. Antivir Ther. 2018;23:229-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Yang ZF, Li J, Ma ZY, Yang XS, Dai B, An JZ, Li HM. Association between expression of LncRNA- HEIH in plasma and hepatocellular carcinoma risk: A case-control study. Xiandai Zhongliu Yixue. 2015;23:80–84. |
| 23. | Mahauad-Fernandez WD, Okeoma CM. The role of BST-2/Tetherin in host protection and disease manifestation. Immun Inflamm Dis. 2016;4:4-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 24. | Medrano J, Barreiro P, Resino S, Tuma P, Rodríguez V, Vispo E, Labarga P, Madejón A, García-Samaniego J, Jiménez-Nácher I, Martín-Carbonero L, Soriano V. Rate and timing of hepatitis C virus relapse after a successful course of pegylated interferon plus ribavirin in HIV-infected and HIV-uninfected patients. Clin Infect Dis. 2009;49:1397-1401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |