Published online May 7, 2020. doi: 10.3748/wjg.v26.i17.2049
Peer-review started: January 31, 2020
First decision: February 24, 2020
Revised: March 25, 2020
Accepted: April 4, 2020
Article in press: April 4, 2020
Published online: May 7, 2020
Processing time: 96 Days and 23.5 Hours
Genetic polymorphism is associated with irritable bowel syndrome (IBS) in terms of susceptibility and clinical manifestations. Previous studies have shown that genetic polymorphism might play a key role in the onset and progression of IBS by modulating components of its pathogenesis such as the gut-brain axis, gastrointestinal motility, inflammatory activity, and immune status. Although underlying pathophysiological mechanisms have not been fully clarified, the potential ethnic differences that are present in worldwide genetic studies of IBS deserve attention. This review surveyed numerous studies focusing on IBS-associated single nucleotide polymorphisms, and investigated the ethnic disparities revealed by them. The results demonstrate the need for more attention on ethnic factors in IBS-related genetic studies. Taking ethnic backgrounds into accounts and placing emphasis on disparities potentially ascribed to ethnicity could help lay a solid and generalized foundation for transcultural, multi-ethnic, or secondary analyses in IBS, for example, a meta-analysis. Broader genetic studies considering ethnic factors are greatly needed to obtain a better understanding of the pathophysiological mechanisms of IBS and to improve the prevention, intervention, and treatment of this disease.
Core tip: The present review focused on the phenomenon of ethnic discrepancies in irritable bowel syndrome-related genetic studies by gathering up-to-date original research and meta-analyses. We discuss the ethnic background and its potential impacts on the inconsistent results of studies, emphasizing the consideration of ethnicity in designing and analyzing irritable bowel syndrome-related genetic investigations, especially for multi-ethnic, transnational, and cross-cultural studies.
- Citation: Xiao QY, Fang XC, Li XQ, Fei GJ. Ethnic differences in genetic polymorphism associated with irritable bowel syndrome. World J Gastroenterol 2020; 26(17): 2049-2063
- URL: https://www.wjgnet.com/1007-9327/full/v26/i17/2049.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i17.2049
Irritable bowel syndrome (IBS) is a functional gastrointestinal (GI) disorder, characterized by recurrent abdominal pain and altered bowel habits[1]. Four clinical subtypes of IBS are indicated in the Rome IV criteria based on the predominant abnormal stool form, namely, constipation-predominant IBS (IBS-C), diarrhea-predominant IBS (IBS-D), IBS with mixed bowel habits (IBS-M), and IBS unclassified[2]. Given that the familial clustering of IBS has been reported[3-5], genetic inheritance is considered to be involved in the pathogenesis of IBS. A Swedish national adoption study showed that the odds ratio (OR) for IBS was 1.67 (95%CI: 1.06-2.62) in adoptees whose biological parents had been diagnosed with IBS, whereas it was only 0.88 (95%CI: 0.48-1.63) in adoptees whose adoptive parents had been diagnosed with IBS[6]. These data indicated the critical role of genetics in the development of IBS.
As functional disorders encountered worldwide, the global prevalence and the symptom of IBS varies from different regions and ethnic groups[7,8]. Attaching importance to different ethnic backgrounds in IBS patients has been encouraged in transnational and cross-cultural studies, as well as being highlighted by the Rome IV Committee in the form of a new chapter “Multicultural Aspects in Functional Gastrointestinal Disorders” included in the published collection of the textbook[9]. While ethnic variation in allele frequency and genotype distribution of polymorphisms has been confirmed genetically[10,11], their role in complex diseases remains controversial. Against the international backgrounds of IBS genetic studies, it is quite necessary to act circumspectly about ethnic factors and their potential impact on the discrepancy between clinical trials, especially in multi-ethnic or transnational multi-center studies.
To date, numerous IBS-associated studies on single nucleotide polymorphisms (SNPs) have been reported[12]. However, given the existence of inaccessible raw data, between-study heterogeneity, and Caucasian- dominated publication bias in IBS genetic studies, it is difficult to precisely figure out the ethnic impact on research results by systematic review with conventional meta-analysis. Instead, a literature review seems more suitable for readers to obtain an overview of the potential ethnic differences in genetic polymorphism associated with IBS. The development and progression of IBS include several different mechanisms such as gut-brain axis, GI dysmotility, inflammation, and immunological activation[13,14]. In this review, we analyzed ethnic differences in genetic polymorphisms associated with IBS from the perspective of pathophysiological mechanisms.
We searched articles addressing IBS-related genetic polymorphisms of different ethnic groups in following databases: MEDLINE (1946-Jan 2020), PubMed (1966–Jan 2020), EMBASE (1947–Jan 2020), Ovid (1950-Jan 2020) and Web of Science (1900–Jan 2020). Literature searches were performed by using “irritable bowel syndrome,” “genetic polymorphism,” “ethnicity,” “pathogenesis,” “single nucleotide polymorphism,” and “genome-wide association” as key phrases in various combinations. The search strategy was modified to suit each database. The reference lists of targeted articles were reviewed and manually searched to obtain additional related studies.
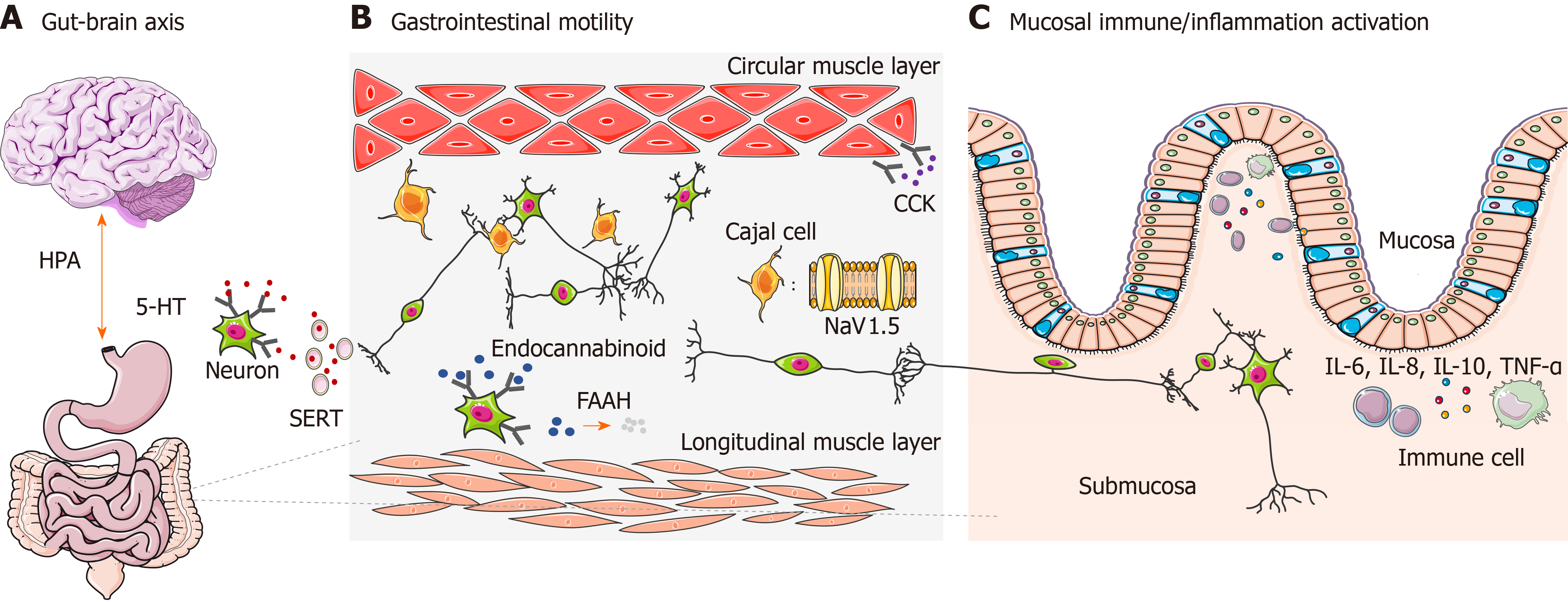
The gut-brain axis refers specifically to a complex reflex circuit that integrates the bidirectional communication between the cortex and the digestive system through afferent and efferent pathways[15]. Information from the GI tract is collected from receptors in the periphery and input to cortical areas. Then a response is generated downstream, further neuromodulating the actions of the enteric nervous system. A dysfunctional gut-brain axis reaction has been observed to contribute to abnormal GI motility and sensitivity, which is closely related to the pathogenesis of IBS[16]. The existence of ethnic differences of genetic polymorphisms associated with the gut-brain axis has already been observed[17] (Figure 1A), among which genetic polymorphism of the 5-hydroxytryptamine (5-HT) system and adrenergic system are the most well studied.
5-HT, also known as serotonin, is a paracrine messenger and neurotransmitter that has been particularly investigated in neuropsychiatric diseases. It is also an important mediator in the gut-brain connection. Notably, it has been estimated that about 95% of serotonin in the body is found in the GI tract instead of the brain and 90% of GI serotonin resides in enterochromaffin cells, while the rest is in enteric neurons[18]. As a crucial signaling molecule within the gut-brain axis, 5-HT stimulates enteric afferent nerve fibers of the vagal nerve and regulates the motility and sensitivity of the GI tract in a 5-HT receptor-dependent manner through submucosal and myenteric neurons that respond to serotonin via various receptors[19]. The genetic polymorphism associated with 5-HT metabolism, such as 5-HT receptors and the 5-HT reuptake transporter, has been studied in detail.
Serotonin receptors: There are at least seven subtypes of receptor (5-HT1-7) within the serotonin receptor superfamily, including 5-HT1–4 and 5-HT7 receptors, which are present in the human GI tract[20]. Results emerging from genetic studies have already shown that genetic polymorphism of 5-HT2, 5-HT3, and 5-HT4 receptors are related to IBS.
5-HT2A receptor, a type of phosphatidylinositol-linked receptor involved in sensitivity to pain, can bind to a G-protein-coupled receptor and directly transduce the signal by coupling to phospholipase C turnover in the cerebral cortex[21]. Rat experiments have confirmed that the 5-HT2A receptor is involved in serotonin-induced hyperalgesia, as intradermal injection of exogenous 5-HT2A receptor agonist (a-methyl 5-HT) into footpad produced a decrease of withdrawal latency to heat stimulation[22]. Significant associations were found between the -1438G/A and 102T/C polymorphisms of the 5-HT2A receptor gene and IBS. -1438G/A polymorphism located in the promoter region regulates the genetic transcription of 5-HT2A receptor. A case-control study among Turkish showed that the A/A genotype of the -1438G/A polymorphism and the C/C genotype of the 102T/C polymorphism conferred a high risk of IBS[23]. Furthermore, patients with T/T genotype of 102T/C polymorphism had much higher scores on visual analog scales reflecting abdominal pain. In Greece, Markoutsaki et al[24] also found that the A allele and the AA genotype of -1438G/A polymorphism were closely related to high risk of IBS. However, the Greek study showed no association between 102C/T polymorphism and IBS, and no polymorphism above was found to significantly correlate with the abdominal pain of IBS patients. Therefore, there might be ethnic disparities in genetic polymorphisms of 5-HT2A receptor.
5-HT3 receptor, a member of the superfamily of ligand-gated ion channels, localizes in numerous neurons of the myenteric and submucosal plexus in the GI tract[25]. It mediates the effect of 5-HT on the parasympathetic ganglia, which results in smooth muscle contraction and increased intestinal secretion[26]. Clinical studies have demonstrated that alosetron, a 5-HT3 receptor antagonist, increases the compliance to colorectal distension[27] and effectively alleviates abdominal pain threshold in IBS patients[28,29]. Ondansetron, another 5-HT3 receptor antagonist, markedly increases fasting small bowel water content by either promoting small bowel secretion or reducing small bowel motility[26]. Together, these studies suggest that 5-HT3 receptor might play a role in the development of IBS by altering the visceral sensitivity and GI motility. Through a series of polymerase chain reaction restriction fragment length polymorphism analyses, Gu et al[30] found that SNP of c.−42C>T in the 5-HT3A receptor gene was associated with a higher risk of IBS-D in Asian women; however, no significant difference was shown in the association between T carrier status and IBS among American cohorts in previous studies[31]. The SNP of c.*76G>A in the 5-HT3E receptor gene, another member of the 5-HT3 receptor family, was proven to have a highly significant association with female IBS-D in two independent Caucasian cohorts from United Kingdom and Germany[32]. Moreover, c.*76G>A was shown to modulate the binding of miR-510 to untranslated regions (UTRs) of the 5-HT3E receptor gene in enterocytes and led to elevated expression of the target gene. This study was the first reported example of a cis-regulatory variant affecting microRNA (miRNA)-related expression in the regulation of serotonin receptor gene. The regulation revoked by the SNP may explain the connection between c.*76G>A and higher risk of IBS. Later, Zhang et al[33] found that the variant of c.*76G>A in the 5-HT3E receptor gene was also significantly associated with IBS-D in Chinese women. Immunohistochemistry in their study suggested that the SNP reduced overexpression of the target gene in human colonic mucosal tissues, which underlines the participation of miRNA-related regulation in IBS as well. Thus, unlike the 5-HT3A receptor, there was no direct evidence supporting the ethnic differences in the polymorphism of 5-HT3E receptor. Meta-analysis reconfirmed that the c.*76G>A was significantly associated with the risk of IBS-D in both Asian and non-Asian populations, while the c.-42C>T was only correlated with that in Asians[34].
5-HT4 receptor, which evokes the 5-HT release of mucosa, degranulation of goblet cell, and Cl− secretion when stimulated, is broadly expressed in the small and large intestines[35]. The role of the 5-HT4 receptor in initiating the peristaltic reflex through myenteric plexus in the human small intestine has already been identified[36]. Wohlfarth et al[37] assessed the genetic heterogeneity of isoform-specific UTRs in British patients (all were of Caucasian origin) and found a relevant SNP of c.*61T>C (rs201253747) residing in the 3′UTR near the stop codon of the gene. This variant was only detected in jejunal biopsy samples from IBS-D patients and proved to increase the risk of IBS-D. To confirm their findings, researchers proceeded to genotype 5 additional cohorts from 4 countries (the United States, Germany, Belgium, and Sweden), and performed a pooled analysis. Strikingly, there was a higher frequency of the variant c.*61T>C in IBS-D patients than in healthy controls and all non-IBS-D patients (P = 0.049, OR = 2.74). It has been further implied that the variant residing in a putative miRNA-binding site in the 3′UTR affects the binding process between the miR-16/miR-103 and 5-HT4 receptor. The disturbance created by the variant putatively impairs the expression of 5-HT4 receptors, which involves in the IBS susceptibility, especially for IBS-D. Caucasian participants from different countries or regions were inclined to present a similar connection between the SNP of c.*61T>C and IBS-D, which might emphasize the importance of ethnic factors in the polymorphism variances rather than geographical discrepancy. Nevertheless, to the best of our knowledge, no related results from other ethnic groups have been reported, and ethnic differences regarding the polymorphisms of 5-HT4 receptor gene warrant further exploration.
Serotonin reuptake transporter: As a transmembrane transport protein, the serotonin reuptake transporter (SERT) modulates the duration and intensity of cumulative impacts from serotonergic neurotransmission by mediating the reuptake of serotonin into the presynaptic neurons[38]. The SERT gene (SLC6A4) locates in chromosome 17q12, and its promoter region contains a genetic polymorphism designated as the “5-HT transporter length polymorphic region” (5HTTLPR), which includes a 44-base-pair deletion or insertion, generating a short (S) or long (L) allele. The short variant of the polymorphism reduces the transcriptional efficiency of the SLC6A4 promoter, further decreases the expression level of the target gene and inhibits serotonin reuptake[39]. Recently, subgroup analyses based on ethnicity in a large-scale meta-analysis proved that the 5HTTLPR insertion/deletion polymorphism was closely related to IBS susceptibility in both Asians and Caucasians[40]. However, a great decrease in the heterogeneity was observed in the Asian subgroup compared to the whole groups, which suggested that there might be an ethnic difference even within the positive correlations between 5HTTLPR polymorphism and IBS risk. Because of limited raw data, the relationship between the 5HTTLPR and the predominant clinical feature of IBS was not statistically analyzed. Previous researches reported that the S/S genotype was predominant in IBS-D patients from Chinese, Indian and Korean[40-42]; furthermore, the serotonin level was higher in rectal biopsy specimens from Indian patients with IBS-D. In contrast, higher frequencies of S/S and L/S genotypes were shared by IBS-C patients from Iran significantly, rather than IBS-D, and even the serotonin levels were proved to be similar in each subtype of IBS[43], which further proved the involvement of ethnic factors related to 5HTTLPR polymorphism in IBS.
The adrenergic receptors, also known as the adrenoceptors, are members of the G-protein-coupled receptor superfamily. The GI tract receives signals from the noradrenaline neuron system via α2-adrenoceptors, which plays an essential role in altering GI mobility and algesthesis. There are three subtypes of α2-adrenoceptors, designated as α2A-, α2B-, and α2C-adrenergic receptors. An earlier study suggested that adrenoceptor polymorphism might be involved in visceral hypersensitivity, as participants with the variant del322–325 in the α2C-adrenergic receptor gene presented a higher pain score in response to the cold presser test than controls[44]. In Caucasians, the polymorphisms of both del322–325 in the α2C-adrenoceptor gene and -1291C>G in α2A-adrenoceptors gene were proven to be associated with IBS-C[45]. Nonetheless, no significant association was identified between the variant -1291C>G and IBS in Turkish cohorts[46]. Moreover, case-control studies conducted in India and South Korea found that the polymorphism of -1291C>G in α2A-adrenergic receptor gene was closely related to IBS-D rather than IBS-C[47,48]. This discrepancy suggested that the development of IBS is possibly directed, at least in part, by unique ethnic variation in the adrenergic system.
Additionally, polymorphisms of the G-protein β3 subunit gene (GNB3), which influences the activity of G-protein-coupled adrenergic receptors, have been extensively investigated concerning its relevance to IBS. Studies performed in South Korea showed that the genotype T/T of 825C/T polymorphism was associated with IBS-C in both children and adults[49,50]. However, other studies in Caucasian populations concluded that there was no significant interaction between the polymorphism of 825C/T and any subtype of IBS[51,52]. Later, in a meta-analysis, Pan et al[53] also failed to show any association between the 825C/T polymorphism and IBS. Nevertheless, after a few years, an updated meta-analysis with a larger sample of researches presents that the SNP of 825C/T was only significantly associated with IBS-C in the Asian population[54], which further suggested the possibility of ethnic differences in the connection of GNB3 polymorphisms and IBS. In fact, scientists have already obtained evidence that ethnic variation exists in the T-allele frequency of 825C/T polymorphism. The prevalence of the T allele is lowest in Caucasian populations (30.1%-31.9%), while it is higher in Asians (42.3%-47.7%), and the highest in Africans (82.3%-84.1%)[55,56]. The diverse distribution of allele frequency may partly contribute to the inconsistency in polymorphism studies on different ethnic groups. Interestingly, statistics have shown a fairly high frequency of the T allele in African populations. Because no relevant studies have been published connecting the 825C/T polymorphism with IBS in African populations, further investigations are warranted.
GI dysmotility is one of the major features leading to IBS. It is not only influenced by spontaneous activities of smooth muscle cells, but is also regulated in multiple ways by neurohumoral and immune systems. Some genetic polymorphisms found in IBS genetic studies are related to GI motility, such as in genes encoding voltage-gated sodium channel NaV1.5, cannabinoid receptor, fatty acid amide hydrolase (FAAH), and cholecystokinin receptor (Figure 1B). Their effects on IBS might vary among different ethnic groups.
The generation of functional neuromuscular movement in the GI tract requires a coordinated operation between the enteric nervous system (ENS), intestinal smooth muscle cells, and interstitial cells of Cajal (ICC)[57]. ICCs are regarded as pacemakers of smooth muscle contractions due to their central role in generating cyclic depolarizations termed slow waves. Depolarizations are transmitted to intestinal smooth muscle cells as electrical stimuli, resulting in GI contraction. The NaV1.5 channel (encoded by SCN5A) is expressed in GI smooth muscle cells and ICCs of the small intestine and colon[58,59]. It has been extensively investigated among cardiac diseases, and its polymorphism of SCN5A is strongly associated with cardiac arrhythmias[60]. An analogous mechanism has also been observed in GI diseases, especially in GI motility disorders. Braak et al[61] found that Caucasians with “gain of function” mutation of SCN5A exhibited a higher risk for IBS. Further genome-wide association (GWA) study in cohorts from the United States, Sweden, Italy and Greece by Beyder et al[62] proved that the SCN5A locus was significantly associated with IBS. It also provided evidence that loss-of-function mutation of SCN5A disrupted NaV1.5 channel function, which might be considered as a potential pathogenic and therapeutic target in IBS. Later, a study on ethnically diverse cohorts including Hispanics, African-Americans, Asians, American-Indians, Pacific Islander, and mixed-racial groups also revealed a significant link between SCN5A variants and IBS, even in mixed-racial groups[63]. Moreover, the frequency of SCN5A mutations in those ethnically diverse groups corresponded to the Caucasian-dominated cohort[62,64], which could partially explain the coordination of genetic polymorphisms of SCN5A and their effects on IBS among different ethnic groups. As above, there was no evidence showing ethnic variance in the genetic polymorphism of SCN5A.
Cannabinoids are natural substances that exert bioactivity in the form of analgesic effects; they can be extracted and synthesized from cannabis plants. Cannabinoids significantly influence GI motility and visceral sensation. A new disease named cannabinoid hyperemesis was added to Rome IV classification and criteria, highlighting the role of cannabinoids in functional gastrointestinal diseases (FGIDs). Endocannabinoids were first discovered by Raphael’s group in 1992, and the composition of the endocannabinoid system (ECS) was then investigated widely, comprising anandamide, 2-arachidonoylglycerol, cannabinoid receptor, and its synthetic and hydrolytic enzymes[65]. The ECS exerts significant anti-propulsive effects in FGIDs, which probably attributes to inhibition of the components in peristaltic reflex[66].
Type 1 cannabinoid receptors (CB1) are abundant in submucosal and myenteric neurons of ENS, accounting for various activities of cannabinoid ligands when activated by cannabis. The length of gene encoding CB1 (CNR1) is affected by a polymorphic (AAT)n triplet. Park et al[67]. first divided the allelic distributions into two groups, shorter alleles (< 10) and longer alleles (> 10). They compared the (AAT)n triplet of the CNR1 gene between IBS patients and healthy controls in the South Koreans, and found the significant correlation between the > 10/> 10 genotype and higher IBS risk. The result was further confirmed by Jiang et al[68] in the Chinese population, which also suggested that the longer the alleles of (AAT)n, the greater the risk of developing IBS. Nevertheless, the connection has not been replicated in predominantly Caucasian IBS patients. Camilleri et al[69] did not find a positive association between the (AAT)n genotype and the symptom of IBS. Meanwhile, an ethnic variation in the allele frequency of (AAT)n triplet repeats was also revealed in contrast to Asian studies, which possibly indicated that the interaction between the polymorphic (AAT)n triplet and IBS may vary among different ethnic groups.
Anandamide, another essential part of the ECS, is inactivated by FAAH in vivo. According to a case-control study conducted by Camilleri et al[70], Caucasians carrying the CA/AA genotype of the +385C/A polymorphism in the FAAH gene were at a higher risk of IBS-D and IBS-M as well as more accelerated colonic transit in IBS-D patients than healthy controls. However, Jiang et al[68] found no significant correlation between the +385 C/A polymorphic region and IBS in Chinese populations, which also suggested the possible existence of ethnic differences in FAAH-encoding gene polymorphism.
Cholecystokinin (CCK) is a type of neuropeptide secreted by endocrine I cells within the duodenum and jejunal mucosa. CCK receptors are widely distributed in both the central nervous system and GI tract. CCK mediates pancreatic enzyme secretion, gallbladder contraction, motor and sensory functions of the GI tract, such as gastric emptying and colonic motility, via two distinct receptors: CCK-1 and CCK-2. Cremonini et al[71] reported that Caucasian IBS-C patients with 779T>C polymorphism of CCK-1 receptor-encoding gene had a significantly lower gastric emptying rate than those with other genotypes. Consistent with the Caucasian study, a case-control study in South Korea found that the variant of 779T>C genotype was more commonly identified in IBS patients, and was significantly associated with IBS-C and IBS-M patients (non-IBS-D)[72], which also referred to a relatively lower level of GI motility.
However, given the relatively small accumulative sample size of studies above, especially in non-Caucasian groups, further investigations with larger scale as well as diverse ethnic backgrounds are greatly needed to shed more lights on the ethnic impact in IBS-associated genetic polymorphism.
Cytokines are essential regulators of immune functions and inflammatory responses. In IBS, abnormal immune regulations secondary to imbalanced cytokine expression have been proven to accelerate the development of IBS. Genetic polymorphisms in cytokines are closely related to intestinal immune activation and inflammatory status[73,74]. IBS-related gene polymorphisms have been identified in interleukin (IL), tumor necrosis factor (TNF), and TNF superfamily (TNFSF) members (Figure 1C).
IL-6, a cytokine secreted by T helper 2 cells, mediates T-cell activation and lymphocytic differentiation by acting as a pro-inflammatory mediator. Previous studies showed that the -174C/G (rs1800795) polymorphic region of the gene encoding IL-6 might be associated with IBS. In Iranians, Barkhordari et al[75] found that the frequency of G/G homozygotes was significantly increased in IBS patients, while the frequency of C/G heterozygotes was considerably decreased, which indicated the role of -174C/G polymorphism in the pathophysiology of IBS. Santhosh et al[76] further verified the genetic association of the -174C/G (rs1800795) polymorphism and IBS in Indians. However, in Caucasian populations, a meta-analysis of case-control studies from the United Kingdom, United States, Belgium, and Sweden showed no association between -174C/G polymorphism and IBS[77].
IL-8, another cytokine secreted by T helper 1 cells, elicits the chemotaxis of inflammatory factors and induces cell proliferation. The Mexican study by Olivo-Diaz et al[78] suggested that +396 G/G and +781C/T genotypes in IL-8 were significantly overrepresented in IBS patients compared with controls. Furthermore, the genotype +396G/G was associated with a higher risk of developing IBS in patients with blastocystosis. Romero-Valdovinos et al[79] confirmed that the proportion of IBS patients carrying the +396 G allele was significantly higher than for other genotypes in Mexicans. However, in a Caucasian meta-analysis, no significant correlation between +781C/T polymorphism and IBS was shown[77]. Although Mexico is geographically adjacent to the United States, with a mixed Indo-European ethnic composition, its ethnic characteristics are remarkably different from those of most Caucasian-dominated countries.
IL-10, a protective cytokine secreted by macrophages and regulatory T cells, inhibits the synthesis of pro-inflammatory mediators and TNF. Patients with IBS had lower serum IL-10 levels and its genetic polymorphism also played a role in the immune response and inflammatory regulation[80]. A meta-analysis demonstrated that carriers of the G allele in the -1082A/G polymorphism of IL-10 had a lower risk of IBS in Caucasian populations, but not in Asians. Carriers with the C allele of the -592A/C polymorphism were particularly susceptible to IBS in Asians, but not in Caucasians[81]. It was also discussed in the meta-analysis that the frequency of A alleles in -1082A/G polymorphism led to greater upregulation of IL-10 expression in Caucasians than in Asians, which further highlighted the various effects of differences in allelic frequency between different ethnic groups.
TNF mainly refers to two closely related cytokines: TNF-α and TNF-β, which are widely involved in immune and inflammatory responses. A significant increase of TNF-α expression was reported in IBS patients[82], and the possession of the A allele (A/A or G/A) of the -308G/A polymorphism in the gene encoding TNF-α was proven to be responsible for increased TNF-α production[83]. Moreover, an increased frequency of the G/A genotype for the -308G/A polymorphism in IBS patients was revealed in a Netherlands study[84], and the association between -308G/A polymorphism and IBS was further confirmed in British patients[85]. However, when characterizing the same polymorphism in a genetically homogeneous South Korean population, Lee et al[50]. found no significant difference between the IBS patients and their healthy counterparts, again indicating the possible existence of ethnic factors in IBS-related genetic polymorphisms.
Members of TNFSF, another essential group of cytokines involved in IBS, share a common TNF homeodomain, which mediates biological effects of TNFSF by connecting TNF ligands to the cysteine-rich domain in corresponding receptors. Based on studies using independent case-control cohorts in Sweden and the United States, the allele rs4263839 G in TNFSF15 has been regarded as a gene conferring susceptibility to IBS in Caucasian populations, especially in IBS-C patients[86]. In the United Kingdom, the other three SNPs of TNFSF15 (rs6478108, rs6478109, and rs7848647) also showed a significant reduction in minor allele frequency of IBS-D patients compared to healthy controls[85], which reconfirms the significance of TNFSF15 polymorphisms in IBS. There are no other studies reported the connection between TNFSF15 polymorphism and IBS in non-Caucasian populations thus far.
Therefore, IBS-associated genetic polymorphisms in IL-6, IL-8, IL-10 may have diverse effects in different ethnic groups. Results from those studies need to be further replicated under the consideration of ethnic factors. Whether ethnicity influences the role of TNFSF genetic polymorphism in IBS remains to be solved.
Besides, considering the mucosal immune functions and inflammatory changes in the pathogenesis of IBS, the gut microbiota has received extensive interests[87,88]. The microbial composition is linked to mucosal lymphocyte phenotypes in post-infectious IBS[89]; intestinal antibacterial gene expression relates to bacterial profiles and immune activity in IBS patients[90]; both the altered profile of the intestinal microbiota and faecal microbial metabolites were evinced to be correlated with the symptom severity of IBS[91,92]. As to the microbiota-related genetic polymorphism, mutations in the mitochondrial genome lead to divergent gut microbial compositions in mice[93]. But since there is no evidence of a particular genetic polymorphism closely related to gut microbial communities in IBS patients yet, the potential ethnic impact of gut microbiota will not be discussed in detail here and it could be explored in the future.
It is worth mentioning that the development of GWA study, which makes the utmost of bio-information technology, has been adding fresh energies to IBS genetic studies in recent years. GWA studies with whole-genome sequencing method in a high-throughput way brought its unique advantages. Ek et al[94] launched a real sense of GWA study within larger-scale IBS cohorts, and included the genotypical and phenotypical information from 14837 individuals in total. They conducted the pilot study in a population sample of 11326 Swedish twins, then replicated in six case-control cohorts from Sweden, Belgium, Italy, Germany, Greece and the United States, and finally delivered a consistent result in 2015 that a locus on chromosome 7p22.1, including genes of KDELR2 (KDEL endoplasmic reticulum protein retention receptor 2) and GRID2IP (glutamate receptor, ionotropic, delta 2 [Grid2] interacting protein) were related to a higher risk of IBS. Meanwhile, an increase of mucosal KDLER2 mRNA expression in IBS patients was also revealed while comparing to healthy controls. Their study demonstrated completely new risk loci in IBS, although the potential roles of those candidate genes remains to be studied, it still represented an incredible start of GWA study in candidate gene hunting for IBS.
Later, Bonfiglio et al[95] resumed their own meta-analysis of five independent IBS GWA studies (Swedish twins mentioned-above and four additional Northern European genotyped cohorts), and reported in 2018 that up to 64 gene candidates associated with IBS risk, and their gene set enrichment analysis further indicated that the function of those risk loci were more closely related to regulations of ion channel activity. Moreover, they included some previously reported candidate genes in their meta-analysis, which replicated that the polymorphism of the genes KDELR2/GRID2IP, SCN5A, TRPM8 (encode transient receptor potential melastatin-8), SI (encoded sucrase-isomaltase), and NPSR1 (encoded neuropeptide S receptor 1) were related to IBS risk. Regretfully, those notable GWA studies were all from Northern European-predominant IBS cohorts, and the ethnic composition of each cohort was not even mentioned, thus could not provide us with clues leading to the ethnic variations. But their studies indeed remind us that large-scale GWA analyses could offer a plausible and effective research approach to identify allele frequencies and evaluate potential ethnic variations of IBS gene polymorphisms between different cohorts with diverse ethnic backgrounds in the future.
Taking together all above findings, the phenomenon of ethnic variations has been recurrently noticed in studies on IBS-related genetic polymorphism (Table 1), which requires more concerns. Factors such as different sample size and research quality could affect the reliability of between-study comparisons to some extent. But after reviewing objectively numerous original researches and up-to-date meta analyses with high-level of evidence, we would like to suggest that the ethnic differences in IBS-associated genetic polymorphisms are more likely to be a worldwide phenomenon needed explaining than the meaningless exception.
| Genetic model | IBS-risk genotype | IBS subtype | Country | Ethnic or demographic group | Research type | Ethnic characteristics | Ref. |
| 5-HT2A receptor | A homozygote in −1438G/A and C homozygote in +102T/C | IBS | Turkey and Greece | Western Asian/Caucasian | Case-control | Consistent in −1438G/A; inconsistent in +102T/C | [23,24] |
| 5-HT3A receptor | c.−42C>T | IBS-D | United States and China | Caucasian/Asian | Case-control; Meta-analysis | Inconsistent | [30,31,34] |
| 5-HT3E receptor | c.*76G>A in female | IBS-D | United Kingdom, Germany, China | Caucasian/Asian | Case-control; Meta-analysis | Consistent | [32-34] |
| 5-HT4 receptor | c.*61 T>C | IBS-D | United States, Germany, Belgium, Sweden | Only Caucasian | Case-control | NA | [37] |
| 5HTTLPR | S/S in short (S) or long (L) allele | IBS-C IBS-D IBS-M | United States, Greece, Italy, Germany, Turkey, China, Japan, South Korea, India, and Iran | NA | Case-control; Meta-analysis | Inconsistent | [40-43] |
| α2A-adrenergic receptor | −1291C/G | IBS-C | United States, Turkey, India, South Korea | Caucasian/Asian | Case-control | Inconsistent | [45-48] |
| GNB3 | T homozygote in 825C/T | IBS-C | United States, Greece, Netherlands, South Korea, China | Caucasian/Asian | Case-control; Meta-analysis | Inconsistent | [49-54] |
| SCN5A | Missense mutations1 | IBS-C IBS-D | United States, Sweden, Italy, Greece | Hispanic/Caucasian/Asian/American Indian/African American | GWA study; Case-control | Consistent | [61-63] |
| CNR1 | (AAT)n > 10 homozygote | IBS | South Korea, China, United States | Asian/Caucasian | Case-control | Inconsistent | [67-69] |
| FAAH | allele carriers in +385C/A | IBS | United States and China | Caucasian/Asian | Case-control | Inconsistent | [68,70] |
| CCK-1 receptor | 779T>C | IBS-C | United States and South Korea | Caucasian/Asian | Case-control | Consistent | [71,72] |
| IL-6 | rs1800795 (−174C/G) | IBS | Iran, India, United Kingdom, United States, Belgium, Sweden | Asian/Caucasian | Case-control; Meta-analysis | Inconsistent | [75-77] |
| IL-8 | rs2227306 (+781C/T) | IBS | Mexico, United Kingdom, United States | Indo-European/ Caucasian | Case-control; Meta-analysis | Inconsistent | [77-79] |
| IL-10 | G allele carriers in −1082A/G; C allele carriers in −592A/C | IBS | United States, United Kingdom, Netherlands, Mexico, Iran, China, South Korea, India | Caucasian/Asian | Meta-analysis | Inconsistent | [81] |
| TNF-α | GA heterozygote in −308G/A | IBS | Netherlands, United Kingdom, South Korea | Caucasian/Asian | Case-control | Inconsistent | [50,82-85] |
| TNFSF | rs4263839 G | IBS-C | United States and Sweden | Only Caucasian | Case-control | NA | [86] |
| TNFSF | rs6478108, rs6478109, rs7848647 | IBS-D | United Kingdom | Only Caucasian | Case-control | NA | [85] |
Actually, non-replication of genetic association results is not rare in case-control studies. Inappropriate population stratification, especially between cohorts with different allele frequencies, which are more common in groups with different ethnic backgrounds, is probably the most often cited reason for the inconsistency of genetic studies[96]. As such, we strongly recommend that ethnic factors should be taken into consideration in large-scale genetic studies as inclusion criteria for the enrollment or the stratification in subgroup analysis. Comparisons of allele frequency between different ethnic groups are also required in multi-ethnic studies, in order to assess whether subgroup analyses based on ethnicity are needed or not.
Limitations and challenges remained in the investigation of potential ethnic differences among genetic studies. First, in contrast to the sex or the age of subjects, few studies have looked at the issue of ethnicity in demographic characteristics of cohorts. Instead of ethnic backgrounds, geographic locations, hospitals, and the institution of cohorts are more likely to be described. Limited original researches with the explicitly-addressed ethnic composition of each individual have conferred huge challenges and limitations for retrospective analysis, especially a meta-analysis or systematic review. Second, no unified standard of ethnic/racial classification has been established for genetic researches. Generally, ethnic groups based on physical similarities are distinguished into Caucasians, Mongoloids, Negroids, and Australoids, while the most of European and Anglo-American studies simply divided ethnic groups into Caucasians, African-Americans, and Asians. Third, due to ancestral patterns of geographical migrations, colonial expansion and the globalization in modern society, individuals with mixed-race also widely exist, making the situation more complex. Without an universal consensus, countries and even geographical regions were used for classifying subgroups in some studies. Furthermore, the specific mechanism in terms of how those genetic polymorphisms influence clinical manifestations and therapeutic responses in IBS patients has not been fully illuminated nowadays. It might interfere the subsequent investigation into the essence of ethnic impacts on IBS-related genetic polymorphisms.
Hence, adequate identification and widely acknowledged classification of the ethnic group are urgently needed to be proposed and verified in further studies, which supposed to be a prerequisite for building a cross-cultural network and conducting better multi-ethnic studies of IBS-related genetic polymorphisms in the future.
In conclusion, genetic polymorphisms with effects on the gut-brain axis, visceral sensitivity, GI motility, and immune and inflammatory responses, have been proven to be correlated with the onset and progression of IBS. IBS-related genetic polymorphisms may potentially vary and have diverse effects in different ethnic groups (Table 1). The different distributions of genotypes and alleles might play a decisive role in disparities among various ethnic groups of IBS cohorts. In-depth discussion and evaluation of ethnicity are thus urgently required to better investigate the genetic polymorphisms associated with IBS worldwide. Further multi-ethnic, transnational and cross-cultural research in this field with larger sample sizes is strongly recommended, which could provide a solid and generalized foundation for clarifying the pathogenesis, identifying candidate genes, and profiling the genetic risk of IBS, as well as further assisting target therapies in precision medicine.
Manuscript source: Unsolicited Manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fagoonee S, Ng QX, Triantafillidis J S-Editor: Wang YQ L-Editor: Filipodia E-Editor: Liu MY
| 1. | Mearin F, Lacy BE, Chang L, Chey WD, Lembo AJ, Simren M, Spiller R. Bowel Disorders. Gastroenterology. 2016;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1781] [Cited by in RCA: 1897] [Article Influence: 210.8] [Reference Citation Analysis (3)] |
| 2. | Lacy BE, Patel NK. Rome Criteria and a Diagnostic Approach to Irritable Bowel Syndrome. J Clin Med. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 320] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 3. | Locke GR 3rd, Zinsmeister AR, Talley NJ, Fett SL, Melton LJ 3rd. Familial association in adults with functional gastrointestinal disorders. Mayo Clin Proc. 2000;75:907-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 180] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 4. | Kalantar JS, Locke GR, Talley NJ, Zinsmeister AR, Fett SL, Melton LJ. Is irritable bowel syndrome more likely to be persistent in those with relatives who suffer from gastrointestinal symptoms? A population-based study at three time points. Aliment Pharmacol Ther. 2003;17:1389-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 5. | Makker J, Chilimuri S, Bella JN. Genetic epidemiology of irritable bowel syndrome. World J Gastroenterol. 2015;21:11353-11361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 6. | Waehrens R, Zöller B, Sundquist J, Sundquist K, Pirouzifard M. A Swedish national adoption study of risk of irritable bowel syndrome (IBS). BMJ Open Gastroenterol. 2017;4:e000156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Sperber AD, Dumitrascu D, Fukudo S, Gerson C, Ghoshal UC, Gwee KA, Hungin APS, Kang JY, Minhu C, Schmulson M, Bolotin A, Friger M, Freud T, Whitehead W. The global prevalence of IBS in adults remains elusive due to the heterogeneity of studies: a Rome Foundation working team literature review. Gut. 2017;66:1075-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 356] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 8. | Gerson CD, Gerson MJ, Awad RA, Chowdhury A, Dancey C, Poitras P, Porcelli P, Sperber A, Wang WA. Irritable bowel syndrome: an international study of symptoms in eight countries. Eur J Gastroenterol Hepatol. 2008;20:659-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Fang X, Francisconi CF, Fukudo S, Gerson MJ, Kang JY, Schmulson W MJ, Sperber AD. Multicultural Aspects in Functional Gastrointestinal Disorders (FGIDs). Gastroenterology. 2016;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Ioannidis JP, Ntzani EE, Trikalinos TA. 'Racial' differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 325] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 11. | Mori M, Yamada R, Kobayashi K, Kawaida R, Yamamoto K. Ethnic differences in allele frequency of autoimmune-disease-associated SNPs. J Hum Genet. 2005;50:264-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 179] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 12. | Cheung CK, Wu JC. Genetic polymorphism in pathogenesis of irritable bowel syndrome. World J Gastroenterol. 2014;20:17693-17698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (1)] |
| 13. | Mayer EA, Labus JS, Tillisch K, Cole SW, Baldi P. Towards a systems view of IBS. Nat Rev Gastroenterol Hepatol. 2015;12:592-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 192] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 14. | Ng QX, Soh AYS, Loke W, Lim DY, Yeo WS. The role of inflammation in irritable bowel syndrome (IBS). J Inflamm Res. 2018;11:345-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 216] [Article Influence: 30.9] [Reference Citation Analysis (2)] |
| 15. | Gaman A, Kuo B. Neuromodulatory processes of the brain-gut axis. Neuromodulation. 2008;11:249-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | Aziz Q, Thompson DG. Brain-gut axis in health and disease. Gastroenterology. 1998;114:559-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 204] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 17. | Chang L, Di Lorenzo C, Farrugia G, Hamilton FA, Mawe GM, Pasricha PJ, Wiley JW. Functional Bowel Disorders: A Roadmap to Guide the Next Generation of Research. Gastroenterology. 2018;154:723-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Kim DY, Camilleri M. Serotonin: a mediator of the brain-gut connection. Am J Gastroenterol. 2000;95:2698-2709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 303] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 19. | González-Arancibia C, Escobar-Luna J, Barrera-Bugueño C, Díaz-Zepeda C, González-Toro MP, Olavarría-Ramírez L, Zanelli-Massai F, Gotteland M, Bravo JA, Julio-Pieper M. What goes around comes around: novel pharmacological targets in the gut-brain axis. Therap Adv Gastroenterol. 2016;9:339-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 20. | Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 988] [Cited by in RCA: 1131] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 21. | Conn PJ, Sanders-Bush E, Hoffman BJ, Hartig PR. A unique serotonin receptor in choroid plexus is linked to phosphatidylinositol turnover. Proc Natl Acad Sci USA. 1986;83:4086-4088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 155] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 22. | Tokunaga A, Saika M, Senba E. 5-HT2A receptor subtype is involved in the thermal hyperalgesic mechanism of serotonin in the periphery. Pain. 1998;76:349-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 85] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Pata C, Erdal E, Yazc K, Camdeviren H, Ozkaya M, Ulu O. Association of the -1438 G/A and 102 T/C polymorphism of the 5-Ht2A receptor gene with irritable bowel syndrome 5-Ht2A gene polymorphism in irritable bowel syndrome. J Clin Gastroenterol. 2004;38:561-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 40] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Markoutsaki T, Karantanos T, Gazouli M, Anagnou NP, Karamanolis DG. 5-HT2A receptor gene polymorphisms and irritable bowel syndrome. J Clin Gastroenterol. 2011;45:514-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Glatzle J, Sternini C, Robin C, Zittel TT, Wong H, Reeve JR, Raybould HE. Expression of 5-HT3 receptors in the rat gastrointestinal tract. Gastroenterology. 2002;123:217-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 130] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 26. | Marciani L, Wright J, Foley S, Hoad CL, Totman JJ, Bush D, Hartley C, Armstrong A, Manby P, Blackshaw E, Perkins AC, Gowland PA, Spiller RC. Effects of a 5-HT(3) antagonist, ondansetron, on fasting and postprandial small bowel water content assessed by magnetic resonance imaging. Aliment Pharmacol Ther. 2010;32:655-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Delvaux M, Louvel D, Mamet JP, Campos-Oriola R, Frexinos J. Effect of alosetron on responses to colonic distension in patients with irritable bowel syndrome. Aliment Pharmacol Ther. 1998;12:849-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 185] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Zheng Y, Yu T, Tang Y, Xiong W, Shen X, Jiang L, Lin L. Efficacy and safety of 5-hydroxytryptamine 3 receptor antagonists in irritable bowel syndrome: A systematic review and meta-analysis of randomized controlled trials. PLoS One. 2017;12:e0172846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 29. | Lacy BE, Nicandro JP, Chuang E, Earnest DL. Alosetron use in clinical practice: significant improvement in irritable bowel syndrome symptoms evaluated using the US Food and Drug Administration composite endpoint. Therap Adv Gastroenterol. 2018;11:1756284818771674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 30. | Gu QY, Zhang J, Feng YC, Dai GR, Du WP. Association of genetic polymorphisms in HTR3A and HTR3E with diarrhea predominant irritable bowel syndrome. Int J Clin Exp Med. 2015;8:4581-4585. [PubMed] |
| 31. | Kilpatrick LA, Labus JS, Coveleskie K, Hammer C, Rappold G, Tillisch K, Bueller JA, Suyenobu B, Jarcho JM, McRoberts JA, Niesler B, Mayer EA. The HTR3A polymorphism c. -42C>T is associated with amygdala responsiveness in patients with irritable bowel syndrome. Gastroenterology. 2011;140:1943-1951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Kapeller J, Houghton LA, Mönnikes H, Walstab J, Möller D, Bönisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F, Gassler N, Fischer C, Whorwell PJ, Atkinson W, Fell C, Büchner KJ, Schmidtmann M, van der Voort I, Wisser AS, Berg T, Rappold G, Niesler B. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 33. | Zhang Y, Li Y, Hao Z, Li X, Bo P, Gong W. Association of the Serotonin Receptor 3E Gene as a Functional Variant in the MicroRNA-510 Target Site with Diarrhea Predominant Irritable Bowel Syndrome in Chinese Women. J Neurogastroenterol Motil. 2016;22:272-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 34. | Guan T, Li T, Cai W, Huang D, Ouyang P, Wang Y, Chen H, Wu K, Ma X. HTR3A and HTR3E gene polymorphisms and diarrhea predominant irritable bowel syndrome risk: evidence from a meta-analysis. Oncotarget. 2017;8:100459-100468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Hoffman JM, Tyler K, MacEachern SJ, Balemba OB, Johnson AC, Brooks EM, Zhao H, Swain GM, Moses PL, Galligan JJ, Sharkey KA, Greenwood-Van Meerveld B, Mawe GM. Activation of colonic mucosal 5-HT(4) receptors accelerates propulsive motility and inhibits visceral hypersensitivity. Gastroenterology. 2012;142:844-854.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 219] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 36. | Grider JR, Foxx-Orenstein AE, Jin JG. 5-Hydroxytryptamine4 receptor agonists initiate the peristaltic reflex in human, rat, and guinea pig intestine. Gastroenterology. 1998;115:370-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 268] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 37. | Wohlfarth C, Schmitteckert S, Härtle JD, Houghton LA, Dweep H, Fortea M, Assadi G, Braun A, Mederer T, Pöhner S, Becker PP, Fischer C, Granzow M, Mönnikes H, Mayer EA, Sayuk G, Boeckxstaens G, Wouters MM, Simrén M, Lindberg G, Ohlsson B, Schmidt PT, Dlugosz A, Agreus L, Andreasson A, D'Amato M, Burwinkel B, Bermejo JL, Röth R, Lasitschka F, Vicario M, Metzger M, Santos J, Rappold GA, Martinez C, Niesler B. miR-16 and miR-103 impact 5-HT4 receptor signalling and correlate with symptom profile in irritable bowel syndrome. Sci Rep. 2017;7:14680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 38. | Bjerregaard H, Severinsen K, Said S, Wiborg O, Sinning S. A dualistic conformational response to substrate binding in the human serotonin transporter reveals a high affinity state for serotonin. J Biol Chem. 2015;290:7747-7755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 39. | Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Müller CR, Hamer DH, Murphy DL. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527-1531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3630] [Cited by in RCA: 3446] [Article Influence: 118.8] [Reference Citation Analysis (0)] |
| 40. | Zhu Y, Zheng G, Hu Z. Association between SERT insertion/deletion polymorphism and the risk of irritable bowel syndrome: A meta-analysis based on 7039 subjects. Gene. 2018;679:133-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 41. | Yuan J, Kang C, Wang M, Wang Q, Li P, Liu H, Hou Y, Su P, Yang F, Wei Y, Yang J. Association study of serotonin transporter SLC6A4 gene with Chinese Han irritable bowel syndrome. PLoS One. 2014;9:e84414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 42. | Park JM, Choi MG, Park JA, Oh JH, Cho YK, Lee IS, Kim SW, Choi KY, Chung IS. Serotonin transporter gene polymorphism and irritable bowel syndrome. Neurogastroenterol Motil. 2006;18:995-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 43. | Mohammadi M, Tahmasebi Abdar H, Mollaei HR, Hajghani H, Baneshi MR, Hayatbakhsh MM. Serotonin Transporter Gene (SLC6A4) Polymorphism and Mucosal Serotonin Levels in Southeastern Iranian Patients with Irritable Bowel Syndrome. Middle East J Dig Dis. 2017;9:26-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 44. | Kohli U, Muszkat M, Sofowora GG, Harris PA, Friedman EA, Dupont WD, Scheinin M, Wood AJ, Stein CM, Kurnik D. Effects of variation in the human alpha2A- and alpha2C-adrenoceptor genes on cognitive tasks and pain perception. Eur J Pain. 2010;14:154-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 45. | Kim HJ, Camilleri M, Carlson PJ, Cremonini F, Ferber I, Stephens D, McKinzie S, Zinsmeister AR, Urrutia R. Association of distinct alpha(2) adrenoceptor and serotonin transporter polymorphisms with constipation and somatic symptoms in functional gastrointestinal disorders. Gut. 2004;53:829-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 147] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 46. | Uğur Kantar F, Simşek İ, Ercal D, Ülgenalp A, Bora E. Alpha-2-adrenergic receptor gene polymorphism in Turkish population with irritable bowel syndrome. Turk J Gastroenterol. 2013;24:483-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 47. | Sikander A, Rana SV, Sharma SK, Sinha SK, Arora SK, Prasad KK, Singh K. Association of alpha 2A adrenergic receptor gene (ADRAlpha2A) polymorphism with irritable bowel syndrome, microscopic and ulcerative colitis. Clin Chim Acta. 2010;411:59-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 48. | Choi YJ, Hwang SW, Kim N, Park JH, Oh JC, Lee DH. Association Between SLC6A4 Serotonin Transporter Gene Lainked Polymorphic Region and ADRA2A -1291C>G and Irritable Bowel Syndrome in Korea. J Neurogastroenterol Motil. 2014;20:388-399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 49. | Park CS, Uhm JH. Polymorphisms of the Serotonin Transporter Gene and G-Protein β3 Subunit Gene in Korean Children with Irritable Bowel Syndrome and Functional Dyspepsia. Gut Liver. 2012;6:223-228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 50. | Lee HJ, Lee SY, Choi JE, Kim JH, Sung IK, Park HS, Jin CJ. G protein beta3 subunit, interleukin-10, and tumor necrosis factor-alpha gene polymorphisms in Koreans with irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:758-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Andresen V, Camilleri M, Kim HJ, Stephens DA, Carlson PJ, Talley NJ, Saito YA, Urrutia R, Zinsmeister AR. Is there an association between GNbeta3-C825T genotype and lower functional gastrointestinal disorders? Gastroenterology. 2006;130:1985-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 52. | Saito YA, Larson JJ, Atkinson EJ, Ryu E, Almazar AE, Petersen GM, Talley NJ. The role of 5-HTT LPR and GNβ3 825C>T polymorphisms and gene-environment interactions in irritable bowel syndrome (IBS). Dig Dis Sci. 2012;57:2650-2657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 53. | Pan ZG, Xiao C, Su DX. No association of G-protein beta polypeptide 3 polymorphism with irritable bowel syndrome: evidence from a meta-analysis. World J Gastroenterol. 2014;20:6345-6352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 54. | Jiang D, Huang D, Cai W, Li T, Wang Y, Chen H, Guan T, Ma X. G protein beta 3(GNβ3) C825T polymorphism and irritable bowel syndrome susceptibility: an updated meta-analysis based on eleven case-control studies. Oncotarget. 2018;9:2770-2781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 55. | Siffert W, Forster P, Jöckel KH, Mvere DA, Brinkmann B, Naber C, Crookes R, Du P Heyns A, Epplen JT, Fridey J, Freedman BI, Müller N, Stolke D, Sharma AM, Al Moutaery K, Grosse-Wilde H, Buerbaum B, Ehrlich T, Ahmad HR, Horsthemke B, Du Toit ED, Tiilikainen A, Ge J, Wang Y, Rosskopf D. Worldwide ethnic distribution of the G protein beta3 subunit 825T allele and its association with obesity in Caucasian, Chinese, and Black African individuals. J Am Soc Nephrol. 1999;10:1921-1930. [PubMed] |
| 56. | Rosskopf D, Manthey I, Siffert W. Identification and ethnic distribution of major haplotypes in the gene GNB3 encoding the G-protein beta3 subunit. Pharmacogenetics. 2002;12:209-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 57. | Beyder A, Farrugia G. Targeting ion channels for the treatment of gastrointestinal motility disorders. Therap Adv Gastroenterol. 2012;5:5-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 58. | Ou Y, Gibbons SJ, Miller SM, Strege PR, Rich A, Distad MA, Ackerman MJ, Rae JL, Szurszewski JH, Farrugia G. SCN5A is expressed in human jejunal circular smooth muscle cells. Neurogastroenterol Motil. 2002;14:477-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 59. | Strege PR, Ou Y, Sha L, Rich A, Gibbons SJ, Szurszewski JH, Sarr MG, Farrugia G. Sodium current in human intestinal interstitial cells of Cajal. Am J Physiol Gastrointest Liver Physiol. 2003;285:G1111-G1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 114] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 60. | Yagihara N, Watanabe H, Barnett P, Duboscq-Bidot L, Thomas AC, Yang P, Ohno S, Hasegawa K, Kuwano R, Chatel S, Redon R, Schott JJ, Probst V, Koopmann TT, Bezzina CR, Wilde AA, Nakano Y, Aiba T, Miyamoto Y, Kamakura S, Darbar D, Donahue BS, Shigemizu D, Tanaka T, Tsunoda T, Suda M, Sato A, Minamino T, Endo N, Shimizu W, Horie M, Roden DM, Makita N. Variants in the SCN5A Promoter Associated With Various Arrhythmia Phenotypes. J Am Heart Assoc. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Braak B, Klooker TK, Scholvinck D, Hofman N, Wilde A, Boeckxstaens GE. Abdominal symptoms in patients with long Qt syndrome and a “gain of function” mutation in the Nav1.5 sodium channel. Gastroenterology. 2008;134:A-683. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 62. | Beyder A, Mazzone A, Strege PR, Tester DJ, Saito YA, Bernard CE, Enders FT, Ek WE, Schmidt PT, Dlugosz A, Lindberg G, Karling P, Ohlsson B, Gazouli M, Nardone G, Cuomo R, Usai-Satta P, Galeazzi F, Neri M, Portincasa P, Bellini M, Barbara G, Camilleri M, Locke GR, Talley NJ, D'Amato M, Ackerman MJ, Farrugia G. Loss-of-function of the voltage-gated sodium channel NaV1.5 (channelopathies) in patients with irritable bowel syndrome. Gastroenterology. 2014;146:1659-1668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 63. | Strege PR, Mazzone A, Bernard CE, Neshatian L, Gibbons SJ, Saito YA, Tester DJ, Calvert ML, Mayer EA, Chang L, Ackerman MJ, Beyder A, Farrugia G. Irritable bowel syndrome patients have SCN5A channelopathies that lead to decreased NaV1.5 current and mechanosensitivity. Am J Physiol Gastrointest Liver Physiol. 2018;314:G494-G503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 64. | Saito YA, Strege PR, Tester DJ, Locke GR, Talley NJ, Bernard CE, Rae JL, Makielski JC, Ackerman MJ, Farrugia G. Sodium channel mutation in irritable bowel syndrome: evidence for an ion channelopathy. Am J Physiol Gastrointest Liver Physiol. 2009;296:G211-G218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 65. | Sharkey KA, Wiley JW. The Role of the Endocannabinoid System in the Brain-Gut Axis. Gastroenterology. 2016;151:252-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 178] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 66. | Pesce M, D'Alessandro A, Borrelli O, Gigli S, Seguella L, Cuomo R, Esposito G, Sarnelli G. Endocannabinoid-related compounds in gastrointestinal diseases. J Cell Mol Med. 2018;22:706-715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 67. | Park JM, Choi MG, Cho YK, Lee IS, Kim SW, Choi KY, Chung IS. Cannabinoid receptor 1 gene polymorphism and irritable bowel syndrome in the Korean population: a hypothesis-generating study. J Clin Gastroenterol. 2011;45:45-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 68. | Jiang Y, Nie Y, Li Y, Zhang L. Association of cannabinoid type 1 receptor and fatty acid amide hydrolase genetic polymorphisms in Chinese patients with irritable bowel syndrome. J Gastroenterol Hepatol. 2014;29:1186-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 69. | Camilleri M, Kolar GJ, Vazquez-Roque MI, Carlson P, Burton DD, Zinsmeister AR. Cannabinoid receptor 1 gene and irritable bowel syndrome: phenotype and quantitative traits. Am J Physiol Gastrointest Liver Physiol. 2013;304:G553-G560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 70. | Camilleri M, Carlson P, McKinzie S, Grudell A, Busciglio I, Burton D, Baxter K, Ryks M, Zinsmeister AR. Genetic variation in endocannabinoid metabolism, gastrointestinal motility, and sensation. Am J Physiol Gastrointest Liver Physiol. 2008;294:G13-G19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 66] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 71. | Cremonini F, Camilleri M, McKinzie S, Carlson P, Camilleri CE, Burton D, Thomforde G, Urrutia R, Zinsmeister AR. Effect of CCK-1 antagonist, dexloxiglumide, in female patients with irritable bowel syndrome: a pharmacodynamic and pharmacogenomic study. Am J Gastroenterol. 2005;100:652-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 72. | Park SY, Rew JS, Lee SM, Ki HS, Lee KR, Cheo JH, Kim HI, Noh DY, Joo YE, Kim HS, Choi SK. Association of CCK(1) Receptor Gene Polymorphisms and Irritable Bowel Syndrome in Korean. J Neurogastroenterol Motil. 2010;16:71-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 73. | Chen J, Zhang Y, Deng Z. Imbalanced shift of cytokine expression between T helper 1 and T helper 2 (Th1/Th2) in intestinal mucosa of patients with post-infectious irritable bowel syndrome. BMC Gastroenterol. 2012;12:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 74. | Bashashati M, Rezaei N, Shafieyoun A, McKernan DP, Chang L, Öhman L, Quigley EM, Schmulson M, Sharkey KA, Simrén M. Cytokine imbalance in irritable bowel syndrome: a systematic review and meta-analysis. Neurogastroenterol Motil. 2014;26:1036-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 75. | Barkhordari E, Rezaei N, Ansaripour B, Larki P, Alighardashi M, Ahmadi-Ashtiani HR, Mahmoudi M, Keramati MR, Habibollahi P, Bashashati M, Ebrahimi-Daryani N, Amirzargar AA. Proinflammatory cytokine gene polymorphisms in irritable bowel syndrome. J Clin Immunol. 2010;30:74-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 76. | Santhosh S, Dutta AK, Samuel P, Joseph AJ, Ashok Kumar J, Kurian G. Cytokine gene polymorphisms in irritable bowel syndrome in Indian population--a pilot case control study. Trop Gastroenterol. 2010;31:30-33. [PubMed] |
| 77. | Czogalla B, Schmitteckert S, Houghton LA, Sayuk GS, Camilleri M, Olivo-Diaz A, Spiller R, Wouters MM, Boeckxstaens G, Bermejo JL, Niesler B. A meta-analysis of immunogenetic Case-Control Association Studies in irritable bowel syndrome. Neurogastroenterol Motil. 2015;27:717-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 78. | Olivo-Diaz A, Romero-Valdovinos M, Gudiño-Ramirez A, Reyes-Gordillo J, Jimenez-Gonzalez DE, Ramirez-Miranda ME, Martinez-Flores WA, Martinez-Hernandez F, Flisser A, Maravilla P. Findings related to IL-8 and IL-10 gene polymorphisms in a Mexican patient population with irritable bowel syndrome infected with Blastocystis. Parasitol Res. 2012;111:487-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 79. | Romero-Valdovinos M, Gudiño-Ramírez A, Reyes-Gordillo J, Martínez-Flores WA, Ramírez-Miranda ME, Maravilla P, Olivo-Díaz A. Interleukin-8 and -10 gene polymorphisms in irritable bowel syndrome. Mol Biol Rep. 2012;39:8837-8843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 80. | Schmulson M, Pulido-London D, Rodriguez O, Morales-Rochlin N, Martinez-García R, Gutierrez-Ruiz MC, López-Alvarenga JC, Robles-Díaz G, Gutiérrez-Reyes G. Lower serum IL-10 is an independent predictor of IBS among volunteers in Mexico. Am J Gastroenterol. 2012;107:747-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 81. | Qin SY, Jiang HX, Lu DH, Zhou Y. Association of interleukin-10 polymorphisms with risk of irritable bowel syndrome: a meta-analysis. World J Gastroenterol. 2013;19:9472-9480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 82. | Liebregts T, Adam B, Bredack C, Röth A, Heinzel S, Lester S, Downie-Doyle S, Smith E, Drew P, Talley NJ, Holtmann G. Immune activation in patients with irritable bowel syndrome. Gastroenterology. 2007;132:913-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 452] [Cited by in RCA: 490] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 83. | Poli F, Boschiero L, Giannoni F, Tonini M, Scalamogna M, Ancona G, Sirchia G. Tumour necrosis factor-alpha gene polymorphism: implications in kidney transplantation. Cytokine. 2000;12:1778-1783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 84. | van der Veek PP, van den Berg M, de Kroon YE, Verspaget HW, Masclee AA. Role of tumor necrosis factor-alpha and interleukin-10 gene polymorphisms in irritable bowel syndrome. Am J Gastroenterol. 2005;100:2510-2516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 146] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 85. | Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, Cox J, Kelly FM, Wilde J, Lennon MG, Neal KR, Whorwell PJ, Hall IP, Spiller RC. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFα. Gut. 2013;62:985-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 86. | Zucchelli M, Camilleri M, Andreasson AN, Bresso F, Dlugosz A, Halfvarson J, Törkvist L, Schmidt PT, Karling P, Ohlsson B, Duerr RH, Simren M, Lindberg G, Agreus L, Carlson P, Zinsmeister AR, D'Amato M. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut. 2011;60:1671-1677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 87. | Adriani A, Ribaldone DG, Astegiano M, Durazzo M, Saracco GM, Pellicano R. Irritable bowel syndrome: the clinical approach. Panminerva Med. 2018;60:213-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 88. | Sundin J, Aziz I, Nordlander S, Polster A, Hu YOO, Hugerth LW, Pennhag AAL, Engstrand L, Törnblom H, Simrén M, Öhman L. Evidence of altered mucosa-associated and fecal microbiota composition in patients with Irritable Bowel Syndrome. Sci Rep. 2020;10:593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 89. | Sundin J, Rangel I, Fuentes S, Heikamp-de Jong I, Hultgren-Hörnquist E, de Vos WM, Brummer RJ. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther. 2015;41:342-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 90. | Bennet SMP, Sundin J, Magnusson MK, Strid H, Tap J, Derrien M, Le Nevé B, Doré J, Törnblom H, Simrén M, Öhman L. Altered intestinal antibacterial gene expression response profile in irritable bowel syndrome is linked to bacterial composition and immune activation. Neurogastroenterol Motil. 2018;30:e13468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 91. | Zhang WX, Zhang Y, Qin G, Li KM, Wei W, Li SY, Yao SK. Altered profiles of fecal metabolites correlate with visceral hypersensitivity and may contribute to symptom severity of diarrhea-predominant irritable bowel syndrome. World J Gastroenterol. 2019;25:6416-6429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 92. | Tap J, Derrien M, Törnblom H, Brazeilles R, Cools-Portier S, Doré J, Störsrud S, Le Nevé B, Öhman L, Simrén M. Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology. 2017;152:111-123.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 444] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 93. | Hirose M, Künstner A, Schilf P, Sünderhauf A, Rupp J, Jöhren O, Schwaninger M, Sina C, Baines JF, Ibrahim SM. Mitochondrial gene polymorphism is associated with gut microbial communities in mice. Sci Rep. 2017;7:15293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 94. | Ek WE, Reznichenko A, Ripke S, Niesler B, Zucchelli M, Rivera NV, Schmidt PT, Pedersen NL, Magnusson P, Talley NJ, Holliday EG, Houghton L, Gazouli M, Karamanolis G, Rappold G, Burwinkel B, Surowy H, Rafter J, Assadi G, Li L, Papadaki E, Gambaccini D, Marchi S, Colucci R, Blandizzi C, Barbaro R, Karling P, Walter S, Ohlsson B, Tornblom H, Bresso F, Andreasson A, Dlugosz A, Simren M, Agreus L, Lindberg G, Boeckxstaens G, Bellini M, Stanghellini V, Barbara G, Daly MJ, Camilleri M, Wouters MM, D'Amato M. Exploring the genetics of irritable bowel syndrome: a GWA study in the general population and replication in multinational case-control cohorts. Gut. 2015;64:1774-1782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 95. | Bonfiglio F, Henström M, Nag A, Hadizadeh F, Zheng T, Cenit MC, Tigchelaar E, Williams F, Reznichenko A, Ek WE, Rivera NV, Homuth G, Aghdassi AA, Kacprowski T, Männikkö M, Karhunen V, Bujanda L, Rafter J, Wijmenga C, Ronkainen J, Hysi P, Zhernakova A, D'Amato M. A GWAS meta-analysis from 5 population-based cohorts implicates ion channel genes in the pathogenesis of irritable bowel syndrome. Neurogastroenterol Motil. 2018;30:e13358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 96. | Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 856] [Cited by in RCA: 836] [Article Influence: 38.0] [Reference Citation Analysis (0)] |