Published online Apr 21, 2020. doi: 10.3748/wjg.v26.i15.1745
Peer-review started: December 16, 2019
First decision: January 19, 2020
Revised: February 7, 2020
Accepted: March 19, 2020
Article in press: March 19, 2020
Published online: April 21, 2020
Processing time: 127 Days and 2.6 Hours
The incidence and mortality rates of pancreatic carcinoma (PC) are rapidly increasing worldwide. Long noncoding RNAs (lncRNAs) play critical roles during PC initiation and progression. Since the lncRNA DNAH17-AS1 is highly expressed in PC, the regulation of DNAH17-AS1 in PC was investigated in this study.
To investigate the expression and molecular action of lncRNA DNAH17-AS1 in PC cells.
The PC expression data for the lncRNA DNAH17-AS1 was downloaded from The Cancer Genome Atlas database and used to examine its profile. Western blot and reverse transcription-quantitative PCR were employed to assess protein and mRNA expression. A subcellular fractionation assay was used to determine the location of DNAH17-AS1 in cells. In addition, the regulatory effects of DNAH17-AS1 on miR-432-5p, PPME1, and tumor activity were investigated using luciferase reporter assay, MTT viability analysis, flow cytometry, and transwell migration analysis.
DNAH17-AS1 was upregulated in PC cells and was associated with aggressive tumor behavior and poor prognosis for patients. Silencing DNAH17-AS1 promoted the apoptosis and reduced the viability, invasion, and migration of PC cells. In addition, DNAH17-AS1 served as a PC oncogene by downregulating miR-432-5p which normally directly targeted PPME1 to downregulate its expression.
DNAH17-AS1 functions in PC as a tumor promoter by regulating the miR-432-5p/PPME1 axis. This finding may provide new insights for PC prognosis and therapy.
Core tip: DNAH17-AS1 is upregulated in pancreatic carcinoma (PC) and is associated with aggressive clinical presentation in PC patients. DNAH17-AS1 promotes PC cell viability, invasion, and migration, and its silencing promotes PC cell apoptosis. DNAH17-AS1 is found primarily in the cytoplasm and participates in the competing endogenous RNA regulatory network. DNAH17-AS1 functions as a tumor promoter in PC by downregulating miR-432-5p and upregulating PPME1.
- Citation: Xu T, Lei T, Li SQ, Mai EH, Ding FH, Niu B. DNAH17-AS1 promotes pancreatic carcinoma by increasing PPME1 expression via inhibition of miR-432-5p. World J Gastroenterol 2020; 26(15): 1745-1757
- URL: https://www.wjgnet.com/1007-9327/full/v26/i15/1745.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i15.1745
Pancreatic carcinoma (PC) is a common digestive system malignancy with serious negative impacts on health, and the seventh leading cause of cancer-related death around the world[1]. Only 15%-20% patients present with resectable disease while the vast majority of patients have either locally advanced or metastatic disease unsuitable for surgical resection[2]. Nonspecific early symptoms, limited methods for detection and diagnosis, and the limited response of PC to chemotherapy and radiotherapy are the main reasons for the serious consequences of PC[3,4]: Overall five-year survival is about 8.3%[5]. It is thus critically important to explore new molecular markers for PC diagnosis and treatment monitoring.
With the increasing use of nuclei acid sequencing technology to understand the multifaceted human genome, more and more new classes of functional molecules have been found. Among these, long noncoding RNAs (lncRNAs) represent a novel and promising class of regulators for tuning tumorigenesis in several human cancers. For example, Chang et al[6] recently reported that the lncRNA NEF restrains non-small-cell lung cancer cell proliferation, while Ma et al[7] found that the lncRNA SPRY4-IT1 promotes proliferation and metastasis in hepatocellular carcinoma via TNF signaling. In addition, some lncRNAs have already been implicated in the development of PC. For example, the lncRNAs MACC1-AS1 and ABHD11-AS1 accelerate the proliferation and metastasis of PC through PAX8/NOTCH1 signaling[8] and PI3K-AKT[9], respectively. In contrast, LINC01111[10] and LINC01197[11] suppress PC.
Numerous studies have revealed that lncRNAs exert their influence in cancer cells by acting as sponges for microRNAs (miRNAs), thereby indirectly regulating the mRNA expression by sequestering miRNAs. MiRNAs are short non-coding RNAs that act as post-transcriptional regulators. They are often involved in tumorigenesis, as they bind target mRNAs and thus reduce the stability and translation of their target genes[12]. LncRNAs and miRNAs and their overall competing endogenous RNA network are increasingly implicated in cancer. For example, the lncRNA CACNA1G-AS1 facilitates hepatocellular carcinoma progression via the miR-2392/C1orf61 pathway[13]. sONE-lncRNA is downregulated in triple-negative breast cancer cells but can act as a tumor suppressor by repressing eNOS-induced NO production, affecting TP53 and c-Myc proteins levels and finally altering the levels of a panel of tumor suppressor miRNAs downstream[14]. There are also similar findings in PC, such as a study reporting that the lncRNA 00976 sponges miR-137 and affects the OTUD7B/EGFR/MAPK pathway to promote PC[15], as well as another demonstrating that lncRNA DLX6-AS1 modulates the miR-497-5p/FZD4/FZD6 axis and Wnt/β-catenin pathway to increase tumorigenesis in PC[16].
The lncRNA DNAH17-AS1 was recently investigated for its potential oncogenic role in colorectal cancer. DNAH17-AS1 was upregulated in colorectal cancer[10]. It has also been reported that lncRNA DLX6-AS1 can promote the progression of liver cancer by targeting miR-424-5p[17]. Interestingly, miR-424-5p also exerts a tumor-suppressive effect in PC[18], but the specific functions of DNAH17-AS1 in PC remain elusive. The interactions between lncRNA DNAH17-AS1 and miRNAs have not yet been investigated thoroughly in PC. Thus, the aim of this work was to investigate the expression of the lncRNA DNAH17-AS1 and its impact on the viability, metastasis, and apoptosis of PC cells, and to unravel its exact mechanistic role in PC by exploring its impact on regulatory proteins and their downstream miRNAs.
Seventy-eight pairs of PC tissue and adjacent normal tissue were obtained from patients with PC at Luoyang Central Hospital Affiliated to Zhengzhou University. Written informed consent was obtained from all patients. The patients did not receive radiotherapy or chemotherapy prior to surgery. Permission for and ethical approval of this research were acquired from the Institutional Ethics Committee of Luoyang Central Hospital affiliated to Zhengzhou University.
Normal human pancreatic duct epithelial cells (HPDE6-C7) and PC cells (Hs766T and SW1990) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States). Cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS) and kept at 37 °C with 5% CO2.
Small interfering RNA (siRNA) oligonucleotides targeting human DNAH17-AS1 (si-DNAH17-AS1), negative control siRNA (si-NC), miR-432-5p mimic, miR-432-5p inhibitor, negative control miR (NC-mimic), the pcDNA3.1 vector targeting PPME1, and an empty vector were constructed by RiboBio (Guangzhou, China). Hs766T PC cells were transfected with the siRNAs (50 nmol), mimics (20 nmol), or inhibitors (20 nmol) using Lipofectamine 2000 (Invitrogen/Thermo Fisher Scientifc). The transfection efficiency was assessed by reverse transcription-quantitative PCR (RT-qPCR).
Total RNA was isolated using Trizol reagent (Invitrogen, United States). cDNA was synthesized using the PrimeScript RT reagent kit (Takara, Dalian, China). RT-qPCR was performed with real-time PCR mixture assays (Takara) according to the manufacturer’s instructions. Expression levels of DNAH17-AS1, miR-432-5p, PPME1, CCNH, and SNRPD2 were normalized to GAPDH and quantified using the 2−△△Cq method.
Transfected Hs766T PC cells (2000 cells/well) were incubated for 24, 48, 72, or 96 h in a 96-well plate. Then, 20 μL of MTT solution (5 mg/mL) was added to each well and then replaced with 150 μL DMSO solution after 4 h of incubation. The absorbance of each suspension at 490 nm was measured using a microplate reader.
Transfected Hs766T PC cells were added into 6-well plates. Artificial wounds for a live cell analysis were made on the cell monolayer using culture inserts. Migratory cells and wound healing were monitored at 0 and 24 h. Three artificial wounds were immediately photographed for each group at the indicated time points following the wound formation. Cell migration was assessed by measuring the difference in the wound area between groups.
Cell invasion was detected in the upper chamber with 60 μL of diluted Matrigel. After 30 min, a Hs766T cell suspension (2 × 103 cells/well) was added to the upper chamber of the transwell plate. RPMI-1640 medium (500 μL, 10% FBS) was then added to the lower chamber. After 24 h, 0.1% crystal violet was used to stain the cells that had invaded the lower chamber. Observations were performed with a light microscope.
Flow cytometry was used to detect apoptosis in PC. First, transfected Hs766T cells (3 × 103 cell/well) were seeded in 6-well plates. After 48 h, trypsin (EDTA-free) digestion was used to collect the cells. We then suspended the collected Hs766T cells in PBS at 4 °C. After resuspension in binding buffer, the Annexin V-FITC probe and propidium iodide (Biovision, K101) were added and the cells were incubated at room temperature for 5 min in the dark, followed by detection using a flow cytometer (BD Biosciences).
Reporter plasmids of DNAH17-AS1 (wt-DNAH17-AS1 and mut-DNAH17-AS1) and PPME1 (wt-PPME1 and mut-PPME1) were purchased from GenePharma (Shanghai, China). The reporter plasmids were transfected into Hs766T cells with the miR-432-5p mimic (RiboBio, Guangzhou, China) for 48 h. Luciferase activity was examined with a dual-luciferase reporter assay system (Promega, United States).
RNA was isolated from the nuclear or cytoplasmic fraction using the Nuclear/Cytosol Fractionation Kit (Biovision, San Francisco Bay, CA, United States) and was measured by qRT-PCR. U1 and GAPDH acted as the identifiers for the nuclear or cytoplasmic fractions, respectively. Nuclear and Cytoplasmic Extraction Reagents were purchased from Thermo Fisher Scientific.
RIPA lysis buffer was used to extract protein from the samples. Next, we used 10% SDS-PAGE to separate proteins. Protein was then transferred to PVDF membranes. After blocking with 5% skim milk for 2 h, the membranes were incubated with primary antibody at 4 °C overnight, using rabbit monoclonal anti-PPME1 (1/2000, ab205956, Abcam, Cambridge, United States), anti-CCNH (1/2000, ab92376, Abcam), anti- SNRPD2 (1/2000, ab251254, Abcam), or rabbit polyclonal anti-GAPDH antibody (1:500, Abcam). The membranes were then washed to remove the primary antibody and incubated with IgG H and L goat anti-rabbit secondary antibody (HRP, 1:1000; Abcam) at 37 °C for 1 h. Finally, ECL (ECL, Pierce) was used to visualize and measure protein expression levels.
Data are expressed as the mean ± SD and were analyzed using SPSS 17.0 or Graphpad Prism 6. The statistical methods employed included Student’s t-test, one-way ANOVA, univariate Kaplan-Meier method with log-rank test, and χ2 test. Differences were considered significant if P < 0.05.
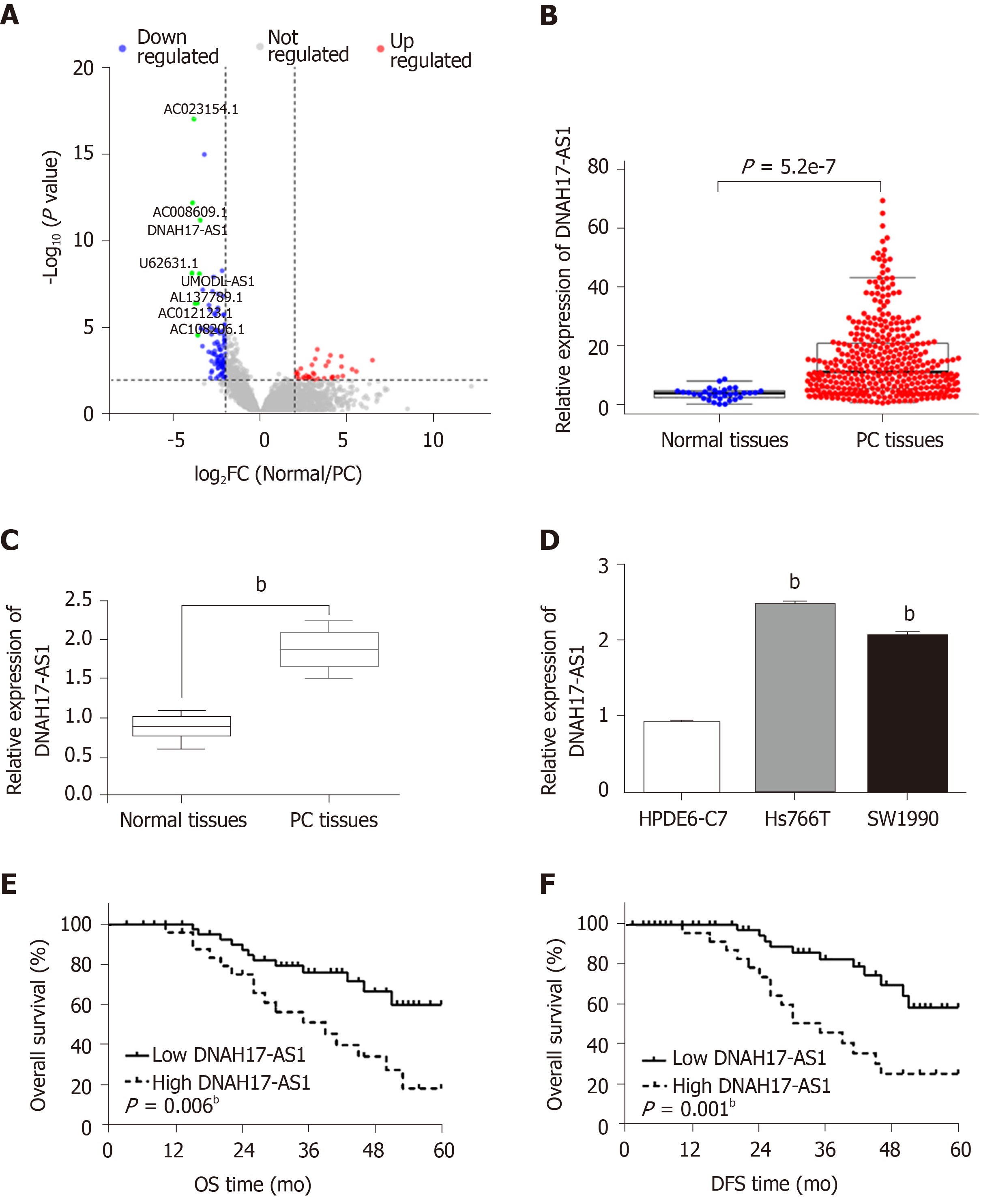
DNAH17-AS1 is an lncRNA that is presumed to be overexpressed in PC patients and tissues, due to its inclusion in the top ten abundant lncRNAs as noted in The Cancer Genome Atlas dataset (Figure 1A and 1B). Indeed, RT-qPCR analysis revealed upregulation of DNAH17-AS1 in PC tissues compared with normal tissues (Figure 1C). Similarly, DNAH17-AS1 expression was higher in pancreatic adenocarcinoma cells (Hs766T and SW1990 cells) than in normal human pancreatic duct epithelial cells (HPDE6-C7; Figure 1D). Since Hs766T cells had the highest DNAH17-AS1 expression, Hs766T cells were selected to perform gain- and loss-of-function experiments.
High DNAH17-AS1 expression was related to aggressive PC tumor behavior, including TNM stage, lymph node metastasis, and differentiation (Table 1). Furthermore, PC patients with DNAH17-AS1 upregulation had shorter overall survival and disease-free survival (Figure 1E and 1F). Based on these results, we suspected that DNAH17-AS1 may be an important regulator of PC progression, and high levels of DNAH17-AS1 expression may play an oncogenic role.
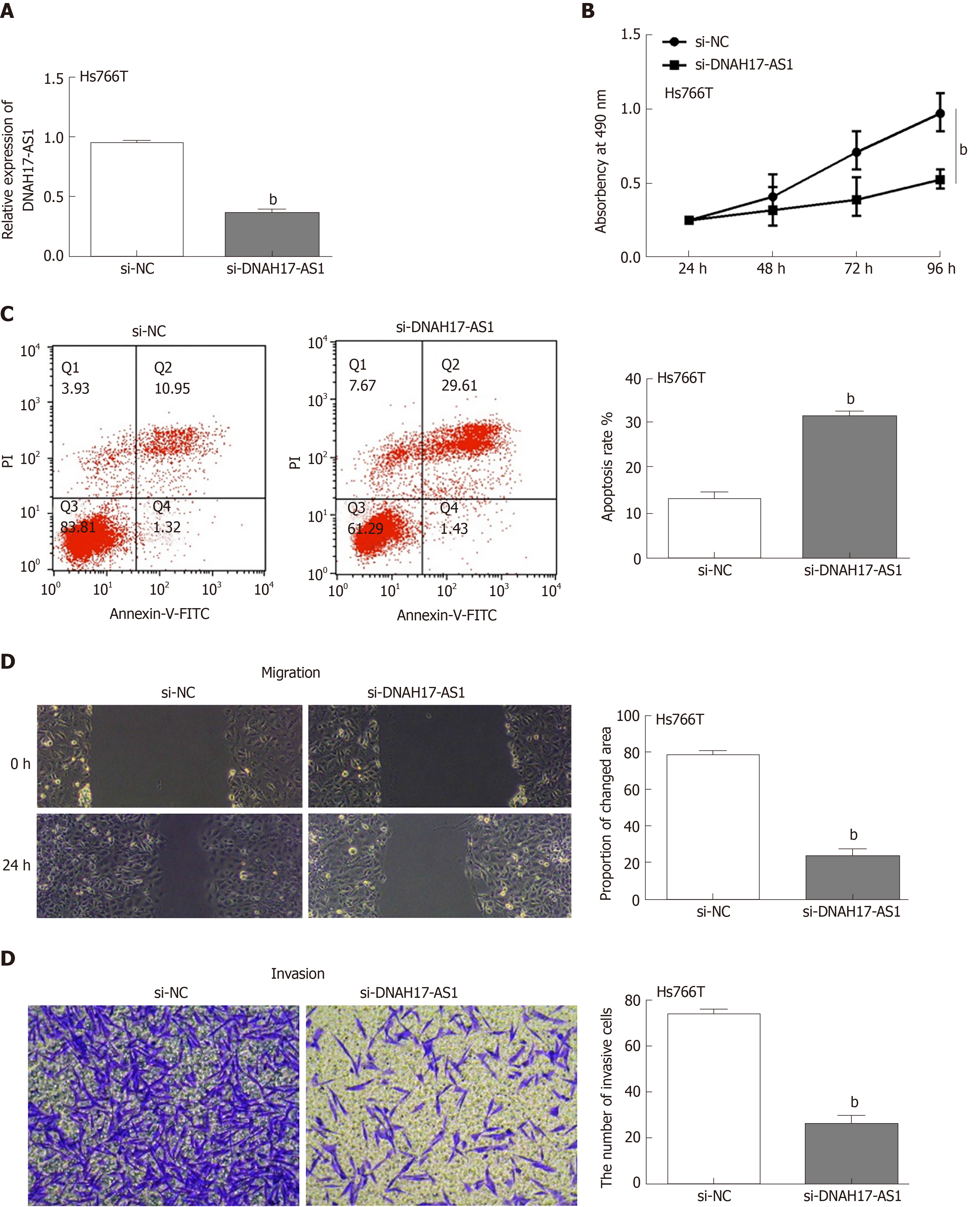
Next, the functions of DNAH17-AS1 were investigated in Hs766T cells transfected with si-DNAH17-AS1 or si-NC. As expected, DNAH17-AS1 was downregulated after transfection with si-DNAH17-AS1 compared with si-NC (Figure 2A). DNAH17-AS1 silencing greatly inhibited Hs766T cell viability compared to control (Figure 2B), with an approximate 50% reduction in cellular viability. Flow cytometry showed that downregulation of DNAH17-AS1 (Figure 2C) promoted apoptosis in PC cells. Furthermore, wound healing and transwell assays indicated that the migration and invasion of Hs766T cells were restrained following DNAHS1-AS1 silencing (Figure 2D and 2E). Taken together, these results showed that DNAH17-AS1 silencing inhibits PC cell viability and metastasis and promotes apoptosis in PC cells.
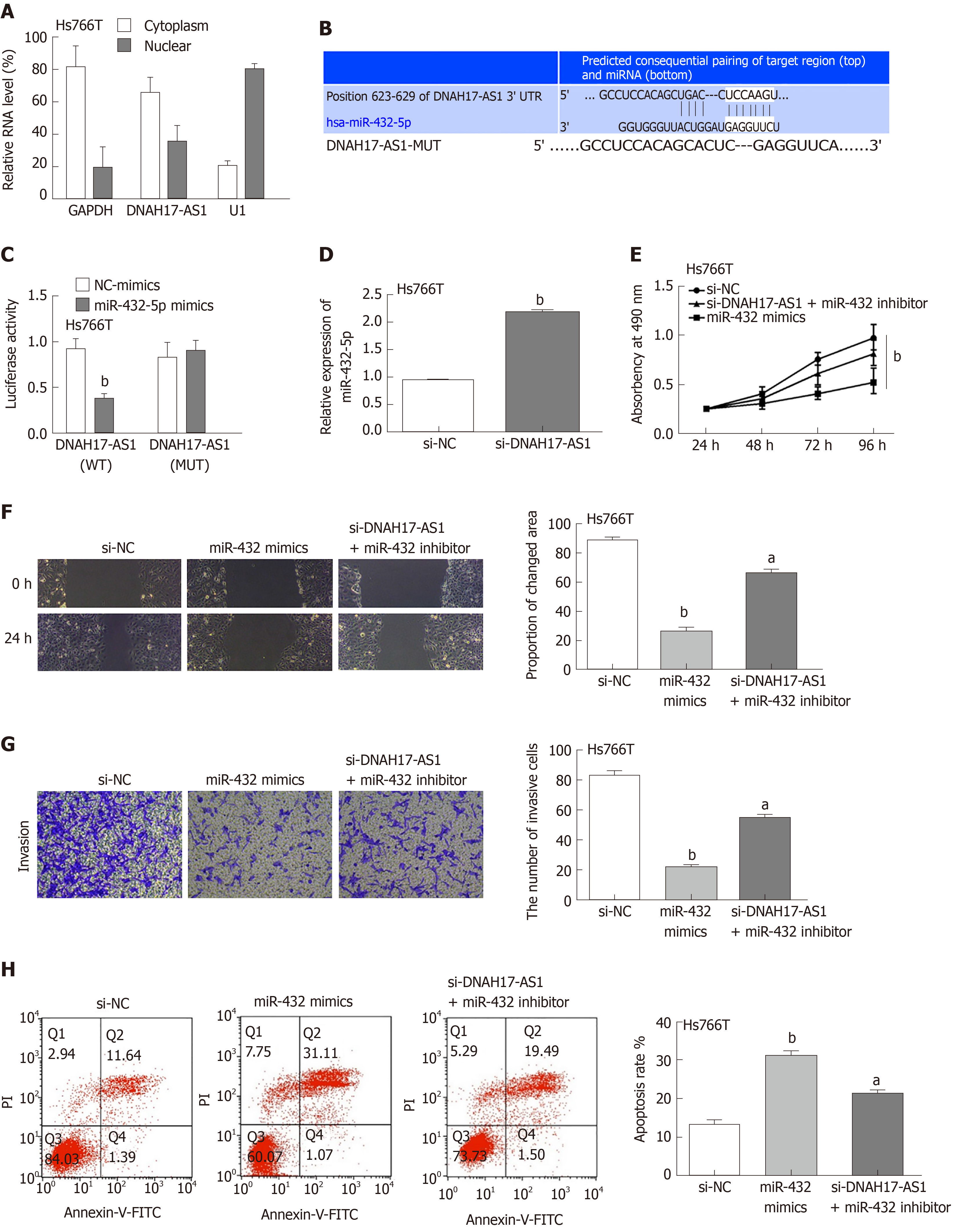
To determine the mechanism by which DNAH17-AS1 exerts its effects in PC, first we examined its cellular location. A subcellular fractionation assay showed that DNAH17-AS1 was predominantly located in the cytoplasm but also occasionally in the nucleus (Figure 3A). We therefore speculated that it is involved in competing endogenous RNA activity in the cytoplasm. A search of the downstream miRNA targets of DNAH17-AS1 in the starBase database (http://starbase.sysu.edu.cn/) and TargetScan database (http://www.targetscan.org) revealed that DNAH17-AS1 can bind to miR-432-5p (Figure 3B). In dual-luciferase reporter assays, miR-432-5p mimic decreased luciferase activity in the DNAH17-AS1-WT vector but had no effect on DNAH17-AS1-MUT (Figure 3C), indicating the specificity of the interaction between DNAH17-AS1 and miR-432-5p. Furthermore, RT-qPCR indicated that miR-432-5p was upregulated following downregulation of DNAH17-AS1 in transfected Hs766T cells (Figure 3D). In addition, two experiments in Hs766T cells showed that miR-432 mimic had similar effects to si-DNAH17-AS1 for regulating PC cell behavior (Figure 2B-E and 3E-H) and the observed consequences of decreased DNAH17-AS1 expression (Figure 2B-E) were rescued by a miR-432-5p inhibitor in cells transfected with si-DNAH17-AS1 (Figure 3E-H). Specifically, the inhibition of cell viability, migration, and invasion was resulted from miR-432-5p increasing and the effect of DNAH17-AS1 silencing on Hs766T cells was reversed by miR-432-5p inhibition (Figure 3E-G). Moreover, the promotion of apoptosis was induced by miR-432-5p mimic and the impact of si-DNAH17-AS1 was also weakened by downregulation of miR-432-5p (Figure 3H). These findings implied that DNAH17-AS1 functions as a tumor promoter in PC by binding to miR-432-5p and inhibiting its level in the cytoplasm.
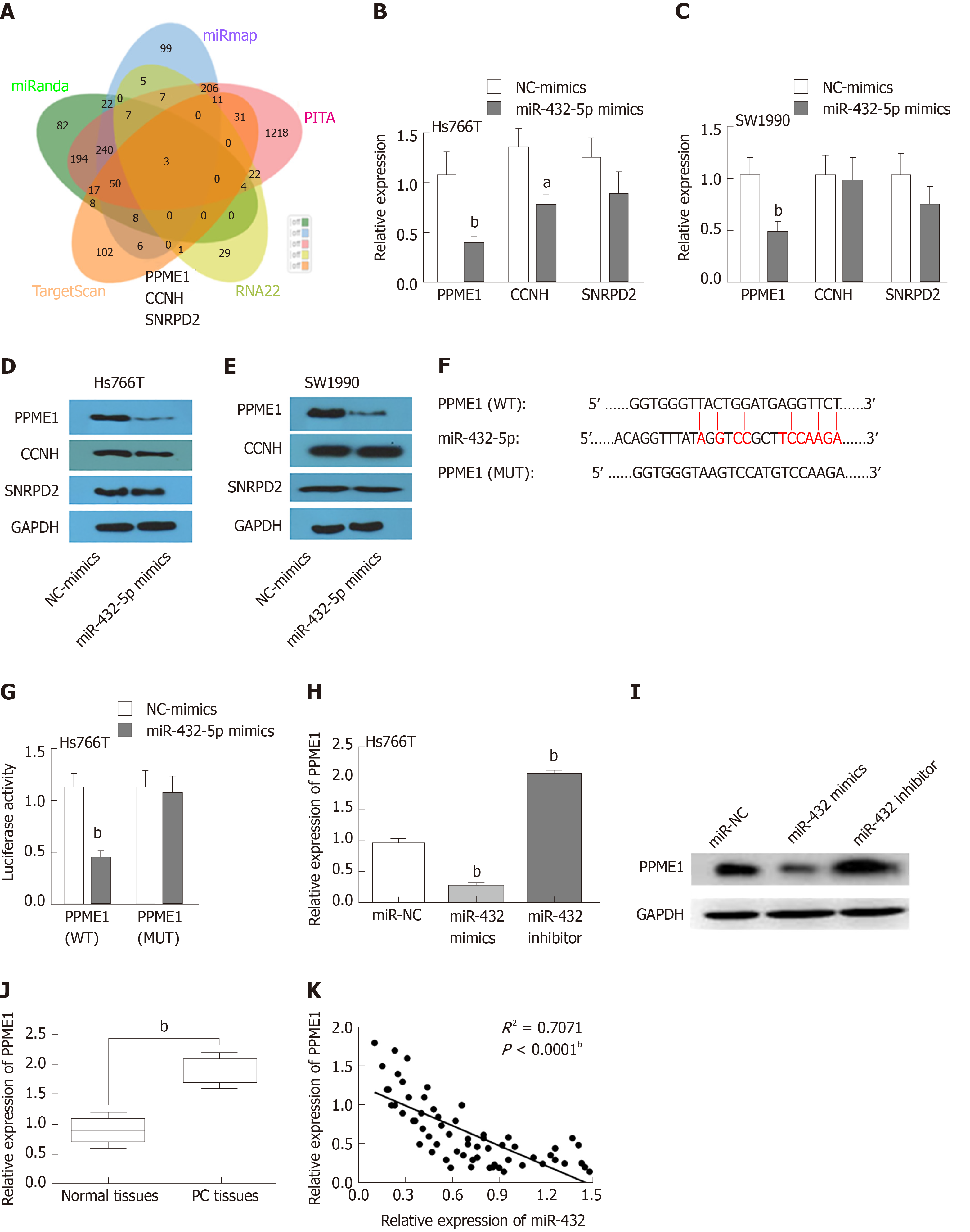
We next investigated the potential downstream mRNAs affected by the interaction of DNAH17-AS1 and miR-432-5p. We used miRanda, miRmap, PITA, RNA22, and TargetScan to identify overlapping targets. A total of three mRNAs were identified as possible targets, including PPME1, CCNH, and SNRPD2 (Figure 4A). In Hs766T and SW1990 cells, however, miR-432-5p mimic reduced PPME1 mRNA and protein levels more than those of CCNH and SNRPD2 (Figure 4B-E). This showed that PPME1 is the most likely one in the three predicted mRNAs regulated by mir-490-3p. A search for targets of miR-432-5p in the TargetScan database revealed that miR-432-5p has a binding site in the 3’-untranslated region (UTR) of PPME1 (Figure 4F). While miR-432-5p mimic suppressed the luciferase activity of PPME1-WT in dual-luciferase reporter assays, PPME1-MUT luciferase activity was unaffected (Figure 4G). Consistent with these results, PPME1 expression was downregulated by overexpression of miR-432-5p and increased by miR-432-5p inhibitor (Figure 4H-I). PPME1 was also upregulated in PC tissues compared with normal tissues (Figure 4J), and miR-432-5p inversely correlated with PPME1 expression in PC tissues (Figure 4K). These results showed that miR-432-5p downregulates PPME1 expression by binding to its 3’-UTR in PC.
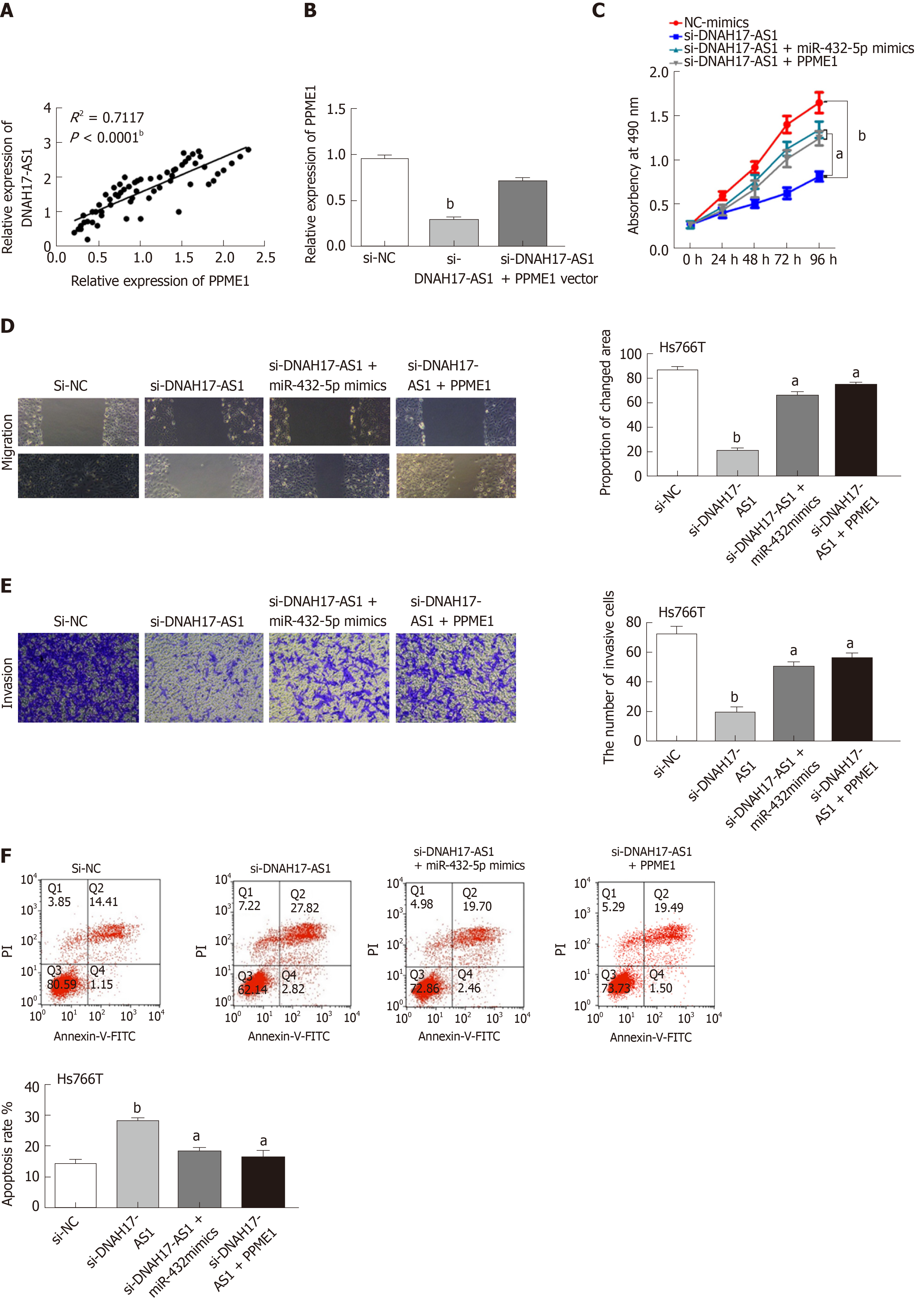
Taking all the above results together, we suspected that DNAH17-AS1 could regulate PPME1 expression through interaction with miR-432-5p. Indeed, DNAH17-AS1 was positively correlated with PPME1 expression in PC (Figure 5A). Moreover, PPME1 expression was downregulated after transfection of si-DNAH17-AS1 vector in Hs766T cells and could be recovered by PPME1 vector (Figure 5B). Upregulation of PPME1 also inhibited the DNAH17-AS1 siRNA-mediated suppression of cell viability and metastasis in Hs766T cells (Figure 5C-E). Similarly, the PPME1 vector weakened the increase in apoptosis typically induced by DNAH17-AS1 siRNA (Figure 5F). Collectively, these data indicated that DNAH17-AS1 promotes the progression of PC through regulation of the miR-432-5p/PPME1 axis.
The incidence, mortality, and morbidity of PC are increasing every year around the world[19]. It is therefore urgent to understand the mechanisms underlying PC tumorigenesis and progression. In addition, the absence of reliable diagnostic and prognostic markers has recently fostered significant research efforts to identify molecular “drivers”. LncRNAs are a promising candidate, and participate in the development of many cancers including PC[10]. Therefore, this study began with mining The Cancer Genome Atlas lncRNA database, focused on a highly-expressed lncRNA DNAH17-AS1 in PC, and deciphered the role of this novel lncRNA in PC.
To date, many lncRNAs have been discovered to be involved in PC tumorigenesis, such as the lncRNAs BANCR and LINC00339[20,21]. Previous reports have indicated that lncRNA DNAH17-AS1 promotes the progression of colorectal cancer[10], but whether DNAH17-AS1 is involved in the regulation of other cancers remains unclear. In this study, we first showed that DNAH17-AS1 is involved in PC progression through suppression of miR-432-5p. In addition, this study is the first to report that PPME1 is a target of miR-432-5p in PC cells. The results of this study will help to elucidate the regulatory mechanisms of DNAH17-AS1 and the miR-432-5p/PPME1 axis in PC.
Consistent with our results, previous studies have reported that DNAH17-AS1 is an lncRNA that is significantly associated with the overall survival of patients with PC[22]. Further, numerous studies have shown that DNAH17-AS1 may play an oncogenic role in both colorectal cancer[10] and head and neck squamous cell carcinoma[23]. All current hypotheses regarding DNAH17-AS1 as a proto-oncogene come from bioinformatic analyses, but to date, none have been verified experimentally. Here we found that DNAH17-AS1 serves as an oncogene in PC by downregulating miR-432-5p using both in silico analysis and experimental data.
LncRNAs can exert various biological effects through both transcriptional and post-transcriptional regulation. The molecular mechanisms of lncRNAs/miRNAs are complicated and can form a complex regulatory network of lncRNA, miRNA, and genes. Consistent with our results, many lncRNAs can interact with miR-432-5p to regulate the occurrence and progression of cancers[24,25]. In addition, some circular RNAs also regulate miR-432-5p to affect the development of cancer[26,27]. These studies showed that ncRNA can promote tumor growth or progression and restrain the apoptosis of tumor stem cells by sponging miR-432-5p. In the current study, we found that miR-432-5p negatively regulated PPME1 expression in PC by binding to its 3’-UTR. Similarly, Hu et al[24] reported that tumor-suppressive miR-432-5p can serve as a prognostic indicator in lung adenocarcinoma by mediating epithelial-mesenchymal transition. Furthermore, Wang et al[28] found that miR-432-5p expression is downregulated in PC. It is clear that miR-432-5p is an important regulator in cancer development and its downregulation predicts poorer prognoses for PC patients.
In this study, we showed that DNAH17-AS1 promoted PC progression by inhibiting miR-432-5p expression. Furthermore, miR-432-5p normally inhibited PPME1 expression to restrain PC progression. Consistent with these findings, the upregulation of PPME1 and resulting carcinogenesis have also been examined in gastric and lung cancer[29]. Hsieh et al[30] demonstrated that the endopeptidase catalytic subunit IMP1 facilitates choriocarcinoma cell migration and invasion via PPME1 upregulation. Taken together, these studies demonstrated that PPME1 plays an important role in the development of cancers including PC and may be a biomarker for PC.
In conclusion, the current study has demonstrated that upregulation of DNAH17-AS1 is related to adverse clinical features and poor prognosis of PC patients. Moreover, DNAH17-AS1 promotes PC progression and serves as a competitive factor to upregulate PPME1 by sequestering miR-432-5p. This study implies that DNAH17-AS1, miR-432-5p, and PPME1 may serve as therapeutic targets and prognostic biomarkers for PC patients and may provide new strategies for PC prevention and treatment.
Among common gastrointestinal malignancies, the annual incidence of pancreatic carcinoma (PC) has dramatically increased in recent years. Many studies have demonstrated that post-transcriptional regulation by long noncoding RNAs (lncRNAs) is important in PC progression. LncRNAs have emerged as pivotal molecules that participate in the initiation and progression of PC, including maintenance of cell growth, evasion of apoptosis, promotion of invasion and metastasis, maintenance of potency, and epithelial-mesenchymal transition.
To discover biomarkers for the diagnosis and treatment of PC.
To investigate the underlying mechanisms of the lncRNA DNAH17-AS1 in PC.
LncRNA DNAH17-AS1 expression was analyzed by Western blot and reverse transcription-quantitative PCR in PC tissue and cell lines, and the clinicopathological significance of DNAH17-AS1 expression in PC patients was also investigated. In vitro experiments were performed to explore the functions of lncRNA DNAH17-AS1 in PC cells. The regulatory effects of DNAH17-AS1/miR-432-5p/PPME1 were also investigated using luciferase reporter assay, MTT assay, flow cytometry, and transwell assay.
We found that the lncRNA DNAH17-AS1 was upregulated in PC tissues and cell lines and had a significant positive relationship with degree of tumor differentiation, TNM stage, and lymph node metastasis. In vitro experiments showed that DNAH17-AS1 increased the proliferation and invasion capacity of PC cells but inhibited their apoptosis. Additionally, DNAH17-AS1 served as an oncogene in PC by downregulating miR-432-5p. Furthermore, miR-432-5p directly targeted PPME1 and normally downregulated its expression. Thus, DNAH17-AS1 increased PPME1 expression to promote PC progression.
This study demonstrates that the lncRNA DNAH17-AS1 can significantly increase the growth, migration, and invasion of PC cells. DNAH17-AS1 also functions by sequestering miR-432-5p, which increases PPME1 expression and ultimately promotes PC progression. Taken together, our study provides the functional involvement of three new possible diagnostic biomarkers for PC.
In the future, additional research will be carried out to further explore the important role of the lncRNA DNAH17-AS1 and whether it can be harnessed to enhance the sensitivity of PC detection and to develop novel anti-cancer treatments. The identification of the lncRNA DNAH17-AS1/miR-432-5p/PPME1 molecular axis may further provide new strategies for PC prevention and treatment.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Youness RA S-Editor: Dou Y L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55815] [Article Influence: 7973.6] [Reference Citation Analysis (132)] |
| 2. | Aier I, Semwal R, Sharma A, Varadwaj PK. A systematic assessment of statistics, risk factors, and underlying features involved in pancreatic cancer. Cancer Epidemiol. 2019;58:104-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 3. | Bach P, Möhring C, Krawzak HW, Goepel M. [Retroperitoneal extravasation as the primary symptom of a pancreatic carcinoma]. Urologe A. 2007;46:1548-1550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 4. | Aronsson L, Bengtsson A, Torén W, Andersson R, Ansari D. Intraductal papillary mucinous carcinoma versus pancreatic ductal adenocarcinoma: A systematic review and meta-analysis. Int J Surg. 2019;71:91-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 5. | Batista IA, Melo SA. Exosomes and the Future of Immunotherapy in Pancreatic Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 6. | Chang L, Xu W, Zhang Y, Gong F. Long non-coding RNA-NEF targets glucose transportation to inhibit the proliferation of non-small-cell lung cancer cells. Oncol Lett. 2019;17:2795-2801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Ma W, Chen X, Wu X, Li J, Mei C, Jing W, Teng L, Tu H, Jiang X, Wang G, Chen Y, Wang K, Wang H, Wei Y, Liu Z, Yuan Y. Long noncoding RNA SPRY4-IT1 promotes proliferation and metastasis of hepatocellular carcinoma via mediating TNF signaling pathway. J Cell Physiol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 8. | Qi C, Xiaofeng C, Dongen L, Liang Y, Liping X, Yue H, Jianshuai J. Long non-coding RNA MACC1-AS1 promoted pancreatic carcinoma progression through activation of PAX8/NOTCH1 signaling pathway. J Exp Clin Cancer Res. 2019;38:344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 9. | Qiao X, Lv SX, Qiao Y, Li QP, Ye B, Wang CC, Miao L. Long noncoding RNA ABHD11-AS1 predicts the prognosis of pancreatic cancer patients and serves as a promoter by activating the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:8630-8639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 10. | Pan S, Shen M, Zhou M, Shi X, He R, Yin T, Wang M, Guo X, Qin R. Long noncoding RNA LINC01111 suppresses pancreatic cancer aggressiveness by regulating DUSP1 expression via microRNA-3924. Cell Death Dis. 2019;10:883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 11. | Ling J, Wang F, Liu C, Dong X, Xue Y, Jia X, Song W, Li Q. FOXO1-regulated lncRNA LINC01197 inhibits pancreatic adenocarcinoma cell proliferation by restraining Wnt/β-catenin signaling. J Exp Clin Cancer Res. 2019;38:179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 12. | Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1868] [Article Influence: 116.8] [Reference Citation Analysis (0)] |
| 13. | Yang J, Li C, Li H, E C. LncRNA CACNA1G-AS1 facilitates hepatocellular carcinoma progression through the miR-2392/C1orf61 pathway. J Cell Physiol. 2019;234:18415-18422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 14. | Youness RA, Hafez HM, Khallaf E, Assal RA, Abdel Motaal A, Gad MZ. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J Cell Physiol. 2019;234:20286-20297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 15. | Lei S, He Z, Chen T, Guo X, Zeng Z, Shen Y, Jiang J. Long noncoding RNA 00976 promotes pancreatic cancer progression through OTUD7B by sponging miR-137 involving EGFR/MAPK pathway. J Exp Clin Cancer Res. 2019;38:470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 16. | Yang J, Ye Z, Mei D, Gu H, Zhang J. Long noncoding RNA DLX6-AS1 promotes tumorigenesis by modulating miR-497-5p/FZD4/FZD6/Wnt/β-catenin pathway in pancreatic cancer. Cancer Manag Res. 2019;11:4209-4221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 17. | Zhao Y, Wang Q, Wang Y, Li J, Lu G, Liu Z. Glutamine protects against oxidative stress injury through inhibiting the activation of PI3K/Akt signaling pathway in parkinsonian cell model. Environ Health Prev Med. 2019;24:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 18. | Fang Y, Liang X, Xu J, Cai X. miR-424 targets AKT3 and PSAT1 and has a tumor-suppressive role in human colorectal cancer. Cancer Manag Res. 2018;10:6537-6547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 19. | Mattiuzzi C, Lippi G. Cancer statistics: a comparison between World Health Organization (WHO) and Global Burden of Disease (GBD). Eur J Public Health. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 116] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 20. | Wu X, Xia T, Cao M, Zhang P, Shi G, Chen L, Zhang J, Yin J, Wu P, Cai B, Lu Z, Miao Y, Jiang K. LncRNA BANCR Promotes Pancreatic Cancer Tumorigenesis via Modulating MiR-195-5p/Wnt/β-Catenin Signaling Pathway. Technol Cancer Res Treat. 2019;18:1533033819887962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 21. | Zhang R, Hao S, Yang L, Xie J, Chen S, Gu G. LINC00339 promotes cell proliferation and metastasis in pancreatic cancer via miR-497-5p/IGF1R axis. J BUON. 2019;24:729-738. [PubMed] |
| 22. | Shi X, Zhao Y, He R, Zhou M, Pan S, Yu S, Xie Y, Li X, Wang M, Guo X, Qin R. Three-lncRNA signature is a potential prognostic biomarker for pancreatic adenocarcinoma. Oncotarget. 2018;9:24248-24259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 23. | Fang XN, Yin M, Li H, Liang C, Xu C, Yang GW, Zhang HX. Comprehensive analysis of competitive endogenous RNAs network associated with head and neck squamous cell carcinoma. Sci Rep. 2018;8:10544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 24. | Hu C, Jiang R, Cheng Z, Lu Y, Gu L, Li H, Li L, Gao Q, Chen M, Zhang X. Ophiopogonin-B Suppresses Epithelial-mesenchymal Transition in Human Lung Adenocarcinoma Cells via the Linc00668/miR-432-5p/EMT Axis. J Cancer. 2019;10:2849-2856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 25. | Yang G, Li J, Cai Y, Yang Z, Li R, Fu W. Glycyrrhizic Acid Alleviates 6-Hydroxydopamine and Corticosterone-Induced Neurotoxicity in SH-SY5Y Cells Through Modulating Autophagy. Neurochem Res. 2018;43:1914-1926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 26. | Liu Y, Lu C, Zhou Y, Zhang Z, Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502:358-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 27. | Liu H, Xue L, Song C, Liu F, Jiang T, Yang X. Overexpression of circular RNA circ_001569 indicates poor prognosis in hepatocellular carcinoma and promotes cell growth and metastasis by sponging miR-411-5p and miR-432-5p. Biochem Biophys Res Commun. 2018;503:2659-2665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 28. | Wang ZX, Deng TX, Ma Z. Identification of a 4-miRNA signature as a potential prognostic biomarker for pancreatic adenocarcinoma. J Cell Biochem. 2019;120:16416-16426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Li J, Han S, Qian Z, Su X, Fan S, Fu J, Liu Y, Yin X, Gao Z, Zhang J, Yu DH, Ji Q. Genetic amplification of PPME1 in gastric and lung cancer and its potential as a novel therapeutic target. Cancer Biol Ther. 2014;15:128-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Hsieh YT, Chou MM, Chen HC, Tseng JJ. IMP1 promotes choriocarcinoma cell migration and invasion through the novel effectors RSK2 and PPME1. Gynecol Oncol. 2013;131:182-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |