Published online Mar 28, 2020. doi: 10.3748/wjg.v26.i12.1365
Peer-review started: December 19, 2019
First decision: January 12, 2020
Revised: March 10, 2020
Accepted: March 22, 2020
Article in press: March 22, 2020
Published online: March 28, 2020
Processing time: 99 Days and 19 Hours
Celiac Disease (CD) is an immune-mediated disorder, in which the HLA immunogenetic background (DQ2 and DQ8 heterodimers) and environmental trigger (gluten) are well established. Indeed, both factors are necessary – but not sufficient – to develop CD. However, it is very likely that CD is underdiagnosed in both developing and developed countries, due to several aspects, including the fact that a lot of patients present mild and/or atypical symptoms, without the presence of any recognized risk factors. Therefore, the possibility and feasibility of widened screening strategies to identify CD patients are debated.
To provide further evidence of the main epidemiological importance of HLA-DQB1*02 allele in the population of CD patients.
We performed a systematic search in PubMed, EMBASE, Cochrane, Web of Science and Scopus databases, in order to produce a systematic review assessing the carrier frequency of HLA-DQB1*02 allele in the celiac population. Following the PRISMA guidelines, we retrieved all the original articles describing CD patients’ HLA-DQB1 genotype in such a way that could allow to assess the HLA-DQB1*02 carrier frequency among CD patients, along with the evidence of the appropriate diagnostic work-up to achieve a correct and final diagnosis of CD.
The final output of this systematic search in the medical literature consisted of 38 studies providing the appropriate HLA-DQB1 genotype information of the respective CD population. According to this systematic review, including a pool of 4945 HLA-DQ genotyped CD patients, the HLA-DQB1*02 carrier frequency was 94.94%, meaning that only 5.06% of CD patients were completely lacking this allelic variant. Interestingly, if we consider only the studies whereby the prevalence of CD patients affected with type 1 diabetes mellitus was supposed or clearly established to be very low, the frequency of non-HLA-DQB1*02 carriers among CD patients dropped to 3.65%.
Such a high carrier frequency of the HLA-DQB1*02 allelic variant (which is > 95%-96% in CD patients without risk factors, like type 1 diabetes mellitus comorbidity) might be exploited to consider a cost-effective and widened screening approach. If a sustainable strategy could be implemented through a low-cost targeted genetic test to detect the individual presence of HLA-DQB1*02 allele, an appropriate algorithm for serological screening in individuals resulting to be genetically predisposed to CD, might be considered.
Core tip: It is well known that HLA-DQ genotyping is useful to assess the individual susceptibility to Celiac Disease (CD) with very high - if not absolute – discriminatory power. Indeed, it is very unlikely that individuals who do not carry specific HLA-DQ alleles coding MHC-DQ2 and MHC-DQ8 heterodimers, may develop CD. Here, we aim at providing further evidence of the specific epidemiological importance of HLA-DQB1*02 allele in the population of CD patients. Briefly, based on 38 original articles that we included in this systematic review (which provided a pool of 4945 HLA-DQ genotyped CD patients, overall), we could find a very high carrier frequency of the HLA-DQB1*02 allelic variant. Indeed, > 95%-96% of CD patients resulted to carry at least one copy of the HLA-DQB1*02 allele. This knowledge might be exploited to consider a cost-effective and widened screening approach: If a sustainable strategy could be implemented through a low-cost targeted genetic test for CD, an appropriate algorithm for serological screening in individuals resulting to be genetically predisposed to CD, might be considered.
- Citation: Poddighe D, Rebuffi C, De Silvestri A, Capittini C. Carrier frequency of HLA-DQB1*02 allele in patients affected with celiac disease: A systematic review assessing the potential rationale of a targeted allelic genotyping as a first-line screening. World J Gastroenterol 2020; 26(12): 1365-1381
- URL: https://www.wjgnet.com/1007-9327/full/v26/i12/1365.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i12.1365
Celiac Disease (CD) is a systemic autoimmune disease triggered by the dietary intake of gluten in a minority of HLA genetically predisposed individuals[1]. The world prevalence of CD in the general population is estimated to be around 1%, despite some geographical and ethnic variations. Recent epidemiological analyses suggested that the CD prevalence in children may be even greater than 1%[2-4].
In the landscape of non-communicable autoimmune diseases, CD presents some peculiar etiological aspects. As mentioned, the necessary environmental trigger for CD is well-known, namely the dietary exposure to gluten, which can lead to CD in a minority of all individuals expressing DQ2 and/or DQ8 heterodimers. Respectively, these antigen-presenting molecules are encoded by the specific alleles DQA1*0501-DQB1*02 and DQA1*0301-DQB1*0302. In North America and Europe, the individuals with such an immunogenetic predisposition to CD are 30%-40% of the general population, but only a minority of them (around 3%) will actually develop CD during life, despite a comparable exposure to gluten foods[1,2].
However, CD is under-diagnosed, even in developed countries. Indeed, according to the so-called “celiac iceberg” epidemiological model, only a minority of cases (the emerged tip of the iceberg) are clinically well-evident, whereas most part (the iceberg mass under the water surface) have atypical and/or pauci-symptomatic clinical presentations, making CD diagnosis be under-considered and/or significantly delayed[2,5]. Case-finding screening strategies helped to diagnose patients with some clinical and family risk factors, but the numerical impact of this approach in terms of number of new CD diagnoses, is quite limited. Indeed, most CD patients do not have any of these specific risk factors[6,7]. Moreover, the onset age of CD is extremely variable, which means that the serological screening should be periodically repeated in at-risk individuals[8].
Therefore, the debate about the opportunity to implement universal and/or widened screening strategies for CD is still open, especially in children, who may experience worse consequences than adults in case of missed or delayed CD diagnosis, due to their longer life expectancy and disease onset during the growth process[6,8-11].
Of course, it is not sustainable to propose a universal CD serological screening; however, the identification of HLA genetically predisposed individuals through a cheap analytic method could exclude that larger part of individuals, who will never develop CD. Through this systematic review, we assessed the percentage of CD patients who are carriers of at least one copy of the HLA-DQB1*02 allele, specifically.
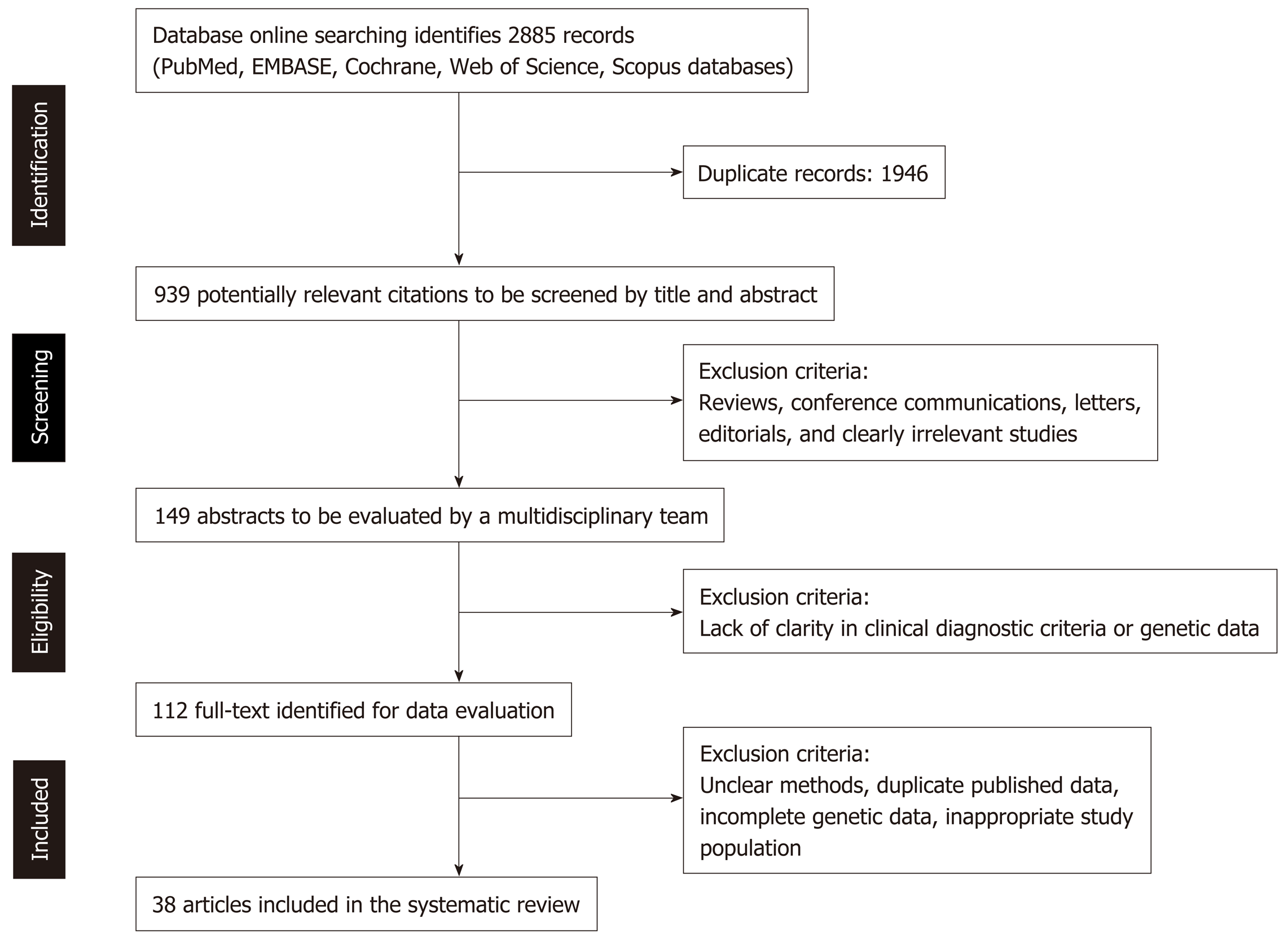
This systematic review was performed according to the PRISMA guidelines, as described in Figure 1, which shows the complete PRISMA flow diagram. Through this systematic review, we aimed at evaluating the carrier frequency of the specific allelic variant HLA-DQB1*02 (coding the β chain of DQ2 heterodimer) in the population of patients diagnosed with CD.
We performed a systematic search in PubMed, EMBASE, Web of Science, Scopus and Cochrane databases, by retrieving all original articles (case series, case–control, cross-sectional, and retrospective cohort studies) describing CD patients’ HLA-DQB1 genotype in detail. We searched all English, French, Spanish, Italian, German, Portuguese articles published up to September 2019.
In detail, an expert librarian performed the following searches in the following medical databases: (1) PubMed: ("celiac disease"[Mesh] OR "celiac disease" OR “coeliac disease” OR CD) AND ("HLA-DQ Antigens" [MeSH] OR "HLA-DQB1" OR "HLA-DQ2" OR "HLA-DQB1*02") AND ("alleles"[MeSH] OR allel* OR "Genes"[MeSH] OR gene OR genes OR variant* OR type* OR genotype[Mesh] OR genotyp*); (2) EMBASE: ('celiac disease'/exp OR 'celiac disease':ti,ab OR 'coeliac disease':ti,ab OR CD:ti,ab) AND ('HLA DQ antigen'/exp OR 'HLA-DQB1':ti,ab OR 'HLA-DQ2:ti,ab OR 'HLA-DQB1*02':ti,ab) AND ('allele'/exp OR allel*:ti,ab OR 'Gene'/exp OR gene:ti,ab OR genes:ti,ab OR variant*:ti,ab OR type*:ti,ab OR 'genotype'/exp OR genotyp*:ti,ab) AND [embase]/lim; (3) Web of Science: TS= ("celiac disease" OR “coeliac disease” OR CD) AND ("HLA-DQ Antigen*" OR "HLA-DQB1" OR "HLA-DQ2" OR "HLA-DQB1*02") AND (allel* OR gene OR genes OR variant* OR type* OR genotyp*); (4) Scopus: ("celiac disease" OR “coeliac disease” OR CD) AND ("HLA-DQ Antigen*" OR "HLA-DQB1" OR "HLA-DQ2" OR "HLA-DQB1*02") AND (allel* OR gene OR genes OR variant* OR type* OR genotyp*): ti,ab; and (5) Cochrane: ("celiac disease"[Mesh] OR "celiac disease":ti,ab,kw OR “coeliac disease”:ti,ab,kw OR CD:ti,ab,kw) AND ("HLA-DQ Antigens" [MeSH] OR "HLA-DQB1":ti,ab,kw OR "HLA-DQ2":ti,ab,kw OR "HLA-DQB1*02":ti,ab,kw) AND ("alleles"[MeSH] OR allel*:ti,ab,kw OR "Genes"[MeSH] OR gene:ti,ab,kw OR genes:ti,ab,kw OR variant*:ti,ab,kw OR type*:ti,ab,kw OR genotype[Mesh] OR genotyp*:ti,ab,kw).
After a critical reading of the articles, two investigators independently performed data extraction according to the following inclusion criteria: Any original articles in which CD patients’ HLA-DQB1 genotype was described in such a way and detail that the number of HLA-DQB1*02 carriers could be clearly defined within the total of the respective CD population. Only articles defining CD cases based on the typical histopathological findings, in addition to the specific serology, were considered. Most of them referred to specific diagnostic criteria, such as those set by Meeuwisse[12], Walker-Smith et al[13] and Husby et al[14], according to the study period. In detail, the following items were extracted from each study: First author's last name, publication date, country of origin, numbers of male/female patients, patient’s age range and HLA-DQB1 genotype (in terms of HLA-DQB1*02 carriers or not).
The process of literature screening and selection according to the PRISMA guidelines, is summarized in Figure 1. As a result, 112 full-text articles have been identified as potentially eligible for this systematic review, because those included some information on the HLA genotype of the respective CD study population. However, as summarized in Table 1, only 38 studies were finally included in the present systematic review, because they only provided enough details of the HLA-DQB1 genotype to appropriately assess the HLA-DQB1*02 carrier frequency among CD patients, along with the evidence of the appropriate diagnostic work-up to achieve a correct and final diagnosis of CD[15-56].
| Ref. | Country | Total number of CD patients | 1Age (yr) | Sex ratio (M/F) | HLA-DQB1*02 carriers | Non-HLA-DQB1*02 carriers | 2Notes | Clinical Background | 3DM1 patients (< 3%) |
| Congia et al[15], 1994 | Italy | 62 | Group I | 24/38 | 61 | 1 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[12] | Symptomatic patients classified as classic (group I) or atypical (group II) CD | + |
| (3.08 ± 2.05) | |||||||||
| Group II | |||||||||
| (5.7 ± 3.1) | |||||||||
| Herrera et al[16],1994 | Argentina | 62 | Children | 12/50 | 51 | 2 | HLA-DQ genotyping of 53 patients previously diagnosed with CD (ESPGHAN criteria)[12] | NA. Patients were diagnosed at the gastroenterology unit | ? |
| Fernández-Arquero et al[17], 1995 | Spain | 70 | 1.7 | NA | 67 | 3 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[12] | NA | ? |
| (NA) | |||||||||
| Polvi et al[18], 1996 | Finland | 49 | Index cases: 8 (1-34) | 25/24 | 49 | 0 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | 35 patients diagnosed on a clinical basis (not specified) and 14 FDR diagnosed by case-finding screening | + |
| FDR: (NA) | |||||||||
| Catassi et al[19], 2001 | Algeria | 79 | 8 | 33/44 | 68 | 1 | HLA-DQ genotyping of 69 patients previously diagnosed with CD (confirmed with intestinal biopsy) | NA | ? |
| (2-37) | |||||||||
| Kaur et al[20], 2002 | India | 35 | 8.42 | 18/17 | 35 | 0 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | Symptomatic patients with consistent GI or extra-GI symptoms | + |
| (1.5-15.6) | |||||||||
| Zubillaga et al[21], 2002 | Spain | 133 | 3.1 | 59/74 | 132 | 1 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | Patients with classic (71.4%) or non-classic (21.8%) CD. Nine asymptomatic patients diagnosed by case-finding screening (5 FRD, 3 DS, 1 DM1) | + |
| (0.5-18) | |||||||||
| Mustalahti et al[22], 2002 | Finland | 56 | NA | 18/38 | 49 | 0 | HLA-DQ genotyping of symptomatic patients previously diagnosed with CD. Seven patients did not undergo the intestinal biopsy (and, thus, have been excluded) | Twenty-eight symptomatic patients with consistent GI or extra-GI symptoms; and 28 asymptomatic siblings by case-finding screening | + |
| Karinen et al[23], 2006 | Finland | 54 | 43.7 ± 14.7 | NA | 53 | 1 | HLA-DQ genotyping of patients previously diagnosed with CD (confirmed with intestinal biopsy) | NA (based on clinical symptoms) | + |
| (1-79) | |||||||||
| Catassi et al[24], 2007 | United States | 22 | (19-83) | 18/4 | 21 | 0 | HLA-DQ genotyping in the context of a multicenter and prospective study (ESPGHAN criteria). HLA-DQ genotyping not done in only one of the 22 new CD diagnoses | Patients enrollment based on specific GI/extra-GI symptoms; or criteria for case-finding screening. Only 1 CD patients with DM1 | ? |
| Murray et al [25], 2007 | United States | 84 | (26-68) | 35/49 | 80 | 4 | HLA-DQ genotyping of patients previously diagnosed with CD (confirmed with intestinal biopsy) | Patients with consistent GI and/or extra-GI symptoms | + |
| Dezsofi et al [26], 2008 | Hungary | 100 | 16 | 47/53 | 96 | 4 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | NA. Anyway, this group was declared as not affected with DM1 | + |
| (3-40) | |||||||||
| Megiorni et al[27], 2009 | Italy | 437 | 5.7 | NA | 392 | 45 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | “Patients were divided on the basis of the clinical manifestations and gastrointestinal symptoms in typical, atypical, and silent forms”. No further numerical information is provided and, thus, it cannot be estimated number of DM1 patients | ? |
| (NA; only children) | |||||||||
| Thomas et al[28], 2009 | United Kingdom | 384 | 44 | 102/282 | 346 | 14 | HLA-DQ genotyping available for 360 patients previously diagnosed with CD (confirmed with intestinal biopsy) | Patients with consistent GI and/or extra-GI symptoms. Only 7 patients were affected with DM1 as well | + |
| (16-84) | |||||||||
| Martins et al[29], 2010 | Brazil | 90 | 15.5 | 35/55 | 84 | 6 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | NA. Patients were diagnosed at the gastroenterology unit | ? |
| (1-55) | |||||||||
| Srivastava et al[30], 2010 | India | 30 | 9.5 | 15/16 | 29 | 1 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | NA. Patients were diagnosed at the gastroenterology unit | ? |
| (3-17) | |||||||||
| El-Akawi et al[31], 2010 | Jordan | 44 | 13.5 | 12/32 | 44 | 0 | HLA-DQ genotyping available for 360 patients previously diagnosed with CD (confirmed with intestinal biopsy) | NA | ? |
| (1-39) | |||||||||
| Alarida et al[32], 2010 | Libya | 31 | 9.2 | 9/22 | 29 | 2 | HLA-DQ genotyping of patients diagnosed with CD (confirmed with intestinal biopsy) | School children undergone mass screening through anti-TG IgA | ? |
| (5-18) | |||||||||
| Castro-Antunes et al[33], 2011 | Brazil | 73 | NA | 37/36 | 60 | 13 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13]; One patient did not undergo HLA analysis | Patients enrollment based on specific GI/extra-GI symptoms; or criteria for case-finding screening (15: FDR or DM1, whose proportion is not specified) | - |
| (Children and adults) | |||||||||
| Mubarak et al[34], 2012 | the Netherlands | 70 | 5.7 | 20/50 | 70 | 0 | HLA-DQ genotyping of patients diagnosed with CD (confirmed with intestinal biopsy) | NA | ? |
| (NA; only children) | |||||||||
| Mubarak et al[34], 2012 | the Netherlands | 85 | 6.2 | 25/60 | 81 | 4 | HLA-DQ genotyping performed in all consecutive CD patients (confirmed with intestinal biopsy) at the time of the diagnosis | NA | ? |
| (NA; only children) | |||||||||
| Piccini et al[35], 2012 | Italy | 89 | NA | 27/62 | 81 | 8 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | NA | ? |
| (< 18) | |||||||||
| Krini et al[36], 2012 | Greece | 118 | NA | NA | 105 | 13 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | NA | ? |
| (< 18) | |||||||||
| Fernández-Cavada-Pollo et al[37], 2013 | Spain | 355 | NA | NA | 335 | 20 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | Patients with consistent GI and/or extra-GI symptoms | + |
| (0.5-76; children: n = 214, adults: n = 141) | |||||||||
| Delgado et al[38], 2014 | Spain | 91 | 6.9 | 23/68 | 88 | 3 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[14] | Patients with consistent GI and/or extra-GI symptoms; or with an affected FDR | + |
| (NA; only children) | |||||||||
| Cilleruelo et al[39], 2014 | Spain | 513 | NA | NA | 496 | 17 | HLA-DQ genotyping in the context of a multicenter and prospective study (ESPGHAN criteria)[13] | Patients enrollment based on classic or atypical symptoms consistent with CD; or criteria for case-finding screening. DM1 is reported in 2.2% (n = 11) of CD patients | + |
| (0.5-15) | |||||||||
| Stanković et al[40], 2014 | Serbia | 73 | 12 | 19/54 | 71 | 2 | HLA-DQ genotyping in the context of a multicenter and prospective study (ESPGHAN criteria)[13] | NA | ? |
| (1-22) | |||||||||
| Uenishi et al[41], 2014 | Brazil | 5 | NA | 1/4 | 5 | 0 | HLA-DQ genotyping of patients diagnosed with CD (confirmed with intestinal biopsy) | FDR case-finding screening | + |
| (7-75) | |||||||||
| Oliveira et al[42], 2014 | Portugal | 39 | 1.8 | 18/21 | 37 | 1 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[14]. One patient did not undergo HLA typing | Patients with consistent GI and/or extra-GI symptoms | + |
| (0.5-17) | |||||||||
| Almeida et al[43], 2016 | Brazil | 237 | 22 | 73/164 | 222 | 15 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | NA | ? |
| (1-75) | |||||||||
| Viken et al[44], 2017 | Norway | 327 | NA | NA | 310 | 17 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | NA. Patients with DM1 have been excluded | + |
| (Both children and adults) | |||||||||
| Murad et al[45], 2018 | Syria | 49 | 9.5 | 14/35 | 45 | 4 | HLA-DQ genotyping of patients diagnosed with CD (confirmed with intestinal biopsy) | NA | ? |
| (1-18) | |||||||||
| Martínez-Ojinaga et al[46], 2019 | Spain | 463 | 2.6 | NA | 454 | 9 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13,14] | Patients with consistent GI/extra-GI symptoms; or criteria for case-finding screening (not specified) | + |
| (0.6-14) | |||||||||
| Poddighe et al[47], 2019 | Italy | 184 | NA | 70/114 | 179 | 5 | HLA-DQ genotyping of 184 patients previously diagnosed with CD (ESPGHAN criteria)[13,14] | Patients with consistent GI (n = 75) or extra-GI (n = 69) symptoms; or criteria for case-finding screening (n = 40) | + |
| (1-16) | |||||||||
| Kauma et al[48], 2019 | Finland | 100 | CD index cases | 19/81 | 64 | 2 | HLA-DQ genotyping was available for 132 patients diagnosed with CD (confirmed with intestinal biopsy) in the context of a research study considering pairs of siblings both affected with CD | Patients with consistent GI/extra-GI symptoms. There is only one patient with IDDM among CD index cases | + |
| (3.08 ± 2.05) | |||||||||
| Kauma et al[48], 2019 | Finland | 100 | CD siblings | 37/63 | 59 | 7 | See previous notes (same study/article) | Patients with consistent GI/extra-GI symptoms. There are 4 patients with DM1 among sibling CD cases | - |
| (5.7 ± 3.1) | |||||||||
| 4Lopes et al[49-51], 2019 | Brazil | 7 | 7.5 | NA | 7 | 0 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13] | FDR case-finding screening. 7 CD cases out of 114 screened FDR | + |
| (6-12) | |||||||||
| 4Bajor et al[52-54], 2019 | Hungary | 105 | 31.2 | 32/73 | 97 | 8 | HLA-DQ genotyping of patients diagnosed with CD (confirmed with intestinal biopsy) | Patients with consistent GI or extra-GI symptoms; or criteria for case-finding screening. No CD patients with DM1 | + |
| (0.5-78) | |||||||||
| Al-Hussaini et al[55], 2019 | Saudi Arabia | 100 | Group 1 | 19/27 | 43 | 3 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13,14] | Patients with consistent GI or extra-GI symptoms (Group 1, n = 46). No data about comorbidity (e.g., DM1) | + |
| (7 ± 3.2) | |||||||||
| Al-Hussaini et al[55], 2019 | Saudi Arabia | 100 | Group 2 | 11/43 | 45 | 9 | Refer to the previous notes (same study/ article) | Patients diagnosed through a mass screening among school children (Group 2, n = 54). No data about comorbidity (e.g., DM1) | - |
| (11.3 ± 2.5) | |||||||||
| Ramosaj-Morina et al[56], 2019 | Kosovo | 60 | 5.5 | 20/40 | 55 | 5 | HLA-DQ genotyping of patients previously diagnosed with CD (ESPGHAN criteria)[13,14] | NA | ? |
| (1.5-18) |
Indeed, as already mentioned, the aim of this systematic review is to assess the frequency of the HLA-DQB1*02 carriers (at least one copy) in the population of CD patients, including both children and adults. This final research output resulted from the analysis of all retrieved full-length original articles, describing the HLA-DQ genotype in CD patients with appropriate resolution and details. Indeed, in order to be considered, the papers had to provide information about the complete HLA-DQB1 genotype for each patient or group of patients affected with CD; or enough information to certainly establish how many CD patients were carriers of at least one copy of the HLA-DQB1*02 allele. Therefore, the original articles reporting only the HLA-DQ allelic frequencies in the study populations were necessarily excluded, since the endpoint is not to assess how frequent is HLA-DQB1*02 in the CD population, but how many of them are carrier of at least one copy of this specific allelic variant.
After this qualitative selection of papers (according to the required information about the HLA-DQ genotype), we excluded all researches including a genetically biased CD population, namely those with a CD population resulting from a preliminary HLA genetic screening. Moreover, according to our main focus on those individuals that are not considered to be at risk in the current clinical recommendations of CD case-finding screening strategies, we have included as far as possible only those articles in which the CD study population was not pre-selected through a specific (autoimmune) comorbidity and, in particular, type 1 diabetes mellitus (DM1). Indeed, it is clearly established that patients with DM1 must be followed-up for the potential CD onset, due to the common background of HLA-DQ genetic predisposition, whereas our analysis aims to provide a preliminary information that could be useful to design a potential strategy of mass screening for that majority of CD patients without any comorbidity, who are actually more likely to have a delayed diagnosis or remain undiagnosed.
The quality of selected studies in terms of laboratory methods, methods description, statistical methodology and clinical features was assessed according to PRISMA standards and resulted to be appropriate.
Based on the 38 selected articles (Table 1), a total of 5065 patients affected with CD were included in this systematic review, but the HLA-DQ genotyping was not available for 120 of them. Among 4945 HLA-DQ genotyped CD patients, 4695 resulted to be carriers of at least one copy of the HLA-DQB1*02 allele, whereas 250 carried different allelic variants at both HLA-DQB1 loci (HLA-DQB1*0302 or others). Therefore, the HLA-DQB1*02 carrier frequency resulted to be 94.94%, meaning that only 5.06% of CD population was completely lacking this allelic variant.
If only the CD pediatric population (n = 2710) is considered, similar figures are obtained: 94.69% of CD children carry the HLA-DQB1*02 allele, whereas only 5.31% is lacking a copy of this allelic variant.
Finally, if we consider only the studies whereby the prevalence of CD patients also affected with DM1 was supposed or clearly established to be very low (< 3%), the frequency of non-HLA-DQB1*02 carriers among CD patients dropped to 3.65%.
Through this systematic review, we demonstrate that around 95% of all patients affected with CD carry the HLA-DQB1*02 allele, at least in one copy. Importantly, this figure raises up to > 96%, if only CD patients without DM1 are considered. This finding may contribute to the implementation of a cost-effective and preliminary genetic test to assess the existence of CD predisposition, through the targeted qualitative analysis of HLA-DQB1*02 allelic presence. This knowledge might help to optimize the use of the serological analyses for CD and, at the same time, extend the screening possibility, at least in the pediatric population.
As described in the introduction, the HLA-DQ genetic background is fundamental to determine the predisposition to develop CD. The coupled alleles DQA1*0501-DQB1*02 and DQA1*0301-DQB1*0302 respectively code the class II MHC heterodimers DQ2 and DQ8, which have been demonstrated to be expressed in almost all patients affected with CD. Indeed, several studies confirmed the very high negative predictive value associated with the absence of any genotypes coding MHC-DQ2 and/or -DQ8[1,2]. Practically, this specific knowledge can be applied to those patients with a suspect of CD, whenever the histopathological findings are not straightforward or the presence of concomitant diseases impairs the reliability of the serological tests for CD (e.g., IgA deficiency, Common Variable Immunodeficiency, et al). Therefore, the high-resolution analysis of HLA-DQ loci resulted to be particularly useful to refine and/or complete the diagnostic work-up for CD in some complex clinical cases, where the absence or presence of DQ2/DQ8 heterodimers can respectively rule out or support the final diagnosis of CD. Of course, the routine use of this expensive HLA analysis is completely inappropriate, because it can only assess the predisposition to develop CD (and not the actual presence of the disease). Indeed, large part of the general population (around 30%-40%, according to some geographical variability) possesses the appropriate HLA background to potentially develop CD[14].
Several studies investigated the relationship between HLA-DQ genotype of CD patients and some clinical characteristics, histopathological features, age of disease onset and even the risk to develop CD[21,25,46,52]. In this specific regard, several original articles reported a risk gradient for CD according to the particular HLA-DQ genotype: People with DQ2 homozygosity and DQ2/DQ8 double heterozygosity resulted to be have the highest risk and, importantly, a comparable risk of CD development was present in individuals carrying a double dose of HLA-DQB1*02 alleles, no matter the paired HLA-DQA1 alleles[27,43,57]. Recently, two meta-analysis by our group supported this finding (OR > 5, compared to the general population) and further emphasized that also a single “dose” of HLA-DQB1*02 is associated with a relatively high risk (OR ≈ 4), as regards pediatric CD[58-59]. In detail, we described that the HLA-DQB1*02 allele was present in 90%-95% of pediatric CD patients[58] and, in our monocentric clinical experience, we found > 95% of CD children who were carriers of at least one copy of HLA-DQB1*02 allele[50]. Therefore, we speculated that such a knowledge might be potentially used to consider the implementation of a widened screening strategy for CD in children, through a qualitative analysis targeting the HLA-DQB1*02 allele exclusively, as a first step, followed by the serological screening, to be applied to positive individuals only[11,50].
In order to further consider this potential idea, we decided to quantitively assess the actual presence of at least one copy of this allele among the CD population and, thus, understand how many patients would be lost if only the HLA-DQB1*02 positive individuals should undergo the serological screening for CD. If the loss of potential CD patients (those negative for HLA-DQB1*02 and, basically, carrying the DQ8 heterodimer only) could result acceptably low, then this approach might prevent the request (and the periodical re-testing) of any serological screening to most children. Anyway, the serological screening could be performed anytime in all those HLA-DQB1*02 negative patients developing symptoms consistent with CD, of course.
The present systematic review estimated that only 5% of the general CD population (including both adults and children) is devoid of HLA-DQB1*02 allele at all. If we could consider a mass screening looking for the carrier status of HLA-DQB1*02 only, we may identify 95% of CD predisposed patients and, concomitantly, rule out (with no more than a 5% error) the lifetime risk of disease in 60%-70% of the general population: These non-predisposed individuals should never receive the serological screening, unless any consistent clinical symptoms appear at some point of the existence without any other explanation.
Actually, according to this last consideration, that error may be even over-estimated. Indeed, if we consider only the articles describing cohorts of CD patients with a low prevalence of concomitant DM1 (Table 1), the percentage of non-HLA-DQB1*02 carriers decrease to 3.65%. Although the genetic predisposition to DM1 relies on both DQ2 and DQ8 heterodimers as well, this disease-genotype association for DM1 is less strong than in CD and, importantly, the DQ8 role and frequency resulted to be more relevant in patients with DM1, as evidenced by several studies. In a pediatric study from northwestern Mexico, including both CD and DM1 patients, Mejía-León et al[60] showed that the HLA-DQ8 combinations with DQ2 or one of its alleles conferred the highest risk for the combination of both diseases. Smigoc Schweiger et al[61] reported that DQ2 and DQ8 were present in 52% and 76% of DM1 patients in their Slovenian cohort, respectively. Mitchell et al[62] described the DQ2/DQ8 analysis of 176 Scottish children with DM1: At least 50%-55% were DQ8 carriers and, importantly, it was not associated with DQ2 in around 20% of patients. In summary, the frequency of the homozygous HLA-DQ8 genotype and, in general, heterozygous HLA-DQ8 without any copy of HLA-DQB1*02 is expected to be higher in DM1 patients than in patients with CD.
Moreover, some studies included in this systematic review paired patients with CD only and cohorts of patients affected with DM1 and DM1+CD. In the study by Dezsofi et al[26], DQ2 negative patients affected with CD only were as few as 4%, whereas DQ2 negative patients were 10.6% in CD patients affected with DM1, and 35% in patients with DM1 only. Viken et al[44] adopted a similar - but larger - study design to investigate the HLA class II alleles in Norwegian patients with coexisting DM1 and CD. Whereas DQ2 negative (all DQ8 homo- or heterozygous) patients do not exceed 5% in the CD-only cohort, they reach 30% in CD+DM1 patients and exceed this percentage in patients with DM1 only. In the study by Kauma et al[48], two groups of CD patients were considered (namely, CD index cases and siblings affected with CD) and the number of patients with concomitant DM1 was quite different (1/66 and 4/66, respectively): Interestingly, the frequency of non-HLA-DQB1*02 carriers was much more prevalent in the latter group (10.6% vs 3%), due to the higher HLA-DQB1*0302 (DQ8) allelic frequency in the DM1 subgroups of CD patients.
This systematic review is limited by the fact that only studies providing enough and appropriate information about the HLA-DQB1 genotype for each patient or groups of patients, could be included in our analyses. However, genetic pre-selection biases were carefully considered and, thus, avoided, in order to provide reliable findings for our analysis purposes.
A potential HLA-DQB1*02-targeted screening approach would be addressed to all those people that does not fall into the ESPGHAN groups at risk for CD (and, thus, receiving a periodical serology screening anyway), because this case-finding strategy leaves the majority of asymptomatic or pauci-symptomatic CD patients undiagnosed[63,64]. Of course, it is not currently sustainable to propose a periodical serology screening for CD to the general population, even if limited to children, who may be more vulnerable to the negative long-term consequences of undiagnosed or belatedly diagnosed CD[6]. If the HLA-DQB1*02 qualitative (present/not present) screening could be feasible after a rigorous economical and ethical assessment, it would allow to identify almost all CD-predisposed children, who should be around 30-40% of the general pediatric population. Therefore, 60-70% should not be eligible to receive the serology screening anymore, and a (periodic) serological test could be proposed to the HLA-DQB1*02 predisposed children only. At the end, this approach might also lead to some savings on the costs currently allocated to the CD serological screening, if we consider that these tests are often over-requested by patients and practitioners[64-66].
The cost of CD diagnostic testing variably affects both patients and the healthcare system. The gold standard for diagnosis uses a combination of methods, which usually begins with a serological testing that includes immunoglobulin A (IgA) and tissue transglutaminase (TTG) IgA[67]. A study (including 250 healthy children) from Europe reported that screening with serum TTG cost €5000 annually, rising to €11250 if anti-endomysium (EMA) testing was additionally performed on every child. Therefore, in Europe, each ELISA-based TTG test costs approximately €20, without considering the total IgA measurement[68]. Identifying that majority of children who do not need the serological screening because they are not genetically predisposed to CD, might result in a significant reduction of costs sustained for CD serology. According to the findings of this systematic review, < 5% of CD-predisposed children may be lost through this screening approach and, if a patient should develop symptoms consistent with CD at any time during own life, the serology test could be performed anyway on a clinical basis.
Low-cost molecular methods for targeted and qualitative HLA-typing can be at the horizon. For instance, Verma et al[69] recently proposed a rapid HLA-DQ typing method to identify subjects genetically susceptible to CD by performing a PCR through a kit containing all four HLA-DQ target alleles only. The cost of such an HLA-DQ genotyping was about €15 and, probably, may be further reduced by using reagents to detect the HLA-DQB1*02 allele only. Children would receive this test only one time and 30%-40% of them (those with positive results) could be identified as CD-predisposed and, then, eligible to serology screening at specific time points to be established.
In conclusion, a cost-effective and widened screening approach may be very helpful in both developed and developing countries, if a sustainable strategy could be implemented through a low-cost targeted genetic test for the HLA-DQB1*02 allelic presence, along with appropriate algorithms for serological screening in the individuals with this specific HLA-DQ predisposition to CD. Of course, specific pharmacoeconomic studies and ethical considerations would be needed, before a specific strategy can be proposed.
Celiac Disease (CD) is an immune-mediated disorder in which the HLA immunogenetic background (DQ2 and DQ8 heterodimers) is well known. This genetic factor is necessary – but not sufficient – to develop CD. Basically, almost 100% of CD patients are carriers of the aforementioned HLA-DQ background and several studies emphasized the main role of the HLA-DQB1*02 allele in such a genetic predisposition.
CD is underdiagnosed in both developing and developed countries, due to several aspects: Indeed, many patients present mild and/or atypical symptoms, without the presence of any recognized risk factors. Therefore, the possibility and feasibility of widened screening strategies to identify CD patients is still debated and this study might provide some additional insights, in order to find novel screening strategies.
Our aim was to define and assess the carrier frequency of the specific allelic variant HLA-DQB1*02 (coding the β chain of DQ2 heterodimer) in the population of patients diagnosed with CD.
In order to achieve our aim, we performed a systematic review, according to the PRISMA guidelines, by retrieving all original articles (case series, case–control, cross-sectional, and retrospective cohort studies) describing CD patients’ HLA-DQB1 genotype in detail. Any original articles, in which CD patients’ HLA-DQB1 genotype was described in such a way and detail that the number of HLA-DQB1*02 carriers could be clearly defined (within the total of the respective CD population), were considered.
As a result of our literature search, 38 studies were finally included in the present systematic review, since those provided details of the HLA-DQB1 genotype in such a way that could allow to assess the HLA-DQB1*02 carrier frequency among CD patients. Among 4945 HLA-DQ genotyped CD patients, the HLA-DQB1*02 carrier frequency resulted to be 94.94%, meaning that only 5.06% of CD population was completely lacking this allelic variant. If only the CD pediatric population is considered, similar figures are obtained: Only 5.31% is lacking a copy of this allelic variant. Finally, if we consider only the studies whereby the prevalence of CD patients also affected with type 1 diabetes mellitus (DM1) was supposed or clearly established to be very low, the frequency of non-HLA-DQB1*02 carriers among CD patients dropped up to 3.65%.
According to the findings of this systematic review, < 4%-5% of CD-predisposed children may be lost through a preliminary evaluation of the presence/absence of HLA-DQB1*02 allele, regardless of the presence of other HLA-DQB1 and HLA-DQA1 CD-predisposing alleles.
A cost-effective and widened screening approach may be very helpful in both developed and developing countries, if a sustainable strategy could be implemented through a low-cost targeted genetic test for the HLA-DQB1*02 allelic presence, along with appropriate algorithms for serological screening in individuals resulting to be genetically predisposed to CD.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Kazakhstan
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ciccone MM, Gabriel S, Jadallah KA S-Editor: Wang YQ L-Editor: A E-Editor: Zhang YL
| 1. | Lindfors K, Ciacci C, Kurppa K, Lundin KEA, Makharia GK, Mearin ML, Murray JA, Verdu EF, Kaukinen K. Coeliac disease. Nat Rev Dis Primers. 2019;5:3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 267] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 2. | Lebwohl B, Sanders DS, Green PHR. Coeliac disease. Lancet. 2018;391:70-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 668] [Article Influence: 95.4] [Reference Citation Analysis (0)] |
| 3. | Poddighe D, Rakhimzhanova M, Marchenko Y, Catassi C. Pediatric Celiac Disease in Central and East Asia: Current Knowledge and Prevalence. Medicina (Kaunas). 2019;55: pii E11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Poddighe D, Turganbekova A, Baymukasheva D, Saduakas Z, Zhanzakova Z, Abdrakhmanova S. Genetic predisposition to celiac disease in Kazakhstan: Potential impact on the clinical practice in Central Asia. PLoS One. 2020;15:e0226546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 5. | Nenna R, Tiberti C, Petrarca L, Lucantoni F, Mennini M, Luparia RP, Panimolle F, Mastrogiorgio G, Pietropaoli N, Magliocca FM, Bonamico M. The celiac iceberg: characterization of the disease in primary schoolchildren. J Pediatr Gastroenterol Nutr. 2013;56:416-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 6. | Ludvigsson JF, Card TR, Kaukinen K, Bai J, Zingone F, Sanders DS, Murray JA. Screening for celiac disease in the general population and in high-risk groups. United European Gastroenterol J. 2015;3:106-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 7. | Catassi C, Lionetti E. Case finding for celiac disease is okay, but is it enough? J Pediatr Gastroenterol Nutr. 2013;57:415-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Ludvigsson JF, Murray JA. Epidemiology of Celiac Disease. Gastroenterol Clin North Am. 2019;48:1-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 9. | US Preventive Services Task Force., Bibbins-Domingo K, Grossman DC, Curry SJ, Barry MJ, Davidson KW, Doubeni CA, Ebell M, Epling JW Jr, Herzstein J, Kemper AR, Krist AH, Kurth AE, Landefeld CS, Mangione CM, Phipps MG, Silverstein M, Simon MA, Tseng CW. Screening for Celiac Disease: US Preventive Services Task Force Recommendation Statement. JAMA. 2017;317:1252-1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 10. | Ludvigsson JF. Mortality and malignancy in celiac disease. Gastrointest Endosc Clin N Am. 2012;22:705-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 11. | Poddighe D. Individual screening strategy for pediatric celiac disease. Eur J Pediatr. 2018;177:1871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Meeuwisse GW. Round table discussion. Diagnostic criteria in coeliac disease. Acta Paediatr Scand. 1970;59:461-464. [RCA] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 178] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Walker-Smith JA, Guandalini S, Schmitz J, Shmerling DH, Visakorpi JK. Revised criteria for diagnosis of coeliac disease. Report of Working Group of European Society of Paediatric Gastroenterology and Nutrition. Arch Dis Child. 1990;65:909-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1014] [Cited by in RCA: 1087] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 14. | Husby S, Koletzko S, Korponay-Szabó IR, Mearin ML, Phillips A, Shamir R, Troncone R, Giersiepen K, Branski D, Catassi C, Lelgeman M, Mäki M, Ribes-Koninckx C, Ventura A, Zimmer KP; ESPGHAN Working Group on Coeliac Disease Diagnosis; ESPGHAN Gastroenterology Committee; European Society for Pediatric Gastroenterology, Hepatology, and Nutrition. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012;54:136-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1708] [Cited by in RCA: 1837] [Article Influence: 141.3] [Reference Citation Analysis (5)] |
| 15. | Congia M, Cucca F, Frau F, Lampis R, Melis L, Clemente MG, Cao A, De Virgiliis S. A gene dosage effect of the DQA1*0501/DQB1*0201 allelic combination influences the clinical heterogeneity of celiac disease. Hum Immunol. 1994;40:138-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | Herrera M, Theiler G, Augustovski F, Chertkoff L, Fainboim L, DeRosa S, Cowan EP, Satz ML. Molecular characterization of HLA class II genes in celiac disease patients of Latin American Caucasian origin. Tissue Antigens. 1994;43:83-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | Fernández-Arquero M, Polanco I, Escobar H, Figueredo MA, de la Concha EG, Clerici-Larradet N. HLA-DQ alleles and susceptibility to celiac disease in Spanish children. Tissue Antigens. 1995;45:145-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Polvi A, Eland C, Koskimies S, Mäki M, Partanen J. HLA DQ and DP in Finnish families with celiac disease. Eur J Immunogenet. 1996;23:221-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | Catassi C, Doloretta Macis M, Rätsch IM, De Virgiliis S, Cucca F. The distribution of DQ genes in the Saharawi population provides only a partial explanation for the high celiac disease prevalence. Tissue Antigens. 2001;58:402-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Kaur G, Sarkar N, Bhatnagar S, Kumar S, Rapthap CC, Bhan MK, Mehra NK. Pediatric celiac disease in India is associated with multiple DR3-DQ2 haplotypes. Hum Immunol. 2002;63:677-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Zubillaga P, Vidales MC, Zubillaga I, Ormaechea V, García-Urkía N, Vitoria JC. HLA-DQA1 and HLA-DQB1 genetic markers and clinical presentation in celiac disease. J Pediatr Gastroenterol Nutr. 2002;34:548-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | Mustalahti K, Holopainen P, Karell K, Mäki M, Partanen J. Genetic dissection between silent and clinically diagnosed symptomatic forms of coeliac disease in multiplex families. Dig Liver Dis. 2002;34:842-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 23. | Karinen H, Kärkkäinen P, Pihlajamäki J, Janatuinen E, Heikkinen M, Julkunen R, Kosma VM, Naukkarinen A, Laakso M. HLA genotyping is useful in the evaluation of the risk for coeliac disease in the 1st-degree relatives of patients with coeliac disease. Scand J Gastroenterol. 2006;41:1299-1304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 24. | Catassi C, Kryszak D, Louis-Jacques O, Duerksen DR, Hill I, Crowe SE, Brown AR, Procaccini NJ, Wonderly BA, Hartley P, Moreci J, Bennett N, Horvath K, Burk M, Fasano A. Detection of Celiac disease in primary care: a multicenter case-finding study in North America. Am J Gastroenterol. 2007;102:1454-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 178] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 25. | Murray JA, Moore SB, Van Dyke CT, Lahr BD, Dierkhising RA, Zinsmeister AR, Melton LJ, Kroning CM, El-Yousseff M, Czaja AJ. HLA DQ gene dosage and risk and severity of celiac disease. Clin Gastroenterol Hepatol. 2007;5:1406-1412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Dezsofi A, Szebeni B, Hermann CS, Kapitány A, Veres G, Sipka S, Körner A, Madácsy L, Korponay-Szabó I, Rajczy K, Arató A. Frequencies of genetic polymorphisms of TLR4 and CD14 and of HLA-DQ genotypes in children with celiac disease, type 1 diabetes mellitus, or both. J Pediatr Gastroenterol Nutr. 2008;47:283-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Megiorni F, Mora B, Bonamico M, Barbato M, Nenna R, Maiella G, Lulli P, Mazzilli MC. HLA-DQ and risk gradient for celiac disease. Hum Immunol. 2009;70:55-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 28. | Thomas HJ, Ahmad T, Rajaguru C, Barnardo M, Warren BF, Jewell DP. Contribution of histological, serological, and genetic factors to the clinical heterogeneity of adult-onset coeliac disease. Scand J Gastroenterol. 2009;44:1076-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 29. | Martins Rde C, Gandolfi L, Modelli IC, Almeida RC, Castro LC, Pratesi R. Serologic screening and genetic testing among brazilian patients with celiac disease and their first degree relatives. Arq Gastroenterol. 2010;47:257-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Srivastava A, Yachha SK, Mathias A, Parveen F, Poddar U, Agrawal S. Prevalence, human leukocyte antigen typing and strategy for screening among Asian first-degree relatives of children with celiac disease. J Gastroenterol Hepatol. 2010;25:319-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | El-Akawi ZJ, Al-Hattab DM, Migdady MA. Frequency of HLA-DQA1*0501 and DQB1*0201 alleles in patients with coeliac disease, their first-degree relatives and controls in Jordan. Ann Trop Paediatr. 2010;30:305-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Alarida K, Harown J, Di Pierro MR, Drago S, Catassi C. HLA-DQ2 and -DQ8 genotypes in celiac and healthy Libyan children. Dig Liver Dis. 2010;42:425-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 33. | Castro-Antunes MM, Crovella S, Brandão LA, Guimaraes RL, Motta ME, Silva GA. Frequency distribution of HLA DQ2 and DQ8 in celiac patients and first-degree relatives in Recife, northeastern Brazil. Clinics (Sao Paulo). 2011;66:227-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | Mubarak A, Spierings E, Wolters VM, Otten HG, ten Kate FJ, Houwen RH. Children with celiac disease and high tTGA are genetically and phenotypically different. World J Gastroenterol. 2013;19:7114-7120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 35. | Piccini B, Vascotto M, Serracca L, Luddi A, Margollicci MA, Balestri P, Vindigni C, Bassotti G, Villanacci V. HLA-DQ typing in the diagnostic algorithm of celiac disease. Rev Esp Enferm Dig. 2012;104:248-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (1)] |
| 36. | Krini M, Chouliaras G, Kanariou M, Varela I, Spanou K, Panayiotou J, Roma E, Constantinidou N. HLA class II high-resolution genotyping in Greek children with celiac disease and impact on disease susceptibility. Pediatr Res. 2012;72:625-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Fernández-Cavada-Pollo MJ, Alcalá-Peña MI, Vargas-Pérez ML, Vergara-Prieto E, Vallcorba-Gómez-Del Valle I, Melero-Ruiz J, Márquez-Armenteros AM, Romero-Albillos JA, Narváez-Rodríguez I, Fernández-de-Mera JJ, González-Roiz C. Celiac disease and HLA-DQ genotype: diagnosis of different genetic risk profiles related to the age in Badajoz, southwestern Spain. Rev Esp Enferm Dig. 2013;105:469-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 38. | Delgado JF, Amengual MJ, Veraguas A, Rodríguez E, de Los Santos MM, Guallarte MP. Paediatric celiac patients carrying the HLA-DR7-DQ2 and HLA-DR3-DQ2 haplotypes display small clinical differences. Acta Paediatr. 2014;103:e238-e242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (1)] |
| 39. | Cilleruelo ML, Roman-Riechmann E, Sanchez-Valverde F, Donat E, Manuel-Ramos J, Martín-Orte E, López MJ, García-Novo D, García S, Pavón P, Martín M, Ortigosa L, Barrio J, Gutierrez C, Espìn B, Castillejo G, Peña-Quintana L, Hualde I, Sebastián M, Calvo C, Fernández S, De Manueles J, Armas H, Urruzuno-Tellerias P, Juste M, Bousoño C, Ribes-Koninckx C. Spanish national registry of celiac disease: incidence and clinical presentation. J Pediatr Gastroenterol Nutr. 2014;59:522-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 40. | Stanković B, Radlović N, Leković Z, Ristić D, Radlović V, Nikčević G, Kotur N, Vučićević K, Kostić T, Pavlović S, Zukic B. HLA genotyping in pediatric celiac disease patients. Bosn J Basic Med Sci. 2014;14:171-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Uenishi RH, Gandolfi L, Almeida LM, Fritsch PM, Almeida FC, Nóbrega YK, Pratesi R. Screening for celiac disease in 1st degree relatives: a 10-year follow-up study. BMC Gastroenterol. 2014;14:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 42. | Oliveira JR, Cabral AJ, Ferreira E, Capelinha F, Spínola H, Gonçalves R. Celiac disease in children from Madeira island and its prevalence in first degree relatives. Arq Gastroenterol. 2014;51:151-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (1)] |
| 43. | Almeida LM, Gandolfi L, Pratesi R, Uenishi RH, de Almeida FC, Selleski N, Nóbrega YK. Presence of DQ2.2 Associated with DQ2.5 Increases the Risk for Celiac Disease. Autoimmune Dis. 2016;2016:5409653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 44. | Viken MK, Flåm ST, Skrivarhaug T, Amundsen SS, Sollid LM, Drivvoll AK, Joner G, Dahl-Jørgensen K, Lie BA. HLA class II alleles in Norwegian patients with coexisting type 1 diabetes and celiac disease. HLA. 2017;89:278-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 45. | Murad H, Jazairi B, Khansaa I, Olabi D, Khouri L. HLA-DQ2 and -DQ8 genotype frequency in Syrian celiac disease children: HLA-DQ relative risks evaluation. BMC Gastroenterol. 2018;18:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 46. | Martínez-Ojinaga E, Fernández-Prieto M, Molina M, Polanco I, Urcelay E, Núñez C. Influence of HLA on clinical and analytical features of pediatric celiac disease. BMC Gastroenterol. 2019;19:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 47. | Poddighe D, Capittini C, Gaviglio I, Brambilla I, Marseglia GL. HLA-DQB1*02 allele in children with celiac disease: Potential usefulness for screening strategies. Int J Immunogenet. 2019;46:342-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Kauma S, Kaukinen K, Huhtala H, Kivelä L, Pekki H, Salmi T, Saavalainen P, Lindfors K, Kurppa K. The Phenotype of Celiac Disease Has Low Concordance between Siblings, Despite a Similar Distribution of HLA Haplotypes. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Sdepanian VL, Lopes LHC, Oliveira RP, Muniz JG. Celiac Disease in First-degree Relatives: Homozygosity of DQB1*02 and At Least One Copy of HLA-DQB1*02 Allele. J Pediatr Gastroenterol Nutr. 2019;69:e149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 50. | Poddighe D. HLA-DQB1*02 Allele in First-degree Relatives of Patients With Celiac Disease. J Pediatr Gastroenterol Nutr. 2019;69:e148-e149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 51. | Lopes LHC, Muniz JG, Oliveira RP, Sdepanian VL. Celiac Disease in Brazilian First-degree Relatives: The Odds Are Five Times Greater for HLA DQ2 Homozygous. J Pediatr Gastroenterol Nutr. 2019;68:e77-e80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 52. | Bajor J, Szakács Z, Juhász M, Papp M, Kocsis D, Szegedi É, Földi I, Farkas N, Hegyi P, Vincze Á. HLA-DQ2 homozygosis increases tTGA levels at diagnosis but does not influence the clinical phenotype of coeliac disease: A multicentre study. Int J Immunogenet. 2019;46:74-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 53. | Poddighe D. Relevance of HLA-DQB1*02 allele in predisposing to coeliac disease. Int J Immunogenet. 2019;46:274-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 54. | Bajor J, Szakács Z, Vincze Á. Response to Letter to the Editor: Relevance of HLA-DQB1*02 allele in predisposing to coeliac disease. Int J Immunogenet. 2019;46:276-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 55. | Al-Hussaini A, Eltayeb-Elsheikh N, Alharthi H, Osman A, Alshahrani M, Sandogji I, Alrashidi S, Bashir MS. HLA-DQ genotypes relative risks for celiac disease in Arabs: A case-control study. J Dig Dis. 2019;20:602-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 56. | Ramosaj-Morina A, Burek Kamenaric M, Azemi M, Spahiu L, Grubic Z, Zunec R. HLA Haplotype Association with Celiac Disease in Albanian Pediatric Patients from Kosovo. Gastroenterol Res Pract. 2019;2019:7369014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 57. | Margaritte-Jeannin P, Babron MC, Bourgey M, Louka AS, Clot F, Percopo S, Coto I, Hugot JP, Ascher H, Sollid LM, Greco L, Clerget-Darpoux F. HLA-DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease. Tissue Antigens. 2004;63:562-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 138] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 58. | De Silvestri A, Capittini C, Poddighe D, Valsecchi C, Marseglia G, Tagliacarne SC, Scotti V, Rebuffi C, Pasi A, Martinetti M, Tinelli C. HLA-DQ genetics in children with celiac disease: a meta-analysis suggesting a two-step genetic screening procedure starting with HLA-DQ β chains. Pediatr Res. 2018;83:564-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 59. | Capittini C, De Silvestri A, Rebuffi C, Tinelli C, Poddighe D. Relevance of HLA-DQB1*02 Allele in the Genetic Predisposition of Children with Celiac Disease: Additional Cues from a Meta-Analysis. Medicina (Kaunas). 2019;55: pii E190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 60. | Mejía-León ME, Ruiz-Dyck KM, Calderón de la Barca AM. HLA-DQ genetic risk gradient for type 1 diabetes and celiac disease in northwestern Mexico. Rev Gastroenterol Mex. 2015;80:135-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 61. | Smigoc Schweiger D, Mendez A, Kunilo Jamnik S, Bratanic N, Bratina N, Battelino T, Brecelj J, Vidan-Jeras B. High-risk genotypes HLA-DR3-DQ2/DR3-DQ2 and DR3-DQ2/DR4-DQ8 in co-occurrence of type 1 diabetes and celiac disease. Autoimmunity. 2016;49:240-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 62. | Mitchell RT, Sun A, Mayo A, Forgan M, Comrie A, Gillett PM. Coeliac screening in a Scottish cohort of children with type 1 diabetes mellitus: is DQ typing the way forward? Arch Dis Child. 2016;101:230-233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 63. | Björck S, Brundin C, Lörinc E, Lynch KF, Agardh D. Screening detects a high proportion of celiac disease in young HLA-genotyped children. J Pediatr Gastroenterol Nutr. 2010;50:49-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 64. | Franceschini E, Lionetti ME, D'Adamo G, D'Angelo E, Gatti S, Naspi Catassi G, Malamisura B, Catassi C. Misuse of serological screening tests for celiac disease in children: A prospective study in Italy. Dig Liver Dis. 2019;51:1547-1550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 65. | Ali M, Danner DJ, Fishman DS, Devaraj S. Utilization of Laboratory Testing Algorithms for Celiac Disease in a Pediatric Hospital. Lab Med. 2020;51:99-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 66. | Huang Y, Don-Wauchope AC, Grey VL, Mansour M, Brill H, Armstrong D. Improving serological test ordering patterns for the diagnosis of celiac disease through clinical laboratory audit of practice. Clin Biochem. 2012;45:455-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 67. | Mearns ES, Taylor A, Boulanger T, Craig KJ, Gerber M, Leffler DA, Drahos J, Sanders DS, Lebwohl B. Systematic Literature Review of the Economic Burden of Celiac Disease. Pharmacoeconomics. 2019;37:45-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 68. | Alessandrini S, Giacomoni E, Muccioli F. Mass population screening for celiac disease in children: the experience in Republic of San Marino from 1993 to 2009. Ital J Pediatr. 2013;39:67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 69. | Verma AK, Singh A, Gatti S, Lionetti E, Galeazzi T, Monachesi C, Franceschini E, Ahuja V, Catassi C, Makharia GK. Validation of a novel single-drop rapid human leukocyte antigen-DQ2/-DQ8 typing method to identify subjects susceptible to celiac disease. JGH Open. 2018;2:311-316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |