Published online Mar 21, 2020. doi: 10.3748/wjg.v26.i11.1113
Peer-review started: December 20, 2019
First decision: January 12, 2020
Revised: March 4, 2020
Accepted: March 9, 2020
Article in press: March 9, 2020
Published online: March 21, 2020
Processing time: 91 Days and 19 Hours
Viruses can alter the expression of host microRNAs (MiRNA s) and modulate the immune response during a persistent infection. The dysregulation of host MiRNA s by hepatitis B virus (HBV) contributes to the proinflammatory and profibrotic changes within the liver. Multiple studies have documented the differential regulation of intracellular and circulating MiRNA s during different stages of HBV infection. Circulating MiRNA s found in plasma and/or extracellular vesicles can integrate data on viral-host interactions and on the associated liver injury. Hence, the detection of circulating MiRNA s in chronic HBV hepatitis could offer a promising alternative to liver biopsy, as their expression is associated with HBV replication, the progression of liver fibrosis, and the outcome of antiviral treatment. The current review explores the available data on miRNA involvement in HBV pathogenesis with an emphasis on their potential use as biomarkers for liver fibrosis.
Core tip: The current review analyses the available data on the role of microRNAs (MiRNA s) in the development and progression of liver fibrosis by focusing on their potential use as diagnostic and prognostic biomarkers for hepatitis B virus-infected patients. Cellular and circulating MiRNA s (in plasma or extracellular vesicles) offer a unique glimpse into the virus-host relationship and the pathogenesis of chronic hepatitis B virus infection. The differential regulation of intracellular and circulating MiRNA s during the natural and on treatment evolution of chronic hepatitis B is discussed.
- Citation: Iacob DG, Rosca A, Ruta SM. Circulating microRNAs as non-invasive biomarkers for hepatitis B virus liver fibrosis. World J Gastroenterol 2020; 26(11): 1113-1127
- URL: https://www.wjgnet.com/1007-9327/full/v26/i11/1113.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i11.1113
MicroRNAs (MiRNA s) are short noncoding RNAs involved in the epigenetic regulation of multiple intracellular and extracellular signalling pathways and in the posttranscriptional regulation of genes across numerous eukaryotic organisms[1-3]. Cellular MiRNA s can modulate viral replication and the immune antiviral response[4,5]. Viruses can encode their own MiRNA s[6] and can alter the cellular miRNome to create a favourable environment for viral replication or latency. Due to these complex roles, MiRNA s have been increasingly evaluated as biomarkers for the diagnosis, prognosis and treatment of viral infections[7,8] as well as for distinct pathologies, including liver fibrosis.
Chronic hepatitis B virus (HBV) infection affects over 257 million people[9] and it is one of the most common causes of liver fibrosis. Liver fibrogenesis is a dynamic process, characterized by an excessive accumulation of extracellular matrix proteins in response to an ongoing liver inflammatory response, with gradual distortion of hepatic architecture and progression to liver cirrhosis[10]. Extracellular matrix is mainly synthesized by hepatic stellate cells (HSCs), which are activated following liver injury, together with proinflammatory cytokines and chemokines. Once activated, HSCs maintain this phenotype through autocrine or paracrine signalling loops[11]. Still, liver fibrosis is a reversible wound-healing process. Experimental studies have shown that an early detection and timely removal of the inciting factor can lead to a complete remission of the fibrotic changes, while interventions performed in later stages are less effective against the already formed architectural changes[12]. Furthermore, given the potential progression of liver fibrosis to hepatocellular carcinoma (HCC), an antifibrotic treatment would ideally need to be started early and would be precisely targeted against the molecular processes occurring at that stage.
Regarding HBV-associated fibrosis, between 8%-20% of untreated patients can progress to liver cirrhosis within 5 years depending on viral characteristics (HBV genotype, viral load, HBV mutations) and host-related factors (age, gender, other comorbidities or coinfections)[13]. Antiviral treatment, with either pegylated interferon-α or nucleoside analogues, halts or attenuates the development of fibrosis[14-18] and the initiation of treatment in the early stages of liver fibrosis is associated with a significant improvement of the histologic scores[17]. However, current treatment options do not ensure a complete cure of the HBV infection (with the elimination of viral reservoirs from hepatocytes) and a persistent activation of fibrotic signalling pathways is possible even in patients with undetectable HBV serum viral loads after treatment[19-21]. Hence, biomarkers which offer additional information on the viral-host interaction could potentially foreshadow new therapeutic agents.
Liver biopsy is currently the gold standard for a complete assessment of liver fibrosis, inflammation, and intrahepatic HBV replication. This technique is nevertheless limited by multiple risks and potential misclassifications, due to examiner variability and sampling[22]. Hence, a series of alternative non-invasive biomarkers have been proposed, including imaging data (elastographic techniques such as transient elastography, acoustic radiation force impulse imaging, two-dimensional shear wave elastography and magnetic resonance elastography), biochemical scores (APRI, Fib-4, Fibrotest), HBV RNA, and HBV core antigen[23-25] or even direct markers (molecules released in the serum following liver fibrogenesis of fibrolysis such as hyalyuronic acid, type IV collagen, matrix metalloproteases or tissue inhibitory metalloprotease-1)[26]. Non-invasive scores are more accessible, which explains why the World Health Organization recommends the use of Fib-4 and APRI for the assessment of liver fibrosis in HBV patients living in low-income countries[27]. Nevertheless, the diagnostic performance of these biomarkers in chronic HBV infection is moderate. Non-invasive methods are less reliable for the prediction of a specific stage of liver fibrosis, yet these can differentiate between an early and an advanced stage of liver fibrosis or even cirrhosis (e.g., F0-F1, ≥ F2 or ≥ F4)[26]. Therefore, combined scores with circulating and cellular MiRNA s could represent an appealing alternative for the diagnosis and monitoring of viral-induced liver fibrosis[28] offering supplementary data on the viral-host interactions and the fibrotic signalling cascades in both the liver and blood.
The current review analyses the available data on the role of MiRNA s in chronic hepatitis B, with an emphasis on their role in liver fibrogenesis and on their potential use as non-invasive biomarkers in the diagnosis, evolution, and treatment of HBV induced-liver fibrosis.
Genes encoding for MiRNA s are transcribed by the RNA polymerase II/III into primary RNA transcripts (pri-miRNA ), further processed in the nucleus by the Drosha ribonuclease to a hairpin loop structure of ~ 60 nucleotides (the pre-miRNA transcript). Pre-MiRNA s are further exported to the cytoplasm, where the Dicer enzyme cleaves the hairpin loop and leads to the mature double-stranded miRNA .
One strand of the mature miRNA is degraded, while the other one (less stable at the 5’ end) becomes the guide strand and is recruited into an RNA-induced silencing complex together with Argonaute proteins, TAR RNA-binding proteins, and other proteins and binds to the 3’ untranslated region of the target mRNAs[29]. Noncanonical interactions can also occur through “seed-like” motifs at the 5’ untranslated region or coding regions[30,31]. This intricate binding mechanism does not imply a perfect complementarity one miRNA can regulate one or more mRNAs, and multiple MiRNA s can bind to the same mRNA. The concentrations of intracellular MiRNA s are extremely variable, depending on the cellular context (cell cycle, metabolism, or differentiation) and concurrent pathologies. This variability could be exploited during viral-host interactions to influence viral tropism and hijack the host transcriptional machinery or to enable the host control on viral infections[32].
HBV modulates miRNA biogenesis by decreasing the expression of Drosha ribonuclease[33]. Novellino et al[34] showed that serum hepatitis B surface antigen (HBsAg) particles transport both Ago2 proteins and a series of MiRNA s (miR-27a, miR-30b, miR-122, miR-126, miR-145, miR-106b, and miR-223) and identified a different miRNA profile in HBsAg particles and plasma[34]. Ago2 interacts with hepatitis B core antigen and HBsAg in various subcellular compartments of infected cells, indicating a potential role of HBV on miRNA packaging into extracellular vesicles (EVs)[35]. The function of extracellular MiRNA s is not well elucidated, yet data on MiRNA s found in EVs (like apoptotic bodies, microvesicles, or exosomes[36,37]) suggest multiple roles in paracrine signalling[38], epigenetic regulation of the recipient cell and regulation of the cellular inflammatory response, through the activation of toll-like receptor signalling pathways[39,40]. Incidentally, the first discovered HBV-encoded miRNA , HBV-miR-3[41] modulates the release of HBV virions and is also incorporated into exosomes and HBV core particles but not into HBsAg subviral particles. It would be interesting to explore if this differential packaging is the result of a viral-host competition and sequestration of host/viral MiRNA s into a certain particle and further exploit these findings for diagnostic or therapeutic purposes.
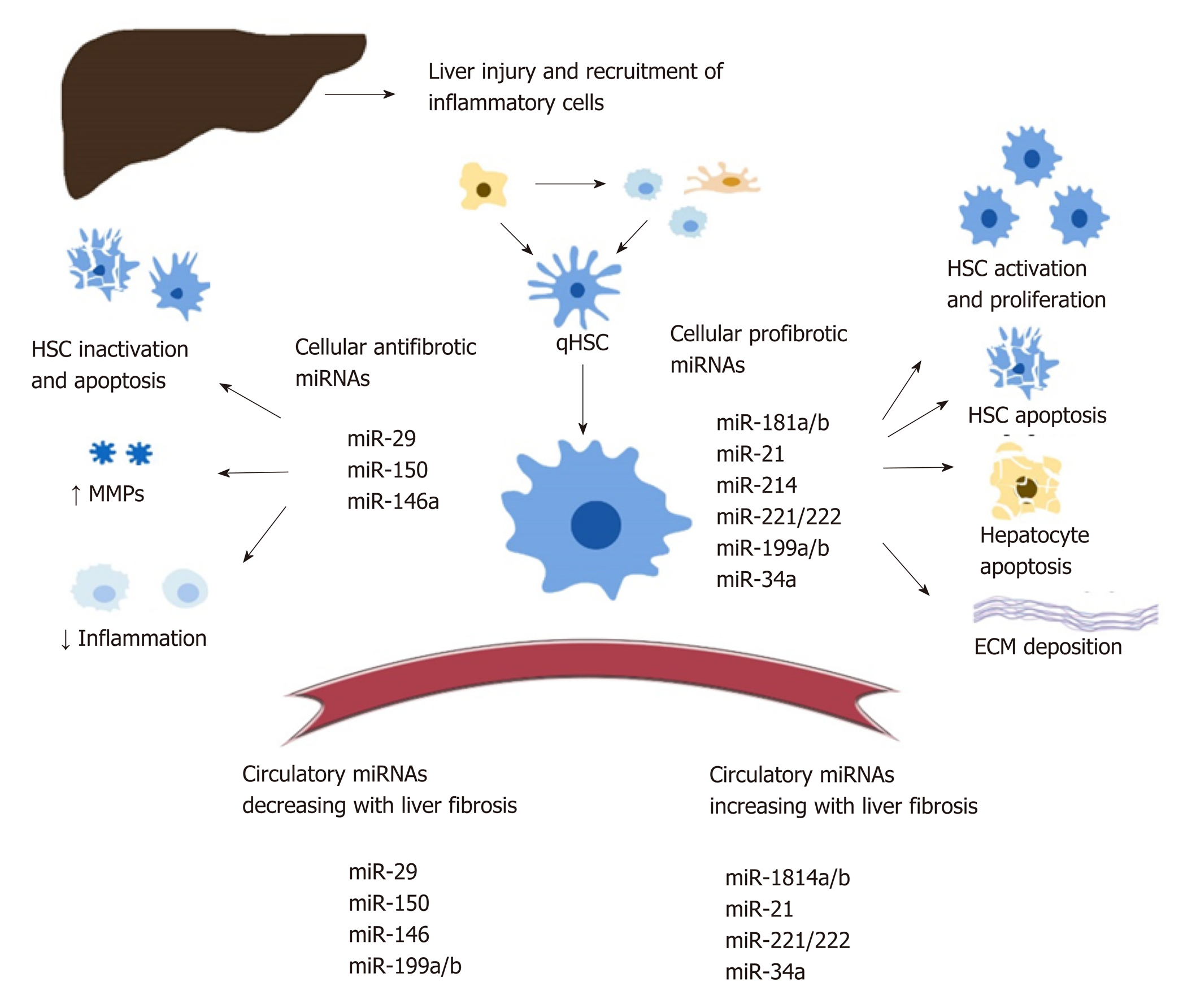
Intracellular miRNA : There is a significant amount of data documenting the role of cellular MiRNA s in liver fibrosis but no consensus on their exact regulatory functions. Various MiRNA s are being proposed as either profibrotic or antifibrotic (Figure 1). This classification is theoretical, based on predicted signalling pathways and reports from various studies[42-44]. Overall, MiRNA s modulate numerous steps during the development of liver fibrosis, including: HSC activation, proliferation, migration, and apoptosis[45-49]; transcription of profibrogenic factors and signalling pathways (such as Col1a1, transforming growth factor beta (TGFβ)-RII[46,47], SPRY2, HNF4a[48], matrix metalloproteinases[49,50], MeCP2[51], retinoid X receptor alpha[52]); modulation of the immune response and intrahepatic recruitment of inflammatory cells, indirectly contributing to the release of profibrotic cytokines such as tumor necrosis factor alpha, Interleukin-6, and Interleukin-1β, regulation of interferon gamma signalling and of the inflammasome pathway[57-60], regulatory activity on the metabolism of lipids, drugs, and alcohol[50,57,58,61]; and the regulation of angiogenesis[63].
The complete characterization of intracellular MiRNA s is nevertheless difficult given that one miRNA can modulate multiple signalling pathways in various tissues. For example, miR-34 mediates both HSC activation through peroxisome proliferator-activated receptor gamma signalling[64] and hepatocyte apoptosis through the miR-34/sirtuin-1/p53 cascade[65]. On the other hand, miR-34a-5p can also display an antifibrotic role within HSC, as its overexpression was correlated with the downregulation of the TGFβ/Smad3 pathway[66].
Both miR-181b and miR-21 favour HSC activation through the inhibition of phosphatase and tensin homolog and activation of the phosphatidylinositol 3-kinase/Akt pathway[67,68]. MiR-29b, a significant antifibrotic cellular miRNA , induces HCS apoptosis, regulates the HSC phenotype, and decreases extracellular matrix synthesis through multiple signalling pathways (TGF-β / Smad, lipopolysaccharide / NF-kB, and oestradiol)[69-72]. HSC activation is also downregulated by multiple antifibrotic MiRNA s, including miR-146 through the suppression of TGF-β / Smad[73], and miR-150 through c-myB and Sp1 signalling pathways[74,75].
Circulating MiRNA s found in the plasma or serum have been extensively studied in the pathogenesis of liver disease due to various aetiologies, including viral hepatitis, nonalcoholic steatohepatitis and alcoholic liver disease, drug-associated liver injury, and HCC[62,76-79].
Circulating MiRNA s correlate with the presence and progression of liver fibrosis and necroinflammation and can be used to predict the survival of patients with cirrhosis or HCC[80-82]. The link between circulating and cellular MiRNA s is still under investigation. In this respect, Table 1 provides a correlation between the regulation of various MiRNA s found in the serum and liver of patients with HBV infection.
| MicroRNA | Plasma level | Liver tissue | Mediated processes involved in liver fibrosis | Ref. |
| miR-34a | ↑ | ↑ | Cell-cycle regulator (cell differentiation, proliferation, metabolism, apoptosis); HSC activation | Guo et al[45]; Singh et al[42] |
| miR-221 miR-222 | ↑ | ↑ | Collagen synthesis; HSC proliferation; Liver fibrosis; Oncogenesis | Singh et al[42] |
| miR 27a/b | ↑ | ↑ | HSC activation, differentiation and proliferation | Zhang et al[131] |
| miR 181a/b | ↑ | ↑ | Cell cycle regulator; HSC activation and proliferation | Yu et al[132] |
| miR 199a/b | ↓ | ↑ | HSC activation | Murakami et al[91] |
| miR-223 | ↓; ↓ EVs | ↑ | Inflammatory response | Akamatsu et al[133]; Bao et al[43]; Ye et al[59]; Wang et al [106] |
| miR-125 (-125a-5p/ -125b) | ↑ | ↑ | HSC activation, proliferation | Zheng et al[134]; You et al[49] |
| miR 21-5p | ↑in total plasma; ↓ in EVs | ↑ | Collagen synthesis; Oncogenesis | Bao et al[43]; Wei et al[68]; Wang et al[135] |
EVs are secreted in multiple body fluids and ensure the transport of various proteins, lipids or RNAs including MiRNA s. Intrahepatic MiRNA s are packaged into EVs and released from injured hepatocytes to further mediate the survival/proliferation or infection in neighbouring cells[83-85]. Additionally, MiRNA s associated to EVs have been shown to play various roles in cell-to-cell communication between parenchymatous and non-parenchymatous cells (such as HSCs, liver sinusoidal endothelial cells, Kupffer cells, or cholangiocytes). Extracellular MiRNA s have been shown to mediate both profibrotic and antifibrotic signalling cascades. For example, hepatitis C virus-infected hepatocytes release EVs containing miR-192 or miR-19a that induce profibrotic TGFβ signalling pathways and activate HSCs[86,87]. Quiescent HSCs release EVs containing miR-214 and miR-199a-5p in order to downregulate fibrogenic pathways in neighbouring activated HSCs and hepatocytes[88,89]. The antifibrotic potential of these EVs is particularly intriguing given that both miR-214 and miR-199a-5p/3p have been known for their profibrotic action[48,90,91].
Compared to the MiRNA s found in the total plasma, EVs can display different miRNA concentrations and even discordant miRNA subsets[92]. Lambrecht et al[93] showed that the same miRNA species can be upregulated in the serum and downregulated in the EVs and suggested that the miRNA signature from circulating EVs reflects the profile found in the vesicles released by activated HSCs in vitro. These discrepancies can also indicate either a higher stability of MiRNA s packaged into EVs against plasma ribonucleases, different intercellular signalling mechanisms[94,95] or could be attributed to the distinct methodology used for miRNA detection and quantification.
Current data on circulating MiRNA s in HBV cirrhosis are limited to host-MiRNA s. The only confirmed HBV-encoded miRNA , HBV-miR-3 is released in the circulation packed in HBV virions and EVs[41]. Given that HBV-miR-3 downregulates the synthesis of HBV virions, it is probable that this miRNA plays a role in the establishment of chronic HBV infection. Hence, the incorporation of HBV-miR-3 into a miRNA diagnostic score could help indicate the contribution of intrahepatic HBV replication to the development of liver inflammation and fibrosis. However, no data are currently available on the role of HBV-miR-3 in HBV-associated liver fibrosis/cirrhosis.
The assessment of miRNA profiles involves the extraction of total RNA and quality control analysis of this purified fraction, followed by their quantification using either reverse-transcription PCR, microarrays, or even next-generation RNA sequencing. Compared to other non-invasive biomarkers, circulating MiRNA s are better able to withstand a low pH, extreme temperatures, ribonucleases, and multiple freeze-thaw cycles[96]. Still, the interpretation of miRNA concentration requires a careful consideration of the methodology, as it depends on the timing of the sample collection, the isolation protocol[97] (e.g., plasma vs serum; MiRNA s in exosomes vs free MiRNA s in plasma or serum) and on the normalization method[98]. Currently, there are various approaches for normalization, including the use of exogenous spike-ins (such as cel-miR-39 from Caenorhabditis elegans), geometrical mean of the quantification cycle for the analysed MiRNA s, and the use of one or more endogenous MiRNA s, small RNAs, or even miRNA /small RNA constructs as reference points[99,100].
One of the most important challenges for the use of circulating MiRNA s in the clinical setting resides in their lack of specificity for a distinct tissue[101]. With few exceptions, such as miR-122, which accounts for an estimated 70% of all MiRNA s in the liver[102], other MiRNA s are less specific for the liver. Moreover, the significance of circulating MiRNA s in chronic liver diseases is complicated by the simultaneous development of the necroinflammatory process and scarring as well as by the potential viral-host interactions. For example serum/plasma miR-122 appears to increase with the progression of the liver necro-inflammatory activity in patients with chronic hepatitis, including those with established liver fibrosis[103], yet it also varies with HBV replication within the liver. Hence, when looking at miRNA expression, a critical interpretation in the clinical context is required.
The circulating MiRNA s associated with liver fibrosis differ between studies, and there is still no consensus on their uses as biomarkers of choice for the diagnosis, staging, or prognosis of liver fibrosis.
The individual expression of MiRNA s in plasma/serum can reach a moderate accuracy for the detection of liver fibrosis. Such an example is miR-29, an antifibrotic miRNA that exhibits an area under the curve between 0.619 to 0.838 in various studies[43,47,69]. The use of multiple MiRNA s or the combination of noncoding RNAs and of other laboratory markers have significantly increased their diagnostic and prognostic accuracy[79,104-106]. Individual MiRNA s and composite scores for liver fibrosis in HBV-infected patients are shown in Table 2 and Table 3. In a meta-analysis by Guo et al[79], the authors identified a panel of eight circulating MiRNA s that could serve as diagnostic markers for liver cirrhosis, irrespective of the viral or nonviral aetiology, displaying an area under the curve of 0.93 (95% confidence interval: 0.91–0.95). Similarly, Murakami et al[91] identified a miRNA score which differentiated between chronic hepatitis B and chronic hepatitis C, non-alcoholic steatohepatitis, and healthy controls with accuracy of 98.35%, 87.5%, or 89.29%, respectively.
| Circulating microRNAs, plasma or serum | Significance for liver disease | Ref. |
| Upregulated microRNAs | ||
| miR-185 | ↑ in advanced (F3-F4) vs early liver fibrosis (F1-F2); and ↑ in early liver fibrosis vs healthy volunteers; No increase with HBV plasma DNA | Li et al[136] |
| miR-2861, miR-345-3p, miR-3620, miR-3656, miR-371a-5p, miR-4646-5p, miR-4651, miR-4695, miR-4800-5p, miR-638 | Individually ↑ in F1-F4 vs F0; Plasma expression differs between each stage of liver fibrosis | Zhang et al[137] |
| miR-1, mR-10b-5p, miR-20b-5p, miR-96b-5p, miR-133b, miR-455-ep, miR-671-5p | Increase in the serum in F3-F4 liver fibrosis | Singh et al[42] |
| miR-499-5p | Increases in the serum in F1-F2 stages | Singh et al[42] |
| miR-106b, miR-181b | Panel for the diagnosis of liver cirrhosis due both HBV and non-HBV associated infection | Chen et al44] |
| Downregulated microRNAs | ||
| miR-29 | ↓ in liver cirrhosis vs healthy controls | Xing et al[138]; Wang et al[106] |
| miR-223 | ↓ with the progression of liver fibrosis from F0-F2 to F3-F4 | Bao et al[43]; Wang et al[106] |
| miR-21 | ||
| miR-143 | ||
| miR-374 | ||
| miR-486-3p, miR-497-5p | Individually ↓ in F1-F4 vs F0; Plasma expression differs between each stage of liver fibrosis | Zhang et al[137] |
| miR-1227-3p | ↓ in the serum in F1-F2 stages | Singh et al[42] |
| Study group | Fibrosis staging/validation method | microRNA detection method/sample1 | Data normalization | microRNA regulation depending on fibrosis staging | microRNA panel for liver fibrosis | Ref. | ||
| 102 treatment naïve CHB | 58 pts F0-F2; 44 pts F3-F4 / liver biopsy and laboratory data | RT-qPCR / serum samples | Spiked-in cel-miR-39 | F3-F4 vs F0-F2 | miR-122, -27b | ↑ | miR-122, -222, PLT, ALP | Appourchaux et al[107], 2016 |
| miR-222, -224 | ↓ | |||||||
| 330 CHB, 165 HC | 165 pts F0-F3; 165 pts F4 / clinical and laboratory data | RT-qPCR / serum samples | Exogenous control using cel-miR-67 | CHB: F4 vs F0-F3 | miR-18a-5p, -21-5p, -29c-3p, -122-5p, -106b-5p, 185-5p | ↓ | Three panels: F4 vs other stages: miR-18a-5p, -21-5p -29c-3p, -122-5p, -106b-5p, 185-5p; F4 vs HC: miR-1, -146a-5p, -451a; CHB vs HC: miR-21-5p, -27a-3p -122-5p, -146a-5p | Jin et al[139], 2015 |
| CHB F4 vs HC | miR-1, -146a-5p | ↑ | ||||||
| miR-451a | ↓ | |||||||
| CHB vs HC | miR-21-5p, -27a-3p, -122-5p, -146a-5p | ↑ | ||||||
| 123 treatment naïve CHB | 69 pts F0-F2 vs 54 staged F3-F4 / liver biopsy | RT-qPCR; Serum samples | Spiked-in cel-miR-39 | F3-F4 vs F0-F2 | miR-29a, -143, -223, -21, -374 | ↓ | miR-29a, -143, -223, PLT | Bao et al[43], 2017 |
| 8 ASC, 8 AVH, 7 HC, 49 treatment naïve CHB | 49 CHB patients: 16 pts F0, 19 pts F1-F2, 14 pts F3-F4 / liver biopsy, clinical and laboratory data | RT-qPCR and microarray analysis; Serum samples | U6 RNA control relative miRNA | F1-F2 | miR-499-5p | ↑ | Analysis of miRNA networks and of individual MiRNA s | Singh et al[42], 2018 |
| miR-1227-3p | ↓ | |||||||
| F3-F4 | miR-34b-3p, -1224-3p, -1, -10b-5p, -20b-5p, -96b-5p, -133, -455-3p, -671-5p | ↑ | ||||||
| 19 CHB, 14 HC | 19 CHB pts with F0-F2 | RT-qPCR total plasma EVs/liver stiffness | Spiked-in cel-miR-39 | Plasma (F0-F2) | miR-192, -200b | ↑ | Expression pattern of each individual miRNA in EVs vs total plasma | Lambrecht et al[93], 2017 |
| EVs (F0-F2) | miR-192, -200b, -92a, -150 | ↓ | ||||||
| 207 CHB, 47 non-HBV-LC, 7 non-CHB, 137 HC | 207 CHB of which: 127 pts F4; 79 pts F0-F3 / liver biopsy | RT-qPCR / plasma samples | miR-1228 control with a log-2 scale transformation | CHB F4 and non-CHB F4; vs other groups; (panel for F4 diagnosis) | miR-106b | ↓ | Panel composed of miR-106 and miR-181b | Chen et al[44], 2013 |
| miR-181b | ↑ | |||||||
| 50 treatment naïve CHB | 10 pts F0, 10 pts F1, 10 pts F2, 10 pts F3, 10 pts F4 / liver biopsy | Microarray analysis / plasma samples | Log standardization of MiRNA s whose target gene expression levels > 2 folds and FDR < 0.05 | F4 vs F0 | miR-2861, -345-3p, -3620-3p, -3656, -371a-5p, -4646-5p, -4651, 4695-5p, -4800-5p, -638, | ↑ | Detailed differential expression of individual MiRNA s for each stage of liver fibrosis F0-F4 | Zhang et al[137], 2015 |
| miR-497-5p, -486-3p | ↓ | |||||||
| 92 CHB | 11 pts F0, 16 pts F1, 12 pts F2, 13 pts F3, 40 pts F4 / liver biopsy and laboratory data | RT-qPCR; Plasma samples | Quanto, EC1, EC2 controls; relative miRNA expression was assessed using 2−ΔΔCq calculation | ≥ F2 | miR-122-5p | ↑ | miR-122-5p, -21-5p, -146a-5p, -223, -29c-3p, -22-3p, -381-3p | Wang et al[106], 2018 |
| miR-223, -29c-3p | ↓ | |||||||
| ≥ F3 | miR-122-5p | ↑ | ||||||
| F4 vs F0 | miR-122-5p, -29c-3p, -146a-5p, -223 | -/↓ | ||||||
Studies on HBV-infected patients have shown that serum/plasma miRNA signatures can assist in the differentiation from other viral or nonviral liver pathologies[93,105,107]. Recently, Shang et al[108] identified a profile of urinary MiRNA s that could serve as diagnostic biomarkers for HBV infection and non-alcoholic steatohepatitis. Nevertheless, there is still insufficient data to recommend a miRNA panel for the specific diagnosis of HBV vs other pathologies. On the other hand, MiRNA s have been associated with a specific HBV immune profile, with the evolution of ne-croinflammatory activity and with the development of chronic liver disease[42,109,110].
MiRNA s can distinguish between early and late fibrosis with a comparable or even higher sensibility and specificity than APRI or Fib-4[43,106,107]. Panels exclusively composed of MiRNA s[106] or panels including both circulating MiRNA s and biological markers (e.g., platelet count and alkaline phosphates) have been evaluated[39,103]. Wang et al[106] showed that a miRNA signature displays a significantly higher accuracy than individual MiRNA s for the detection of moderate and advanced liver fibrosis (area under the curve of 0.90 for stages beyond F2, 0.88 for F3-F4, and 0.83 for F4).
Serum/plasma MiRNA s precede the increase of liver transaminases in studies on acute liver injury[111,112]. Translating this result in patients with chronic hepatitis is nevertheless challenging, due to the persistent elevation of laboratory markers in chronic liver diseases. In HBV-associated liver fibrosis diagnosis, it is important to distinguish between MiRNA s that signal the presence of liver inflammation vs fibrosis, a challenge in practice because both can be present in the progression of chronic livery injury. Examples of circulating MiRNA s that correlate with either liver necroinflammation and/or fibrosis are presented in Table 4. Still, circulating MiRNA s could be used as prognostic markers for survival as well as for the developing risk of HCC in cirrhotic patients, including those with chronic hepatitis B[113,114].
| Circulating microRNA | microRNA regulation | Clinical significance in HBV infection | Ref. |
| miR-122 | ↑ | Correlates with the necroinflammatory activity, HBsAg and HBV DNA; Also correlated with ≥ F2 stage of liver fibrosis | Waidmann et al[103], Ji et al[109], Wang et al[106] |
| miR-210 | ↑ | Marker of necroinflammation; Varies with the severity of HBV hepatitis | Song et al[140] |
| miR-125 (-125a-5p/ -125b) | ↑ | Correlates with HBV intrahepatic replication and necroinflammatory activity | Li et al[141], Zheng et al[134] |
| miR-124 | ↑ | Marker of HBV-associated necroinflammation | Wang et al[142] |
| miR-29 | ↓ | Marker of liver fibrosis irrespective of aetiology | Xing et al[139] |
| miR-223 | ↓ | Marker of liver fibrosis, decreases with the progression to cirrhosis | Bao et al[43] |
| miR-185 | ↑ | Increases in advanced HBV fibrosis; Could play a therapeutic role in HBV gene suppression in tumoral cells | Li et al[136], Fan et al[143] |
miR-122 gradually decreases in the serum of patients with decompensated liver cirrhosis and its value is independently associated with the survival and MELD score[115], while miR-34 indicates the degree of portal hypertension in patients with liver cirrhosis[116]. miRNA scores also yield a satisfactory sensitivity and sensibility for the detection of HCC in patients with cirrhosis due to viral and non-viral aetiologies[117-120]. Various profibrotic MiRNA s, such as miR-21 and miR-221/222, are also well-known oncogenic MiRNA s and regulate tumoral signalling pathways. Circulating miR-21 is associated with the detection of HCC and with the presence of distant metastasis[49,77,121-123]. Similarly, serum miR-221 plays an important role in the growth and proliferation of tumoral cells[124] and appears to be regulated by HBx[125]. In this respect, Huang et al[117] devised a miRNA score that differentiated between HBV- or hepatitis C virus-associated HCC.
Plasma miRNA expression varies in response to antiviral treatment and could provide a promising tool for treatment selection. Van der Ree et al[126] found higher pretreatment levels of miR-301a-3p and miR-145-5p in patients with HBsAg loss, while Yang et al[127] devised a combination of miR-3960 and miR-126-3p that correlated with the clearance of HBsAg.
miRNA panels have also been studied in patients receiving either interferon or nucleoside analogues. Zhang et al[128] constructed a model of 11 MiRNA s for the prediction of an early virological response to an interferon-based regimen, while Brunetto et al[129] defined a miR-B index (combining serum miR-122, miR-99, miR-192, miR-335, miR-126, miR-320) for the prediction of a sustained virological response. Li et al[130] used a miRNA panel composed of miR-22, miR-210, and alanine aminotransaminase to predict the early and sustained virological response but did not find any correlations with HBsAg or HBeAg clearance during a regimen with interferon-alpha.
Further studies on the serum miRNA dynamics during treatment could help establish the correlation between a specific pretreatment miRNA profile and the outcome of the treatment measured as both viral suppression and fibrosis regression.
Cellular and circulating MiRNA s offer a unique glimpse into the intrahepatic development of liver fibrosis and intrahepatic viral replication. Diagnostic and prognostic panels that combine different serum MiRNA s alone or with other biological parameters display a moderately high sensibility and sensitivity compared to validated non-invasive scores. Although current data remain heterogenous, there is growing proof that serum MiRNA s correlate with virologic, immunologic, and fibrotic changes in liver and could become powerful biomarkers during HBV infection.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: European Society of Clinical Microbiology and Infectious Diseases (Fellow).
Specialty type: Gastroenterology and hepatology
Country of origin: Romania
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Esmat SM, Komatsu H, Rezaee-Zavareh MS, Said ZNA S-Editor: Tang JZ L-Editor: A E-Editor: Zhang YL
| 1. | Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2408] [Cited by in RCA: 2409] [Article Influence: 100.4] [Reference Citation Analysis (0)] |
| 2. | Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3501] [Cited by in RCA: 3545] [Article Influence: 147.7] [Reference Citation Analysis (0)] |
| 3. | Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1723] [Cited by in RCA: 1727] [Article Influence: 69.1] [Reference Citation Analysis (0)] |
| 4. | Li C, Hu J, Hao J, Zhao B, Wu B, Sun L, Peng S, Gao GF, Meng S. Competitive virus and host RNAs: the interplay of a hidden virus and host interaction. Protein Cell. 2014;5:348-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Bruscella P, Bottini S, Baudesson C, Pawlotsky JM, Feray C, Trabucchi M. Viruses and MiRNA s: More Friends than Foes. Front Microbiol. 2017;8:824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 158] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 6. | Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1206] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 7. | Auvinen E. Diagnostic and Prognostic Value of MicroRNA in Viral Diseases. Mol Diagn Ther. 2017;21:45-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 8. | Rosca A, Anton G, Botezatu A, Temereanca A, Ene L, Achim C, Ruta S. miR-29a associates with viro-immunological markers of HIV infection in treatment experienced patients. J Med Virol. 2016;88:2132-2137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 9. | World Health Organization. Global hepatitis report, 2017. World Health Organization. Apr 2017. Available from: https://apps.who.int/iris/handle/10665/255016. License: CC BY-NC-SA 3.0 IGO. |
| 10. | Baiocchini A, Montaldo C, Conigliaro A, Grimaldi A, Correani V, Mura F, Ciccosanti F, Rotiroti N, Brenna A, Montalbano M, D'Offizi G, Capobianchi MR, Alessandro R, Piacentini M, Schininà ME, Maras B, Del Nonno F, Tripodi M, Mancone C. Extracellular Matrix Molecular Remodeling in Human Liver Fibrosis Evolution. PLoS One. 2016;11:e0151736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 166] [Article Influence: 18.4] [Reference Citation Analysis (1)] |
| 11. | Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1369] [Article Influence: 97.8] [Reference Citation Analysis (0)] |
| 12. | Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat Rev Immunol. 2014;14:181-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 777] [Cited by in RCA: 1001] [Article Influence: 91.0] [Reference Citation Analysis (0)] |
| 13. | European Association for the Study of the Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2323] [Cited by in RCA: 2400] [Article Influence: 184.6] [Reference Citation Analysis (0)] |
| 14. | Dienstag JL, Goldin RD, Heathcote EJ, Hann HW, Woessner M, Stephenson SL, Gardner S, Gray DF, Schiff ER. Histological outcome during long-term lamivudine therapy. Gastroenterology. 2003;124:105-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 574] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 15. | Hadziyannis SJ, Tassopoulos NC, Heathcote EJ, Chang TT, Kitis G, Rizzetto M, Marcellin P, Lim SG, Goodman Z, Ma J, Brosgart CL, Borroto-Esoda K, Arterburn S, Chuck SL; Adefovir Dipivoxil 438 Study Group. Long-term therapy with adefovir dipivoxil for HBeAg-negative chronic hepatitis B for up to 5 years. Gastroenterology. 2006;131:1743-1751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 680] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 16. | Chang TT, Lai CL, Kew Yoon S, Lee SS, Coelho HS, Carrilho FJ, Poordad F, Halota W, Horsmans Y, Tsai N, Zhang H, Tenney DJ, Tamez R, Iloeje U. Entecavir treatment for up to 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2010;51:422-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 463] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 17. | Marcellin P, Gane E, Buti M, Afdhal N, Sievert W, Jacobson IM, Washington MK, Germanidis G, Flaherty JF, Aguilar Schall R, Bornstein JD, Kitrinos KM, Subramanian GM, McHutchison JG, Heathcote EJ. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet. 2013;381:468-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1228] [Cited by in RCA: 1363] [Article Influence: 113.6] [Reference Citation Analysis (0)] |
| 18. | Boyd A, Bottero J, Miailhes P, Lascoux-Combe C, Rougier H, Girard PM, Serfaty L, Lacombe K. Liver fibrosis regression and progression during controlled hepatitis B virus infection among HIV-HBV patients treated with tenofovir disoproxil fumarate in France: a prospective cohort study. J Int AIDS Soc. 2017;20:21426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 689] [Article Influence: 68.9] [Reference Citation Analysis (0)] |
| 20. | Mu D, Yuan FC, Chen Y, Jiang XY, Yan L, Jiang LY, Gong JP, Zhang DZ, Ren H, Liao Y. Baseline value of intrahepatic HBV DNA over cccDNA predicts patient's response to interferon therapy. Sci Rep. 2017;7:5937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Zhang X, Lu W, Zheng Y, Wang W, Bai L, Chen L, Feng Y, Zhang Z, Yuan Z. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J Clin Invest. 2016;126:1079-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | Bravo AA, Sheth SG, Chopra S. Liver biopsy. N Engl J Med. 2001;344:495-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 1732] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 23. | Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, Zhang R, Chen R, Li T, Zhang T, Yuan Q, Li PC, Huang Q, Colonno R, Jia J, Hou J, McCrae MA, Gao Z, Ren H, Xia N, Zhuang H, Lu F. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. 2016;65:700-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 337] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 24. | Giersch K, Allweiss L, Volz T, Dandri M, Lütgehetmann M. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol. 2017;66:460-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 130] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 25. | Chen EQ, Feng S, Wang ML, Liang LB, Zhou LY, Du LY, Yan LB, Tao CM, Tang H. Serum hepatitis B core-related antigen is a satisfactory surrogate marker of intrahepatic covalently closed circular DNA in chronic hepatitis B. Sci Rep. 2017;7:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 26. | Castera L. Noninvasive methods to assess liver disease in patients with hepatitis B or C. Gastroenterology. 2012;142:1293-1302.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 451] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 27. | World Health Organization. Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. World Health Organization. 2015. Available from: https://apps.who.int/iris/handle/10665/186275. |
| 28. | Lambrecht J, Verhulst S, Mannaerts I, Reynaert H, van Grunsven LA. Prospects in non-invasive assessment of liver fibrosis: Liquid biopsy as the future gold standard? Biochim Biophys Acta Mol Basis Dis. 2018;1864:1024-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 29. | O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne). 2018;9:402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2111] [Cited by in RCA: 3313] [Article Influence: 473.3] [Reference Citation Analysis (0)] |
| 30. | Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol Cell. 2016;64:320-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 332] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 31. | Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 919] [Cited by in RCA: 997] [Article Influence: 83.1] [Reference Citation Analysis (0)] |
| 32. | Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 544] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 33. | Ren M, Qin D, Li K, Qu J, Wang L, Wang Z, Huang A, Tang H. Correlation between hepatitis B virus protein and microRNA processor Drosha in cells expressing HBV. Antiviral Res. 2012;94:225-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 34. | Novellino L, Rossi RL, Bonino F, Cavallone D, Abrignani S, Pagani M, Brunetto MR. Circulating hepatitis B surface antigen particles carry hepatocellular microRNAs. PLoS One. 2012;7:e31952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 35. | Hayes CN, Akamatsu S, Tsuge M, Miki D, Akiyama R, Abe H, Ochi H, Hiraga N, Imamura M, Takahashi S, Aikata H, Kawaoka T, Kawakami Y, Ohishi W, Chayama K. Hepatitis B virus-specific MiRNA s and Argonaute2 play a role in the viral life cycle. PLoS One. 2012;7:e47490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 36. | Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003-5008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2629] [Article Influence: 187.8] [Reference Citation Analysis (0)] |
| 37. | Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating MiRNA s: cell-cell communication function? Front Genet. 2013;4:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 288] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 38. | Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442-17452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1582] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 39. | Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110-E2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1064] [Cited by in RCA: 1253] [Article Influence: 96.4] [Reference Citation Analysis (0)] |
| 40. | Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, Habbel P, Kälin R, Franzoni E, Rybak A, Nguyen D, Veh R, Ninnemann O, Peters O, Nitsch R, Heppner FL, Golenbock D, Schott E, Ploegh HL, Wulczyn FG, Lehnardt S. An unconventional role for miRNA : let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 602] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 41. | Yang X, Li H, Sun H, Fan H, Hu Y, Liu M, Li X, Tang H. Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication. J Virol. 2017;91:pii: e01919-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 42. | Singh AK, Rooge SB, Varshney A, Vasudevan M, Bhardwaj A, Venugopal SK, Trehanpati N, Kumar M, Geffers R, Kumar V, Sarin SK. Global microRNA expression profiling in the liver biopsies of hepatitis B virus-infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology. 2018;67:1695-1709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 43. | Bao S, Zheng J, Li N, Huang C, Chen M, Cheng Q, Yu K, Chen S, Zhu M, Shi G. Serum MicroRNA Levels as a Noninvasive Diagnostic Biomarker for the Early Diagnosis of Hepatitis B Virus-Related Liver Fibrosis. Gut Liver. 2017;11:860-869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 44. | Chen YJ, Zhu JM, Wu H, Fan J, Zhou J, Hu J, Yu Q, Liu TT, Yang L, Wu CL, Guo XL, Huang XW, Shen XZ. Circulating microRNAs as a Fingerprint for Liver Cirrhosis. PLoS One. 2013;8:e66577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 45. | Guo CJ, Pan Q, Cheng T, Jiang B, Chen GY, Li DG. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009;276:5163-5176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 92] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 46. | Coll M, El Taghdouini A, Perea L, Mannaerts I, Vila-Casadesús M, Blaya D, Rodrigo-Torres D, Affò S, Morales-Ibanez O, Graupera I, Lozano JJ, Najimi M, Sokal E, Lambrecht J, Ginès P, van Grunsven LA, Sancho-Bru P. Integrative miRNA and Gene Expression Profiling Analysis of Human Quiescent Hepatic Stellate Cells. Sci Rep. 2015;5:11549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 47. | Wang J, Chu ES, Chen HY, Man K, Go MY, Huang XR, Lan HY, Sung JJ, Yu J. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget. 2015;6:7325-7338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 148] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 48. | Ma L, Yang X, Wei R, Ye T, Zhou JK, Wen M, Men R, Li P, Dong B, Liu L, Fu X, Xu H, Aqeilan RI, Wei YQ, Yang L, Peng Y. MicroRNA-214 promotes hepatic stellate cell activation and liver fibrosis by suppressing Sufu expression. Cell Death Dis. 2018;9:718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 49. | You K, Li SY, Gong J, Fang JH, Zhang C, Zhang M, Yuan Y, Yang J, Zhuang SM. MicroRNA-125b Promotes Hepatic Stellate Cell Activation and Liver Fibrosis by Activating RhoA Signaling. Mol Ther Nucleic Acids. 2018;12:57-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 50. | Gupta P, Sata TN, Yadav AK, Mishra A, Vats N, Hossain MM, Sanal MG, Venugopal SK. TGF-β induces liver fibrosis via miRNA -181a-mediated down regulation of augmenter of liver regeneration in hepatic stellate cells. PLoS One. 2019;14:e0214534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 51. | Zhang Y, Ghazwani M, Li J, Sun M, Stolz DB, He F, Fan J, Xie W, Li S. MiR-29b inhibits collagen maturation in hepatic stellate cells through down-regulating the expression of HSP47 and lysyl oxidase. Biochem Biophys Res Commun. 2014;446:940-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 52. | Zhao J, Tang N, Wu K, Dai W, Ye C, Shi J, Zhang J, Ning B, Zeng X, Lin Y. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLoS One. 2014;9:e108005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 53. | Iizuka M, Ogawa T, Enomoto M, Motoyama H, Yoshizato K, Ikeda K, Kawada N. Induction of microRNA-214-5p in human and rodent liver fibrosis. Fibrogenesis Tissue Repair. 2012;5:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 54. | Meng F, Glaser SS, Francis H, Yang F, Han Y, Stokes A, Staloch D, McCarra J, Liu J, Venter J, Zhao H, Liu X, Francis T, Swendsen S, Liu CG, Tsukamoto H, Alpini G. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am J Pathol. 2012;181:804-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 55. | Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705-714, 714.e1-714.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 321] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 56. | Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 252] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 57. | Wang Y, Zhu P, Qiu J, Wang J, Zhu H, Zhu Y, Zhang L, Zhu J, Liu X, Dong C. Identification and characterization of interferon signaling-related microRNAs in occult hepatitis B virus infection. Clin Epigenetics. 2017;9:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 58. | Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 529] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 59. | Ye D, Zhang T, Lou G, Liu Y. Role of miR-223 in the pathophysiology of liver diseases. Exp Mol Med. 2018;50:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 60. | Yang Z, Peng Y, Yang S. MicroRNA-146a regulates the transformation from liver fibrosis to cirrhosis in patients with hepatitis B via interleukin-6. Exp Ther Med. 2019;17:4670-4676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 61. | Li LM, Wang D, Zen K. MicroRNAs in Drug-induced Liver Injury. J Clin Transl Hepatol. 2014;2:162-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 62. | Pirola CJ, Fernández Gianotti T, Castaño GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, Mirshahi F, Sanyal AJ, Sookoian S. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 442] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 63. | Yang L, Dong C, Yang J, Yang L, Chang N, Qi C, Li L. MicroRNA-26b-5p Inhibits Mouse Liver Fibrogenesis and Angiogenesis by Targeting PDGF Receptor-Beta. Mol Ther Nucleic Acids. 2019;16:206-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 64. | Li X, Chen Y, Wu S, He J, Lou L, Ye W, Wang J. microRNA-34a and microRNA-34c promote the activation of human hepatic stellate cells by targeting peroxisome proliferator-activated receptor γ. Mol Med Rep. 2015;11:1017-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 65. | Tian XF, Ji FJ, Zang HL, Cao H. Activation of the miR-34a/SIRT1/p53 Signaling Pathway Contributes to the Progress of Liver Fibrosis via Inducing Apoptosis in Hepatocytes but Not in HSCs. PLoS One. 2016;11:e0158657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 66. | Feili X, Wu S, Ye W, Tu J, Lou L. MicroRNA-34a-5p inhibits liver fibrosis by regulating TGF-β1/Smad3 pathway in hepatic stellate cells. Cell Biol Int. 2018;42:1370-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 67. | Zheng J, Wu C, Xu Z, Xia P, Dong P, Chen B, Yu F. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol Cell Biochem. 2015;398:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 68. | Wei J, Feng L, Li Z, Xu G, Fan X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother. 2013;67:387-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 69. | Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M, Tacke F, Trautwein C, Luedde T. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 656] [Cited by in RCA: 658] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 70. | Zhang Y, Wu L, Wang Y, Zhang M, Li L, Zhu D, Li X, Gu H, Zhang CY, Zen K. Protective role of estrogen-induced miRNA -29 expression in carbon tetrachloride-induced mouse liver injury. J Biol Chem. 2012;287:14851-14862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 71. | Jing F, Geng Y, Xu XY, Xu HY, Shi JS, Xu ZH. MicroRNA29a Reverts the Activated Hepatic Stellate Cells in the Regression of Hepatic Fibrosis through Regulation of ATPase H⁺ Transporting V1 Subunit C1. Int J Mol Sci. 2019;20:pii: E796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 72. | Matsumoto Y, Itami S, Kuroda M, Yoshizato K, Kawada N, Murakami Y. MiR-29a Assists in Preventing the Activation of Human Stellate Cells and Promotes Recovery From Liver Fibrosis in Mice. Mol Ther. 2016;24:1848-1859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 73. | Zou Y, Cai Y, Lu D, Zhou Y, Yao Q, Zhang S. MicroRNA-146a-5p attenuates liver fibrosis by suppressing profibrogenic effects of TGFβ1 and lipopolysaccharide. Cell Signal. 2017;39:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 74. | Zheng J, Lin Z, Dong P, Lu Z, Gao S, Chen X, Wu C, Yu F. Activation of hepatic stellate cells is suppressed by microRNA-150. Int J Mol Med. 2013;32:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 75. | Venugopal SK, Jiang J, Kim TH, Li Y, Wang SS, Torok NJ, Wu J, Zern MA. Liver fibrosis causes downregulation of miRNA -150 and miRNA -194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am J Physiol Gastrointest Liver Physiol. 2010;298:G101-G106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 76. | Ding W, Xin J, Jiang L, Zhou Q, Wu T, Shi D, Lin B, Li L, Li J. Characterisation of peripheral blood mononuclear cell microRNA in hepatitis B-related acute-on-chronic liver failure. Sci Rep. 2015;5:13098. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 77. | Mirzaei HR, Sahebkar A, Mohammadi M, Yari R, Salehi H, Jafari MH, Namdar A, Khabazian E, Jaafari MR, Mirzaei H. Circulating microRNAs in Hepatocellular Carcinoma: Potential Diagnostic and Prognostic Biomarkers. Curr Pharm Des. 2016;22:5257-5269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 127] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 78. | Torres JL, Novo-Veleiro I, Manzanedo L, Alvela-Suárez L, Macías R, Laso FJ, Marcos M. Role of microRNAs in alcohol-induced liver disorders and non-alcoholic fatty liver disease. World J Gastroenterol. 2018;24:4104-4118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 65] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (2)] |
| 79. | Guo L, Li W, Hu L, Zhou H, Zheng L, Yu L, Liang W. Diagnostic value of circulating microRNAs for liver cirrhosis: a meta-analysis. Oncotarget. 2018;9:5397-5405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 80. | Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol. 2015;21:9853-9862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 58] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 81. | Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, Cai F, Ma L, Yu Y. The Role of MicroRNAs in Hepatocellular Carcinoma. J Cancer. 2018;9:3557-3569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 82. | Loosen SH, Schueller F, Trautwein C, Roy S, Roderburg C. Role of circulating microRNAs in liver diseases. World J Hepatol. 2017;9:586-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 83. | Nojima H, Freeman CM, Schuster RM, Japtok L, Kleuser B, Edwards MJ, Gulbins E, Lentsch AB. Hepatocyte exosomes mediate liver repair and regeneration via sphingosine-1-phosphate. J Hepatol. 2016;64:60-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 227] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 84. | Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014;10:e1004424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 277] [Cited by in RCA: 339] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 85. | Chen L, Chen R, Kemper S, Brigstock DR. Pathways of production and delivery of hepatocyte exosomes. J Cell Commun Signal. 2018;12:343-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 86. | Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB. Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 131] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 87. | Kim JH, Lee CH, Lee SW. Exosomal Transmission of MicroRNA from HCV Replicating Cells Stimulates Transdifferentiation in Hepatic Stellate Cells. Mol Ther Nucleic Acids. 2019;14:483-497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 88. | Chen L, Chen R, Kemper S, Charrier A, Brigstock DR. Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1. Am J Physiol Gastrointest Liver Physiol. 2015;309:G491-G499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 118] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 89. | Chen L, Chen R, Velazquez VM, Brigstock DR. Fibrogenic Signaling Is Suppressed in Hepatic Stellate Cells through Targeting of Connective Tissue Growth Factor (CCN2) by Cellular or Exosomal MicroRNA-199a-5p. Am J Pathol. 2016;186:2921-2933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 90. | Lino Cardenas CL, Henaoui IS, Courcot E, Roderburg C, Cauffiez C, Aubert S, Copin MC, Wallaert B, Glowacki F, Dewaeles E, Milosevic J, Maurizio J, Tedrow J, Marcet B, Lo-Guidice JM, Kaminski N, Barbry P, Luedde T, Perrais M, Mari B, Pottier N. miR-199a-5p Is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS Genet. 2013;9:e1003291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 194] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 91. | Murakami Y, Toyoda H, Tanaka M, Kuroda M, Harada Y, Matsuda F, Tajima A, Kosaka N, Ochiya T, Shimotohno K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One. 2011;6:e16081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 238] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 92. | Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA. 2014;111:14888-14893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 680] [Cited by in RCA: 860] [Article Influence: 78.2] [Reference Citation Analysis (0)] |
| 93. | Lambrecht J, Jan Poortmans P, Verhulst S, Reynaert H, Mannaerts I, van Grunsven LA. Circulating ECV-Associated MiRNA s as Potential Clinical Biomarkers in Early Stage HBV and HCV Induced Liver Fibrosis. Front Pharmacol. 2017;8:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 94. | Yang J, Li C, Zhang L, Wang X. Extracellular Vesicles as Carriers of Non-coding RNAs in Liver Diseases. Front Pharmacol. 2018;9:415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 95. | Szabo G, Momen-Heravi F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat Rev Gastroenterol Hepatol. 2017;14:455-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 219] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 96. | Ishikawa H, Yamada H, Taromaru N, Kondo K, Nagura A, Yamazaki M, Ando Y, Munetsuna E, Suzuki K, Ohashi K, Teradaira R. Stability of serum high-density lipoprotein-microRNAs for preanalytical conditions. Ann Clin Biochem. 2017;54:134-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 97. | Ramón-Núñez LA, Martos L, Fernández-Pardo Á, Oto J, Medina P, España F, Navarro S. Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS One. 2017;12:e0187005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 98. | Binderup HG, Madsen JS, Heegaard NHH, Houlind K, Andersen RF, Brasen CL. Quantification of microRNA levels in plasma - Impact of preanalytical and analytical conditions. PLoS One. 2018;13:e0201069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 99. | Faraldi M, Gomarasca M, Sansoni V, Perego S, Banfi G, Lombardi G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci Rep. 2019;9:1584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 119] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 100. | Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data Normalization Strategies for MicroRNA Quantification. Clin Chem. 2015;61:1333-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 364] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 101. | Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E, Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865-3877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 593] [Cited by in RCA: 756] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 102. | Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 657] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 103. | Waidmann O, Bihrer V, Pleli T, Farnik H, Berger A, Zeuzem S, Kronenberger B, Piiper A. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J Viral Hepat. 2012;19:e58-e65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 104. | Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, Kosaka N, Ochiya T, Taguchi YH. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS One. 2012;7:e48366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 105. | Yamaura Y, Tatsumi N, Takagi S, Tokumitsu S, Fukami T, Tajiri K, Minemura M, Yokoi T, Nakajima M. Serum microRNA profiles in patients with chronic hepatitis B, chronic hepatitis C, primary biliary cirrhosis, autoimmune hepatitis, nonalcoholic steatohepatitis, or drug-induced liver injury. Clin Biochem. 2017;50:1034-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 106. | Wang TZ, Lin DD, Jin BX, Sun XY, Li N. Plasma microRNA: A novel non-invasive biomarker for HBV-associated liver fibrosis staging. Exp Ther Med. 2019;17:1919-1929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 107. | Appourchaux K, Dokmak S, Resche-Rigon M, Treton X, Lapalus M, Gattolliat CH, Porchet E, Martinot-Peignoux M, Boyer N, Vidaud M, Bedossa P, Marcellin P, Bièche I, Estrabaud E, Asselah T. MicroRNA-based diagnostic tools for advanced fibrosis and cirrhosis in patients with chronic hepatitis B and C. Sci Rep. 2016;6:34935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 108. | Shang JW, Yan XL, Zhang H, Su SB. Expression and significance of urinary microRNA in patients with chronic hepatitis B. Medicine (Baltimore). 2019;98:e17143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 109. | Ji F, Yang B, Peng X, Ding H, You H, Tien P. Circulating microRNAs in hepatitis B virus-infected patients. J Viral Hepat. 2011;18:e242-e251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 110. | Xing T, Xu H, Yu W, Wang B, Zhang J. Expression profile and clinical significance of MiRNA s at different stages of chronic hepatitis B virus infection. Int J Clin Exp Med. 2015;8:5611-5620. [PubMed] |
| 111. | Antoine DJ, Dear JW, Lewis PS, Platt V, Coyle J, Masson M, Thanacoody RH, Gray AJ, Webb DJ, Moggs JG, Bateman DN, Goldring CE, Park BK. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58:777-787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 321] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 112. | Park HK, Jo W, Choi HJ, Jang S, Ryu JE, Lee HJ, Lee H, Kim H, Yu ES, Son WC. Time-course changes in the expression levels of miR-122, -155, and -21 as markers of liver cell damage, inflammation, and regeneration in acetaminophen-induced liver injury in rats. J Vet Sci. 2016;17:45–51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 113. | Wen Y, Han J, Chen J, Dong J, Xia Y, Liu J, Jiang Y, Dai J, Lu J, Jin G, Han J, Wei Q, Shen H, Sun B, Hu Z. Plasma MiRNA s as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137:1679-1690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 172] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 114. | Jin Y, Wong YS, Goh BKP, Chan CY, Cheow PC, Chow PKH, Lim TKH, Goh GBB, Krishnamoorthy TL, Kumar R, Ng TP, Chong SS, Tan HH, Chung AYF, Ooi LLPJ, Chang JPE, Tan CK, Lee CGL. Circulating microRNAs as Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Sci Rep. 2019;9:10464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 115. | Waidmann O, Köberle V, Brunner F, Zeuzem S, Piiper A, Kronenberger B. Serum microRNA-122 predicts survival in patients with liver cirrhosis. PLoS One. 2012;7:e45652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 116. | Jansen C, Eischeid H, Goertzen J, Schierwagen R, Anadol E, Strassburg CP, Sauerbruch T, Odenthal M, Trebicka J. The role of miRNA -34a as a prognostic biomarker for cirrhotic patients with portal hypertension receiving TIPS. PLoS One. 2014;9:e103779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 117. | Huang YH, Liang KH, Chien RN, Hu TH, Lin KH, Hsu CW, Lin CL, Pan TL, Ke PY, Yeh CT. A Circulating MicroRNA Signature Capable of Assessing the Risk of Hepatocellular Carcinoma in Cirrhotic Patients. Sci Rep. 2017;7:523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 118. | Giray BG, Emekdas G, Tezcan S, Ulger M, Serin MS, Sezgin O, Altintas E, Tiftik EN. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol Biol Rep. 2014;41:4513-4519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (1)] |
| 119. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 120. | Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, Lin D. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 449] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 121. | Bandopadhyay M, Banerjee A, Sarkar N, Panigrahi R, Datta S, Pal A, Singh SP, Biswas A, Chakrabarti S, Chakravarty R. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer. 2014;14:721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 122. | Guo X, Lv X, Lv X, Ma Y, Chen L, Chen Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget. 2017;8:44050-44058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 123. | Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K, Kanto T, Doki Y, Mori M. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 284] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 124. | Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406:70-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 125. | Chen JJ, Tang YS, Huang SF, Ai JG, Wang HX, Zhang LP. HBx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBVrelated hepatocellular carcinoma by targeting estrogen receptor-α. Oncol Rep. 2015;33:792-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 126. | van der Ree MH, Jansen L, Kruize Z, van Nuenen AC, van Dort KA, Takkenberg RB, Reesink HW, Kootstra NA. Plasma MicroRNA Levels Are Associated With Hepatitis B e Antigen Status and Treatment Response in Chronic Hepatitis B Patients. J Infect Dis. 2017;215:1421-1429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 127. | Yang Y, Liu M, Deng Y, Guo Y, Zhang X, Xiang D, Jiang L, You Z, Wu Y, Li M, Mao Q. Pretreatment microRNA levels can predict HBsAg clearance in CHB patients treated with pegylated interferon α-2a. Virol J. 2018;15:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 128. | Zhang X, Chen C, Wu M, Chen L, Zhang J, Zhang X, Zhang Z, Wu J, Wang J, Chen X, Huang T, Chen L, Yuan Z. Plasma microRNA profile as a predictor of early virological response to interferon treatment in chronic hepatitis B patients. Antivir Ther. 2012;17:1243-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 129. | Brunetto MR, Cavallone D, Oliveri F, Moriconi F, Colombatto P, Coco B, Ciccorossi P, Rastelli C, Romagnoli V, Cherubini B, Teilum MW, Blondal T, Bonino F. A serum microRNA signature is associated with the immune control of chronic hepatitis B virus infection. PLoS One. 2014;9:e110782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 130. | Li J, Zhang X, Chen L, Zhang Z, Zhang J, Wang W, Wu M, Shi B, Zhang X, Kozlowski M, Hu Y, Yuan Z. Circulating miR-210 and miR-22 combined with ALT predict the virological response to interferon-alpha therapy of CHB patients. Sci Rep. 2017;7:15658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 131. | Zhang H, Yan XL, Guo XX, Shi MJ, Lu YY, Zhou QM, Chen QL, Hu YY, Xu LM, Huang S, Su SB. MiR-27a as a predictor for the activation of hepatic stellate cells and hepatitis B virus-induced liver cirrhosis. Oncotarget. 2018;9:1075-1090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 132. | Yu F, Zhou G, Li G, Chen B, Dong P, Zheng J. Serum miR-181b Is Correlated with Hepatitis B Virus Replication and Disease Progression in Chronic Hepatitis B Patients. Dig Dis Sci. 2015;60:2346-2352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 133. | Akamatsu S, Hayes CN, Tsuge M, Miki D, Akiyama R, Abe H, Ochi H, Hiraga N, Imamura M, Takahashi S, Aikata H, Kawaoka T, Kawakami Y, Ohishi W, Chayama K. Differences in serum microRNA profiles in hepatitis B and C virus infection. J Infect. 2015;70:273–287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 134. | Zheng J, Zhou Z, Xu Z, Li G, Dong P, Chen Z, Lin D, Chen B, Yu F. Serum microRNA-125a-5p, a useful biomarker in liver diseases, correlates with disease progression. Mol Med Rep. 2015;12:1584-1590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 135. | Wang W, Liu R, Su Y, Li H, Xie W, Ning B. MicroRNA-21-5p mediates TGF-β-regulated fibrogenic activation of spinal fibroblasts and the formation of fibrotic scars after spinal cord injury. Int J Biol Sci. 2018;14:178-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 136. | Li BB, Li DL, Chen C, Liu BH, Xia CY, Wu HJ, Wu CQ, Ji GQ, Liu S, Ni W, Yao DK, Zeng ZY, Chen DG, Qin BD, Xin X, Yan GL, Dan Tang, Liu HM, He J, Yan H, Zhu WJ, Yu HY, Zhu L. Potentials of the elevated circulating miR-185 level as a biomarker for early diagnosis of HBV-related liver fibrosis. Sci Rep. 2016;6:34157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 137. | Zhang Q, Xu M, Qu Y, Li Z, Zhang Q, Cai X, Lu L. Analysis of the differential expression of circulating microRNAs during the progression of hepatic fibrosis in patients with chronic hepatitis B virus infection. Mol Med Rep. 2015;12:5647-5654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 138. | Xing TJ, Jiang DF, Huang JX, Xu ZL. Expression and clinical significance of miR-122 and miR-29 in hepatitis B virus-related liver disease. Genet Mol Res. 2014;13:7912-7918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 139. | Jin BX, Zhang YH, Jin WJ, Sun XY, Qiao GF, Wei YY, Sun LB, Zhang WH, Li N. MicroRNA panels as disease biomarkers distinguishing hepatitis B virus infection caused hepatitis and liver cirrhosis. Sci Rep. 2015;5:15026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 140. | Song G, Jia H, Xu H, Liu W, Zhu H, Li S, Shi J, Li Z, He J, Chen Z. Studying the association of microRNA-210 level with chronic hepatitis B progression. J Viral Hepat. 2014;21:272-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 141. | Li F, Zhou P, Deng W, Wang J, Mao R, Zhang Y, Li J, Yu J, Yang F, Huang Y, Lu M, Zhang J. Serum microRNA-125b correlates with hepatitis B viral replication and liver necroinflammation. Clin Microbiol Infect. 2016;22:384.e1-384.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 142. | Wang JY, Mao RC, Zhang YM, Zhang YJ, Liu HY, Qin YL, Lu MJ, Zhang JM. Serum microRNA-124 is a novel biomarker for liver necroinflammation in patients with chronic hepatitis B virus infection. J Viral Hepat. 2015;22:128-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 143. | Fan HX, Feng YJ, Zhao XP, He YZ, Tang H. MiR-185-5p suppresses HBV gene expression by targeting ELK1 in hepatoma carcinoma cells. Life Sci. 2018;213:9-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |