Published online Sep 7, 2019. doi: 10.3748/wjg.v25.i33.4921
Peer-review started: April 25, 2019
First decision: May 30, 2019
Revised: June 29, 2019
Accepted: July 19, 2019
Article in press: July 19, 2019
Published online: September 7, 2019
Processing time: 135 Days and 20.4 Hours
The potential role of chronic inflammation in the development of cancer has been widely recognized. However, there has been little research fully and thoroughly exploring the molecular link between hepatitis B virus (HBV) and hepatocellular carcinoma (HCC).
To elucidate the molecular links between HBV and HCC through analyzing the molecular processes of HBV-HCC using a multidimensional approach.
First, maladjusted genes shared between HBV and HCC were identified by disease-related differentially expressed genes. Second, the protein-protein interaction network based on dysfunctional genes identified a series of dysfunctional modules and significant crosstalk between modules based on the hypergeometric test. In addition, key regulators were detected by pivot analysis. Finally, targeted drugs that have regulatory effects on diseases were predicted by modular methods and drug target information.
The study found that 67 genes continued to increase in the HBV-HCC process. Moreover, 366 overlapping genes in the module network participated in multiple functional blocks. It could be presumed that these genes and their interactions play an important role in the relationship between inflammation and cancer. Correspondingly, significant crosstalk constructed a module level bridge for HBV-HCC molecular processes. On the other hand, a series of non-coding RNAs and transcription factors that have potential pivot regulatory effects on HBV and HCC were identified. Among them, some of the regulators also had persistent disorders in the process of HBV-HCC including microRNA-192, microRNA-215, and microRNA-874, and early growth response 2, FOS, and Kruppel-like factor 4. Therefore, the study concluded that these pivots are the key bridge molecules outside the module. Last but not least, a variety of drugs that may have some potential pharmacological or toxic side effects on HBV-induced HCC were predicted, but their mechanisms still need to be further explored.
The results suggest that the persistent inflammatory environment of HBV can be utilized as an important risk factor to induce the occurrence of HCC, which is supported by molecular evidence.
Core tip: The potential role of chronic inflammation in the development of cancer has been widely recognized. However, the molecular link between hepatitis B virus (HBV) and hepatocellular carcinoma (HCC) has not been fully and thoroughly explored. Therefore, this study analyzed the molecular processes of HBV-HCC using a multidimensional approach to elucidate the molecular links between the two groups. The results suggest that the persistent inflammatory environment of HBV can be used as an important risk factor to induce the occurrence of HCC, which is supported by molecular evidence.
- Citation: Huang XB, He YG, Zheng L, Feng H, Li YM, Li HY, Yang FX, Li J. Identification of hepatitis B virus and liver cancer bridge molecules based on functional module network. World J Gastroenterol 2019; 25(33): 4921-4932
- URL: https://www.wjgnet.com/1007-9327/full/v25/i33/4921.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i33.4921
Epidemiological research has shown that chronic low levels of inflammation can significantly increase the risk of cancer[1]. A series of genes including inflammatory molecules and transcription factors (TFs), adhesion molecules, AP-1, chemokines, C-reactive protein and enzymes are involved in inflammation, which have crucial impacts on inflammatory-mediated tumors[2]. In the process of chronic inflammation caused by virus infection, abnormal long-term expression of related proteins may induce physiological disorders such as oxidative stress and inflammation in tissues and organs. Thereby, a potential carcinogenic microenvironment has been formed within it, and different functions are exerted in different stages of cancer development[3]. On the other hand, the occurrence and development of tumors also affect inflammatory response processes. Many types of cancer can change the secretion levels of chemokines and inflammatory cytokines in the microenvironment, which is conducive to promoting immune escape in cancer[4,5]. Specifically, chronic hepatitis B virus (HBV) infection seriously threatens human health, which is one of the most common infectious diseases in the world, and has become a public health problem worldwide[6]. Long-term infection of HBV has the possibility of inducing liver failure, cirrhosis, and liver cancer[7]. The key mechanism is that viral DNA is integrated into the genome of host cells to alter the genetic mechanism and gene expression of host cells[8]. Studies have shown that the large surface of HBV surface antigen can induce DNA damage and polo-like kinase 1-mediated cell cycle G2/M cell division failure, which leads to unstable reproductive cycle of chromatin to drive the development of hepatocellular carcinoma (HCC)[9]. In addition, protein 4 (VSIG4) with immunoglobulin domain contains VSIG4 has poor prognosis in patients with HBV-positive HCC, but has no predictive significance in patients with HBV-negative HCC[10]. This indicates that HBV infection not only affects the occurrence of HCC, but also affects its development, and has a negative effect on the prognosis of patients. Therefore, a systematic and in-depth understanding of the potential molecular links between HBV and HCC is essential for the exploration of the mechanism of HBV-induced HCC process and the development of targeted therapies. On the other hand, HCC is one of the most common cancers and has a higher mortality. Although the treatment of HCC has improved in the past few decades, the survival rate of patients is still very low[11]. Accumulating evidence has indicated that liver cancer is a complex disease with multiple factors and steps. In terms of risk factors, chronic persistent infection of hepatitis C virus or HBV, chronic untreated hepatitis inflammation with different etiologies, oxidative stress, and fatty liver disease may lead to the occurrence of HCC[12]. From the molecular mechanism, the increased expression of A-Raf and fatty acid 2-hydrolase (FA2H) in HCC cells leads to lipid metabolism disorder and promotes the development of cancer[13]. However, in drug sensitivity tests, overexpression of FA2H also increases the drug sensitivity of human colorectal and cervical cancer cells, while silencing FA2H makes the cells resistant to the drug[14]. Furthermore, studies have suggested that arginase 1 (ARG1) can participate in the proliferation of HBV-specific CD8 (+) T cells and regulate the occurrence of HBV[15]. At the same time, ARG1 may also promote the epithelial to mesenchymal transition process by upregulating Vimentin, N-cadherin, and beta-catenin, thus mediating the development and invasion of HCC[16]. Therefore, it is speculated that it is the key molecule in the HBV-HCC process, which needs further exploration. On the other hand, NOP7 interacts with beta-catenin to activate the inflammatory signaling pathway of beta-catenin/TCF, and its upregulation promotes the proliferation and migration of HCC cancer cells[17]. To some extent, these results indicate that HBV may mediate the occurrence and development of HCC, and guide a comprehensive and in-depth discussion on the bridge mechanism between them.
The study explored the co-imbalance bridging molecules between HBV and HCC and their potential drugs based on the dysfunction module. The results not only help to clarify the potential molecular links between HBV and HCC, but also provide biologists with abundant candidate resources for further research.
The National Center for Biotechnology Information Gene contains numerous published results on HBV. To systematically analyze the molecular links between HBV and HCC, 128 expression profiles of HBV-related RNA (GSE83148) and 16 microRNAs (miRNAs) (GSE33857) were collected from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). All of these were assessed using Affymetrix Human Genome U133 Plus 2.0 Array, including normal and disease samples. Subsequently, 424 RNA-seq data (original count) and 850 miRNA expression profile data of HCC-related genes were collected from The Cancer Genome Atlas (TCGA) database.
In this study, differences in the expression of RNA and miRNAs in both disease and normal samples were calculated using the R-language limma package. For chip data, we first used the background correct function for background correction and standardization. Then the control probes and low-expression probes were filtered out to obtain high-quality standardized data based on the quantile normalization method of normalizing Between Array function. For RNA-seq gene expression data, the voom function was utilized to standardize reads counts. Finally, these standardized chips and RNA-seq data were analyzed by using lmFit and eBayes functions with default parameters, and the differentially expressed genes (DEGs) of HBV and HCC were screened by R language limma package, with a screening threshold P value < 0.01. The DEGs of hepatitis B and HCC were screened for logFC > 1 and logFC < 1, respectively.
The database STRING (a search tool for retrieving interacting genes/proteins) is specially designed for protein-protein interaction (PPI). It provides the most comprehensive view of the current most complete PPI, so it can be used as a metadata base for extensive PPI analysis. All human protein interaction data in this study were derived from STRING data, involving 405916 interaction pairs of 10514 proteins. Then the inflammation and cancer DEGs were mapped onto the PPI network, and the maximum connected component was obtained. Based on the maximum connected component generated above, we used the perfect MCODE method with default parameters to identify the functional modules related to inflammation and cancer. Cytoscape 3.6.1 network visualization software and ClusterONE algorithm were used to select modules with node degree > 50, and 16 modules were obtained
The roles of HBV- and HCC-related genes in the pathogenesis of HBV and HCC are intricate, and there are innumerable links between them. Correspondingly, the functions of the modules are also rich and colorful, and the interactions between modules are intricate as well. In order to clarify the interactions between modules and build a bridge between HBV and HCC at the module level, we used human PPI information as a background set to conduct comprehensive crosstalk analysis of all modules to further understand the interaction mechanism of co-expression modules between HBV and HCC diseases. First, based on the hypothesis that the crosstalk between functional modules is significant when the number of interactions is significantly greater than the random distribution, we constructed 1000 random PPI networks with a network size and degree of each node unchanged. Subsequently, for each pair of modules between HBV and HCC, we compared the actual number of interactions with the random distribution extracted from 1000 random PPI networks. According to the computational rules, the number of interaction pairs between modules is larger than the interaction pairs under random background. These interactions are called crosstalk. The method of calculating significant crosstalk was as follows: First, under the background of random network, the number of interaction pairs between modules in N random networks was larger than that in real networks, and the number of interaction pairs between modules was counted as n. Then the formula for calculating p value was P = n/N (in this study, N = 1000). When P ≤ 0.05, it can be considered that these crosstalk modules are more significant than random ones. Finally, Cytoscape was utilized to elucidate the significant crosstalk to intuitively observe the complex regulatory relationship between co-expression modules.
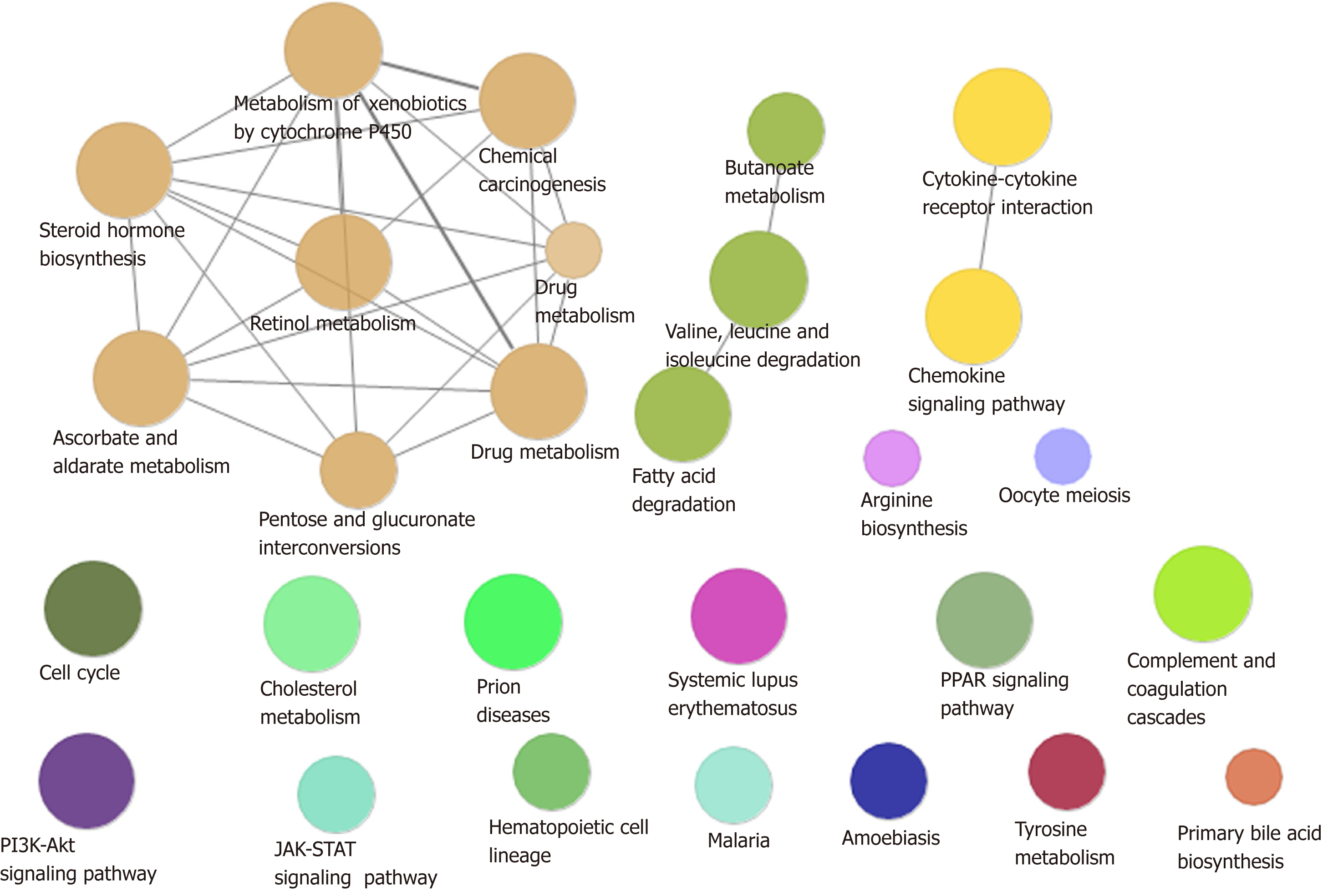
Functions and signaling pathways are often important mediators of genes and diseases, and the study of them is often an effective means to explore the molecular pathways and potential mechanisms of diseases. Therefore, enrichment analysis of Gene Ontology (GO) function (P value cutoff = 0.01, q value cutoff = 0.01) and KEGG pathway (P value cutoff = 0.01, q value cutoff = 0.01) was carried out for all modules related to HBV and HCC using R language Cluster profiler package, respectively. Subsequently, we extracted the functions and pathways involved in both HBV and HCC, and considered them to be the molecular bridges between the two diseases at the levels of function and pathway.
Pivot node is a node that not only interacts with two modules but also has at least two pairs of interactions with each module. The hypergeometric test significance analysis of the interaction between the node and each module is P ≤ 0.05. Python program was written to find the pivot node of the interaction module for further analysis. Gene transcription and post-transcriptional regulation are often driven by non-coding RNA (ncRNA) and TFs. Hence, we scientifically predicted and detected their role in HBV- and HCC-related dysfunction modules. Pivot is defined as a regulator that has significant regulatory effects on modules in the pathogenesis of HBV and HCC including ncRNA, TFs, and potential drugs. More than two control links between each regulator and each module were required, and the significance of enriched targets in each module based on the hypergeometric test calculation was P < 0.01. In addition, we examined the overlap of DEGs between HBV and HCC in these significant pivot regulators.
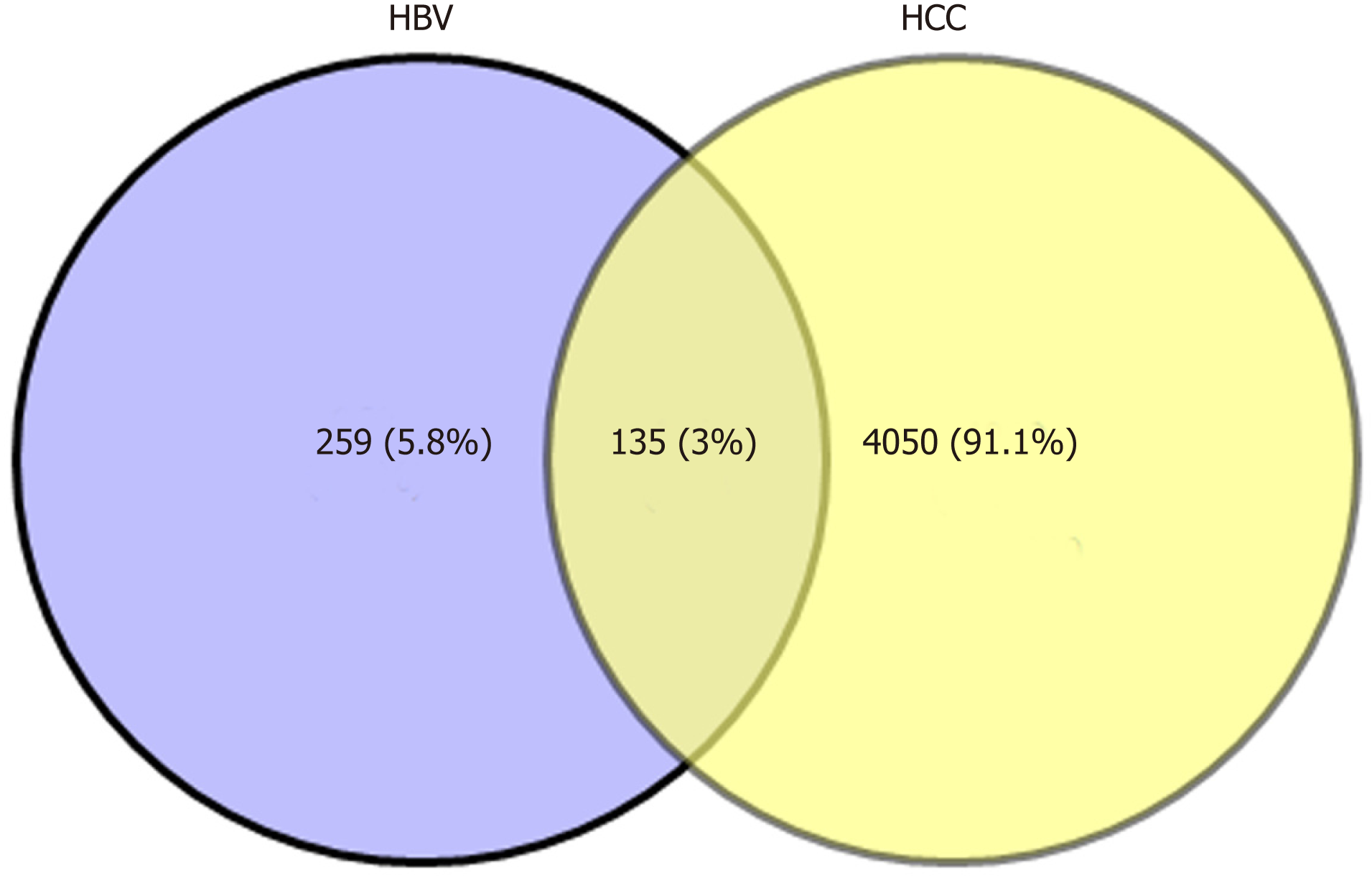
Biologists have conducted many experiments and studies on the relationship between HBV and HCC, and determined that HBV infection is a key factor that induces HCC. However, the complex interaction mechanism between them remains unclear. Therefore, molecular links and functional effects of HBV and HCC were explored during the course of disease. We integrated related genes of the samples and screened the DEGs of HBV and HCC. Through significant screening of DEG, 394 HBV significant DEGs and 4185 HCC significant DEGs were obtained. After screening of HBV- and HCC-related DEGs, 135 common genes were obtained (Figure 1).
In order to determine the functional clusters of hepatitis B and HCC DEGs, we searched 4444 differential gene interactions based on the PPI network. In our results, 16 dysfunction modules were obtained including 1585 nodes and 145616 edges. As shown in the table, inflammation and cancer DEGs were closely clustered together in the modules. We also observed that butyrylcholinesterase had the largest connectivity (16) in the modular network among the common genes of HBV and HCC, while lipoprotein metabolism and fatty acid-binding protein 5 were linked to 15 other genes respectively, which means that the 3 genes played a central role in their modules. At the same time, their modules were significantly involved in hepatitis B and HCC, which could be a bridge molecule between HBV and HCC. Therefore, we inferred that HBV and HCC may promote their own proliferation through some common function module-related genes, and are closely related to the microenvironment of liver diseases.
The expression level of the same gene in two related diseases is different, which may represent the progressive bridge between diseases. Therefore, to identify key bridge molecules between HBV and HCC, we screened 135 common DEGs of diseases to identify key molecules that could characterize the process of HBV to HCC. Sixty-seven persistent dysregulated genes were obtained. Interestingly, these genes were upregulated, and most of the genes in HCC were significantly higher than those in HBV. Thus, these significantly elevated genes could characterize the progression of disease from HBV to HCC and are key genes for bridging the two diseases. On the other hand, overlapping screening of pathogenic module genes clustered by DEGs clarified that the same genes existed among multiple modules. A total of 366 overlapping genes were screened, which indicated that these genes could be associated with the disease process of HBV-related HCC at the same time. Subsequently, the connectivity of module genes was calculated and analyzed. The results suggested that the highest connectivity of PIK3CD was 670, and there were 15 genes larger than 600. The higher the connectivity, the more significant the role of the gene in the whole regulatory network, and the more important influence it has in the process of two diseases.
A total of 54% of all HCC cases are associated with HBV, making it the most common cause of cancer worldwide[18]. In addition to directly overlapping nodes as the most direct bridge between HBV and HCC, we also analyzed other possible links between inflammation and cancer. In other words, PPI was used to find the crosstalk interaction among modules, and 40 significant crosstalk connections were obtained by screening the significant crosstalk. Because Module 7 had the highest connectivity among them, focusing on the genes of Module 7 allowed us to further understand the bridge mechanism. Inflammatory mediators play an important role in the microenvironment of tumors, which can affect all stages of tumorigenesis and development, especially the initial stage of formation. Based on GO functional analysis, we found that DEGs of the central dysfunction module tended to significantly enrich multiple disease functions (Figure 2). These pathways included positive regulation of lipid kinase activity, protein kinase B signal transduction, phosphorylation of phosphatidylinositol, acute inflammatory response, and myocyte proliferation. The modules not only shared some DEGs, but also participated in the same or similar functions and paths through crosstalk interaction. In conclusion, exploration of bridge mechanism at the module level suggested that the connections of HBV and HCC could communicate and transit through module bridging to a certain extent, demonstrating the process of disease under the global effect. Therefore, exploring the potential processes of crosstalk and molecular linkage through crosstalk may further our understanding of the detailed pathogenesis of HBV-related HCC.
Although the regulation of HBV-related HCC by single or several TFs and ncRNA has been extensively studied, little attention has been paid to their comprehensive regulation of dysfunctional modules. Therefore, in order to explore these transcriptional regulators, we applied the predictive analysis of regulators to the dysfunction module based on the relationship between transcription and post-transcriptional regulation. We obtained 496 ncRNAs and 158 TFs involving 739 ncRNA-Module interaction pairs and 213 TF-Module interaction pairs. Statistical analysis of the predicted results showed that there were five regulatory modules, which were targeted by long-chain ncRNA MALAT1, and three modules were targeted by mi410-3p. Other ncRNAs also regulated multiple dysfunction modules to varying degrees, and had potential regulatory effects on HBV and HCC. According to statistics, TF PPARA could regulate five modules, and NFKB1 and RELA also had significant regulatory effect on the four modules. These TFs may mediate the occurrence and development of HBV-related HCC and play a crucial role in the process of disease.
Three of the same miRNAs in HBV and HCC were identical to the predicted ncRNA including miR-192, miR-215, and miR-874. At the same time, it was predicted that three genes in the TFs of the regulatory module were identical to those of the persistent disorder including EGR2, FOS, and KLF4 involved in modules 1 and 9. According to the analysis of GO function, these two modules mainly play a role in regulating the JAK-STAT and MAPK signaling pathways. Therefore, we presume that these six TFs and ncRNAs are key regulatory factors and key components of connecting HBV and HCC bridges. Generally speaking, it is convenient for us to understand the potential mechanism of disease by exploring the regulatory role of pivot regulators in dysfunction modules. The pivot regulators can also be used as candidates for further experimental studies by other biologists.
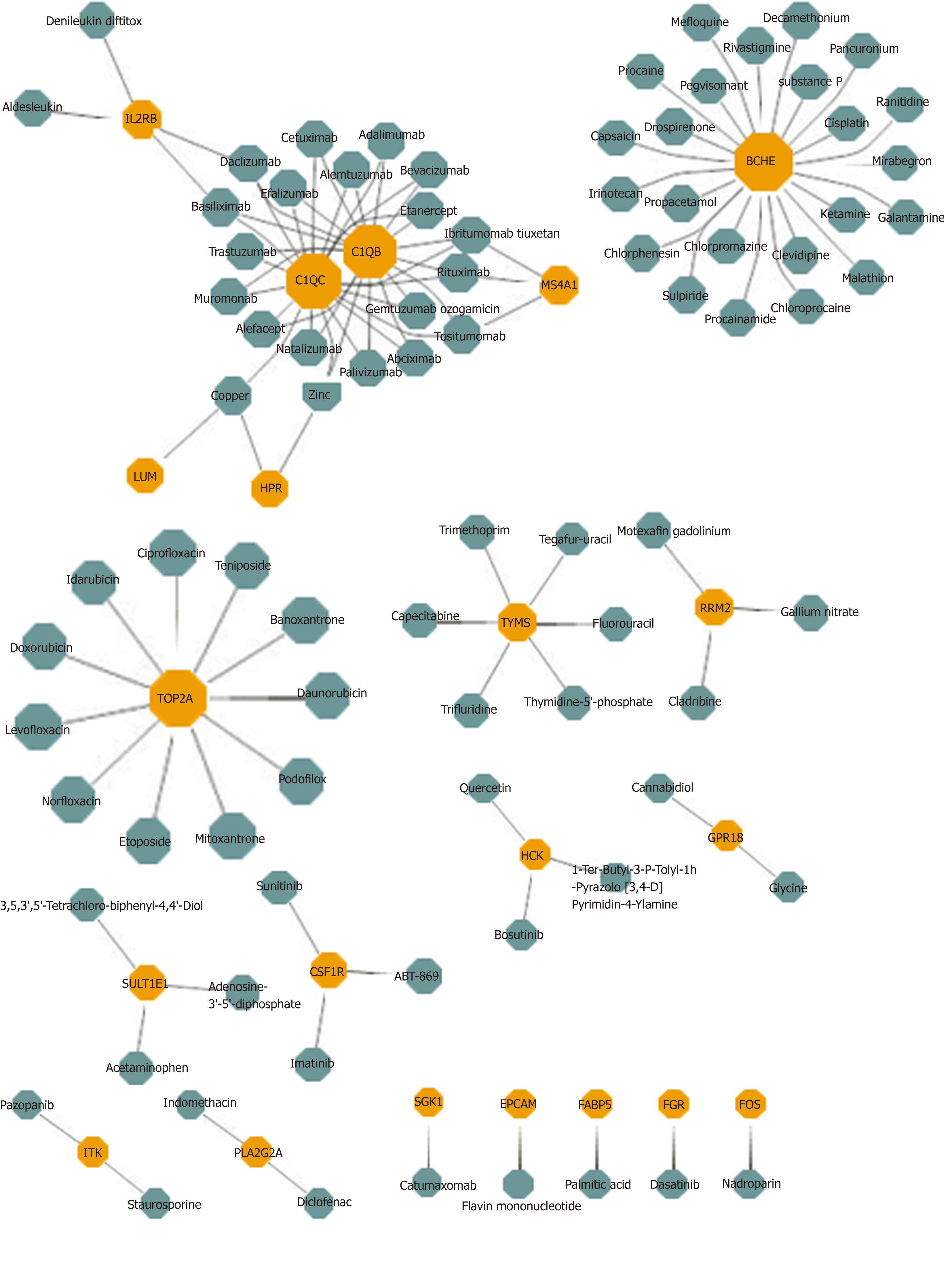
Potential drug prediction was made based on the bridge mechanism and drug target information between HBV and HCC explored previously. The results reported that 1,633 drug-module drug target pairs of 953 drugs may represent the potential therapeutic mechanism of the disease. In the statistical results, Sarilumab had significant pharmacological effects on six modules, while Capsaicin, Imipramine, and Mirtazapine had potential therapeutic or side effects on five modules. Other drugs also had different degree of targeting dysfunction modules, which had a certain regulatory effect on HBV and HCC. After screening the same DEGs of the two diseases, 21 drug target genes were found, and each gene corresponded to multiple drugs (Figure 3). In conclusion, these targeted drug predictions of bridge molecules and functional dysfunction modules provide references and inspirations for biologists in the treatment of diseases and the analysis of pharmacodynamics, and it can be used as candidate drugs as well. Potential target drug prediction based on dysfunction module has become an important research method for personalized treatment and drug use.
HCC is the most difficult end-stage liver disease to cure. A total of 60%-80% of HCC patients worldwide are potential liver diseases caused by HCV or HBV[19]. Although scientists have done extensive research on the close relationship between hepatitis and HCC, there has been a lack of exploration of molecular bridges based on functional modules of HBV and HCC. Therefore, resources from several databases were integrated including gene transcription and miRNA level changes in normal and disease patients, PPI network, transcriptional and post-transcriptional regulation, and other related data to study the potential molecular bridge of HBV-mediated HCC. The combination of PPI and crosstalk analysis showed that the functional module-based method can provide abundant resources for potential candidate genes, interactions, ncRNA, and TFs of molecular bridges between the two diseases.
In our analysis, there were 135 identical genes in the DEGs of HBV and HCC and 67 genes were assumed to be persistent dysfunctional genes among them with the increased expression of TOP5, GRHL2, VIPR1, CHST4, SLC25A47, and FXYD1 from hepatitis to HCC. We postulate that these genes play an important role in the occurrence and development of HCC induced by HBV, which has been confirmed in some previous studies. GRHL2 levels in alcoholic liver patients and model mice increased significantly among them, which seems to increase the level of hepatic inflammation by targeting the inhibition of the transcription of microRNA122, while HIF1 alpha can promote the metastasis of cancer cells and angiogenesis[20,21]. Inhibitory effect on miRNA 122 can also affect the differentiation potential of hepatic stem/progenitor cells and aggravate the occurrence of liver diseases[22]. GRHL2 can also promote cell proliferation in a variety of HCC cell lines and is significantly associated with early recurrence of HCC[23]. In addition, the binding of VIP to receptors can participate in neutrophil recruitment, adhesion molecule expression, and fibrinogen synthesis in different target organs to regulate inflammation[24]. VIPR1 is expressed in the majority of most common human tumors including breast cancer, prostate cancer, pancreatic cancer, lung cancer, colon cancer, gastric cancer, liver and bladder cancers, lymphoma, and meningioma[25]. In addition, Jinawath et al[26] identified a significant increase in CHST4 expression in intrahepatic cho-langiocarcinoma disease samples by gene expression profile. As an organ with metabolic function, the liver plays a major role in metabolism-related proteins in tissues and cells, and the imbalance of metabolism-related proteins may cause liver dysfunction, even the occurrence of diseases. SLC transporters, as the "metabolic gates" of cells, mediate the transport of many essential nutrients and metabolites. Human genome studies have identified SLC transporters as susceptible or pathogenic genes in various diseases such as cancer, cardiovascular disease, metabolic disorders, autoimmune diseases, and neurological dysfunction[27]. Finally, FXYD proteins can act as Na, K-ATPase functional regulators by reducing the affinity of the system to potassium and sodium. The expression level of FXYD proteins in normal liver tissues is low, but it has a significant increase in the detection data in this study, de-monstrating that FXYD is also a key gene causing liver diseases[28].
Through in-depth analysis of HBV-related HCC dysfunction module, it was found that overlapping genes existed among multiple modules, including a variety of chemokines that have the ability to chemoattract white blood cells to the site of infection, thereby regulating the inflammatory response. CCL21 also participated in five modules. CCL21 chemokines bind to CR7 receptors and T cells of mature DCs regulated DC migration to the white pulp of the spleen, where physical contacts with lymphocytes triggers immune cell responses and regulates tumor-mediated immunosuppression[29,30]. Another chemokine CCL20 participated in four modules simultaneously, and its expression level in HBV-infected cells was markedly increased. CCL20/CCR6 chemokine/receptor axis is able to recruit CCR6-positive white blood cells into the tumor microenvironment and promote the initiation and progression of HCC[31,32]. While some chemokine receptors also existed in several modules. The knockdown of CCR1 results in the reduction of HCC metastasis promoter osteopontin in vitro and in vivo induced liver cancer migration, invasion, and lung metastasis[33]. In addition, PIK3CD had the highest 670 connectivity among all modules, which is a key regulatory gene with one-stop and whole-body effects. The high expression of PIK3CD can promote the proliferation and migration of HCC cells, and also participates in acute liver injury model in mice. Long-term inflammation of liver injury is an important factor leading to liver fibrosis and even cirrhosis and HCC[34,35]. Later, interesting module pairs were observed and module 4 and module 6 showed significant crosstalk, including the most common DEGs of which most were related to chemokines and receptors. Functional analysis showed that they may regulate pivot regulators by regulating inflammation, cell cycle regulation, and cell adhesion, thus completing the potential relationship between HBV and HCC.
Transcriptional and post-transcriptional regulation are regarded as key factors in the occurrence and development of diseases. Evaluating the transcriptional regulation of dysfunction module has become an important means to explore the bridge molecules of HBV-mediated HCC pathogenesis in a comprehensive manner. To elucidate the transcriptional regulatory factors associated with the molecular links between the two diseases, pivot regulators were analyzed based on transcriptional and post-transcriptional regulatory relationships. The results showed that MALAT1, ANCR, and BANCR were the main long-chain ncRNAs, miRNAs dominated by miRNA-410-3p, TFs dominated by PPARA, NFKB1, and RELA had significant regulatory effects on dysfunction modules. For common DEGs of HBV and HCC persistent disorder genes and miRNAs, the same genes were found with these pivot regulators including EGR2, FOS, and KLF4, as well as miR-192, miR-215, and miR-874. These genes exist in two disease-related modules and play a regulatory role in these modules, so they can be presumed to be key bridge molecules between diseases. These genes regulated activation of T cell, production of cytokine, change of cell cycle, activation of inflammatory and cancer-related signaling pathways by targeting multiple genes in the module. EGR plays a crucial role in the expression of FasL mediated by HBx, thus affecting the occurrence of HBV-related HCC[36]. Inhibition of EGR2 in HCC cell lines reduces the expression of SOCS-1 and the phosphorylation of JAK2 and STAT3, thus affecting cell proliferation[37]. FOS signal transduction is associated with TLR9-mediated IFN production in plasma-like dendritic cells, and the gene expression level of it is also significantly changed in HCC[38,39]. KLF4 affects inflammation by regulating M1/M2 macrophage polarization, and can also be used as a candidate marker for HCC development[40,41]. The regulation of small RNA is the focus of biological mechanism research. Among them, miRNA-192 not only affects the replication of HBV, but also affects the proliferation of HCC cell lines through the regulation of apoptotic proteins and ER stress[42]. MiRNA-215 is significantly correlated with hepatitis grade, fibrosis stage, and tumor tissue differentiation[43]. MiRNA-874 can inhibit the angiogenesis of endothelial cells derived from tumors. Overexpression of miRNA-874-3p in HCC cell lines can significantly inhibit cell growth and colony formation, and promote cell apoptosis[44,45]. Based on the functions of these transcriptional and post-transcriptional regulators, it is believed that they may represent key linkages in the development of HBV to HCC. TFs mediated modules 1 and 9, which is an important mechanism of dysfunction. All pivot regulators mediated dysfunction modules and played an overall regulatory role including the recombinant genes, indicating the potential pathogenesis of HBV-related HCC.
Drug prediction results based on multi-regulator-driven dysfunction module and drug target information showed that Sarilumab had significant regulatory effects on six dysfunction modules. Sarilumab is a human monoclonal antibody against IL-6 receptor-alpha, which has the ability to reduce neutrophils, showing that the drug has a certain effect on inflammation[46]. 26 DEGs results were obtained with DEGs targeting HBV and HCC. Among them, butyrylcholinesterase targeted predictive drug Mefloquine acts on the beta-catenin pathway and plays a role in the treatment of HCC[47]. Sulpiride induces fatty liver in rats by phosphorylating IRS-1 in Ser 307-mediated adipose tissue insulin resistance, so the drug may have potential toxic side effects on the liver[48]. Many drugs need to be further explored for their treatment or side effects. However, this study provides a new method for choosing common drugs for HBV and HCC. This is not just helpful for drug research and development personnel to conduct drug screening, but also provides theoretical guidance for clinical medical personnel to conduct personalized treatment. Generally speaking, the functional module-based approach can not only comprehensively and thoroughly explore the mechanism of the occurrence and development of disease, but also predict its potential therapeutic methods and mechanisms.
The potential role of chronic inflammation in the development of cancer has been widely recognized. However, there has been little research fully and thoroughly exploring the molecular link between hepatitis B virus (HBV) and hepatocellular carcinoma (HCC).
To conduct a comprehensive and in-depth discussion on the bridge mechanism between HBV and HCC.
The purpose of this study was to explore the co-imbalance bridging molecules between HBV and HCC and their potential drugs based on the dysfunction module.
First, maladjusted genes shared between HBV and HCC were identified by disease-related DEGs. Second, the PPI network based on dysfunctional genes identified a series of dysfunctional modules and significant crosstalk between modules based on the hypergeometric test. In addition, key regulators were detected by pivot analysis. Finally, targeted drugs that have regulatory effects on diseases were predicted by modular methods and drug target information.
The study found that 67 genes continued to increase in the HBV-HCC process. Moreover, 366 overlapping genes in the module network participated in multiple functional blocks. It could be presumed that these genes and their interactions play an important role in the relationship between inflammation and cancer. Correspondingly, significant crosstalk constructed a module level bridge for HBV-HCC molecular processes. On the other hand, a series of ncRNAs and TFs that have potential pivot regulatory effects on HBV and HCC were identified. Among them, some of the regulators also had persistent disorders in the process of HBV-HCC including miRNA-192, miRNA-215, and miRNA-874, and EGR2, FOS, and KLF4. Therefore, the study concluded that these pivots are the key bridge molecules outside the module. Last but not least, a variety of drugs that may have some potential pharmacological or toxic side effects on HBV-induced HCC were predicted, but their mechanisms need to be further explored.
The results suggest that the persistent inflammatory environment of HBV can be utilized as an important risk factor to induce the occurrence of HCC, which is supported by molecular evidence.
In the future, research may comprehensively and thoroughly explore the mechanism of HCC occurrence and development and predict the potential therapeutic methods and mechanisms.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dourakis SP, Rostami-Nejad M, Yakoot M S-Editor: Yan JP L-Editor: Filipodia E-Editor: Ma YJ
| 1. | Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis. 2009;30:1073-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1802] [Cited by in RCA: 2085] [Article Influence: 130.3] [Reference Citation Analysis (1)] |
| 2. | Gupta SC, Kunnumakkara AB, Aggarwal S, Aggarwal BB. Inflammation, a Double-Edge Sword for Cancer and Other Age-Related Diseases. Front Immunol. 2018;9:2160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 168] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 3. | Virzì A, Roca Suarez AA, Baumert TF, Lupberger J. Oncogenic Signaling Induced by HCV Infection. Viruses. 2018;10:pii: E538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Susek KH, Karvouni M, Alici E, Lundqvist A. The Role of CXC Chemokine Receptors 1-4 on Immune Cells in the Tumor Microenvironment. Front Immunol. 2018;9:2159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 5. | Zang M, Li Y, He H, Ding H, Chen K, Du J, Chen T, Wu Z, Liu H, Wang D, Cai J, Qu C. IL-23 production of liver inflammatory macrophages to damaged hepatocytes promotes hepatocellular carcinoma development after chronic hepatitis B virus infection. Biochim Biophys Acta Mol Basis Dis. 2018;1864:3759-3770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 6. | Mysore KR, Leung DH. Hepatitis B and C. Clin Liver Dis. 2018;22:703-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Maddrey WC. Hepatitis B: An important public health issue. J Med Virol. 2000;61:362-366. [PubMed] |
| 8. | Yang L, Ye S, Zhao X, Ji L, Zhang Y, Zhou P, Sun J, Guan Y, Han Y, Ni C, Hu X, Liu W, Wang H, Zhou B, Huang J. Molecular Characterization of HBV DNA Integration in Patients with Hepatitis and Hepatocellular Carcinoma. J Cancer. 2018;9:3225-3235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 9. | Musa J, Li J, Grünewald TG. Hepatitis B virus large surface protein is priming for hepatocellular carcinoma development via induction of cytokinesis failure. J Pathol. 2019;247:6-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Zhu S, Tan W, Li W, Zhou R, Wu X, Chen X, Li W, Shang C, Chen Y. Low expression of VSIG4 is associated with poor prognosis in hepatocellular carcinoma patients with hepatitis B infection. Cancer Manag Res. 2018;10:3697-3705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, Cai F, Ma L, Yu Y. The Role of MicroRNAs in Hepatocellular Carcinoma. J Cancer. 2018;9:3557-3569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 12. | Abdel-Hamid NM, Abass SA, Mohamed AA, Muneam Hamid D. Herbal management of hepatocellular carcinoma through cutting the pathways of the common risk factors. Biomed Pharmacother. 2018;107:1246-1258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Ranjpour M, Wajid S, Jain SK. Elevated expression of A-Raf and FA2H in hepatocellular carcinoma is associated with lipid metabolism dysregulation and cancer progression. Anticancer Agents Med Chem. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Herrero AB, Astudillo AM, Balboa MA, Cuevas C, Balsinde J, Moreno S. Levels of SCS7/FA2H-mediated fatty acid 2-hydroxylation determine the sensitivity of cells to antitumor PM02734. Cancer Res. 2008;68:9779-9787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 15. | Kong X, Sun R, Chen Y, Wei H, Tian Z. γδT cells drive myeloid-derived suppressor cell-mediated CD8+ T cell exhaustion in hepatitis B virus-induced immunotolerance. J Immunol. 2014;193:1645-1653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 16. | You J, Chen W, Chen J, Zheng Q, Dong J, Zhu Y. The Oncogenic Role of ARG1 in Progression and Metastasis of Hepatocellular Carcinoma. Biomed Res Int. 2018;2018:2109865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 17. | Wu N, Zhao J, Yuan Y, Lu C, Zhu W, Jiang Q. NOP7 interacts with β-catenin and activates β-catenin/TCF signaling in hepatocellular carcinoma cells. Onco Targets Ther. 2018;11:6369-6376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Pazgan-Simon M, Simon KA, Jarowicz E, Rotter K, Szymanek-Pasternak A, Zuwała-Jagiełło J. Hepatitis B virus treatment in hepatocellular carcinoma patients prolongs survival and reduces the risk of cancer recurrence. Clin Exp Hepatol. 2018;4:210-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 19. | Park JW, Chen M, Colombo M, Roberts LR, Schwartz M, Chen PJ, Kudo M, Johnson P, Wagner S, Orsini LS, Sherman M. Global patterns of hepatocellular carcinoma management from diagnosis to death: The BRIDGE Study. Liver Int. 2015;35:2155-2166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 569] [Cited by in RCA: 945] [Article Influence: 94.5] [Reference Citation Analysis (0)] |
| 20. | Satishchandran A, Ambade A, Rao S, Hsueh YC, Iracheta-Vellve A, Tornai D, Lowe P, Gyongyosi B, Li J, Catalano D, Zhong L, Kodys K, Xie J, Bala S, Gao G, Szabo G. MicroRNA 122, Regulated by GRLH2, Protects Livers of Mice and Patients From Ethanol-Induced Liver Disease. Gastroenterology. 2018;154:238-252.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 21. | Kim A, Ma JY. Rhaponticin decreases the metastatic and angiogenic abilities of cancer cells via suppression of the HIF‑1α pathway. Int J Oncol. 2018;53:1160-1170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Tanimizu N, Kobayashi S, Ichinohe N, Mitaka T. Downregulation of miR122 by grainyhead-like 2 restricts the hepatocytic differentiation potential of adult liver progenitor cells. Development. 2014;141:4448-4456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Tanaka Y, Kanai F, Tada M, Tateishi R, Sanada M, Nannya Y, Ohta M, Asaoka Y, Seto M, Shiina S, Yoshida H, Kawabe T, Yokosuka O, Ogawa S, Omata M. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J Hepatol. 2008;49:746-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 24. | Martínez C, Juarranz Y, Abad C, Arranz A, Miguel BG, Rosignoli F, Leceta J, Gomariz RP. Analysis of the role of the PAC1 receptor in neutrophil recruitment, acute-phase response, and nitric oxide production in septic shock. J Leukoc Biol. 2005;77:729-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Reubi JC. In vitro evaluation of VIP/PACAP receptors in healthy and diseased human tissues. Clinical implications. Ann N Y Acad Sci. 2000;921:1-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 70] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Jinawath N, Chamgramol Y, Furukawa Y, Obama K, Tsunoda T, Sripa B, Pairojkul C, Nakamura Y. Comparison of gene expression profiles between Opisthorchis viverrini and non-Opisthorchis viverrini associated human intrahepatic cholangiocarcinoma. Hepatology. 2006;44:1025-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 27. | Zhang Y, Zhang Y, Sun K, Meng Z, Chen L. The SLC transporter in nutrient and metabolic sensing, regulation, and drug development. J Mol Cell Biol. 2019;11:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 28. | Floyd RV, Wray S, Martín-Vasallo P, Mobasheri A. Differential cellular expression of FXYD1 (phospholemman) and FXYD2 (gamma subunit of Na, K-ATPase) in normal human tissues: A study using high density human tissue microarrays. Ann Anat. 2010;192:7-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Nico D, Martins Almeida F, Maria Motta J, Soares Dos Santos Cardoso F, Freire-de-Lima CG, Freire-de-Lima L, de Luca PM, Maria Blanco Martinez A, Morrot A, Palatnik-de-Sousa CB. NH36 and F3 Antigen-Primed Dendritic Cells Show Preserved Migrating Capabilities and CCR7 Expression and F3 Is Effective in Immunotherapy of Visceral Leishmaniasis. Front Immunol. 2018;9:967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 30. | Zhou S, Chen L, Qin J, Li R, Tao H, Zhen Z, Chen H, Chen G, Yang Y, Liu B, She Z, Zhong C, Liang C. Depletion of CD4+ CD25+ regulatory T cells promotes CCL21-mediated antitumor immunity. PLoS One. 2013;8:e73952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Liu Y, Li L, Liu J, She WM, Shi JM, Li J, Wang JY, Jiang W. Activated hepatic stellate cells directly induce pathogenic Th17 cells in chronic hepatitis B virus infection. Exp Cell Res. 2017;359:129-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Benkheil M, Van Haele M, Roskams T, Laporte M, Noppen S, Abbasi K, Delang L, Neyts J, Liekens S. CCL20, a direct-acting pro-angiogenic chemokine induced by hepatitis C virus (HCV): Potential role in HCV-related liver cancer. Exp Cell Res. 2018;372:168-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 33. | Viallat JR, Rey F, Farisse P, Henric A, Gastaut JA. [The frequency of Kaposi's bronchial sarcoma in AIDS. Personal experience in 1987]. Rev Mal Respir. 1989;6:71-73. [PubMed] |
| 34. | Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS One. 2016;11:e0158347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 267] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 35. | Xu G, Han X, Yuan G, An L, Du P. Screening for the protective effect target of deproteinized extract of calf blood and its mechanisms in mice with CCl4-induced acute liver injury. PLoS One. 2017;12:e0180899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 36. | Yoo YG, Lee MO. Hepatitis B virus X protein induces expression of Fas ligand gene through enhancing transcriptional activity of early growth response factor. J Biol Chem. 2004;279:36242-36249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 37. | Lu L, Ye X, Yao Q, Lu A, Zhao Z, Ding Y, Meng C, Yu W, Du Y, Cheng J. Egr2 enhances insulin resistance via JAK2/STAT3/SOCS-1 pathway in HepG2 cells treated with palmitate. Gen Comp Endocrinol. 2018;260:25-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Janovec V, Aouar B, Font-Haro A, Hofman T, Trejbalova K, Weber J, Chaperot L, Plumas J, Olive D, Dubreuil P, Nunès JA, Stranska R, Hirsch I. The MEK1/2-ERK Pathway Inhibits Type I IFN Production in Plasmacytoid Dendritic Cells. Front Immunol. 2018;9:364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Liu S, Yao X, Zhang D, Sheng J, Wen X, Wang Q, Chen G, Li Z, Du Z, Zhang X. Analysis of Transcription Factor-Related Regulatory Networks Based on Bioinformatics Analysis and Validation in Hepatocellular Carcinoma. Biomed Res Int. 2018;2018:1431396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 40. | Li B, Sheng Z, Liu C, Qian L, Wu Y, Wu Y, Ma G, Yao Y. Kallistatin Inhibits Atherosclerotic Inflammation by Regulating Macrophage Polarization. Hum Gene Ther. 2019;30:339-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 41. | Zhang Y, Liu Z, Li JS. Identifying Biomarkers of Hepatocellular Carcinoma Based on Gene Co-Expression Network from High-Throughput Data. Stud Health Technol Inform. 2017;245:667-671. [PubMed] |
| 42. | Nielsen KO, Jacobsen KS, Mirza AH, Winther TN, Størling J, Glebe D, Pociot F, Hogh B. Hepatitis B virus upregulates host microRNAs that target apoptosis-regulatory genes in an in vitro cell model. Exp Cell Res. 2018;371:92-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 43. | Mamdouh S, Khorshed F, Aboushousha T, Hamdy H, Diab A, Seleem M, Saber M. Evaluation of Mir-224, Mir-215 and Mir-143 as Serum Biomarkers for HCV Associated Hepatocellular Carcinoma. Asian Pac J Cancer Prev. 2017;18:3167-3171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 44. | Lopatina T, Grange C, Fonsato V, Tapparo M, Brossa A, Fallo S, Pitino A, Herrera-Sanchez MB, Kholia S, Camussi G, Bussolati B. Extracellular vesicles from human liver stem cells inhibit tumor angiogenesis. Int J Cancer. 2019;144:322-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 45. | Leong KW, Cheng CW, Wong CM, Ng IO, Kwong YL, Tse E. miR-874-3p is down-regulated in hepatocellular carcinoma and negatively regulates PIN1 expression. Oncotarget. 2017;8:11343-11355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 46. | Lee EB. A review of sarilumab for the treatment of rheumatoid arthritis. Immunotherapy. 2018;10:57-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 47. | Li YH, Yang SL, Zhang GF, Wu JC, Gong LL, Ming-Zhong, Lin RX. Mefloquine targets β-catenin pathway and thus can play a role in the treatment of liver cancer. Microb Pathog. 2018;118:357-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 48. | Zhou X, Ren L, Yu Z, Huang X, Li Y, Wang C. The antipsychotics sulpiride induces fatty liver in rats via phosphorylation of insulin receptor substrate-1 at Serine 307-mediated adipose tissue insulin resistance. Toxicol Appl Pharmacol. 2018;345:66-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |