Published online Nov 21, 2018. doi: 10.3748/wjg.v24.i43.4880
Peer-review started: August 3, 2018
First decision: October 8, 2018
Revised: October 22, 2018
Accepted: November 2, 2018
Article in press: November 2, 2018
Published online: November 21, 2018
Processing time: 110 Days and 14.1 Hours
To establish patient-individual tumor models of rectal cancer for analyses of novel biomarkers, individual response prediction and individual therapy regimens.
Establishment of cell lines was conducted by direct in vitro culturing and in vivo xenografting with subsequent in vitro culturing. Cell lines were in-depth characterized concerning morphological features, invasive and migratory behavior, phenotype, molecular profile including mutational analysis, protein expression, and confirmation of origin by DNA fingerprint. Assessment of chemosensitivity towards an extensive range of current chemotherapeutic drugs and of radiosensitivity was performed including analysis of a combined radio- and chemotherapeutic treatment. In addition, glucose metabolism was assessed with 18F-fluorodeoxyglucose (FDG) and proliferation with 18F-fluorothymidine.
We describe the establishment of ultra-low passage rectal cancer cell lines of three patients suffering from rectal cancer. Two cell lines (HROC126, HROC284Met) were established directly from tumor specimens while HROC239 T0 M1 was established subsequent to xenografting of the tumor. Molecular analysis classified all three cell lines as CIMP-0/ non-MSI-H (sporadic standard) type. Mutational analysis revealed following mutational profiles: HROC126: APCwt, TP53wt, KRASwt, BRAFwt, PTENwt; HROC239 T0 M1: APCmut, P53wt, KRASmut, BRAFwt, PTENmut and HROC284Met: APCwt, P53mut, KRASmut, BRAFwt, PTENmut. All cell lines could be characterized as epithelial (EpCAM+) tumor cells with equivalent morphologic features and comparable growth kinetics. The cell lines displayed a heterogeneous response toward chemotherapy, radiotherapy and their combined application. HROC126 showed a highly radio-resistant phenotype and HROC284Met was more susceptible to a combined radiochemotherapy than HROC126 and HROC239 T0 M1. Analysis of 18F-FDG uptake displayed a markedly reduced FDG uptake of all three cell lines after combined radiochemotherapy.
These newly established and in-depth characterized ultra-low passage rectal cancer cell lines provide a useful instrument for analysis of biological characteristics of rectal cancer.
Core tip: Ultra-low passage and in-depth characterized tumor models are highly desirable for basic research and assessment of individual response prediction to current or novel therapy regimens. Here, for the first time, we describe three patient-derived rectal cancer cell lines established either directly from patient’s tumor samples or after xenografting. These tumor models were characterized according to phenotype, molecular-, as well as growth and morphological features and sensitivity to chemotherapeutic drugs and radiation, including radiochemotherapy. In addition, glucose metabolism was assessed with 18F-fluorodeoxyglucose and proliferation with 18F-fluorothymidine. These cell lines provide excellent tools for basic and translational research of rectal cancers’ biological characteristics.
- Citation: Gock M, Mullins CS, Bergner C, Prall F, Ramer R, Göder A, Krämer OH, Lange F, Krause BJ, Klar E, Linnebacher M. Establishment, functional and genetic characterization of three novel patient-derived rectal cancer cell lines. World J Gastroenterol 2018; 24(43): 4880-4892
- URL: https://www.wjgnet.com/1007-9327/full/v24/i43/4880.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i43.4880
Since two decades our understanding of colorectal carcinoma (CRC) as a heterogeneous disease entity in terms of both molecular carcinogenesis and morphologic multistep pathways has steadily grown[1]. Meanwhile, three major molecular carcinogenesis pathways have been identified: (1) Chromosomal instability (CIN); (2) microsatellite instability (MSI); and (3) CpG island methylator phenotype (CIMP) or epigenetic instability pathways[2,3]. In conclusion, Jass et al[4] classified CRCs into 5 molecular subtypes primarily by underlying types of genetic instability and presence of DNA methylation: (1) CIMP-H/MSI-H; (2) CIMP-H/non-MSI-H; (3) CIMP-L/non-MSI-H; (4) CIMP-0/non-MSI-H (spStd); and (5) CIMP-0/MSI-H. But these molecular subtypes are not distributed equally along the large bowel. Yamauchi et al[5] could show that clinicopathological and molecular features of CRCs differed depending on the bowel subsite of the tumor.
Recent algorithms for treatment of locally advanced rectal cancer consist of neoadjuvant chemoradiation (nCRT) followed by rectal resection and total mesorectal excision (TME). Currently, beside clinical evaluation, imaging modalities including endorectal ultrasound, computed tomography (CT), magnetic resonance imaging (MRI) as well as 18F-fluorodeoxyglucose (FDG) positron emission tomography-computed tomography (18F-FDG-PET/CT) are used for staging, assessment of response to nCRT, and restaging after nCRT[6,7]. However, accuracy of parameters derived from these imaging modalities, especially for predicting a pathological complete response (pCR) after nCRT, is currently limited due to their low sensitivity and specificity[6,7].
In the last decade, many patient-individual tumor models of CRC could be generated by us and others[8,9]. But no novel rectal cancer models have been published up to now.
In this study, we describe the establishment and functional characterization of three patient-derived rectal cancer cell lines along with corresponding patient-derived xenografts (PDX). A broad analysis of tumor biology, genetic alterations, protein expression, and assessment of chemosensitivity towards a range of chemotherapeutic drugs and of radiosensitivity was performed.
In addition, an analysis of the metabolism of the glucose analogue 18F-FDG was carried out. Due to their higher glucose metabolism, 18F-FDG is differentially taken up by malignant cells[10]. This phenomenon can be assessed by molecular imaging with 18F-FDG-PET/CT and may be used to detect tumors’ therapy responses, that are either not apparent with other morphological imaging modalities or may precede a significant tumor shrinkage by weeks or months[11]. Furthermore 3-deoxy-3-18F-fluorothymidine (18F-FLT) uptake was analyzed. 18F-FLT is intracellularly trapped when monophosphorylated by thymidine kinase 1 and 18F-FLT-PET uptake reflects activity of thymidine kinase 1 which is elevated during the S phase of the cell cycle. Thus, 18F-FLT-PET uptake mirrors tumor cell proliferation[12].
Considering these aspects, our characterized matched in vitro and in vivo tumor models represent excellent tools for further development of individual response prediction, therapy regimens, and might prove especially valuable to gain additional insights in the tumor biology of rectal cancer.
Primary rectal cancer resection specimens of HROC126, HROC239 and resection specimens of a rectal cancer liver metastasis (HROC284Met) were received fresh from surgery, with informed written patient consent. Tumor samples cut into small pieces (3 mm × 3 mm × 3 mm) were vitally frozen in freezing medium [fetal calf serum (FCS) containing 10% DMSO] at -80 °C for subsequent xenografting. Snap-frozen pieces were stored at -160 °C for subsequent molecular analysis. Cell line establishment was adapted according to Maletzki et al[13].
Six-week-old female NMRI nu/nu mice were used as recipients for subcutaneous tumor in vivo engraftment as described before[14]. Established xenograft tumors (max. 1.500 mm3) were removed and taken into culture as described above.
Procedures involving patient material were in accordance with generally accepted guidelines for the use of human material approved by the Ethics Committee of the Medical faculty, University of Rostock (reference number II HV 43/2004) only after informed patient consent was obtained in written. In vivo experimental procedures were carried out according to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The experimental protocol was approved by the Committee on the Ethics of Animal Experiments of the University of Rostock (Landesamt für Landwirtschaft, Lebensmittelsicherheit und Fischerei Mecklenburg-Vorpommern; Thierfelder Str. 18, 18059 Rostock, Germany; permit number: LALLF M-V/TSD/7221.3-1.1-071-10).
The rectal cancer cell lines HROC126, HROC239 T0 M1 and HROC284Met as well as the control CRC cell line HCT116 were cultured in T75 culture flasks using Dulbecco’s modified Eagle medium (DMEM) /Ham’s F12 supplement with 2 mmol/L L-glutamine and 10% FCS (all cell culture reagents were obtained from Pan Biotech, Aidenbach, Germany).
Hematoxylin and eosin (HE)-stained primary tumors and corresponding PDX were clinicopathologically staged[15], and additional information was extracted from clinical charts.
Molecular classification was performed as described before[3]. Mutation status of the genes APC, TP53, KRAS, PIK3CA, PTEN and BRAFV600E were analyzed. DNA-methylation was checked using a modified marker panel originally published by Ogino et al[16]. CIN was assessed using the SNP Array 6.0 from Affymetrix (Cleveland, OH, United States).
Genomic DNA was isolated from cell lines, matched tumor and normal tissue, PDX tissue as well as corresponding B cells using the Wizard® Genomic DNA Purification Kit (Promega Madison, WI, United States). Highly polymorphic short tandem repeat (STR) DNA marker (CSF1PO, TPOX, THO1, vWA, D16S539, D13S317, and D7S820) and the marker amelogenin for gender determination were used by taking advantage of published PCR primers[17].
Peripheral blood mononuclear cells were isolated by density-gradient centrifugation. B-lymphoid cell lines (B-LCLs) were generated via Epstein-Barr virus (EBV)-transformation[18]. Outgrowing B-LCL cultures were harvested, expanded, characterized by flow cytometry, and frozen down in a master cell bank.
Doubling times of HROC126, HROC239 T0 M1 and HROC284Met cells were determined from serial passages. Five times 105 cells were seeded into 25-cm2 flasks and viable cells (defined by trypan blue exclusion) were subsequently counted for seven consecutive days. Cultures were fed when needed. Cell cycle and ploidy were determined by flow cytometry (FACSCalibur, BD Biosciences, Heidelberg, Germany) using fixed (70% ethanol) and RNase A digested (100 μg/mL; Sigma Aldrich, Munich, Germany) and propidium iodide (10 μg/mL) stained cells. For each sample, at least 10000 events were measured. Cell cycle analysis was done by taking advantage of the Modfit software (Verity Software House, Topsham, ME, United States) using matched B-LCLs as diploid controls.
Levels of markers expressed on the cells’ surface were determined by flow cytometry with and without interferon (IFN)-γ pre-treatment using a panel of FITC-, PE- or APC-conjugated antibodies: CD26, CD29, CD44, CD49a, CD50, CD56, CD58, CD66acde, CD71, CD73,CD90, CD102, CD166, human leukocyte antigen (HLA)-ABC (Immunotools, Friesoythe, Germany); CD152, CD275, CD278, CD326, β2-M, HLA-DR, HLA-E, HLA-G (Miltenyi Biotec, Bergisch- Gladbach, Germany), Ki-67 (Biolegend, San Diego, United States) and HLA-A2 (cell culture supernatant clone BB7.2). For HLA-A2, a polyclonal, secondary FITC-conjugated anti-mouse serum was used (Dako, Hamburg, Germany). Sample analysis was done by CellQuest (BD Biosciences).
Presence or absence of mycoplasma as well as potential polyomavirus infection (JC/BK and SV40) was tested as described before[19].
Tumor cell invasion capacity was tested using the classical Boyden chamber test (8-μm pore size in a 24-well plate format) with Matrigel-coating (both BD Biosciences). Two times 105 cells were seeded in 500 μL serum-free medium per upper Boyden chamber. Medium supplemented with 10% heat-inactivated FCS served as chemo-attractant in the lower Boyden chamber. Three days later, the non-invading cells on the surface of the upper inserts were removed and viability of cells on the lower surface was measured by the 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,6-benzene disulfonate (WST-1) test (Roche Diagnostics, Mannheim, Germany). In parallel experiments, for cells’ capacity to migrate were tested using uncoated upper Boyden chambers.
For chemosensitivity, cells were seeded into 96-well microtiter plates at 5 × 103 or 1 × 104 cells/well. When cells reached 30%-40% confluency, cultures were exposed to increasing concentrations of 5-FU, oxaliplatin, irinotecan, combination of 5-FU and oxaliplatin (FOLFOX) and combination of 5-FU and irinotecan (FOLFIRI) (freshly provided by pharmacy of the University Medical Center Rostock). After 72 h, media and therapeutics were refreshed. Following another 72 h, plates were carefully washed and stained with crystal violet (0.2%, 10 min). Finally, drug effects from triplicate wells were determined at the level of 50 % inhibition (IC50) in comparison to control, measured at 570 nm (reference wavelength: 620 nm). For radiosensitivity analysis, cells radiated with different doses up to 60 Gy using a 137Cs-source were seeded into 96-well microtiter plates in triplicates (1 × 105 cells per well and six serial two-fold dilutions). Control cells were not radiated. After 4 and 7 d, triplicate plates were analyzed for total cell growth using crystal violet as described above.
For analysis of combined radio- and chemosensitivity, cells were seeded and exposed to increasing drug concentrations as described above. After three days of exposure, media were replaced and cells were radiated with 50 Gy. Following another three days, triplicate plates were analyzed for total cell growth.
Western blot was done as previously described[20]. Antibodies specific for the following targets were from Santa Cruz (Heidelberg, Germany): BAX, histone deacetylase 2 (HDAC2), HDAC1, and p53. Anti-survivin was obtained from Novus Biologicals (Cambridge, United Kingdom). HSP90 antibody was provided by Enzo Life Sciences (Lörrach, Germany).
For analysis of 18F-FDG and 18F-FLT uptake, rectal cancer cells were seeded into 24-well microtiter plates at 1 × 105 cells/well in complete culture medium. At day 1, cells were either exposed to FOLFOX at an IC25 concentration alone or radiated (50 Gy) alone or exposed to combinations thereof, whereas control cultures were treated with solvent. After three days of exposure, media were removed and substituted by DMEM (Fisher Scientific, Schwerte, Germany) without FCS, glucose, glutamine, and incubation continued for 1 h before 18F-labeled tracers (0.5 MBq/mL culture medium) were added. Thirty minutes later, incubation was terminated by aspirating the medium and rinsing the cell layer three times with ice-cold PBS. The cells were solubilized with 0.1 mol/L NaOH, and incorporated 18F activity was determined using a gamma counter (WIZARD2 10-Detector Gamma Counter, Perkinelmer, Waltham, MA, United States). Counts per minute were normalized on total protein level of the cells using a commercial Bradford assay (Bio-Rad Laboratories, Munich, Germany).
18F-FDG and 18F-FLT uptake of treated cells is expressed as percent of controls exposed to the solvent only (n = 10 per cell line and experimental condition).
Values are reported as mean ± SEM from at least three measurements. After proving the assumption of normality, differences were determined by the unpaired Student’s t-test. Mean group differences were checked by the Kruskal-Wallis test before for multiple comparisons subgroups were tested with post hoc Dunn’s test. All statistics were performed using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, United States).
We report on three female patients suffering from rectal cancer with an age of 58, 72, and 67 years. The first patient (HROC126) suffered from a carcinoma of the middle rectal third. Preoperative staging revealed an Union for International Cancer Control (UICC) stage II (cT2 cN0 cM0) tumor and a low anterior resection with complete TME was performed. Interestingly, pathological examination showed an UICC stage IIIB tumor [pT3 pN1b (4/13) cM0], therefore a regular adjuvant radio-chemo-therapy (RCT) was conducted. After five years of follow up, the patient is still in complete remission.
The second patient (HROC239) suffered from a rectal cancer of the upper third and preoperative staging revealed an UICC stage IIIB tumor (cT3 cN+). A regular anterior rectal resection was performed. Pathological analysis revealed an UICC stage IIIC [pT4 pN2b (7/20) cM0] tumor and a regular adjuvant RCT was added. After five years of follow up, this patient is likewise in complete remission.
The third patient (HROC284) suffered from a rectal cancer of the middle third and preoperative staging revealed an UICC IIIB stage (cT3 cN1 cM0). A regular preoperative neoadjuvant RCT consisting of 50.4 Gy and 5-FU was performed; thereafter, a low anterior resection with complete TME was conducted. Postoperative pathological examination showed an UICC IIIB [ypT2 pN2a (4/13) cM0] tumor stage. The patient developed after 6 mo a singular liver metastasis that could be resected completely. Subsequently, she developed further irresectable liver metastases and a chemotherapy consisting of FOLFIRI and bevacizumab was administered. Unfortunately, the tumor showed only a partial remission with the additional diagnosis of lung metastases. The patient died 22 mo after the operation due to tumor progression with liver and lung metastases.
In vitro and in vivo approaches were combined as described previously[13] for establishment of matching cell lines and PDX. With this method, direct cell line establishment could be achieved in two out of three cases (HROC126 and HROC284Met) with HROC239 T0 M1 originating from a parallel PDX (Table 1).
| Tumor ID | Direct cell line establishment | Cell line from xenograft | Corresponding xenograft | Paired B-LCL |
| HROC126 | + | - | + | + |
| HROC239 T0 M1 | - | + | + | + |
| HROC284Met | + | - | + | + |
Outgrowth of cells in culture occurred immediately. Doubling times of the cell lines were 34.8 h for HROC126, 28.8 h for HROC239 T0 M1 and 38.1 h for HROC284Met. Tumor formation in immunodeficient NMRI nu/nu mice could be observed as fast as 1-3 mo after the tumor engraftment. Histological analysis showed that tumor architecture was preserved in the PDX compared to the original patient tumor architecture (data not shown).
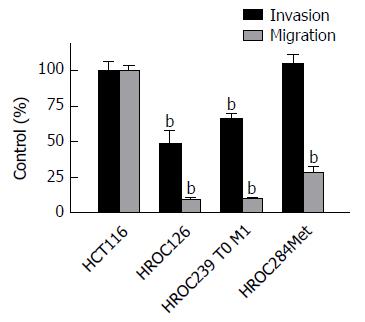
Analysis of invasion and migration revealed a significant reduced infiltrative activity of HROC126 and of HROC239 T0 M1 compared to the reference line HCT116 (t-test, P < 0.05), while infiltrative activity of HROC284Met was comparable to HCT116.
Regarding the migratory activity, all three cell lines (HROC126, HROC239 T0 M1 and HROC284Met) were significantly less migratory active (t-test, P < 0.01) through uncoated Boyden chambers then the HCT116 reference cell line (Figure 1).
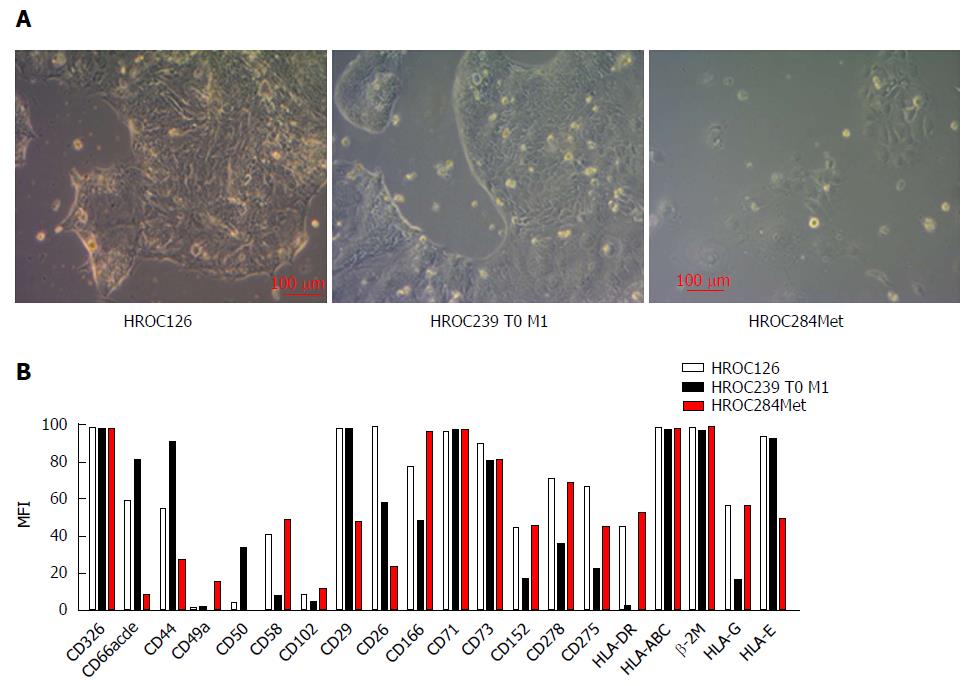
As determined by phase contrast microscopy, cells of all three HROC lines adhered tightly to the cell culture flasks. The cell lines were growing as monolayers on conventional tissue culture plastic and showed a stable outgrowth as defined by passaging > 40 times. HROC284Met cells proliferated as polygonal cell clusters with more regular dimensions, while HROC126 and HROC239 T0 M1 cells formed tightly packed multicellular islands (Figure 2). Morphology did not change during long term passage (up to 40 passages) (data not shown). As determined by semi-quantitative PCR, HROC126, HROC239 T0 M1 and HROC284Met cell lines were free of mycoplasma and several other potential contaminants (JC/BK and SV40) which have been described for (colorectal) cell lines (data not shown).
The epithelial phenotype of all three cell lines was confirmed by high positive immunoreactivity for CD326 (EpCAM) while expression of CD66abcd (CEACAM) differed. HROC126 and HROC239 T0 M1 showed moderate to high expression of CD66abcd, while HROC284Met showed no expression (Figure 2).
A more detailed characterization revealed very heterogeneous expression of adhesion and co-stimulatory factors (moderate to high expression of CD44, CD58 and CD166 but low to absent expression of CD49a, CD50 and CD102) (Figure 2). Further analysis showed a high to moderate expression of CD26 and CD29, which have been described as stem cell and metastasis-promoting surface receptors[21,22] (Figure 2).
The proliferation marker CD71 was highly expressed by all three cell lines (Figure 2) reflecting high proliferative activity of the tumor cells.
Regarding HLA molecules that play an important role in specific immune recognition and tumor cell defense, all three cell lines showed a high expression of HLA class I (β2M and pan-HLA-ABC) molecules. HROC126 and HROC284Met showed a moderate expression of HLA class II (HLA-DR), while HROC239 T0 M1 showed no expression. In addition, all three tumor models presented a distinct expression of HLA-E and HLA-G molecules which are involved in immune suppression (Figure 2).
Additional immune evasion molecules analyzed included CD73 which was highly expressed by all three cell lines. Lower levels were observed for CD278 [inducible co-stimulator (ICOS)], CD275 (B7-H2) and CD152 [cytotoxic T-lymphocyte-associated protein 4 (CTLA-4)] (Figure 2).
To verify the genetic identity of our established tumor models to the parental tumors, a DNA fingerprint was performed. Several highly polymorphic STR DNA markers that covered nine validated STR loci on different chromosomal locations were included into this analysis. We could confirm identity to the parental tumors in all established cell lines and PDX from the three clinical cases included, using this approach. Also, cross contamination of any of the tumor cell lines with other tumor cells could be excluded (data not shown).
Furthermore, the molecular features of the cell lines were determined in direct comparison to the original tumors as well as to PDX tissues. All three cases showed a distinct degree of aneuploidy, absent MSI and no methylation in CIMP-sensitive promotors. Thus, they can all be classified as CIMP-0/ non-MSI-H (spStd) type. HROC126 presented wild type APC, TP53, KRAS, and BRAF genes, while HROC239 T0 M1 presented mutations in the APC, KRAS, TP53, and PTEN genes with a BRAF wild type. HROC284Met presented with a mutation in the KRAS and PTEN gene and in addition a BRAF wild type (Table 2).
| Cell line | CIMP-number | MSI-status | Ploidy status | TP53 | APC | K-Ras | PIK3CA | BRAF | PTEN |
| HROC126 | 0 | Mss | Aneuploid | Wt | Wt | Wt | Wt | Wt | Wt |
| HROC239 T0 M1 | 0 | Mss | Aneuploid | Mut | Mut | Mut | Wt | Wt | Mut |
| HROC284Met | 0 | Mss | Aneuploid | Mut | Wt | Mut | Wt | Wt | Mut |
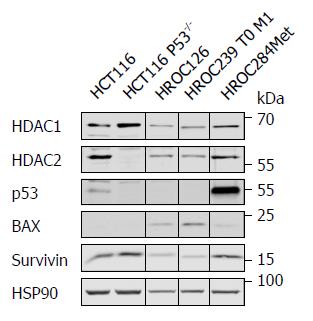
To confirm that HROC126 cells have wild type p53, HROC239 T0 M1 cells have mutant p53, and to gain insight into the p53 status of HROC284Met cells, we performed Western blot analyses for p53 and some of its target genes. Since epigenetic regulators of the histone deacetylase family are frequently dysregulated in colon cancer cells, we also tested for their expression. P53-positive and p53-negative HCT116 cells served as control.
All new HROC cell lines were positive for HDAC1 and HDAC2 (Figure 3). HROC126 and HROC239 T0 M1 carried undetectable levels of p53, which can be a marker for active wild type p53 or a loss of p53[23]. While HROC239 T0 M1 cells expressed the p53 target gene BAX avidly, HROC126 expressed very low levels of this protein. HROC284Met showed high p53 expression, which is a typical feature of mutant p53. This ties in with its low level of BAX and increased level of survivin (Figure 3).
These data verify our genetic analyses and suggest that the p53 status of our novel cellular models reflects p53 activity.
Response testing of individual tumor cell lines to current chemotherapeutic drugs and concomitant radiation therapy has become more and more valuable due to huge heterogeneity of individual tumor responses to established therapy regimens. Thus, sensitivity of the three cell lines to a panel of current chemotherapeutic drugs and their combination was assessed using standard proliferation and cytotoxicity assays.
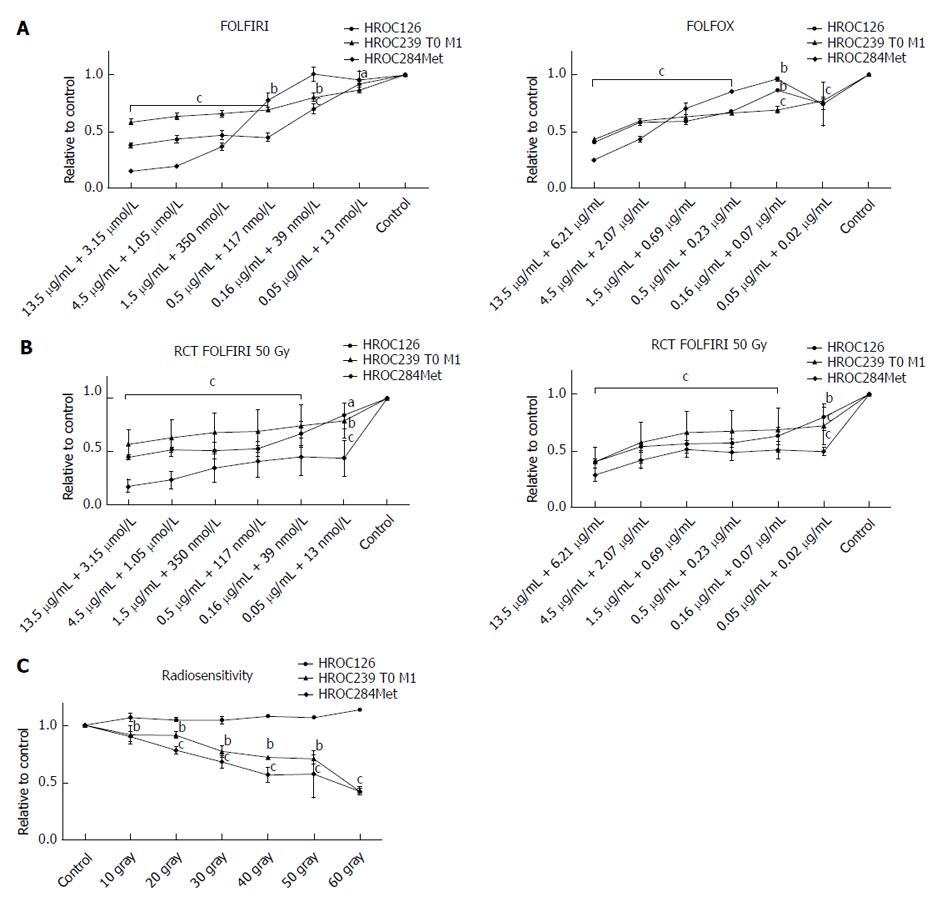
Analysis of drug sensitivity to single substances of 5-FU, irinotecan and oxaliplatin showed an individual response of all three cell lines, with HROC239 T0 M1 being more resistant to oxaliplatin (IC50 = 2.4 μg/mL) (Table 3). Doses were comparable or lower than achievable plasma concentrations in patients (Table 3).
| Cell line | 5-FU (μg/mL) | Irinotecan (μmol/L) | Oxaliplatin (μg/mL) |
| HROC126 | 0.42 | 0.72 | 0.3 |
| HROC239 T0 M1 | 21 | 4.4 | 2.4 |
| HROC284Met | 7 | 4.3 | 0.68 |
| Plasma levels (pharmacokinetic) | 50 | 10 | 2 |
Combination of drugs, currently administered in intensified therapy regimens, FOLFIRI and FOLFOX showed again a good response of all three cell lines, with HROC284Met being less susceptible to FOLFIRI and FOLFOX than HROC126 and HROC239 T0 M1 (Figure 4).
Next step was evaluating the resistance of the cell lines to γ-radiation. Here, all cell lines showed a remarkable resistance to radiation and even radiation with 50 Gy could not prevent further growth of the cell lines; particularly HROC126 displayed a highly radio resistant phenotype (Figure 4).
At last, as actual neoadjuvant regimens for advanced rectal cancer consist of a combined radiochemotherapy (FOLFOX or FOLFIRI with 50 Gy), this was administered to the cell lines. Furthermore, an individual response of all three cell lines could be observed, with HROC284Met being more susceptible than HROC126 and HROC239 T0 M1. Again, when patient plasma levels are set for reference, measured IC50 concentrations were below those plasma concentrations (Figure 4).
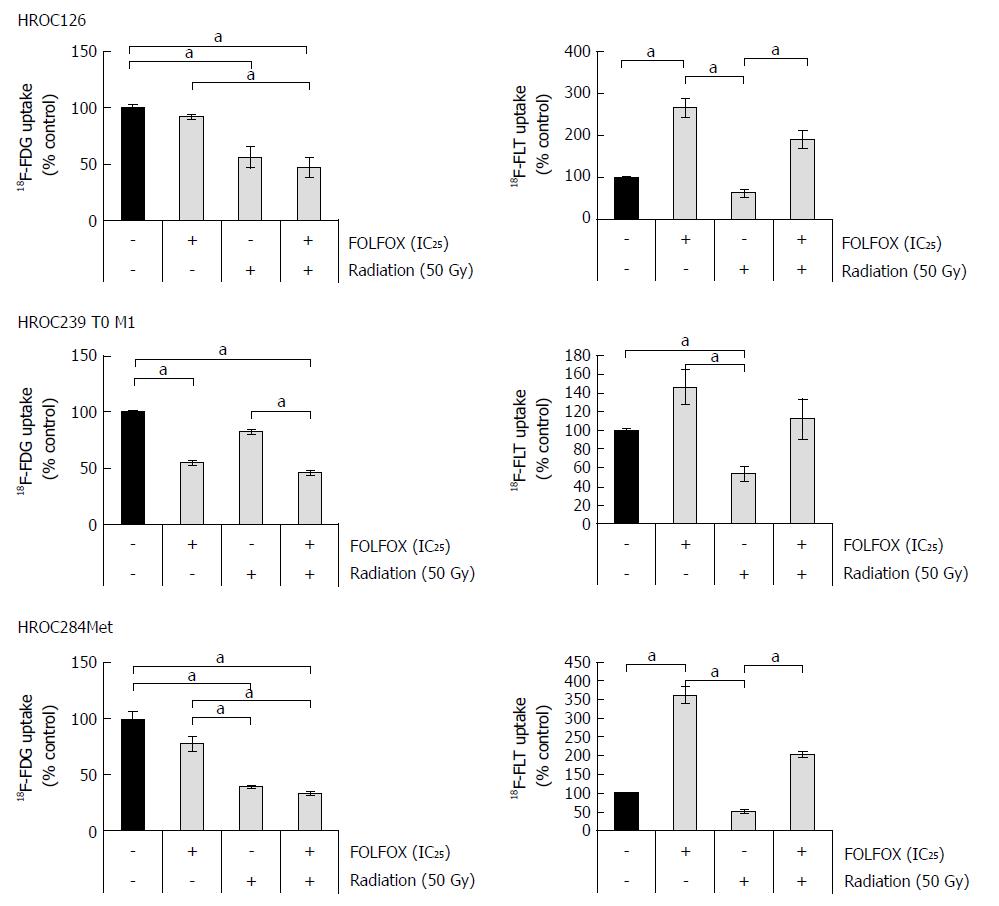
Analysis of 18F-FDG uptake showed an individual response of all three cell lines.
HROC239 T0 M1 is the only cell line that presented a reduced glucose uptake after chemotherapy, whereas HROC126 and HROC284Met were sensitive to single radiation treatment (Figure 5). Only after radiation, a significant reduction of tracer uptake was determined in these two cell lines. In HROC239 T0 M1 cells, the application of FOLFOX at an IC25 dose led to a reduced 18F-FDG uptake. All three tumor models displayed a reduced 18F-FDG uptake after a combined radiochemotherapy.
Regarding uptake of 18F-FLT, which might reflect higher tumor cell proliferation activity, we found mixed results. Interestingly, HROC126 and HROC284Met displayed a similar response pattern as shown in the FDG trials. In both cell lines, FLT uptake was elevated after FOLFOX treatment, whereas a single radiation therapy showed a lower thymidine uptake in comparison with single chemotherapy or combination with FOLFOX. In HROC239 T0 M1 cells, a markedly but not significantly higher uptake of FOLFOX treated cultures compared to control was observed. Again, radiation alone displayed a lower number of counts in comparison to FOLFOX treatment or controls.
To gain additional information on the proliferation status of the cell lines under treatment, a flow cytometric analysis of Ki-67 expression was performed. In all treatment groups (FOLFOX, radiation, combined FOLFOX and radiation), HROC126 and HROC239 T0 M1 showed a significantly reduced expression of Ki-67 compared to untreated controls. HROC284Met revealed a significantly reduced expression of Ki-67 in the radiation and combination group (FOLFOX and radiation) and a markedly reduced activity in the FOLFOX group (Supplementary Figure 1).
In the last decade, many patient-individual tumor models of colonic cancer were established by our lab and others[8,9]. These models have shown to be extremely helpful in decoding colorectal cancers’ molecular heterogeneity[9,24] and identification of novel biomarkers[25], druggable targets and novel treatment strategies[26]. But no novel rectal cancer models have been published up to now. Here, we describe the establishment and functional characterization of three patient-derived rectal cancer cell lines that could be established from fresh patients’ tumor samples for the first time.
These established cell lines reproduce the original molecular signature of the tumor in a perfect way. In addition, PDX models of the original tumors could be established. PDX models show high morphologic and molecular correspondence with the analogous patient tumor, facilitating pharmacologic studies to predict clinical response[27,28].
While HROC126 and HROC239 T0 M1 were obtained from locally advanced rectal cancer primaries, HROC284Met was acquired from a liver metastasis. Thus, the established rectal cancer cell lines may represent valuable models for advanced rectal cancer that requires multimodal therapy according to international guidelines[29].
Regarding growth kinetics and to some aspect morphology, the three cell lines displayed some variations. While migratory activity was comparable between all three cell lines, HROC284Met showed a significantly higher infiltrative activity compared to HROC126 and HROC239 T0 M1. This fact might reflect the worse clinical outcome of HROC284Met in terms of a metastatic and rapid progressive disease.
The epithelial phenotype of all three lines was confirmed by high positive immunoreactivity for CD326 (EpCAM); while expression of CD66abcd (CEACAM) was heterogeneous with HROC284Met showing even no expression of this tumor antigen. Further characterization revealed very heterogeneous expression of adhesion and co-stimulatory factors (moderate to high expression of CD44, CD58 and CD166 but low to absent expression of CD49a, CD50, and CD102). Interestingly, a high to moderate expression of CD26 and CD29, which have been described as stem cell and metastasis-promoting surface receptors[21,22] could be shown. Reflecting the high proliferative activity of the tumor cells, the proliferation marker CD71 was highly expressed in all three cell lines.
As tumor immune escape mechanisms play a more and more important role in clinical oncology, we analyzed several key molecules of immune escape. All three cell lines express high levels of CD73, which has been described as immune-evasion molecule and overexpression was observed in various tumor tissues[30]. Interestingly, a moderate expression of CD152 (CTLA-4) could be demonstrated, especially in HROC126 and HROC284Met. CTLA-4 is an immunoglobulin superfamily receptor and was the first immune-checkpoint receptor that was clinically targeted using blocking antibodies[31]. This finding suggests that the rectal cancer tumor models could also help to develop immune checkpoint inhibiting strategies and to optimize immune-therapeutic applications.
Regarding HLA molecules that play an important role in specific immune recognition and tumor cell defense, expression of HLA class I molecules was preserved in all three cell lines. HROC126 and HROC284Met cells were additionally found to be positive for HLA class II molecules. This provides a basis for further immunological analysis aiming at identification of immunogenic epitopes from shared or individual neo-antigens.
Lately, tumor sidedness has emerged as an important prognostic and predictive element in the treatment of CRC[32]. Clinicopathological and molecular features of CRCs differ depending on the bowel subsite of the tumor[5]. The molecular profiles of all three rectal cancer cell lines were associated with the CIMP-0/ non-MSI-H (spStd) type[33]. This finding is not surprising as approximately 70% of all sigmoid and rectal cancers show the CIMP-0/ non-MSI-H (spStd) subtype[34]. This fact may account for the differences in the clinicopathological features of right sided and left sided or rectal cancers[35]
Regarding the tumor specific mutational analysis, a BRAF wild type could be detected in all three cell lines. This is not unexpected as Salem et al[32]. analyzed 1445 rectal tumors and found a BRAF mutation in only 3.2% of all examined tumor samples. Likewise, the observed mutations of KRAS, TP53 and APC in HROC239 T0 M1 and HROC284Met correlate with the frequent mutation of these genes in rectal tumors ranging from 50%- 66%. Of interest, a PTEN mutation was found in HROC239 T0 M1 and in HROC284Met, while several studies could show that PTEN mutations are rare in rectal tumors [35].
Current guidelines recommend nCRT in locally advanced rectal cancer[29]. Hence, cell lines were tested against a panel of current chemotherapeutical regimens. In addition, analysis of radiosensitivity and sensitivity of combined radio- and chemotherapy was evaluated, particularly with regard to the known differing clinical response rates to nCRT. In principle, all cell lines were susceptible towards these regimens as well as to the single substances (5-FU, irinotecan, oxaliplatin) or combinations of drugs (FOLFOX, FOLFIRI) which are currently administered in intensified therapy regimens. Of note, doses were comparable or lower than plasma concentrations measurable in patients.
Analysis of sensitivity to sole radiation showed a remarkable resistance of HROC126 even at high radiation doses, while HROC239 T0 M1 and HROC284Met displayed a moderate sensitivity. As shown above, HROC239 T0 M1 and HROC284Met exhibited a PTEN mutation. It is known that the PI3K/PTEN/AKT/mTOR signaling pathway participates in drug resistance, tumorigenesis and progression of cancer[36]. These alterations have been implicated as modulators of clinical outcomes in patients with esophageal cancer who underwent CRT[37]. In particular, a loss of PTEN decreases CHK1 and TP53 activity by regulating their protein levels and promoting genomic instability in tumor cells[38]. In rectal cancer, a recent study by Peng et al. could show that patients with a PTEN mutation displayed a better response to CRT than those with a wild-type genotype[39]. This might contribute to a more distinct sensitivity to radiation of HROC239 T0 M1 and HROC284Met in contrast to the PTEN wild type HROC126.
Concerning p53 and its target gene induction, we see no clear correlation between p53 status and the sensitivity of our cell models to drugs and γ-irradiation. This is not due to a lack of correlation between p53 expression and activity. The low to undetectable levels of p53 in HROC126 and HROC239 T0 M1 cells seems to be a marker for active wild type TP53 or a loss of TP53, respectively[22]. While HROC126 cells express various p53 target genes, HROC239 T0 M1 carry very low levels thereof (Figure 3). The mutation of TP53 in HROC284Met agrees with TP53 mutations in advanced cancers and metastases[40]. The high levels of survivin, which is a target of mutant p53 in combination with nuclear factor kappa B (NF-кB) p65, and a suppressor of chemosensitivity[40] is not reflected by our assays assessing cellular sensitivity to drugs and γ-irradiation. Thus, additional factors need to be considered when molecular markers are used to predict the responsiveness of cancer cells to drugs and γ-irradiation. Although the class I HDAC HDAC2 is frequently dysregulated in MSI colorectal cells[23], we see no loss of HDAC2 in our systems. This might be due to the fact that a loss of HDAC2 only affects a portion of HDAC2 positive cells in culture (Krämer et al, unpublished observation).
Analyzing the combination of chemo- and radiotherapy, once more an individual response of all three cell lines became evident. Identification of novel mechanisms leading to radiation-resistance might help to overcome this and might thus improve therapy efficacy[41] particularly as different responses to nCRT are associated with differences in long-term outcomes including disease-free survival[42]. Therefore, this novel set of rectal cancer models represents an ideal tool for further research also in this area.
At last, we analyzed uptake of 18F-FDG and 18F-FLT in the cell lines. Rosenberg et al[43] reported that the metabolic response to nCRT in rectal cancer can be correlated with histopathological response using 18F-FDG PET. Analysis of glucose uptake revealed a good response of all three cell lines. While HROC126 and HROC284Met showed a significantly reduced 18F-FDG uptake in radiated cultures, HROC239 T0 M1 showed a significantly reduced uptake after FOLFOX treatment and in the chemoradiation group. In addition, the combination of FOLFOX and radiation resulted in all three cell lines in a significantly reduced 18F-FDG uptake compared to solitary FOLFOX treatment. This finding emphasizes the possible use of 18F-FDG PET for detection of early tumor responses and restaging after nCRT.
Many anti-cancer drugs target cell proliferation as primary or secondary target. These changes can be investigated using 18F-FLT PET after chemotherapeutic treatment. However, 18F-FLT changes following chemotherapy are inconstant and depend on the tumors and specific treatments[44]. In two out of the three novel rectal cancer cell lines, FLT could be predictor for response to a chemotherapy regimen, where an increased FLT uptake was determined.
After radiation, all three cell lines showed a significantly reduced uptake of 18F-FLT in comparison to FOLFOX, reflecting an expected decline of cell proliferation after radiation. The combined RCT treatment displayed again a significantly higher uptake of 18F-FLT for two of the three cell lines. This marked retention of 18F-FLT might be triggered by 5-FU that interferes with endogenous thymidine synthesis and can cause rapid accumulation of thymidine kinase 1 levels[45]. Of note, Hong et al[46] reported that the 18F-FLT flare observed during 5-FU infusion was associated with poor treatment response in patients with mCRC. Hence, more clinical studies are needed to define the role of 18F-FLT PET after FOLFOX treatment.
In summary, the enormous strength of the present study lies in the establishment and characterization of the first pairs of matching in vitro and in vivo patient-individual rectal cancer models (cell lines + PDX). They represent ideal tools for further development of personalized medicine concepts of rectal cancer. However, given the small sampling size of n = 3, the number of rectal cancer cases successfully transferred into models must be considered as a limitation. Similarly, the molecular and functional data provided could be expanded.
It is well known that ultra-low passage and in-depth characterized patient-derived tumor models are highly desirable for basic research and for predicting individual responses to current or novel therapy regimens.
To establish individual tumor models of rectal cancer from patient-derived tumor samples to gain further insights into the biological behavior of rectal cancer.
Main objective of the study was the establishing and profound characterization of new patient-derived rectal cancer cell lines with corresponding patient-derived xenograft models that allow testing of drug response, translational and basic research.
Establishment of cell lines could be achieved by direct in vitro culturing and in vivo xenografting with following in vitro culturing. Profound analysis of morphological features, invasive and migratory behavior, phenotype, molecular profile including mutational analysis, and protein expression was done. Responsiveness to current chemotherapeutic drugs was evaluated and sensitivity to radiation and combined radio-chemotherapy was examined. At last the positron emission tomography (PET) tracers 18F-fluorodeoxyglucose (FDG) and 18F-fluorothymidine were used to assess glucose metabolism and proliferation activity respectively.
Three individual ultra-low passage rectal cancer cell lines could be established. In vitro and in vivo experiments demonstrated that all cell lines retained their malignant properties. Molecular analysis classified all three cell lines as sporadic type (CIMP-0/non-MSI-H). Mutational analysis revealed an individual mutational profile of each cell line (HROC126: APCwt, TP53wt, KRASwt, BRAFwt, PTENwt; HROC239 T0 M1: APCmut, P53wt, KRASmut, BRAFwt, PTENmut and HROC284Met: APCwt, P53mut, KRASmut, BRAFwt, PTENmut). The cell lines demonstrated a heterogeneous response to chemotherapy, radiation and combined radio-chemotherapy. Interestingly, analysis of glucose metabolism showed a markedly reduced uptake of the PET tracer 18F-FDG after combined radio-chemotherapy of all three cell lines.
Taken together, this study describes the development and in-depth characterization of three patient-derived rectal cancer cell lines that could be established from fresh patients´ tumor samples for the first time. These powerful matched in vitro and in vivo models provide useful tools not only to perform basic research to better understand the biology of rectal cancer, but also to test and establish novel therapy regimens.
This descriptive study exemplifies the methodology and characterization of rectal cancer cell lines obtained directly from patients´ tumor material. This is an important step to extend the abilities of personalized tumor therapy in the near future.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Biondi A, Costi R S- Editor: Ma RY L- Editor: A E- Editor: Bian YN
| 1. | Bae JM, Kim JH, Kang GH. Molecular Subtypes of Colorectal Cancer and Their Clinicopathologic Features, With an Emphasis on the Serrated Neoplasia Pathway. Arch Pathol Lab Med. 2016;140:406-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 2. | Jass JR. Molecular heterogeneity of colorectal cancer: Implications for cancer control. Surg Oncol. 2007;16 Suppl 1:S7-S9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 3. | Ostwald C, Linnebacher M, Weirich V, Prall F. Chromosomally and microsatellite stable colorectal carcinomas without the CpG island methylator phenotype in a molecular classification. Int J Oncol. 2009;35:321-327. [PubMed] |
| 4. | Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 955] [Cited by in RCA: 1004] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 5. | Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 493] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 6. | van der Paardt MP, Zagers MB, Beets-Tan RG, Stoker J, Bipat S. Patients who undergo preoperative chemoradiotherapy for locally advanced rectal cancer restaged by using diagnostic MR imaging: a systematic review and meta-analysis. Radiology. 2013;269:101-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 279] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 7. | Joye I, Deroose CM, Vandecaveye V, Haustermans K. The role of diffusion-weighted MRI and (18)F-FDG PET/CT in the prediction of pathologic complete response after radiochemotherapy for rectal cancer: a systematic review. Radiother Oncol. 2014;113:158-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 139] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 8. | Maletzki C, Gock M, Randow M, Klar E, Huehns M, Prall F, Linnebacher M. Establishment and characterization of cell lines from chromosomal instable colorectal cancer. World J Gastroenterol. 2015;21:164-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (1)] |
| 9. | van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1313] [Cited by in RCA: 1728] [Article Influence: 192.0] [Reference Citation Analysis (0)] |
| 10. | Zaniboni A, Savelli G, Pizzocaro C, Basile P, Massetti V. Positron Emission Tomography for the Response Evaluation following Treatment with Chemotherapy in Patients Affected by Colorectal Liver Metastases: A Selected Review. Gastroenterol Res Pract. 2015;2015:706808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Gauthé M, Richard-Molard M, Cacheux W, Michel P, Jouve JL, Mitry E, Alberini JL, Lièvre A; Fédération Francophone de Cancérologie Digestive (FFCD). Role of fluorine 18 fluorodeoxyglucose positron emission tomography/computed tomography in gastrointestinal cancers. Dig Liver Dis. 2015;47:443-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Bollineni VR, Collette S, Liu Y. Functional and molecular imaging in cancer drug development. Chin Clin Oncol. 2014;3:17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 13. | Maletzki C, Stier S, Gruenert U, Gock M, Ostwald C, Prall F, Linnebacher M. Establishment, characterization and chemosensitivity of three mismatch repair deficient cell lines from sporadic and inherited colorectal carcinomas. PLoS One. 2012;7:e52485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Linnebacher M, Maletzki C, Ostwald C, Klier U, Krohn M, Klar E, Prall F. Cryopreservation of human colorectal carcinomas prior to xenografting. BMC Cancer. 2010;10:362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Fielding LP, Arsenault PA, Chapuis PH, Dent O, Gathright B, Hardcastle JD, Hermanek P, Jass JR, Newland RC. Clinicopathological staging for colorectal cancer: an International Documentation System (IDS) and an International Comprehensive Anatomical Terminology (ICAT). J Gastroenterol Hepatol. 1991;6:325-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 284] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 16. | Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90-96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 633] [Cited by in RCA: 640] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 17. | Goodenough DJ. The use of ROC curves in testing the proficiency of individuals in classifying pneumoconiosis. Radiology. 1975;114:472-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 18. | Klier U, Maletzki C, Klar E, Linnebacher M. Generation of highly pure fusions of colorectal carcinoma and antigen-presenting cells. Langenbecks Arch Surg. 2010;395:365-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 19. | Gock M, Mullins CS, Harnack C, Prall F, Ramer R, Göder A, Krämer OH, Klar E, Linnebacher M. Establishment, functional and genetic characterization of a colon derived large cell neuroendocrine carcinoma cell line. World J Gastroenterol. 2018;24:3749-3759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 20. | Rauch A, Carlstedt A, Emmerich C, Mustafa AM, Göder A, Knauer SK, Linnebacher M, Heinzel T, Krämer OH. Survivin antagonizes chemotherapy-induced cell death of colorectal cancer cells. Oncotarget. 2018;9:27835-27850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Davies S, Beckenkamp A, Buffon A. CD26 a cancer stem cell marker and therapeutic target. Biomed Pharmacother. 2015;71:135-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Vassilopoulos A, Chisholm C, Lahusen T, Zheng H, Deng CX. A critical role of CD29 and CD49f in mediating metastasis for cancer-initiating cells isolated from a Brca1-associated mouse model of breast cancer. Oncogene. 2014;33:5477-5482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Wagner T, Brand P, Heinzel T, Krämer OH. Histone deacetylase 2 controls p53 and is a critical factor in tumorigenesis. Biochim Biophys Acta. 2014;1846:524-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, Arango D, Strausberg RL, Buchanan D, Wormald S. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238-3247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 292] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 25. | Falzone L, Scola L, Zanghì A, Biondi A, Di Cataldo A, Libra M, Candido S. Integrated analysis of colorectal cancer microRNA datasets: identification of microRNAs associated with tumor development. Aging (Albany NY). 2018;10:1000-1014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 26. | Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, Buscarino M, Isella C, Lamba S, Martinoglio B. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 240] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 27. | Rosfjord E, Lucas J, Li G, Gerber HP. Advances in patient-derived tumor xenografts: from target identification to predicting clinical response rates in oncology. Biochem Pharmacol. 2014;91:135-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 28. | William D, Mullins CS, Schneider B, Orthmann A, Lamp N, Krohn M, Hoffmann A, Classen CF, Linnebacher M. Optimized creation of glioblastoma patient derived xenografts for use in preclinical studies. J Transl Med. 2017;15:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Glynne-Jones R, Wyrwicz L, Tiret E, Brown G, Rödel C, Cervantes A, Arnold D; ESMO Guidelines Committee. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28:iv22-iv40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1112] [Cited by in RCA: 1190] [Article Influence: 148.8] [Reference Citation Analysis (0)] |
| 30. | Jin D, Fan J, Wang L, Thompson LF, Liu A, Daniel BJ, Shin T, Curiel TJ, Zhang B. CD73 on tumor cells impairs antitumor T-cell responses: a novel mechanism of tumor-induced immune suppression. Cancer Res. 2010;70:2245-2255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 354] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 31. | Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9936] [Cited by in RCA: 10302] [Article Influence: 792.5] [Reference Citation Analysis (34)] |
| 32. | Salem ME, Weinberg BA, Xiu J, El-Deiry WS, Hwang JJ, Gatalica Z, Philip PA, Shields AF, Lenz HJ, Marshall JL. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget. 2017;8:86356-86368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 157] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 33. | Prall F, Ostwald C, Linnebacher M. Tubular invasion and the morphogenesis of tumor budding in colorectal carcinoma. Hum Pathol. 2009;40:1510-1512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 34. | Bae JM, Kim JH, Cho NY, Kim TY, Kang GH. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer. 2013;109:1004-1012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 35. | Loree JM, Pereira AAL, Lam M, Willauer AN, Raghav K, Dasari A, Morris VK, Advani S, Menter DG, Eng C. Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular Subtypes. Clin Cancer Res. 2018;24:1062-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 218] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 36. | Costa C, Pereira S, Lima L, Peixoto A, Fernandes E, Neves D, Neves M, Gaiteiro C, Tavares A, Gil da Costa RM. Abnormal Protein Glycosylation and Activated PI3K/Akt/mTOR Pathway: Role in Bladder Cancer Prognosis and Targeted Therapeutics. PLoS One. 2015;10:e0141253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 37. | Hildebrandt MA, Yang H, Hung MC, Izzo JG, Huang M, Lin J, Ajani JA, Wu X. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol. 2009;27:857-871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 142] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 38. | Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, Mansukhani M, Murty VV, Gaciong Z, Meek SE. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 259] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 39. | Peng J, Ma W, Zhou Z, Gu Y, Lu Z, Zhang R, Pan Z. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway predict tumor response and disease-free survival in locally advanced rectal cancer patients receiving preoperative chemoradiotherapy and radical surgery. J Cancer. 2018;9:1067-1077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 40. | Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1319] [Cited by in RCA: 1278] [Article Influence: 106.5] [Reference Citation Analysis (0)] |
| 41. | Alonzi R. Functional Radiotherapy Targeting using Focused Dose Escalation. Clin Oncol (R Coll Radiol). 2015;27:601-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 42. | Maas M, Nelemans PJ, Valentini V, Das P, Rödel C, Kuo LJ, Calvo FA, García-Aguilar J, Glynne-Jones R, Haustermans K. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11:835-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1189] [Cited by in RCA: 1454] [Article Influence: 96.9] [Reference Citation Analysis (0)] |
| 43. | Rosenberg R, Herrmann K, Gertler R, Künzli B, Essler M, Lordick F, Becker K, Schuster T, Geinitz H, Maak M. The predictive value of metabolic response to preoperative radiochemotherapy in locally advanced rectal cancer measured by PET/CT. Int J Colorectal Dis. 2009;24:191-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 70] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 44. | Soloviev D, Lewis D, Honess D, Aboagye E. [(18)F]FLT: an imaging biomarker of tumour proliferation for assessment of tumour response to treatment. Eur J Cancer. 2012;48:416-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 45. | Lee SJ, Kim SY, Chung JH, Oh SJ, Ryu JS, Hong YS, Kim TW, Moon DH. Induction of thymidine kinase 1 after 5-fluorouracil as a mechanism for 3’-deoxy-3’-[18F]fluorothymidine flare. Biochem Pharmacol. 2010;80:1528-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 46. | Hong YS, Kim HO, Kim KP, Lee JL, Kim HJ, Lee SJ, Lee SJ, Oh SJ, Kim JS, Ryu JS. 3’-Deoxy-3’-18F-fluorothymidine PET for the early prediction of response to leucovorin, 5-fluorouracil, and oxaliplatin therapy in patients with metastatic colorectal cancer. J Nucl Med. 2013;54:1209-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |