Published online Oct 28, 2018. doi: 10.3748/wjg.v24.i40.4536
Peer-review started: July 4, 2018
First decision: August 1, 2018
Revised: August 31, 2018
Accepted: October 5, 2018
Article in press: October 5, 2018
Published online: October 28, 2018
Processing time: 118 Days and 14 Hours
Hepatocellular carcinoma (HCC) is the 3rd leading cause of cancer-related death worldwide. More than 80% of HCCs arise within chronic liver disease resulting from viral hepatitis, alcohol, hemochromatosis, obesity and metabolic syndrome or genotoxins. Projections based on Western lifestyle and its metabolic consequences anticipate a further increase in incidence, despite recent breakthroughs in the management of viral hepatitis. HCCs display high heterogeneity of molecular phenotypes, which challenges clinical management. However, emerging molecular classifications of HCCs have not yet formed a unified corpus translatable to the clinical practice. Thus, patient management is currently based upon tumor number, size, vascular invasion, performance status and functional liver reserve. Nonetheless, an impressive body of molecular evidence emerged within the last 20 years and is becoming increasingly available to medical practitioners and researchers in the form of repositories. Therefore, the aim this work is to review molecular data underlying HCC classifications and to organize this corpus into the major dimensions explaining HCC phenotypic diversity. Major efforts have been recently made worldwide toward a unifying “clinically-friendly” molecular landscape. As a result, a consensus emerges on three major dimensions explaining the HCC heterogeneity. In the first dimension, tumor cell proliferation and differentiation enabled allocation of HCCs to two major classes presenting profoundly different clinical aggressiveness. In the second dimension, HCC microenvironment and tumor immunity underlie recent therapeutic breakthroughs prolonging patients’ survival. In the third dimension, metabolic reprogramming, with the recent emergence of subclass-specific metabolic profiles, may lead to adaptive and combined therapeutic approaches. Therefore, here we review recent molecular evidence, their impact on tumor histopathological features and clinical behavior and highlight the remaining challenges to translate our cognitive corpus into patient diagnosis and allocation to therapeutic options.
Core tip: Recent work revealed substantial steps toward a unifying molecular classification of human hepatocellular carcinomas. The expected translation of high-throughput assays to the clinical practice will further refine evidence-based patient management in terms of prognosis and response to treatment.
- Citation: Désert R, Nieto N, Musso O. Dimensions of hepatocellular carcinoma phenotypic diversity. World J Gastroenterol 2018; 24(40): 4536-4547
- URL: https://www.wjgnet.com/1007-9327/full/v24/i40/4536.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i40.4536
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide[1] and its incidence has doubled over the past 20 years in Western countries. More than 80% of HCCs emerge within chronic inflammatory liver diseases resulting from chronic viral hepatitis, alcohol abuse, hemochromatosis, obesity and metabolic syndrome or DNA-damaging food or environmental toxins. Based on the current epidemics of obesity and metabolic syndrome in the Western world, projections anticipate an increase in HCC incidence, despite recent breakthroughs in the management of viral hepatitis[2]. HCCs display a high heterogeneity of molecular phenotypes, which raises major challenges in clinical management[3]. Currently, patient management is based on tumor number, size, vascular invasion, performance status and functional reserve of the liver[2]. A further obstacle complicating the understanding of basic human HCC biology (and of patient management) is that liver biopsy cannot routinely be used for HCC diagnosis because of the risk of complications[4,5]. Thus, surveillance for HCC emergence in patients with chronic liver diseases relies on biannual ultrasound or magnetic resonance imaging (MRI). As a surrogate for tumor liver biopsy, functional MRI-based metabolic imaging will become increasingly relevant for diagnosis and allocation to treatments in early-stage HCCs[6-8]. Surveillance programs currently detect small early-stage tumors that are candidates for potentially curative therapies (local ablation, resection or transplantation) with 5-year survival rates of approximately 50%-70%. However, high 5-year recurrence rates (up to 70%) after HCC resection or even higher after percutaneous ablation make liver transplantation the best possible treatment, with a recurrence rate of 10%[2]. As a result, the prediction of early recurrence and of the potential for cancer aggressiveness are key factors leading patient eligibility for liver transplantation[4]. Therefore, gaining insight into the diversity of HCC phenotypes and deepening our understanding of the mechanisms of progression from low-grade HCCs to aggressive tumors, that will ultimately kill the patient, will shed basic knowledge to improve patient management[4,7,9]. Systemic therapies for intermediate and advanced HCC patients not eligible for surgical approaches have made encouraging progress in the past 10 years. Sorafenib was the first systemic therapy to be approved in HCC[10-12]. Then, regorafenib and nivolumab were approved in the second-line setting after sorafenib, significantly improving the original survival benefit[13]. Importantly, HCC patients presenting beyond transplant or resection eligibility may benefit from locoregional therapies [tumor ablation, transarterial chemoembolization (TACE) and radioembolization with yttrium-90 microspheres (Y90)], according to recent Clinical Practice Guidelines[14]. Whereas tumor ablation is recommended as a first-line therapy for early-stage HCC[14]; TACE has been recommended for intermediate-stage HCC[14]. Y90 has been investigated as an alternative to TACE, with a good safety profile in delaying tumor progression[15] and has been proposed as a treatment of choice for down-staging HCC patients as a bridging strategy toward liver transplantation[16]. However, Y90 has not shown overall survival benefit over sorafenib in intermediate and locally advanced HCC patients after unsuccessful TACE[14,17].
The well-recognized phenotypic and genetic heterogeneity of HCCs occurs after a marathon of cellular changes driven by chronic inflammation, which progressively leads to severe fibrosis and profound remodeling of the tissue architecture. The two major consequences of this process are impaired liver function and the emergence of HCC[18]. In fact, chronic liver inflammation and progressive liver fibrosis generate a pro-tumorigenic microenvironment known as “the field effect”[19,20], whereby the diseased liver turns into a “minefield” riddled with pre-neoplastic and neoplastic foci. This longtime process can explain the relatively important number of mutations within each tumor, approximately 40[21-23], and the high heterogeneity between patients, as well as the frequent intra-tumor heterogeneity[3,24,25].
HCC heterogeneity is also amplified by the potential multiplicity of cell origin. One possibility for hepatocellular carcinogenesis is a well-established multistep process defined by a precise sequence of lesions, from cirrhosis to low-grade, then high-grade dysplastic nodules followed by early and advanced HCC[9]. This process is strongly enhanced by the TERT promoter mutation[26] and by MYC activation[27] and may involve retro-differentiation of hepatocytes to liver progenitor cells in an inflammatory environment[28-31]. In a different pathological context, HCCs can result from the malignant transformation of a hepatocellular adenoma carrying exon 3 mutations of CTNNB1, the gene encoding β-catenin[32]. Regardless of the case, a parallel can be stablished between the diversity of histopathological patterns of HCCs observed in the routine Anatomic Pathology laboratory and specific molecular programs[33-35].
HCC classifications (Table 1) have revealed multidimensional mechanisms leading to HCC heterogeneity. Here, we provide an overview of the major dimensions explaining the HCC phenotypic diversity with the aim of providing a unifying perspective of HCC classifications.
Pioneering transcriptomics studies classified 91 HCCs into two groups: Cluster A and Cluster B[36]. Cluster A showed high alpha fetoprotein (AFP), high Edmondson-Steiner’s scores indicating low cyto-architectural differentiation and unfavorable survival. These clinical features correlated with high expression of genes associated with cell proliferation and low hepatocyte differentiation. By contrast, genes typical of cluster B were liver-specific, indicating that these tumors were composed of well-differentiated, hepatocyte-like cells. Subsequent studies integrated the gene expression program of the 91-HCC dataset with the orthologous genes from several mouse models[37], whereby poorly differentiated human HCCs matched Myc-Tgfα transgenic mice. These mice typically showed early and high incidence rates of HCC development with ensuing high mortality. Their HCCs showed high rates of genomic instability and of expression of transcripts indicating poor prognosis[38]. Poorly differentiated human HCCs also matched a MET activation signature in genetically modified mice[39]. Moreover, integration of gene expression data from fetal hepatoblasts and adult hepatocytes with 61 cases of human HCCs revealed a group sharing gene expression patterns with fetal hepatoblasts[40]. These tumors fell into the category of proliferative poorly differentiated HCCs (cluster A). These pioneering studies shed light on a first layer of HCC heterogeneity and set the basis for the well-established classification of HCCs into two classes: non-proliferative and proliferative. Proliferative tumors are in general poorly differentiated, highly aggressive and associated with unfavorable patient outcome. On the other hand, non-proliferative HCCs tend to preserve a certain degree of hepatocyte differentiation and they are associated with more favorable patient outcome[2,9].
Further work searched for underlying dimensions that could explain phenotypic diversity within proliferative and non-proliferative HCCs by combining analysis of tumor transcriptomics’ programs and genetic mutations. Fifty-seven HCCs were classified into six groups (G1 to G6) and the expression of genes of interest was confirmed by real time qPCR in an independent collection of 63 HCCs. The aim of this approach was two-fold: first, to gain insight into the mechanisms leading to HCC heterogeneity; second, to find molecular markers enabling researchers, surgeons and oncologists to identify patients who may benefit from adaptive therapeutic approaches. However, one of the major pitfalls of biomarker identification is that the number of features (genes) entering the analysis is exceedingly higher than the number of observations (patients/tumors), which involves the risk of generating overfitting models. Overfitting may lead the classifiers to better describe a particular set of observations, but may not be equally performant to describe and classify sets of observations collected in different contexts. To circumvent this pitfall, statistical methods such as partial least squares are used to reduce the number of discriminant genes[41-43]. Importantly, the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria are applied to warrant that a set of markers performs consistently[44,45]. These include, among other recommendations, the use of training and validation sets to ensure consistency of results, followed by reproducibility studies in external validation cohorts.
Applying these procedures, studies revealed group G1, expressing high AFP levels in young women; group G2 associated with hemochromatosis and both groups G1 and G2 showing AKT activation as well as frequent AXIN1 mutation. G2 and G3 tumors were enriched in TP53 mutations, G3 containing the most poorly differentiated HCCs. Together, G1, G2 and G3 HCCs shared activation of biological pathways leading to cell proliferation and chromosome instability. G4 was a heterogeneous group of HCCs. Finally, groups G5 and G6 were associated with CTNNB1 mutation, which occurs in 30%-40% of HCCs, and showed strong expression of β-catenin target genes[9,42]. These tumors fit within the non-proliferative HCC class[9] because they proliferate at lower rates than G1-G3 tumors and tend to show higher cyto-architectural differentiation. However, two studies analyzing three different cohorts totaling 819 patients demonstrated that HCCs with mutated CTNNB1 do not differ in clinical outcome from HCCs with wild-type CTNNB1[43,46]. In fact, the survival curves and clinical features of patients undergoing resection of HCCs with mutated CTNNB1 suggest that the intrinsic aggressiveness of these tumors is intermediate between well-differentiated, non-proliferative HCCs with wild-type CTNNB1 and poorly-differentiated, proliferative HCCs[43,47].
As AXIN1 is part of the molecular scaffold involved in β-catenin inactivation, it was first suggested that AXIN1 mutations, which occur in 10% of HCCs, would lead to activation of the β-catenin pathway in HCCs[48,49]. However, later studies showed no association between AXIN1 mutations and activation of the β-catenin pathway in HCCs[50]. Indeed, AXIN1 deficiency induces HCCs in mice in the absence of activation of the β-catenin pathway[51]. HCCs with mutated AXIN1 differ in histology, genomic signature and outcome from those with CTNNB1 mutations[51]. Indeed, AXIN1-mutated HCCs impact the Notch and YAP pathways[51]. By contrast, although APC mutations are infrequent in HCCs (1%-2%), they are associated with activation of the β-catenin pathway[52,53].
The analysis of the clinical features associated with G1 to G6 subgroups in a cohort of 82 HCC patients from Singapore[54] confirmed the significant correlation between high AFP and groups G1 and 2, and showed an association between microvascular invasion and group G3. In a large cohort of 343 HCCs, associations were confirmed between the groups G1-G6 and clinical and pathological features as well as genetic aberrations[34]. G1 tumors were associated will female gender and high AFP, showed strong levels of KRT19, EPCAM and phospho-ERK protein expression. Also, groups G1 and G2 were enriched in AXIN1 mutations. Group G3 confirmed a high rate of macrovascular invasion and were associated with macrotrabecular massive subtype and poor outcome[33,34]. Groups G1, G2 and G3 had increased AFP serum levels and high TP53 mutation rates. On the other hand, group G4 was negatively associated with tumor size, vascular invasion, CTNNB1 and TP53 mutations and positively associated with the steatohepatitic subtype, inflammatory infiltrates and CRP protein expression. Groups G5 and G6 showed a high CTNNB1 mutation rate (80%), were positively associated with tumor differentiation and, by immunohistochemistry, were positive for β-catenin in the nuclei and cytoplasm and for GLUL (glutamine synthetase), a β-catenin target gene expressed in well-differentiated hepatocytes[33,34,50]. Finally, patients with G3 HCCs showed early tumor recurrence after resection and poor overall survival by univariate and multivariate analysis in a large cohort of 244 HCCs[55].
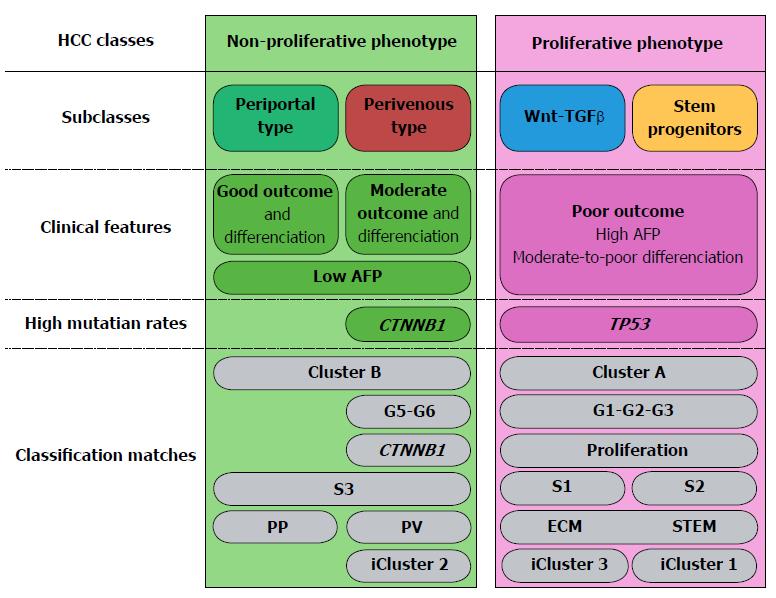
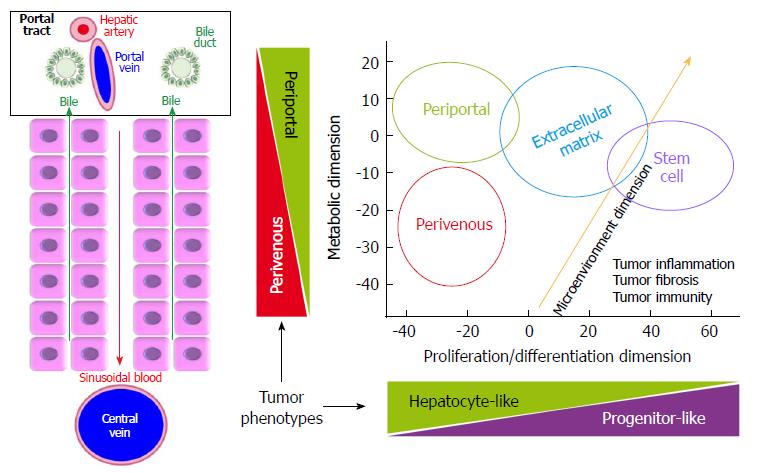
Taken together, a vast body of evidence confirms that HCCs can be grouped in two large classes: non-proliferative and proliferative (Figure 1), which are, respectively, well-differentiated and poorly differentiated tumors[2,9,31,47,56-58]. Proliferative and non-proliferative HCCs are respectively characterized by almost mutually exclusive mutational patterns. Indeed, CTNNB1 and TP53 mutations rarely coexist; they are respectively associated with the non-proliferative and proliferative HCC classes[31,52,56,57]. Non-proliferative, well-to-moderately differentiated HCCs, with low AFP serum levels match groups G5-G6[42]; S3[59,60] and B[36,40]. Our recent work showed that the class of non-proliferative HCCs can be split into two subclasses: Periportal-type (wild-type CTNNB1) and Perivenous-type (mutated CTNNB1), according to their respective metabolic liver zonation programs (Figure 1). Of note, patients undergoing resection of Periportal-type HCCs showed the lowest early (< 2 years) recurrence and the highest survival rates among all HCC patients treated by tumor resection in two cohorts of 247 and 210 subjects[43,47]. At the opposite, proliferative HCCs are poorly differentiated and highly aggressive. They match the G1-G2-G3[42], S1-S2[59,60], A[36,40] and ECM-STEM[43] subclasses and form a consensual class of tumors sharing a common background of large tumor size, high TP53 mutation rates, with loss of the hepatocyte-like phenotype, high AFP serum levels, extracellular matrix remodeling and angiogenesis[61-63]. Of note, the relationships between tumor size, differentiation, proliferation and development of a rich vascular network enabling tumor growth had been illustrated before the advent of molecular HCC classifications. In fact, modeling of HCC growth revealed two curves which were anti-parallel, but had similar slopes[64]. The first represented the loss of the hepatocyte-like phenotype through the expression of a liver-specific gene marker. The second represented the increase in tumor size and in the density of the vascular network[64]. Further studies revealed that this pattern was concomitant with an increase in extracellular matrix remodeling[65,66], suggesting plasticity of the tumor microenvironment across HCC progression.
HCC classification based on three publicly available transcriptome datasets (90, 82 and 60 HCCs, respectively) applying three unsupervised clustering methods (hierarchical clustering, non-negative matrix factorization and k-means clustering) defined HCC subclasses independently in the training sets before integration by a subclass mapping algorithm[59]. Three robust subclasses (S1, S2 and S3) were validated in six datasets (totaling 371 HCCs). Further work using whole genome sequencing and transcriptomics in 88 human HCCs confirmed that the outcome of the S1 and S2 subclasses was less favorable than that of the S3 subclass[67]. The S1 subclass was associated with signatures of Wnt/β-catenin, transforming growth factor beta (TGF-β) activation, epithelial-to-mesenchymal transition, vascular invasion and early recurrence by univariate and multivariate analyses.
It is interesting to note that both the S1 and S3 subclasses showed β-catenin pathway activation[59]. However, the mechanisms of pathway activation and the target genes responding to such stimuli were shown to be quite different. In the S3 subclass of non-proliferating, well-differentiated HCCs, grossly 50% of HCCs carried CTNNB1 mutations and expressed the so-called “liver-specific” β-catenin target genes, involved in hepatocyte metabolism, such as GLUL[59]. By contrast, the S1 subclass was not enriched in CTNNB1 mutations and expressed high levels of “classical Wnt genes”, i.e., Wnt targets involved in tumor cell proliferation, angiogenesis, epithelial-to-mesenchymal transition and extracellular matrix remodeling, such as CCND1, VEGF and MMP7[60]. In HCCs of the S3 subclass, activation of the β-catenin pathway results from CTNNB1 mutations in exon 3. By contrast, in HCCs of the S1 subclass activation of the β-catenin pathway was associated with upregulation in the expression of Wnt ligands, FZD receptors and TGFB1 target genes, that suggested Wnt/TGFB1 pathway cross-talks[60]. In vitro, well-differentiated hepatocyte-like HepaRG human HCC cells (wild-type CTNNB1), when plated at low density, spontaneously retro-differentiate to liver progenitors through a transient phase of epithelial-to-mesenchymal transition. This process involves translocation of β-catenin to the cell nuclei, indicating activation of the Wnt/β-catenin pathway[68]. These cells express a transcriptomic program that matches the S1 subclass of HCCs[28,29]. In vitro modeling of the microenvironment of the S1 subclass of HCC by incubating HepaRG liver progenitor cells with soluble Wnt3a ligand enhances and perpetuates the S1-like HCC phenotype, increasing HCC cell proliferation, invasive activity and driving the expression of extracellular matrix remodeling genes, as well as progenitor/stem cell markers associated with signatures of unfavorable HCC outcome[69]. This molecular phenotype can be reverted in vitro with small interfering RNAs targeting β-catenin, as well as with the soluble Wnt inhibitors FZD7_CRD and FZD8_CRD, that block the interaction of Wnt ligands with their cognate FZD receptors[69].
Although HCC are soft, cellular tumors with very scant extracellular matrix, some HCCs contain intra-tumor foci enriched in extracellular matrix glycoproteins, myofibroblasts and stem/progenitor cell markers that we called “fibrous nests”[69]. HCCs containing fibrous nests express high levels of the Wnt2 ligand, the FZD1 and FZD7 receptors, the Wnt inhibitors SFRP1, 2, 5; DKK1, the β-catenin target genes CD44, LEF1, LGR5, SOX9, GPC3 and, in particular, a minimal signature composed of COL4A1, LAMC1, DKK1 and SFRP1, associated with poor HCC outcome[69]. Of note, although the Wnt3 ligand is up-regulated in HCCs[70], HCCs containing fibrous nests are enriched in Wnt2 (and not Wnt3)[35]. In fact, Wnt2 is expressed by liver endothelial cells and stimulates liver regeneration from progenitor cells upon tissue damage[71]. In addition, Wnt2 secreted by tumor fibroblasts promotes progression of esophageal cancer[72]. These mechanisms are in line with the evidence that progenitor cell markers predicted outcome in 132 HCC patients undergoing liver transplantation[73].
The above body of evidence points to the clinical relevance of inflammation and the immune response of the host to the emergence and progression of HCCs. Transcriptomics analysis of a training set of 228 and a validation set of 728 HCCs revealed that approximately 25% of the tumors exhibit an immune profile compatible with a favorable response to immunotherapy, i.e., expression of the immune checkpoint modulators Programmed Death-Ligand 1 (CD274, a.k.a., PDL1) and Programmed Cell Death 1 (PDCD1, a.k.a., PD1)[74]. Thus, 25% of HCCs can be considered as the Immune HCC class, which contains two subtypes, exhibiting markers of adaptive T-cell response (active immune response) or of T-cell exhaustion (exhausted immune response). The latter may result from TGFB1 signaling mediating immunosuppression and is associated with S1 (i.e., Wnt/TGFB1-type) HCCs. The potential clinical relevance of these findings resides in the identification of an HCC class that may predict responses to checkpoint inhibitors in terms of survival benefits[75,76]. At least 80% of HCCs worldwide arise in a background of chronic inflammation and immune reactivity; thus, inflammatory mediators in the underlying non-tumor liver impact cancer HCC aggressiveness[19,77]. Cross-talk between the HCC cells and their microenvironment affect numerous biological functions. Therefore, it is not surprising that a wealth of transcriptome-based studies have identified signatures reflecting vascular invasion[78], metastasis[79] or angiogenesis[80] in patients with HCC with poor outcome. HCCs expressing cholangiocarcinoma traits[81], as well as those expressing the stem/biliary epithelial marker EPCAM[82,83] had also poor outcome, as well as a subset of HCCs expressing a signature reflecting late Tgfb1 activation in mice[84]. Most of these signatures were compared in a large cohort of 287 HCCs[55]. The study showed that most of the signatures fell within the same group of tumors, matching G3[42], proliferation class[58], and S1-S2[59] HCC subclasses. Therefore, inflammation, fibrosis and immunity are important components highly related to HCC phenotypic diversity.
Cancer cells proliferate and modify their microenvironment, which leads to extracellular matrix degradation, tissue invasion and metastases. These cancer cell functions call for increased energy production and macromolecule synthesis, which explains why malignant cells display a profound reprogramming of their metabolic pathways[85,86]. Non-targeted metabolic profiling on liquid chromatography-mass spectrometry of 50 HCCs revealed increased glycolysis, β-oxidation and gluconeogenesis; with reduced tricarboxylic acid (TCA) cycle activity. These changes were accompanied by increased levels of antioxidant molecules such as glutathione, as well as by lower levels of inflammatory-related polyunsaturated fatty acids. In particular, betaine and propionyl carnitine were proposed as markers to distinguish HCC from chronic hepatitis and cirrhosis[87]. A large-scale multicenter serum metabolite biomarker study identified a metabolite panel of interest in the detection of early HCC in patients at risk[88]. Combined transcriptomics and metabolomics in two tumor collections of 31 and 59 HCCs by gas chromatography-mass spectrometry-based metabolomics similarly showed increased glycolysis over mitochondrial oxidative phosphorylation and, in particular, increased lipid catabolism in the subgroup G1 of HCCs[89]. A transcriptomics study of 2761 metabolic genes in eight microarray datasets gathering 521 human HCCs confirmed down-downregulation of genes involved in physiologic hepatocyte metabolic functions, such as xenobiotic detoxification, fatty acid and amino acid metabolism[90]. The same study identified up-regulation of genes involved in glycolysis, pentose phosphate pathway, nucleotide biosynthesis, TCA cycle, oxidative phosphorylation and glycan metabolism; with several metabolic genes being associated with patient outcome and tumor progression markers[90]. These data fit the Warburg model of energy metabolism in cancer cells, whereby they bypass the TCA cycle and utilize glycolysis as the primary source of energy, that enables a less efficient but faster production of ATP[85,86]. However, up-regulation of genes involved in the TCA cycle[90] suggests that the metabolic landscape in HCCs is much more complex than a simple shift from oxidative phosphorylation to glycolysis, because cancer cells may rely on the TCA cycle for macromolecule synthesis[85]. Also, β-catenin-activated HCCs are not glycolytic, but oxidize fatty acids at high rates as an energy source, under the control of the transcription factor PPARα[91].
The Cancer Genome Atlas (TCGA) research network integrated data from multiple platforms, comprising whole exome sequencing, copy number analysis, RNA sequencing, microRNA sequencing, methylomics and proteomics[57,92]. This network analyzed an international cohort of 363 HCC cases by whole-exome sequencing and DNA copy number and 196 cases by DNA methylation, RNA, miRNA and proteomics analysis (Table 1). This comprehensive work confirmed previous HCC classifications (Figure 1) into two large non-proliferative and proliferative classes and subclasses corresponding to the S1, S2 and S3 subclasses[57,59,60] and identified gene expression changes resulting from mutation or down-regulation by hypermethylation in genes likely to participate in metabolic reprogramming, such as ALB, APOB and CSP1. In particular, isocitrate dehydrogenase (IDH) mutations are associated with poor patient outcome in the S2 HCC subclass[92], in line with the previous demonstration that mutant IDH inhibits HNF4α, thus blocking hepatocyte differentiation[93].
Well-differentiated HCCs, i.e., groups B[36,40], G5-G6[2,42,54], S3[59,60], iCluster2[57,92] (Figure 1) and morphologically low-grade tumors, as defined by the Edmondson-Steiner’s score and other histological classification systems[94-96] are composed of hepatocyte-like cells, which are easily recognizable by routine microscopic analysis of standard hematoxylin-eosin stained tumor tissue slides. Well-differentiated, hepatocyte-like HCCs in these groups were classically described as enriched in CTNNB1 mutations, because approximately 50% of them presented mutations in the third exon of this gene in humans, thus expressing a perivenous-type metabolic program comprising β-catenin-regulated, liver-specific genes.
In normal liver, the interplay between HNF4α and β-catenin governs the differential distribution of metabolic functions along the periportal to perivenous axis, which is known as liver zonation (Figure 2). Periportal HNF4α-regulated networks include such functions as amino-acid catabolism and gluconeogenesis. At the opposite, perivenous β-catenin-regulated networks include, for example, glycolysis and glutamine synthesis[97-100]. Thus, parenchymal cells near the portal triads are called periportal (PP) hepatocytes; whereas those close to the centrilobular vein are known as perivenous (PV) hepatocytes. We recently showed that well-differentiated HCCs display mutually exclusive liver zonation programs, i.e., they express either a periportal or a perivenous metabolic program[43] and they respectively display periportal (wild-type β-catenin) or perivenous (mutant β-catenin) metabolic phenotypes. Periportal-type HCCs show the highest 2-year recurrence-free survival rates by multivariate analysis, suggesting that these tumors have the lowest potential for early recurrence among all HCCs. They can be identified because they express high levels of an 8-gene signature composed of genes involved in periportal metabolic functions[43]. Periportal, extracellular matrix (ECM) and stem cell (STEM) HCC subclasses seem to be distributed in a continuum across the spectrum of proliferation/differentiation ratios (Figure 2). In addition, orthogonally to the hepatocyte proliferation/differentiation dimension, well-differentiated HCCs distribute across the metabolic zonation dimension[43,47]. This body of evidence indicates that HCC subclasses may show specific metabolic reprogramming profiles.
Over the past decade, transcriptome-based classifications increased our knowledge on the molecular heterogeneity of HCCs. They demonstrated that HCCs could be divided into two subtypes of less aggressive tumors (PP and PV) and two subtypes of more aggressive tumors (S1 and S2 or ECM and STEM). They also showed that TP53 mutation was associated with the most aggressive HCC subtypes and that CTNNB1 mutation defined a homogenous subtype displaying intermediate outcomes (Figure 1).
We can imagine a future where HCC patients would benefit from high-throughput technologies. However, a major obstacle complicating further understanding of basic human HCC biology and patient management is that liver tumor biopsy cannot routinely be used for HCC diagnosis and follow-up because of the risk of complications. As a surrogate for tumor liver biopsy, MRI-based metabolic imaging may become increasingly relevant for diagnosis and allocation to treatments in early-stage HCCs[6-8]. Future challenges will call for validation of circulating protein markers and molecular studies on liquid biopsies. Data management will call for further bio-statistical refinements, such as defining standard methods enabling the classification of a single sample by itself, without requiring an entire cohort. But the effort could pay off.
It is however important to bear in mind three major features in the natural history of HCCs. First, these tumors arise in more than 80% of the cases in severely fibrotic livers, with impaired liver function. Second, HCCs show high intra-tumor heterogeneity[24,25] despite a limited number of trunk mutational events[56]. Third, despite their metastatic capacity, HCC may develop locally advanced disease given the vascular anatomy of the liver. The natural history of HCC explains why tumor diagnosis, staging and treatment allocation is based upon tumor size and number, vascular invasion, location with respect to main vascular structures, underlying functional liver reserve and patient’s performance status. As a consequence, liver and HCC imaging are thriving fields of research and development. They will benefit from statistical refinements in HCC texture analyses by MRI[7,101,102], in the light of molecular tumor profiles. This body of cognitive data will spur translational efforts toward evidence-based patient management.
The authors thank the following members of the Institut NuMeCan, at INSERM in Rennes, France: Anne Corlu, for helpful discussion and inspiring remarks; Bruno Clément, for critical reading of the manuscript and helpful suggestions; Michèle Le Guennec, Patricia Jouas and Thomas Poussou for efficient secretarial assistance. We thank Christelle Reynès and Robert Sabatier, Biostatistics and Informatics laboratory, Pharmacy School, University of Montpellier, France and Pierre-Antoine Eliat, Small Animal Imaging Core Facility, Rennes 1 University, INSERM, CNRS, for enlightening discussions.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: France
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Liedtke C, Tomizawa M S- Editor: Ma RY L- Editor: A E- Editor: Bian YN
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21359] [Article Influence: 2135.9] [Reference Citation Analysis (3)] |
| 2. | Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1933] [Cited by in RCA: 1867] [Article Influence: 207.4] [Reference Citation Analysis (4)] |
| 3. | Nault JC, Villanueva A. Intratumor molecular and phenotypic diversity in hepatocellular carcinoma. Clin Cancer Res. 2015;21:1786-1788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 4. | Sapisochin G, Bruix J. Liver transplantation for hepatocellular carcinoma: outcomes and novel surgical approaches. Nat Rev Gastroenterol Hepatol. 2017;14:203-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 331] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 5. | Bruix J, Reig M, Sherman M. Evidence-Based Diagnosis, Staging, and Treatment of Patients With Hepatocellular Carcinoma. Gastroenterology. 2016;150:835-853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 1274] [Article Influence: 141.6] [Reference Citation Analysis (2)] |
| 6. | Yamashita T, Kitao A, Matsui O, Hayashi T, Nio K, Kondo M, Ohno N, Miyati T, Okada H, Yamashita T. Gd-EOB-DTPA-enhanced magnetic resonance imaging and alpha-fetoprotein predict prognosis of early-stage hepatocellular carcinoma. Hepatology. 2014;60:1674-1685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 7. | Hectors SJ, Wagner M, Bane O, Besa C, Lewis S, Remark R, Chen N, Fiel MI, Zhu H, Gnjatic S. Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci Rep. 2017;7:2452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 8. | Ueno A, Masugi Y, Yamazaki K, Komuta M, Effendi K, Tanami Y, Tsujikawa H, Tanimoto A, Okuda S, Itano O. OATP1B3 expression is strongly associated with Wnt/β-catenin signalling and represents the transporter of gadoxetic acid in hepatocellular carcinoma. J Hepatol. 2014;61:1080-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 9. | Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology. 2015;149:1226-1239.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 744] [Cited by in RCA: 949] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 10. | Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 390] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 11. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9016] [Cited by in RCA: 10244] [Article Influence: 602.6] [Reference Citation Analysis (2)] |
| 12. | Roberts LR. Sorafenib in liver cancer--just the beginning. N Engl J Med. 2008;359:420-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 88] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 13. | Raoul JL, Kudo M, Finn RS, Edeline J, Reig M, Galle PR. Systemic therapy for intermediate and advanced hepatocellular carcinoma: Sorafenib and beyond. Cancer Treat Rev. 2018;68:16-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 117] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 14. | European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu.; European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5593] [Cited by in RCA: 6023] [Article Influence: 860.4] [Reference Citation Analysis (3)] |
| 15. | Salem R, Gordon AC, Mouli S, Hickey R, Kallini J, Gabr A, Mulcahy MF, Baker T, Abecassis M, Miller FH. Y90 Radioembolization Significantly Prolongs Time to Progression Compared With Chemoembolization in Patients With Hepatocellular Carcinoma. Gastroenterology. 2016;151:1155-1163.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 448] [Cited by in RCA: 486] [Article Influence: 54.0] [Reference Citation Analysis (30)] |
| 16. | Garin E, Pallard X, Edeline J. Does Y90 Radioembolization Prolong Overall Survival Compared With Chemoembolization in Patients With Hepatocellular Carcinoma? Gastroenterology. 2017;152:1624-1625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 17. | Vilgrain V, Pereira H, Assenat E, Guiu B, Ilonca AD, Pageaux GP, Sibert A, Bouattour M, Lebtahi R, Allaham W. Efficacy and safety of selective internal radiotherapy with yttrium-90 resin microspheres compared with sorafenib in locally advanced and inoperable hepatocellular carcinoma (SARAH): an open-label randomised controlled phase 3 trial. Lancet Oncol. 2017;18:1624-1636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 620] [Cited by in RCA: 593] [Article Influence: 74.1] [Reference Citation Analysis (0)] |
| 18. | Wallace MC, Friedman SL. Hepatic fibrosis and the microenvironment: fertile soil for hepatocellular carcinoma development. Gene Expr. 2014;16:77-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 19. | Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995-2004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1044] [Cited by in RCA: 1004] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 20. | Hernandez-Gea V, Toffanin S, Friedman SL, Llovet JM. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 591] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 21. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5906] [Cited by in RCA: 5588] [Article Influence: 465.7] [Reference Citation Analysis (0)] |
| 22. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1339] [Article Influence: 133.9] [Reference Citation Analysis (0)] |
| 23. | Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 535] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 24. | Friemel J, Rechsteiner M, Frick L, Böhm F, Struckmann K, Egger M, Moch H, Heikenwalder M, Weber A. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res. 2015;21:1951-1961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 232] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 25. | Xue R, Li R, Guo H, Guo L, Su Z, Ni X, Qi L, Zhang T, Li Q, Zhang Z. Variable Intra-Tumor Genomic Heterogeneity of Multiple Lesions in Patients With Hepatocellular Carcinoma. Gastroenterology. 2016;150:998-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 170] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 26. | Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, Laurent A, Cherqui D, Balabaud C, Zucman-Rossi J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 414] [Cited by in RCA: 511] [Article Influence: 46.5] [Reference Citation Analysis (1)] |
| 27. | Nault JC, Calderaro J, Di Tommaso L, Balabaud C, Zafrani ES, Bioulac-Sage P, Roncalli M, Zucman-Rossi J. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology. 2014;60:1983-1992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 258] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 28. | Dubois-Pot-Schneider H, Fekir K, Coulouarn C, Glaise D, Aninat C, Jarnouen K, Le Guével R, Kubo T, Ishida S, Morel F. Inflammatory cytokines promote the retrodifferentiation of tumor-derived hepatocyte-like cells to progenitor cells. Hepatology. 2014;60:2077-2090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 29. | Cabillic F, Corlu A. Regulation of Transdifferentiation and Retrodifferentiation by Inflammatory Cytokines in Hepatocellular Carcinoma. Gastroenterology. 2016;151:607-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 30. | He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, Shalapour S, Seki E, Yost SE, Jepsen K. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 367] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 31. | Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 831] [Article Influence: 103.9] [Reference Citation Analysis (2)] |
| 32. | Pilati C, Letouzé E, Nault JC, Imbeaud S, Boulai A, Calderaro J, Poussin K, Franconi A, Couchy G, Morcrette G. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer Cell. 2014;25:428-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 199] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 33. | Ziol M, Poté N, Amaddeo G, Laurent A, Nault JC, Oberti F, Costentin C, Michalak S, Bouattour M, Francoz C. Macrotrabecular-massive hepatocellular carcinoma: A distinctive histological subtype with clinical relevance. Hepatology. 2018;68:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (1)] |
| 34. | Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouzé E, Blanc JF, Laurent C, Hajji Y, Azoulay D, Bioulac-Sage P. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67:727-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 534] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 35. | Désert R, Mebarki S, Desille M, Sicard M, Lavergne E, Renaud S, Bergeat D, Sulpice L, Perret C, Turlin B. “Fibrous nests” in human hepatocellular carcinoma express a Wnt-induced gene signature associated with poor clinical outcome. Int J Biochem Cell Biol. 2016;81:195-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 36. | Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 677] [Cited by in RCA: 689] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 37. | Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306-1311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 377] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 38. | Sargent LM, Zhou X, Keck CL, Sanderson ND, Zimonjic DB, Popescu NC, Thorgeirsson SS. Nonrandom cytogenetic alterations in hepatocellular carcinoma from transgenic mice overexpressing c-Myc and transforming growth factor-alpha in the liver. Am J Pathol. 1999;154:1047-1055. [PubMed] |
| 39. | Kaposi-Novak P, Lee JS, Gòmez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582-1595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 296] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 40. | Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 747] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 41. | Wold H. Estimation of principal components and related models by iterative least squares. Multivariate analysis. New York: Academic Press 1966; 391-420. |
| 42. | Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, Hérault A, Saric J, Belghiti J, Franco D. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 827] [Cited by in RCA: 925] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 43. | Désert R, Rohart F, Canal F, Sicard M, Desille M, Renaud S, Turlin B, Bellaud P, Perret C, Clément B. Human hepatocellular carcinomas with a periportal phenotype have the lowest potential for early recurrence after curative resection. Hepatology. 2017;66:1502-1518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 44. | McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM; Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97:1180-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1072] [Cited by in RCA: 1149] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 45. | Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 545] [Cited by in RCA: 633] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 46. | Rebouissou S, Franconi A, Calderaro J, Letouzé E, Imbeaud S, Pilati C, Nault JC, Couchy G, Laurent A, Balabaud C. Genotype-phenotype correlation of CTNNB1 mutations reveals different β-catenin activity associated with liver tumor progression. Hepatology. 2016;64:2047-2061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 237] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 47. | Ng CKY, Piscuoglio S, Terracciano LM. Molecular classification of hepatocellular carcinoma: The view from metabolic zonation. Hepatology. 2017;66:1377-1380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 48. | Clevers H. Axin and hepatocellular carcinomas. Nat Genet. 2000;24:206-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 49. | Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 703] [Cited by in RCA: 722] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 50. | Zucman-Rossi J, Benhamouche S, Godard C, Boyault S, Grimber G, Balabaud C, Cunha AS, Bioulac-Sage P, Perret C. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 185] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 51. | Abitbol S, Dahmani R, Coulouarn C, Ragazzon B, Mlecnik B, Senni N, Savall M, Bossard P, Sohier P, Drouet V. AXIN deficiency in human and mouse hepatocytes induces hepatocellular carcinoma in the absence of β-catenin activation. J Hepatol. 2018;68:1203-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 52. | Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 962] [Cited by in RCA: 1148] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 53. | Colnot S, Decaens T, Niwa-Kawakita M, Godard C, Hamard G, Kahn A, Giovannini M, Perret C. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc Natl Acad Sci USA. 2004;101:17216-17221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 253] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 54. | Allen JC Jr, Nault JC, Zhu G, Khor AY, Liu J, Lim TK, Zucman-Rossi J, Chow PK. The transcriptomic G1-G6 signature of hepatocellular carcinoma in an Asian population: Association of G3 with microvascular invasion. Medicine (Baltimore). 2016;95:e5263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 55. | Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501-1512.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 334] [Cited by in RCA: 334] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 56. | Torrecilla S, Sia D, Harrington AN, Zhang Z, Cabellos L, Cornella H, Moeini A, Camprecios G, Leow WQ, Fiel MI. Trunk mutational events present minimal intra- and inter-tumoral heterogeneity in hepatocellular carcinoma. J Hepatol. 2017;67:1222-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 120] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 57. | Sia D, Llovet JM. Liver cancer: Translating ‘-omics’ results into precision medicine for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2017;14:571-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 58. | Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Solé M. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779-6788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 570] [Cited by in RCA: 564] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 59. | Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385-7392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 956] [Cited by in RCA: 940] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 60. | Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y, Villanueva A, Minguez B, Newell P, Tsai HW. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res. 2012;18:4997-5007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 244] [Article Influence: 18.8] [Reference Citation Analysis (1)] |
| 61. | Yuan RH, Jeng YM, Hu RH, Lai PL, Lee PH, Cheng CC, Hsu HC. Role of p53 and β-catenin mutations in conjunction with CK19 expression on early tumor recurrence and prognosis of hepatocellular carcinoma. J Gastrointest Surg. 2011;15:321-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 62. | Mise K, Tashiro S, Yogita S, Wada D, Harada M, Fukuda Y, Miyake H, Isikawa M, Izumi K, Sano N. Assessment of the biological malignancy of hepatocellular carcinoma: relationship to clinicopathological factors and prognosis. Clin Cancer Res. 1998;4:1475-1482. [PubMed] |
| 63. | Hayashi H, Sugio K, Matsumata T, Adachi E, Takenaka K, Sugimachi K. The clinical significance of p53 gene mutation in hepatocellular carcinomas from Japan. Hepatology. 1995;22:1702-1707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 64. | Musso O, Rehn M, Théret N, Turlin B, Bioulac-Sage P, Lotrian D, Campion JP, Pihlajaniemi T, Clément B. Tumor progression is associated with a significant decrease in the expression of the endostatin precursor collagen XVIII in human hepatocellular carcinomas. Cancer Res. 2001;61:45-49. [PubMed] |
| 65. | Théret N, Musso O, Turlin B, Lotrian D, Bioulac-Sage P, Campion JP, Boudjéma K, Clément B. Increased extracellular matrix remodeling is associated with tumor progression in human hepatocellular carcinomas. Hepatology. 2001;34:82-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 147] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 66. | Le Pabic H, Bonnier D, Wewer UM, Coutand A, Musso O, Baffet G, Clément B, Théret N. ADAM12 in human liver cancers: TGF-beta-regulated expression in stellate cells is associated with matrix remodeling. Hepatology. 2003;37:1056-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 161] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 67. | Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422-1433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 355] [Cited by in RCA: 411] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 68. | Cerec V, Glaise D, Garnier D, Morosan S, Turlin B, Drenou B, Gripon P, Kremsdorf D, Guguen-Guillouzo C, Corlu A. Transdifferentiation of hepatocyte-like cells from the human hepatoma HepaRG cell line through bipotent progenitor. Hepatology. 2007;45:957-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 285] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 69. | Mebarki S, Désert R, Sulpice L, Sicard M, Desille M, Canal F, Dubois-Pot Schneider H, Bergeat D, Turlin B, Bellaud P. De novo HAPLN1 expression hallmarks Wnt-induced stem cell and fibrogenic networks leading to aggressive human hepatocellular carcinomas. Oncotarget. 2016;7:39026-39043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 70. | Bengochea A, de Souza MM, Lefrançois L, Le Roux E, Galy O, Chemin I, Kim M, Wands JR, Trepo C, Hainaut P. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br J Cancer. 2008;99:143-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 175] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 71. | Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180-185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 600] [Cited by in RCA: 539] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 72. | Fu L, Zhang C, Zhang LY, Dong SS, Lu LH, Chen J, Dai Y, Li Y, Kong KL, Kwong DL. Wnt2 secreted by tumour fibroblasts promotes tumour progression in oesophageal cancer by activation of the Wnt/β-catenin signalling pathway. Gut. 2011;60:1635-1643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 73. | Miltiadous O, Sia D, Hoshida Y, Fiel MI, Harrington AN, Thung SN, Tan PS, Dong H, Revill K, Chang CY. Progenitor cell markers predict outcome of patients with hepatocellular carcinoma beyond Milan criteria undergoing liver transplantation. J Hepatol. 2015;63:1368-1377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 74. | Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, Putra J, Camprecios G, Bassaganyas L, Akers N. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology. 2017;153:812-826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 690] [Cited by in RCA: 659] [Article Influence: 82.4] [Reference Citation Analysis (0)] |
| 75. | Sim HW, Knox J. Hepatocellular carcinoma in the era of immunotherapy. Curr Probl Cancer. 2018;42:40-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 76. | Iñarrairaegui M, Melero I, Sangro B. Immunotherapy of Hepatocellular Carcinoma: Facts and Hopes. Clin Cancer Res. 2018;24:1518-1524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 77. | Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19:222-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 735] [Article Influence: 105.0] [Reference Citation Analysis (0)] |
| 78. | Mínguez B, Hoshida Y, Villanueva A, Toffanin S, Cabellos L, Thung S, Mandeli J, Sia D, April C, Fan JB. Gene-expression signature of vascular invasion in hepatocellular carcinoma. J Hepatol. 2011;55:1325-1331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 79. | Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202-10212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 761] [Cited by in RCA: 786] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 80. | Villa E, Critelli R, Lei B, Marzocchi G, Cammà C, Giannelli G, Pontisso P, Cabibbo G, Enea M, Colopi S. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. Results from a prospective study. Gut. 2016;65:861-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 198] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 81. | Woo HG, Lee JH, Yoon JH, Kim CY, Lee HS, Jang JJ, Yi NJ, Suh KS, Lee KU, Park ES. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034-3041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 158] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 82. | Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451-1461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 545] [Cited by in RCA: 593] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 83. | Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012-1024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 936] [Cited by in RCA: 956] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 84. | Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059-2067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 283] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 85. | Anderson NM, Mucka P, Kern JG, Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018;9:216-237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 365] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 86. | Shang RZ, Qu SB, Wang DS. Reprogramming of glucose metabolism in hepatocellular carcinoma: Progress and prospects. World J Gastroenterol. 2016;22:9933-9943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 66] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 87. | Huang Q, Tan Y, Yin P, Ye G, Gao P, Lu X, Wang H, Xu G. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013;73:4992-5002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 336] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 88. | Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, Yin Z, Xie X, Wang X, Chen W. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2017; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 284] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 89. | Beyoğlu D, Imbeaud S, Maurhofer O, Bioulac-Sage P, Zucman-Rossi J, Dufour JF, Idle JR. Tissue metabolomics of hepatocellular carcinoma: tumor energy metabolism and the role of transcriptomic classification. Hepatology. 2013;58:229-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 179] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 90. | Nwosu ZC, Megger DA, Hammad S, Sitek B, Roessler S, Ebert MP, Meyer C, Dooley S. Identification of the Consistently Altered Metabolic Targets in Human Hepatocellular Carcinoma. Cell Mol Gastroenterol Hepatol. 2017;4:303-323.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 91. | Senni N, Savall M, Cabrerizo Granados D, Alves-Guerra MC, Sartor C, Lagoutte I, Gougelet A, Terris B, Gilgenkrantz H, Perret C. β-catenin-activated hepatocellular carcinomas are addicted to fatty acids. Gut. 2018; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 92. | Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327-1341.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1721] [Article Influence: 215.1] [Reference Citation Analysis (1)] |
| 93. | Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS, Gurumurthy S, Akbay EA, Sia D, Cornella H. Mutant IDH inhibits HNF-4α to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 372] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 94. | Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 95. | Bosman FT, World Health Organization. International Agency for Research on Cancer. Who classification of tumours of the digestive system. Lyon: International Agency for Research on Cancer 2010; . |
| 96. | Goodman ZD. Neoplasms of the liver. Mod Pathol. 2007;20 Suppl 1:S49-S60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 81] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 97. | Berasain C, Avila MA. Deciphering liver zonation: new insights into the β-catenin, Tcf4, and HNF4α triad. Hepatology. 2014;59:2080-2082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 98. | Braeuning A, Ittrich C, Köhle C, Hailfinger S, Bonin M, Buchmann A, Schwarz M. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006;273:5051-5061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 183] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 99. | Holloway MG, Miles GD, Dombkowski AA, Waxman DJ. Liver-specific hepatocyte nuclear factor-4alpha deficiency: greater impact on gene expression in male than in female mouse liver. Mol Endocrinol. 2008;22:1274-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 84] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 100. | Gougelet A, Torre C, Veber P, Sartor C, Bachelot L, Denechaud PD, Godard C, Moldes M, Burnol AF, Dubuquoy C. T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344-2357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 101. | De Certaines JD, Larcher T, Duda D, Azzabou N, Eliat P-A, Escudero LM, Pinheiro AM, Yang G, Coatrieux J-L, Snezkho E. Application of texture analysis to muscle mri: 1-what kind of information should be expected from texture analysis? EPJ Nonlinear Biomed Phys. 2015;3:3. [RCA] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 102. | Lerski RA, de Certaines JD, Duda D, Klonowski W, Yang G, Coatrieux JL, Azzabou N, Eliat P-A. Application of texture analysis to muscle mri: 2 - technical recommendations. EPJ Nonlinear Biomed Phys. 2015;3:2. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |