Published online Sep 7, 2018. doi: 10.3748/wjg.v24.i33.3695
Peer-review started: May 4, 2018
First decision: May 16, 2018
Revised: June 28, 2018
Accepted: July 16, 2018
Article in press: July 16, 2018
Published online: September 7, 2018
Processing time: 125 Days and 18.2 Hours
Hepatocellular carcinoma (HCC) is the fifth most common cancer and is the second leading cause of cancer death. Since the diagnosis of HCC is difficult, in many cases patients with HCC are diagnosed advanced stage of development. Hepatocyte growth factor (HGF)/c-mesenchymal-epithelial transition receptor (c-Met) axis is a key signaling pathway in HCC, either via canonical or non-canonical pathways. Available treatments against HCC based upon HGF/c-Met inhibition can increase patient lifespan, but do not reach the expected therapeutic benefits. In HCC, c-Met monomers can bind other receptor monomers, activating several noncanonical signaling pathways, leading to increased cell proliferation, invasion, motility, and drug resistance. All of these processes are enhanced by the tumor microenvironment, with stromal cells contributing to boost tumor progression through oxidative stress, angiogenesis, lymphangiogenesis, inflammation, and fibrosis. Novel treatments against HCC are being explored to modulate other targets such as microRNAs, methyltransferases, and acetyltransferases, which are all involved in the regulation of gene expression in cancer. This review compiles basic knowledge regarding signaling pathways in HCC, and compounds already used or showing potential to be used in clinical trials.
Core tip: Hepatocellular carcinoma (HCC) is a tumor usually arising from previous hepatic diseases as cirrhosis and chronic hepatitis B and C infections. Several studies have shown that a key factor for HCC oncogenesis is chronic inflammation. Inflammation induces changes in the gene expression pattern in surrounding cells. These changes provide an environment with a high level of cytokines, promoting hepatocyte transformation to tumor cells. New therapies against HCC are focused on regulating stromal cells within the tumor microenvironment to avoid HCC progression.
- Citation: García-Vilas JA, Medina MÁ. Updates on the hepatocyte growth factor/c-Met axis in hepatocellular carcinoma and its therapeutic implications. World J Gastroenterol 2018; 24(33): 3695-3708
- URL: https://www.wjgnet.com/1007-9327/full/v24/i33/3695.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i33.3695
Liver is a vital organ responsible for hundreds of chemical reactions. Among them, it is involved in metabolizing many toxins. These reactions are carried out by the hepatocytes, which represent around 80% of the hepatic tissue cell population. Since the liver is the main detoxifying organ, hepatocytes are exposed to many toxins[1] which may cause insults inducing several anomalies, such as primary liver cancer. There are two types of adult primary liver cancers: cholangiocarcinoma and hepatocellular carcinoma (HCC), the latter being responsible for 85% to 90%[2] of total primary liver cancer instances. HCC is one of the deadliest malignancies worldwide[3]. It can be preceded by chronic inflammation due to cirrhosis, hepatitis B virus or hepatitis C virus infections, all of which increase 20-fold the risk of liver cancer[4].
Hepatocytes have a high regeneration rate, being controlled by multiple growth factors. The first molecule discovered with the ability to stimulate hepatocyte division was hepatocyte growth factor (HGF). HGF is expressed and released by specialized non-parenchymal cells called hepatocyte stellate cells. These cells release HGF into the extracellular space, where it acts in a paracrine manner on its receptor, known as c-Met, which is located on the surface of hepatocytes. HGF was characterized as a potent mitogen due to its ability to induce c-Met dimerization. This activates a canonical signal transduction pathway including effector molecules such as RAS-ERK and PI3K-AKT, which increase DNA synthesis and increase cell cycle progression.
The first therapies developed to treat HCC were focused on inhibiting the HGF/c-Met axis, thus stopping hepatocytes in the G1 phase of the cell cycle[5]. Studies in mice have shown that HGF or c-Met deletion are lethal[6]. Moreover, some tumors overexpress these proteins. Therapies targeting HGF or c-Met have been used for years. However, patients receiving these therapies still presented high rates of mortality, as well resistance to chemotherapy, radiotherapy, immunotherapy or hormonal therapy[7].
Recently, novel studies have elucidated other noncanonical signaling pathways that are modified in HCC. It is also now known that the fate of hepatocytes is determined by the interaction with nearby stromal cells. New therapies are being developed targeting the tumor microenvironment, including endothelial cells, immune cells, fibroblasts and the extracellular matrix.
Nonetheless, HGF/c-Met levels are currently being used to predict tumor aggressiveness and the prognosis of HCC patients.
In this review, we have analyzed HCC literature to generate a comprehensive view about the molecular processes already known and important discoveries that remain to be made.
The HGF gene is located on chromosome 7q21. It contains 20 exons and is expressed by mesenchymal cells. Hepatocyte growth factor (HGF) is a member of the peptidase S1 family of serine proteases, although it lacks peptidase activity. This protein is synthesized as an inactive pro-peptide generating an alpha/beta heterodimer linked by a disulfide bond. Proteolytic conversion of pro-HGF to HGF can be mediated by three enzymes present in the tumor environment: matriptase, hepsin and HGF activator (HGFA). However, there is evidence that urokinase plasminogen activator (uPA), transmembrane protease, serine 13 (TMPRSS13)[8] may also activate it. Although HGF was originally identified as a hepatocyte mitogen, it is now known to be a cytokine with pleiotropic effects. It has roles in enhancing angiogenesis, immune response, cell motility, and cell differentiation.
The gene for c-Met is located in chromosome 7q21-31 and contains 24 exons. Its promoter region, however, is located in chromosome 1. c-Met is expressed in epithelial cells. c-Met is a single pass tyrosine kinase receptor made up of an alpha and a beta subunit linked by disulfide bonds. The beta subunit is a transmembrane monomer that contains 5 catalytic tyrosines in its cytoplasmic tail. Y1003 negatively regulates c-Met by linking it to the ubiquitin ligase casitas beta-lineage lymphoma (c-CBL)[9]. In contrast, Y1234; Y1235; Y1349 and Y1356 positively regulate c-Met. Furthermore, S985 in c-Met can be phosphorylated by protein kinase-C, inducing c-Met degradation (ubiquitination and endocytosis) [10].
c-Met activation has pleiotropic effects because its cytoplasmic domain can interact with multiple proteins involved in several cellular signaling pathways. Because of this, c-Met is considered an oncogene involved in cell proliferation, invasion, motility, angiogenesis and apoptosis.
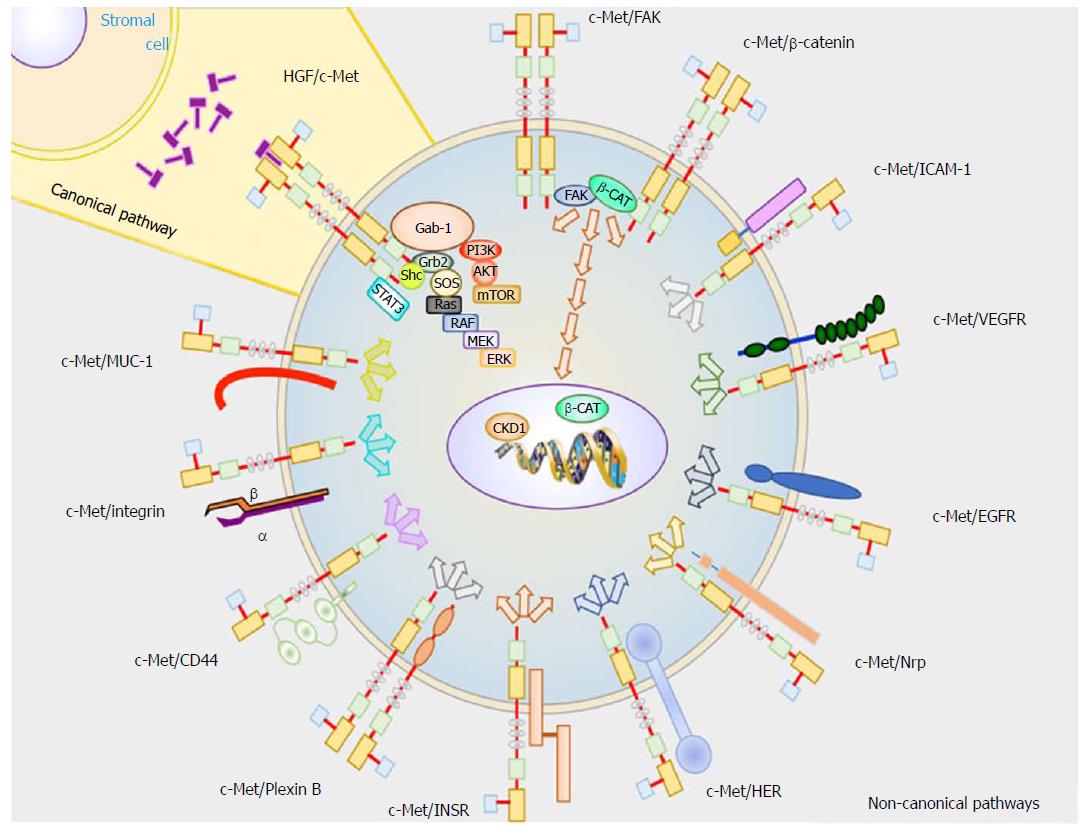
The pathophysiology of hepatocellular carcinoma at a cellular level is complex and it is very possible that there are many unknown c-Met interactions with others signaling pathways. Multiple cell pathways are aberrant in HCC, but this review just focuses on the signaling pathways related to c-Met. The activation of c-Met can take place by the canonical pathway, which involves HGF binding to c-Met resulting in c-Met homodimerization. It can also take place through non-canonical pathways, where c-Met dimerizes with different receptors.
Heterodimers of receptor proteins and c-Met are involved in overstimulation and dysregulation of c-Met signaling pathways. This occurs during hypoxia, which can cause c-Met overexpression, mutations on tyrosine kinase domain or HGF gene amplification[11]. However, this latter event rarely occurs in HCC[12].
These signaling pathways involve proteins with SH2 domains or phosphotyrosine-binding domains that are able to interact with phosphorylated tyrosine residues[13] that, in turn, interact physically with the cytoplasmic domain of c-Met.
Growth factor receptor-bound protein 2 (Grb-2): Grb-2 interacts with Y1356 of c-Met to transduce HGF signaling to the cytoplasm. Grb-2 is considered a key protein in HGF/c-Met axis because it connects to several signaling transducers, such as Ras, SOS, and Gab1. Grb-2 is involved in cell motility, cycle progression, angiogenesis, amongst other.
GRB2-associated binding protein 1 (Gab1): Activated c-Met is phosphorylated on Y1349 and Y1356 residues which specifically interact and phosphorylate to Gab1. However, Gab1 can also be phosphorylated by Grb-2. Gab1 is involved in many signal transduction pathways by binding to effector proteins that have a role in cell motility and extracellular matrix invasion, such as Shp2, Shc, PLCγ1, p120[14].
Phosphoinositide 3 kinase (PI3K): PI3K is an enzyme able to phosphorylate proteins downstream of c-Met thereby linking oncogenes and many receptors essential for cellular functions. The phospho Y1356 in c-Met can phosphorylate PI3K, inducing cell mobility[9] by activating focal adhesion kinase (FAK). However, PI3K can also be activated by Gab1 where it promotes cell survival[11].
Signal transducer and activator of transcription 3 (STAT3): HGF binds to c-Met inducing the phosphorylation on Y1356. This phosphorylated amino acid interacts and actives STAT3, as was shown by Boccaccio et al[15]. When it is activated, it translocates to the nucleus where it binds to DNA and promotes gene expression (related with angiogenesis, and long-term response)[15].
Shc-transforming protein 1 (Shc): SHC is an adaptor protein involved in the mitogenic signal transduction from tyrosine receptors. On the other hand, experiments carried out in fibroblast showed that Shc is highly stimulated by VEGF and that activation correlated with the angiogenic response[16].
c-Met activation by non-canonical pathways takes place when this receptor is over-expressed and dimerizes with other receptor subunits, or may bind to ligands other than HGF. Non-canonical pathways are usually associated with c-Met gene amplification, and are common in treatment resistant cancers[17,18], tumor progression, and metastasis, as shown in in vivo experiments using mice[19,20]. It has also been reported that c-Met dimerization takes place in the absence of ligand binding[21] when it interacts with the following proteins:
Epithelial growth factor receptor (EGFR): Physical interaction between EGFR and c-Met was found in A431 cells[22]. In HCC, the transactivation between these two receptors takes place, inducing the common downstream signaling effectors PI3K and Ras[23].
Human epidermal growth factor receptor (HER): The dimerization between c-Met and HER increases activation of PI3K/AKT signaling[18], which is associated with resistance to EGFR inhibitors[24] as well as cancer progression.
Integrin α6β4: Trusolino et al[25] determined that integrin α6β4 physically interacts with c-Met on the membrane surface of carcinoma cells. This protein is necessary for cancer invasion because the cytosolic domain of β4 induces c-Met activation. In this case, the signaling transduction is performed by Shc and PI3K[14].
β-catenin (β-CAT): Phosphorylated β-CAT may bind to c-Met, activating its downstream signaling. Phosphorylation of Y654 in β-CAT actives FAK, which induces cyclin D1 (CKD1) expression[26]. At the same time, β-catenin is translocated to the nucleus, and promotes c-myc gene expression[27].
Receptor for hyaluronic acid (CD44): The CD44v3 splice variant is the CD44 isoform with high affinity for heparin domains. This v3 may be activated by different growth factors with heparin domains, such as fibroblast growth factor (FGF) and HGF. CD44 may act as a concentrator of HGF to present it to c-Met resulting in downstream signaling transduction[28]. On the other hand, Olaku et al[29] reported that CD44v6 splice variant is necessary for c-Met activation. It is thought that three specific amino acid residues (RWH in human) in v6 are necessary for complete c-Met activation.
ICAM-1: This protein can substitute for the role of CD44v6 in c-Met activation, as shown in hepatocytes from Cd44 null mice[29].
Plexin B1: Receptor with high similarity to c-Met, which is also expressed in the same tissues as c-Met. After mutation and expression of exogenous c-Met in cells, Giordano et al[30] shown that plexin B1 links to c-Met when it is activated by semaphorin 4D. This interaction was reported in invasive cancer cells growing in response to semaphorin 4D.
Vascular endothelial growth factor A (VEGF-A): HGF can induce VEGF expression[31] by phosphorylation of a key transcription factor called Sp1. This characteristic of HGF increases the expression of Bcl-2[32], which acts as an antiapoptotic protein.
Insulin receptor (INSR) tyrosine kinase: This receptor has an extracellular α-chain and a transmembrane β-chain. This protein has a very similar structure compared to c-Met. Furthermore, it has been reported that insulin and HGF can phosphorylate INSR in its Y1146 and Y1150 or Y1151 residues. In HCC cells, Y1322 is also phosphorylated, thus activating PI3K. HGF-stimulated hepatocytes have shown to form a INSR-c-Met complex. There is evidence that c-Met can also phosphorylate insulin receptor substrates (IRS) on Y895. Likewise, INSR phosphorylates IRS on Y612[33]. In summary, both c-Met and IRS increase downstream signaling through PI3K-AKT, promoting cell growth, cell survival and cell motility.
Fas: This protein is one of the surface death receptors and triggers apoptosis signaling when binds its ligand (FasL). Wang et al[34] showed that Fas and c-Met associate with each other using coimmunoprecipitation experiments in Hep G2 cells. These authors proposed that c-Met promotes cell survival by two different pathways. (1) When there are low levels of FasL in the microenvironment, Fas binds to c-Met avoiding to trigger its intracellular signaling pathway. (2) However, in the presence of high levels of HGF, c-Met releases Fas activating death receptor-mediated apoptosis. Nevertheless, the c-Met/Fas signaling pathway ratio is so high that cells activate antiapoptotic signals through PI3K/AKT/Bad axis to prevent Fas-mediated apoptosis.
Mucin 1 (MUC1): MUC1 expression is increased during transformation from the normal liver to HCC, as was described by Bozkaya et al[35] MUC1 silencing in HCC cells leads to β-catenin activation and c-Myc expression. Under this condition, high levels of HGF in the microenvironment increase cellular motility and invasiveness[35]. However, there are also contradicting studies. For instance, Singh et al[36] reported that MUC1-induced c-Met activation by physical interaction decreased MMP-1 transcription and cell motility[36].
Neuropilin-1 and -2 (Nrp-1, -2): Neuropilins are a family of transmembrane glycoproteins involved in several processes, including axonal guidance, angiogenesis, tumorigenesis, and immunologic response. Nrp-1 can bind VEGF-A165, VEGF-B, VEGF-E, and placental growth factor (PIGF). On the other hand, Nrp-2 can bind class III semaphorins and VEGF proteins (VEGF-A165, VEGF-A145, and VEGF-C). Nrp-2 binds VEGF proteins, and increases the VEGFR-2 phosphorylation threshold, promoting migration, and sprouting cells[37]. Nrp-2 and VEGFR2 can bind each other enhancing the signaling initiated by the HGF/c-Met axis. Moreover, Nrp-1 and Nrp-2 interact with other receptor tyrosine kinase, such as VEGFRs[37]. Neuropilins have a short cytoplasmic domain to act as catalytic domain. This evidence suggests that the intracellular domain may present a binding site involved in kinase signal transduction[37].
Focal adhesion kinase (FAK): Studies carried out in MEFs and HEK293 cells showed that FAK interacts directly with c-Met[38]. FAK is a non-receptor tyrosine kinase involved in several cell signaling pathways. Notably, it is well characterized for its role in formation and disassembly of focal adhesions, as well as cell protrusions[26]. However, FAK is also intimately involved in the regulation of cell proliferation because it is able to phosphorylate PI3K and ERK. Experiments in FAK knockout mice revealed suppressed hepatocarcinogenesis due to decreased PI3K and ERK signaling pathways[26].
Collectively, these pathways are responsible for promoting initiation and progression of HCC (Figure 1).
The expression of HGF is decreased in HCC, but it is increased in the surrounding tissue. On the other hand, c-Met is expressed in HCC at higher levels than in the surrounding tissue. These observations suggest that the overexpression of c-Met, together with additional oncogenes, is responsible of HCC aggressiveness[39].
Microenvironment is created by extracellular matrix (ECM) and stromal cells. Stromal cells, such as endothelial cells, fibroblasts, and immune cells, increase the gene expression and release of chemokines, cytokines, proteases and growth factors, such as HGF[40]. HGF released from stromal cells interacts with hepatocytes enhancing HGF/c-Met signaling transduction. Subsequently, hepatocytes intensify cell survival and cell proliferation. HGF may also modify cell-cell interaction, and cell-extracellular matrix interactions through activating proteolytic networks[41,42]. Furthermore, stromal cells increase the gene expression of HGF. Furthermore, HGF promotes angiogenesis from endothelial cells around the chronic liver disease.
The initial stages of liver tumor are characterized by hypoxia, a condition leading to the release of VEGF to increase tumor vascularization. In parallel, immune cells contribute to the activation of the vascularization process by releasing inflammatory mediators, including interleukin-1α (IL-1α), IL-1β, tumor necrosis factor-α, and prostaglandin E2. HCC is a highly vascularized tumor. However, the new blood vessels formed to irrigate the tumor mass present abnormalities, such as tortuous vessels and sinusoidal capillarization. Tumor blood vessels have incomplete basal membrane and incomplete pericyte coverage. Simultaneously, lymphangiogenesis, proliferation and sprouting of new lymphatic vessels from preexisting ones, also takes places. Lymphangiogenesis is tightly regulated by lymphoangiogenic growth factors, such as VEGF-C and VEGF-D, released from tumor and stromal cells. VEGF-C and VEGF-D interact with VEGFR-2 and VEGFR-3, respectively, in lymphatic cells. VEGFR-2 is responsible for vessel enlargement, whereas VEGFR-3 is critical for lymphangiogenic sprouting[43]. However, VEGF-A may interact with VEGFR-3. Further, venous and lymphatic endothelial cells express neuropilin-2 (Nrp-2), a protein that it has recently been associated to lymphatic cell survival and migration[37]. On the other hand, Nrp-2 is not involved in venous or lymphatic cell proliferation[44]. Tumor-associated lymphatic vessels express stromal cell-derived factor 1 (CXCL12), and tumor cells express the receptor for CXCL12, called C-X-C chemokine receptor 4 (CXCR4). Tumor cells have upregulated CXCR4 under hypoxia conditions. However, hypoxia promotes migration of metastatic cells into the tumor-draining lymph node. Endothelial and lymphatic cell survival and cell proliferation promote tumor metastasis. In fact, high densities of vascular and lymphatic vessels are correlated with high patient lethality[44].
The role of immune cells in cancer is controversial, because they can eliminate tumor cells in their initial stages, but, due to their generation of oxidative stress and DNA damage, they may also promote cancer development.
A study carried out in a small cohort showed that patients with viral hepatitis presented high neutrophil levels and low lymphocyte levels. This has been proposed as a substitute parameter to determine initial development HCC stages, because neutrophils are the first kind of immune cells to interact with tumor cells[45]. Neutrophils located in the tumor mass are responsible for promoting inflammation and facilitating tumor progression. From the same study, the authors detected high platelet levels in circulation. This association might be caused because platelets are responsible for activating neutrophils, enabling neutrophils to migrate through blood vessels. A lower risk of HCC was shown with anti-platelet therapy from another independent study[46]. Additionally, platelets are key effectors to enhance the accumulation of CD8+ T lymphocytes and mediate immune injury, intensifying the micro-inflammation.
Tumor-infiltrating T cells are regulated by VEGF-A and VEGF-C, as well as their release into the tumor microenvironment, cell proliferation phenotype, and activity[47].
Macrophages are responsible for digesting all damaged cells in the tissues including cancer cells. However, macrophages activated by inflammatory cytokines from the tumor microenvironment, express the PD-L1 protein. This results in them adopting a suppressive macrophage phenotype, avoiding tumor-specific T cell immunity and increasing HCC progression[48]. These macrophages activated by the tumor microenvironment express tumor necrosis factor α (TNF-α), which induces c-Met-expression on their surface. These macrophages also express MMP-9, which increases the remodeling of the tumor microenvironment of HCC[49].
Fibroblasts are an important cell population in the microenvironment, and have a prominent role in tumor cell progression and metastasis. Fibroblasts have key functions in the tumor microenvironment: (1) synthesis and remodeling of the ECM; and (2) release of multiple cytokines and chemokines to promote inflammation, angiogenesis and epithelial differentiation. Tumor-associated fibroblasts acquire a modified phenotype induced by transforming growth factor-β (TGFβ), and become hepatic progenitor cancer cells (HPCs)[50].
HPCs have the ability to differentiate to several kinds of cells depending on the stimulus from their microenvironment. Under the influence of a tumor microenvironment, HPCs acquire genetic and epigenetic mutations, inducing their transformation from HPCs to cancer stem cells (CSCs)[51]. CSCs are able to perpetuate themselves through self-renewal, and generate mature cells of a particular tissue through differentiation, supporting tumor cell proliferation[52]. Likewise, CSCs have been proposed to be the clonogenic core of HCC, sustain the primary tumor, confer drug resistance, promote metastasis, and promote angiogenesis.
All of the stages described above in the present section, cell transvasation, cell-cell interaction, oxidative stress, cytokines and proteases released into the tumor microenvironment, provoke changes and deregulation in the ECM. The deregulation of ECM is characterized by increasing deposition of fibronectin, fibrillar collagen types I and II into the liver. This ECM deregulation decreases liver plasticity, increasing ECM stiffness, cell survival and cell proliferation of tumor cells. These processes are mediated by integrin signaling pathways, such as α1β1 and α2β1. Moreover, the ECM increases immune cell activation and differentiation, promotes angiogenesis, and activates tissue invasion[42].
Dapito et al[53] showed that translocation of components of intestinal bacterial, and Toll-like receptor 4 (TLR4), can reach the liver by the portal circulation. Both components promote liver inflammation, increasing the secretion of growth factor, such as epiregulin from hepatic stellate cells, and promoting the synthesis of ECM. This scenario enhances fibrosis and HCC development.
The microenvironment not only creates favorable conditions to develop the unregulated cell proliferation characteristic of cancer cells, but also influences their sensitivity to drugs and promotes metastasis. HGF delivered by stromal cells has been described as a key factor to confer resistance to molecular targeted drugs[54].
Only 2% of the human genome encodes proteins, and the majority of the transcriptome contains non-coding RNAs (ncRNAs). ncRNAs have regulatory functions and may be classified by their sequence length, in micro RNAs (miRNAs), small nucleolar RNAs (snoRNAs), small interfering RNAS (siRNAs), long non-coding RNAs (lncRNAs), and very long intergenic ncRNAs (vlincRNAs)[55,56]. However, there are other classifications based on association with annotated protein-coding genes, association with other DNA elements of known function, protein-coding RNA resemblance, association with repeats, with a biochemical pathway or stability, sequence and structure conservation, expression in different biological states, subcellular structures, and based on function[56]. Independently of the classification criteria, the ncRNAs have physiological relevance in genetic and epigenetic regulation, such as control of chromatin remodeling, gene transcription, protein transport and metabolism[57].
Dysregulation of miRNAs has been associated with alterations in cell signaling pathways playing important roles in the control of cell invasion, proliferation, and metastasis, among many other pathophysiologic processes. For instance, miR-26a. behaves as a tumor suppressor in hepatocellular carcinoma (HCC), but it shows oncogenic properties in lung cancer. These observations suggest that microenvironment also determines the roles of miRNAs in cancer (Table 1). A RNA-sequencing study performed in 23 liver biopsies, between tumor and adjacent non-tumor tissue, showed 57 lncRNA with differential expression[58].
| miRNA | Functions | Effect of miR in HGF/c-Met axis | Levels in HCC | Ref. |
| miR-34 | Cell invasion, proliferation | Inhibits c-Met | Downregulated | [83] |
| miR-199 | Proliferation, cell motility, | Tumor-suppressor | Downregulated | [84] |
| cell invasion | ||||
| miR-340 | Cell invasion, cell migration | Inhibits c-Met | Downregulated | [85] |
| miR-126 | Cell proliferation, | Inhibits c-Met | Downregulated | [86] |
| cell invasion, inhibits angiogenesis | ||||
| miR181-a | Cell motility and invasion | Inhibits c-Met | Upregulated | [87] |
| let-7 family | Represses cell proliferation, invasion, metastasis and resistance therapy | Inhibits c-Met signaling downstream | Downregulated | [88,89] |
| miR-148 | Promotes apoptosis, suppress cell invasion | Tumor-suppressor | Downregulated | [90] |
| miR-1 | Cell migration, cell proliferation | Tumor-suppressor | Downregulated | [91] |
| miR-26a | Cell proliferation, invasion, and migration | Inhibits c-Met signaling downstream | Downregulated | [92] |
| miR-122 | Induces apoptosis | Inhibition of c-Met | Downregulated | [93] |
| miR-145 | Cell viability, cell migration | Inhibits c-Met signaling downstream | Downregulated | [94] |
| miR-449 | Promotes apoptosis, reduces proliferation | Decreases c-Met levels | Downregulated | [95] |
| miR-200 | Cell migration and invasion | Decreases HGF-synthesis in fibroblasts | Upregulated | [96] |
| miR-101 | Cell proliferation, migration and invasion | Inhibits c-Met signaling downstream | Downregulated | [97] |
Histones acetylation, methylation, phosphorylation, sumoylation, and ubiquitylation, are post-translational modifications (PTMs) intimately related to epigenetic processes. These PTMs are responsible for the induction or repression of genes through modification in the level of DNA compaction or the recruitment of transcriptional machinery. Histones deacetylases (HDACs) are a family of proteins that remove acetyl groups from histone tail amino acids, decreasing gene expression. This family of proteins is overexpressed in HCC[59]. In fact, human HCC hallmarks are the loss of acetylation in H4K16[60], and increased methylation in H3K27 by EZH2 methylase[61]. Furthermore, EZH2 can inhibit the expression of miR-622, thus increasing the severity of HCC[62]. SET8 is a specific H4K20 methylase required for S phase progression by coupling to PCNA[63]. SET8 dysregulation has been described in human HCC[55]. Histone hypermethylation has also been related with decreased gene expression.
On the other hand, histones acetyltransferases (HAT) are a family of proteins that add acetyl groups to histone amino acids, inducing the relaxation of DNA strains, and subsequently up-regulating gene expression.
In the liver, xenobiotics such as alcohol can alter the epigenetic state by generating reactive oxygen species (ROS) and depleting S-adenosylmethionine (SAM) levels. This promotes H3K9 acetylation and alters the expression of several miRNAs[64]. Changes in the microbiota and viral liver infections can also modify DNA methylation, altering gene expression[65,66].
HCC is difficult to diagnose by current methods with biopsy being the most validated method. However, it is difficult to obtain a representative biopsy sample because open tumor biopsies of HCC are not allowed[67]. This scenario makes difficult to perform an accurate diagnostic.
Chronic liver diseases caused by hepatitis virus B and/or C virus are the most prevalent causes of HCC. However, other factors have been related to increase the prevalence of HCC such alcohol-related liver diseases, obesity, and type 2 diabetes mellitus-related non-alcoholic fatty liver[45].
The HGF/c-Met axis has been proposed as a key target for clinical intervention. This is due to its relevance in cellular processes such as 3D morphogenesis, cell survival and metastasis. Due to their key role in HCC, HGF and c-Met have been proposed as essential therapeutic targets (Table 2). A recent review on the therapeutic targeting of the HGF/c-Met signaling in HCC has focused on small c-Met kinase inhibitors[68]. This same review also comments problems as resistance to c-Met targeting drugs and side effects of c-Met targeting.
| Agent | Targets | Phase | Activity | HCC stage | Ref. |
| Sorafenib | Raf, MAPK, VEGFR, PDGFRβ | III | Anti-tumor | Advanced | [98] |
| Anti-angiogenesis | |||||
| Cabozanitinib | VEGFR2, KIT, RET, AXL | III | Anti-tumor | Advanced | [99] |
| Brivanib | FGFR, VEGFR | II | Anti-angiogenesis | Advanced | [100] |
| Foretinib | FLT1, PDGFRβ, c-Met, VEGFR-2, Tie-2 | II | Anti-tumor | Advanced | [101] |
| FLT4, RON, FLT3, KIT | Anti-angiogenesis | ||||
| Everolimus | mTOR | III | Anti-tumor | Advanced | [102] |
| Cobazitinib | c-Met, VEGFR-2, RET | II | Anti-tumor | Advanced | [103] |
| Anti-angiogenesis | |||||
| Ramucirumab | VEGFR-2 | III | Anti-angiogenesis | Advanced | [104] |
| MSC2156119J | c-Met | Ib/II | Anti-tumor, anti-mestastasis | Advanced | [105] |
| Gefitinib | EGFR, c-Met, HGF, | II | Anti-tumor | Advanced | [106] |
| Bevacizumab | VEGFRs | II | Anti-angiogenesis | Advanced | [107] |
| AZD6244 | MEK1/2 | Ib | Anti-tumor | Advanced | [108] |
| AZD4547 | p-FGFR-1, p-FGFR-2 | I | Anti-tumor, anti-angiogenesis | Advanced | [109] |
| p-c-Met, | |||||
| p-AKT, | |||||
| p-ERK | |||||
| MK2461 | c-Met, Ftl-1 | I | Anti-tumor | Advanced | [110] |
| Crizotinib | p-c-Met | Ib | Anti-angiogenesis | Advanced | [111] |
| ALK | |||||
| Bortezomib | Proteasome inhibitor | II | Anti-tumor | Advanced | [102] |
| Docetaxel | EGFR | II | Anti-tumor | Advanced | |
| INC280 | p-c-Met | II | Anti-tumor | Advanced | [111] |
Although some improvements have been obtained in HCC therapy, this cancer still remains largely incurable. Nowadays, there is palliative therapy using a multikinase inhibitor, such as sorafenib, which limits cell proliferation, and tumor angiogenesis[69]. Many clinical trials have shown that sorafenib improves overall survival of patients with advanced HCC. However, there is no effective treatment for HCC based on conventional monotherapies using tyrosine kinase inhibitors. For example, a monotherapy with Tivantinib, a small molecule tyrosine kinase inhibitor exhibiting a high selectivity against c-Met, was clinically tested. Unfortunately, in a phase III clinical trial, this compound failed to meet the primary endpoint.
HCC develops resistance to the conventional chemotherapy and radiotherapy treatments[7]. Moreover, the tumor microenvironment blocks drug effects[70]. HGF confers resistance of HCC to inhibitors of EGFR. On the other hand, HGF decreases the expression of E-cadherin[71] and increases the expression of Snail 1, to protect cancer cells from apoptosis[72].
Another difficulty in detecting HCC is the lack of a clinically relevant circulating biomarker. Current investigations are focused on discovering circulating biomarkers[45]. Sitia et al[73] showed that the CD8+ T cell/platelet ratio could be a sign of HCC progression. Moreover, Sitia et al[73] verified that aspirin or clopidogrel, two anti-platelet drugs, decreased the number of CD8+ T cells in liver infected with hepatitis B virus. After treatment with anti-platelet drugs, they observed a decrease in inflammation, fibrosis severity, and progression to HCC in a transgenic mouse model[73]. The effect was enhanced when mice were treated with aspirin and clopidogrel. The authors concluded that anti-platelet drugs diminished the amount of CD8+ T cells into the liver, avoiding hepatocellular necrosis, hepatocyte regeneration, and inflammation. These events prevent or delay HCC in the mouse model used. On the other hand, this treatment does not eradicate the viral infection.
Preclinical studies in animals have shown that anti-lymphangiogenic strategies, sequestering VEGF-C and VEGF-D, or blocking VEGFR-3 with antibodies, are feasible therapies against HCC tumors[74]. Experimental therapy against Nrp-2 reduced tumor lymphangiogenesis, apparently delaying the departure of cells from the primary tumor[44]. Lymphatic cells release CXCL12, which can be blocked with antibodies and may provide therapeutics benefits[75]. Inhibiting the expression of Nrp-1revealed that Nrp-1 is required to activate VEGFR-2 signaling-dependent mitogenic[76].
Novel anticancer strategies are emerging based on ncRNA expression, because they have a myriad of cell functions, such as chromatin remodeling, protein transport, genes transcription and metabolism[57,77]. MicroRNAs (miRs) are being studied in cancer for their key role as post-transcriptional regulators of gene expression, and for their potential to be used to classify tumors[78]. Recently studies are revealing that some miRNAs could be useful as prognostic biomarkers based on their presence in serum. They could be promising therapeutic targets as well.
Combinations of current drugs used for clinical treatments, such as sorafenib, tivantinib, 5-fluorouracil (5-FU), and potential miRs inhibitors, such as miR-93 inhibitor, are proposed as promising anti-HCC therapies[79]. Other studies demonstrated that miR26[80], miR-499, miR-30a, miR-122, and miR-148, among others, play a role in the development of normal physiological function in liver.
Some anti-cancer therapies against HCC are focused on HDACs as key targets. HDAC inhibitors (either miRNAs or small molecule HDAC inhibitors) have been shown to increase apoptosis and decrease cell proliferation. HCC treated with trichostatin A (TSA), a compound described as HDAC inhibitor, as well as with siRNA against HDACs (-1, -2, and -3) have increased apoptosis and decreased cell proliferation[60]. Sirtuins, vorinostat, romidepsin, belinostat, panobinastat, valproate and ITF2357 are HDAC inhibitors that have shown promising anti-cancer effects in clinical trials[55].
Histone methyltransferases, such as SET8, SUV39H1 and EZH2, are also promising targets for HCC therapy. Experiments performed in HCC cells showed that the silencing of EZH2 decreased the expression of CDKN2A, FOXO3, E2F1 and NOTCH2[81]. The silencing of EZH2 also repressed the expression of miR-622, increasing CXCR4 levels[61]. Small molecule inhibitors against EZH2, such as EPZ011989, may be potentially useful for the treatment of HCC patients[82].
The discovery of novel molecules related with HGF/c-Met axis signaling pathways presents a promising clinically avenue for predicting individual susceptibility and designing better specific personal therapies (Table 3).
| Agent | Target | Activity | Ref. |
| LZ8 | Inhibits the expression of c-Met, ERK, AKT | Induces apoptosis | [112] |
| Inhibits the phosphorylation of JNK, AKT, ERK, p-AKT | |||
| Stabilizes p53 | |||
| Damnacanthal | Inhibits the phosphorylation of c-Met | Inhibits cell proliferation and cell invasion | [113] |
| Decreases MMP-2 activity | |||
| GEN-203 | Inhibits the phosphorylation of c-Met | Blocks cell proliferation | [114] |
| JNJ38877605 | Inhibits phosphorylation of c-Met | Induces apoptosis | [70] |
| SRI31215 | Blocks pro-HGF activation | Inhibits cell proliferation | [70] |
| Madecassoside | Inhibits the phosphorylation of ERK1/2 and | Blocks cell proliferation | [115] |
| PKC | |||
| PHA665752 | Inhibits phosphorylation of c-Met and FGFR | Blocks cell proliferation | [116] |
| EPZ011989 | Inhibits histone methylation | Blocks cell proliferation | [82] |
| YC-1 | Inhibits the DNA synthesis | Arrests cell cycle and induce apoptosis | [117] |
HCC is a complex pathology with interconnected regulatory networks involved. This cancer is difficult to diagnose and the current treatments only produce modest therapeutic benefits. Moreover, the lack of biomarkers for early diagnosis contributes to making HCC one of the cancer types with the poorest prognosis. Novel approaches to elucidate the molecular etiology of HCC have shown that, in addition to the HGF/c-Met axis signaling, there are other receptors and ligands involved. Dysregulation in protein interactions, lncRNAs, post-translational modifications in histones, and DNA methylation levels can be responsible for cell proliferation, migration, invasion, and mestastasis. Novel treatments are being developed against HDACs, methyltransferases, and miRs, among other molecular targets. Therapies combining HGF/c-Met signaling pathway inhibitors with epigenetic inhibitors, such as histone methyltransferase or HDAC inhibitors, seem to be the most promising therapies to date. Additional experimental efforts will be needed to identify useful predictive biomarkers and to suggest new personal therapies.
We thank Dr. Michael J Hendzel (Univerity of Alberta, Canada) for carefully reading the manuscript for English grammar and having made editing suggestions.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Aravinthan AD, Elalfy H, Roohvand F, Vento S, Zhu X S- Editor: Gong ZM L- Editor: A E- Editor: Huang Y
| 1. | Waller LP, Deshpande V, Pyrsopoulos N. Hepatocellular carcinoma: A comprehensive review. World J Hepatol. 2015;7:2648-2663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4267] [Article Influence: 237.1] [Reference Citation Analysis (2)] |
| 3. | Lau EY, Lo J, Cheng BY, Ma MK, Lee JM, Ng JK, Chai S, Lin CH, Tsang SY, Ma S. Cancer-Associated Fibroblasts Regulate Tumor-Initiating Cell Plasticity in Hepatocellular Carcinoma through c-Met/FRA1/HEY1 Signaling. Cell Rep. 2016;15:1175-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 260] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 4. | Donato F, Boffetta P, Puoti M. A meta-analysis of epidemiological studies on the combined effect of hepatitis B and C virus infections in causing hepatocellular carcinoma. Int J Cancer. 1998;75:347-354. [PubMed] |
| 5. | Mohammed FF, Khokha R. Thinking outside the cell: proteases regulate hepatocyte division. Trends Cell Biol. 2005;15:555-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 6. | Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1076] [Cited by in RCA: 1034] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 7. | Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989-5005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 608] [Cited by in RCA: 680] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 8. | Kawaguchi M, Kataoka H. Mechanisms of hepatocyte growth factor activation in cancer tissues. Cancers (Basel). 2014;6:1890-1904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Bradley CA, Salto-Tellez M, Laurent-Puig P, Bardelli A, Rolfo C, Tabernero J, Khawaja HA, Lawler M, Johnston PG, Van Schaeybroeck S; MErCuRIC consortium. Targeting c-MET in gastrointestinal tumours: rationale, opportunities and challenges. Nat Rev Clin Oncol. 2017;14:562-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 10. | Nakayama M, Sakai K, Yamashita A, Nakamura T, Suzuki Y, Matsumoto K. Met/HGF receptor activation is regulated by juxtamembrane Ser985 phosphorylation in hepatocytes. Cytokine. 2013;62:446-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Ghiso E, Giordano S. Targeting MET: why, where and how? Curr Opin Pharmacol. 2013;13:511-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Seruca R, Suijkerbuijk RF, Gärtner F, Criado B, Veiga I, Olde-Weghuis D, David L, Castedo S, Sobrinho-Simões M. Increasing levels of MYC and MET co-amplification during tumor progression of a case of gastric cancer. Cancer Genet Cytogenet. 1995;82:140-145. [PubMed] |
| 13. | Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027-1047. [PubMed] |
| 14. | Viticchiè G, Muller PAJ. c-Met and Other Cell Surface Molecules: Interaction, Activation and Functional Consequences. Biomedicines. 2015;3:46-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 61] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 15. | Boccaccio C, Andò M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio PM. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 410] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 16. | Saucier C, Khoury H, Lai KM, Peschard P, Dankort D, Naujokas MA, Holash J, Yancopoulos GD, Muller WJ, Pawson T. The Shc adaptor protein is critical for VEGF induction by Met/HGF and ErbB2 receptors and for early onset of tumor angiogenesis. Proc Natl Acad Sci USA. 2004;101:2345-2350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Migliore C, Giordano S. Molecular cancer therapy: can our expectation be MET? Eur J Cancer. 2008;44:641-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 99] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 18. | Scagliotti GV, Novello S, von Pawel J. The emerging role of MET/HGF inhibitors in oncology. Cancer Treat Rev. 2013;39:793-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 19. | Jeffers M, Fiscella M, Webb CP, Anver M, Koochekpour S, Vande Woude GF. The mutationally activated Met receptor mediates motility and metastasis. Proc Natl Acad Sci USA. 1998;95:14417-14422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 172] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 20. | Rong S, Segal S, Anver M, Resau JH, Vande Woude GF. Invasiveness and metastasis of NIH 3T3 cells induced by Met-hepatocyte growth factor/scatter factor autocrine stimulation. Proc Natl Acad Sci USA. 1994;91:4731-4735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 276] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 21. | Ferracini R, Di Renzo MF, Scotlandi K, Baldini N, Olivero M, Lollini P, Cremona O, Campanacci M, Comoglio PM. The Met/HGF receptor is over-expressed in human osteosarcomas and is activated by either a paracrine or an autocrine circuit. Oncogene. 1995;10:739-749. [PubMed] |
| 22. | Jo M, Stolz DB, Esplen JE, Dorko K, Michalopoulos GK, Strom SC. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem. 2000;275:8806-8811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 275] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 23. | Fischer OM, Giordano S, Comoglio PM, Ullrich A. Reactive oxygen species mediate Met receptor transactivation by G protein-coupled receptors and the epidermal growth factor receptor in human carcinoma cells. J Biol Chem. 2004;279:28970-28978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 97] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 24. | Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3442] [Cited by in RCA: 3669] [Article Influence: 203.8] [Reference Citation Analysis (0)] |
| 25. | Trusolino L, Bertotti A, Comoglio PM. A signaling adapter function for alpha6beta4 integrin in the control of HGF-dependent invasive growth. Cell. 2001;107:643-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 328] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 26. | Shang N, Arteaga M, Zaidi A, Stauffer J, Cotler SJ, Zeleznik-Le NJ, Zhang J, Qiu W. FAK is required for c-Met/β-catenin-driven hepatocarcinogenesis. Hepatology. 2015;61:214-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 27. | Monga SP, Mars WM, Pediaditakis P, Bell A, Mulé K, Bowen WC, Wang X, Zarnegar R, Michalopoulos GK. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res. 2002;62:2064-2071. [PubMed] |
| 28. | van der Voort R, Taher TE, Wielenga VJ, Spaargaren M, Prevo R, Smit L, David G, Hartmann G, Gherardi E, Pals ST. Heparan sulfate-modified CD44 promotes hepatocyte growth factor/scatter factor-induced signal transduction through the receptor tyrosine kinase c-Met. J Biol Chem. 1999;274:6499-6506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 170] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 29. | Olaku V, Matzke A, Mitchell C, Hasenauer S, Sakkaravarthi A, Pace G, Ponta H, Orian-Rousseau V. c-Met recruits ICAM-1 as a coreceptor to compensate for the loss of CD44 in Cd44 null mice. Mol Biol Cell. 2011;22:2777-2786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 30. | Giordano S, Corso S, Conrotto P, Artigiani S, Gilestro G, Barberis D, Tamagnone L, Comoglio PM. The semaphorin 4D receptor controls invasive growth by coupling with Met. Nat Cell Biol. 2002;4:720-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 341] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 31. | Corso S, Comoglio PM, Giordano S. Cancer therapy: can the challenge be MET? Trends Mol Med. 2005;11:284-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 32. | Xin X, Yang S, Ingle G, Zlot C, Rangell L, Kowalski J, Schwall R, Ferrara N, Gerritsen ME. Hepatocyte growth factor enhances vascular endothelial growth factor-induced angiogenesis in vitro and in vivo. Am J Pathol. 2001;158:1111-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 302] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 33. | Fafalios A, Ma J, Tan X, Stoops J, Luo J, Defrances MC, Zarnegar R. A hepatocyte growth factor receptor (Met)-insulin receptor hybrid governs hepatic glucose metabolism. Nat Med. 2011;17:1577-1584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Wang X, DeFrances MC, Dai Y, Pediaditakis P, Johnson C, Bell A, Michalopoulos GK, Zarnegar R. A mechanism of cell survival: sequestration of Fas by the HGF receptor Met. Mol Cell. 2002;9:411-421. [PubMed] |
| 35. | Bozkaya G, Korhan P, Cokaklı M, Erdal E, Sağol O, Karademir S, Korch C, Atabey N. Cooperative interaction of MUC1 with the HGF/c-Met pathway during hepatocarcinogenesis. Mol Cancer. 2012;11:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 36. | Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, Gendler SJ, Bennett EP, Hollingsworth MA. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283:26985-26995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 37. | Sulpice E, Plouët J, Bergé M, Allanic D, Tobelem G, Merkulova-Rainon T. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood. 2008;111:2036-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 133] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 38. | Chen SY, Chen HC. Direct interaction of focal adhesion kinase (FAK) with Met is required for FAK to promote hepatocyte growth factor-induced cell invasion. Mol Cell Biol. 2006;26:5155-5167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 116] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 39. | Tward AD, Jones KD, Yant S, Cheung ST, Fan ST, Chen X, Kay MA, Wang R, Bishop JM. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc Natl Acad Sci USA. 2007;104:14771-14776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 173] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 40. | Matsumoto K, Nakamura T. Hepatocyte growth factor: renotropic role and potential therapeutics for renal diseases. Kidney Int. 2001;59:2023-2038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 254] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 41. | Kadono Y, Shibahara K, Namiki M, Watanabe Y, Seiki M, Sato H. Membrane type 1-matrix metalloproteinase is involved in the formation of hepatocyte growth factor/scatter factor-induced branching tubules in madin-darby canine kidney epithelial cells. Biochem Biophys Res Commun. 1998;251:681-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Hernandez-Gea V, Toffanin S, Friedman SL, Llovet JM. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 593] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 43. | Wirzenius M, Tammela T, Uutela M, He Y, Odorisio T, Zambruno G, Nagy JA, Dvorak HF, Ylä-Herttuala S, Shibuya M. Distinct vascular endothelial growth factor signals for lymphatic vessel enlargement and sprouting. J Exp Med. 2007;204:1431-1440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 139] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 44. | Caunt M, Mak J, Liang WC, Stawicki S, Pan Q, Tong RK, Kowalski J, Ho C, Reslan HB, Ross J. Blocking neuropilin-2 function inhibits tumor cell metastasis. Cancer Cell. 2008;13:331-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 263] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 45. | Margetts J, Ogle LF, Chan SL, Chan AWH, Chan KCA, Jamieson D, Willoughby CE, Mann DA, Wilson CL, Manas DM. Neutrophils: driving progression and poor prognosis in hepatocellular carcinoma? Br J Cancer. 2018;118:248-257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 46. | Lee M, Chung GE, Lee JH, Oh S, Nam JY, Chang Y, Cho H, Ahn H, Cho YY, Yoo JJ. Antiplatelet therapy and the risk of hepatocellular carcinoma in chronic hepatitis B patients on antiviral treatment. Hepatology. 2017;66:1556-1569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 47. | Ziogas AC, Gavalas NG, Tsiatas M, Tsitsilonis O, Politi E, Terpos E, Rodolakis A, Vlahos G, Thomakos N, Haidopoulos D. VEGF directly suppresses activation of T cells from ovarian cancer patients and healthy individuals via VEGF receptor Type 2. Int J Cancer. 2012;130:857-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 48. | Kuang DM, Zhao Q, Peng C, Xu J, Zhang JP, Wu C, Zheng L. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med. 2009;206:1327-1337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 591] [Cited by in RCA: 724] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 49. | Zhao L, Wu Y, Xie XD, Chu YF, Li JQ, Zheng L. c-Met identifies a population of matrix metalloproteinase 9-producing monocytes in peritumoural stroma of hepatocellular carcinoma. J Pathol. 2015;237:319-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 50. | Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3347] [Cited by in RCA: 3531] [Article Influence: 185.8] [Reference Citation Analysis (1)] |
| 51. | Yagci T, Cetin M, Ercin PB. Cancer Stem Cells in Hepatocellular Carcinoma. J Gastrointest Cancer. 2017; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 52. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6844] [Cited by in RCA: 6917] [Article Influence: 288.2] [Reference Citation Analysis (0)] |
| 53. | Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 854] [Cited by in RCA: 1029] [Article Influence: 79.2] [Reference Citation Analysis (0)] |
| 54. | Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500-504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1432] [Cited by in RCA: 1457] [Article Influence: 112.1] [Reference Citation Analysis (0)] |
| 55. | Wilson CL, Mann DA, Borthwick LA. Epigenetic reprogramming in liver fibrosis and cancer. Adv Drug Deliv Rev. 2017;121:124-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 56. | St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 880] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 57. | Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 264] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 58. | Esposti DD, Hernandez-Vargas H, Voegele C, Fernandez-Jimenez N, Forey N, Bancel B, Le Calvez-Kelm F, McKay J, Merle P, Herceg Z. Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget. 2016;7:31862-31877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 59. | Quint K, Agaimy A, Di Fazio P, Montalbano R, Steindorf C, Jung R, Hellerbrand C, Hartmann A, Sitter H, Neureiter D. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Arch. 2011;459:129-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 60. | Buurman R, Gürlevik E, Schäffer V, Eilers M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kühnel F, Skawran B. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 2012;143:811-820.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 61. | Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J, Hui CK, Lau GK, He ML, Sung J. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology. 2007;46:200-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 128] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 62. | Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 601] [Cited by in RCA: 627] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 63. | Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073-11077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 64. | Shukla SD, Lim RW. Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Res. 2013;35:47-55. [PubMed] |
| 65. | Wijetunga NA, Pascual M, Tozour J, Delahaye F, Alani M, Adeyeye M, Wolkoff AW, Verma A, Greally JM. A pre-neoplastic epigenetic field defect in HCV-infected liver at transcription factor binding sites and polycomb targets. Oncogene. 2017;36:2030-2044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 66. | Thaiss CA, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell. 2016;167:1495-1510.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 566] [Article Influence: 62.9] [Reference Citation Analysis (0)] |
| 67. | Thelen A, Jonas S, Benckert C, Weichert W, Schott E, Bötcher C, Dietz E, Wiedenmann B, Neuhaus P, Scholz A. Tumor-associated lymphangiogenesis correlates with prognosis after resection of human hepatocellular carcinoma. Ann Surg Oncol. 2009;16:1222-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 68. | Hu CT, Wu JR, Cheng CC, Wu WS. The Therapeutic Targeting of HGF/c-Met Signaling in Hepatocellular Carcinoma: Alternative Approaches. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 69. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9016] [Cited by in RCA: 10271] [Article Influence: 604.2] [Reference Citation Analysis (2)] |
| 70. | Owusu BY, Galemmo R, Janetka J, Klampfer L. Hepatocyte Growth Factor, a Key Tumor-Promoting Factor in the Tumor Microenvironment. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 71. | Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006;66:944-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 410] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 72. | Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J. 2006;25:3534-3545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 278] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 73. | Sitia G, Aiolfi R, Di Lucia P, Mainetti M, Fiocchi A, Mingozzi F, Esposito A, Ruggeri ZM, Chisari FV, Iannacone M. Antiplatelet therapy prevents hepatocellular carcinoma and improves survival in a mouse model of chronic hepatitis B. Proc Natl Acad Sci USA. 2012;109:E2165-E2172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 261] [Article Influence: 20.1] [Reference Citation Analysis (1)] |
| 74. | Pytowski B, Goldman J, Persaud K, Wu Y, Witte L, Hicklin DJ, Skobe M, Boardman KC, Swartz MA. Complete and specific inhibition of adult lymphatic regeneration by a novel VEGFR-3 neutralizing antibody. J Natl Cancer Inst. 2005;97:14-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 194] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 75. | Phillips RJ, Burdick MD, Lutz M, Belperio JA, Keane MP, Strieter RM. The stromal derived factor-1/CXCL12-CXC chemokine receptor 4 biological axis in non-small cell lung cancer metastases. Am J Respir Crit Care Med. 2003;167:1676-1686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 356] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 76. | Murga M, Fernandez-Capetillo O, Tosato G. Neuropilin-1 regulates attachment in human endothelial cells independently of vascular endothelial growth factor receptor-2. Blood. 2005;105:1992-1999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 102] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 77. | Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220-2222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 193] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 78. | Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390-7394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 787] [Cited by in RCA: 812] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 79. | Ohta K, Hoshino H, Wang J, Ono S, Iida Y, Hata K, Huang SK, Colquhoun S, Hoon DS. MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. 2015;6:3211-3224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 144] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 80. | Yang X, Zhang XF, Lu X, Jia HL, Liang L, Dong QZ, Ye QH, Qin LX. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology. 2014;59:1874-1885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 170] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 81. | Gao SB, Xu B, Ding LH, Zheng QL, Zhang L, Zheng QF, Li SH, Feng ZJ, Wei J, Yin ZY. The functional and mechanistic relatedness of EZH2 and menin in hepatocellular carcinoma. J Hepatol. 2014;61:832-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 82. | Campbell JE, Kuntz KW, Knutson SK, Warholic NM, Keilhack H, Wigle TJ, Raimondi A, Klaus CR, Rioux N, Yokoi A. EPZ011989, A Potent, Orally-Available EZH2 Inhibitor with Robust in Vivo Activity. ACS Med Chem Lett. 2015;6:491-495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 103] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 83. | Zhou JY, Chen X, Zhao J, Bao Z, Chen X, Zhang P, Liu ZF, Zhou JY. MicroRNA-34a overcomes HGF-mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer Lett. 2014;351:265-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 84. | Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J Biol Chem. 2008;283:18158-18166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 172] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 85. | Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J, Zhao JJ, Mao SS, Zhang GH, Xu XC, Zhang N. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer. 2011;117:2842-2852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 167] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 86. | Hu MH, Ma CY, Wang XM, Ye CD, Zhang GX, Chen L, Wang JG. MicroRNA-126 inhibits tumor proliferation and angiogenesis of hepatocellular carcinoma by down-regulating EGFL7 expression. Oncotarget. 2016;7:66922-66934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 87. | Meng F, Glaser SS, Francis H, DeMorrow S, Han Y, Passarini JD, Stokes A, Cleary JP, Liu X, Venter J. Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med. 2012;16:160-173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 104] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 88. | Shi W, Zhang Z, Yang B, Guo H, Jing L, Liu T, Luo Y, Liu H, Li Y, Gao Y. Overexpression of microRNA let-7 correlates with disease progression and poor prognosis in hepatocellular carcinoma. Medicine (Baltimore). 2017;96:e7764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 89. | Jin B, Wang W, Meng XX, Du G, Li J, Zhang SZ, Zhou BH, Fu ZH. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer. 2016;16:863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 90. | Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, Liu L, Jia D, Tian Q, Wu J. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574-7583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 233] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 91. | Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394-33405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 293] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 92. | Yang X, Liang L, Zhang XF, Jia HL, Qin Y, Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 237] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 93. | Yang YM, Lee CG, Koo JH, Kim TH, Lee JM, An J, Kim KM, Kim SG. Gα12 overexpressed in hepatocellular carcinoma reduces microRNA-122 expression via HNF4α inactivation, which causes c-Met induction. Oncotarget. 2015;6:19055-19069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 94. | Ding W, Tan H, Zhao C, Li X, Li Z, Jiang C, Zhang Y, Wang L. MiR-145 suppresses cell proliferation and motility by inhibiting ROCK1 in hepatocellular carcinoma. Tumour Biol. 2016;37:6255-6260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 95. | Korhan P, Erdal E, Atabey N. MiR-181a-5p is downregulated in hepatocellular carcinoma and suppresses motility, invasion and branching-morphogenesis by directly targeting c-Met. Biochem Biophys Res Commun. 2014;450:1304-1312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 96. | Chen Y, Du M, Wang J, Xing P, Zhang Y, Li F, Lu X. MiRNA-200a expression is inverse correlation with hepatocyte growth factor expression in stromal fibroblasts and its high expression predicts a good prognosis in patients with non-small cell lung cancer. Oncotarget. 2016;7:48432-48442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 97. | Zhu Z, Zhang X, Wang G, Zheng H. Role of MicroRNAs in Hepatocellular Carcinoma. Hepat Mon. 2014;14:e18672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 98. | Runge A, Hu J, Wieland M, Bergeest JP, Mogler C, Neumann A, Géraud C, Arnold B, Rohr K, Komljenovic D. An inducible hepatocellular carcinoma model for preclinical evaluation of antiangiogenic therapy in adult mice. Cancer Res. 2014;74:4157-4169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 99. | Xiang Q, Chen W, Ren M, Wang J, Zhang H, Deng DY, Zhang L, Shang C, Chen Y. Cabozantinib suppresses tumor growth and metastasis in hepatocellular carcinoma by a dual blockade of VEGFR2 and MET. Clin Cancer Res. 2014;20:2959-2970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 179] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 100. | Chan SL, Yeo W. Targeted therapy of hepatocellular carcinoma: present and future. J Gastroenterol Hepatol. 2012;27:862-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 101. | Yau TC, Sukeepaisarnjaroen W, Chao Y, Yen CJ, Lausoontornsiri W, Chen PJ, Sanpajit T, Lencioni R, Camp AC, Cox DS. A phase I/II study of foretinib, an oral multikinase inhibitor targeting MET, RON, AXL, TIE-2, and VEGFR in advanced hepatocellular carcinoma (HCC). J Clin Oncol. 2012;30:4108-4108. |
| 102. | Zhu AX, Kudo M, Assenat E, Cattan S, Kang YK, Lim HY, Poon RT, Blanc JF, Vogel A, Chen CL. Effect of everolimus on survival in advanced hepatocellular carcinoma after failure of sorafenib: the EVOLVE-1 randomized clinical trial. JAMA. 2014;312:57-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 488] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 103. | Goyal L, Muzumdar MD, Zhu AX. Targeting the HGF/c-MET pathway in hepatocellular carcinoma. Clin Cancer Res. 2013;19:2310-2318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 252] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 104. | Zhu AX, Baron AD, Malfertheiner P, Kudo M, Kawazoe S, Pezet D, Weissinger F, Brandi G, Barone CA, Okusaka T. Ramucirumab as Second-Line Treatment in Patients With Advanced Hepatocellular Carcinoma: Analysis of REACH Trial Results by Child-Pugh Score. JAMA Oncol. 2016;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 105. | Bladt F, Friese-Hamim M, Ihling C, Wilm C, Blaukat A. The c-Met Inhibitor MSC2156119J Effectively Inhibits Tumor Growth in Liver Cancer Models. Cancers (Basel). 2014;6:1736-1752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 106. | Schiffer E, Housset C, Cacheux W, Wendum D, Desbois-Mouthon C, Rey C, Clergue F, Poupon R, Barbu V, Rosmorduc O. Gefitinib, an EGFR inhibitor, prevents hepatocellular carcinoma development in the rat liver with cirrhosis. Hepatology. 2005;41:307-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 251] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 107. | Kaseb AO, Morris JS, Iwasaki M, Al-Shamsi HO, Raghav KPS, Girard L, Cheung S, Nguyen V, Elsayes KM, Xiao L. Phase II trial of bevacizumab and erlotinib as a second-line therapy for advanced hepatocellular carcinoma. OncoTargets and therapy. 2016;773-780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 108. | Tai WM, Yong WP, Lim C, Low LS, Tham CK, Koh TS, Ng QS, Wang WW, Wang LZ, Hartano S. A phase Ib study of selumetinib (AZD6244, ARRY-142886) in combination with sorafenib in advanced hepatocellular carcinoma (HCC). Ann Oncol. 2016;27:2210-2215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 109. | Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, Rooney C, Coleman T, Baker D, Mellor MJ. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045-2056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 424] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 110. | Sun CY, Zhu Y, Li XF, Tang LP, Su ZQ, Wang XQ, Li CY, Yang HM, Zheng GJ, Feng B. Norcantharidin alone or in combination with crizotinib induces autophagic cell death in hepatocellular carcinoma by repressing c-Met-mTOR signaling. Oncotarget. 2017;8:114945-114955. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 111. | Tanwandee T, Sukeepaisarnjaroen W, Chan SL, Choo SP, Han GH, Sriuranpong V, Pan HM, Yau TC, Ren ZG, Xu JM. A phase (Ph) II study of the efficacy and safety of the cMET inhibitor capmatinib (INC280) in patients (pts) with advanced hepatocellular carcinoma (HCC). J Clin Oncol. 2016;34:4074-4074. |
| 112. | Li Z, You K, Li J, Wang Y, Xu H, Gao B, Wang J. Madecassoside suppresses proliferation and invasiveness of HGF-induced human hepatocellular carcinoma cells via PKC-cMET-ERK1/2-COX-2-PGE2 pathway. Int Immunopharmacol. 2016;33:24-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 113. | García-Vilas JA, Quesada AR, Medina MA. Damnacanthal, a noni anthraquinone, inhibits c-Met and is a potent antitumor compound against Hep G2 human hepatocellular carcinoma cells. Sci Rep. 2015;5:8021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 114. | Diaz D, Ford KA, Hartley DP, Harstad EB, Cain GR, Achilles-Poon K, Nguyen T, Peng J, Zheng Z, Merchant M. Pharmacokinetic drivers of toxicity for basic molecules: strategy to lower pKa results in decreased tissue exposure and toxicity for a small molecule Met inhibitor. Toxicol Appl Pharmacol. 2013;266:86-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 115. | Qu Z, Wu J, Wu J, Luo D, Jiang C, Ding Y. Exosomes derived from HCC cells induce sorafenib resistance in hepatocellular carcinoma both in vivo and in vitro. J Exp Clin Cancer Res. 2016;35:159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 126] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 116. | Jo JC, Choi EK, Shin JS, Moon JH, Hong SW, Lee HR, Kim SM, Jung SA, Lee DH, Jung SH. Targeting FGFR Pathway in Human Hepatocellular Carcinoma: Expressing pFGFR and pMET for Antitumor Activity. Mol Cancer Ther. 2015;14:2613-2622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 117. | Wang SW, Pan SL, Guh JH, Chen HL, Huang DM, Chang YL, Kuo SC, Lee FY, Teng CM. YC-1 [3-(5’-Hydroxymethyl-2’-furyl)-1-benzyl Indazole] exhibits a novel antiproliferative effect and arrests the cell cycle in G0-G1 in human hepatocellular carcinoma cells. J Pharmacol Exp Ther. 2005;312:917-925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |