Published online Jul 7, 2018. doi: 10.3748/wjg.v24.i25.2647
Peer-review started: March 29, 2018
First decision: May 9, 2018
Revised: May 18, 2018
Accepted: June 16, 2018
Article in press: June 16, 2018
Published online: July 7, 2018
Processing time: 98 Days and 17.4 Hours
Hepatocellular carcinoma (HCC) is the most common primary liver malignant neoplasia. HCC is characterized by a poor prognosis. The need to find new molecular markers for its diagnosis and prognosis has led to a progressive increase in the number of scientific studies on this topic. MicroRNAs (miRNAs) are small non-coding RNA that play a role in almost all main cellular pathways. miRNAs are involved in the regulation of expression of the major tumor-related genes in carcinogenesis, acting as oncogenes or tumor suppressor genes. The aim of this review was to identify papers published in 2017 investigating the role of miRNAs in HCC tumorigenesis. miRNAs were classified according to their role in the main molecular pathways involved in HCC tumorigenesis: (1) mTOR; (2) Wnt; (3) JAK/STAT; (4) apoptosis; and (5) MAPK. The role of miRNAs in prognosis/response prediction was taken into consideration. Bearing in mind that the analysis of miRNAs in serum and other body fluids would be crucial for clinical management, the role of circulating miRNAs in HCC patients was also investigated. The most represented miRNA-regulated pathway in HCC is mTOR, but apoptosis, Wnt, JAK/STAT or MAPK pathways are also influenced by miRNA expression levels. These miRNAs could thus be used in clinical practice as diagnostic, prognostic or therapeutic targets for HCC treatment.
Core tip: Hepatocellular carcinoma (HCC) is the most common primary liver neoplasia and is characterized by a poor prognosis. MicroRNAs (miRNAs) are involved in the regulation of expression of the major tumor-related pathways in carcinogenesis and may act as oncogenes or tumor suppressor genes. mTOR is the most represented miRNA-regulated pathway in HCC. miRNAs found to be deregulated in HCC could be used in clinical practice as diagnostic, prognostic or therapeutic targets.
- Citation: Vasuri F, Visani M, Acquaviva G, Brand T, Fiorentino M, Pession A, Tallini G, D’Errico A, de Biase D. Role of microRNAs in the main molecular pathways of hepatocellular carcinoma. World J Gastroenterol 2018; 24(25): 2647-2660
- URL: https://www.wjgnet.com/1007-9327/full/v24/i25/2647.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i25.2647
Hepatocellular carcinoma (HCC), the most common primary liver malignant neoplasia, is a very diffuse malignancy, with variable incidence according to geography, and represents the fifth cause of cancer-related death worldwide[1,2]. Generally, HCC is characterized by a poor prognosis, mainly due to the limited treatment choices[3]: prognosis after surgery - including liver transplantation - depends on the tumor stage, the association with cirrhosis and on liver function[4]. Histology can be of little help in predicting the response to surgery, since the classic well-established histological features of HCC are seldom evaluable on liver needle biopsy: microvascular invasion (MVI) is rarely seen on biopsy, and tumor grade and architecture are too heterogeneous in HCC to be assessed on the basis of small sampling. Up to now, the most reliable prognostic markers after surgery are clinical and surgical, e.g., the complete resection of the lesion, liver function tests, and alpha-fetoprotein[5]. For liver transplantation, the inclusion within the Milan criteria remains a cornerstone for the good outcome of the recipients[6].
As for the systemic therapy for HCC, no serious options have been available until 2007, when the neoangiogenesis agent Sorafenib was introduced. Sorafenib gave one more chance to those patients with high-stage and MVI-positive HCC, not suitable for surgery[7,8]. Again, no predictive tests are available to assess which patients will benefit most from Sorafenib therapy.
The need to find new tissue and/or serum markers for HCC diagnosis and prognosis progressively increased the number of studies on the molecular mechanism behind liver carcinogenesis. The issue, however, still remains tangled, due to the high heterogeneity of HCC (not only in phenotype), and to the complex multistep carcinogenesis occurring differently in cirrhotic and non-cirrhotic livers[2].
MicroRNAs (miRNAs) are small non-coding RNA (20-25 nucleotides) that play a role in almost all main cellular pathways[9]. miRNAs contribute to a variety of physiological and pathological events, including several types of tumors[10-15]. Thus, miRNAs are involved in the regulation of expression of the major tumor-related genes in carcinogenesis, acting as oncogenes or tumor suppressor genes[16].
The aim of this review was to identify papers published in the last 12 mo (from January 2017 to December 2017), investigating the possible role of miRNAs in HCC tumorigenesis. miRNAs were classified according to their role in the main molecular pathways involved in HCC: (1) mTOR; (2) Wnt; (3) JAK/STAT; (4) apoptosis; and (5) MAPK. Moreover, the possible role of miRNAs in prognosis/response prediction and the level of circulating miRNAs in HCC patients were also investigated.
mTOR (mammalian target of rapamycin) is a well conserved serine-threonine kinase that plays a fundamental role in the signaling network that controls growth and cell metabolism. mTOR is the physical target of rapamycin. mTOR exists in two different multi-protein complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). mTORC1 is composed of five components: mTOR, Raptor, PRAS40, GβL, and DEPTOR[17]. mTORC1 is directly inhibited by rapamycin but, at the same time, it is modulated by genotoxic stress, growth factors, oxygen and energy status, resulting in modulation of cell growth and proliferation[18]. These inputs indirectly regulate mTORC1 by controlling the activation status of TSC1-TSC2. For example, growth factors block TSC1-TSC2 and mTORC1 is consequently activated. On the contrary, energy deficit, genotoxic stress or oxygen deprivation are positive signals on TSC1-TSC2, which inhibit mTORC1. Activated mTORC1 promotes protein synthesis and lipid biogenesis, it controls mitochondrial metabolism and biogenesis; it also inhibits catabolism by blocking autophagy and it inhibits growth factor signaling by activating negative feedback loops that block the PI3K pathway[18-20].
The second complex, mTORC2, is composed of six components: mTOR, Rictor, GβL, Sin1, PRR5/Protor-1, and DEPTOR. mTORC2 regulates cellular survival, cytoskeletal organization and metabolism. Compared to mTORC1, the mechanisms of mTORC2 are less understood: It is insensitive to acute treatment with rapamycin, but it was reported that a long-term treatment with rapamycin reduces mTORC2 signaling, suppressing the assembly of the complex[19,21]. This complex is activated by growth factors with a PI3K-dependent mechanism and the work of Zinzalla et al[22] suggested a possible role of ribosomes for mTORC2 complex activation[20,22].
Due to its key role in regulating cell growth, survival and metabolism, the mTOR pathway is aberrantly activated in many diseases, including cancer, cardiovascular disease and diabetes[20]. In up to 50% of HCC cases, an aberrant activation of mTOR was reported, mainly downstream of the insulin growth factor (IGF) or epidermal growth factor (EGF) signaling cascades[23].
Many studies showed tumor suppressor miRNAs, the down-regulation of which lead to the mTOR pathway activation in HCC cells.
miR-758-3p: Jiang et al[24] showed that the restoration of miR-758-3p in HCC cell line could suppress cell proliferation, migration, and invasion. miR-758-3p markedly down-regulates the expression of MDM2 and mTOR and, at the same time, the expression of p53, AKT and PRAS40 resulted up-regulated. mTOR can be regulated by its upstream effector AKT, which can suppress PRAS40 so as to eliminate the inhibition it exerts on mTORC1[24].
miR-142: Yu et al[25] demonstrated that miR-142 expression was reduced in 50 tumor tissues in comparison to correspondent normal tissues and in two HCC cell lines compared to human normal liver cell line. This lower expression was linked to poor clinical parameters like high TNM stage and distant metastasis. miR-142 was identified to directly target the transforming growth factor β (TGF-β), which controls cell vitality, proliferation, epithelial-mesenchymal transition (EMT) and neo-angiogenesis. mTOR is one of the effector pathways of TGF-β signaling. These findings imply that miR-142 is a tumour suppressor gene in HCC and that it increases the TGF-β-induced development of hepatocellular carcinoma[25].
miR-199b-5p: In 100 pairs of HCC patients’ tumor tissues and adjacent liver tissues a significant down-regulation of miR-199b-5p was observed and associated to poor clinical outcome. N-cadherin was the demonstrated target of miR-199b-5p and it promoted EMT in HCC cells. The restoration of miR-199b-5p suppressed cell migration, invasion and metastasis in xenograft tumors. It was demonstrated that the miR-199b-5p overexpression lead to suppression of TGF-β1-induced Akt phosphorylation. Moreover, inhibition of the PI3K/Akt signaling pathway blocked TGF-β1-induced N-cadherin overexpression in HCC cells. The inhibitory effects on EMT and on the TGF-β1 signaling pathway support the potential use of miR-199b-5p as a promising strategy to treat HCC[26].
miR-187, miR-497, miR-99a, miR-592: IGF-1R activation, through the PI3K/Akt/mTOR axis, is responsible for cell proliferation, migration and invasion in HCC[27]. IGF-1R is a target of miR-187, miR-497, miR-99a and miR-592[28-30]. miR-187 was found downregulated in HCC tissues and cell lines: as reported by Han et al[29], the restoration of miR-187 leads to a significant arrest of HCC growth. miR-497 and miR-99a target the 3′-UTR of both IGF-1R and mTOR and were shown to be down-regulated in HCC human tissues and cell lines: the co-transfection with both miRNAs slowed cell proliferation and the tumor growth in HCC cell lines and in xenograft models[28]. Wang and colleagues demonstrated that miR-592 was significantly downregulated in HCC tissues and cell lines and that its low expression was associated with lymph node metastases[30]. These results indicated that miR-187, miR-497, miR-99a, miR-592 could be investigated as potential therapeutic targets for HCC in the future.
miR-296-5p: The low expression of miR-296-5p is directly linked to the activation of the mTOR pathway in HCC growth. Gainoffunction experiments demonstrated that miR2965p inhibited HCC cell proliferation, migration and invasion in vitro, by targeting AKT2. These findings indicated that the miR2965p/AKT2 axis plays important roles in HCC carcinogenesis and progression, and that miR2965p/AKT2 could be considered a potential target for HCC therapy[31].
miR-139-5p: PDK1/AKT/mTOR axis activation could lead to hepatocellular carcinoma cell proliferation. PDK1 is a known target of miR-139-5p, found down-regulated in HCC tissues and cell lines. Mo et al[32] also observed that miR139-5p/PDK1 expression was regulated by long-coding RNA XIST, which was found over-expressed in HCC.
miR-15b-5p: Opa interacting protein 5 (OIP5) was found up-regulated in HCC, inducing tumor growth and metastasis in vitro and in vivo. OIP5 induces mTORC1 and GSK-3β/β-catenin signaling activation, through AKT. miR-15b-5p was found down-regulated in HCC cells and OIP5 was found to be its direct target. These findings suggest that the restoration of miR-15b-5p could inhibit OIP5-mediated oncogenic signaling in HCC[33].
miR-345: Yu and colleagues found that the expression of miR-345 was significantly down-regulated in 65 HCC cases, and matching tumor-adjacent tissues, and in HCC cell lines. They also reported a clinical correlation between the low expression of miR-345 and venous infiltration, multiple lymph node metastases, and advanced TNM stage. The restoration of miR-345 inhibited migration and invasion ability of HCC cells. It was demonstrated that interferon regulatory factor 1 (IRF1) was a direct target of miR-345 and that IRF-1 mediated the oncogenic effects triggering mTOR/STAT3/AKT signaling[34].
miR-223: Dong and colleagues showed that miR-223 was able to suppress cell growth and to promote apoptosis in HCC cell lines (HepG2 and Bel-7402). Ras-related protein Rab-1(Rab1) is specifically regulated by miR-223. These data suggested that, in HCC cells, the anti-tumor effects due to miR-223 restoration may be due to the inactivation of the mTOR pathway, caused by the suppression of Rab1 when miR-223 is over-expressed. According to these results, miR-223 may be a potential therapeutic target for treating HCC, mediating mTOR signaling silencing[35].
Other studies showed oncogenic miRNAs, the upregulation of which leads to the mTOR pathway activation in HCC cells.
miR-33a: The levels of miR-33a were observed as significantly higher in HCC tissues than in adjacent non-tumor tissues. This elevated expression of miR-33a correlated with adverse clinical features and poor prognosis. It was demonstrated that miR-33a could promote cell growth by modulating the proliferation and apoptosis of HCC cells. Its direct target is PPARα, one of the targets of mTORC1[36].
miR-302d: Chen and colleagues demonstrated that the overexpression of miR-302d promoted cell growth and migration and suppressed apoptosis in HCC cell lines and that it promotes xenograft tumor growth in vivo. These mechanisms were found to be mediated by TGFbR2-signaling, a target of miR-302d[37].
miR-23b: miR-23b was found to be significantly up-regulated in tumor tissues of HCC patients. It was demonstrated that this miRNA regulated ST7L, a suppressor of the AKT/GSK3β/β-catenin pathway in HCC cells. MiR-23b thus acts as an oncomir in HCC, stimulating proliferation and metastasis through the mTOR and β-catenin signaling cascades[38].
miR-181a, miR-155-5p, miR-25: miR-181a was found to be up-regulated in HCC tissues compared to adjacent tissues; moreover, its levels were dramatically higher in metastatic HCC tissues than in non-metastatic HCC tissues. miR-181a regulates the proliferation and invasion of HCC cells by targeting PTEN, the reduction of which activates the PI3K/Akt pathway[39]. Another miRNA was found to be over-expressed in HCC and plays an oncogenic role in HCC by targeting PTEN: miR-155-5p promotes cell growth, migration and invasion, but inhibits apoptosis in vitro and promoted HCC progression in vivo[40]. PTEN is a known target also of miR-25, which is over-expressed in HCC cell lines and in liver cancer stem cells (LCSCs)[41].
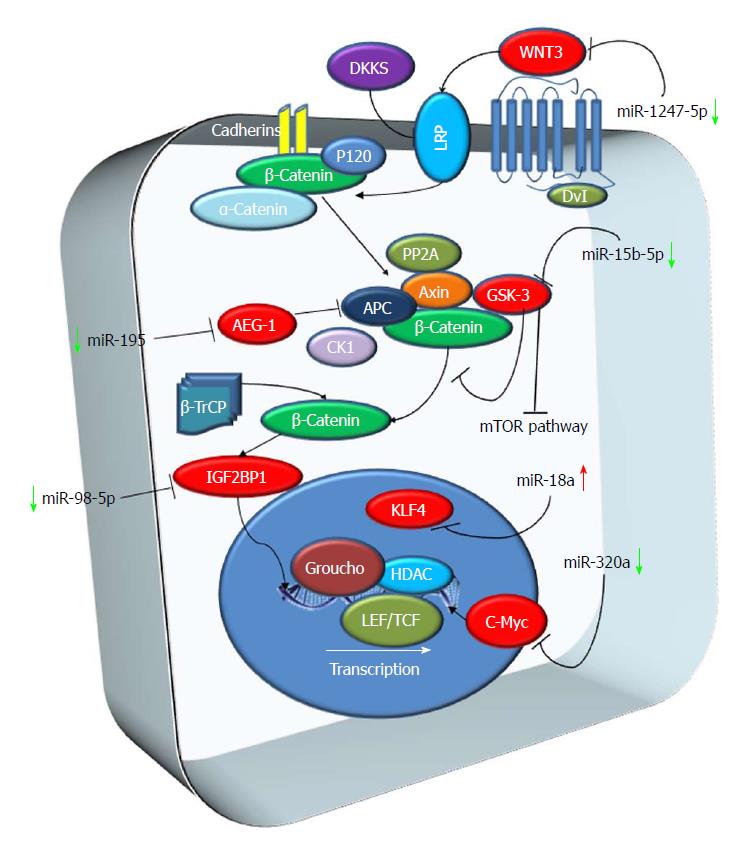
β-catenin phosphorylation and degradation and its regulation by Wnt are the essence of the Wnt pathway. Signaling by the Wnt family proteins is one of the fundamental mechanisms that induce cell proliferation, cell polarity and cell fate during development and tissue homeostasis (Logan and Nusse, 2004). Canonical Wnt signaling functions by regulating the amount of β-catenin. In the absence of Wnt, cytoplasmic β-catenin protein is degraded by the Axin complex. This non-stop degradation prevents β-catenin from reaching the nucleus, and Wnt target genes are thereby repressed[42-44]. Inhibitors of Wnt signaling might be effective in HCC, where mutations in the Wnt pathway components are quite common. In the HepG2 cell line, knockdown of β-catenin, mediated by RNA interference, decreased proliferation and growth in vitro[45,46] (Figure 1).
miRNA-10a, miR-30e, miR-215, miR-125b and miR-148a: Ashmawy and colleagues observed that 11 miRNAs (miR-10a, miR-106b, miR-99a, miR-148a, miR-125b, miR-30e, miR-199a, miR-199a3p, miR-24, miR-122 and miR-215) were down-regulated in HCC patients. Five of these miRNAs (miRNA-10a, miR-30e, miR-215, miR-125b and miR-148a) were also associated with the expression of genes involved in the Wnt/β-catenin pathway, such as β-catenin, APC and c-myc[47].
miR-155 and miR-183: In the same study, the authors detected that miR-155 and miR-183 were up-regulated in HCC patients if compared to controls and that miR-155 was correlated with liver cirrhosis[47].
miR-18a: Other than miR-155 and miR-183, other miRNAs involved in the Wnt/b-catenin pathway were observed up-regulated in HCC. Liu et al[48] identified that miR-18a expression was upregulated in human HCC if compared to the adjacent non-tumoral liver tissue. This up-regulation promotes the proliferation and migration of HCC cell lines by inhibiting KLF4, a factor that negatively regulates β-catenin expression. This data led the authors to hypothesize that miR-18 could be a therapeutic target for HCC treatment[48].
miR-195: Yan and colleagues showed a potential application of miR-195 in the cancer therapy of HCC. They observed that miR-195 was markedly down-regulated both in HCC cell lines and in 36 HCCs compared to adjacent non-tumoral liver tissues. The expression of miR-195 was inversely correlated with AEG-1 expression, which was demonstrated to be a target of miR-195. Overexpression of AEG-1 activates the PI3K/Akt, nuclear factor-κB, and Wnt/β-catenin signaling pathways stimulating proliferation, metastasis, angiogenesis and chemoresistance[49]. An analysis performed using miR-195 mimics showed how miR195 inhibited liver cancer cell growth and induced apoptosis in HCC cell lines. The overexpression of miR-195 also decreased tumor growth of hepatoma xenografts in nude mice[50].
miR-320a: miR-320a was observed to be down-regulated in HCC tissues if compared to paired adjacent non-tumoral liver tissues. In samples with miR-320a inhibition, an up-regulation of the expression levels of β-catenin, c-myc, cyclin D1 and DKK-1 was observed. According to this data miR-320a may be considered as a tumor-suppressive microRNA in human HCC, through the down-regulation of the β-catenin pathway[51]. miR320a was observed to be down-regulated also in a cohort of 50 HCC tissues by Xie and colleagues. The authors also identified c-Myc as a direct target of miR-320a and observed that inducing upregulation of miR-320a in HCC leads to inhibition of HCC cell proliferation and invasion capability through c-Myc silencing. These data support the role of miR320a as a tumor-suppressive microRNA in HCC and provide evidence that miR-320a may be used as a potential target for HCC treatment[52].
miR-98-5p: Another miRNA observed to be down-regulated in HCC tissues is miR-98-5p. Down-regulation of miR-98-5p correlates with tumor size, lymph node metastasis, and clinical stage. In addition, HCC patients with low expression of miR-98-5p had a shorter survival time compared to those with high miR-98-5p levels. miR-98-5p down-regulation in HCC has been associated with up-regulation of Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in HCC, which induces cell proliferation while inhibiting cell apoptosis[53].
miR-15b-5p: As reported above (see mTOR pathway paragraph), miR-15b-5p is involved in AKT/mTOR pathway. However, its expression also inhibits GSK-3β/β-catenin signaling in HCC[33].
miR-1247-5p: miR-1247-5p levels are down-regulated in patients with HCC and in HCC cell lines. This downregulation is probably due to hypermethylation of miR-1247-5p gene. Also, the overexpression of miR1247-5p inhibits the invasion and proliferation of HepG2 cells, induces cell apoptosis in vitro, and suppresses the growth of transplanted tumors in vivo. Wnt3 is a target of miR-1247-5p and overexpression of miR-1247-5p significantly down-regulated its expression. The evidence that the expression of miR-1247-5p can be regulated by methylation indicates that miR-1247-5p may be a potential therapeutic target for HCC[54].
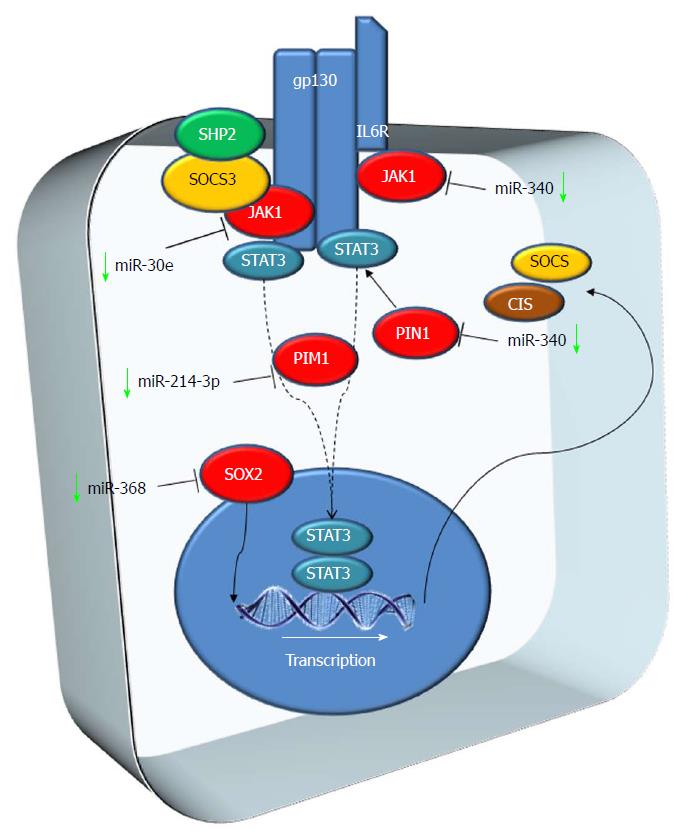
The JAK/STAT pathway regulates development and it is involved in stem cell maintenance, hematopoiesis and inflammatory response. Active JAKs recruit STAT (signal transducers and activators of transcription) proteins that form dimers that translocate to the nucleus when phosphorylated. These dimers modulate transcription of genes involved in differentiation, proliferation and apoptosis. The JAK/STAT pathway is regulated at multiple levels. Many cancers and neoplastic cells employ several strategies to activate the JAK/STAT pathway (e.g., activating mutations in STATs or reduced expression of negative regulators[55-57]) (Figure 2).
miR-214-3p: miR-214-3p is expressed at low levels in HCC[58]. PIM-1 is an oncogene encoding for a serine/threonine kinase protein involved in several human cancers. PIM-1 transcription is initiated by STAT proteins and plays a key role in signal transduction, contributing to both cell proliferation and survival, and thus providing an advantage in tumorigenesis[59]. PIM-1 is a miR-214-3p target and PIM-1 expression is enhanced in HCC[58].
miR-30e: miR-30e is down-regulated in the majority of HCC tissues. Restoration of its expression down-regulates JAK1 expression levels. Silencing JAK1 inhibits migration, proliferation and invasion of HCC cells. For this reason, miR-30e might be a prognostic marker of HCC and a putative therapeutic target[60].
miR-340: JAK-1 was also identified as a direct target of miR-340. miR-340 was found to be significantly down-regulated in HCC tissues and cell lines and, in vitro, its overexpression inhibited migration, cell proliferation and invasion[61].
miR-140-5p and miR-200: miR-140-5p and miR-200 were down-regulated in HCC and predicted to target Pin1[62]. Pin-1 is an independent factor for poor prognosis in HCC, it is overexpressed in ~70% of human HCCs[63], and it is correlated with larger tumor size, higher incidence of MVI and poor prognosis in HCC[64]. The over-expression of miR-140-5p inhibits human HCC cell growth, colony formation and migration[62].
miR-638: miR-638 expression in HCC tissues is down-regulated if compared to the paired non-tumoral tissues. Low expression of miR-638 was linked to venous infiltration and TNM stage. Moreover, low levels of miR-638 are associated with a lower E-cadherin and vimentin expression if compared to cells showing high miR-638 levels. miR-638 down-regulation increases SOX2 expression, a gene overexpressed in HCC and involved in oncogenesis and in the progression of various cancers[65,66]. Moreover, SOX2 expression is associated with overall poor survival in HCC patients and it promotes cancer cell invasion[67]. These data lead to considering miR-638-SOX2 as a putative target for repressing the development and metastasis of HCC[68].
Apoptosis occurs normally during development to maintain cell populations in tissues and as a defense mechanism in immune reactions or when cells are damaged[69,70]. TP53 was the first tumor suppressor gene to be linked to apoptosis and it is well established that TP53 mutations occur in the vast majority of human cancers[71]. In fact, if on one hand wild type p53 promotes apoptosis, cell-cycle arrest and senescence, the loss of p53 function increases viability, chromosomal instability and cellular lifespan. Disruption of the apoptotic pathway correlates with the progression of several tumors[72].
miR-30a: Anoikis is a form of cell death due to loss of contact of the cells with the extracellular matrix. Low levels of miR-30a were observed in several HCC cell lines (Hep3B, HepG2, SMMC-7721, MHCC97-L, MHCC97-H and HCCLM3) and in HCC tumor tissue compared to adjacent non-neoplastic tissue. miR-30a silencing is accompanied by an increase in the expression of Beclin 1 and Atg5 proteins and by a decreased number of cells undergoing anoikis[73].
miR-365: miR-365 expression was significantly lower in HCC cells (SMC7721, HepG2, Bel7404 and Bel7402) compared to a normal hepatocellular cell line (LO2). Inducing up-regulation of miR-365 leads to a significant decrease in cellular activity and to the inhibition of tumor growth. It has also been observed that in cells with an up-regulation of miR-365, the expression of Bax, cyto C and cleaved caspase 3, which is downstream of Bcl-2, were also markedly up-regulated[74].
miR526a: miR526a is down-regulated in HCC tissues. In vitro, the introduction of miR526a into HCC cell lines significantly decreased HCC proliferation, migration and invasion, via p21 inactivation[75].
miR-377: miR-377 was down-regulated in HCC tumors if compared to the adjacent non-neoplastic tissue. In vitro experiments with HCC cell lines demonstrated that a gain of miR-377 function inhibited colony formation, suggesting that miR-377 plays a key role as a tumor suppressor in HCC. In cells with high levels of miR-377 the apoptotic rate was significantly higher than in the controls. Bcl-xL is an anti-apoptotic protein and it is overexpressed in about 33% of HCC[76], conferring resistance to apoptosis. The higher level of apoptotic rate observed in cell lines with miR-377 over-expression was associated with a concomitant down-regulation of Bcl-xL mRNA levels, suggesting that Bcl-xL is a target of miR-377[77].
miR-199a-5p: miR-199a-5p was down-regulated in HCC tissues compared to pair-matched non-neoplastic hepatic tissues[78], and the same was the case for let-7c expression[79]. miR-199a-5p down-regulation was correlated with tumor size and invasion. Moreover, the low expression of miR-199a-5p and let-7c was associated with higher metastatic capability in HCC cell lines. MAP4K3 is a pro-apoptotic kinase that activates the Intrinsic Apoptosis Pathway[80]. MAP4K3 gene was predicted as a possible target of miR-199a-5p and let-7c and the up-regulation of both miRNAs leads to a significant decrease in MAP4K3 protein level, resulting also in a decrease in HCC cell migration and invasion[78].
miR-330: miR-330 level was higher in HCC tissues if compared to adjacent non-neoplastic specimens. The up-regulation of miR-330 was associated with shorter survival in HCC patients. ING genes have been reported to be implicated in apoptosis, cell cycle regulation, and DNA repair. ING4 plays important roles in many cancer-related processes, such as apoptosis, cell proliferation and growth, angiogenesis and migration. ING4 expression is decreased in several cancers[81]. Overexpression of miR-330 reduced the expression of ING4 in HCC cells promoting HCC cell proliferation and invasion[82].
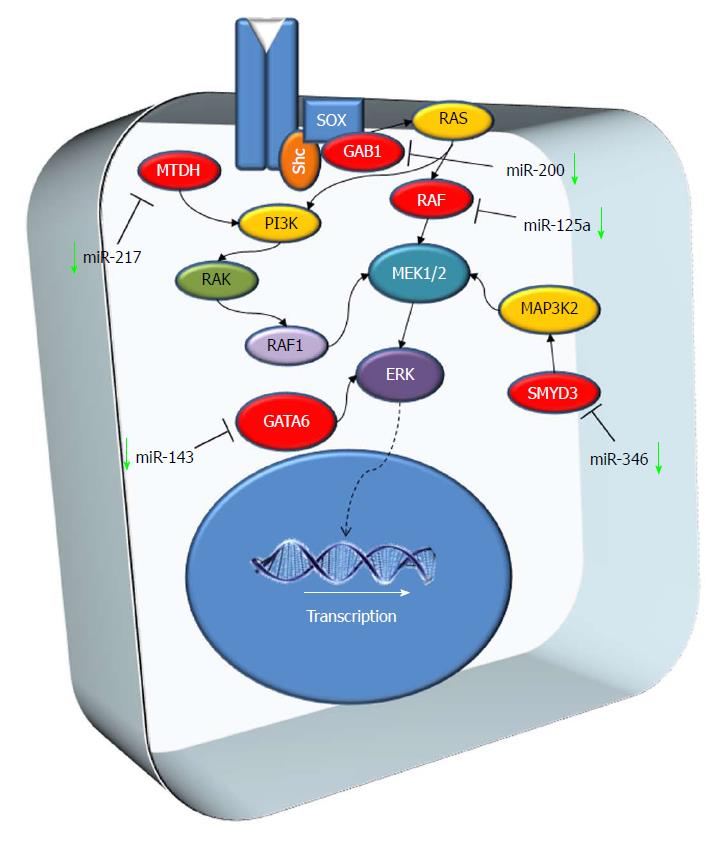
The mitogen-activated protein kinase (MAPK) pathway is characterized by different kinase proteins that link extracellular signals to the machinery that controls physiological cellular processes such as growth, differentiation, proliferation, migration and apoptosis. Alterations in the MAPK cascade impinge on almost all previously listed physiological processes and play a critical role in the development and progression of cancer[83] (Figure 3).
miR-346: miR-346 was down-regulated in HCC tissues and its expression levels are associated with tumor size and TNM stage. An in vitro study revealed that the loss of miR-346 leads to the up-regulation of S phase in HCC cell lines. One of the putative targets of miR-346 is SMYD3. SMYD3 is an oncogene up-regulated in several tumors, including HCC[84]. SMYD3 expression leads to methylation of MAP3K2 by increasing MAP kinase signaling and promoting the formation of Ras-driven carcinomas[85]. Restoring miR-346 levels in HCC cell lines prevented proliferation through the suppression of SMYD3 expression[86].
miR-143: miR-143 expression was reduced in HCC tumor tissues and human liver cancer cell lines (SMMC-7721, Hep3B, HepG2, Huh7, Bel7402, MHCC97-H and SK-Hep1) compared to non-neoplastic adjacent tissues and normal liver cell line (L02), respectively[87]. GATA-binding factor 6 (GATA6) is a transcriptional factor with an oncogenic role in various types of tumor, favoring cancer progression[88]. GATA6 is a potential target for miR-143 and its expression is downregulated by miR-143 overexpression in HepG2 and Bel7402 cells[87].
miR-125a: miR-125a was detected as down-regulated in 80% of HCC biopsies if compared with the adjacent non-tumor liver tissue. When grouping patients according to HCC etiology, miR-125a was downregulated in 80% of HBV patients, 78% of HCV patients, and in all 4 patients with non-alcoholic steatohepatitis. In vitro analysis revealed that MMP11, SIRT7 and c-Raf were the main miR-125a targets. In support of this, MMP11, SIRT7 and c-Raf were up-regulated in about 80% patients with miR-125a down-regulation, hinting at an oncosuppressor effect of the microRNA through the regulation of MMP11, c-Raf and SIRT7 expression[89].
miR-217: miR-217 expression levels in HCC tissues were significantly decreased, while MTDH levels were significantly upregulated. miR-217 up-regulation in HCC cell lines lead to a remarkable downregulation of MTDH mRNA expression, demonstrating that MTDH is a target of miR-217. Metadherin (MTDH) is a transmembrane protein overexpressed in several cancers[90] and it is associated with tumor development and with aggressive course[49]. In cell lines with miR-217 up-regulation and MTDH down-regulation, cell apoptosis notably increased, indicating that miRNA expression promotes apoptosis of HCC cells[91].
miR-200a: miR-200a was down-regulated in HCC cell lines and tissues and it was correlated with the metastatic ability of HCC[92]. GAB1 is one of the molecules targeted by miR-200a. The overexpression of miR-200a suppressed the levels of GAB1 in HCC cell lines, inhibiting cell migration and invasion[92].
In a wide-array miRNA analysis on resected patients with low-stage HCC (within the Milan criteria), Sato et al[93] found that the deregulation of 13 intratumoral miRNAs (miR-100, miR-99a, miR-99b, miR-125b, miR-378, miR-129-5p, , miR-125a-5p, miR-497, miR-22, miR-140-3p, miR-145, miR-221, miR-195) significantly correlated with post-surgical (Table 1). The same is applies to the deregulation of more than 50 miRNAs in the non-tumoral tissue, among which the most significant was miR-96. miR-125b downregulation, in combination with more typical prognostic factors, was also correlated with low disease-free survival (DFS) after resection in later works[94] (Table 1).
| miRNAs (regulation) | Pathway | Clinical |
| 99a | mTOR | Recurrence after resection |
| 497 | Recurrence after resection, recurrence after transplantation | |
| 195 | Recurrence after resection | |
| 140-5p | Recurrence after resection | |
| 23b | Recurrence after resection | |
| 223 | Recurrence after resection | |
| 199a-5p | Recurrence after transplantation | |
| 181a-5p | Resistance to Sorafenib/CHT | |
| 33a-5p | Resistance to Sorafenib/CHT | |
| 125b | Wnt | Recurrence after resection, recurrence after transplantation |
| 195 | Recurrence after resection | |
| 18 (up) | Recurrence after transplantation | |
| 140-5p | JAK-STAT | Recurrence after resection |
| 365 | Apoptosis | Recurrence after resection |
| 125a | MAPK | Recurrence after resection, resistance to Sorafenib/CHT |
In a more recent study on resected HCC, Lin et al[3] identified 16 miRNAs related to MVI, the hierarchical clustering of which was able to predict survival after resection: miR-452-5p, miR-378, miR-9-5p, miR-550a-5p, miR-15a-5p, miR-140-5p, let7g, miR-152-3p, miR-122-5p, miR-212-3p, miR-23b, miR-365a, miR-629-5p, miR-1270, miR-659-3p and miR-3941 (Table 1).
Other miRNAs whose down-regulation was correlated with poor prognosis after resection were miR342-3p[95], miR-655-3p[96], miR-105-1 via NCOA1 deregulation[97], miR-223 associated with an increased Stathmin-1 expression[98], and miR-483-3p in “histologically advanced” HCC (high-grade and/or MVI)[99]. Upregulation of miR-19b was correlated with good prognosis after resection in resected patients with advanced HCC[100]. Albeit miRNA down-regulation is often associated to worse prognosis, up-regulation is not always a good prognostic sign: for example, the up-regulation of miR-135a, miR-29a5p and miR-221 was significantly associated with early HCC recurrence[101-103] (Table 1).
The search for molecular predictors for HCC recurrence after orthotopic liver transplantation (OLT) is even more tangled than after resection, due to the particular immune status of the recipients and the intrinsic capability of HCC tumor cells not only to give metastases to distant organs, but also to implant in the graft. A wide microarray profiling by Barry et al[104] found more than 60 miRNAs to be deregulated in recurrent HCC after OLT. Interestingly, miR-125b and miR-497 - already mentioned above in post-resection recurrence - are among the most significant[93,104]. Common mechanisms beyond HCC recurrence after both resection and liver transplantation are likely to indicate that a more aggressive tumor biology leads to a higher risk of recurrence or dissemination.
Morita et al[105] demonstrated that the concomitant upregulation of miR-18a and down-regulation of miR-199a-5p correlated with the worst disease-free survival in a population of 70 transplanted patients. The most represented sites of recurrence were lymph nodes, lung and bone. The mechanisms proposed by the authors included the link between miR-18a and TNFα, as well as the regulation of the HIF1α, the VEGF-A, and the IGF pathways by miR-199a-5p[105] (Table 1).
The role of miRNAs in the response to chemotherapy is still largely unresolved, especially for HCCs, which are malignancies with an extreme molecular heterogeneity. Due to this heterogeneity, no target therapies specific for HCC have been available since the introduction of Sorafenib in 2005. Sorafenib changed the natural history of patients with advanced HCC not suitable for surgery[7], but no molecular markers for the prediction of the response to this therapy are available yet. Only few in vitro studies focusing on the role of miRNAs in the response to Sorafenib exist, e.g., miR-137[106] and miR-125a-5p[107]. A very interesting recent study found that the deregulation of miR-181a-5p in patients’ serum was correlated with a worst disease control after Sorafenib therapy[108] (Table 1).
As for the resistance to classic chemotherapic drugs, other in vitro studies showed that miR-205-5p and miR-503 were involved in the resistance to 5-Fuorouracil[109,110], miR-33a-5p was involved in the resistance to Cisplatin[111], and miR-31 was involved in the resistance to Adriamycin[112]. An exhaustive in vitro and in vivo study by Jin et al[113] showed that the tissue levels of miR-26a/b regulated the mechanism of autophagy, thus influencing the cell’s resistance to drugs. So, in spite of the many contributions in the literature about miRNAs and HCC, there is a lack of in vivo studies on the issue of the response to systemic therapy.
The analysis of miRNAs in serum and other body fluids (i.e., urines) would be crucial for clinical management, since it would allow diagnosis and/or prognosis of HCC patients before surgery. However, the study of serum miRNAs is still a complicated issue, due to the high tumor/patient variability and the lack of a standard control among laboratories. miR-122 - the miRNA most represented quantitatively in the human liver - is the most promising in early HCC diagnosis, albeit all authors generally agree that miR-122 serum levels also increase in non-neoplastic liver diseases[114]. The first proposed serum panel for HCC detection was miR-122 associated with miR-21 and miR-223[115]. A recent meta-analysis by Ding et al[116] showed that difficulties exist also in the comparison of scientific results among centers: the “high-frequency expression miRNAs” best suited for HCC diagnosis from serum were miR-122, miR-21, and miR-199, and generally a panel of multiple serum miRNAs is advisable. For example, a panel composed by three serum miRNAs (miR-92-3p, miR-3126-5p and miR-107) together with serum alpha-fetoprotein (AFP) showed higher sensitivity and specificity in the early diagnosis of HCC compared to AFP alone[117]. Another study found that serum miR-939, miR-595, miR-519d, and miR-494 were able to differentiate cirrhotic patients with and without HCC better than AFP[118]. Other serum miRNAs are likely to be useful in the diagnosis of local or distant HCC recurrence after surgery, like miR-486-5p[119] and miR-34a[120].
The extreme heterogeneity of HCC, in both its morphological picture (as assessed by radiologists and pathologists) and in its clinical course and outcome, reflects the heterogeneity of its bio-molecular status. Several molecular pathways are involved in hepatocarcinogenesis, as well as in the regenerative-dysplastic-neoplastic progression observed in cirrhotic nodules. As a consequence, the up- or down-regulation of several miRNAs is involved. As evidenced by the present review, the most represented miRNA-regulated pathway in HCC is mTOR, but other pathways, such as as apoptosis, Wnt or MAPK, are also influenced by miRNA expression levels. Moreover, as shown in Table 1, some miRNAs involved in at least one of the main molecular pathways of hepatocarcinogenesis are likely to have a clinical significance (recurrence after surgery, response to systemic therapy).
The identification of specific tissue and serum miRNAs, able to predict the arising of HCC in cirrhosis, to predict HCC recurrence after surgery, or to predict the response to systemic therapy, might lead to a drastic improvement in the management of these patients. Anyhow, the clinical application of miRNAs has always been complicated, especially because of inter-laboratory variability, due to the choice of control to be used for normalization. A recent study of our group showed how the miRNA profile of HCCs changed using a pool of cirrhotic tissues or a pool of healthy livers as non-tumor controls[121]. Other authors suggested to employ stable miRNAs as controls for the study of the expression of other miRNAs[122]. In the light of the available data, it would be useful to elaborate, based on the most representative miRNA in in vivo model (e.g., rat, mouse), to better understand the possible role of these molecules as theragnostic markers in HCC. The issue is still open, and the standardization of miRNA analysis among laboratories is crucial for the development of a miRNA-based diagnosis of liver nodules, as well as of a miRNA-regulatory therapy.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Xu CF S- Editor: Gong ZM L- Editor: A E- Editor: Huang Y
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12135] [Cited by in RCA: 12991] [Article Influence: 1443.4] [Reference Citation Analysis (2)] |
| 2. | Loda M, Mucci LA, Mittelstadt ML, Hemelrijck MV, Cotter MB, Springerlink (Online Service). Pathology and Epidemiology of Cancer. XIV. 2017;. |
| 3. | Lin Z, Cai YJ, Chen RC, Chen BC, Zhao L, Xu SH, Wang XD, Song M, Wu JM, Wang YQ. A microRNA expression profile for vascular invasion can predict overall survival in hepatocellular carcinoma. Clin Chim Acta. 2017;469:171-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Pang TC, Lam VW. Surgical management of hepatocellular carcinoma. World J Hepatol. 2015;7:245-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 5. | Bosman FT, World Health Organization, International Agency for Research on Cancer. WHO classification of tumours of the digestive system. (4th). IARC, Lyon. 2010;. |
| 6. | Mazzaferro V, Regalia E, Doci R, Andreola S, Pulvirenti A, Bozzetti F, Montalto F, Ammatuna M, Morabito A, Gennari L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5110] [Cited by in RCA: 5313] [Article Influence: 183.2] [Reference Citation Analysis (0)] |
| 7. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6573] [Article Influence: 469.5] [Reference Citation Analysis (1)] |
| 8. | Kudo M. Systemic Therapy for Hepatocellular Carcinoma: 2017 Update. Oncology. 2017;93 Suppl 1:135-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 87] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 9. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7919] [Cited by in RCA: 8601] [Article Influence: 409.6] [Reference Citation Analysis (0)] |
| 10. | de la Chapelle A, Jazdzewski K. MicroRNAs in thyroid cancer. J Clin Endocrinol Metab. 2011;96:3326-3336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 11. | Wang W, Luo YP. MicroRNAs in breast cancer: oncogene and tumor suppressors with clinical potential. J Zhejiang Univ Sci B. 2015;16:18-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 128] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 12. | Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 113] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 13. | Inamura K, Ishikawa Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment. J Clin Med. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 138] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 14. | Ishiguro H, Kimura M, Takeyama H. Role of microRNAs in gastric cancer. World J Gastroenterol. 2014;20:5694-5699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 67] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 15. | Visani M, de Biase D, Marucci G, Cerasoli S, Nigrisoli E, Bacchi Reggiani ML, Albani F, Baruzzi A, Pession A; PERNO study group. Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I-III. Mol Oncol. 2014;8:417-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 16. | Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 278] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 17. | Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 966] [Cited by in RCA: 952] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 18. | Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589-3594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1542] [Cited by in RCA: 1729] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 19. | Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5794] [Cited by in RCA: 6609] [Article Influence: 508.4] [Reference Citation Analysis (1)] |
| 20. | Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2855] [Cited by in RCA: 3175] [Article Influence: 211.7] [Reference Citation Analysis (0)] |
| 21. | Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1994] [Cited by in RCA: 2110] [Article Influence: 111.1] [Reference Citation Analysis (0)] |
| 22. | Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 517] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 23. | Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972-1983, 1983.e1-1983.11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 591] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 24. | Jiang D, Cho W, Li Z, Xu X, Qu Y, Jiang Z, Guo L, Xu G. MiR-758-3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomed Pharmacother. 2017;96:535-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 25. | Yu Q, Xiang L, Yin L, Liu X, Yang D, Zhou J. Loss-of-function of miR-142 by hypermethylation promotes TGF-β-mediated tumour growth and metastasis in hepatocellular carcinoma. Cell Prolif. 2017;50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Zhou SJ, Liu FY, Zhang AH, Liang HF, Wang Y, Ma R, Jiang YH, Sun NF. MicroRNA-199b-5p attenuates TGF-β1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Br J Cancer. 2017;117:233-244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 27. | Tovar V, Alsinet C, Villanueva A, Hoshida Y, Chiang DY, Solé M, Thung S, Moyano S, Toffanin S, Mínguez B. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. J Hepatol. 2010;52:550-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 187] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 28. | Cheng H, Xue J, Yang S, Chen Y, Wang Y, Zhu Y, Wang X, Kuang D, Ruan Q, Duan Y. Co-targeting of IGF1R/mTOR pathway by miR-497 and miR-99a impairs hepatocellular carcinoma development. Oncotarget. 2017;8:47984-47997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Han X, Wang X, Zhao B, Chen G, Sheng Y, Wang W, Teng M. MicroRNA-187 inhibits tumor growth and metastasis via targeting of IGF-1R in hepatocellular carcinoma. Mol Med Rep. 2017;16:2241-2246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | Wang W, Zhang H, Tang M, Liu L, Zhou Z, Zhang S, Wang L. MicroRNA-592 targets IGF-1R to suppress cellular proliferation, migration and invasion in hepatocellular carcinoma. Oncol Lett. 2017;13:3522-3528. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Ma X, Zhuang B, Li W. MicroRNA2965p downregulated AKT2 to inhibit hepatocellular carcinoma cell proliferation, migration and invasion. Mol Med Rep. 2017;16:1565-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Mo Y, Lu Y, Wang P, Huang S, He L, Li D, Li F, Huang J, Lin X, Li X. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317690999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 33. | Li H, Zhang J, Lee MJ, Yu GR, Han X, Kim DG. OIP5, a target of miR-15b-5p, regulates hepatocellular carcinoma growth and metastasis through the AKT/mTORC1 and β-catenin signaling pathways. Oncotarget. 2017;8:18129-18144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 34. | Yu M, Xue H, Wang Y, Shen Q, Jiang Q, Zhang X, Li K, Jia M, Jia J, Xu J. miR-345 inhibits tumor metastasis and EMT by targeting IRF1-mediated mTOR/STAT3/AKT pathway in hepatocellular carcinoma. Int J Oncol. 2017;50:975-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 35. | Dong Z, Qi R, Guo X, Zhao X, Li Y, Zeng Z, Bai W, Chang X, Hao L, Chen Y. MiR-223 modulates hepatocellular carcinoma cell proliferation through promoting apoptosis via the Rab1-mediated mTOR activation. Biochem Biophys Res Commun. 2017;483:630-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 36. | Chang W, Zhang L, Xian Y, Yu Z. MicroRNA-33a promotes cell proliferation and inhibits apoptosis by targeting PPARα in human hepatocellular carcinoma. Exp Ther Med. 2017;13:2507-2514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 37. | Chen YL, Xu QP, Guo F, Guan WH. MicroRNA-302d downregulates TGFBR2 expression and promotes hepatocellular carcinoma growth and invasion. Exp Ther Med. 2017;13:681-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Zhuang L, Wang X, Wang Z, Ma X, Han B, Zou H, Wu Z, Dong S, Qu Z, Zang Y. MicroRNA-23b functions as an oncogene and activates AKT/GSK3β/β-catenin signaling by targeting ST7L in hepatocellular carcinoma. Cell Death Dis. 2017;8:e2804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 39. | Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17:248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 111] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 40. | Fu X, Wen H, Jing L, Yang Y, Wang W, Liang X, Nan K, Yao Y, Tian T. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 2017;108:620-631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 41. | Feng X, Jiang J, Shi S, Xie H, Zhou L, Zheng S. Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int J Oncol. 2016;49:2600-2610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 42. | Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429-22433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 969] [Cited by in RCA: 1030] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 43. | Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68-75. [PubMed] |
| 44. | MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4613] [Cited by in RCA: 4510] [Article Influence: 281.9] [Reference Citation Analysis (0)] |
| 45. | Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4 pii:a008052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 683] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 46. | Zeng G, Apte U, Cieply B, Singh S, Monga SP. siRNA-mediated beta-catenin knockdown in human hepatoma cells results in decreased growth and survival. Neoplasia. 2007;9:951-959. [PubMed] |
| 47. | Ashmawy AM, Elgeshy KM, Abdel Salam ET, Ghareeb M, Kobaisi MH, Amin HAA, Sharawy SK, Abdel Wahab AHA. Crosstalk between liver-related microRNAs and Wnt/β-catenin pathway in hepatocellular carcinoma patients. Arab J Gastroenterol. 2017;18:144-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 48. | Liu L, Cai X, Liu E, Tian X, Tian C. MicroRNA-18a promotes proliferation and metastasis in hepatocellular carcinoma via targeting KLF4. Oncotarget. 2017;8:68263-68269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 49. | Sarkar D, Fisher PB. AEG-1/MTDH/LYRIC: clinical significance. Adv Cancer Res. 2013;120:39-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 50. | Yan JJ, Chang Y, Zhang YN, Lin JS, He XX, Huang HJ. miR-195 inhibits cell proliferation via targeting AEG-1 in hepatocellular carcinoma. Oncol Lett. 2017;13:3118-3126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 51. | Lu C, Liao Z, Cai M, Zhang G. MicroRNA-320a downregulation mediates human liver cancer cell proliferation through the Wnt/β-catenin signaling pathway. Oncol Lett. 2017;13:573-578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 52. | Xie F, Yuan Y, Xie L, Ran P, Xiang X, Huang Q, Qi G, Guo X, Xiao C, Zheng S. miRNA-320a inhibits tumor proliferation and invasion by targeting c-Myc in human hepatocellular carcinoma. Onco Targets Ther. 2017;10:885-894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 53. | Jiang T, Li M, Li Q, Guo Z, Sun X, Zhang X, Liu Y, Yao W, Xiao P. MicroRNA-98-5p Inhibits Cell Proliferation and Induces Cell Apoptosis in Hepatocellular Carcinoma via Targeting IGF2BP1. Oncol Res. 2017;25:1117-1127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 54. | Chu Y, Fan W, Guo W, Zhang Y, Wang L, Guo L, Duan X, Wei J, Xu G. miR-1247-5p functions as a tumor suppressor in human hepatocellular carcinoma by targeting Wnt3. Oncol Rep. 2017;38:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 55. | Haddad BR, Gu L, Mirtti T, Dagvadorj A, Vogiatzi P, Hoang DT, Bajaj R, Leiby B, Ellsworth E, Blackmon S. STAT5A/B gene locus undergoes amplification during human prostate cancer progression. Am J Pathol. 2013;182:2264-2275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 56. | He B, You L, Uematsu K, Zang K, Xu Z, Lee AY, Costello JF, McCormick F, Jablons DM. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc Natl Acad Sci U S A. 2003;100:14133-14138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 323] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 57. | Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113:365-371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 343] [Cited by in RCA: 445] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 58. | Huang PS, Lin YH, Chi HC, Chen PY, Huang YH, Yeh CT, Wang CS, Lin KH. Thyroid hormone inhibits growth of hepatoma cells through induction of miR-214. Sci Rep. 2017;7:14868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 59. | Xie Y, Xu K, Dai B, Guo Z, Jiang T, Chen H, Qiu Y. The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene. 2006;25:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 97] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 60. | Mao J, Hu X, Pang P, Zhou B, Li D, Shan H. miR-30e acts as a tumor suppressor in hepatocellular carcinoma partly via JAK1/STAT3 pathway. Oncol Rep. 2017;38:393-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 61. | Yuan J, Ji H, Xiao F, Lin Z, Zhao X, Wang Z, Zhao J, Lu J. MicroRNA-340 inhibits the proliferation and invasion of hepatocellular carcinoma cells by targeting JAK1. Biochem Biophys Res Commun. 2017;483:578-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 62. | Yan X, Zhu Z, Xu S, Yang LN, Liao XH, Zheng M, Yang D, Wang J, Chen D, Wang L. MicroRNA-140-5p inhibits hepatocellular carcinoma by directly targeting the unique isomerase Pin1 to block multiple cancer-driving pathways. Sci Rep. 2017;7:45915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 63. | Pang R, Yuen J, Yuen MF, Lai CL, Lee TK, Man K, Poon RT, Fan ST, Wong CM, Ng IO. PIN1 overexpression and beta-catenin gene mutations are distinct oncogenic events in human hepatocellular carcinoma. Oncogene. 2004;23:4182-4186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 79] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 64. | Shinoda K, Kuboki S, Shimizu H, Ohtsuka M, Kato A, Yoshitomi H, Furukawa K, Miyazaki M. Pin1 facilitates NF-κB activation and promotes tumour progression in human hepatocellular carcinoma. Br J Cancer. 2015;113:1323-1331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 65. | Huang P, Qiu J, Li B, Hong J, Lu C, Wang L, Wang J, Hu Y, Jia W, Yuan Y. Role of Sox2 and Oct4 in predicting survival of hepatocellular carcinoma patients after hepatectomy. Clin Biochem. 2011;44:582-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 66. | Wen W, Han T, Chen C, Huang L, Sun W, Wang X, Chen SZ, Xiang DM, Tang L, Cao D. Cyclin G1 expands liver tumor-initiating cells by Sox2 induction via Akt/mTOR signaling. Mol Cancer Ther. 2013;12:1796-1804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 67. | Sun C, Sun L, Li Y, Kang X, Zhang S, Liu Y. Sox2 expression predicts poor survival of hepatocellular carcinoma patients and it promotes liver cancer cell invasion by activating Slug. Med Oncol. 2013;30:503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 68. | Zhang Y, Zhang D, Jiang J, Dong L. Loss of miR-638 promotes invasion and epithelial-mesenchymal transition by targeting SOX2 in hepatocellular carcinoma. Oncol Rep. 2017;37:323-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 69. | Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495-516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10243] [Cited by in RCA: 9604] [Article Influence: 533.6] [Reference Citation Analysis (0)] |
| 70. | Norbury CJ, Hickson ID. Cellular responses to DNA damage. Annu Rev Pharmacol Toxicol. 2001;41:367-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 381] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 71. | Wallace-Brodeur RR, Lowe SW. Clinical implications of p53 mutations. Cell Mol Life Sci. 1999;55:64-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 150] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 72. | Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485-495. [PubMed] |
| 73. | Fu XT, Shi YH, Zhou J, Peng YF, Liu WR, Shi GM, Gao Q, Wang XY, Song K, Fan J. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 2018;412:108-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 74. | Li M, Yang Y, Kuang Y, Gan X, Zeng W, Liu Y, Guan H. miR-365 induces hepatocellular carcinoma cell apoptosis through targeting Bcl-2. Exp Ther Med. 2017;13:2279-2285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 75. | Zhan L, Pan Y, Chen L, Chen Z, Zhang H, Sun C. MicroRNA-526a targets p21-activated kinase 7 to inhibit tumorigenesis in hepatocellular carcinoma. Mol Med Rep. 2017;16:837-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 76. | Shigematsu S, Fukuda S, Nakayama H, Inoue H, Hiasa Y, Onji M, Higashiyama S. ZNF689 suppresses apoptosis of hepatocellular carcinoma cells through the down-regulation of Bcl-2 family members. Exp Cell Res. 2011;317:1851-1859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 77. | Ge H, Zou D, Wang Y, Jiang H, Wang L. MicroRNA-377 Downregulates Bcl-xL and Increases Apoptosis in Hepatocellular Carcinoma Cells. Oncol Res. 2017;25:29-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 78. | Liu L, Lu L, Zheng A, Xie J, Xue Q, Wang F, Wang X, Zhou H, Tong X, Li Y. MiR-199a-5p and let-7c cooperatively inhibit migration and invasion by targeting MAP4K3 in hepatocellular carcinoma. Oncotarget. 2017;8:13666-13677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Zhu XM, Wu LJ, Xu J, Yang R, Wu FS. Let-7c microRNA expression and clinical significance in hepatocellular carcinoma. J Int Med Res. 2011;39:2323-2329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 80. | Lam D, Dickens D, Reid EB, Loh SH, Moisoi N, Martins LM. MAP4K3 modulates cell death via the post-transcriptional regulation of BH3-only proteins. Proc Natl Acad Sci U S A. 2009;106:11978-11983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 81. | Li M, Zhu Y, Zhang H, Li L, He P, Xia H, Zhang Y, Mao C. Delivery of inhibitor of growth 4 (ING4) gene significantly inhibits proliferation and invasion and promotes apoptosis of human osteosarcoma cells. Sci Rep. 2014;4:7380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 82. | Hu X, Feng Y, Sun L, Qu L, Sun C. Roles of microRNA-330 and Its Target Gene ING4 in the Development of Aggressive Phenotype in Hepatocellular Carcinoma Cells. Dig Dis Sci. 2017;62:715-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 83. | Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279-3290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1960] [Cited by in RCA: 2233] [Article Influence: 124.1] [Reference Citation Analysis (0)] |
| 84. | Sarris ME, Moulos P, Haroniti A, Giakountis A, Talianidis I. Smyd3 Is a Transcriptional Potentiator of Multiple Cancer-Promoting Genes and Required for Liver and Colon Cancer Development. Cancer Cell. 2016;29:354-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 85. | Mazur PK, Reynoird N, Khatri P, Jansen PW, Wilkinson AW, Liu S, Barbash O, Van Aller GS, Huddleston M, Dhanak D. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283-287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 267] [Cited by in RCA: 308] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 86. | Zhu W, Qian J, Ma L, Ma P, Yang F, Shu Y. MiR-346 suppresses cell proliferation through SMYD3 dependent approach in hepatocellular carcinoma. Oncotarget. 2017;8:65218-65229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 87. | Xue F, Yin J, Xu L, Wang B. MicroRNA-143 inhibits tumorigenesis in hepatocellular carcinoma by downregulating GATA6. Exp Ther Med. 2017;13:2667-2674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 88. | Kwei KA, Bashyam MD, Kao J, Ratheesh R, Reddy EC, Kim YH, Montgomery K, Giacomini CP, Choi YL, Chatterjee S. Genomic profiling identifies GATA6 as a candidate oncogene amplified in pancreatobiliary cancer. PLoS Genet. 2008;4:e1000081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 92] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 89. | Coppola N, de Stefano G, Panella M, Onorato L, Iodice V, Minichini C, Mosca N, Desiato L, Farella N, Starace M. Lowered expression of microRNA-125a-5p in human hepatocellular carcinoma and up-regulation of its oncogenic targets sirtuin-7, matrix metalloproteinase-11, and c-Raf. Oncotarget. 2017;8:25289-25299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 90. | Shi X, Wang X. The role of MTDH/AEG-1 in the progression of cancer. Int J Clin Exp Med. 2015;8:4795-4807. [PubMed] |
| 91. | Zhang M, Li M, Li N, Zhang Z, Liu N, Han X, Liu Q, Liao C. miR-217 suppresses proliferation, migration, and invasion promoting apoptosis via targeting MTDH in hepatocellular carcinoma. Oncol Rep. 2017;37:1772-1778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 92. | Wang J, Song W, Shen W, Yang X, Sun W, Qu S, Shang R, Ma B, Pu M, Tao K. MicroRNA-200a Suppresses Cell Invasion and Migration by Directly Targeting GAB1 in Hepatocellular Carcinoma. Oncol Res. 2017;25:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 93. | Sato F, Hatano E, Kitamura K, Myomoto A, Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S, Shimizu K. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One. 2011;6:e16435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 94. | Shimagaki T, Yoshizumi T, Harimoto N, Yoshio S, Naito Y, Yamamoto Y, Ochiya T, Yoshida Y, Kanto T, Maehara Y. MicroRNA-125b expression and intrahepatic metastasis are predictors for early recurrence after hepatocellular carcinoma resection. Hepatol Res. 2018;48:313-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 95. | Gao Y, Zhang SG, Wang ZH, Liao JC. Down-regulation of miR-342-3p in hepatocellular carcinoma tissues and its prognostic significance. Eur Rev Med Pharmacol Sci. 2017;21:2098-2102. [PubMed] |
| 96. | Zhao XQ, Liang B, Jiang K, Zhang HY. Down-regulation of miR-655-3p predicts worse clinical outcome in patients suffering from hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2017;21:748-752. [PubMed] |
| 97. | Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie RT, Yang HQ, Chang ZY, Sun R, Chai L. High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. 2017;8:11896-11905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 98. | Imura S, Yamada S, Saito YU, Iwahashi S, Arakawa Y, Ikemoto T, Morine Y, Utsunomiya T, Shimada M. miR-223 and Stathmin-1 Expression in Non-tumor Liver Tissue of Patients with Hepatocellular Carcinoma. Anticancer Res. 2017;37:5877-5883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 99. | Vasuri F, Fittipaldi S, De Pace V, Gramantieri L, Bertuzzo V, Cescon M, Pinna AD, Fiorentino M, D’Errico A, Ravaioli M. Tissue miRNA 483-3p expression predicts tumor recurrence after surgical resection in histologically advanced hepatocellular carcinomas. Oncotarget. 2018;9:17895-17905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 100. | Hung CL, Yen CS, Tsai HW, Su YC, Yen CJ. Upregulation of MicroRNA-19b predicts good prognosis in patients with hepatocellular carcinoma presenting with vascular invasion or multifocal disease. BMC Cancer. 2015;15:665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 101. | Chen F, Li XF, Fu DS, Huang JG, Yang SE. Clinical potential of miRNA-221 as a novel prognostic biomarker for hepatocellular carcinoma. Cancer Biomark. 2017;18:209-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 102. | von Felden J, Heim D, Schulze K, Krech T, Ewald F, Nashan B, Lohse AW, Wege H. High expression of micro RNA-135A in hepatocellular carcinoma is associated with recurrence within 12 months after resection. BMC Cancer. 2017;17:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 103. | Zhu HT, Dong QZ, Sheng YY, Wei JW, Wang G, Zhou HJ, Ren N, Jia HL, Ye QH, Qin LX. MicroRNA-29a-5p is a novel predictor for early recurrence of hepatitis B virus-related hepatocellular carcinoma after surgical resection. PLoS One. 2012;7:e52393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 104. | Barry CT, D’Souza M, McCall M, Safadjou S, Ryan C, Kashyap R, Marroquin C, Orloff M, Almudevar A, Godfrey TE. Micro RNA expression profiles as adjunctive data to assess the risk of hepatocellular carcinoma recurrence after liver transplantation. Am J Transplant. 2012;12:428-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 105. | Morita K, Shirabe K, Taketomi A, Soejima Y, Yoshizumi T, Uchiyama H, Ikegami T, Yamashita Y, Sugimachi K, Harimoto N. Relevance of microRNA-18a and microRNA-199a-5p to hepatocellular carcinoma recurrence after living donor liver transplantation. Liver Transpl. 2016;22:665-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 106. | Lu AQ, Lv B, Qiu F, Wang XY, Cao XH. Upregulation of miR-137 reverses sorafenib resistance and cancer-initiating cell phenotypes by degrading ANT2 in hepatocellular carcinoma. Oncol Rep. 2017;37:2071-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 107. | Potenza N, Mosca N, Zappavigna S, Castiello F, Panella M, Ferri C, Vanacore D, Giordano A, Stiuso P, Caraglia M. MicroRNA-125a-5p Is a Downstream Effector of Sorafenib in Its Antiproliferative Activity Toward Human Hepatocellular Carcinoma Cells. J Cell Physiol. 2017;232:1907-1913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 108. | Nishida N, Arizumi T, Hagiwara S, Ida H, Sakurai T, Kudo M. MicroRNAs for the Prediction of Early Response to Sorafenib Treatment in Human Hepatocellular Carcinoma. Liver Cancer. 2017;6:113-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (1)] |
| 109. | Shao P, Qu WK, Wang CY, Tian Y, Ye ML, Sun DG, Sui JD, Wang LM, Fan R, Gao ZM. MicroRNA-205-5p regulates the chemotherapeutic resistance of hepatocellular carcinoma cells by targeting PTEN/JNK/ANXA3 pathway. Am J Transl Res. 2017;9:4300-4307. [PubMed] |
| 110. | Yang X, Zang J, Pan X, Yin J, Xiang Q, Yu J, Gan R, Lei X. miR-503 inhibits proliferation making human hepatocellular carcinoma cells susceptible to 5fluorouracil by targeting EIF4E. Oncol Rep. 2017;37:563-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 111. | Meng W, Tai Y, Zhao H, Fu B, Zhang T, Liu W, Li H, Yang Y, Zhang Q, Feng Y. Downregulation of miR-33a-5p in Hepatocellular Carcinoma: A Possible Mechanism for Chemotherapy Resistance. Med Sci Monit. 2017;23:1295-1304. [PubMed] |
| 112. | Du Z, Niu S, Xu X, Xu Q. MicroRNA31-NDRG3 regulation axes are essential for hepatocellular carcinoma survival and drug resistance. Cancer Biomark. 2017;19:221-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 113. | Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, Wang F, Zhang CY, Zen K, Li L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 176] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 114. | Loosen SH, Schueller F, Trautwein C, Roy S, Roderburg C. Role of circulating microRNAs in liver diseases. World J Hepatol. 2017;9:586-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 115. | Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 450] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 116. | Ding Y, Yan JL, Fang AN, Zhou WF, Huang L. Circulating miRNAs as novel diagnostic biomarkers in hepatocellular carcinoma detection: a meta-analysis based on 24 articles. Oncotarget. 2017;8:66402-66413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 117. | Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma X, Wei Q, Han L. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore). 2017;96:e5642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 118. | Fornari F, Ferracin M, Trerè D, Milazzo M, Marinelli S, Galassi M, Venerandi L, Pollutri D, Patrizi C, Borghi A. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, Identify Cirrhotic Patients with HCC. PLoS One. 2015;10:e0141448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 116] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 119. | Wang L, Liu M, Zhu H, Rong W, Wu F, An S, Liu F, Feng L, Wu J, Xu N. Identification of recurrence-related serum microRNAs in hepatocellular carcinoma following hepatectomy. Cancer Biol Ther. 2015;16:1445-1452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 120. | Xiang ZL, Zhao XM, Zhang L, Yang P, Fan J, Tang ZY, Zeng ZC. MicroRNA-34a expression levels in serum and intratumoral tissue can predict bone metastasis in patients with hepatocellular carcinoma. Oncotarget. 2016;7:87246-87256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 121. | Fittipaldi S, Vasuri F, Bonora S, Degiovanni A, Santandrea G, Cucchetti A, Gramantieri L, Bolondi L, D’Errico A. miRNA Signature of Hepatocellular Carcinoma Vascularization: How the Controls Can Influence the Signature. Dig Dis Sci. 2017;62:2397-2407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 122. | Shen J, Wang Q, Gurvich I, Remotti H, Santella RM. Evaluating normalization approaches for the better identification of aberrant microRNAs associated with hepatocellular carcinoma. Hepatoma Res. 2016;2:305-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |