Published online Jun 28, 2018. doi: 10.3748/wjg.v24.i24.2537
Peer-review started: March 25, 2018
First decision: April 27, 2018
Revised: May 4, 2018
Accepted: June 9, 2018
Article in press: June 9, 2018
Published online: June 28, 2018
Processing time: 94 Days and 15.5 Hours
Pancreatic cancer (PaC) shows a clear tendency to increase in the next years and therefore represents an important health and social challenge. Currently, there is an important need to find biomarkers for PaC early detection because the existing ones are not useful for that purpose. Recent studies have indicated that there is a large window of time for PaC early detection, which opens the possibility to find early biomarkers that could greatly improve the dismal prognosis of this tumor. The present manuscript reviews the state of the art of the existing PaC biomarkers. It focuses on the anomalous glycosylation process and its role in PaC. Glycan structures of glycoconjugates such as glycoproteins are modified in tumors and these modifications can be detected in biological fluids of the cancer patients. Several studies have found serum glycoproteins with altered glycan chains in PaC patients, but they have not shown enough specificity for PaC. To find more specific cancer glycoproteins we propose to analyze the glycan moieties of a battery of glycoproteins that have been reported to increase in PaC tissues and that can also be found in serum. The combination of these new candidate glycoproteins with their aberrant glycosylation together with the existing biomarkers could result in a panel, which would expect to give better results as a new tool for early diagnosis of PaC and to monitor the disease.
Core tip: There is an urgent need to find new biomarkers for pancreatic cancer (PaC) diagnosis. The review focuses on the field of glycoproteomics and describes serum glycoproteins that have been identified up to date as potential biomarkers and their limitations basically due to the fact that they are neither pancreatic specific nor cancer specific. The review proposes new glycoprotein candidates that have been described to be overexpressed in PaC tissues and that are secreted into serum. The combination of the candidate protein levels and their glycan moieties, which could be altered in cancer, could improve their potential as PaC biomarkers.
- Citation: Llop E, Guerrero PE, Duran A, Barrabés S, Massaguer A, Ferri MJ, Albiol-Quer M, de Llorens R, Peracaula R. Glycoprotein biomarkers for the detection of pancreatic ductal adenocarcinoma. World J Gastroenterol 2018; 24(24): 2537-2554
- URL: https://www.wjgnet.com/1007-9327/full/v24/i24/2537.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i24.2537
Cancer still represents both a tremendous social and healthcare problem and its incidence is increasing in developed and developing countries. In pancreatic ductal adenocarcinoma (PaC), total cancer deaths are projected to increase dramatically to become the second leading cause of cancer-related deaths before 2030[1-4].
Nowadays, PaC presents the lowest five-year survival rate, being surgery the only possible curative option, but this is only possible for 20% of patients. The current therapeutic treatments for PaC are not very effective, extending the overall survival by only 8.5 mo as much[2].
PaC has a poor prognosis due to delayed diagnosis. In most cases, more than 90% of patients with PaC are diagnosed at stages III and IV[5]. Symptoms of PaC are often unnoticed as they are not very specific for the disease. The most common symptoms are abdominal unspecific pain, back pain, nausea, diarrhea and jaundice[6]. New-onset diabetes mellitus (DM) and acute exacerbation of DM can also be signs of PaC. Thus, in all these cases a detailed examination is needed[7].
It is particularly disturbing that there are no effective detection assay systems and treatment protocols for PaC, advances that have improved the management of other important carcinomas. It is therefore necessary an outstanding effort to study this tumor, to analyze its complexity, and to stablish efficient diagnostic systems and treatment options. Today there is a big improvement in the knowledge of the biology of PaC, but the disease still remains difficult to analyze, to detect and to treat[2]. PaC is a very aggressive tumor presenting a great desmoplastic area that creates a very dense stroma that prevents the arrival of therapeutic drugs. Genetically, PaC is a very complicated disease with at least 63 genetic alterations that promote resistance to the therapies[2,8,9].
Fortunately, a long period of time before the establishment of metastatic PaC has been indicated by Yachida et al[10]. Further, Poruk et al[11] have indicated that “a reasonable amount of time exits that allow for an early diagnosis of PaC” which indicates that it is possible to early detect the tumor. It is also worthy to mention that an incidental diagnosis of PaC is synonymous with an improved prognosis[12].
Early detection remains one of the most promising approaches to improve the long-term survival of cancer patients, and this may be achieved by efficient screening of biomarkers in biological fluids. The objective of a screening program is to detect cancer at a curable stage, before symptoms develop[13,14].
For many malignancies, biomarkers have become an established part of patient management, and are included in many clinical guidelines. As a consequence, the search for new and better biomarkers has become an integral component of contemporary cancer research. Biomarkers serve multiple purposes, as early detection, therapeutic monitoring, predicting responses, side-effects, surrogate endpoints, and they guide critical treatment decisions.
The term tumor marker (TM), or biomarker, traditionally has referred to molecules, mainly proteins, which are either directly produced by malignant cells, or are produced by other cells in response to certain malignant or other non-malignant conditions. Most clinically relevant biomarkers have been discovered serendipitously, or by trial and error.
Current tumor markers may be grouped into a variety of categories, including proteins, glycoproteins, onco-fetal antigens, hormones, receptors, genetic markers, RNA molecules and metabolites. Despite advances in laboratory technology and an enormous expansion in relevant literature (768000 papers indexed in PubMed in 2016 related to biomarkers), we are far from widespread clinical use of biomarkers. In fact, only a few dozen clinical relevant biomarkers useful for cancer exist nowadays (Table 1). The number of biomarkers receiving FDA approval has declined substantially over the last ten years to less than one protein biomarker per year. After almost two decades of intensive research using advanced genomics and proteomics technologies, only a handful of tumor markers have been translated into patient care. Of the proteins that could be differentially expressed in human cancers, only nine have been approved as tumor associated antigens[15]. All of these cancer biomarkers are glycosylated proteins[16-20]. Despite their clinical usefulness, they present limitations in terms of sensitivity and specificity as the case of carbohydrate antigen 19-9 (CA 19-9) for the diagnosis of PaC, as discussed later.
| Biomarker | Tumor class | Glycosylated protein |
| Alpha-feto protein (AFP) | Testicular, hepatocellular | Yes |
| Human chorionic gonadotropin | Testicular | Yes |
| CA 15-3 | Breast | Yes |
| CA 19-9 | Pancreatic | Yes |
| CA 27-29 | Breast | Yes |
| CA 125 | Ovarian | Yes |
| Carcinoembryonic antigen | Colon | Yes |
| Human epidermal growth factor receptor 2 (HER-2) | Breast | Yes |
| Prostate specific antigen (PSA) | Prostate | Yes |
| Thyroglobulin | Thyroid | Yes |
TMs are minimally invasive and convenient, and their associated costs are low. Single TMs were traditionally used but these have come under scrutiny due to their low sensitivity and specificity. Recent research has shown superior performance using a combination of multiple TMs as a panel that also incorporates other clinical factors as imaging systems[21,22]. In the past few years it has become increasingly clear that no single biomarker can be reliably used for cancer diagnosis.
Serum CA 19-9 was described after obtaining monoclonal antibodies against human colorectal cancer cells. It is a carbohydrate antigen called sialyl-Lewis a (SLea), which is embedded on cell surface molecules, as gangliosides and mucins[23]. CA 19-9 is routinely used as a biomarker of PaC, with an 82% of sensitivity and a 90% of specificity. But CA 19-9 is also elevated in other situations as pancreatitis, jaundice, hepatic and pancreatic cysts, and other cancers as colorectal and breast. CA 19-9 is not expressed in the 10%-20% of the Caucasian population due to a genetic deficiency of a fucosyltransferase. The marker was ineffective for general population screening and diagnosis, but CA 19-9 has been extensively studied in PaC, and it is continuously requested by clinicians, especially for monitoring. Recently, it has been shown that CA 19-9 is upregulated up to 2 years before PaC diagnosis[24], and it seems to be a good companion with other markers improving the diagnostic accuracy of PaC, as for instance, when used together with albumin and IGF, it allows the distinction between PaC and chronic pancreatitis (ChrP) with high sensitivity and specificity[25].
Detection of asymptomatic PaC has only been achieved incidentally by imaging techniques[11,26]. Significant progress has been made over the past two decades in imaging technology and at present, available techniques for pancreas imaging have a key role in PaC diagnosis as well as in cancer staging, assessment of the treatment response and detection of metastatic lesions. Pancreatic imaging tests include several modalities: Ultrasonography (US), computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), and endoscopic ultrasonography (EUS)[27-31].
Ultrasonography: Ultrasonography (US) may have some limitations in the early diagnosis of PaC. The entire pancreas is difficult to visualize clearly on ultrasonography especially for small tumors located in the head or tail[32].
Multidetector computed tomography: After the direct suspicion of PaC, the initial diagnosis and staging of PaC mainly relies on a multi-detector row CT[22]. Overall, CT is reported to have a sensitivity of 89%-97% for PaC diagnosis, although it is less effective in diagnosing small (< 2 cm) tumors with a sensitivity of 65%-75%[33,34].
Since CT has a wide anatomic coverage, it also shows a high performance for the evaluation of vascular structures invasion (100% sensitivity and 72% specificity), which is a key factor for predicting the tumor resectability[35,36]. CT is also a primary imaging modality for monitoring the response to treatment[34].
Magnetic resonance imaging: MRI can be useful in imaging for PaC in patients when EUS or CT findings are not diagnostic[37].
Diffusion-weighted MRI (DWI) helps to detect solid pancreatic neoplasms with extremely dense cellularity[38,39]. However, DWI may not be capable of definitively distinguishing between inflammatory/neoplastic lesions.
Endoscopic ultrasonography: CT of the pancreas can be complemented by Endoscopic ultrasonography (EUS), which is the most sensitive imaging method for the detection of malignant pancreatic lesions. EUS sensitivity has been reported to be higher than 95%[40,41].
EUS is particularly useful for identification of small tumors (< 3 cm diameter) that are not visualized by other imaging modalities (93% sensitivity)[42]. Besides providing important information for tumor detection and staging, EUS allows to obtain tissue samples for histological diagnosis using fine-needle aspiration (FNA) during the same examination. EUS-guided FNA has been shown to have a sensitivity and specificity for PaC of 85% and 98%, respectively[43,44]. In addition, EUS is equivalent to CT at determining the surgical resectability of PaC[45].
Positron emission tomography: PaC assessment can be improved using PET techniques. PET relies on the enhanced metabolism of glucose in cancer cells, which overexpress glucose transporter 1, leading to glucose accumulation in tumors. Fluorine 18-fluorodeoxyglucose (FDG), a glucose analogue, is normally used as radiotracer. The reported sensitivity and specificity of FDG-PET for the identification of PaC are low[46]. The main limitations of PET for PaC diagnosis are low-false negative results in hyperglycemia and false-positive results caused by a physiologic FDG uptake by normal tissues or inflammatory masses (pancreatitis)[47].
However, FDG-PET has been described to be more sensitive than CT for monitoring the response to treatment and for detecting tumor recurrence after resection[48,49]. One of the main advantages of PET is its wide anatomic coverage, which allows the detection of possible metastases in the entire body[35,50].
Although imaging techniques have provided significant advances in the diagnosis and follow up of PaC patients, they still have limitations in the early diagnose of small primary pancreatic tumors and metastases, which is crucial for the effectiveness of therapeutic interventions. Further improvement could be made with a combination of imaging techniques and biomarkers, since the presence of biomarkers in serum could be a very early finding that would increase the ability of imaging techniques to diagnose pancreatic carcinoma.
Currently, the only FDA-approved blood-based biomarker for PaC is CA 19-9 which is recommended for monitoring the disease or following up. Therefore, discovering new biomarkers to diagnose PaC, especially in early stages, is mandatory to improve PaC patients live expectancy.
In this context, apart from glycoproteomic strategies to identify novel PaC biomarkers based on altered glycosylation of particular proteins, which will be discussed in depth later on, real efforts have been made to identify new specific biomarkers for the early detection of PaC[12,51-54]. Harsha et al[55] have collected an extensive list of potential new protein PaC biomarkers (207 proteins over-expressed), but many candidates have not worked differentiating between benign and malignant situations. Proteomic analysis of exosomes and tissues of PaC patients have also been investigated[56-58].
Other approaches including molecules different than proteins are also under investigation. Detection of specific PaC mutations on cell-free circulating tumor DNA (ctDNA) or circulating tumor cells (CTC) has been explored[59-63]. Some commonly mutated genes in PaC are KRAS, TP53, SMAD4 and CDKN2A[64-66]; which allow classifying PaC into four different genetic subtypes that might have potential clinical relevance[65]. These mutations have also been detected in ctDNA or CTCs[62-63]. The study of Bettegowda et al[59] has shown that about 40% of patients with stages I to III of PaC have detectable ctDNA, a value that increases up to 90% for patients with stage IV tumor. On the other hand, Kinugasa et al[60] have reported the detection of mutated KRAS in ctDNA of 63% PaC patients (all stages), 20% of patients with ChrP and 5% of healthy controls (HC). Thus, these approaches could be interesting for developing an early detection PaC biomarker, although nowadays liquid biopsies still have some drawbacks, such as low diagnostic sensitivity or limitations on the detection techniques, and are not yet suitable to substitute tissue biopsies[61,67-68]. Other genetic changes such as epigenetic alterations[62,69-70] or non-coding RNAs expression[71-75] have also been detected in circulating free nucleic acids. Several miRNAs present better performance for PaC diagnosis than CA 19-9[76]. A recent study analyzing the blood levels of a panel of microRNAs previously associated with PaC did not show differential miRNA expression levels among patients with localized PaC and metastatic PaC; however, some miRNAs (miR-10b, miR-21-5p and miR-30c) present a consistent association between over-expression in plasma collected years before PaC diagnosis and risk, interesting for follow-up purposes[74]. The detection of miRNAs in circulating exosomes has also been explored and showed diagnostic potential for PaC, since the level of miR-10b, miR-20a, miR-21, miR-30c, miR-106b and miR-let7a (significantly different in PaC patients plasma compared to HC and ChrP patients) normalized after tumor resection[77]. Also, another especially interesting mRNA is miR-107, which might be useful not only as a diagnostic biomarker (AUC = 0.851, PaC patients compared with HC) but also as a target for PaC treatment[75].
Marker combinations: The combination of these novel biomarkers among them or with those already in use (mainly CA 19-9) might achieve the sensitivity and specificity required for early PaC diagnosis for high-risk population screening[25,78-81]. The recent study of Sefrioui et al[80] compares the diagnostic performance of CA 19-9, CTCs and KRAS mutational status detected on ctDNA in patients who also underwent endoscopic ultrasound-guided fine needle aspiration (EUS-FNA). The detection of CA 19-9 in combination with ctDNA and CTC analysis improved the sensitivity and specificity (78% and 91% respectively) in comparison with EUS-FNA alone (73% and 88% respectively). Moreover, unlike CA 19-9, the combined markers improved the diagnosis accuracy compared with CA 19-9 alone in cholestasis cases. Another study performed with plasma from PaC patients, ChrP patients and HC showed that the combination of relative abundances of miR-16, miR-196a and CA 19-9 levels (AUC = 0.979) improved in comparison with the analysis of CA 19-9 alone (AUC = 0.903) and the miRNA panel alone (AUC = 0.895). Similar results were obtained when comparing PaC patients against ChrP patients, and PaC patients against ChrP patients plus HC. The same study shows that the miR panel is more effective than CA 19-9 for early diagnosis of PaC[78]. Some combinatory analyses have been patented[82]. For example, the combination of plasma tissue factor pathway inhibitor (TFPI), tenascin C (TNC-FN III-C) and CA 19-9 levels, which presented a strong capacity in distinguishing all early-stage cancer from both HC and ChrP[81].
The value of all these findings is still uncertain since they lack proper robust clinical trials and the subsequent multicenter trials with larger cohorts to validate the data obtained. Moreover, it has to be taken into account that the methodologies used to discover novel PaC biomarkers would need to be simplified to apply them into clinic laboratories[82].
Anomalous glycosylation associated to cancer development was already described 50 years ago. During the last decades the field of glycobiology and our understanding of the central importance of glycans in biology have grown dramatically. Glycans are directly involved in virtually every disease affecting humankind, including inflammation, infectious diseases, cancer, diabetes and neurodegeneration[20,83-85].
Protein glycosylation is one of the most common protein post translational modifications. Approximately 50% of cell proteins are glycosylated. A number of N-linked glycoprotein changes have been identified in association with different diseases using glycoproteomic approaches. These glycoproteins have been shown to have aberrant glycosylation patterns in malignancy, but only their total protein levels are clinically monitored (Table 1).
Changes in glycosylation are believed to be a main feature in oncogenic transformation as glycans are known to be continuously involved in several processes, such as protein folding and clearance rates, cell signaling, angiogenesis, differentiation, cell growth, cell-matrix interactions, immune modulation, tumor cell dissociation, invasion, epithelial-mesenchymal transition (EMT), and metastasis.
Glycans expressed in several types of glycoconjugates are known to change during cancer genesis and progression[84,86]. Glycosylation changes that are commonly associated with cancer transformation include sialylation, fucosylation, increased GlcNAc-branching of N-glycans and over-expression of truncated mucin-type O-glycans[87]. These changes increase the structural glycan heterogeneity and alter the function of cells. These alterations become more marked as the tumor acquires a more aggressive phenotype. In a malignant cell, almost every glycoprotein will be affected by aberrant glycosylation, and cells usually synthesize an array of glycoforms of every protein.
Changes in glycosylation tend to be more pronounced than alterations in protein expression. Glycan modifications change rapidly and dramatically in response to a disease. This makes glycan alterations more reliable qualitative biomarkers in terms of the predictive value[88]. Some of the glycoproteins with altered glycosylation in cancer cells can be shed into the bloodstream so that their tumor associated glycoforms can potentially be specific cancer biomarkers[89,90]. Therefore, new tumor markers based on the detection of altered glycoforms of a particular glycoprotein, regardless of their levels, may have a much higher specificity for the detection of cancer than variable levels of the proteins themselves, which up to date have shown limited clinical utility. That is why glycoproteomics technologies are of particular interest to identify glycoproteins with altered glycoforms, caused by tumor transformation, as potential tumor markers.
The feasibility of this approach has been demonstrated for α-fetoprotein (AFP), which is a glycoprotein used as a marker for the diagnosis of hepatocellular carcinoma (HCC). AFP serum levels are increased in hepatocellular carcinoma, but also in other non-malignant chronic hepatic diseases such as chronic hepatitis or liver cirrhosis, which limits AFP specificity for HCC detection. The detection of an AFP fucosylated glycoform produced by HCC cells, which corresponded to an N-glycan with a disialylated biantennary structure with internal fucose (core-fucose), called AFP-L3, has been shown to improve the specificity of AFP as a marker of HCC. Thus, the ratio of fucosylated AFP to total AFP (AFP-L3%) has been approved by the FDA as a biomarker for the risk assessment of patients with chronic liver disease for the development of HCC[91,92].
In the same context, we have described changes in glycosylation at the level of core fucose and the type of sialic acid linkage of the prostate-specific antigen (PSA) in prostate cancer[93,94]. The increase of α2,3-sialylated PSA glycoforms and the decrease of core fucosylated PSA glycoforms have been reported in cohorts of patient sera, and have proven useful in differentiating high-risk or aggressive prostate cancer from indolent prostate cancer and benign prostatic hyperplasia[95,96], which is not possible to achieve with the current PSA tests. The fact that PSA is a specific protein of the prostate facilitates that these glycosylation changes on PSA are specific to the prostate tumor, and their detection can be transferred into the clinic, in a similar way to that described for AFP.
Identification and detection of abnormal glycosylated proteins in bodily fluids of PaC patients may allow the discovery of new markers for early diagnosis. A large number of proteomic studies in PaC have been reported. Also, many glycomic studies have revealed an aberrant N-glycosylation profile on human pancreatic carcinomas, and human PaC cells show large variation in their N-glycosylation status[53,97-100].
Zhao et al[101] observed increased branching of N-linked oligosaccharides, as well as an increasing in protein fucosylation and sialylation in the sera of PaC patients. The increase in Lewis and blood group glycans is a near universal feature of PaC[102,103]. Truncated O-linked glycosylation resulting in the Tn and sialyl-Tn antigens occurs in almost all epithelial cancers, and in PaC in particular[104].
In this review we will describe the scientific advances and the forthcoming challenges to identify glycoproteins with altered glycosylation in PaC that could be useful as biomarkers, either alone or in combination with other biomarkers.
Proteomic studies have led to identification of panels of proteins with potential as PaC biomarkers. However, in recent years, glycoproteomic strategies in biomarker discovery have become more important than just proteomics because they combine both the protein expression levels and glycan changes associated with PaC, thus increasing the protein potential for cancer diagnosing[105].
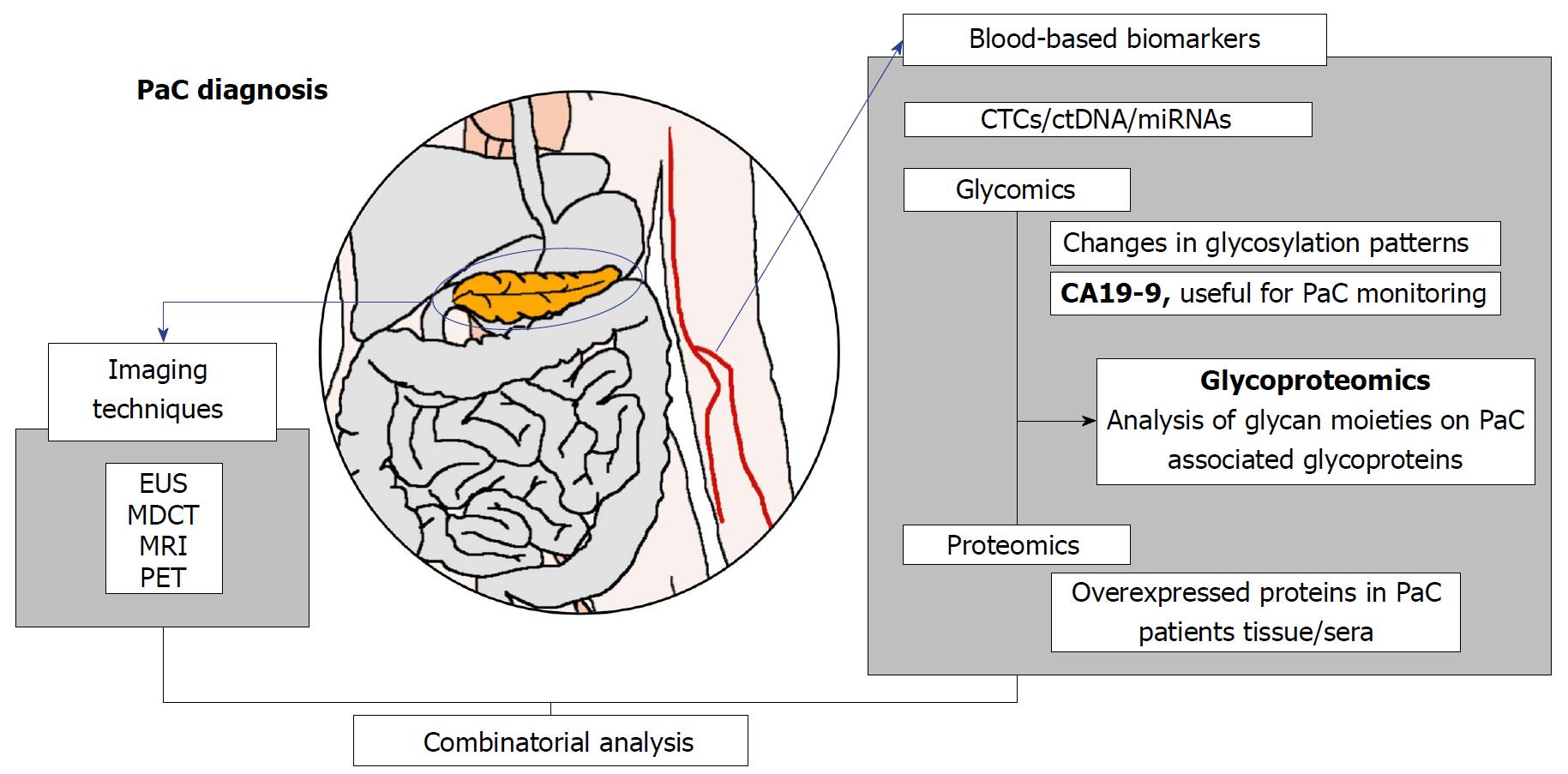
Although it is well-known that glycosylation plays an important role in epithelial cancers, the glycoproteome of the human pancreas and the aberrant glycosylation associated to PaC has not been deeply addressed. It should be noted that analysis of the glycoproteome is much more complex than the proteome because contrary to proteins, in which the amino acid sequences are unique and coded in the genome, no template exists for glycans. Glycosylation comprises a big heterogeneity of oligosaccharide residues in different sequences and linkages giving rise to a high amount of existing structures. In the present review, we focus on the strategies that have been proposed for the detection and/or quantification of glycobiomarkers in biological fluids and tissues from PaC patients. The summary of the approaches and/or glycoproteins proposed in this article intended to provide glycoproteins candidates with tumor associated glycans that should be explored for the development of a highly PaC sensitive and specific test (Figure 1).
The gold standard biomarker for diagnosing cancer should be ideally identified in easily accessible biological fluids, being for this reason, plasma and sera the samples most analyzed. Several plasma glycoproteins display altered N-glycan patterns in tumors. This was shown by N-glycan analysis of whole serum proteins (total serum N-glycome) from several cancer types[106] and of individual proteins shed or secreted from cancer tissues into the blood[89]. Some of these serum glycoproteins may also present glycan alterations in chronic inflammatory diseases. For this reason, finding a cancer specific glycoform in a serum glycoprotein, which was not present in benign diseases or in healthy individuals, would improve pancreatic cancer diagnosis.
In the last decade, several studies have identified serum glycoproteins with altered glycosylation in PaC patients using different glycoproteomic approaches. Most authors have usually taken the serum as a starting point and have used either lectins or antibodies against specific glycan structures, or mass-spectrometry (MS) techniques to identify the differential glycosylation of serum glycoproteins in PaC patients (summarized in Table 2). However, candidate biomarkers are expected to exist in a very low concentration and serum samples are very abundant in others glycoproteins like immunoglobulins and other high-abundance proteins (acute-phase proteins) that can be removed before the analysis with multiple available commercial options (Table 2).
| Techniques | Serum cohort (n) | Serum depletion (Yes/No) | Glycosylation change | Main glycoproteins identified | Ref. |
| ConA chromatography. Tryptic digestion. N-glycan MS analysis of the N-glycopeptides | 2 PaC | No | Increased branching, fucosylation and sialylation | [101] | |
| ConA and WGA chromatography. Glycoprotein separation and array with lectins (SNA, PNA, MAL, AAL). | 6 PaC 8 ChrP 10 HC | Yes (IgY 12) (albumin, IgG and major acute-phase proteins) | Increased fucosylation and sialylation | Hemopexin Serum amyloid P component Antithrombin-III Haptoglobin (HPT)-related protein β-2-glycoprotein 1 | [110] |
| Antibody microarray to α1-β-glycoprotein, amyloid P and antithrombin followed by lectin detection (SNA). MS glycoprotein identification. | 22 PaC (III/IV) 35 ChrP 89 HC 37 diabetic | Increase affinity to SNA (α2,6-sialic acid) by 69% in PaC patients | α1-β-glycoprotein. ROC curve (cancer vs non cancer samples), 100% sensitivity 98% specificity. AUC = 0.998 | [111] | |
| Bead-based antibody-lectin (SNA, Con A) multiplex assay-for determining SNA and Con A reactivity of α1-β-glycoprotein, and amyloid P component. | 20 PaC (III/IV) 20 ChrP 20 HC | SNA affinity (α2,6-sialic acid) | α2,6-sialic glycoforms of α1-β-glycoprotein Differentiation of ChrP vs PaC (P = 0.035) | [112] | |
| SNA affinity chromatography to enrich sialylated glycopeptides and compared their relative abundance by ultra performance LC-MS | 10 PaC (II-III) 5 Acute Pancreatitis 16 HC | Yes (albumin depleted) | SNA (α2,6-sialic acid) | Sialylated glycopeptides of HPT, α-1-antitrypsin (A1AT), transferrin, ceruloplasmin, α1-acid-glycoprotein (AGP), fetuin A and Igs. Change in acute pancreatitis and PaC | [97] |
| 2DE followed by N-glycan sequencing | 9 PaC (I-IV) 3 ChrP 5HC | SLex Fucosylation | Increase in SLex on AGP, HPT and transferrin in advanced PaC and ChrP Increase in core fucosylation of HPT and AGP in PaC vs ChrP and HC | [113] | |
| Electrophoresis (1DE) followed by WB with anti SLex. Immunoprecipitation of ceruloplasmin and SLex detection | 20 PaC (IIa-IV) 14 ChrP 13 HC | Yes (IgY 12) (albumin, IgG and major acute-phase proteins | SLex | Ceruloplasmin Tendency to an increase of SLex on ceruloplasmin in PaC vs HC and ChrP | [114] |
| Lectin (AAL)-antibody ELISA | 72 PaC 22 HC 63 pancreatitis | AAL (fucosylation). | Increase of fucosylated HPT in advanced PaC | [115] | |
| AGP purification MS analysis of AGP N-glycans and AAL ELISA | 19 PaC (I-IV) 6 ChrP 6 HC | α1,3 fucosylation | Increase of fucosylated AGP in advanced PaC | [116-118] | |
| N-glycan sequencing of human serum ribonuclease (RNase 1). | 2 PaC 2 HC | Core fucosylation | Increase of core fucosylation in RNase 1 in PaC | [120] | |
| ELISA to measure N-glycosylation Asn-88 site occupancy of serum RNase 1 | 91 PaC 60 HC | Asn-88 N-glycosylation | Increase in N-glycosylated Asn-88 of RNase 1 (normalized to RNase 1) in PaC. | [121] | |
| AAL to enrich fucosylated glycoproteins LC-MS/MS analyses ELISA/lectin ELISAs | 20 IPMN 10 MCN 37 PaC (I-IV) 30 HC 30 ChrP 22 OJ 30 Type II DM | IgY-14 LC10 columns | Fucosylation (AAL) | ACT trombospondin-1 HPT High diagnostic potential combined with CA 19-9 | [119] |
| nanoLC-MS/MS analysis of iTRAQ labelled glycopeptides. | 13 HC 13 ChrP 13 PaC 1 Std | IgY-14 LC10 column | Core-fucosylation | One core fucosylated peptide from ACT different between groups | [108] |
| PHA-L lectin to enrich complex N-glycoproteins 2D nanoLC-MS/MS analyses Western Blot with biotinylated PHA-L nanoLC-MS/MS of tryptic digested gel bands that corresponded to specific lectin interactions on Western blot | 26 HC (include ChrP + pseudo cysts) 76 PaC | Albumin/IgG depletion | Increased fucosylation N211 Novel glycosylation site N64 New N-glycosylated side at N2336 in PaC N-glycosylation N877 | HPT Leukemia inhibitory factor receptor LIFR Centrosome-associate protein 350 CE350 Vacuolar protein sorting-associated protein 13A VP13A | [122] |
| 2D-LC-MS/MS | 231 serum women samples pooled in groups: time-to-diagnosis | Yes | N-glycosylation occupancy | A1AT HPT AGP | [123] |
| Lectin (CCL2)-antibody ELISA | 109 PaC 91 control (plasma) | No | CCL2 (3’ fucosylation) | MUC5AC | [99] |
| Antibody lectin sandwitch array | 156 PaC (I-IV) 160 control (plasma) | No | SLea related | MUC5AC MUC16 | [127] |
| Antibody microarray capture of proteins. Glycan analysis with lectins (AAL, WGA) and CA 19-9 | 23 PaC (I-IV) 23 HC | No | CA 19-9 | MUC1 CEA | [128] |
| Antibody lectin sandwich array | 23 PaC (I-IV) 23 HC | No | SLea(CA 19-9) | MUC1 MUC5AC | [129] |
| Antibody array | 285 PaC (I-IV) 102 ChrP 144 HC (serum & plasma) | No | CA 19-9 | MUC1 MUC5AC MUC16 | [130] |
Although the molecular mechanisms underlying PaC-associated glycosylation events have not been perfectly understood, the most common glycan methodologies developed are focused on the analysis of alterations in branching, fucosylation[107,108] and sialylation[97]. It has also been reported that specific N-glycosylation occupancy in certain glycoproteins can be significantly altered[109].
Using glycoprotein microarrays with multi-lectin detection techniques, an increase in both fucosylation and sialylation of some serum glycoproteins was described for PaC patients compared to HC and pancreatitis patients[110,111]. Serum depletion to remove the most abundant proteins (albumin, IgGs and acute-phase proteins) to identify specific PaC glycoproteins with changes in glycosylation in PaC patients did not proportionate any pancreatic specific glycoprotein. However, several acute-phase proteins liver-derived, such as α1-β-glycoprotein, showed an increase in the affinity to Sambucus nigra agglutinin (SNA) lectin, which binds α2,6-sialic acid, in most of the PaC serum samples, which were stage III/IV, and allowed to discriminate them from ChrP patients[111,112].
More recently, Kontro et al[97] analyzed N-glycopeptides from albumin-depleted serum of PaC, ChrP, and HC. They showed significant changes in the concentration of the α2,6 sialylated N-glycopeptides in both PaC and ChrP. The glycopeptides corresponded to acute-phase proteins [haptoglobin (HPT), α-1-antitrypsin (A1AT), transferrin, ceruloplasmin, α-1-acid-glycoprotein (AGP), fetuin A] and immunoglobulins.
Using electrophoresis and western blotting of albumin and IgG-depleted serum samples, we identified serum glycoproteins with an increased sialyl-Lewis × (SLex) level in advanced PaC. However, the glycoproteins identified: AGP, HPT and transferrin, which are acute-phase proteins, also showed a SLex increase in ChrP patients, suggesting that the increase in SLex was related to inflammation rather than cancer itself[113]. In the search for more specific PaC glycoproteins with an increased SLex level, further serum depletion that included the twelve most abundant serum proteins was performed in order to remove the previously described acute-phase proteins. Other glycoproteins with SLex increase were identified, but turned out to be acute-phase proteins. Among them, ceruloplasmin was further analyzed in a bigger cohort of PaC serum samples. The ratio of SLex on ceruloplasmin related to ceruloplasmin levels showed an increased tendency in the sera from PaC compared to HC and ChrP patients[114].
Looking for other potential cancer associated glycoforms, our group performed the N-glycan sequencing of some major serum acute-phase proteins. An increase in core fucosylation was found in HPT and AGP in PaC patients compared to the ChrP and HC groups[113]. Other authors also analyzed HPT fucosylation in a larger cohort of patients. Although the increase of fucosylated HPT did not show significant differences in PaC vs pancreatitis patients, its combination with CA 19-9 gave higher diagnostic potential compared to CA 19-9 alone[115].
Recently, we have addressed the analysis of AGP fucosylation using several complementary analytical methodologies, including lectin and MS analyses, and have shown an increase in AGP α1,3-fucose glycoforms in advanced PaC compared to ChrP and HC patients[116-118].
The expression of fucosylated glycoproteins in PaC depleted serum samples was analyzed using Aleuria aurantia lectin (AAL)[119]. α1-Antichymotrypsin (ACT), trombospondin-1 and HPT were proposed as the best fucosylated candidates and were further validated with ELISAs or lectin ELISAs. Their combination with CA 19-9 resulted to be very promising for distinguishing PaC from other conditions with or without obstructive jaundice, indicating a high diagnostic potential[119].
The study of N-linked core fucosylated glycopeptides with potential use in PaC diagnosis has also been addressed performing nanoLC-MS/MS analysis of iTRAQ labelled glycopeptides. One core fucosylated peptide, belonging again to an acute phase glycoprotein, ACT, presented significant differences between healthy, ChrP and PaC groups[108].
In a previous study, our group addressed the analysis of serum ribonuclease 1 (RNase 1), which is an acinar pancreatic protein found in serum. However, this protein is also secreted by endothelial cells into serum. Serum pancreatic RNase 1 was found to be much more core fucosylated in PaC than in HC[120], which suggests that the quantification of core fucosylation in some PaC serum glycoproteins might be useful as a PaC biomarker. Nakata et al[121] showed that N-glycosylation at Asn-88 in serum RNase 1 was significantly increased in patients with PaC and proposed the serum RNase 1 N-glycosylation occupancy at Asn-88 site as a novel diagnostic marker for PaC.
Besides fucosylation, other authors focused on N-glycan branching of serum glycoproteins, which is one of the most common altered glycosylation features found in most types of cancers. After albumin/IgG depletion, Drabik et al[122] performed a glycoprotein enrichment using Phaseolus vulgaris leucoagglutinin (PHA-L). Then, using two-dimensional capillary chromatography combined with tandem mass spectrometry, they found four glycoproteins (LIFR, CE350, VP13A and HPT) that carried an altered N-glycan structure in PaC compared to HC. Recently, Krishnan et al[123] carried out a pilot proteomic study to evaluate changes in serum N-glycoproteins that could be potential biomarkers prior to clinical presentation of PaC being inflammatory response, coagulation and immune-related proteins the most promising ones. One of the top-scoring glycoproteins identified, A1AT, was quantified by ELISA. After analyzing 120 control and 92 PaC cases they concluded that whilst A1AT levels performed reasonably well for discriminating prediagnosis cancer cases, it is unlikely to be specific for detecting PaC.
Another group of highly glycosylated proteins with high potential as novel clinical PaC biomarkers are mucins. In this regard, altered glycosylation pattern of glycoproteins belonging to members of mucin (MUC) families have also been associated with PaC in several studies using PaC tissues as well as serum samples (Table 2). MUC4 altered glycosylation was identified in PaC by immunohistochemistry, as only one out of two antibodies directed towards different MUC4-Tn glycoforms showed staining in PaC tissues[104]. These authors also identified changes in MUC1 glycosylation, as the glycoform MUC1-T observed in normal pancreas became MUC1-STn in neoplastic stages. Recently, the study of immunoprecipitated MUC1 glycoforms using lectins has exposed its potential to discriminate between PaC and cholangiocarcinoma patients[124]. As mucins can be cleaved from cells and released to the bloodstream, all the specific glycoforms detected in tissues could be potentially detected in serum to be exploited as biomarkers. In addition, in cyst fluid samples, MUC5AC specific glycosylation detected with the Wheat germ agglutinin (WGA) lectin was reported to discriminate malignant from benign cases in pre-cancerous lesions as pancreatic cysts[125]. We also reported recently the increased expression of MUC1-SLex and MUC5AC-SLex in PaC tissues compared to benign controls[126].
Some of these mucin glycoforms described as PaC specific in tissues were also analyzed in sera (see Table 2). Although most of the well-established serological biomarkers are based on immunological methods to detect mucin glycoproteins such as CA125 assay detecting MUC16 in ovarian cancer or CA15-3 assay detecting MUC1 in breast cancer, the exact structural epitope recognized is unknown. Moreover, the structural characterization of O-glycans in mucins have been overlooked due to the analytical limitations (high molecular weight (MW) proteins with high number of glycosylation sites, high glycan heterogeneity in each individual site and lack of enzymes able to release O-glycans).
Regarding MUC5AC, increased levels of specific glycoforms carrying 3’ fucose (for instance SLex or Ley) were observed in serum from PaC patients compared to benign controls[99]. Further, a panel of serum glycans and different MUC5AC glycoforms (including SLea and SLex) showed improved accuracy over the standard CA 19-9 assay for detecting PaC[127].
Chen et al[128], using antibody microarrays to capture multiple serum glycoproteins followed by detection with lectins or glycan-binding antibodies, described cancer-associated glycan alterations on MUC1 and CEA in serum of PaC patients. Using antibody-lectin sandwich arrays, MUC1 and MUC5AC were shown to carry distinct glycan alterations in PaC patients, in particular MUC1 carried the CA 19-9 antigen (SLea) in a majority of PaC patients compared to HC[129]. Using antibody arrays[130], they noticed that the detection of CA 19-9 on individual carriers, like MUC5AC or MUC16, did not exceed the performance of the CA 19-9 assay. Notwithstanding, these analyses could discriminate between PaC and ChrP patients in low CA 19-9 individuals, so the combined analysis of CA 19-9 plus its detection on specific mucins could yield improved sensitivity for PaC detection.
The studies above reported (summarized in Table 2) consisted on preclinical exploratory works to compare tumor and non-tumor serum samples, to generate potential biomarker candidates for PaC detection. Using serum or depleted serum as starting material to find glycoproteins with altered glycosylation in PaC patients has resulted mainly in the identification of acute-phase glycoproteins mostly liver derived, as well as mucins such as MUC1 and MUC5AC. The glycosylation analysis of some of these reported individual glycoproteins, mainly focused on changes in sialic acid, fucose, or SLex/a, in PaC patients, showed usually significant changes in advanced PaC, but not in low PaC stages, and in many cases in inflammatory conditions, which limits their use as biomarkers of PaC. The high levels in serum of these acute-phase proteins, which are not tumor specific, have hindered the identification of minor proteins with altered glycosylation from a pancreatic tumor origin that could be more PaC specific, which should be one of the challenges of next investigations.
Some of the best and most widely recognized cancer bio-markers are highly tissue-specific, such as PSA, hCG and AFP. Although proteins with such characteristics are quite rare, we consider that pancreatic tissues are the ideal biological sample for studying the tumor-specific glycoproteins. Once the pancreatic tissue-specific glycoproteins or glycoforms can be found, their presence in serum could be explored.
Although the high number of biomarker studies reported in the last years, no valuable marker for this lethal cancer has been translated into clinics. The detection of the tumor associated glycans on selected glycoproteins, overexpressed or neo expressed specifically in PaC tissues, could improve the biomarker performance of the glycoprotein.
Although the search of potential biomarkers in PaC cultured cells is a good starting sample, PaC stromal components represent up to 90% of the pancreatic tumor volume, which implies that cell line models are not completely representative of the physiological environment. For that, proteomic analysis of tumor tissues is proposed as starting point to provide glycoprotein candidates that could be secreted or shed into blood for further biomarker analysis in serum or plasma. In this review we have focused on those glycoproteins that have been described in the literature to be differentially expressed and/or oversecreted in PaC cells, tissues, or pancreatic juice and that can also be found in plasma or serum.
The main protein carriers of CA 19-9 described are MUC1, MUC5AC and MUC16 and as we have mentioned previously, the study of their altered glycosylation is of interest for improving the performance of the CA 19-9 test. However, the analysis of these high MW glycoproteins (MW over 200 KDa with a glycan component that contributes to more than 50% of the MW) in serum presents a big challenge. Despite being one of the main families of proteins with potential as cancer biomarkers, in this study we will concentrate on other non-mucin glycoproteins that may be analyzed easily.
Among the large number of proteins described in different proteomic approaches, we have focused on glycoproteins different from acute-phase proteins because these last ones are usually increased in inflammation conditions as discussed previously. In our selection, the proteins associated to inflammatory processes have also been discarded because their abnormal expression may be associated with more than one type of cancer (lack of specificity). The most promising glycoprotein candidates are listed in Table 3 and may be analyzed in detail to determine whether a specific glycoform could be PaC associated and be used alone or together with CA 19-9 for the detection of early-stage PaC.
| Glycoprotein | Glycosylation sites | Reported glycosylation | Serum auto-antibodies | Ref. |
| Mesothelin (cleaved form) | 3 N-glycosylation sites | Reduction of MW after PNGase F digestion of A431 cancer cells | Yes | [133-144] |
| IGFBP-3 | 3 N-glycosylation sites O-glycosylation | Increase of N-glycosylation levels in tumor tissues Increase of biantennary complex type N-glycans having more mannose, fucose, bisecting GlcNAc and terminal sialic acid in breast cancer serum | Not described | [109,145] |
| IGFBP-2 | O-glycosylation | Not described | Not described | [141,146] |
| REG 1A REG 1B REG 3A REG 4 | O-glycosylation O-glycosylation 1 potential N-glycosylation site 1 N-glycosylation site | Increase protein glycoform diversity in pancreatic ductal fluid of PaC by western blot | Not described | [147-151] |
| TIMP-1 | 2 N-glycosylation sites | Core fucosylated, biantennary N-glycans with Gal or GalNAc in HEK293. Some of the glycans are sialylated, and many have outer arm fucosylation. Aberrant N-glycosylation in colon cancer | Not described | [152-156] |
| HER-2 | 7 N-glycosylation sites O-glycosylation | Altered N-glycosylation in breast cancer cells. Altered O-glycans (Tn and T) in PaC cells | Yes | [157-162] |
It has been described that the immune system reacts to developing tumors and generates auto-antibodies against tumor-associated antigens. These serum auto-antibodies can be detected in early stages and have recently emerged as early stage bio-markers for different types of cancer, including PaC. Possibly auto-antibodies signatures might therefore be best used as a screening tool to detect the presence of cancer in general, to be followed by more specific diagnostic measures in case of positive results[131].
Cancer patients produce auto-antibodies to proteins that are either mutated, misfolded, over-expressed, or to proteins that show altered post-translational modifications like glycosylation. The mechanisms of auto-antibody production are not fully clear. Secretion of abnormal glycoforms into blood induces the immune system to produce antibodies which results in the amplification of an early malignant alteration. Therefore, the detection of these antibodies indicates that the affected tissue shed these glycoproteins in the blood and may allow the diagnosis in an early stage of the disease, when there are no clinical signs yet, and the amount of anomalous glycan antigens is still very low. Some studies have evaluated serological auto-antibodies in PaC and the combination of four auto-antibodies including anti-mesothelin showed a reasonable diagnostic performance[132]. The finding of serum auto-antibodies recognizing some of the proposed glycoprotein candidates reinforce them as potential PaC biomarkers (Table 3).
Mesothelin, one of the potential candidates, is highly expressed in some common epithelial cancers. Its expression by immunohistochemistry has been reported in approximately 100% of malignant mesotheliomas[133], and in other cancers such as ovarian[134] and lung adenocarcinomas[135]. Regarding PaC, Zheng et al[136], reported mesothelin expression by western blot and RT-PCR assay in human PaC cell lines, and Argani et al[137], reported its expression in the majority of the PaC tissues analyzed by immunohistochemistry. Interestingly, mesothelin is present on normal mesothelial cells, peritoneum and pericardium, but it is not expressed either in normal pancreatic tissues or in ChrP[138]. Bournet et al[139] also demonstrated the overexpression of the mesothelin gene in biopsies of PaC samples when compared to ChrP. Although mesothelin is attached to the cell membrane, it can be shed like many other cell membrane proteins. Some authors have identified an increase in mesothelin serum levels when compared with ChrP and HC samples[140,141]. However, this protein has been also detected in sera from patients with ovarian carcinoma[142] and malignant mesothelioma[143]. Mature mesothelin presents three potential glycosylation sites and although mesothelin glycosylation has not been analyzed yet, Onda et al[144] observed a change in the SDS-PAGE mobility of mesothelin from cell extracts after being treated with PNGase F.
Some members of the insulin-like growth factor binding protein (IGFBP) family have also been proposed as potential candidates. It is known that six IGF-binding proteins interact with IGFs and modulate their tissue distribution and access to cell receptors. Serum levels of IGFBP-3 are associated with several carcinomas. IGFBP-3 incorporates altered glycan chains in breast cancer (containing increased amounts of bi-antennary chains, fucose and sialic acid residues)[145], as well as in hepatocellular carcinoma and in PaC. In this regard, Pan et al[109], using a quantitative glycoproteomics approach with tissue proteins labelled with stable isotope and N-glycopeptide enrichment with hydrazide beads, observed that IGFBP-3, among other proteins, had elevated N-glycosylation level in PaC compared with healthy pancreas.
Chen et al[52] performed a quantitative proteomic approach in pancreatic juice and identifyed IGFBP-2 as a novel PaC target. The IGFBP-2 overexpression was further validated by western blotting in pancreatic juice and pancreatic tissues compared to normal and ChrP samples. IGFBP-2 has also been studied as a serum PaC biomarker in combination to mesothelin[141]. In particular, this secreted glycoprotein does not contain any N-glycosylation site but the presence of O-glycosylation has been reported[146].
Searching for a more organ-specific glycoprotein we included lithostathine, also known as regenerating protein (REG) in the biomarkers’ list. It is a glycosylated protein which is normally found in the exocrine pancreas and its glycan chains have already been characterized[147,148]. It is the major non-enzymatic component of pancreatic juice, and its presumed role is to prevent stone formation in pancreatic ducts. It is expressed in pancreatic ductal carcinoma, as well as in colon cancer[149]. REG1 is only present in acinar pancreatic cells and malignant ductal cells and is not expressed in normal ducts or islets cells. REG1 is upregulated in PaC working as a growth factor, and is increased in serum and urine levels in PaC patients[150,151]. REG1 presents an altered glycosylation pattern in PaC ductal fluid, so its potential as a glycobiomarker should be considered in future investigations[148].
Mammalian tissue inhibitors of metalloproteinases (MMPs) are also considered biomarker candidates. They are endogenous secreted proteins that inhibit all metalloproteinases and are crucial cancer regulators[152]. TIMP-1 is a MMP inhibitor and it is expressed in many organs. TIMP-1 levels are altered in many cancers, as lung, prostate, breast, colon and pancreas, and its serum and urine levels are increased in more advanced tumors[21,153,154]. TIMP-1 contains two N-glycosylation sites that present an altered pattern in colon, lung and prostate cancer and some of these aberrant glycan chains have already been sequenced[155,156].
Another proposed candidate is HER-2, a transmembrane protein belonging to the EGFR family that possesses intrinsic tyrosine kinase activity. Its expression has been described in normal and cancerous human pancreas. However, its overexpression has been associated with PaC malignant phenotype and bad prognosis[157,158]. In regard to glycosylation, some authors analyzed HER-2 N-glycans from human breast cancer cell lines using a mass spectrometry based approach[159-161]. Although several reports have described N-glycosylation structures of HER-2 from breast cell lines, the N-glycans attached to this protein in PaC cells or tissues remain to be elucidated. In regards to O-glycosylation, Chugh et al[162] described that the loss of GALNT3 in poorly differentiated PaC cells produced altered O-glycans (Tn and T) on EGFR and HER-2, resulting in an increase in phosphorylation and tumor aggressiveness. The glycosylation of HER-2 could influence in the signalling pathway of PaC cells. The search of aberrant HER-2 glycoforms in PaC sera may be used as a biomarker and also as a target for PaC treatment.
Although there is no single ideal method for the detection of new cancer biomarkers in complex bodily fluids and tissues, we propose the combination of a large variety of analytical methods to develop new approaches based on the glycan analysis of the candidate glycoproteins in a similar way that the ones that have been stablished for the detection of AFP and PSA in HCC and prostate cancer respectively. The most promising approaches include lectin based methods and mass spectrometry technology. The lectin-based methods commonly adopted in cancer biomarker discovery research are lectin affinity chromatography, enzyme-linked lectin assays, lectin histochemistry, lectin blotting and new lectin arrays[163]. Some efforts have also been made to achieve glycan-specific antibodies and/or to produce antibodies that can recognize a specific glycoform in a glycopeptide or glycoprotein. However, glycan-specific antibodies are rare, compared with antibodies recognizing peptide epitopes, because carbohydrates have been shown to be poor immunogens.
Early detection of PaC using glyco-biomarkers could provide a solution and future directions for PaC management. Simultaneous measurement of specific glycoforms combined with protein levels might increase the diagnostic potential of the candidate glycoproteins. However, we have to consider that the aberrant glycosylation is disease specific but not tissue specific, which means that some alterations such as core fucosylation levels or sialylation degree of the candidate glycoprotein could be a common epithelial cancer feature. Furthermore, the use of a multimarker panel composed of the candidate glycoproteins combined with CA 19-9, as well as other medical approaches (as imaging), could enhance the lack of discriminatory power of a single marker.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Guo XZ, Nakai Y S- Editor: Wang XJ L- Editor: A E- Editor: Yin SY
| 1. | Smith BD, Smith GL, Hurria A, Hortobagyi GN, Buchholz TA. Future of cancer incidence in the United States: burdens upon an aging, changing nation. J Clin Oncol. 2009;27:2758-2765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1195] [Cited by in RCA: 1318] [Article Influence: 82.4] [Reference Citation Analysis (0)] |
| 2. | Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913-2921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5379] [Cited by in RCA: 5138] [Article Influence: 467.1] [Reference Citation Analysis (0)] |
| 3. | Malvezzi M, Carioli G, Bertuccio P, Boffetta P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2017, with focus on lung cancer. Ann Oncol. 2017;28:1117-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 4. | Lucas AL, Malvezzi M, Carioli G, Negri E, La Vecchia C, Boffetta P, Bosetti C. Global Trends in Pancreatic Cancer Mortality From 1980 Through 2013 and Predictions for 2017. Clin Gastroenterol Hepatol. 2016;14:1452-1462.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 5. | Brierley JD, Gospodarowicz MK, Wittekind C, editors . UICC. TNM Classification of malignant tumors. ; Wiley-Blackwell: Hoboken, NJ, USA 2017; . |
| 6. | Kanno A, Masamune A, Hanada K, Maguchi H, Shimizu Y, Ueki T, Hasebe O, Ohtsuka T, Nakamura M, Takenaka M, Kitano M, Kikuyama M, Gabata T, Yoshida K, Sasaki T, Serikawa M, Furukawa T, Yanagisawa A, Shimosegawa T; Japan Study Group on the Early Detection of Pancreatic Cancer (JEDPAC). Multicenter study of early pancreatic cancer in Japan. Pancreatology. 2018;18:61-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 168] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 7. | Kikuyama M, Kamisawa T, Kuruma S, Chiba K, Kawaguchi S, Terada S, Satoh T. Early Diagnosis to Improve the Poor Prognosis of Pancreatic Cancer. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 8. | O'Neill CB, Atoria CL, O’Reilly EM, LaFemina J, Henman MC, Elkin EB. Costs and trends in pancreatic cancer treatment. Cancer. 2012;118:5132-5139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 9. | Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185-203.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1247] [Cited by in RCA: 1389] [Article Influence: 173.6] [Reference Citation Analysis (0)] |
| 10. | Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114-1117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2041] [Cited by in RCA: 1944] [Article Influence: 129.6] [Reference Citation Analysis (0)] |
| 11. | Poruk KE, Firpo MA, Adler DG, Mulvihill SJ. Screening for pancreatic cancer: why, how, and who? Ann Surg. 2013;257:17-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 183] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 12. | Zhou B, Xu JW, Cheng YG, Gao JY, Hu SY, Wang L, Zhan HX. Early detection of pancreatic cancer: Where are we now and where are we going? Int J Cancer. 2017;141:231-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 13. | Selleck MJ, Senthil M, Wall NR. Making Meaningful Clinical Use of Biomarkers. Biomark Insights. 2017;12:1177271917715236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 14. | Holdenrieder S, Pagliaro L, Morgenstern D, Dayyani F. Clinically Meaningful Use of Blood Tumor Markers in Oncology. Biomed Res Int. 2016;2016:9795269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 15. | Polanski M, Anderson NL. A list of candidate cancer biomarkers for targeted proteomics. Biomark Insights. 2007;1:1-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Füzéry AK, Levin J, Chan MM, Chan DW. Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin Proteomics. 2013;10:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 296] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 17. | Schiess R, Wollscheid B, Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Mol Oncol. 2009;3:33-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 275] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 18. | Pavlou MP, Diamandis EP, Blasutig IM. The long journey of cancer biomarkers from the bench to the clinic. Clin Chem. 2013;59:147-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 19. | Diamandis EP. Cancer biomarkers: can we turn recent failures into success? J Natl Cancer Inst. 2010;102:1462-1467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 270] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 20. | Kuzmanov U, Kosanam H, Diamandis EP. The sweet and sour of serological glycoprotein tumor biomarker quantification. BMC Med. 2013;11:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Capello M, Bantis LE, Scelo G, Zhao Y, Li P, Dhillon DS, Patel NJ, Kundnani DL, Wang H, Abbruzzese JL. Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer. J Natl Cancer Inst. 2017;109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 22. | Chang JC, Kundranda M. Novel Diagnostic and Predictive Biomarkers in Pancreatic Adenocarcinoma. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 23. | Goh SK, Gold G, Christophi C, Muralidharan V. Serum carbohydrate antigen 19-9 in pancreatic adenocarcinoma: a mini review for surgeons. ANZ J Surg. 2017;87:987-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | O'Brien DP, Sandanayake NS, Jenkinson C, Gentry-Maharaj A, Apostolidou S, Fourkala EO, Camuzeaux S, Blyuss O, Gunu R, Dawnay A. Serum CA19-9 is significantly upregulated up to 2 years before diagnosis with pancreatic cancer: implications for early disease detection. Clin Cancer Res. 2015;21:622-631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 25. | Ferri MJ, Saez M, Figueras J, Fort E, Sabat M, López-Ben S, de Llorens R, Aleixandre RN, Peracaula R. Improved Pancreatic Adenocarcinoma Diagnosis in Jaundiced and Non-Jaundiced Pancreatic Adenocarcinoma Patients through the Combination of Routine Clinical Markers Associated to Pancreatic Adenocarcinoma Pathophysiology. PLoS One. 2016;11:e0147214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 26. | Winter JM, Cameron JL, Lillemoe KD, Campbell KA, Chang D, Riall TS, Coleman J, Sauter PK, Canto M, Hruban RH. Periampullary and pancreatic incidentaloma: a single institution’s experience with an increasingly common diagnosis. Ann Surg. 2006;243:673-80; discussion 680-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 123] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 27. | Morana G, Cancian L, Pozzi Mucelli R, Cugini C. Staging cancer of the pancreas. Cancer Imaging. 2010;10:S137-S141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Appel BL, Tolat P, Evans DB, Tsai S. Current staging systems for pancreatic cancer. Cancer J. 2012;18:539-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 29. | Fusaroli P, Kypraios D, Caletti G, Eloubeidi MA. Pancreatico-biliary endoscopic ultrasound: a systematic review of the levels of evidence, performance and outcomes. World J Gastroenterol. 2012;18:4243-4256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Conrad C, Fernández-Del Castillo C. Preoperative evaluation and management of the pancreatic head mass. J Surg Oncol. 2013;107:23-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Raman SP, Horton KM, Fishman EK. Multimodality imaging of pancreatic cancer-computed tomography, magnetic resonance imaging, and positron emission tomography. Cancer J. 2012;18:511-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 32. | Hanada K, Okazaki A, Hirano N, Izumi Y, Teraoka Y, Ikemoto J, Kanemitsu K, Hino F, Fukuda T, Yonehara S. Diagnostic strategies for early pancreatic cancer. J Gastroenterol. 2015;50:147-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 33. | Wong JC, Lu DS. Staging of pancreatic adenocarcinoma by imaging studies. Clin Gastroenterol Hepatol. 2008;6:1301-1308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 101] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 34. | Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1397] [Cited by in RCA: 1732] [Article Influence: 192.4] [Reference Citation Analysis (1)] |
| 35. | Lee ES, Lee JM. Imaging diagnosis of pancreatic cancer: a state-of-the-art review. World J Gastroenterol. 2014;20:7864-7877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 247] [Cited by in RCA: 250] [Article Influence: 22.7] [Reference Citation Analysis (4)] |
| 36. | Karmazanovsky G, Fedorov V, Kubyshkin V, Kotchatkov A. Pancreatic head cancer: accuracy of CT in determination of resectability. Abdom Imaging. 2005;30:488-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 91] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 37. | Tummala P, Junaidi O, Agarwal B. Imaging of pancreatic cancer: An overview. J Gastrointest Oncol. 2011;2:168-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 42] [Reference Citation Analysis (0)] |
| 38. | De Robertis R, Tinazzi Martini P, Demozzi E, Dal Corso F, Bassi C, Pederzoli P, D’Onofrio M. Diffusion-weighted imaging of pancreatic cancer. World J Radiol. 2015;7:319-328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 46] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 39. | Wang Y, Miller FH, Chen ZE, Merrick L, Mortele KJ, Hoff FL, Hammond NA, Yaghmai V, Nikolaidis P. Diffusion-weighted MR imaging of solid and cystic lesions of the pancreas. Radiographics. 2011;31:E47-E64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 40. | Agarwal B, Abu-Hamda E, Molke KL, Correa AM, Ho L. Endoscopic ultrasound-guided fine needle aspiration and multidetector spiral CT in the diagnosis of pancreatic cancer. Am J Gastroenterol. 2004;99:844-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 221] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 41. | Volmar KE, Vollmer RT, Jowell PS, Nelson RC, Xie HB. Pancreatic FNA in 1000 cases: a comparison of imaging modalities. Gastrointest Endosc. 2005;61:854-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 151] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 42. | Wang W, Shpaner A, Krishna SG, Ross WA, Bhutani MS, Tamm EP, Raju GS, Xiao L, Wolff RA, Fleming JB. Use of EUS-FNA in diagnosing pancreatic neoplasm without a definitive mass on CT. Gastrointest Endosc. 2013;78:73-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 43. | Hewitt MJ, McPhail MJ, Possamai L, Dhar A, Vlavianos P, Monahan KJ. EUS-guided FNA for diagnosis of solid pancreatic neoplasms: a meta-analysis. Gastrointest Endosc. 2012;75:319-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 510] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 44. | Mohammad Alizadeh AH, Shahrokh S, Hadizadeh M, Padashi M, Zali MR. Diagnostic potency of EUS-guided FNA for the evaluation of pancreatic mass lesions. Endosc Ultrasound. 2016;5:30-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 45. | Dewitt J, Devereaux BM, Lehman GA, Sherman S, Imperiale TF. Comparison of endoscopic ultrasound and computed tomography for the preoperative evaluation of pancreatic cancer: a systematic review. Clin Gastroenterol Hepatol. 2006;4:717-725; quiz 664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 168] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 46. | Kauhanen SP, Komar G, Seppänen MP, Dean KI, Minn HR, Kajander SA, Rinta-Kiikka I, Alanen K, Borra RJ, Puolakkainen PA. A prospective diagnostic accuracy study of 18F-fluorodeoxyglucose positron emission tomography/computed tomography, multidetector row computed tomography, and magnetic resonance imaging in primary diagnosis and staging of pancreatic cancer. Ann Surg. 2009;250:957-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 219] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 47. | Matsumoto I, Shirakawa S, Shinzeki M, Asari S, Goto T, Ajiki T, Fukumoto T, Kitajima K, Ku Y. 18-Fluorodeoxyglucose positron emission tomography does not aid in diagnosis of pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol. 2013;11:712-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 48. | Sperti C, Pasquali C, Bissoli S, Chierichetti F, Liessi G, Pedrazzoli S. Tumor relapse after pancreatic cancer resection is detected earlier by 18-FDG PET than by CT. J Gastrointest Surg. 2010;14:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 49. | Kinupe AB, Ung Y, Ko Y-J, Berry SR. FDG PET/CT in pancreatic cancer staging and management: A retrospective study. Journal of Clin Oncol. 2017;35:464-464. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 50. | Dibble EH, Karantanis D, Mercier G, Peller PJ, Kachnic LA, Subramaniam RM. PET/CT of cancer patients: part 1, pancreatic neoplasms. AJR Am J Roentgenol. 2012;199:952-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 51. | Makawita S, Smith C, Batruch I, Zheng Y, Rückert F, Grützmann R, Pilarsky C, Gallinger S, Diamandis EP. Integrated proteomic profiling of cell line conditioned media and pancreatic juice for the identification of pancreatic cancer biomarkers. Mol Cell Proteomics. 2011;10:M111.008599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 52. | Chen R, Pan S, Yi EC, Donohoe S, Bronner MP, Potter JD, Goodlett DR, Aebersold R, Brentnall TA. Quantitative proteomic profiling of pancreatic cancer juice. Proteomics. 2006;6:3871-3879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 84] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 53. | Pan S, Brentnall TA, Chen R. Glycoproteins and glycoproteomics in pancreatic cancer. World J Gastroenterol. 2016;22:9288-9299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 48] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (2)] |
| 54. | Brand RE, Nolen BM, Zeh HJ, Allen PJ, Eloubeidi MA, Goldberg M, Elton E, Arnoletti JP, Christein JD, Vickers SM. Serum biomarker panels for the detection of pancreatic cancer. Clin Cancer Res. 2011;17:805-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 190] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 55. | Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, Balakrishnan L, Dwivedi SB, Telikicherla D, Selvan LD. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 239] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 56. | Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2108] [Cited by in RCA: 2203] [Article Influence: 220.3] [Reference Citation Analysis (0)] |
| 57. | Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329-335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2697] [Cited by in RCA: 3761] [Article Influence: 376.1] [Reference Citation Analysis (0)] |
| 58. | Bauden M, Kristl T, Sasor A, Andersson B, Marko-Varga G, Andersson R, Ansari D. Histone profiling reveals the H1.3 histone variant as a prognostic biomarker for pancreatic ductal adenocarcinoma. BMC Cancer. 2017;17:810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 59. | Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2770] [Cited by in RCA: 3552] [Article Influence: 322.9] [Reference Citation Analysis (0)] |
| 60. | Kinugasa H, Nouso K, Miyahara K, Morimoto Y, Dohi C, Tsutsumi K, Kato H, Matsubara T, Okada H, Yamamoto K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121:2271-2280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 181] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 61. | Pishvaian MJ, Joseph Bender R, Matrisian LM, Rahib L, Hendifar A, Hoos WA, Mikhail S, Chung V, Picozzi V, Heartwell C. A pilot study evaluating concordance between blood-based and patient-matched tumor molecular testing within pancreatic cancer patients participating in the Know Your Tumor (KYT) initiative. Oncotarget. 2016;8:83446-83456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 62. | Yi JM, Guzzetta AA, Bailey VJ, Downing SR, Van Neste L, Chiappinelli KB, Keeley BP, Stark A, Herrera A, Wolfgang C. Novel methylation biomarker panel for the early detection of pancreatic cancer. Clin Cancer Res. 2013;19:6544-6555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 63. | Kulemann B, Rösch S, Seifert S, Timme S, Bronsert P, Seifert G, Martini V, Kuvendjiska J, Glatz T, Hussung S. Pancreatic cancer: Circulating Tumor Cells and Primary Tumors show Heterogeneous KRAS Mutations. Sci Rep. 2017;7:4510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 64. | Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1690] [Cited by in RCA: 1647] [Article Influence: 126.7] [Reference Citation Analysis (0)] |
| 65. | Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2130] [Cited by in RCA: 1993] [Article Influence: 199.3] [Reference Citation Analysis (1)] |
| 66. | Roberts NJ, Norris AL, Petersen GM, Bondy ML, Brand R, Gallinger S, Kurtz RC, Olson SH, Rustgi AK, Schwartz AG. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. Cancer Discov. 2016;6:166-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 278] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 67. | Imamura T, Komatsu S, Ichikawa D, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Arita T, Konishi H, Shiozaki A. Liquid biopsy in patients with pancreatic cancer: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2016;22:5627-5641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 68. | Riva F, Dronov OI, Khomenko DI, Huguet F, Louvet C, Mariani P, Stern MH, Lantz O, Proudhon C, Pierga JY. Clinical applications of circulating tumor DNA and circulating tumor cells in pancreatic cancer. Mol Oncol. 2016;10:481-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (1)] |
| 69. | Syren P, Andersson R, Bauden M, Ansari D. Epigenetic alterations as biomarkers in pancreatic ductal adenocarcinoma. Scand J Gastroenterol. 2017;52:668-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 70. | Henriksen SD, Madsen PH, Larsen AC, Johansen MB, Pedersen IS, Krarup H, Thorlacius-Ussing O. Promoter hypermethylation in plasma-derived cell-free DNA as a prognostic marker for pancreatic adenocarcinoma staging. Int J Cancer. 2017;141:2489-2497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 71. | Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Nagata H, Konishi H, Shiozaki A, Ikoma H, Okamoto K. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105:1733-1740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 213] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 72. | Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 183] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 73. | Miyamae M, Komatsu S, Ichikawa D, Kawaguchi T, Hirajima S, Okajima W, Ohashi T, Imamura T, Konishi H, Shiozaki A. Plasma microRNA profiles: identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br J Cancer. 2015;113:1467-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 74. | Duell EJ, Lujan-Barroso L, Sala N, Deitz McElyea S, Overvad K, Tjonneland A, Olsen A, Weiderpass E, Busund LT, Moi L. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int J Cancer. 2017;141:905-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 75. | Imamura T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Ohashi T, Kiuchi J, Nishibeppu K, Konishi H, Shiozaki A. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep. 2017;7:5708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 76. | Li Y, Sarkar FH. MicroRNA Targeted Therapeutic Approach for Pancreatic Cancer. Int J Biol Sci. 2016;12:326-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 77. | Lai X, Wang M, McElyea SD, Sherman S, House M, Korc M. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 261] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 78. | Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 234] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 79. | Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, Schmidt CM, Yip-Schneider MT, Allen PJ, Schattner M. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci USA. 2017;114:10202-10207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 435] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 80. | Sefrioui D, Blanchard F, Toure E, Basile P, Beaussire L, Dolfus C, Perdrix A, Paresy M, Antonietti M, Iwanicki-Caron I. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br J Cancer. 2017;117:1017-1025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 81. | Balasenthil S, Huang Y, Liu S, Marsh T, Chen J, Stass SA, KuKuruga D, Brand R, Chen N, Frazier ML. A Plasma Biomarker Panel to Identify Surgically Resectable Early-Stage Pancreatic Cancer. J Natl Cancer Inst. 2017;109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 82. | Boulaiz H, Ramos MC, Griñán-Lisón C, García-Rubiño ME, Vicente F, Marchal JA. What’s new in the diagnosis of pancreatic cancer: a patent review (2011-present). Expert Opin Ther Pat. 2017;27:1319-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 83. | Kailemia MJ, Park D, Lebrilla CB. Glycans and glycoproteins as specific biomarkers for cancer. Anal Bioanal Chem. 2017;409:395-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 260] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 84. | Munkley J, Elliott DJ. Hallmarks of glycosylation in cancer. Oncotarget. 2016;7:35478-35489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 382] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 85. | Hart GW. Myriad Roles of Glycans in Biology. J Mol Biol. 2016;428:3147-3149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 86. | Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1664] [Cited by in RCA: 2187] [Article Influence: 218.7] [Reference Citation Analysis (0)] |
| 87. | Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol. 2015;10:473-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 636] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 88. | Silva ML. Cancer serum biomarkers based on aberrant post-translational modifications of glycoproteins: Clinical value and discovery strategies. Biochim Biophys Acta. 2015;1856:165-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 89. | Peracaula R, Barrabés S, Sarrats A, Rudd PM, de Llorens R. Altered glycosylation in tumours focused to cancer diagnosis. Dis Markers. 2008;25:207-218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 126] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 90. | Meany DL, Chan DW. Aberrant glycosylation associated with enzymes as cancer biomarkers. Clin Proteomics. 2011;8:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 182] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 91. | Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, Block TM, Mehta AS. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18:1914-1921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 129] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 92. | Li D, Mallory T, Satomura S. AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin Chim Acta. 2001;313:15-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 258] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 93. | Tabarés G, Radcliffe CM, Barrabés S, Ramírez M, Aleixandre RN, Hoesel W, Dwek RA, Rudd PM, Peracaula R, de Llorens R. Different glycan structures in prostate-specific antigen from prostate cancer sera in relation to seminal plasma PSA. Glycobiology. 2006;16:132-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 133] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 94. | Sarrats A, Comet J, Tabarés G, Ramírez M, Aleixandre RN, de Llorens R, Peracaula R. Differential percentage of serum prostate-specific antigen subforms suggests a new way to improve prostate cancer diagnosis. Prostate. 2010;70:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 95. | Llop E, Ferrer-Batallé M, Barrabés S, Guerrero PE, Ramírez M, Saldova R, Rudd PM, Aleixandre RN, Comet J, de Llorens R. Improvement of Prostate Cancer Diagnosis by Detecting PSA Glycosylation-Specific Changes. Theranostics. 2016;6:1190-1204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 96. | Ferrer-Batallé M, Llop E, Ramírez M, Aleixandre RN, Saez M, Comet J, de Llorens R, Peracaula R. Comparative Study of Blood-Based Biomarkers, α2,3-Sialic Acid PSA and PHI, for High-Risk Prostate Cancer Detection. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 97. | Kontro H, Joenväärä S, Haglund C, Renkonen R. Comparison of sialylated N-glycopeptide levels in serum of pancreatic cancer patients, acute pancreatitis patients, and healthy controls. Proteomics. 2014;14:1713-1723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 98. | Mann BF, Goetz JA, House MG, Schmidt CM, Novotny MV. Glycomic and proteomic profiling of pancreatic cyst fluids identifies hyperfucosylated lactosamines on the N-linked glycans of overexpressed glycoproteins. Mol Cell Proteomics. 2012;11:M111.015792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 99. | Singh S, Pal K, Yadav J, Tang H, Partyka K, Kletter D, Hsueh P, Ensink E, Kc B, Hostetter G. Upregulation of glycans containing 3’ fucose in a subset of pancreatic cancers uncovered using fusion-tagged lectins. J Proteome Res. 2015;14:2594-2605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 100. | Holst S, Belo AI, Giovannetti E, van Die I, Wuhrer M. Profiling of different pancreatic cancer cells used as models for metastatic behaviour shows large variation in their N-glycosylation. Sci Rep. 2017;7:16623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 101. | Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6:1126-1138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 136] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 102. | Pour PM, Tempero MM, Takasaki H, Uchida E, Takiyama Y, Burnett DA, Steplewski Z. Expression of blood group-related antigens ABH, Lewis A, Lewis B, Lewis X, Lewis Y, and CA 19-9 in pancreatic cancer cells in comparison with the patient’s blood group type. Cancer Res. 1988;48:5422-5426. [PubMed] |
| 103. | Pérez-Garay M, Arteta B, Llop E, Cobler L, Pagès L, Ortiz R, Ferri MJ, de Bolós C, Figueras J, de Llorens R. α2,3-Sialyltransferase ST3Gal IV promotes migration and metastasis in pancreatic adenocarcinoma cells and tends to be highly expressed in pancreatic adenocarcinoma tissues. Int J Biochem Cell Biol. 2013;45:1748-1757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 104. | Remmers N, Anderson JM, Linde EM, DiMaio DJ, Lazenby AJ, Wandall HH, Mandel U, Clausen H, Yu F, Hollingsworth MA. Aberrant expression of mucin core proteins and o-linked glycans associated with progression of pancreatic cancer. Clin Cancer Res. 2013;19:1981-1993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 136] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 105. | Kailemia MJ, Xu G, Wong M, Li Q, Goonatilleke E, Leon F, Lebrilla CB. Recent Advances in the Mass Spectrometry Methods for Glycomics and Cancer. Anal Chem. 2018;90:208-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 106. | Arnold JN, Saldova R, Hamid UM, Rudd PM. Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Proteomics. 2008;8:3284-3293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 259] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 107. | Terao N, Takamatsu S, Minehira T, Sobajima T, Nakayama K, Kamada Y, Miyoshi E. Fucosylation is a common glycosylation type in pancreatic cancer stem cell-like phenotypes. World J Gastroenterol. 2015;21:3876-3887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 39] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 108. | Tan Z, Yin H, Nie S, Lin Z, Zhu J, Ruffin MT, Anderson MA, Simeone DM, Lubman DM. Large-scale identification of core-fucosylated glycopeptide sites in pancreatic cancer serum using mass spectrometry. J Proteome Res. 2015;14:1968-1978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 109. | Pan S, Chen R, Tamura Y, Crispin DA, Lai LA, May DH, McIntosh MW, Goodlett DR, Brentnall TA. Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. J Proteome Res. 2014;13:1293-1306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 110. | Zhao J, Patwa TH, Qiu W, Shedden K, Hinderer R, Misek DE, Anderson MA, Simeone DM, Lubman DM. Glycoprotein microarrays with multi-lectin detection: unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res. 2007;6:1864-1874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 121] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 111. | Li C, Simeone DM, Brenner DE, Anderson MA, Shedden KA, Ruffin MT, Lubman DM. Pancreatic cancer serum detection using a lectin/glyco-antibody array method. J Proteome Res. 2009;8:483-492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 112. | Li C, Zolotarevsky E, Thompson I, Anderson MA, Simeone DM, Casper JM, Mullenix MC, Lubman DM. A multiplexed bead assay for profiling glycosylation patterns on serum protein biomarkers of pancreatic cancer. Electrophoresis. 2011;32:2028-2035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 113. | Sarrats A, Saldova R, Pla E, Fort E, Harvey DJ, Struwe WB, de Llorens R, Rudd PM, Peracaula R. Glycosylation of liver acute-phase proteins in pancreatic cancer and chronic pancreatitis. Proteomics Clin Appl. 2010;4:432-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 114. | Balmaña M, Sarrats A, Llop E, Barrabés S, Saldova R, Ferri MJ, Figueras J, Fort E, de Llorens R, Rudd PM. Identification of potential pancreatic cancer serum markers: Increased sialyl-Lewis X on ceruloplasmin. Clin Chim Acta. 2015;442:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 115. | Matsumoto H, Shinzaki S, Narisada M, Kawamoto S, Kuwamoto K, Moriwaki K, Kanke F, Satomura S, Kumada T, Miyoshi E. Clinical application of a lectin-antibody ELISA to measure fucosylated haptoglobin in sera of patients with pancreatic cancer. Clin Chem Lab Med. 2010;48:505-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 116. | Giménez E, Balmaña M, Figueras J, Fort E, de Bolós C, Sanz-Nebot V, Peracaula R, Rizzi A. Quantitative analysis of N-glycans from human alfa-acid-glycoprotein using stable isotope labeling and zwitterionic hydrophilic interaction capillary liquid chromatography electrospray mass spectrometry as tool for pancreatic disease diagnosis. Anal Chim Acta. 2015;866:59-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 117. | Balmaña M, Giménez E, Puerta A, Llop E, Figueras J, Fort E, Sanz-Nebot V, de Bolós C, Rizzi A, Barrabés S. Increased α1-3 fucosylation of α-1-acid glycoprotein (AGP) in pancreatic cancer. J Proteomics. 2016;132:144-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 118. | Mancera-Arteu M, Giménez E, Barbosa J, Peracaula R, Sanz-Nebot V. Zwitterionic-hydrophilic interaction capillary liquid chromatography coupled to tandem mass spectrometry for the characterization of human alpha-acid-glycoprotein N-glycan isomers. Anal Chim Acta. 2017;991:76-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 119. | Nie S, Lo A, Wu J, Zhu J, Tan Z, Simeone DM, Anderson MA, Shedden KA, Ruffin MT, Lubman DM. Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis. J Proteome Res. 2014;13:1873-1884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 120. | Barrabés S, Pagès-Pons L, Radcliffe CM, Tabarés G, Fort E, Royle L, Harvey DJ, Moenner M, Dwek RA, Rudd PM. Glycosylation of serum ribonuclease 1 indicates a major endothelial origin and reveals an increase in core fucosylation in pancreatic cancer. Glycobiology. 2007;17:388-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 121. | Nakata D. Increased N-glycosylation of Asn88 in serum pancreatic ribonuclease 1 is a novel diagnostic marker for pancreatic cancer. Sci Rep. 2014;4:6715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 122. | Drabik A, Bodzon-Kulakowska A, Suder P, Silberring J, Kulig J, Sierzega M. Glycosylation Changes in Serum Proteins Identify Patients with Pancreatic Cancer. J Proteome Res. 2017;16:1436-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 123. | Krishnan S, Whitwell HJ, Cuenco J, Gentry-Maharaj A, Menon U, Pereira SP, Gaspari M, Timms JF. Evidence of Altered Glycosylation of Serum Proteins Prior to Pancreatic Cancer Diagnosis. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 124. | Matsuda A, Higashi M, Nakagawa T, Yokoyama S, Kuno A, Yonezawa S, Narimatsu H. Assessment of tumor characteristics based on glycoform analysis of membrane-tethered MUC1. Lab Invest. 2017;97:1103-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 125. | Haab BB, Porter A, Yue T, Li L, Scheiman J, Anderson MA, Barnes D, Schmidt CM, Feng Z, Simeone DM. Glycosylation variants of mucins and CEACAMs as candidate biomarkers for the diagnosis of pancreatic cystic neoplasms. Ann Surg. 2010;251:937-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 126. | Balmaña M, Duran A, Gomes C, Llop E, López-Martos R, Ortiz MR, Barrabés S, Reis CA, Peracaula R. Analysis of sialyl-Lewis x on MUC5AC and MUC1 mucins in pancreatic cancer tissues. Int J Biol Macromol. 2018;112:33-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 127. | Tang H, Partyka K, Hsueh P, Sinha JY, Kletter D, Zeh H, Huang Y, Brand RE, Haab BB. Glycans related to the CA19-9 antigen are elevated in distinct subsets of pancreatic cancers and improve diagnostic accuracy over CA19-9. Cell Mol Gastroenterol Hepatol. 2016;2:201-221.e15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 128. | Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4:437-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 168] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 129. | Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, Brand RE, Haab BB. The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays. Mol Cell Proteomics. 2009;8:1697-1707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 130. | Yue T, Maupin KA, Fallon B, Li L, Partyka K, Anderson MA, Brenner DE, Kaul K, Zeh H, Moser AJ. Enhanced discrimination of malignant from benign pancreatic disease by measuring the CA 19-9 antigen on specific protein carriers. PLoS One. 2011;6:e29180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 131. | Mirus JE, Zhang Y, Li CI, Lokshin AE, Prentice RL, Hingorani SR, Lampe PD. Cross-species antibody microarray interrogation identifies a 3-protein panel of plasma biomarkers for early diagnosis of pancreas cancer. Clin Cancer Res. 2015;21:1764-1771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 132. | Dumstrei K, Chen H, Brenner H. A systematic review of serum autoantibodies as biomarkers for pancreatic cancer detection. Oncotarget. 2016;7:11151-11164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 133. | Ordóñez NG. Value of mesothelin immunostaining in the diagnosis of mesothelioma. Mod Pathol. 2003;16:192-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 181] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 134. | Hassan R, Kreitman RJ, Pastan I, Willingham MC. Localization of mesothelin in epithelial ovarian cancer. Appl Immunohistochem Mol Morphol. 2005;13:243-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 145] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 135. | Miettinen M, Sarlomo-Rikala M. Expression of calretinin, thrombomodulin, keratin 5, and mesothelin in lung carcinomas of different types: an immunohistochemical analysis of 596 tumors in comparison with epithelioid mesotheliomas of the pleura. Am J Surg Pathol. 2003;27:150-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 137] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 136. | Zheng C, Jia W, Tang Y, Zhao H, Jiang Y, Sun S. Mesothelin regulates growth and apoptosis in pancreatic cancer cells through p53-dependent and -independent signal pathway. J Exp Clin Cancer Res. 2012;31:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 137. | Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, Murugesan SR, Leach SD, Jaffee E, Yeo CJ. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE). Clin Cancer Res. 2001;7:3862-3868. [PubMed] |
| 138. | Hassan R, Laszik ZG, Lerner M, Raffeld M, Postier R, Brackett D. Mesothelin is overexpressed in pancreaticobiliary adenocarcinomas but not in normal pancreas and chronic pancreatitis. Am J Clin Pathol. 2005;124:838-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 155] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 139. | Bournet B, Pointreau A, Souque A, Oumouhou N, Muscari F, Lepage B, Senesse P, Barthet M, Lesavre N, Hammel P. Gene expression signature of advanced pancreatic ductal adenocarcinoma using low density array on endoscopic ultrasound-guided fine needle aspiration samples. Pancreatology. 2012;12:27-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 140. | Johnston FM, Tan MC, Tan BR Jr, Porembka MR, Brunt EM, Linehan DC, Simon PO Jr, Plambeck-Suess S, Eberlein TJ, Hellstrom KE, Hellstrom I, Hawkins WG, Goedegebuure P. Circulating mesothelin protein and cellular antimesothelin immunity in patients with pancreatic cancer. Clin Cancer Res. 2009;15:6511-6518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 141. | Kendrick ZW, Firpo MA, Repko RC, Scaife CL, Adler DG, Boucher KM, Mulvihill SJ. Serum IGFBP2 and MSLN as diagnostic and prognostic biomarkers for pancreatic cancer. HPB (Oxford). 2014;16:670-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 142. | Scholler N, Fu N, Yang Y, Ye Z, Goodman GE, Hellström KE, Hellström I. Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in sera from patients with ovarian carcinoma. Proc Natl Acad Sci USA. 1999;96:11531-11536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 242] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 143. | Hassan R, Remaley AT, Sampson ML, Zhang J, Cox DD, Pingpank J, Alexander R, Willingham M, Pastan I, Onda M. Detection and quantitation of serum mesothelin, a tumor marker for patients with mesothelioma and ovarian cancer. Clin Cancer Res. 2006;12:447-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 210] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 144. | Onda M, Nagata S, Ho M, Bera TK, Hassan R, Alexander RH, Pastan I. Megakaryocyte potentiation factor cleaved from mesothelin precursor is a useful tumor marker in the serum of patients with mesothelioma. Clin Cancer Res. 2006;12:4225-4231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 145. | Baricević I, Masnikosa R, Lagundzin D, Golubović V, Nedić O. Alterations of insulin-like growth factor binding protein 3 (IGFBP-3) glycosylation in patients with breast tumours. Clin Biochem. 2010;43:725-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 146. | Roghani M, Segovia B, Whitechurch O, Binoux M. Purification from human cerebrospinal fluid of insulin-like growth factor binding proteins (IGFBPs). Isolation of IGFBP-2, an altered form of IGFBP-3 and a new IGFBP species. Growth Regul. 1991;1:125-130. [PubMed] |
| 147. | De Reggi M, Capon C, Gharib B, Wieruszeski JM, Michel R, Fournet B. The glycan moiety of human pancreatic lithostathine. Structure characterization and possible pathophysiological implications. Eur J Biochem. 1995;230:503-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 148. | Porterfield M, Zhao P, Han H, Cunningham J, Aoki K, Von Hoff DD, Demeure MJ, Pierce JM, Tiemeyer M, Wells L. Discrimination between adenocarcinoma and normal pancreatic ductal fluid by proteomic and glycomic analysis. J Proteome Res. 2014;13:395-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 149. | Zhang YW, Ding LS, Lai MD. Reg gene family and human diseases. World J Gastroenterol. 2003;9:2635-2641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 112] [Cited by in RCA: 112] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 150. | Makawita S, Dimitromanolakis A, Soosaipillai A, Soleas I, Chan A, Gallinger S, Haun RS, Blasutig IM, Diamandis EP. Validation of four candidate pancreatic cancer serological biomarkers that improve the performance of CA19.9. BMC Cancer. 2013;13:404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 151. | Radon TP, Massat NJ, Jones R, Alrawashdeh W, Dumartin L, Ennis D, Duffy SW, Kocher HM, Pereira SP, Guarner posthumous L. Identification of a Three-Biomarker Panel in Urine for Early Detection of Pancreatic Adenocarcinoma. Clin Cancer Res. 2015;21:3512-3521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 142] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 152. | Jackson HW, Defamie V, Waterhouse P, Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer. 2017;17:38-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 341] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 153. | Slater EP, Fendrich V, Strauch K, Rospleszcz S, Ramaswamy A, Mätthai E, Chaloupka B, Gress TM, Langer P, Bartsch DK. LCN2 and TIMP1 as Potential Serum Markers for the Early Detection of Familial Pancreatic Cancer. Transl Oncol. 2013;6:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 154. | Roy R, Zurakowski D, Wischhusen J, Frauenhoffer C, Hooshmand S, Kulke M, Moses MA. Urinary TIMP-1 and MMP-2 levels detect the presence of pancreatic malignancies. Br J Cancer. 2014;111:1772-1779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 155. | Kim YS, Kim SH, Kang JG, Ko JH. Expression level and glycan dynamics determine the net effects of TIMP-1 on cancer progression. BMB Rep. 2012;45:623-628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 156. | Kim HIe, Saldova R, Park JH, Lee YH, Harvey DJ, Wormald MR, Wynne K, Elia G, Kim HJ, Rudd PM, Lee ST. The presence of outer arm fucose residues on the N-glycans of tissue inhibitor of metalloproteinases-1 reduces its activity. J Proteome Res. 2013;12:3547-3560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 157. | Yamanaka Y, Friess H, Kobrin MS, Büchler M, Kunz J, Beger HG, Korc M. Overexpression of HER2/neu oncogene in human pancreatic carcinoma. Hum Pathol. 1993;24:1127-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 138] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 158. | Komoto M, Nakata B, Amano R, Yamada N, Yashiro M, Ohira M, Wakasa K, Hirakawa K. HER2 overexpression correlates with survival after curative resection of pancreatic cancer. Cancer Sci. 2009;100:1243-1247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 92] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 159. | Kaneshiro K, Watanabe M, Terasawa K, Uchimura H, Fukuyama Y, Iwamoto S, Sato TA, Shimizu K, Tsujimoto G, Tanaka K. Rapid quantitative profiling of N-glycan by the glycan-labeling method using 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid. Anal Chem. 2012;84:7146-7151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 160. | Watanabe M, Terasawa K, Kaneshiro K, Uchimura H, Yamamoto R, Fukuyama Y, Shimizu K, Sato TA, Tanaka K. Improvement of mass spectrometry analysis of glycoproteins by MALDI-MS using 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid. Anal Bioanal Chem. 2013;405:4289-4293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 161. | Lee LY, Thaysen-Andersen M, Baker MS, Packer NH, Hancock WS, Fanayan S. Comprehensive N-glycome profiling of cultured human epithelial breast cells identifies unique secretome N-glycosylation signatures enabling tumorigenic subtype classification. J Proteome Res. 2014;13:4783-4795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 162. | Chugh S, Meza J, Sheinin YM, Ponnusamy MP, Batra SK. Loss of N-acetylgalactosaminyltransferase 3 in poorly differentiated pancreatic cancer: augmented aggressiveness and aberrant ErbB family glycosylation. Br J Cancer. 2016;114:1376-1386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 163. | Hashim OH, Jayapalan JJ, Lee CS. Lectins: an effective tool for screening of potential cancer biomarkers. PeerJ. 2017;5:e3784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |