Published online Mar 21, 2018. doi: 10.3748/wjg.v24.i11.1206
Peer-review started: December 10, 2017
First decision: December 21, 2017
Revised: December 25, 2017
Accepted: January 16, 2018
Article in press: January 16, 2018
Published online: March 21, 2018
Processing time: 97 Days and 2 Hours
To identify multiple microRNAs (miRNAs) for predicting the prognosis of gastric cancer (GC) patients by bioinformatics analysis.
The original microarray dataset GSE93415, which included 20 GC and 20 tumor adjacent normal gastric mucosal tissues, was downloaded from the Gene Expression Omnibus database and used for screening differentially expressed miRNAs (DEMs). The cut-off criteria were P < 0.05 and fold change > 2.0. In addition, we acquired the miRNA expression profiles and clinical information of 361 GC patients from The Cancer Genome Atlas database to assess the prognostic role of the DEMs. The target genes of miRNAs were predicted using TargetScan, miRDB, miRWalk, and DIANA, and then the common target genes were selected for functional enrichment analysis.
A total of 110 DEMs including 19 up-regulated and 91 down-regulated miRNAs were identified between 20 pairs of GC and tumor adjacent normal tissues, and the Kaplan-Meier survival analysis found that a three-miRNA signature (miR-145-3p, miR-125b-5p, and miR-99a-5p) had an obvious correlation with the survival of GC patients. Furthermore, univariate and multivariate Cox regression analyses indicated that the three-miRNA signature could be a significant prognostic marker in GC patients. The common target genes of the three miRNAs are added up to 108 and used for Gene Functional Enrichment analysis. Biological Process and Molecular Function analyses showed that the target genes are involved in cell recognition, gene silencing and nucleic acid binding, transcription factor activity, and transmembrane receptor activity. Cellular Component analysis revealed that the genes are portion of nucleus, chromatin silencing complex, and TORC1/2 complex. Biological Pathway analysis indicated that the genes participate in several cancer-related pathways, such as the focal adhesion, PI3K, and mTOR signaling pathways.
This study justified that a three-miRNA signature could play a role in predicting the survival of GC patients.
Core tip: We identified 110 differentially expressed miRNAs through mining the datasets of Gene Expression Omnibus database and acquired the miRNA expression profiles and clinical information of 361 gastric cancer (GC) patients from The Cancer Genome Atlas database. Multiple miRNAs together acting as biomarkers may have a stronger reliability in survival prediction. Our study found that a novel three-miRNA signature could be used for predicting the prognosis of GC patients.
- Citation: Zhang C, Zhang CD, Ma MH, Dai DQ. Three-microRNA signature identified by bioinformatics analysis predicts prognosis of gastric cancer patients. World J Gastroenterol 2018; 24(11): 1206-1215
- URL: https://www.wjgnet.com/1007-9327/full/v24/i11/1206.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i11.1206
Gastric cancer (GC) is the fourth most common cancer in incidence and the second in mortality among all cancers worldwide[1]. In 2008, a total of 989600 individuals were newly diagnosed with GC and 738,000 deaths occurred, therefore, this disease is a serious public health issue worldwide[2]. Research studies that explore the cellular and molecular mechanisms of GC development and the validation of novel biomarkers are urgently needed to achieve early diagnosis and treatment.
MicroRNAs (miRNAs), which are endogenous small noncoding RNAs (20-22 nt), have been identified as the key regulators of genes at the post-transcriptional level[3]. Increasing studies have found that miRNAs are associated with the development and progression of GC, and can act as important biomarkers in diagnosis[4,5], therapy[6], and prognosis[7,8]. Thus, the identification of differentially expressed miRNAs (DEMs) may contribute to the early diagnosis and the prediction of survival prognosis in GC.
Several studies have found that a number of miRNAs are differentially expressed in GC and are associated with survival prognosis. However, these studies lack a large sample size or an appropriate proportion of samples. A reliable survival prediction requires large-scale samples that include detailed clinical characteristics. The Gene Expression Omnibus (GEO) database is a public functional genomics data repository that includes array- and sequence-based data and allows users to query and download experiments or curated gene expression profiles[9]. The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) project is one of the most useful cancer genomics programs and has generated, analyzed, and made available genomic sequence, expression, methylation, and copy number variation data on over 11000 individuals who represent over 30 different types of cancer[10]. In the present study, we identified DEMs between GC and adjacent normal tissues by analyzing the miRNA data of GSE93415 from GEO. In addition, the associations between DEMs and survival prognosis were analyzed using the expression profiles and clinical features downloaded from TCGA.
The microarray data of GSE93415 were downloaded from the GEO database (https://http://www.ncbi.nlm.nih.gov/geo/) and the miRNA expression data were processed with the limma package in R. Statistically significant DEMs between GC and adjacent normal samples were identified with the cut-off criterion P < 0.05 and fold change > 2.0.
TCGA (https://cancergenome.nih.gov/) stomach adenocarcinoma and adjacent normal tissue miRNA sequencing data and clinical information were downloaded for analysis. The inclusion criteria included: (1) samples with completed data for analysis; (2) patients had not received preoperative chemoradiation; and (3) overall survival time less than 80 mo. Consequently, 361 GC samples were included in the present study. The Kaplan-Meier method and log-rank test were conducted to test the prognostic value of DEMs. When P < 0.05, miRNAs were considered significantly associated with the prognosis of patients. Then, we ranked prognosis-related miRNAs according to the median expression level. Subsequently, we scored each GC patient in accordance with a high or low level of expression, and a risk grade was defined by the total scores. Finally, GC patients were sorted into high and low risk groups by the risk-score rank. The prognosis-related miRNA signature was used to analyze overall survival between high and low risk group patients using a Kaplan-Meier curve.
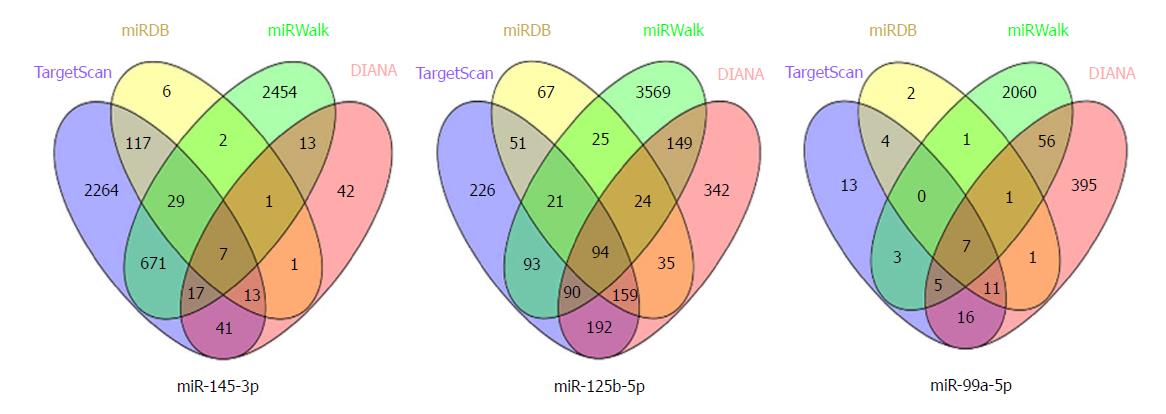
We used four online tools to predict the potential target genes of the prognostic related DEMs, including TargetScan (http://www.targetscan.org/vert_71/), miRDB (http://www.mirdb.org/), miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html), and DIANA (http://www.microrna.gr/microT-CDS). In order to obtain the more reliable target genes, the Venn plot was performed to acquire the consensus genes of the four online tools.
FunRich [Functional Enrichment analysis tool (http://www.funrich.org/)] is a stand-alone software used for functional enrichment and interaction network analysis of genes and proteins[11]. Enrichment analysis was conducted on the consensus genes using the FunRich tool in the following categories: Biological Process, Cellular Component, Molecular Function, and Biological Pathways. P < 0.05 was considered statistically significant.
The data of miRNA expression in GC and adjacent normal samples were performed by unpaired t-test. The association between DEMs expression and clinical characteristics was analyzed by the chi-square and t-tests. Kaplan-Meier survival analysis and the univariate/multivariate Cox regression analysis were used to assess the expression levels of DEMs and prognostic features. All the statistical analyses were performed with IBM SPSS version 19.0 and P < 0.05 was considered statistically significant.
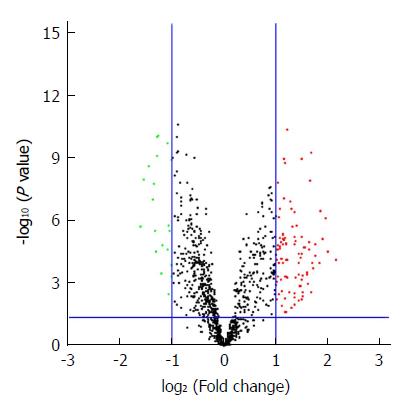
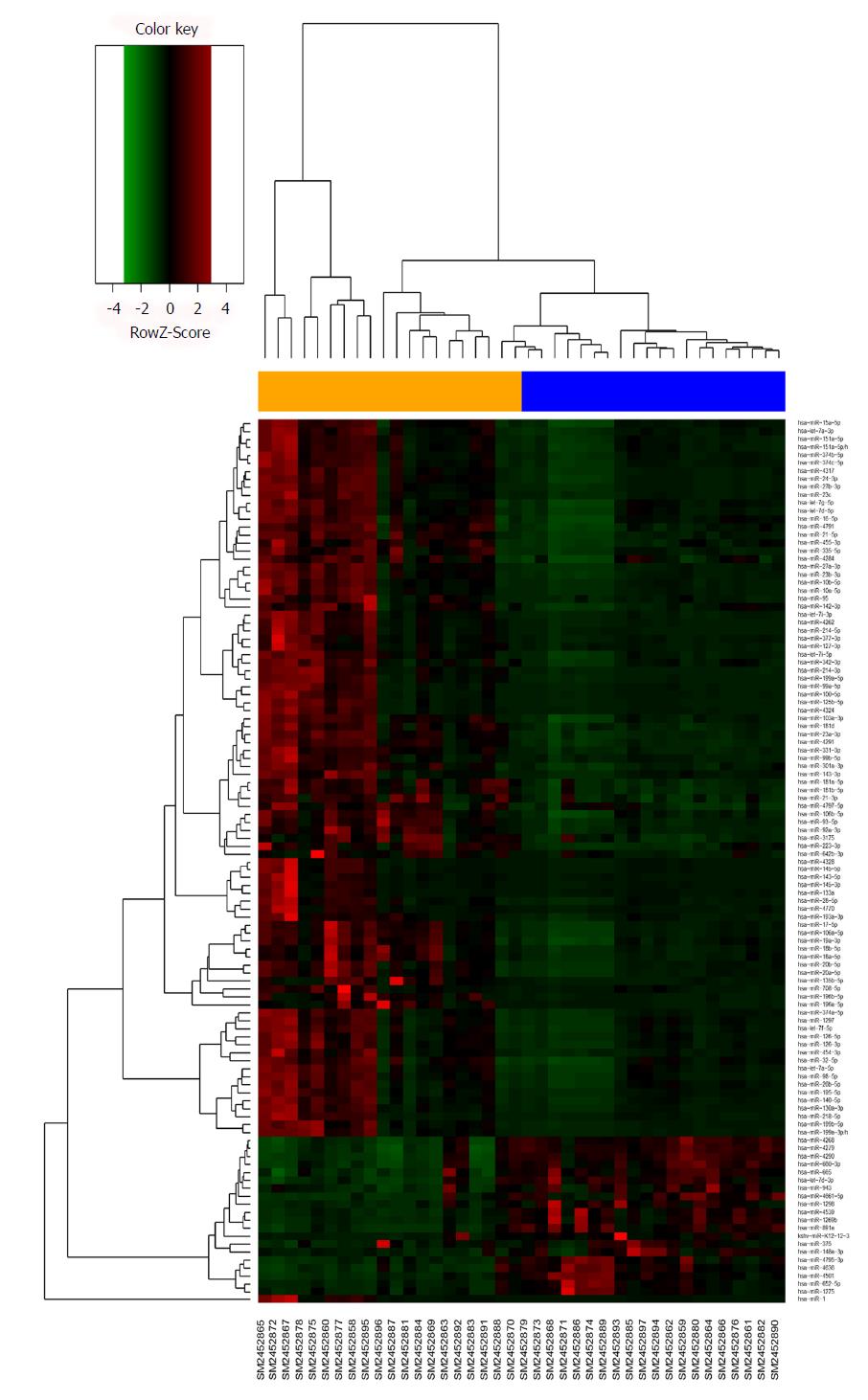
The microarray data of GSE93415, including 20 pairs of GC and adjacent normal tissue samples, were obtained from the NCBI-GEO database. After applying cut-off criteria of P < 0.05 and fold change > 2.0, a total of 110 DEMs were identified between GC and adjacent normal tissues (Table 1). The results of 19 downregulated miRNAs and 91 upregulated miRNAs are displayed in the volcano plot (Figure 1). A heat map of hierarchic cluster analysis showed that DEMs could be discriminated between GC and normal tissues (Figure 2).
| Upregulated DEMs1 | P value | Downregulated DEMs | P value |
| hsa-miR-199a-3p/hsa-miR-199b-3p | 7.10E-05 | hsa-miR-652-5p | 0.000135 |
| hsa-miR-125b-5p | 2.99E-05 | hsa-miR-1269b | 1.13E-09 |
| hsa-miR-199a-5p | 7.65E-07 | hsa-miR-665 | 2.86E-06 |
| hsa-miR-223-3p | 7.49E-06 | hsa-miR-375 | 0.003304 |
| hsa-miR-196a-5p | 3.22E-07 | hsa-miR-4501 | 1.64E-06 |
| hsa-miR-27a-3p | 0.000112 | hsa-miR-4279 | 1.94E-10 |
| hsa-miR-23b-3p | 4.71E-05 | hsa-miR-943 | 2.46E-05 |
| hsa-miR-21-5p | 1.36E-05 | hsa-miR-148a-3p | 1.53E-05 |
| hsa-miR-100-5p | 0.000197 | hsa-miR-1275 | 0.000349 |
| hsa-miR-20a-5p | 0.000102 | hsa-miR-4290 | 8.91E-11 |
| hsa-miR-23a-3p | 5.15E-10 | hsa-miR-4268 | 9.16E-11 |
| hsa-miR-1 | 0.002694 | hsa-miR-891a | 7.29E-10 |
| hsa-miR-214-3p | 1.23E-08 | hsa-miR-4795-3p | 3.07E-05 |
| hsa-miR-10a-5p | 2.33E-05 | hsa-miR-1298 | 3.00E-06 |
| hsa-miR-135b-5p | 1.04E-05 | hsa-miR-660-3p | 1.75E-08 |
| hsa-miR-99a-5p | 0.001109 | hsa-miR-4661-5p | 9.22E-08 |
| hsa-miR-20b-5p | 0.000365 | hsa-miR-4539 | 2.37E-09 |
| hsa-miR-199b-5p | 0.000291 | hsa-let-7d-3p | 1.10E-08 |
| hsa-miR-10b-5p | 1.83E-05 | hsa-miR-4636 | 1.94E-06 |
| hsa-miR-27b-3p | 1.95E-05 | ||
| hsa-miR-126-3p | 0.004079 | ||
| hsa-miR-130a-3p | 0.000442 | ||
| hsa-miR-142-3p | 0.002661 | ||
| hsa-miR-4291 | 1.12E-09 | ||
| hsa-miR-24-3p | 6.14E-06 | ||
| hsa-let-7a-5p | 0.0059 | ||
| hsa-miR-145-5p | 0.00066 | ||
| hsa-miR-17-5p | 3.84E-05 | ||
| hsa-miR-143-5p | 6.07E-05 | ||
| hsa-let-7f-5p | 0.001364 | ||
| hsa-miR-4328 | 0.001281 | ||
| hsa-miR-4324 | 3.91E-05 | ||
| hsa-miR-145-3p | 0.000389 | ||
| hsa-miR-143-3p | 0.006402 | ||
| hsa-miR-95 | 0.000106 | ||
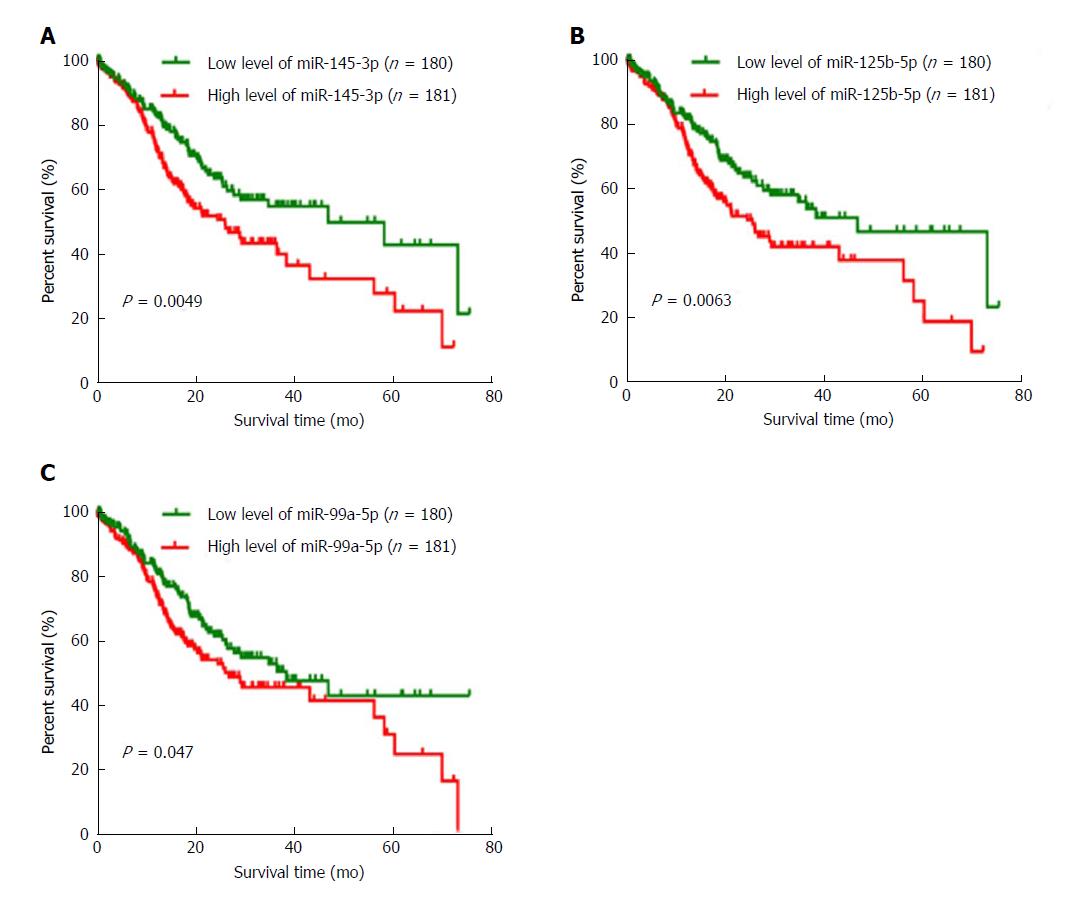
To identify the DEMs which could be used to predict the overall survival of GC patients, we collected 361 samples from TCGA to assess the relationship between DEMs and the overall survival of GC patients. The patients’ clinical characteristics including age at diagnosis, gender, race, TNM stage, and histologic grade are shown in Table 2. By using a log-rank test and Kaplan-Meier curve, we found that three DEMs (miR-145-3p, miR-125b-5p, and miR-99a-5p) were negatively associated with overall survival (Figure 3). The association analysis between the three DEMs and clinical characteristics indicated that miR-145-3p, miR-125b-5p, and miR-99a-5p were all significantly associated with histologic grade (P < 0.05). The detailed results are shown in Table 3.
| Variables | Case, n (%) |
| Age at diagnosis (yr) | |
| < 60 | 113 (31.3) |
| ≥ 60 | 248 (68.7) |
| Gender | |
| Male | 241 (66.8) |
| Female | 120 (33.2) |
| T stage | |
| T1 + T2 | 88 (24.4) |
| T3 + T4 | 273 (75.6) |
| Histologic grade | |
| G1 + G2 | 134 (37.1) |
| G3 + G4 | 227 (62.9) |
| Race | |
| White | 234 (64.8) |
| Asian | 83 (23.0) |
| Black or African American | 12 (3.3) |
| NA | 32 (8.9) |
| Pathologic stage | |
| I | 44 (12.2) |
| II | 117 (32.4) |
| III | 162 (44.9) |
| IV | 30 (8.3) |
| NA | 8 (2.2) |
| Node status | |
| N0 | 111 (30.7) |
| N1-3 | 249 (69.0) |
| NA | 1 (0.3) |
| Metastasis | |
| M0 | 325 (90.0) |
| M1 | 22 (6.1) |
| Mx | 14 (3.9) |
| Variables | miR-145-3p expression | P value | miR-125b-5p expression | P value | miR-99a-5p expression | P value | |||
| Low | High | Low | High | Low | High | ||||
| Age at diagnosis (yr) | |||||||||
| < 60 | 51 | 62 | 0.225 | 42 | 71 | 0.001a | 49 | 64 | 0.096 |
| ≥ 60 | 129 | 119 | 138 | 110 | 131 | 117 | |||
| T stage | |||||||||
| T1 + T2 | 40 | 48 | 0.342 | 51 | 37 | 0.081 | 46 | 42 | 0.603 |
| T3 + T4 | 140 | 133 | 129 | 144 | 134 | 139 | |||
| N stage | |||||||||
| N0 | 58 | 53 | 0.567 | 57 | 54 | 0.731 | 57 | 54 | 0.731 |
| N1-3 | 119 | 124 | 120 | 123 | 120 | 123 | |||
| M stage | |||||||||
| M0 | 163 | 162 | 0.670 | 165 | 160 | 0.191 | 164 | 161 | 0.386 |
| M1 | 10 | 12 | 8 | 14 | 9 | 13 | |||
| Histologic grade | |||||||||
| G1 + G2 | 77 | 57 | 0.026a | 87 | 47 | < 0.001a | 80 | 54 | 0.004a |
| G3 + G4 | 103 | 124 | 93 | 134 | 100 | 127 | |||
| Pathologic stage | |||||||||
| I + II | 81 | 80 | 0.876 | 86 | 75 | 0.221 | 80 | 81 | 0.954 |
| III + IV | 95 | 97 | 90 | 102 | 96 | 96 | |||
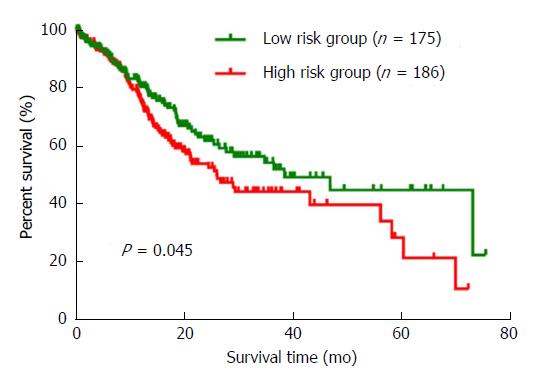
We ranked the three DEMs by the median of expression and then scored each GC patient in accordance with high or low-level expression. A risk grade was defined by the total scores. As a result, all the 361 GC patients were sorted into a high or low risk group. A survival analysis with the Kaplan-Meier method and log-rank test was conducted. The results indicated that the overall survival between the high risk and low risk groups was significantly different (P = 0.045). Interestingly, compared to patients in the high risk group, the low risk patients tended to have a better prognosis (Figure 4). Furthermore, we performed univariate and multivariate Cox regression analyses to verify the prognostic role of the three-DEM signature according to clinical features. The univariate analysis showed that pathologic stage (HR = 1.825, P < 0.001), T stage (HR = 1.864, P = 0.006), N stage (HR = 2.005, P = 0.001), and the three-DEM signature (HR = 1.422, P = 0.039) were significantly associated with the prognostic outcome of GC patients. The multivariate analysis revealed that T stage (HR = 1.623, P = 0.044) and the three-DEM signature (HR = 1.451, P = 0.032) were all independent factors in predicting the prognosis of GC patients (Table 4).
| Variables | Univariate analysis | Multivariate analysis | ||
| HR (95%CI) | P value | HR (95%CI) | P value | |
| Age at diagnosis (≥ 60 vs < 60) | 1.373 (0.948-1.988) | 0.094 | 1.642 (1.122-2.401) | 0.011a |
| Pathologic stage (III + IV vs I + II) | 1.825 (1.332-2.499) | < 0.001a | 1.252 (0.812-1.929) | 0.309 |
| T stage (T3 + T4 vs T1 + T2) | 1.864 (1.197-2.902) | 0.006a | 1.623 (1.012-2.603) | 0.044a |
| N stage (N1-2 vs N0) | 2.005 (1.328-3.026) | 0.001a | 1.602 (0.935-2.744) | 0.086 |
| M stage (M1 vs M0) | 1.368 (0.964-1.941) | 0.080 | 1.313 (0.919-1.875) | 0.134 |
| Three-DEM signature (high vs low risk) | 1.442 (1.018-1.988) | 0.039a | 1.451 (1.033-2.040) | 0.032a |
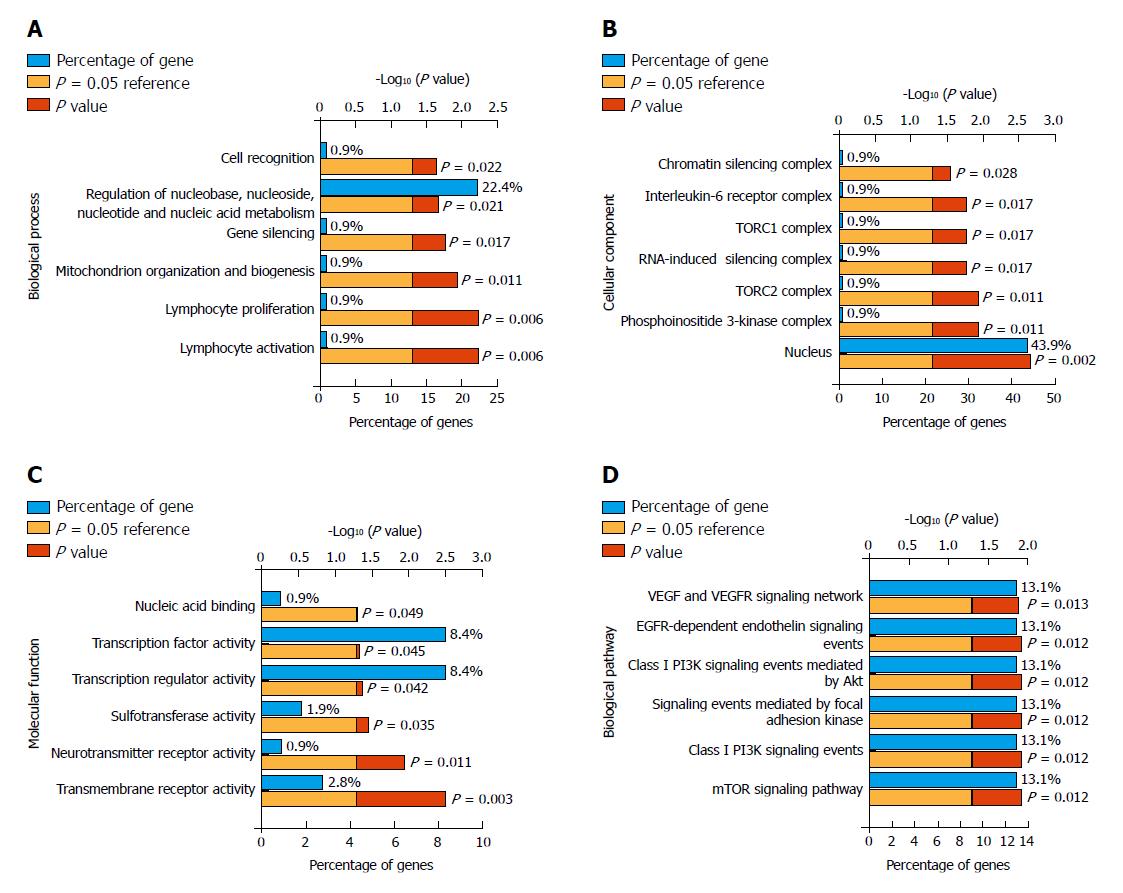
The online target prediction tools TargetScan, miRDB, miRWalk, and DIANA were used to predict the targets genes of miR-145-3p, miR-125b-5p, and miR-99a-5p. We then obtained the consensus genes of each DEM from the four online predictions (Figure 5). As a result, we identified a 108 consensus target genes. Furthermore, we conducted gene enrichment analysis to identify the biological function of common target genes (Figure 6). The Biological Process analysis indicated that the genes were mostly enriched in cell recognition, regulation of nucleic acid, and gene silencing. Cellular Component analysis indicated that genes were enriched in the nucleus, RNA-induced silencing complex, chromatin silencing complex, and phosphoinositide 3-kinase complex. Molecular Function analysis showed that genes were enriched in transmembrane receptor, transcription regulator, and transcription factor activity. Biological Pathways were mainly enriched in the VEGFR, PI3K/Akt, and mTOR signaling pathways.
Due to the reduction in chronic Helicobacter pylori infection and improvement of sanitation, the incidence and mortality rates of GC have declined in recent years[12]. However, there are still almost 460,000 new GC cases and 350000 GC deaths each year in China[13]. The prognosis of GC patients is poor and the five-year survival rate is 5%-20% despite advances in GC therapy[14]. Thus, to improve the clinical treatment and management of GC patients, it is urgent to identify reliable prognostic biomarkers. In this study, we identified a total of 110 DEMs by analyzing the GSE93415 data and discovered that three miRNAs (miR-145-3p, miR-125b-5p, and miR-99a-5p) were negatively associated with overall survival. Additionally, we constructed a three-miRNA signature to predict the prognosis of GC patients.
For decades, a large number of studies have reported that miRNAs can play oncogenic or tumor-suppressing roles in regulating cell biological behavior of cells[15-18]. At present, several miRNAs are known to be useful in the early diagnosis of cancers, including miR-21[19], miR-486[20], miR-24[21], and miR-125a-5p[22]. In addition, miR-191[23], miR-1908[24], miR-200c[25], and miR-217[26] were found to be potential prognostic indictors in cancer. However, these studies only used a single indictor or a limited number of patients for survival analysis. In this study, we identified DEMs by analyzing the array data from the GEO database and found that three highly expressed miRNAs (miR-145-3p, miR-125b-5p, and miR-99a-5p) may be potential prognostic indictors in GC. Kaplan-Meier and Log-rank test survival analysis indicated that the three-miRNA signature can be used to predict the prognosis of GC patients.
We then searched present publications online to compare and test our findings. Chang et al[27] showed that miR-125b-5p was overexpressed in GC patients and promoted invasion and metastasis of GC by targeting STARD13 and NEU1. It was also indicated that miR-125b-5p could be a potential biomarker for predicting prognoses and clinical outcomes in patients with HER2-positive GC that receive trastuzumab treatment[28]. Wu et al[29] found that miR-125b-5p promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways and acts as an independent prognostic factor in GC. Furthermore, Zhang et al[30] demonstrated that miR-99a-5p might function as a novel molecule to regulate cisplatin resistance by directly targeting the calpain small subunit 1 (CAPNS1)-associated pathway in GC. Interestingly, there are no studies describing relations between miR-145-3p and GC. However, in non-small cell lung cancer, miR-145-3p was found to inhibit cancer cell migration and invasion by targeting PDK1 via the mTOR signaling pathway[31]. Moreover, miR-145-3p was also identified to be down-regulated in metastatic castration-resistant prostate cancer and target four molecules which can significantly predict survival in prostate cancer[32]. These results may suggest that miR-145-3p has a complicated effect in different cancers and, in future research, we will investigate the role of miR-145-3p in GC.
Dysregulated genes may participate in tumorigenesis and progression by aberrant signaling pathways. In this study, we predicted the target genes of the three miRNAs and performed gene functional enrichment analysis. The results showed that these target genes were associated with the process of gene silencing and cell recognition, as well as focal adhesion, EGFR, PI3K/Akt, and mTOR signaling pathways. Xu et al[33] suggested that the EGFR-Akt signaling pathway regulates drug resistance in GC patients. The PI3K/Akt pathway was demonstrated to be associated with poor prognosis, tumor progression, and resistance to systematic therapy in many cancers including GC[34,35]. In addition, the PI3K/Akt/mTOR pathway is a key signaling pathway that is reported to be involved in GC[36]. Thus, the results of our functional enrichment analysis are in accordance with present studies.
Above all, we identified a three-miRNA signature for predicting the prognosis of patients with GC and analyzed potential signaling pathways in the development and progression of GC. However, to determine the genesis and development mechanism of GC, more large-scale and systematic investigations are required.
Increasing studies have reported that microRNAs (miRNAs) play an important role in the development and progression of cancers, including gastric cancer (GC). Furthermore, miRNAs can also act as accurate biomarkers in diagnosis and prognosis prediction. In this study, we found that a three-miRNA signature could be used for predicting the prognosis of GC patients and multiple miRNAs together acting as biomarkers may have a stronger reliability in survival prediction.
The worldwide incidence and mortality rates of GC are fairly high. Most of GC patients have been in the advanced stage when diagnosed and endure a poor prognosis. Identifying accurate biomarkers in predicting prognosis of patients is an urgent issue to be solved, so that patients could have an individualized treatment and an improvement in prognosis.
We aimed to identify multiple miRNAs for predicting the prognosis of patients with gastric cancer. In the present study, we found that a three-miRNA (miR-145-3p, miR-125b-5p, and miR-99a-5p) signature could be used for predicting the prognosis of patients with gastric cancer. This objective could be applied to clinical practice and have a guidance role in improving the prognosis of patients with gastric cancer.
We obtained the differentially expressed miRNAs by analyzing a microarray dataset from the Gene Expression Omnibus database with the limma package in R. The Kaplan-Meier method and log-rank test were used for describing the survival curve. The target genes of the three miRNAs (miR-145-3p, miR-125b-5p, and miR-99a-5p) were predicted with the online tools of TargetScan, miRDB, miRWalk, and DIANA. Venn plot was performed to obtain the common target genes from these four online tools. Enrichment analysis was conducted on the consensus genes using the FunRich tool.
In the present study, we found that a three-miRNA (miR-145-3p, miR-125b-5p, and miR-99a-5p) signature could be used for predicting the prognosis of patients with gastric cancer. Multiple miRNAs together acting as prognosis-related biomarkers may have a stronger reliability and this finding could be useful in clinical treatment according to gastric cancer patients with different prognoses. However, the role of miR-145-3p in the tumorigenesis and progression of gastric cancer remains unclear. Thus, this problem remains to be solved in our future study.
Our study identified that the three miRNAs (miR-145-3p, miR-125b-5p, and miR-99a-5p) were up-regulated in gastric cancer patients by analyzing a microarray dataset. Besides, the novel three-miRNA signature could be used for predicting the prognosis of patients with gastric cancer. Multiple miRNAs together acting as prognosis-related biomarkers may have a stronger reliability, so that our finding could be useful in clinical treatment according to gastric cancer patients with different prognoses.
This study provides us with a new insight that multiple miRNAs can be together used for predicting the prognosis of patients with gastric cancer. In order to further confirm the prognostic value of the three-miRNA signature, our future research may focus on exploring the relation between miR-145-3p and gastric cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Kimura A, Matowicka-Karna J S- Editor: Gong ZM L- Editor: Wang TQ E- Editor: Huang Y
| 1. | Piazuelo MB, Correa P. Gastric cáncer: Overview. Colomb Med (Cali). 2013;44:192-201. [PubMed] |
| 2. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25545] [Article Influence: 1824.6] [Reference Citation Analysis (7)] |
| 3. | Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3661] [Cited by in RCA: 3994] [Article Influence: 234.9] [Reference Citation Analysis (0)] |
| 4. | Wang D, Fan Z, Liu F, Zuo J. Hsa-miR-21 and Hsa-miR-29 in Tissue as Potential Diagnostic and Prognostic Biomarkers for Gastric Cancer. Cell Physiol Biochem. 2015;37:1454-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Liu X, Kwong A, Sihoe A, Chu KM. Plasma miR-940 may serve as a novel biomarker for gastric cancer. Tumour Biol. 2016;37:3589-3597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Merhautova J, Demlova R, Slaby O. MicroRNA-Based Therapy in Animal Models of Selected Gastrointestinal Cancers. Front Pharmacol. 2016;7:329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Hou CG, Luo XY, Li G. Diagnostic and Prognostic Value of Serum MicroRNA-206 in Patients with Gastric Cancer. Cell Physiol Biochem. 2016;39:1512-1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 8. | Zhang L, Huang Z, Zhang H, Zhu M, Zhu W, Zhou X, Liu P. Prognostic value of candidate microRNAs in gastric cancer: A validation study. Cancer Biomark. 2017;18:221-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991-D995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4527] [Cited by in RCA: 6787] [Article Influence: 522.1] [Reference Citation Analysis (0)] |
| 10. | Wang Z, Jensen MA, Zenklusen JC. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol. 2016;1418:111-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 435] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 11. | Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597-2601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 968] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 12. | Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030-3044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1946] [Cited by in RCA: 1974] [Article Influence: 103.9] [Reference Citation Analysis (0)] |
| 13. | Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, He J. Report of incidence and mortality in China cancer registries, 2009. Chin J Cancer Res. 2013;25:10-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 167] [Reference Citation Analysis (0)] |
| 14. | Wang Z, Cai Q, Jiang Z, Liu B, Zhu Z, Li C. Prognostic role of microRNA-21 in gastric cancer: a meta-analysis. Med Sci Monit. 2014;20:1668-1674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1981] [Article Influence: 104.3] [Reference Citation Analysis (0)] |
| 16. | Nohata N, Hanazawa T, Kinoshita T, Okamoto Y, Seki N. MicroRNAs function as tumor suppressors or oncogenes: aberrant expression of microRNAs in head and neck squamous cell carcinoma. Auris Nasus Larynx. 2013;40:143-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Chen Y, Fu LL, Wen X, Liu B, Huang J, Wang JH, Wei YQ. Oncogenic and tumor suppressive roles of microRNAs in apoptosis and autophagy. Apoptosis. 2014;19:1177-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 18. | Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer. 2011;47:1127-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 19. | Zeng Z, Wang J, Zhao L, Hu P, Zhang H, Tang X, He D, Tang S, Zeng Z. Potential role of microRNA-21 in the diagnosis of gastric cancer: a meta-analysis. PLoS One. 2013;8:e73278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 20. | Li W, Wang Y, Zhang Q, Tang L, Liu X, Dai Y, Xiao L, Huang S, Chen L, Guo Z. MicroRNA-486 as a Biomarker for Early Diagnosis and Recurrence of Non-Small Cell Lung Cancer. PLoS One. 2015;10:e0134220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 21. | Fang Z, Tang J, Bai Y, Lin H, You H, Jin H, Lin L, You P, Li J, Dai Z. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. 2015;34:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 22. | Wang P, Yang D, Zhang H, Wei X, Ma T, Cheng Z, Hong Q, Hu J, Zhuo H, Song Y. Early Detection of Lung Cancer in Serum by a Panel of MicroRNA Biomarkers. Clin Lung Cancer. 2015;16:313-319.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 23. | Gao X, Xie Z, Wang Z, Cheng K, Liang K, Song Z. Overexpression of miR-191 Predicts Poor Prognosis and Promotes Proliferation and Invasion in Esophageal Squamous Cell Carcinoma. Yonsei Med J. 2017;58:1101-1110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Teng C, Zheng H. Low expression of microRNA-1908 predicts a poor prognosis for patients with ovarian cancer. Oncol Lett. 2017;14:4277-4281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Si L, Tian H, Yue W, Li L, Li S, Gao C, Qi L. Potential use of microRNA-200c as a prognostic marker in non-small cell lung cancer. Oncol Lett. 2017;14:4325-4330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Yang J, Zhang HF, Qin CF. MicroRNA-217 functions as a prognosis predictor and inhibits pancreatic cancer cell proliferation and invasion via targeting E2F3. Eur Rev Med Pharmacol Sci. 2017;21:4050-4057. [PubMed] |
| 27. | Chang S, He S, Qiu G, Lu J, Wang J, Liu J, Fan L, Zhao W, Che X. MicroRNA-125b promotes invasion and metastasis of gastric cancer by targeting STARD13 and NEU1. Tumour Biol. 2016;37:12141-12151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 28. | Sui M, Jiao A, Zhai H, Wang Y, Wang Y, Sun D, Li P. Upregulation of miR-125b is associated with poor prognosis and trastuzumab resistance in HER2-positive gastric cancer. Exp Ther Med. 2017;14:657-663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Wu JG, Wang JJ, Jiang X, Lan JP, He XJ, Wang HJ, Ma YY, Xia YJ, Ru GQ, Ma J. MiR-125b promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways in gastric cancer, resulting in a poor prognosis. Gastric Cancer. 2015;18:729-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 30. | Zhang Y, Xu W, Ni P, Li A, Zhou J, Xu S. MiR-99a and MiR-491 Regulate Cisplatin Resistance in Human Gastric Cancer Cells by Targeting CAPNS1. Int J Biol Sci. 2016;12:1437-1447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 31. | Chen GM, Zheng AJ, Cai J, Han P, Ji HB, Wang LL. microRNA-145-3p inhibits non-small cell lung cancer cell migration and invasion by targeting PDK1 via the mTOR signaling pathway. J Cell Biochem. 2018;119:885-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 32. | Goto Y, Kurozumi A, Arai T, Nohata N, Kojima S, Okato A, Kato M, Yamazaki K, Ishida Y, Naya Y. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117:409-420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 33. | Xu H, Miao ZF, Wang ZN, Zhao TT, Xu YY, Song YX, Huang JY, Zhang JY, Liu XY, Wu JH. HCRP1 downregulation confers poor prognosis and induces chemoresistance through regulation of EGFR-AKT pathway in human gastric cancer. Virchows Arch. 2017;471:743-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Lin HL, Yang MH, Wu CW, Chen PM, Yang YP, Chu YR, Kao CL, Ku HH, Lo JF, Liou JP. 2-Methoxyestradiol attenuates phosphatidylinositol 3-kinase/Akt pathway-mediated metastasis of gastric cancer. Int J Cancer. 2007;121:2547-2555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455-7464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 977] [Cited by in RCA: 1047] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 36. | Liu M, Li CM, Chen ZF, Ji R, Guo QH, Li Q, Zhang HL, Zhou YN. Celecoxib regulates apoptosis and autophagy via the PI3K/Akt signaling pathway in SGC-7901 gastric cancer cells. Int J Mol Med. 2014;33:1451-1458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |