Published online Jul 14, 2017. doi: 10.3748/wjg.v23.i26.4712
Peer-review started: January 22, 2017
First decision: March 16, 2017
Revised: April 5, 2017
Accepted: May 19, 2017
Article in press: May 19, 2017
Published online: July 14, 2017
Processing time: 173 Days and 7 Hours
To evaluate the frequency of Helicobacter pylori (H. pylori) CagA antibodies in H. pylori infected subjects and to identify potential histopathological and bacterial factors related to H. pylori CagA-immune response.
Systematic data to H. pylori isolates, blood samples, gastric biopsies for histological and molecular analyses were available from 99 prospectively recruited subjects. Serological profile (anti-H. pylori, anti-CagA) was correlated with H. pylori isolates (cagA, EPIYA, vacA s/m genotype), histology (Sydney classification) and mucosal interleukin-8 (IL-8) mRNA and protein expression. Selected H. pylori strains were assessed for H. pylori CagA protein expression and IL-8 induction in co-cultivation model with AGS cells.
Thirty point three percent of microbiologically confirmed H. pylori infected patients were seropositive for CagA. Majority of H. pylori isolates were cagA gene positive (93.9%) with following vacA polymorphisms: 42.4% vacA s1m1, 23.2% s1m2 and 34.3% s2m2. Anti-CagA-IgG seropositivity was strongly associated with atrophic gastritis, increased mucosal inflammation according to the Sydney score, IL-8 and cagA mRNA expression. VacA s and m polymorphisms were the major determinants for positive (vacA s1m1) or negative (vacA s2m2) anti-CagA serological immune response, which also correlated with the in vitro inflammatory potential in AGS cells. In vitro co-cultivation of representative H. pylori strains with AGS cells confirmed functional CagA translocation, which showed only partial correlation with CagA seropositivity in patients, supporting vacA as major co-determinant of the immune response.
Serological immune response to H. pylori cagA+ strain in H. pylori infected patients is strongly associated with vacA polymorphism, suggesting the crucial role of bacterial factors in immune and clinical phenotype of the infection.
Core tip:Helicobacter pylori (H. pylori) related diseases are commonly associated with cagA+ strains, although seropositivity against CagA varies among different studies. In this prospective study, we evaluated potential factors related to the H. pylori CagA-immune response. We show that anti-CagA-IgG seropositivity was strongly associated with histopathological and inflammatory factors. Most importantly, we identified H. pylori vacA polymorphism as one of the main determinants of immune response to CagA and inflammatory potential of H. pylori strains ex vivo and in vitro. Our data support the crucial role of bacterial factors that co-determine the complex interaction with H. pylori and define the immune and clinical phenotypes of the infection.
- Citation: Link A, Langner C, Schirrmeister W, Habendorf W, Weigt J, Venerito M, Tammer I, Schlüter D, Schlaermann P, Meyer TF, Wex T, Malfertheiner P. Helicobacter pylori vacA genotype is a predominant determinant of immune response to Helicobacter pylori CagA. World J Gastroenterol 2017; 23(26): 4712-4723
- URL: https://www.wjgnet.com/1007-9327/full/v23/i26/4712.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i26.4712
Infection with Helicobacter pylori (H. pylori) causes chronic inflammation of the gastric mucosa with progression to severe complications in a subset of patients[1-3]. The determinants for the magnitude of inflammation and progression to complication include H. pylori with its bacterial virulence factors, host genetic background and environmental factors. H. pylori virulence factors facilitate colonization (urease, flagella and catalase) and induce inflammation (OipA, NapA, DupA, IceA, VacA and CagA) of the gastric mucosa[4,5]. CagA and VacA are the most relevant pro-inflammatory factors and are closely related to peptic ulcer disease (PUD) as well as gastric cancer (GC)[6-9].
CagA is the principal protein encoded in the complex of the cytotoxin associated gene pathogenicity island (cag PAI), which is shuttled from H. pylori into gastric epithelial cells through the type IV bacterial secretion system[7,8]. Intracellularly, CagA undergoes tyrosine phosphorylation by Src and Abl kinases to interact with several host proteins, influence their activity and subsequently alter morphological properties of the host cells[10-13]. CagA protein stimulates expression of inflammatory cytokine interleukin-8 (IL-8) in gastric epithelial cells by activating nuclear factor-κB and leads to increased inflammation of the gastric mucosa[14]. Overall, H. pylori cagA+ strains are associated with an increased risk of gastric cancer compared to cagA- strains[15]. The oncogenic role of CagA is further supported by in vivo experiments in mice, where transgenic cagA expression in stomach leads to gastric epithelial hyperplasia, adenocarcinoma, myeloid leukemia and B-cell lymphoma[16].
One of the interesting features related to CagA is the induction of a systemic immune response to CagA and this in fact led to the discovery of this protein[17]. Infection with cagA+ strains and serological detection of anti-CagA antibodies have been associated with increased risk for PUD as well as for GC[18,19]. A meta-analysis of 16 studies concludes that seropositivity for anti-CagA-IgG is associated with a 2.87-fold higher risk for gastric cancer development[20]. In earlier studies, Ando et al[21] found a significant correlation between anti-CagA-IgG and IL-8 expression in biopsy culture supernatant and described an association of anti-CagA-IgG with increased neutrophil infiltration and H. pylori density. Therefore, it has been suggested that screening for the cagA status of H. pylori may provide an additional advantage for identifying patients at high risk for gastric cancer development[20]. However, low levels of anti-CagA-IgG in subjects infected with cagA+ strains have been reported[22,23]. H. pylori IgG seroprevalence in a large study in our center was 44.4%, and proportion anti-CagA-IgG positive was 43.3%[22]. In another prospective study on patients undergoing screening colonoscopy, we observed an even lower proportion (36.6%) of anti-CagA-IgG positivity[23]. In studies performed in various geographic regions of the world the CagA-seropositivity ranges from 35% to 80%[22-24]. The low number of CagA-seropositivity in spite of the high prevalence of H. pylori cagA+ strains has not been explained. At present only few studies addressed this observation, however, systematic data are not yet available[25-27]. In the present prospective study, we aimed to identify the factors related to serological reactivity or immune response to CagA.
In a prospective study 413 patients were recruited between July 2011 and April 2014. Among those, 99 patients (98 patients of European descent) in total fulfilled the inclusion criteria such as microbiologically confirmed H. pylori infection with successful isolation and characterization of H. pylori strains and known H. pylori anti-CagA status (Figure S1). Patients, with current or past history of non-gastric cancers or stomach surgery, acute bleeding, oral anticoagulation, immunosuppressive or antibiotic therapy (within the last 2 wk before entering the study) were excluded. The study was conducted according to the “World Medical Association Declaration of Helsinki - Ethical Principles for Medical Research Involving Human Subjects” and approved by the ethical board of the Otto-von-Guericke University (Study Number 80/11). All patients provided written informed consent. Blood samples were drawn and systematic biopsy protocol was completed during upper GI endoscopy at the Department of Gastroenterology, Hepatology and Infectious Diseases at the Otto-von-Guericke University of Magdeburg, Germany.
During upper GI endoscopy, biopsies from antrum and corpus were collected for microbiology cultivation of H. pylori, rapid urease test (CLOtest, Kimberly Clark, United States), histological assessment and further molecular analyses. Histological evaluation was performed according to the updated Sydney protocol from five biopsies (two from each antrum and corpus and one from incisura angularis)[28,29]. Following fixation, slides were stained with hematoxilin, eosin, PAS and modified Giemsa stain for H. pylori detection. Gastric cancer tumor tissues were characterized according to the International Classification of Diseases for Oncology and Lauren criteria.
Serological assessment of anti-H. pylori IgG and anti-CagA-IgG
Serological assessment for H. pylori was performed using H. pylori IgG ELISA Kit (Biohit, Helsinki, Finland) and CagA IgG ELISA Kit (GENESIS Diagnostics, Cambridgeshire, Great Britain). Both tests exhibited a high sensitivity for detection of H. pylori infection in our region and have been validated in multiple studies in the past[22,23]. All tests were performed according to manufacturer’s instructions with internal and external validation. Cut-off values for positive testing were ≥ 30.0 EIU or ≥ 6.25 U/mL for H. pylori IgG ELISA and CagA IgG ELISA, respectively. To confirm the data on anti-CagA-IgG we performed immunoblot testing using Helicobacter ViraStripe® IgG immunoblot (Viramed Biotech AG, Planegg, Germany). The test result was considered positive if following criteria were fulfilled: quantitative evaluation of the blots using an automated scanning system provided by the manufacturer (positivity values ≥ 80% in comparison to control), and two researchers independently and blinded to results, confirmed the positivity.
Gastric biopsies were collected in 1.5 mL 0.9 vol% isotonic sodium chloride solution (Berlin-Chemie AG, Berlin, Germany) and immediately transported to the Institute of Medical Microbiology for further cultivation. Cultivation and identification of H. pylori was performed as described previously[30]. Positive cultures were harvested in 0.9 vol% isotonic sodium chloride solution, centrifuged at 13.000 rpm for 3 min and cell pellets were stored at -30 °C until further analysis.
Six days before the experiment, frozen stocks of several H. pylori isolates from patients were inoculated on Columbia-agar-based medium that contained 10 vol% washed human erythrocytes and 10 vol% heat inactivated horse serum (purchased from the NRZ, Nationales Referenzzentrum Helicobacter Freiburg, Germany). Bacteria were cultivated under microaerophilic conditions at 37 °C. The strain H. pylori ATCC® BAA-1606™ (BCM300) was cultivated on selective agar plates (bioMérieux, Marcy l’Etoile, France) under the same conditions. After 3 d bacteria were removed into PBS and cultivated on fresh agar plates for another three days under the same conditions. For the experiments, bacteria were re-suspended in PBS (with Ca2+ and Mg2+) and concentration (bacteria/mL) was determined by measuring the optical density (λ = 580 nm). To check bacteria for viability, suspensions were inspected microscopically for motility and shape.
AGS cells (CRL-1739; American Type Culture Collection-ATCC) were maintained in RPMI 1640 (Life Technologies, Carlsbad, CA, United States) with 10% Fetal Calf Serum, 100 U/mL Penicillin, 100 μg/mL streptomycin, and 100 µg/mL gentamycin (PAA, Cölbe, Germany) at 37 °C and 5% CO2. Twenty-four hours prior to infection experiments, cells were seeded in 6 well plates at a concentration of 300000 cells/mL in the same medium as mentioned above. Four hours prior infection, medium was removed, cells were washed twice with PBS without Ca2+ and Mg2+ (Life Technologies, Carlsbad, CA, United States) and fresh antibiotic free medium was added. One well was harvested by trypsination (5 min, 37 °C) and cell number was determined. Cells were infected with H. pylori at a “multiplicity of infection” of 100 for 24 h. Cell culture supernatant was removed, centrifuged at 13.000 rpm for 5 min and transferred into a new reaction tube. After cells were washed twice with PBS, cells were harvested, washed with PBS and cell pellet was stored at -80 °C until further analysis.
DNA extraction of H. pylori was performed using DNA Mini Kit (Qiagen, Hilden, Germany) following manufacturer’s recommendations. Amplification of DNA was done in a T3 Thermocycler machine (Biometra, Goettingen, Germany) with 15 μL HotStar Taq Plus DNA Polymerase Mix (Qiagen, Hilden, Germany), 11.6 μL RNase-free water, 0.2 μL of each forward and reverse primer (50 μmol/L) and 3 μL H. pylori DNA. Seven primer sets were used for the study: cagA, EPIYA, vacA s, vacA m, glmM, cagE and virB11. The primer sequences and size of product are shown in Table S1. The reactions were carried out as follows: enzyme activation at 95 °C for 15 min, 40 cycles of denaturation at 95 °C for 30 s, annealing for 30 s, extension at 72 °C for 1 min followed by final extension at 72 °C for 10 min PCR products were analyzed by agarose gel electrophoresis, ethidium bromide staining and Hyperladder IV (Bioline, Luckenwalde, Germany) as molecular weight marker. E.A.S.Y RH system (Herolab, Wiesloch, Germany) was used for gel imaging.
H. pylori total RNA was extracted using RNeasy Protect Bacteria Reagent and RNeasy Mini Kit (QIAGEN, Hilden, Germany) following manufacturer’s recommendations. Total RNA of gastric specimens and AGS cells was isolated using RNeasy Plus Universal Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s recommendations (without gDNA Eliminator Solution). RNA concentration was determined spectrophotometrically by measuring absorbance at 260/280 nm (Biophotometer, Eppendorf, Hamburg, Germany). cDNA synthesis was performed in a 40 μL reaction volume with 500 ng of total RNA of H. pylori or 1 μg RNA of antrum biopsies. CagA and glmM mRNA of H. pylori and β-actin with IL-8 of gastric tissue and AGS cells was determined with quantitative real-time PCR (qRT-PCR) using the CDX96-Cycler (BioRAD, Munich, Germany). A single 30 μL reaction contained 15 μL QuantiTect SYBR Green PCR Master Mix (QIAGEN, Hilden, Germany), 13.4 μL RNase-free water, 0.2 μL of each forward and reverse primer (50 μmol/L) and 1.2 μL H. pylori or antrum cDNA. For qRT-PCR programs see above (qualitative PCR program). Annealing temperature and primers are shown in supplementary data (Table S1). Quality of qRT-PCR products was verified by melt curve analysis and agarose gel electrophoresis (see above). Expression data were analyzed using the 2-ΔCt method.
Cell pellets were mixed with 2x Laemmli buffer (4% SDS, 20% glycerol, 120 mmol/L Tris-Cl (pH 6.8) and 0.02% bromphenol blue) and boiled for 10 min at 95 °C. Thereafter, samples were separated using 10% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. Membranes were blocked with TBS buffer and incubated with antibodies as previously described[31].
Interleukin 8 (IL-8) concentration in AGS co-culture supernatants was determined with quantitative sandwich enzyme-linked immunoassay (Quantikine® ELISA, R and D Systems, Abingdon, United Kingdom) according to manufacturer´s recommendations. Results are displayed in pg/mL.
Statistical analysis was performed using GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA, United States). All data are presented as mean ± SD. χ2 test and Fisher´s exact test were used for contingency tables. The Mann-Whitney U-test and the Kruskal-Wallis analyses of variance were used to analyze the statistical significance for two unpaired groups or multiple groups, respectively. Post hoc analyses were performed using Dunn’s multiple comparison tests. Correlation analyses were performed using Spearman’s test. Two-sided P-values < 0.05 were considered as statistically significant.
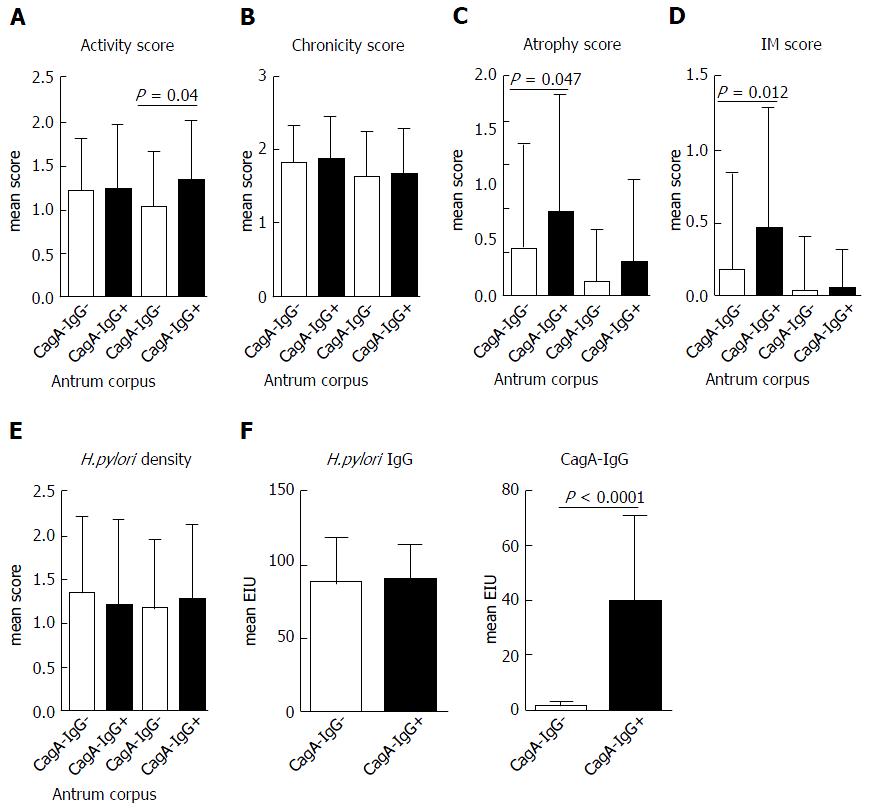
From 99 patients with successful cultivation of H. pylori from the stomach, 30 (30.3%) patients had positive anti-CagA-IgG serology. First, we questioned if CagA-IgG seropositive and seronegative groups may have a difference in clinical phenotype. Clinical and demographical data are presented in Table 1. Among different histological conditions, corpus predominant gastritis and chronic atrophic gastritis were more frequently found in the group of patients with seropositivity for anti-CagA-IgG. More patients with chronic non-active gastritis or patients without any inflammation were found in the anti-CagA-IgG negative group, suggesting the weaker Inflammation related to H. pylori infection. With further focus on the clinical phenotype, we observed a slightly higher polymophonuclear neutrophil infiltration in corpus and a more severe atrophy with intestinal metaplasia in antrum of patients with anti-CagA-IgG based on the mean Sydney Scores for corpus and antrum separately (Figure 1). No difference in H. pylori-IgG antibody titer or H. pylori density was found histologically between those groups.
| Total | H. pylori + CagA-IgG- | H. pylori + CagA-IgG+ | P value | |
| Total | 99 | 69 (69.7) | 30 (30.3) | |
| Gender | NS | |||
| Female | 72 | 51 (73.9) | 21 (70) | |
| Male | 27 | 18 (26.1) | 9 (30) | |
| Age | ||||
| mean ± SD | 54.1 ± 14.1 | 53.7 ± 13.7 | 55.1 ± 15.0 | NS |
| H. pylori status | ||||
| Anti-H.pylori-IgG+ | 93 | 64 (92.8) | 29 (96.7) | NS |
| Anti-CagA-IgG+ | 30 | - | 30 (100) | |
| mean CagA-IgG EIU | 1.5 ± 1.7 | 39.6.5 ± 31.1 | < 0.0001 | |
| Culture+ | 99 | 69 (100) | 30 (100) | NS |
| Histology+ | 79 | 55 (79.7) | 24 (80) | NS |
| Clinical phenotype | ||||
| Chronic active Gastritis (any severity) | 92 | 63 (91.3) | 29 (96.7) | NS |
| Chronic non-active Gastritis (grade > 2)1 | 7 | 6 (8.7) | 1 (3.3) | NS |
| Corpus predominant gastritis | 5 | 0 (0) | 5 (16.7) | 0.0020 |
| Antrum-/pangastritis | 87 | 63 (91.3) | 24 (80) | NS |
| Chronic atrophic gastritis (any severity) | 46 | 25 (36.2) | 21 (70) | 0.0023 |
| Chronic atrophic gastritis (> 2/3)1 | 28 | 14 (20.3) | 14 (46.7) | 0.014 |
| Intestinal metaplasia (any) | 23 | 12 (17.4) | 11 (36.7) | 0.068 |
| Gastric cancer | 6 | 2 (2.9) | 4 (13.3) | 0.056 |
| PUD or MALT-Lymphoma (any) | 5 | 4 (5.8) | 1 (3.3) | NS |
| Normal mucosa (no PMNs and ≤ 1 chronicity)1 | 4 | 4 (5.8) | 0 (0) | NS |
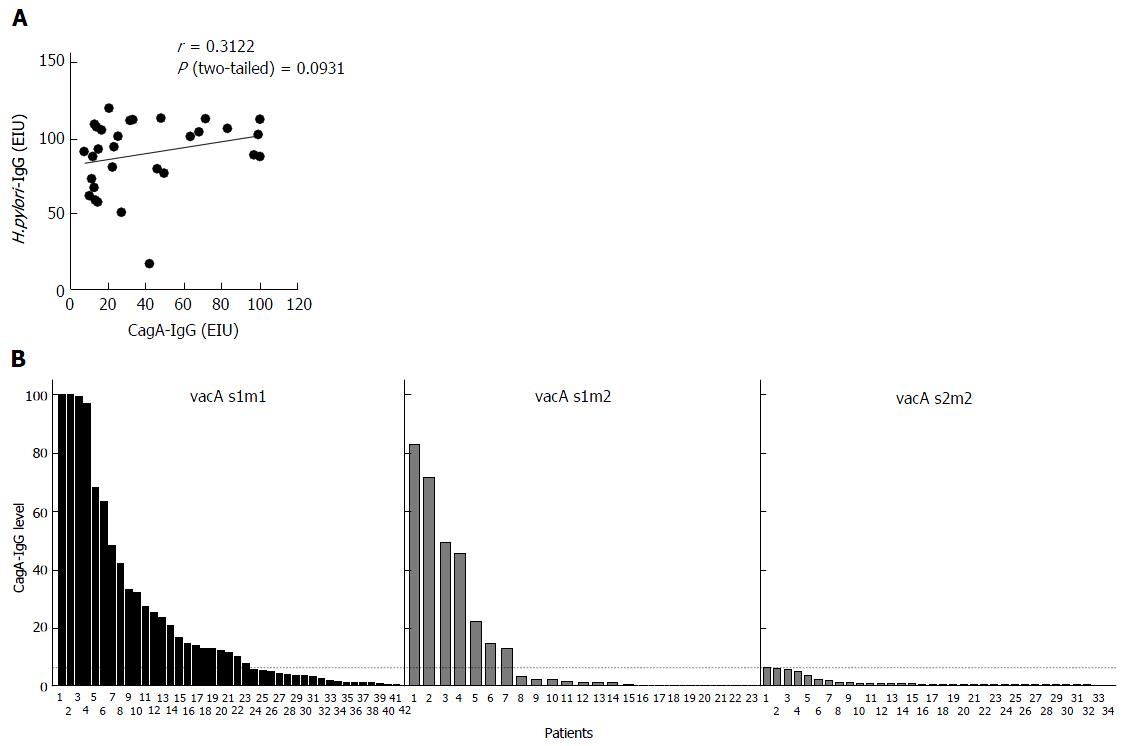
It has been previously suggested that immune response to CagA may be dependent on H. pylori strain characteristics and its virulence factors. As expected all six patients in CagA-IgG- group had cagA-strains. All strains from patients with CagA-IgG+ had cagA+ strains (Table 2). To evaluate if studied patients show immune response to H. pylori we compared CagA-IgG in both groups. We found that seropositivity against H. pylori was present in most of cases in 64 (92.8%) and 29 (96.7%) patients without and with anti-CagA-IgG, respectively, suggesting that the majority of patients are immunologically capable of showing the serological response to H. pylori or its virulence factors. Correlation analyses between the H.pylori-IgG and CagA-IgG titers did not reveal any correlation (Figure 2A).
| Total | H. pylori +CagA-IgG- | H. pylori +CagA-IgG+ | P value | |
| Total | 99 | 69 (69.7) | 30 (30.3) | |
| cagA gene1 | NS | |||
| Positive | 93 | 63 (91.3) | 30 (100) | |
| Negative | 6 | 6 (8.7) | 0 | |
| cagA mRNA2 | 87 | 60 | 27 | < 0.0001 |
| Positive | 61 | 34 (56.7) | 27 (100) | |
| Negative | 26 | 26 (43.3) | 0 (0) | |
| EPIYA motifs | NS | |||
| Negative | 5 | 5 (7.2) | 0 | |
| AB | 4 | 2 (2.9) | 2 (6.7) | |
| ABC | 33 | 21 (30.4) | 12 (40) | |
| ABCC | 7 | 6 (8.7) | 1 (3.3) | |
| ABCCC | 2 | 1 (14.5) | 1 (3.3) | |
| Mixed | 48 | 34 (49.3) | 14 (46.7) | |
| VacA-IgG1,3 | 8 (13.1) | 5 (18.5) | NS | |
| vacA subtype1 | ||||
| s1 | 65 | 35 (50.7) | 30 (100) | < 0.0001 |
| s2 | 34 | 34 (49.3) | 0 | |
| m1 | 42 | 19 (27.5) | 23 (76.7) | < 0.0001 |
| m2 | 57 | 50 (72.5) | 7 (23.3) | |
| s1m1 | 42 | 19 (27.5) | 23 (76.7) | < 0.0001 |
| s1m2 | 23 | 16 (23.2) | 7 (23.3) | |
| s2m2 | 34 | 34 (49.3) | 0 |
Next, we speculated that CagA immune response may be further dependent on successful transcription of cagA mRNA. All strains in CagA-IgG+ group showed moderate or high cagA mRNA expression. At the same time 34 (56.7%) patients of the CagA-IgG-group had also positive cagA mRNA expression. We questioned if differences in EPIYA motifs or a missing Type IV secretion system could have an impact on production of CagA-IgG. A large proportion of the patients had an evidence for H. pylori with mixed EPIYA motifs and no specific differences were observed among CagA-IgG positive and negative groups (Table 2). As a surrogate for the presence of cagA pathogenicity island and type IV secretion system, we examined cagE (cagPAI marker) and virB11 (T4SS marker) expression in 54 patients of the cagA+ and CagA IgG negative group. CagE was detectable in all tested H. pylori isolates, while only one strain was negative for virB11 (data not shown) further excluding the potentially missing T4SS.
It is well known that VacA and CagA are the main pro-inflammatory bacterial factors. It has been earlier hypothesized that vacA polymorphism may also be related to CagA seropositivity[25]. As shown in Table 2, all of the strains from patients with immune response had H. pylori with vacA s1 subtype (with m1 76.7% and m2 23.3%). None of the patients with s2m2 showed CagA seropositivity. In support, the level of anti-CagA-IgG were higher and more frequent positive in vacA s1m1 (50%) and vacA s1m2 (36.8%) compared to vacA s2m2 (0%), further suggesting the importance of H. pylori vacA virulence factor in immune response (Figure 2B).
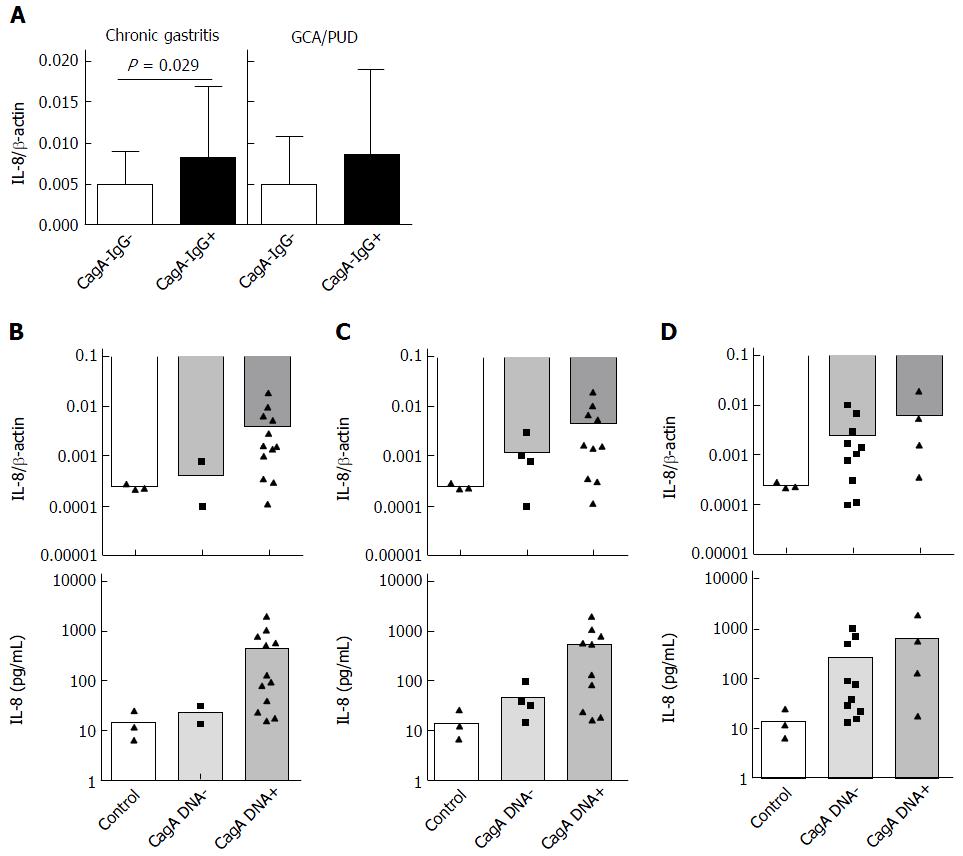
CagA with functional TSS4 is known to induce IL-8 in vitro and in vivo. Having shown increased histological inflammation in subjects with anti-CagA-IgG, we questioned if this may correlate with H. pylori-related cytokine IL-8 in antrum mucosa. Independently of histological phenotype, IL-8 was significantly higher (about 2 fold) in patients with anti-CagA-IgG+ compared to anti-CagA-IgG- patients with CG (0.0082 ± 0.009 vs 0.0048 ± 0.004, P = 0.026) (Figure 3A). This, however, was not the case in mucosa from patients with GC and PUD, although the number of the patients was very small. We observed no correlation between IL-8 expression in mucosa and the level of anti-CagA-IgG. To confirm those strain-dependent observations, we performed in vitro analyses using H. pylori co-culture with AGS cell line. We randomly selected H. pylori strains with different strain characteristics including cagA mRNA expression, CagA-IgG and vacA polymorphisms (Table 3). As expected, cagA+ stains and strains with cagA mRNA expression induced slightly higher IL-8 mRNA expression compared to controls (AGS without H. pylori) and cagA- strains (Figure 3B and C). However, anti-CagA-IgG positivity did not correlate with IL-8 expression suggesting that host serological immunotype/phenotype does not correlate with in vitro potential of H. pylori to induce inflammation (Figure 3D). IL-8 mRNA expression in AGS cells correlated significantly with IL-8 expression in supernatant (Figure S2A), and we observed identical pattern for IL-8 release in supernatant of AGS cells in confirmation of the results (Figure 3B-D).
| Strain characterization | In vitro | ||||||
| ID | cagA DNA | cagA RNA | CagA-IgG | vacA | H.pylori CagA | AGS + H.pylori CagA | AGS + H.pylori p-CagA |
| BCM300 | + | + | s1m1 | + | + | + | |
| 117 | + | + | + | s1m1 | + | + | + |
| 6 | + | + | - | s1m1 | + | + | + |
| 255 | + | + | - | s1m1 | + | + | + |
| 13/1 | + | + | + | s1m1 | + | + | - |
| 46 | + | + | - | s1m1 | + | + | - |
| 89 | + | + | - | s1m2 | + | + | - |
| 424 | + | + | + | s1m2 | - | - | - |
| 21 | + | - | - | s1m1 | - | - | - |
| 321 | + | + | - | s2m2 | - | - | - |
| 374 | + | + | - | s2m2 | - | - | - |
| 342 | + | - | - | s2m2 | - | - | - |
| 314 | - | - | - | s1m2 | - | - | - |
| 450 | - | - | - | s1m2 | - | - | - |
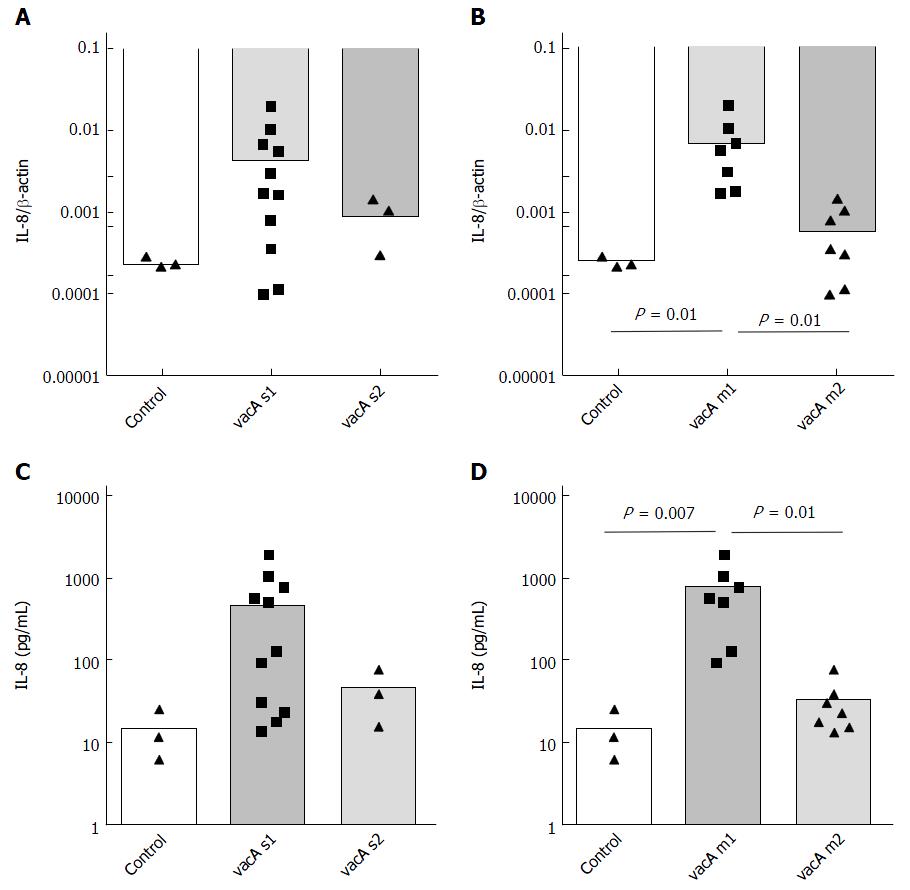
The inflammatory potential of H. pylori cagA+ strains showed relatively high distribution suggesting other bacterial factors potentially responsible for the observation. Therefore, we questioned whether vacA s and m polymorphisms may correlate with inflammatory potential of H. pylori in vitro. Strains with vacA s1 induced higher IL-8 mRNA (Figure 4) and IL-8 expression in supernatant; however, the highest difference was related to vacA m polymorphism with highest values for vacA m1 compared to vacA m2. This data further confirms the highest inflammatory potential defined by IL-8 expression of vacA s1m1 compared to vacA s1m2 or s2m2 (Figure S2B and C).
Having shown that multiple factors may be related to seropositivity to CagA, we questioned if the H. pylori strains indeed a capable of expression of functional CagA protein (including its phosphorylated form) in AGS cells. For this purpose, we performed CagA Western blotting using bacterial pellets and AGS cells co-cultivated with H. pylori (Table 3). As expected, we found that the majority of H. pylori cagA+ strains with vacA s1m1 polymorphism indeed were capable of CagA protein expression independently to anti-CagA-IgG positivity in host (Table 3). This provides an additional level of evidence that anti-CagA-IgG is dependent on various bacterial and probably host factors but may not be useful as a biomarker for lesser pathogenic H. pylori infection.
For the analysis of IgG response against CagA, we used well established ELISA-based method[22,23]. To confirm these results and to further evaluate seropositivity, we performed an independent analysis using Immunoblot based method to evaluate the seropositivity. Helicobacter ViraStripe® IgG Kit includes, besides CagA, also various other Antigen-preparations such as VacA, p90, UreA, etc. Overall, there was a strong correlation between the two tests [r = 0.722 (95%CI: 0.6-0.81), P < 0.0001] (Figure S3A). All samples with positivity in anti-CagA-IgG ELISA test (Omega Genesis) showed very strong signal in immunoblot with values above 200 (Figure S3B). However, there were also several samples with positive signal in immunoblot and low values in ELISA, suggesting that certain samples with anti-CagA-IgG could be probably missed due to methodological issues (Figure S3C). However, the immunoblot-based method (ViraStripe CagA-IgG Blot) was positive in some patients without evidence for past or present H. pylori infection and the lower specificity could be at least in part be the explanation for the higher detection rate (data not shown).
A substantial number of patients infected with H. pylori cagA-positive strains do not develop systemic immune response to CagA. In this study, we performed prospective and systematic analysis of H. pylori and its virulence factors CagA and VacA to find the explanation for the missing CagA-seropositivity. We confirm that the seroprevalence of CagA in unselected population with microbiologically confirmed H. pylori infected patients is low despite the high prevalence of H. pylori cagA+ strains. Following multilevel analyses, we found that among various potential factors vacA polymorphism is the most important factor associated with anti-CagA-IgG seropositivity.
The anti-CagA-seropositivity varies between different regions with highest prevalence in Asian countries and lowest in Europe. While earlier data suggested correlation between cagA gene and seropositivity against CagA, our data showed that only 36%-43% patients had anti-CagA-IgG[22,23,32]. In a recent work from Blaser´s group, the prevalence of anti-CagA-IgG in a large cohort of children in Europe was 32%[33]. The data from those studies confirm the low seropositivity in a European population with seropositivity for anti-CagA-IgG of microbiologically confirmed H. pylori cagA+ infected subjects.
Experience from H. pylori vaccine trials suggests that an immune response to CagA is a common event. In the phase-I vaccine trials, intramuscular application of CagA, VacA and NAP induced strong systemic immune reactions measured via anti-CagA-IgG[34]. So, basically any contact of inflammatory cells with CagA leads to antibody production in B-lymphocytes following antigen presentation. The failure in CagA presentation may happen during various steps of infection such as defective CagA expression, missing translocation due to T4SS system or missing or low cell death related to H. pylori infection and according low antigen presentation to immune cells. Indeed, the majority of H. pylori strains from subjects with anti-CagA-IgG exhibited mRNA expression in vitro, while a subgroup of bacteria showed no or very low cagA mRNA expression which further correlated with CagA protein expression the in vitro AGC cell model. However, multiple factors related to host and environment (for example low acidity, predisposition to inflammation) may play very important role[35]. Using in vitro model, we confirmed that the inflammatory potential of cagA positive strain was confirmed in vitro using the classical co-cultivation model of AGS cells and using CagA expression analysis in AGS cells (Table 3). To the first, direct analyses of strains with anti-CagA-IgG seropositivity did not reveal significant difference in inflammatory potential measured by IL-8 in vitro, suggesting that other bacterial factors could contribute to immune reaction. Second, co-cultivation analysis using AGC cells confirmed from mRNA expression showing that the multiple H. pylori strains from patients with negative anti-CagA-IgG have fully functional CagA and TSS4 (Table 3).
Increasing evidence highlights the role of vacA polymorphisms in gastric diseases[26]. Assessment of H. pylori vacA and cagA genotypes and serological host response earlier revealed the association with vacA s1[25]. Systematic analyses of vacA subtypes in background of anti-CagA-IgG have revealed crucial dependency of seropositivity on H. pylori vacA s1m1 polymorphism in our cohort. The in vitro data highlight the inflammatory potential of H. pylori strains with vacA s1m1 polymorphism. This observation is further supported by data showing the dependency of apoptotic activity of H. pylori on vacA[36]. This led us to believe that the immune response to cagA may be at least in part triggered by the effect of VacA on the gastric mucosa. Therefore, the amount of inflammation related to cell toxicity and apoptosis through VacA may influence the interaction of cellular CagA with the immune system and ultimately determine the immune response. The interaction between VacA and CagA has been in focus of several recent studies providing evidence for complex interaction and showing that VacA and CagA can counter-regulate or antagonize each other and affect the host-bacteria interaction[37,38].
Whether a host will develop an immune response to an infection may be influenced by multiple factors. The seroprevalence may typically change during the course of infection, however, it only partially true for H. pylori infection that shows relatively similar pattern during the life-time starting with early infection to death. We have previously shown that anti-CagA-IgG seropositivity was similar in different age groups (above or below 30 years) from H. pylori positive subjects[22]. Recently, the similar serological pattern was shown in children, where anti-CagA-IgG was positive in 32% despite the very young age[33]. Based on this observation, we speculate that the initial Infection with H. pylori and according very first contact to CagA may determine the serological status of the host, which will then remain stable through the whole life until H. pylori treatment, disappearance or death. In this regard, the host factors and especially genetic predisposition may play the very important role. Certain host factors such as genetic polymorphism (exp. HLA) have previously been suggested to be associated with susceptibility or resistance to H. pylori infection[39]. Furthermore, nonfunctional TLR1 SNP 602S/S has been associated with a reduced risk of H.pylori-induced gastritis[40]. Also, a genome-wide association study identified an association between TLR1 and H. pylori seroprevalence that could potentially explain the variation[41]. However, TLR1 is not the only candidate gene, and IL1-beta should be also considered as a potential determinant[42].
From the clinical perspective, our data support significant association of H. pylori CagA seropositivity and corpus predominant gastritis, atrophic alterations in gastric mucosa. However, the absence of anti-CagA-IgG does not preclude the infection of individual with the more virulent CagA positive H. pylori strain. Based on the current data, the knowledge of individual anti-CagA-IgG status does not allow any specific prognostic clinically-relevant management in support of existing recommendation[43,44].
One of the limitations of our study is that due to the low number of patients, we were unable to suitably address the host related genetic factors. In the present study, we focused on the systemic anti-CagA-IgG production and the locally produced IgA response may be an interesting target for evaluation. Even though we could correlate CagA-IgG data from two different tests, there still may be some difference related to different techniques[25]. Nevertheless, we observed the best specificity with the ELISA kit while immunoblot although had slightly higher sensitivity, it was also associated with high number of false positive results (data not shown). Furthermore, even though the H. pylori CagA-IgG positive and negative groups were well balanced, the higher number of subjects were female and potential gender specificity cannot be fully excluded.
In summary, we show that seropositivity for CagA in subjects with H. pylori infection is positive in one third of H. pylori infected European population despite the presence of CagA positive strain. The immune response to CagA was associated with various bacterial factors and most importantly with H. pylori vacA gene polymorphisms. Our data support a crucial role of bacterial and probably host-related factors that co-determine the complex interaction with H. pylori and define the immunologic and clinical phenotypes of the infection.
We thank the endoscopy team for their assistance during the study. We thank further the team from Institute of Pathology specifically Prof. Dr. Albert Roessner for support to the study. We are also grateful to Marion Holley, Ursula Stolz (from the GI Research Laboratory of the Department of Gastroenterology, Hepatology and Infectious Diseases), Andrea Carl and Bettina Neitzel (Institute of Medical Microbiology) for their excellent technical assistance during the experimental work in this study.
Helicobacter pylori (H. pylori) -related peptic ulcer disease and gastric adenocarcinoma are commonly associated with cagA+ H. pylori strains. However, seropositivity against CagA varies among different studies with positivity below 50% in multiple studies from Europe.
Infection with H. pylori induces strong and sustained inflammation in mucosa and triggers immune positivity against H.pylori. Although similar immune response to CagA is expected for H. pylori cagA+ strains, the positivity is substantially lower. The mechanism responsible for the seropositivity to CagA is not sufficiently understood. This knowledge may be helpful to identify the factors responsible for the differences in clinical phenotype of H. pylori infection. Furthermore, it may also facilitate the preventive and treatment strategies.
In this well-characterized cohort of patients, we demonstrated a low anti-CagA-IgG positivity in H. pylori infected patients, which was independent to the high rate of H. pylori cagA positive strains. Immune response to H. pylori CagA was strongly associated with atrophic gastritis, increased mucosal inflammation and IL-8 expression. Most importantly, we observed a strong association of anti-CagA positivity to H. pylori vacA s and m polymorphisms, which also correlated with the inflammatory potential in vitro in AGS cell lines. Altogether, our data suggest that H. pylori vacA polymorphism may determine the immune response to CagA through modulation of mucosal inflammation.
These data strengthens the role of H. pylori vacA polymorphisms and immune response to CagA and in H. pylori infection. Whether H. pylori vacA may become a clinical tool for risk stratification of H. pylori-related diseases needs further evaluation.
The results in this manuscript have demonstrated that seropositivity for CagA in subjects with CagA positive H. pylori status is present in one third of H. pylori infected European population. The immune response to CagA is associated with various bacterial factors and most importantly with vacA gene polymorphisms. The data supported that both bacterial and host-related factors determined the complex interaction of H. pylori with the immunologic system and clinical phenotypes of the infection.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Allaix ME, Tian YP S- Editor: Qi Y L- Editor: A E- Editor: Li D
| 1. | Malfertheiner P, Link A, Selgrad M. Helicobacter pylori: perspectives and time trends. Nat Rev Gastroenterol Hepatol. 2014;11:628-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 118] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 2. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3126] [Cited by in RCA: 3179] [Article Influence: 132.5] [Reference Citation Analysis (0)] |
| 3. | Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64:1353-1367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1322] [Cited by in RCA: 1173] [Article Influence: 117.3] [Reference Citation Analysis (0)] |
| 4. | Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11:385-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 476] [Article Influence: 39.7] [Reference Citation Analysis (2)] |
| 5. | Shiota S, Suzuki R, Yamaoka Y. The significance of virulence factors in Helicobacter pylori. J Dig Dis. 2013;14:341-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 6. | Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu Rev Pathol. 2006;1:63-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 410] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 7. | Odenbreit S. Translocation of Helicobacter pylori CagA into Gastric Epithelial Cells by Type IV Secretion. Science. 2000;287:1497-1500. [RCA] [DOI] [Full Text] [Cited by in Crossref: 954] [Cited by in RCA: 965] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 8. | Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 508] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 9. | Müller A. Multistep activation of the Helicobacter pylori effector CagA. J Clin Invest. 2012;122:1192-1195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 755] [Cited by in RCA: 782] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 11. | Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, Neish AS, Collier-Hyams L, Perez-Perez GI, Hatakeyama M. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102:10646-10651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 412] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 12. | Mueller D, Tegtmeyer N, Brandt S, Yamaoka Y, De Poire E, Sgouras D, Wessler S, Torres J, Smolka A, Backert S. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J Clin Invest. 2012;122:1553-1566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 202] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 13. | Tammer I, Brandt S, Hartig R, König W, Backert S. Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology. 2007;132:1309-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 190] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 14. | Brandt S, Kwok T, Hartig R, König W, Backert S. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci USA. 2005;102:9300-9305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 420] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 15. | Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111-2115. |
| 16. | Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, Higashi H, Musashi M, Iwabuchi K, Suzuki M. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105:1003-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 465] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 17. | Apel I, Jacobs E, Kist M, Bredt W. Antibody response of patients against a 120 kDa surface protein of Campylobacter pylori. Zentralbl Bakteriol Mikrobiol Hyg A. 1988;268:271-276. [PubMed] |
| 18. | Blaser MJ, Perez-perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori Strains Possessing cagA Is Associated with an Increased Risk of Developing Adenocarcinoma of the Stomach Infection with Helicobacter pylori Strains Possessing cagA Is Associated with an Increased Risk of Developing Adeno. Cancer Res. 1995;55:2111-2115. |
| 19. | Cover TL, Glupczynski Y, Lage AP, Burette A, Tummuru MK, Perez-Perez GI, Blaser MJ. Serologic detection of infection with cagA+ Helicobacter pylori strains. J Clin Microbiol. 1995;33:1496-1500. [PubMed] |
| 20. | Huang JQ, Zheng GF, Sumanac K, Irvine EJ, Hunt RH. Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology. 2003;125:1636-1644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 380] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 21. | Ando T, Perez-Perez GI, Kusugami K, Ohsuga M, Bloch KC, Blaser MJ. Anti-CagA immunoglobulin G responses correlate with interleukin-8 induction in human gastric mucosal biopsy culture. Clin Diagn Lab Immunol. 2000;7:803-809. [PubMed] |
| 22. | Wex T, Venerito M, Kreutzer J, Götze T, Kandulski A, Malfertheiner P. Serological prevalence of Helicobacter pylori infection in Saxony-Anhalt, Germany, in 2010. Clin Vaccine Immunol. 2011;18:2109-2112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Selgrad M, Bornschein J, Kandulski A, Hille C, Weigt J, Roessner A, Wex T, Malfertheiner P. Helicobacter pylori but not gastrin is associated with the development of colonic neoplasms. Int J Cancer. 2014;135:1127-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Yamaoka Y, Kodama T, Kashima K, Graham DY. Antibody against Helicobacter pylori CagA and VacA and the risk for gastric cancer. J Clin Pathol. 1999;52:215-218. [PubMed] |
| 25. | Figueiredo C, Quint W, Nouhan N, van den Munckhof H, Herbrink P, Scherpenisse J, de Boer W, Schneeberger P, Perez-Perez G, Blaser MJ. Assessment of Helicobacter pylori vacA and cagA genotypes and host serological response. J Clin Microbiol. 2001;39:1339-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, Rhead JL, Schiavon S, Guariso G, Ceroti M, Nitti D. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135:91-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 294] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 27. | Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, Atherton JC. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 312] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 28. | Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-1181. [PubMed] |
| 29. | Link A, Schirrmeister W, Langner C, Varbanova M, Bornschein J, Wex T, Malfertheiner P. Differential expression of microRNAs in preneoplastic gastric mucosa. Sci Rep. 2015;5:8270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 30. | Selgrad M, Meissle J, Bornschein J, Kandulski A, Langner C, Varbanova M, Wex T, Tammer I, Schlüter D, Malfertheiner P. Antibiotic susceptibility of Helicobacter pylori in central Germany and its relationship with the number of eradication therapies. Eur J Gastroenterol Hepatol. 2013;25:1257-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 31. | Schlaermann P, Toelle B, Berger H, Schmidt SC, Glanemann M, Ordemann J, Bartfeld S, Mollenkopf HJ, Meyer TF. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65:202-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 177] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 32. | Miehlke S, Go MF, Kim JG, Graham DY, Figura N. Serologic detection of Helicobacter pylori infection with cagA-positive strains in duodenal ulcer, gastric cancer, and asymptomatic gastritis. J Gastroenterol. 1998;33 Suppl 10:18-21. [PubMed] |
| 33. | den Hollander WJ, Holster IL, van Gilst B, van Vuuren a. J, Jaddoe VW V., Hofman a., Perez-Perez GI, Kuipers EJ, Moll H a., Blaser MJ. Intergenerational reduction in Helicobacter pylori prevalence is similar between different ethnic groups living in a Western city. Gut. 2014;1200-1208. [RCA] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Malfertheiner P, Schultze V, Rosenkranz B, Kaufmann SH, Ulrichs T, Novicki D, Norelli F, Contorni M, Peppoloni S, Berti D. Safety and immunogenicity of an intramuscular Helicobacter pylori vaccine in noninfected volunteers: a phase I study. Gastroenterology. 2008;135:787-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 35. | Karita M, Tummuru MK, Wirth HP, Blaser MJ. Effect of growth phase and acid shock on Helicobacter pylori cagA expression. Infect Immun. 1996;64:4501-4507. [PubMed] |
| 36. | Cover TL, Krishna US, Israel DA, Peek RM. Induction of gastric epithelial cell apoptosis by Helicobacter pylori vacuolating cytotoxin. Cancer Res. 2003;63:951-957. [PubMed] |
| 37. | Abreu MT, Peek RM. Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146:1534-1546.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 235] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 38. | Backert S, Tegtmeyer N. the versatility of the Helicobacter pylori vacuolating cytotoxin vacA in signal transduction and molecular crosstalk. Toxins (Basel). 2010;2:69-92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 39. | González CA, Sala N, Capellá G. Genetic susceptibility and gastric cancer risk. Int J Cancer. 2002;100:249-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 234] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 40. | Yang CA, Scheibenbogen C, Bauer S, Kleinle C, Wex T, Bornschein J, Malfertheiner P, Hellmig S, Schumann RR, Hamann L. A frequent Toll-like receptor 1 gene polymorphism affects NK- and T-cell IFN-γ production and is associated with Helicobacter pylori-induced gastric disease. Helicobacter. 2013;18:13-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 41. | Mayerle J, den Hoed CM, Schurmann C, Stolk L, Homuth G, Peters MJ, Capelle LG, Zimmermann K, Rivadeneira F, Gruska S. Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA. 2013;309:1912-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 42. | El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1690] [Cited by in RCA: 1673] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 43. | Malfertheiner P, Megraud F, O‘Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1719] [Cited by in RCA: 1587] [Article Influence: 122.1] [Reference Citation Analysis (5)] |
| 44. | Fock KM, Talley N, Moayyedi P, Hunt R, Azuma T, Sugano K, Xiao SD, Lam SK, Goh KL, Chiba T. Asia-Pacific consensus guidelines on gastric cancer prevention. J Gastroenterol Hepatol. 2008;23:351-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 261] [Article Influence: 15.4] [Reference Citation Analysis (0)] |