Published online Jan 14, 2017. doi: 10.3748/wjg.v23.i2.286
Peer-review started: September 1, 2016
First decision: September 20, 2016
Revised: October 19, 2016
Accepted: November 16, 2016
Article in press: November 16, 2016
Published online: January 14, 2017
Processing time: 136 Days and 16.6 Hours
To understand the interference of carbohydrates absorbance in nucleic acids signals during diagnosis of malignancy using Fourier transform infrared (FTIR) spectroscopy.
We used formalin fixed paraffin embedded colonic tissues to obtain infrared (IR) spectra in the mid IR region using a bruker II IR microscope with a facility for varying the measurement area by varying the aperture available. Following this procedure we could measure different regions of the crypt circles containing different biochemicals. Crypts from 18 patients were measured. Circular crypts with a maximum diameter of 120 μm and a lumen of about 30 μm were selected for uniformity. The spectral data was analyzed using conventional and advanced computational methods.
Among the various components that are observed to contribute to the diagnostic capabilities of FTIR, the carbohydrates and nucleic acids are prominent. However there are intrinsic difficulties in the diagnostic capabilities due to the overlap of major absorbance bands of nucleic acids, carbohydrates and phospholipids in the mid-IR region. The result demonstrates colonic tissues as a biological system suitable for studying interference of carbohydrates and nucleic acids under ex vivo conditions. Among the diagnostic parameters that are affected by the absorbance from nucleic acids is the RNA/DNA ratio, dependent on absorbance at 1121 cm-1 and 1020 cm-1 that is used to classify the normal and cancerous tissues especially during FTIR based diagnosis of colonic malignancies. The signals of the nucleic acids and the ratio (RNA/DNA) are likely increased due to disappearance of interfering components like carbohydrates and phosphates along with an increase in amount of RNA.
The present work, proposes one mechanism for the observed changes in the nucleic acid absorbance in mid-IR during disease progression (carcinogenesis).
Core tip: Techniques like infrared (IR) spectroscopy have been used in different research areas and their potential has been established in the field of biomedicine. However studies connecting the basic spectral data to histological features are very few. The connection between diagnosis of the disease pathology and spectral features often needs a correlation. This study address one such issue pertaining to the varying histology and the spectral signatures in mid IR region by trying to understand the contribution of one type of bio molecules in presence and absence of another using biopsies rather than synthetically created combinations.
- Citation: Sahu RK, Salman A, Mordechai S. Tracing overlapping biological signals in mid-infrared using colonic tissues as a model system. World J Gastroenterol 2017; 23(2): 286-296
- URL: https://www.wjgnet.com/1007-9327/full/v23/i2/286.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i2.286
In fourier transform infrared (FTIR) based diagnosis of cancer and other abnormalities, the signals from nucleic acids form a vital part and occur due to changes in the composition of the DNA[1-5] and RNA. For example, changes occur in DNA composition and structure in biopsies which are routinely used in pathology laboratories. Biopsied tissues are subjected to various procedures like paraffin embedding, deparaffinization with hot xylol and ethanol and thus, many smaller molecules are removed and what remains essentially are the nucleic acids, glycogen, proteins and other macromolecules. It is well known that nucleic acids undergo changes in all types of cancers arising due to the composition of DNA[6] that is altered due to mutations or other effects and the transcription and translation is a consequence of these alterations as well as cause of disease manifestation. Both the nucleic acids, RNA and DNA have been found to absorb in the wave numbers region 900-1300 cm-1 in the mid-infrared (IR) that are of diagnostic potential[2]. However, this region is also the region where carbohydrates and phosphates/phospholipids prominently absorb. Thus, it becomes important to study how the absorbance of nucleic acids is affected by other compounds which are present in the tissues and absorb at overlapping wave numbers.
Many approaches have been undertaken to identify the contribution of nucleic acids by measuring the intensities at wave numbers where nucleic acids absorb[7-10]. However, most of these cases dealt with in vitro conditions where lipids and proteins were not affected by the fixation procedures. Moreover in these cases, the signals were measured from intact cells and not from tissues, while during preparation of sections from biopsies, even the nucleus has a possibility of being sliced, leading to the reality of exposure of the nucleic acids to histopathological processing.
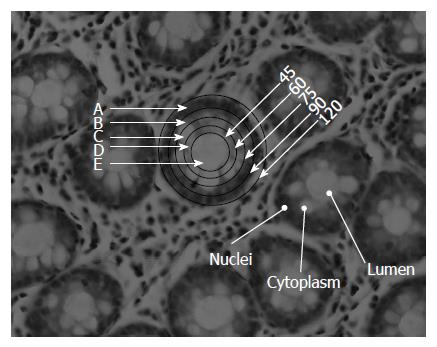
We under took the study in colonic tissues which provide an ideal condition for studying the absorbance bands arising from the nuclei (nucleic acids) by measuring different zones across the transverse section of the crypt. The colonic crypts in transverse section display the epithelial cells arranged around the lumen with the nuclei located at the periphery of the crypt. Thus, by gradually increasing the (circular) aperture of the FTIR-microscope, circular areas of various diameters can be measured that include and exclude the nuclei. In this system at least three different regions can be measured. The lumen that is filled with secretions, consisting of mostly glycoproteins with negligible contribution from lipids and nucleic acids is the inner most and measured by the smallest aperture. The outer circle that includes the nuclei along with other cellular structures gets included in measurements at the maximum aperture. Contribution from the central region that has mostly cellular structures especially the endoplasmic reticulum (ER) and Golgi bodies that would be required for synthesis of the glycoproteins and their packing (rich in lipids due to the folding nature of the ER) as well as organelles filled with mucin is varied with the aperture. We assume that migrations of extra material from the surrounding areas into the crypt during all histopathological procedures is minimum. Since one region of the crypt is measured with respect to another region in the same crypt, using difference of the normalized spectra, the variability in the thickness of the sample, and presence of cell debris and other factors that are usually encountered in biological samples are not expected to affect the study. Similarly while doping of compounds can be used as model system to study interference, the presence of multiple components makes the designing of such doping experiments complicated and may not mimic what occurs in the real biological systems. We utilized the advantage of the microscopic setup that allows the selection of regions of the crypts suitable for the study, and focusing on the various crypt zones as detailed in the methods section. The present work thus allows us to look into the contribution of the nucleus that contains mainly nucleic acids and proteins besides lipids that form the structure around it without much interference from carbohydrates which have a significant overlapping absorbance with these biomolecules in the mid-IR region. We propose at least one mechanism by which the RNA/DNA parameter plays a role in diagnosis of colonic abnormality, namely due to the decreasing interference from carbohydrates/glycoproteins.
Formalin-fixed, paraffin-embedded colonic tissue from biopsies (taken from regions far from the location of cancer) of colon cancer patients were retrieved with their consent from the histopathology files of Soroka University Medical Center, Beer-Sheva (SUMC). Identical sections, 10 μm thick of colonic mucosa were taken[11] and used for FTIR-MSP or for parallel hematoxylin eosin staining. A pathologist examined the tissue histology under microscope to ascertain that there was no influence of stromal material in the measurement sites in the biopsy and the region where measurement on crypts was undertaken was equivalent to what is observed in a normal colonic mucosa with regards to morphology, when the data were being collected for establishing the principle. For the study using samples of IBD patients, measurements were made on crypts that were in the region considered as having normal morphology as well as in regions where the disease was diagnosed by the expert pathologist. All diseased spectra presented are the average of spectra of that category of disease. Sub categories were not evaluated in this study and are a future prospectus.
The measurements were made only on circular crypts that had no contaminating material and were of the required diameter. The crypts were selected such that they represented the measurements along the middle of the height of a crypt as seen by their proximity to the stromal layers. The FTIR measurements on biopsies were performed using the FTIR microscope, IR scope II, with liquid nitrogen cooled mercury-cadmium-telluride detector, coupled to the FTIR spectrometer (Bruker Equinox model 55, OPUS software). After focusing on a crypt, the lowest diameter was selected (45 μm) and the moving stage so adjusted that the entire lumen was covered by the circular area of measurement (area E in Figure 1). Once this was established, the slide was not moved on the microscope stage, but only the aperture was increased to cover the entire crypt regions gradually measuring regions D, C, B and A. Only those crypts where the entire crypt was enclosed in the circular aperture of diameter of 120 μm without elimination of any crypt structure or inclusion of stromal substance, were selected. Then measurements were made from the lowest to the highest diameter aperture sequentially. A background measurement was made separately for each slit diameter and was used to calculate the corrected absorbance for each aperture separately. To achieve a high signal to noise ratio 128 co-added scans were collected in the wavenumber region 600 to 4000 cm-1 for Fourier transform processing. The spectra were baseline corrected using OPUS software and normalized to amide I band in the region 800-1800 cm-1 after smoothing, using the software to further exclude noise arising due to the effect of decreasing apertures. This method was carefully followed to allow a relative comparison of various zones. The intensities obtained at various wave numbers were used for subsequent analysis. Additional normalization methods, such as amide II or vector normalization, have also been tested and gave negligible changes in the results.
We used 19 crypts of human colonic tissues from patients that were of similar morphology, size and spectral features away from any abnormality to establish the initial spectral variation pattern. The spectra were collected from different crypts using different apertures as represented in Figure 1. All the spectra were cut in the region 800-1800 cm-1. The spectra in each region were manipulated separately using OPUS (7) software before computational analyses.
Principal component analysis: Five categories of crypt spectra were considered here according to the microscope aperture used during the measurements. Our aim was to find differences among the spectral categories by analyzing its IR spectrum and established that measurements at each aperture gives rise to spectra which can be grouped together. This indicated that a spatial variation exists in the distribution of biological components across the crypt which can be monitored by FTIR spectroscopy. Each spectral measurement includes D points, which may be represented as one point (or vector) in a D-dimensional space. The number of representing points (i.e., number of different spectra) is denoted by N. In order to find the features characteristic of each category, two methods were considered: principal component analysis (PCA), and linear discriminant analysis (LDA)[12-16]. PCA is was used for dimensionality reduction[12,14,17-20], as well as for a rough classification of the measured data into the five categories. In PCA, the data is represented in a reduced “principal” subspace, defined according to the variance between the measurements, with orthogonal basis vectors ν1, ν2, ..., νk, arranged according to the variance, in increasing order (ν1 is the direction of maximal variance, etc.). The corresponding components are the 1st, 2nd, …, kth principal components (PCs). The data points can also be represented graphically, in a two-dimensional subspace, defined by only two chosen components. This alone can provide some order of differentiation between measurements belonging to different categories. It is usually a rather poor method of differentiation, because it uses only partial knowledge regarding the category features. Nevertheless, it is instructive to investigate the differentiation graphically, even at this early stage of data analysis.
LDA: A more comprehensive method of differentiation is the LDA, a linear model which separates data into categories by defining a linear combination of category features[21]. In our case, the features were the measured data points, but it is computationally better to use the data after having performed the PCA procedure. Data classification is obtained by analyzing a training set (a subset of the total data) in order to examine the model’s efficiency.
The LDA analysis was carried out using the “Leave one out” algorithm (LOO)[12,14]. In the LOO algorithm, the training set contained all of the measured spectra, except for one. The category of this left-out measurement was then calculated, and compared to the known category. This procedure was repeated N times; in each repetition a different measured spectrum was left out. LOO usually applied with small amount of data was used when k = N, the number of data points[22,23].
Cluster analysis: Using cluster analysis[24,25] which is an unsupervised technique, it is possible to cluster the spectra based on the distances between them. For this manner, each spectrum is considered as a place vector in N dimensional space. The results are shown in dendrogram which is a 2D plot of the date for example name of file vs heterogeneity[25,26]. Unsupervised cluster analysis was performed on spectra of the aperture 45 μm and 90 μm samples at different regions 900-1800 cm-1.
Histological section of a colonic biopsy, displaying the anatomy of the crypts in transverse section is shown in Figure 1. Most crypts which are tubular structures have a circular to oval cross section as seen in the figure and vary in their outer and luminal diameters. The most frequent size encountered in crypts is a diameter of 120 μm and a lumen diameter of 45 μm and these crypts were mainly selected for measurements. It is observed that the lumen is largely unstained in the histological section under H&E staining. This zone consists mainly of secretions of glycoprotein in the crypt. On the contrary, Zone A contains nuclei in a peripheral alignment to the lumen is densely stained. Zone B has some portions of nuclei, while Zones C and D mostly contain cell organelles and cytoplasm. The FTIR spectra of Zone E largely reflect contributions from glycoproteins (and other crypt secretions) while the spectra obtained from Zone A reflect absorbance from component of nucleic acids, especially DNA in the presence of other components. Lipids contribute towards absorbance in all zones and minimally to Zone E and can be traced with the absorbance bands between 1700-1800 cm-1 (Esters) and 1345-1469 (CH2, CH3) cm-1. DNA being a prominent contributor to nucleic acid absorbance would be found mostly in Zones A and B while RNA absorbance would be found in Zones A-D. Thus, this system presents a unique opportunity to study contribution of different cellular components in the IR spectra from a variety of de-paraffinized tissue sections under ex vivo conditions.
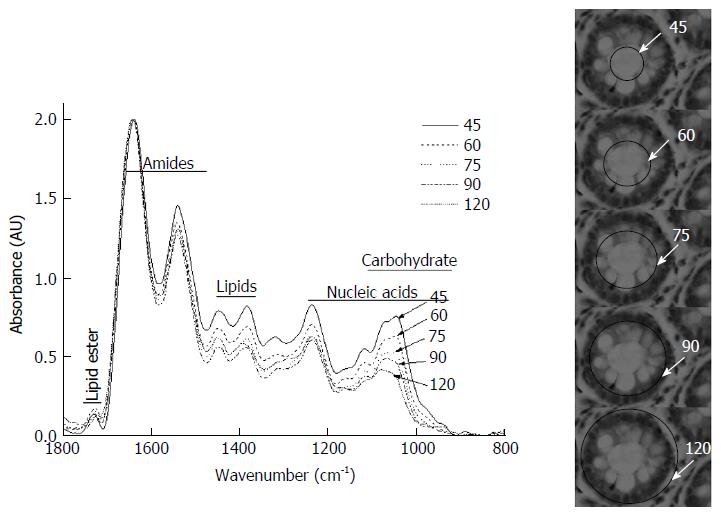
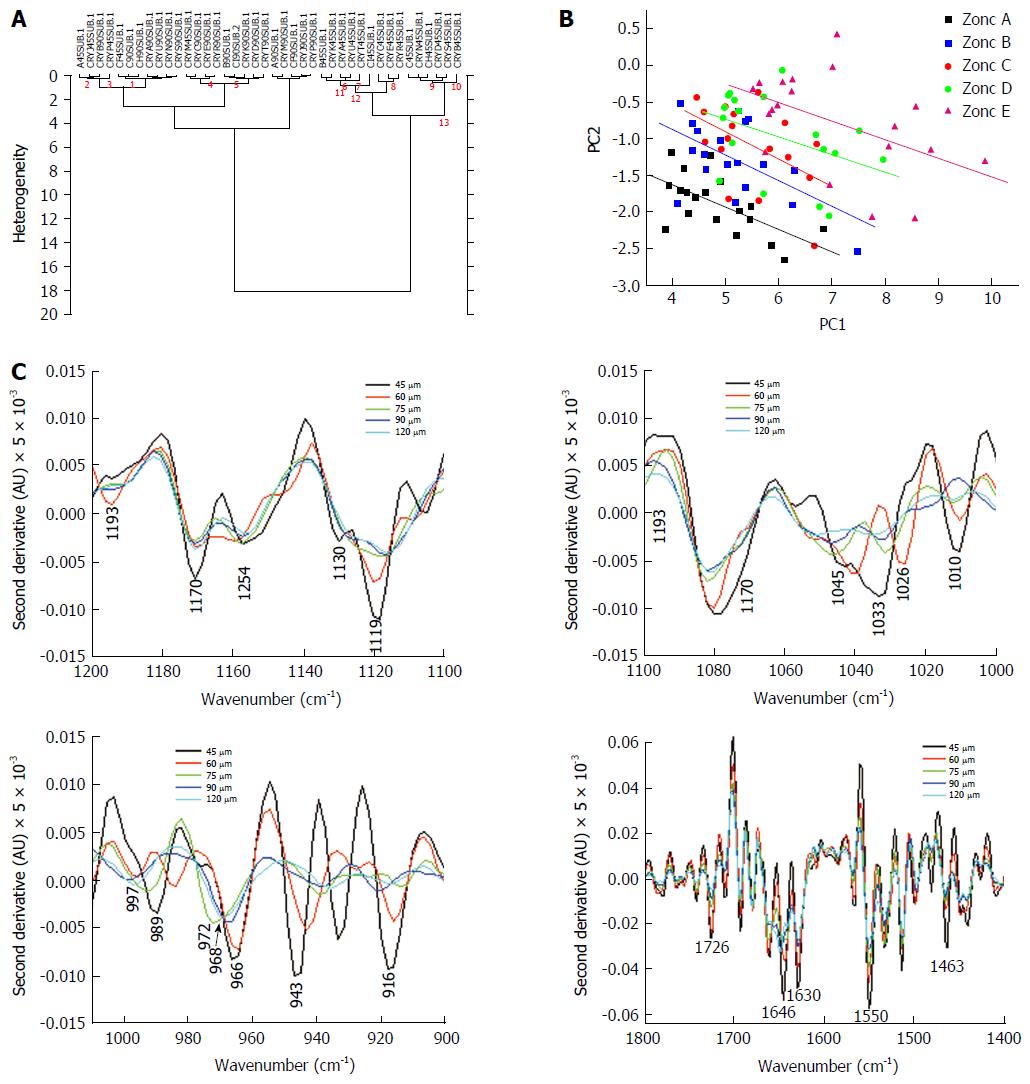
Figure 2 shows typical spectra of the above five zones in the mid-IR region of 800-1800 cm-1, which has been used for diagnosis of cancer and contains relevant information from all metabolites in the cells/tissues. There is a progressive decrease in the intensities in the region between 900-1200 cm-1vs aperture diameter. This systematic decrease is attributed mainly to glucose (carbohydrates) and glycoprotein absorbance. To further understand the distribution and gradient of various biochemicals along the crypt diameter, cluster analysis was performed. The data are presented in Figure 3A, which shows that the spectra obtained with the same aperture cluster together due to unique combination of the components in that zone of the crypt. Although the main goal of PCA is dimensionality reduction by choosing the maximum variance directions, sometimes, it is possible to detect classification potential when looking in the PCs domain. Figure 3B shows the scores of PC1 and PC2 in a two dimensional figure generated using the calculated scores derived using PCA calculation. As can be seen from this figure, there is a total separation between the data points of the far zones (A and E, A and D), while the data points of near zones are overlapped (D and C). To further establish that neighboring zones had variations in spectra and composition we used a more sophisticated LDA classifier. LDA analysis followed the PCA calculations performed spectra in the range 900-1800 cm-1 show clear differentiation among spectra obtained with different apertures (Table 1). These results establish that we have a biochemical variation along the crypt diameter and this is reflected in the mid-IR spectra.
| Zone A | 20 | 0 | 0 | 0 | 0 |
| Zone B | 1 | 16 | 3 | 0 | 0 |
| Zone C | 0 | 4 | 14 | 2 | 0 |
| Zone D | 1 | 0 | 2 | 16 | 1 |
| Zone E | 0 | 0 | 2 | 5 | 13 |
We next examined the spectra to see which regions exhibited the maximum variation in the diagnostic parameters. The most striking variation seen in the region 900-1200 cm-1 is due to decreasing glucose (carbohydrate) and glycoprotein absorbance when the aperture was increased gradually from 45 to 120 μm. This indicates that as the aperture increases contribution from other macromolecules like proteins, lipids and nucleic acids becomes significant, causing a relative lowering of signals from the carbohydrate/glycoprotein absorbance. Second derivative analysis of the spectra showed variation of metabolites absorbing in different regions obtained due to aperture variation (Figure 3C). The lumen secretions originate from the secretory vesicles that are concentrated in Zones B and C. Thus, monitoring the glucose/glycosylation variation would establish the validity of the model. (Zone E containing glycoproteins in a higher proportion than other zones, exhibits a deeper minima at this wavenumber (approximately 1030, 1045 cm-1) in the second derivative spectra (Top right panel Figure 3C).
The pattern obtained in Zone E shows bands that correspond to absorbance from glycosylated proteins (increased absorbance at amide II, and at wave numbers between 900-1300 cm-1) which are known to be present in the crypt lumen. Thus, we establish that there is gradual decrease of this component using our model which correlates with the histological features. Glycoproteins are secreted into the crypt lumen and as expected a decrease in protein level is reflected also in the increasing minima (in the second derivative spectra) at the amide II due to decreasing proportion of protein absorbance with the inclusion of additional cell components and nuclei into the spectra as we move towards the periphery (Figure 3C lower right panel).
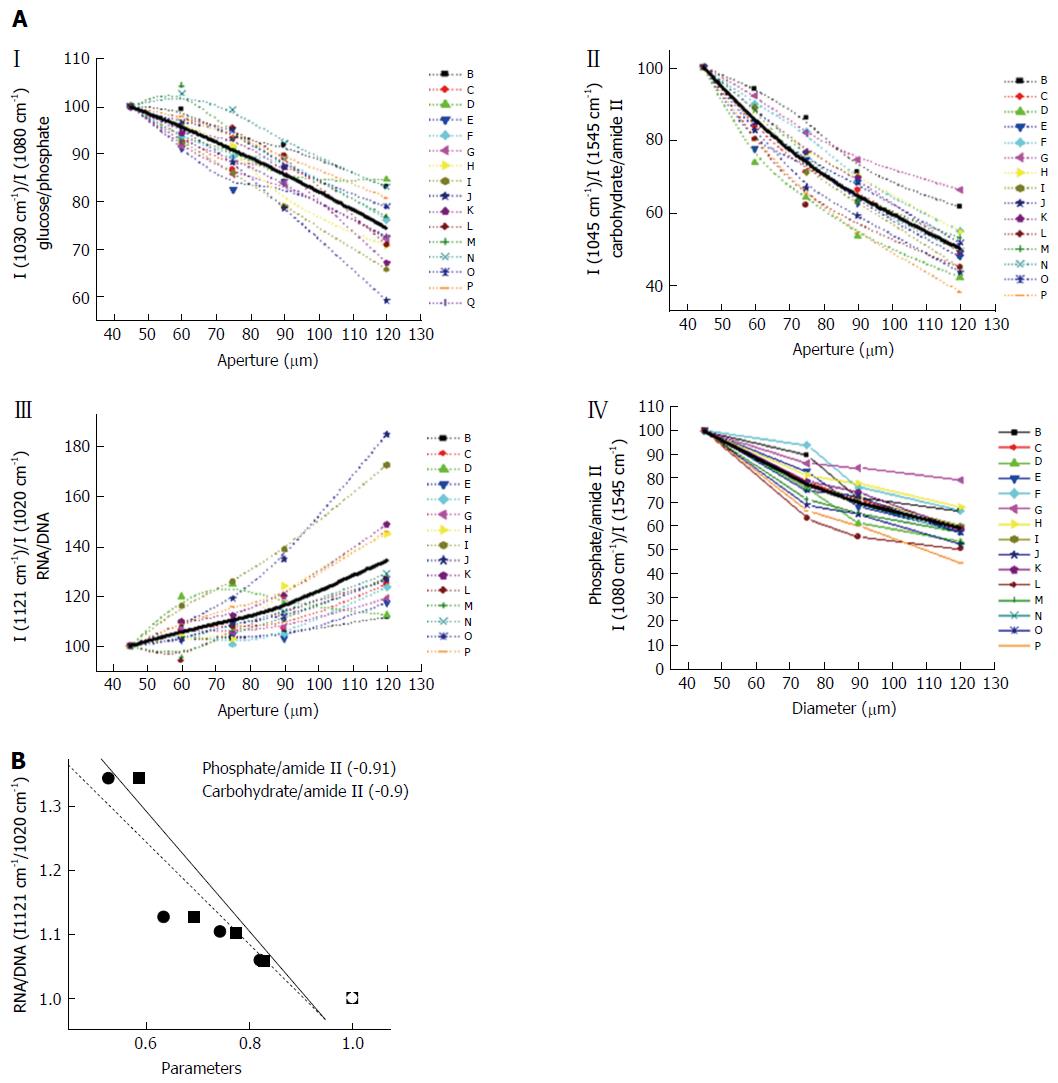
Figure 4A shows the variation in different metabolites across the crypt. We evaluated the variation of carbohydrates/Amide II (I (1045 cm-1)/I (1545 cm-1), which is expected to be highest in the lumen and progressively decrease with increasing aperture as most carbohydrates and glycoproteins are secreted into the lumen and hence are concentrated in that region (Figure 4A). It is observed that as expected, the level of glucose and phosphates decrease when the aperture increases, indicating that the bulk of the carbohydrate absorbance arises from the luminal components (Figure 4A). The RNA/DNA ratio which decreases with aperture shows that DNA signals arise mainly from components localized at the periphery further confirming that the absorbance from nucleic acids and carbohydrates can be spatially partitioned in this system (Figure 4A). Other parameters that indicate levels of carbohydrates and nucleic acids display similar trends (data not shown).
Under normal conditions DNA and RNA are present in the nucleus while the cytoplasm contains mostly RNA. As the aperture increases signals from DNA get included and the signals from RNA have a relative lowering. The classical ratio I (1121 cm-1)/I (1020 cm-1) seems to best denote the inclusion of nucleic acid absorbance in the spectra as this increases considerably with aperture in the larger apertures (Zones B and A) in the spectral measurement. Inclusion of the nuclei results in inclusion of signals from DNA which is mostly confined to the nuclei. Both RNA and DNA signals are expected to increase with aperture. However our results indicate that the RNA/DNA ratio increases. Therefore RNA signals seem to increase more rapidly than DNA. This observation may be due to the fact that several types of RNA exist in the cells and nuclei and most of the DNA is usually in compact form with less contribution to IR absorbance. However the region where these two nucleic acids prominently absorb IR is also the region of considerable absorption by sugars and their derivatives.
To understand the variation of the nucleic acid contribution in presence and absence of carbohydrates and other interfering components, the relation of the parameter representing the RNA/DNA ratio was plotted against the parameters representing concentration of carbohydrates/phosphates (Figure 4B). It can be seen that the parameter shows an inverse relation as indicated by the negative correlation coefficient between RNA/DNA ratio and those representing the carbohydrates or phosphates. This shows that if there is a decrease in carbohydrates due to any disease condition, an increase in signals from RNA/DNA may be expected and becomes relevant when the diagnosis is based on FTIR spectral measurements.
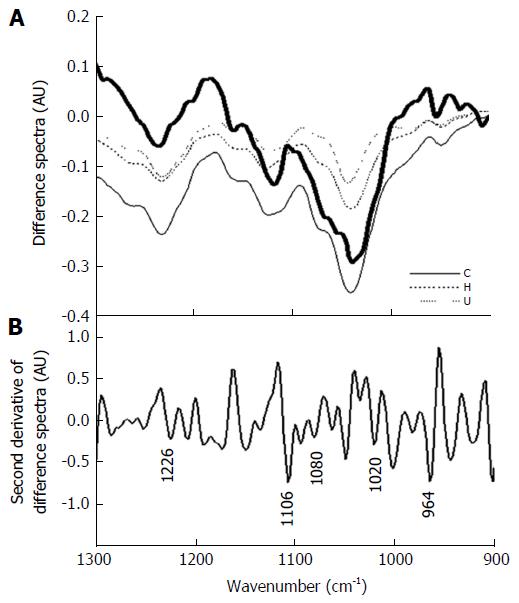
To further verify the proposition that disappearing carbohydrate/glycoprotein signals can lead to increasing signals from nucleic acids, we subjected the spectra obtained from different grades of colonic malignancies to a qualitative analysis where data were obtained by measuring absorbance in the entire crypt circle and a difference spectra of the malignant with the normal was plotted. As can be seen (Figure 5A), there is similarity between the difference spectra obtained for colonic malignancies with the difference spectra obtained between Zones A and B. This indicates that the observed diagnostic potential of the technique owes its origin to the phenomenon, i.e., loss of luminal structures and components that are present in normal histology but are absent or disturbed during malignancy. The nucleic acid signature is obtained under both these conditions is presence or absence of the interfering components.
To further confirm the absorbance of nucleic acids, we analyzed the second derivative of the difference spectra of the total crypt area (Zone A) and Zone B containing some portions of nuclei should result in identifying the signatures exclusive to DNA. Figure 5B shows the second derivative spectra where minima are observed at wave numbers that correspond to nucleic acid absorbance bands. A statistical analysis of the parameters showed significant difference in the ratio obtained with the inclusion of absorbance from Zone A when compared to the parameter obtained from absorbance in the lumen alone (Table 2). This shows that the RNA/DNA ratio though susceptible to interference from the carbohydrate moieties can still be a significant parameter when used to detect changes occurring due to variation in nucleic acid content even in tissues rich in glycoproteins. We further studied this correlation between the average parameters and the RNA/DNA ratio to see the interference of the substances. Histologically studies of the cross sections of crypts, indicate that the lumen is shrunken and there is a collapsing structure with nuclei of the cells occurring inner to their normal occurrence[27]. Thus, this indicates that indeed the nuclei contribute to the absorbance.
| Apertures | 1121/1020 | 1244/1230 | Glucose/phosphate | 996/966 |
| Compared | P values | P values | P values | P values |
| 45, 60 | 0.00164 | 0.000501 | 0.008998 | 0.267273 |
| 60, 75 | 0.00376 | 0.0003 | 0.187822 | 0.002968 |
| 75, 90 | 0.235976 | 0.319264 | 0.123108 | 0.003836 |
| 90, 120 | 4.26E-05 | 0.199106 | 1.55E-05 | 0.016475 |
| 45, 120 | 1.10E-05 | 0.000104 | 1.07E-10 | 0.007219 |
| 75, 120 | 0.000104 | 0.037284 | 9.58E-09 | 0.000418 |
FTIR spectroscopy has been increasingly seen as a potential technique for analysis of metabolic profiles of cells and tissues during diagnostics in biomedicine[19,20]. However, it has an intrinsic complexity associated with overlapping signals from different metabolites that can simultaneously absorb making it difficult to often assign variations in absorbance at a particular wavenumber to a single metabolite. To overcome this problem other than taking very specific intensities corresponding to known metabolites that are expected to vary due to biological variations, advanced methodologies for spectral analyses have been adopted[15,18]. These have been useful to partly increase the sensitivity and specificity of spectral analyses especially in cancer diagnostics.
A question of considerable implications in this field is how the variations of chemical composition from one group of compounds may affect the signals arising due to another group absorbing at similar wave numbers and how such interference may result in affecting the diagnostic features of the FTIR spectroscopy. While the effect of contaminating extraneous materials[28] and confounding variables[30] have been studied as a part of the basic research into spectral interference, no study about how the actual interference from components intrinsic to tissues has been undertaken.
Phospholipids are known to absorb at overlapping wave numbers with nucleic acids and can influence the absorbance due of nucleic acids. We confirmed that the variation of the phospholipids along the crypt diameter was minimum and did not contribute significantly to the observed variations of the nucleic acids by monitoring the changes at 1737 cm-1 where lipids have absorbance with least interference from other macromolecules (Figure 4A). Thus a decreasing level of carbohydrates coupled with an increasing signal from nucleic acids can help confirm the absorbance due to nucleic acids at their specific wave numbers.
The correlation between the average parameters and the RNA /DNA ratio was plotted to see the interference of the substances. Figure 4B shows the negative correlation of the two components, carbohydrates, phosphates with the RNA/DNA ratio showing that these substances decrease while the RNA/DNA (1121/1020) ratio increases with correlation around -0.9. Thus, our initial hypothesis that a decreasing carbohydrate and phosphate absorbance could contribute to increasing signals of RNA/DNA is verified.
Our groups had previously demonstrated that utilization of simple ratios of intensities of nucleic acids in the mid-IR region from normal and cancerous samples of different tissues can be quite effective. Implementing such simplistic analysis is important when the technique is to be used in clinical set ups as most personnel dealing with such preliminary diagnosis may not have the required expertise to undertake more complicated analysis as well as to avoid dependence on more complex software programs and personnel. Interestingly, the absorbance of nucleic acids is also affected by the presence of carbohydrates or carbohydrate containing macromolecules, which are known to vary from the normal levels in tissues due to carcinogenesis[31]. This has been observed in several tissues like colon, cervix and cell lines[2]. Thus, it became important to undertake a study to understand how the variation in carbohydrates affects the signals of nucleic acids.
The colonic crypts in cross section provide a unique opportunity to understand this phenomenon of how increasing or decreasing levels of absorbance of one component affects the signals associated with a diagnostic variable. Under situations where permissible, therefore developing techniques to eliminate the contribution of carbohydrates would render the technique more efficient when diagnosis is based on nucleic acid absorbance. It may also lead to identification of changes in nucleic acids composition of content using FTIR spectroscopy due to enhancement of signals from DNA or RNA. The RNA/DNA ratio is one of the parameters that has a near universal diagnostic potential across different types of malignancy. During malignancy accumulated glycogen or carbohydrates decrease in both Cervical intraepithelial neoplasia and cervical cancer and during this condition the RNA/DNA ratio is an effective parameter[31]. Thus, lowered interference from carbohydrates may indeed lead to improved nucleic acid signatures. We show cases of IBD in the present study and the variation. Observations with different grades of colonic malignancy previously had shown that RNA/DNA is indeed an effective ratio and histopathological features indicated the shrinking of the lumen or distortion of the lumen in crypt circle evident of disappearing or altered carbohydrate metabolism[11].
With increasing evidence from different studies indicating common variations during malignancies, it is feasible to identify and classify the disease stage and grade based on spectral data. However the pathologist would still be needed for ascertaining that the measurements are made in the right region. While a pathologist’s classification would still be subjective, a spectral data would help further lend a quantitative approach to the diagnosis. This has been the objective behind using computational methods that are free from human bias. Previously studies have indicated the potential of predicting diseases from biopsies[32]. While the disappearance of carbohydrates (Glycogen) an decrease in its signal in the FTIR spectra is a function of the differentiation process in the cervical epithelium[33] and is elucidated by measurements in different zones , in the crypt the same analogy may be observed by varying the measurement using different aperture in the crypt circle. This establishes that biological variations can be manifested as chemical signatures and are the basis for the diagnostics.
We have traced the variation in the interfering components along the colon crypt diameter along with the nucleic acid variation and shown that decreasing carbohydrates from interfering substances may indeed lead to improvement of signals from nucleic acids leading to its effectiveness as a diagnostic variable.
In conclusion, we utilize the unique features present in the cross sections of crypts in the colonic biopsies and advanced computational methods to establish the contribution of nucleic acids in presence of carbohydrates. We demonstrate the validity of using the RNA/DNA ratio as a diagnostic parameter for detecting colonic malignancies (abnormalities) by studying how the nucleic acid absorbance may be effective in spite of presence of confounding variables like carbohydrates and phosphates in tissues. The present study indicates that one has to be careful when comparing results obtained from different apertures and utilizing the system for diagnosis. Different apertures strongly affect the extracted biological markers due to overlap between the IR signals at different zones.
Fourier transform infrared (FTIR) microscopy principles for diagnostics are still being evaluated. This study looks at the role of interference from absorbance of compounds in the same wave numbers.
The authors established the effect of removal or reduction of one component on the efficacy or potential of another being used as a diagnostic parameter.
The authors used the complex architecture of the colonic crypt transverse section to study variation of metabolites and the ensuing spectral features.
These findings are used to diagnose the diseases from evaluation of biopsies in an objective method.
The authors focus on differentiating normal and cancerous tissues by using FTIR spectroscopy. The overlap of major absorbance bands of nucleic acids, carbohydrates and phospholipids in the mid-infrared region would decrease the diagnostic potential of this technique. The authors used the colonic tissues as a model system to study the interference of macromolecules and eliminate RNA/DNA ratio as an improved diagnostic parameter for detecting colonic malignancies. This article is well written with good readability.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Israel
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Hazard FK, Li YQ S- Editor: Gong ZM L- Editor: A E- Editor: Zhang FF
| 1. | Dovbeshko GI, Chegel VI, Gridina NY, Repnytska OP, Shirshov YM, Tryndiak VP, Todor IM, Solyanik GI. Surface enhanced IR absorption of nucleic acids from tumor cells: FTIR reflectance study. Biopolymers. 2002;67:470-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 104] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 2. | Sahu RK, Argov S, Salman A, Huleihel M, Grossman N, Hammody Z, Kapelushnik J, Mordechai S. Characteristic absorbance of nucleic acids in the Mid-IR region as possible common biomarkers for diagnosis of malignancy. Technol Cancer Res Treat. 2004;3:629-638. [PubMed] |
| 3. | Pevsner A, Diem M. IR spectroscopic studies of major cellular components. III. Hydration of protein, nucleic acid, and phospholipid films. Biopolymers. 2003;72:282-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | Bhatt AN, Mathur R, Farooque A, Verma A, Dwarakanath BS. Cancer biomarkers - current perspectives. Indian J Med Res. 2010;132:129-149. [PubMed] |
| 5. | Lewis PD, Lewis KE, Ghosal R, Bayliss S, Lloyd AJ, Wills J, Godfrey R, Kloer P, Mur LA. Evaluation of FTIR spectroscopy as a diagnostic tool for lung cancer using sputum. BMC Cancer. 2010;10:640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 6. | Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog. 2006;5:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 919] [Cited by in RCA: 963] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 7. | Lasch P, Pacifico A, Diem M. Spatially resolved IR microspectroscopy of single cells. Biopolymers. 2002;67:335-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 84] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 8. | Benedetti E, Bramanti E, Papineschi F, Rossi I, Benedetti E. Determination of the relative amount of nucleic acids and proteins in leukemic and normal lymphocytes by means of Fourier transform infrared microspectroscopy. Applied Spectroscopy. 1997;51:792-797. |
| 9. | Mourant JR, Yamada YR, Carpenter S, Dominique LR, Freyer JP. FTIR spectroscopy demonstrates biochemical differences in mammalian cell cultures at different growth stages. Biophys J. 2003;85:1938-1947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 105] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | Andrus PG, Strickland RD. Cancer grading by Fourier transform infrared spectroscopy. Biospectroscopy. 1998;4:37-46. [PubMed] |
| 11. | Argov S, Ramesh J, Salman A, Sinelnikov I, Goldstein J, Guterman H, Mordechai S. Diagnostic potential of Fourier-transform infrared microspectroscopy and advanced computational methods in colon cancer patients. J Biomed Opt. 2002;7:248-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 101] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Salman A, Lapidot I, Pomerantz A, Tsror L, Shufan E, Moreh R, Mordechai S, Huleihel M. Identification of fungal phytopathogens using Fourier transform infrared-attenuated total reflection spectroscopy and advanced statistical methods. J Biomed Opt. 2012;17:017002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 13. | Salman A, Pomerantz A, Tsror L, Lapidot I, Moreh R, Mordechai S, Huleihel M. Utilizing FTIR-ATR spectroscopy for classification and relative spectral similarity evaluation of different Colletotrichum coccodes isolates. Analyst. 2012;137:3558-3564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Salman A, Shufan E, Zeiri L, Huleihel M. Detection and identification of cancerous murine fibroblasts, transformed by murine sarcoma virus in culture, using Raman spectroscopy and advanced statistical methods. Biochim Biophys Acta. 2013;1830:2720-2727. [PubMed] |
| 15. | Bishop CM. Pattern recognition and machine learning, Springer, New York, 2006. . |
| 16. | Sahu RK, Salman A, Mordechai S, Manor E. Study of plasma-induced peripheral blood mononuclear cells survival using Fourier transform infrared microspectroscopy. J Biomed Opt. 2013;18:115004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Camastra F, Vinciarelli A. Machine learning for audio, image and video analysis: theory and applications, Springer, London, 2008. . |
| 18. | Zwielly A, Gopas J, Brkic G, Mordechai S. Discrimination between drug-resistant and non-resistant human melanoma cell lines by FTIR spectroscopy. Analyst. 2009;134:294-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Travo A, Desplat V, Barron E, Poychicot-Coustau E, Guillon J, Déléris G, Forfar I. Basis of a FTIR spectroscopy methodology for automated evaluation of Akt kinase inhibitor on leukemic cell lines used as model. Anal Bioanal Chem. 2012;404:1733-1743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Diem M, Griffiths P, Chalmers JM. Vibrational Spectroscopy for Medical Diagnosis. Wiley, 2008. . |
| 22. | Huberty CJ. Applied discriminant analysis. Wiley, 1994. . |
| 23. | Fukunaga K. Introduction to statistical pattern recognition. Boston: Academic Press, 1990. . |
| 24. | Otto M. Chemometrics: statistics and computer application in analytical chemistry. Wiley-VCH, Weinheim, New York, 1999. . |
| 25. | Everitt B. Cluster analysis, E. Arnold. Halsted Press, London, New York, 1993. . |
| 26. | Flury B, Riedwyl H. Multivariate Statistics: A Practical Approach, CRC Press LLC, 1988. . |
| 27. | Argov S, Sahu RK, Bernshtain E, Salman A, Shohat G, Zelig U, Mordechai S. Inflammatory bowel diseases as an intermediate stage between normal and cancer: a FTIR-microspectroscopy approach. Biopolymers. 2004;75:384-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Sahu RK, Argov S, Salman A, Zelig U, Huleihel M, Grossman N, Gopas J, Kapelushnik J, Mordechai S. Can Fourier transform infrared spectroscopy at higher wavenumbers (mid IR) shed light on biomarkers for carcinogenesis in tissues? J Biomed Opt. 2005;10:054017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Sahu RK, Zelig U, Huleihel M, Brosh N, Talyshinsky M, Ben-Harosh M, Mordechai S, Kapelushnik J. Continuous monitoring of WBC (biochemistry) in an adult leukemia patient using advanced FTIR-spectroscopy. Leuk Res. 2006;30:687-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | Romeo MJ, Wood BR, Quinn MA, McNaughton D. Removal of blood components from cervical smears: implications for cancer diagnosis using FTIR spectroscopy. Biopolymers. 2003;72:69-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Podshyvalov A, Sahu RK, Mark S, Kantarovich K, Guterman H, Goldstein J, Jagannathan R, Argov S, Mordechai S. Distinction of cervical cancer biopsies by use of infrared microspectroscopy and probabilistic neural networks. Appl Opt. 2005;44:3725-3734. [PubMed] |
| 32. | Sahu RK, Argov S, Walfisch S, Bogomolny E, Moreh R, Mordechai S. Prediction potential of IR-micro spectroscopy for colon cancer relapse. Analyst. 2010;135:538-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Chiriboga L, Xie P, Yee H, Vigorita V, Zarou D, Zakim D, Diem M. Infrared spectroscopy of human tissue. I. Differentiation and maturation of epithelial cells in the human cervix. Biospectroscopy. 1998;4:47-53. [PubMed] |