Published online Nov 21, 2016. doi: 10.3748/wjg.v22.i43.9506
Peer-review started: July 16, 2016
First decision: August 19, 2016
Revised: September 10, 2016
Accepted: October 10, 2016
Article in press: October 10, 2016
Published online: November 21, 2016
Processing time: 126 Days and 12.1 Hours
To identify common copy number alterations on gastric cancer cell lines.
Four gastric cancer cell lines (ACP02, ACP03, AGP01 and PG100) underwent chromosomal comparative genome hybridization and array comparative genome hybridization. We also confirmed the results by fluorescence in situ hybridization analysis using the bacterial artificial chromosome clone and quantitative real time PCR analysis.
The amplification of 9p13.3 was detected in all cell lines by both methodologies. An increase in the copy number of 9p13.3 was also confirmed by fluorescence in situ hybridization analysis. Moreover, the interleukin 11 receptor alpha (IL11RA) and maternal embryonic leucine zipper kinase (MELK) genes, which are present in the 9p13.3 amplicon, revealed gains of the MELK gene in all the cell lines studied. Additionally, a gain in the copy number of IL11RA and MELK was observed in 19.1% (13/68) and 55.9% (38/68) of primary gastric adenocarcinoma samples, respectively.
The characterization of a small gain region at 9p13.3 in gastric cancer cell lines and primary gastric adenocarcinoma samples has revealed MELK as a candidate target gene that is possibly related to the development of gastric cancer.
Core tip: While the presence of alterations in the DNA copy number is one of the key hallmarks of carcinogenesis, in gastric cancer, the chromosomal regions with frequent gain and loss are still poorly defined. Array comparative genome hybridization is a high resolution tool that allows the simultaneous detection of sub-microscopic copy number changes across the genome. The characterization of a small gain or loss region in gastric cancer cell lines and primary gastric adenocarcinoma samples could reveal a candidate target gene that may possibly be linked to the development of gastric cancer.
- Citation: Calcagno DQ, Takeno SS, Gigek CO, Leal MF, Wisnieski F, Chen ES, Araújo TMT, Lima EM, Melaragno MI, Demachki S, Assumpção PP, Burbano RR, Smith MC. Identification of IL11RA and MELK amplification in gastric cancer by comprehensive genomic profiling of gastric cancer cell lines. World J Gastroenterol 2016; 22(43): 9506-9514
- URL: https://www.wjgnet.com/1007-9327/full/v22/i43/9506.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i43.9506
Gastric cancer (GC) remains a major public health issues, as it is the fifth most common malignancy and the third leading cause of cancer death in both sexes worldwide[1]. The most common type of GC is adenocarcinoma, which can be further categorized into two main types, intestinal type and diffuse type, which are biologically different with distinct clinical and epidemiological profiles[2]. The difference in the clinicopathological characteristics between the histological types of gastric cancer indicate that gastric tumor development occurs through the progressive accumulation of distinct genetic alterations[2-5]. Thus, the characterization of these genomic abnormalities in gastric cancer may help to clarify the molecular pathogenesis of the disease and may unveil genetic markers of progression and for predicting treatment response or survival.
Genomic instability with frequent DNA copy number variations (CNVs) is one of the key hallmarks of gastric carcinogenesis[6]. Tumor progression seems to depend on the successive acquisition of chromosomal aberrations, leading to gains or losses of parts of the genome. However, there is no clear agreement on the genetic changes underlying gastric carcinogenesis.
In the last decades, chromosomal comparative genome hybridization (cCGH) and array CGH (aCGH) analyses of gastric tumors and gastric cell lines have revealed recurrent DNA CNVs[7-11]. Using cCGH, Burbano et al[3] showed that the copy number gain of 8q24.1, the locus containing the MYC oncogene, is a frequent alteration in GC. Further investigations by our group demonstrated that MYC amplification is a common finding in preneoplastic gastric lesions and tumors[4,5,12-15].
Moreover, Takeno et al[10] stated that diffuse-type GC shows a complex pattern of chromosomal alterations, especially chromosome region losses. Recently, Liang et al[16] suggested that the detection of DNA CNVs from tissue or blood samples may be a useful tool for guiding individualized treatment strategies and for identifying new drug targets in patients with GC.
In the current study, we analyzed the chromosomal abnormalities of four GC cell lines by cCGH and aCGH. The occurrence of the amplification of chromosomal region 9p13 in GC cell lines was validated by fluorescence in situ hybridization (FISH) and confirmed in primary gastric adenocarcinoma samples by quantitative polymerase chain reaction (qPCR). Among the genes within the 9p13 region, we chose two genes for validation in primary GC samples, interleukin 11 receptor alpha (IL11RA) and maternal embryonic leucine zipper kinase (MELK).
The ACP02, ACP03 and AGP01 gastric adenocarcinoma cell lines, which were previously established and characterized by our research group, were used in the present study[17,18]. Additionally, we used the GC cell line, PG100, obtained from the Rio de Janeiro Cell Bank (Rio de Janeiro, RJ, Brazil), which was previously characterized cytogenetically by our group[19]. All cell lines were cultured according to Lima et al[20].
Quantitative gene copy number measurements were performed on 68 primary gastric adenocarcinoma samples that were obtained from patients who underwent surgery resection in João de Barros Barreto University Hospital (HUJBB), Belém, Pará, Brazil. In Pará, Brazil, the human population is composed of interethnic crosses between three main origin groups, European (mainly represented by Portuguese), Africans, and Amerindians[21].
All the patients had negative histories of exposure to either chemotherapy or radiotherapy before surgery, and there were no other diagnosed cancers. Signed informed consent, with the approval of the ethics committee of HUJBB, was obtained from all patients prior to the collection of samples.
DNA from the GC cells lines and gastric tumors were isolated using the QiAmp DNA isolation kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommended protocol. DNA concentration and purity were evaluated by Nanodrop (NanoDrop Technologies, Houston, TX, United States) and agarose gel electrophoresis. All DNA samples used had an A260/280 ratio of 1.8-2.0 and an A260/A230 ratio of > 1.5 and were visualized as a high molecular weight band on an agarose gel.
DNA samples from GC cell lines were labeled using the CGH Nick Translation Kit (Abbott Laboratories, IL, United States) with Control DNA (Promega, Madison, United States) according to the manufacturer’s instructions. Hybridization was performed with CGH Metaphase Target Slides (Abbott Laboratories, Illinois, United States), following the manufacturer’s protocols. The slides were analyzed by Corel Photo-Paint - Version 5.00 - Isis Zeiss® software, using an Axioskop Zeiss microscope (Carl Zeiss Inc. Canada, Don Mills, ON, Canada) equipped with an epi-illuminator and fluorochrome-specific optical filters.
The three-color images with red, green, and blue were acquired from 15 metaphases. Chromosome imbalances were detected on the basis of the deviation of the fluorescence ratio profile from the balanced value (FITC:rhodamine = 1). For each chromosome, the final ratio values were prepared from the mean values of at least ten chromosome homologues from separate metaphase spreads. The CGH results were plotted as a series of green to red ratio profiles.
To evaluate the complete genome of all the four cell lines studied, high density microarray analysis was performed using the AffymetrixR CytoScan™ HD Array platform (Affymetrix, Santa Clara, CA, United States). First, genomic DNA was digested by the NspI restriction enzyme, and the digested samples were ligated using the NspI adaptor. The fragments were amplified by PCR and run on a 2% agarose gel to verify that the PCR product size distribution was between 150 bp and 2000 bp. After PCR product purification and dilution, we performed the quantification of each sample using a NanodropR 1000 Spectophotometer (NanoDrop Technologies, Houston, TX, United States). The average purification yield for each sample was ≥ 3.0 μg/μL.
The purified samples were then fragmented using DNAse I enzyme, and the products were run on a 4% agarose gel to verify that the majority of fragments had a size distribution between 25 and 125 bp.
Labeling was performed using terminal deoxynucleotidyl transferase enzyme, which adds biotinylated nucleotides at the 3’ end of fragmented samples.
During the hybridization step, each sample was hybridized onto a CytoScan® HD Array (Affymetrix, Santa Clara, CA, United States) and placed in a GeneChip® Hybridization Oven 640 (Affymetrix, Santa Clara, CA, United States) at 50 °C and 60 rpm for 16 to 18 h. The processes prior to scanning of arrays, washing and staining, were carried out at a Fluidics Station 450 (Affymetrix, Santa Clara, CA, United States). The arrays were scanned using GeneChip® Scanner 3000 7G (Affymetrix, Santa Clara, CA, United States).
The copy number was deduced from the weighted log2 ratio and the aberration type was identified and confirmed using allelic plots.
FISH was performed on nuclei and metaphase spreads of the cell lines, ACP02, ACP03 and AGP01. Metaphase spreads of lymphocytes from a healthy donor were used as a control. The bacterial artificial chromosome (BAC) clone, RP11-165H19, was obtained from BAC/PAC Resources (http://bacpac.chori.org/). Bacterial cultures and DNA isolation was performed using Qiagen Plasmid Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Alu-PCR products of the BAC were used as probes and were biotinylated using nick translation, as described previously[22].
For the validation of 9p13 amplification, we evaluated the copy number of two genes within this locus, IL11RA and MELK. For this, we used the same DNA samples from GC cell lines that were used for cCGH and aCGH and from GC tissues. qPCR was performed using the FAM/MGB-labeled TaqMan probes (Life Technologies, Foster City, CA, USA) for IL11RA (Hs01842695_cn) or MELK (Hs05076287_cn). VIC/TAMRA-labeled TaqManCopy Number Reference Assay RNAse P (#4403326; Life Technologies, Foster City, CA, United States) was used as an internal control. All the real-time qPCR reactions were performed in quadruplicate with gDNA using a 7500 Fast Real-Time PCR system (Life Technologies, Foster City, CA, United States) as described previously[13]. The copy number of each sample was estimated by CNV analysis using Copy Caller Software V1.0 (Life Technologies, Foster City, CA, United States). Known Human Genomic DNA, G1471 and G1521 (Promega, Madison, United States), were used for calibration.
The data on clinical features were compared by the χ2 test or two-tailed Fisher’s exact test for categorical variables. All statistical analyses were performed with the statistical package SPSS for Windows (V.17.0, SPSS Inc, Chicago, IL, United States). P values of ≤ 0.05 were considered significant.
The ACP02, ACP03, AGP01 and PG100 cell lines showed multiple gains and losses by cCGH and aCGH. Most chromosomal aberrations detected in these cell lines by cCGH were confirmed by aCGH (Table 1), although aCGH analysis enabled the identification of many additional chromosomal gains and losses. On the other hand, the gain of 16p21-p23 in ACP03 and the gains of 6p11-p12, 12p11.1 and 18p11.2-p11.3 in AGP01 were detected only by cCGH.
| Cell line | cCGH | aCGH |
| ACP02 | +8p21-pter, +8q24, +9p12-p22, +9q21.1-q21.3, +15q11.1-q14, +16p, +16q, +17p11, +17q11.2, +17q23, +22q11.1-q12.3 | +1p13.2, +1p21.3, +1q21.2, +1q21.3, +2p11.2, +4p12, +4p13, +4q12, +5q12.1, +7q11.22, +7q11.23, +8p11.23, +8p21.3, +8q24.11, +8q24.22, -8q24.22, +8q24.23, +8q24.3, +9p13.2, +9p13.3, +9p11.3, +9q21.12, +10p11.22, +10p12.1, +10p12.31, +10p13, +10p14, +10q11.21, -11p11.12, +11p11.2, , -11q12.1, -11q12.2, -11q12.3, +12q12-q15, +13q12.11, +14q11.2, +15q11.2, +15q12, +15q14, +15q15.3, +15q24.1, +15q25.1, +15q26.1, +16p11.2, +16p12.3, +16q11.2, +16q13, +16q21, +16q22.1, +17p11.2, +17q11.2, +17q23.1, +18q11.2, +19q13.11, +20p11.1, +20p11.23, +20p12.2, +20q11.21, +20q11.23, +22q11.21,+ 22q11.23, +22q12.2, +22q12.3 |
| ACP03 | +4p15.1-pter, +6p22.3-p24, +6q25.1,q26, +8p22-pter, -8q11.1-q11.2, +9p12-p22, +10p12-p14, -11p11.1, -11q12, +15q11.1-q15, +15q23-q26.1, +16p12-p13.1, +16p21-p23, +22q11.1-q12.1 | +1p13.2, +1p13.3, +1p21.3, +1q21.1, +1q21.2, 1q21.3, +2p12, +4p12, +4p14-p13, +4p15.1, +5q12.1, +6p24.2, +7q11.21, +7q11.22, +7q11.23, +8p11.21, +8p11.23-p11.22, +8p21.2, +8p21.3, -8q11.21, +8q24.11, -8q24.12, +8q24.13, +8q24.21, +8q24.22, -8q24.22, +8q24.23, +8q24.3, +9p13.2, +9p13.3, +9q21.13, +9p23, +10p11.21, +10p11.22, +10p12.1, +10p12.2, +10p12.31, +10p12.33, +10p13, +10p14, +10q11.21, -11p11.12, +11q11.2, -11q12.1-q12.2, -11q12.3, +12q12-q15, +14q11.2, +14q13.2, +15q11.2, +15q12, +15q13.1, +15q14, +15q24.1, +15q25.1, +15q25.2, +15q26.1, +16p11.2, +16p12.3, +16q11.2, +16q12.1, +16q12.2, +16q21, +16q22.1, +17p11.2, +17q11.2, +17q23.1, +18q11.2, +18q12.1, +19q13.11, +20p11.1, +20p11.23, +20p12.2, +20q11.21, +20q11.22, +20q11.23, +22q11.21, +22q11.23, +22q12.2, +22q12.3 |
| AGP01 | +1p13-21, +1q12-q21.3, +2p11.2-p12, +4p11-p12, +4q12-q13.1, +5p11-p12, +5q11.2-q12, +6p11-p12, +6q12-q16.1, +7q11.1-q11.2, +9p12-p13, +9q13-q21.3, +10p11.2-p12.3, +10q11.1-q21.1, +11p11-p11.2, +12p11.1, +12q12, +13q11-q12, +14q11.1-q13, +15q11-q14, +16p11.2, +16q12, +17p11.2, +17q11.2, +18p11.2-p11.3, +18q11-q12, +19q12-q13.1, +20p11.2-p12, +20q11.1-q11.2 | +1p13.3, +1p21.3, +1q21.2, +1q21.3, +2p11.2, +2p12, +4p12, +4p14-p13, +5p12, +5q12.1, +6p24.2, +6q13, +7q11.21, +7q11.22, +7q11.23, +8p11.21, +8p11.23-p11.22, +8p21.3, -8q11.21, +8q24.11, -8q24.12, +8q24.21, +8q24.22, +8q24.23, +8q24.3, +9p13.2, +9p13.3, +9p22.3, +9q21.12, +9q21.13, +10p11.22, +10p12.1, +10p12.31, +10p13, +10p14, +10q11.21, -11p11.12, +11p11.2, -11q12.1, -11q12.2, -11q12.3, +12q12-q15, +13q12.11, +14q11.2, +15q12, +15q14, +15q15.1, +15q15.3, +15q24.1, +15q25.1, +15q25.2, +15q25.3, +15q26.1, +16p11.2, +16p12.3, +16q11.2, +16q12.1, +16q13, +16q21, +16q22.1, +17p11.2, +17q11.2, +17q23.1, +18q11.2, +19q13.11, +20p11.21, +20p11.1, +20p11.23, +20p12.2, 20q11.21, +20q11.23, +22q11.21, +22q11.23, +22q12.2, +22q12.3 |
| PG100 | +9p12-p23 | +1p13.2, +1p13.3, +1p21.3, +1q21.1, +1q21.2, +1q21.3, +2p11.2, +4p15.1, +6q13, +7q11.21, +7q11.22, +7q11.23, +8p11.21, +8p11.23-p11.22, +8p21.2, +8p21.3, -8q11.21, +8q24.11, +8q24.13, +8q24.22, -8q24.22, +8q24.23, +8q24.3, +9p13.2, +9p13.3, +9p22.3, +9q21.12, +9q21.13, +10p11.22, +10p12.1, +10p12.2, +10p12.31, +10p14, +10q11.21, +11p11.2, -11q12.1, -11q12.2, -11q12.3, +12q12-q15, +13q12.11, +14q11.2, +15q11.2, +15q12, +15q14, +15q15.1, +15q15.3, +15q24.1, +15q25.2, +15q25.3, +15q26.1, +16p11.2, +16p12.3, +16q11.2, +16q12.1, +16q21, +16q22.1, +17p11.2, +17q11.2, +17q23.1, +18q11.2, +19q13.11, +20p11.1, +20p11.23, +20p12.2, +20q11.21, +20q11.23, +22q11.21, +22q11.23, +22q12.1-q12.2, +22q12.2, +22q12.3 |
Notably, the gain of chromosome region 9p13 was common in all cell lines and as such, this locus was selected for further investigation.
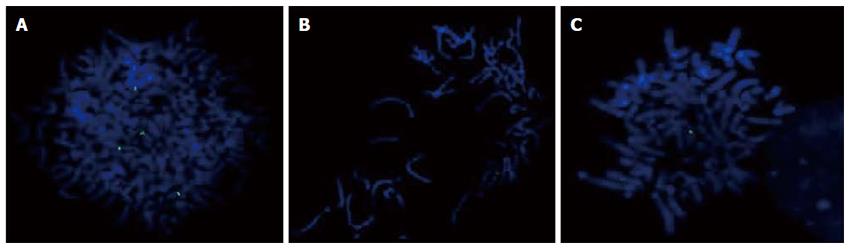
The presence of the 9p13 amplification in the GC cell lines was confirmed by metaphase FISH using a BAC clone (Figure 1). We observed signal gain in all cell lines, and only ACP02 showed high amplification of this region (Table 2).
| Cell line | 0 signal | 1 signal | 2 signals | 3 signals | 4 signals | ≥5 signals |
| ACP02 | 12 (6.0) | 26 (13.0) | 112 (56.0) | 25 (12.5) | 22 (11.0) | 3 (1.5) |
| ACP03 | 22 (11.0) | 40 (20.0) | 99 (49.5) | 22 (11.0) | 6 (3.0) | 1 (0.5) |
| AGP01 | 19 (9.5) | 45 (22.5) | 99 (49.5) | 27 (13.5) | 10 (5.0) | - |
| PG100 | 18 (9.0) | 55 (27.5) | 87 (43.5) | 32 (16.0) | 7 (3.5) | 1 (0.5) |
| Control | 34 (17.0) | 68 (34.0) | 97 (48.5) | 1 (0.5) | - | - |
Based on gene location and annotated gene function, we selected the MELK and IL11RA genes for validation in GC cell lines and in 68 primary gastric adenocarcinoma by qPCR. We detected two copies of IL11RA and three copies of MELK in all GC cell lines. By analyzing the CNV of these two genes in gastric tumors, we observed that 19.1% (13/68) and 55.9% (38/68) of gastric tumors had ≥ 3 copies of IL11RA and MELK, respectively. No association was found between the clinicopathological characteristics of patients and the number of copies of the studied genes (Table 3).
| MELK | IL11RA | |||||
| 2 copies (n = 38) | ≥3 copies (n = 30) | P value | 2 copies (n = 55) | ≥3 copies (n = 13) | P value | |
| Age (yr) (mean ± SD) | ||||||
| > 50 (64.5 ± 6.9) | 23 | 25 | 0.0748 | 39 | 9 | 0.7461 |
| ≤ 50 (42.5 ± 5.2) | 15 | 5 | 16 | 4 | ||
| Gender | ||||||
| Male | 23 | 21 | 0.5781 | 34 | 9 | 0.7544 |
| Female | 15 | 9 | 21 | 4 | ||
| Histopathology | ||||||
| Intestinal | 23 | 20 | 0.7886 | 32 | 11 | 0.1110 |
| Diffuse | 15 | 10 | 23 | 2 | ||
| Depth of tumor invasion | ||||||
| pT1-pT2 | 10 | 11 | 0.5137 | 18 | 3 | 0.4076 |
| pT3-pT4 | 28 | 19 | 36 | 10 | ||
| Lymph node metastasis | ||||||
| Absent | 10 | 7 | 1.0000 | 11 | 6 | 0.1091 |
| Present | 28 | 23 | 44 | 7 | ||
| Stage | ||||||
| I-II | 25 | 17 | 0.6049 | 33 | 9 | 0.7525 |
| III-IV | 13 | 13 | 22 | 4 | ||
aCGH is a high resolution tool that allows the simultaneous detection of sub-microscopic copy number changes across the genome, thus overcoming the several limitations of cCHG[23]. In this study, most of the copy number changes observed in ACP02, ACP03, AGP01 and PG100 by cCGH were confirmed by aCGH. ACP02, ACP03 and AGP01 are gastric adenocarcinoma cell lines from diffuse and intestinal types and cancerous ascitic fluid and were previously established and characterized by our research group[17,18], while PG100 is a commercially available primary gastric adenocarcinoma cell line[19]. Furthermore, aCGH analysis enabled the identification of many additional chromosomal gains and losses. On the other hand, the gain of the 16p21-p23 region in ACP03 and the gains of the 6p11-p12, 12p11.1 and 18p11.2-p11.3 regions in AGP01 were only detected by cCGH. This may be due to technical reasons, as cCGH is more sensitive than aCGH for detecting large chromosome regions, as previously discussed by Kamradt et al[24].
When comparing the GC cell lines, only a few differences in cytogenetic composition were found by cCGH and aCGH. The gain on 9p13.3 was found in all cell lines, and the presence of this amplicon in these gastric cell lines was confirmed by metaphase FISH, using a BAC clone for the amplified region. It is noteworthy that high levels of this amplification were only found in ACP02.
Genetic alterations in the short arm of chromosome 9 are commonly observed in different cancer types[25]. In GC, losses of 9p have been frequently described[26-29]. Fan et al[29] (2012) observed a homozygous deletion at 9p21, which encompasses the P16INK4A tumor suppressor gene, in 11% (8/72) of the gastric tumors studied. To our knowledge, this is the first study that describes gains at 9p in GC.
Amplifications on 9p have been reported in esophageal cancer[30], lung sarcomatoid carcinoma[31] and breast cancer[32]. Towle et al[33] found that 16.6% (36/217) of the cell lines carried regions of genomic gain spanning part of chromosome 9p13. Additionally, 1.8% (4/217) harbored high-level DNA amplification of this region, including a ductal breast carcinoma line (B0T-474), a tongue squamous cell carcinoma line (SCC-9), a melanoma line (WM-115), and an osteosarcoma line (MG-63).
Because this region harbors several tumor-related genes, several studies in the literature have correlated gene copy number alterations of 9p13 with cancer[24,34]. Sarhadi et al[34] observed that the gain of chromosome 9p13 encompasses many genes, such as KIAA1161, C9orf24, C9orf25, DNAI1, ENHO, CNTFR, LOC415056, C9orf23, DCTN3, ARID3C, SIGMAR1, GALT, IL11RA, CCL27, CCL19, CCL21 and FAM205A, in different types of cancer.
In this study, we selected the IL11RA and MELK genes to validate this amplification region in GC cell lines and primary gastric adenocarcinoma. The results showed an increase in the copy number of the MELK gene in ACP02, ACP03, AGP01 and PG100. Moreover, 19.1% (13/68) and 55.9% (38/68) of gastric tumors showed ≥ 3 copies of IL11RA and MELK, respectively.
Kamradt et al[24] analyzed a small amplicon at 9p13.3 in prostate cancer cell lines and validated IL11RA copy number gain in 75% (15/20) of prostate tumors. In addition, it has been demonstrated that IL11RA is overexpressed in GC, colon cancer, breast cancer, prostate cancer and osteosarcoma[35-41]. IL11RA encodes a specific receptor for IL11, and the IL11/IL11RA signaling pathway is involved in the regulation of several biological activities, such as adipogenesis, osteoclastogenesis, neurogenesis, and megakaryocyte maturation and platelet production[42,43].
With regard to MELK, the other gene that was selected for validation, this study describes, for the first time, that the copy number gain of the MELK gene occurs in cancer. To our knowledge, only one previous study on astrocytoma samples has investigated MELK amplification, and they did not find any MELK copy number gain[44].
MELK is a highly conserved serine/threonine kinase that was first found to be expressed in a wide range of early embryonic cellular stages, and as a result, it has been implicated in embryogenesis and cell cycle control[45]. Additionally, several studies have identified MELK overexpression in stem cell populations and several human cancers, including aggressive astrocytoma, breast cancer, prostate cancer, melanoma and GC[44-49].
Preclinical studies have suggested MELK as a potential therapeutic target for multiple cancers. Since then, novel therapeutics that selectively inhibit MELK have been developed, such as OTSSP167, which is currently in a Phase I trial for patients with solid tumors and who have not responded to treatment[45,50-53].
Li et al[54] observed MELK overexpression more frequently in GC lesions than in the corresponding noncancerous mucosa and that higher MELK levels were associated with lymph node involvement, distant metastasis, and poor prognosis in patients with GC. In addition, these authors demonstrated that reducing MELK expression or inhibiting its kinase activity resulted in growth inhibition, G2/M arrest, apoptosis and the suppression of the invasive capability of GC cells in vitro and in vivo. MELK knockdown also led to alterations in the levels of epithelial mesenchymal transition (EMT)-associated proteins. Furthermore, in GC patient-derived xenograft models, targeted treatment with OTSSP167 showed anticancer effects. These results suggest that MELK may be a promising target for GC treatment.
In conclusion, our results from generating genome wide DNA copy number profiles in GC cell lines and validation in primary gastric adenocarcinoma specimens revealed genomic aberrations redundancies, indicating that the cell lines retain the gross genomic architecture of primary tumors. Moreover, the characterization of a small gained region at 9p13.3 in GC cell lines and primary gastric adenocarcinoma samples revealed MELK as a candidate target gene this region that may possibly be linked to the development of GC. Therefore, we hypothesize that the copy number gain of MELK may be a mechanism of gene overexpression and may represent an interesting therapeutic target in gastric carcinogenesis.
Despite alterations in DNA copy number is one of the key hallmarks of carcinogenesis, the chromosomal regions with frequent gain and loss are still poorly defined in gastric cancer. The characterization of a small gain or loss region in gastric cancer cell lines and primary gastric adenocarcinoma samples could reveal a candidate target gene that may possibly be linked to the development of gastric cancer.
DNA copy number profiles in gastric cancer cell lines and validation in primary gastric adenocarcinoma specimens revealed genomic aberrations redundancies, indicating that the cell lines retain the gross genomic architecture of primary tumors. Moreover, the characterization of a small gained region at 9p13.3 in gastric cancer cell lines and primary gastric adenocarcinoma samples revealed MELK as a candidate target gene that is possibly related to the development of gastric cancer.
Several studies in the literature have correlated gene copy number alterations of 9p13 region. A study described a small amplicon at 9p13.3 in prostate cancer cell lines and validated IL11RA copy number gain in 75% (15/20) of prostate tumors. However, this is the first time that the copy number gain of the MELK gene was described in tumor. Furthermore, in gastric cancer patient-derived xenograft models, targeted treatment with OTSSP167 (a MELK inhibitor) showed anticancer effects. These results suggest that MELK may be a promising target for gastric cancer treatment.
The authors suggested that the copy number gain of MELK may be a mechanism of gene overexpression and may represent an interesting therapeutic target in gastric carcinogenesis in the future.
Copy number variation (CNV) is a type of structural variation characterized by duplication or deletion of sections of the genome, which in turn can result in phenotypic alterations. Array comparative genomic hybridization (aCGH) is a technology developed for a high-resolution evaluation of DNA copy number alterations associated with chromosome abnormalities.
The authors tried to identify common copy number alterations by using chromosomal comparative genome hybridization and array comparative genome hybridization in four gastric cancer cell lines. They concluded MELK as a candidate target gene that is possibly related to the development of gastric cancer.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Kupeli S S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 1. | Globocan 2012. Available from: http://globocan.iarc.fr. |
| 2. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] |
| 3. | Burbano RR, Assumpção PP, Leal MF, Calcagno DQ, Guimarães AC, Khayat AS, Takeno SS, Chen ES, De Arruda Cardoso Smith M. C-MYC locus amplification as metastasis predictor in intestinal-type gastric adenocarcinomas: CGH study in Brazil. Anticancer Res. 2006;26:2909-2914. [PubMed] |
| 4. | Calcagno DQ, Leal MF, Taken SS, Assumpção PP, Demachki S, Smith Mde A, Burbano RR. Aneuploidy of chromosome 8 and C-MYC amplification in individuals from northern Brazil with gastric adenocarcinoma. Anticancer Res. 2005;25:4069-4074. [PubMed] |
| 5. | Calcagno DQ, Guimarães AC, Leal MF, Seabra AD, Khayat AS, Pontes TB, Assumpção PP, De Arruda Cardoso Smith M, Burbano RR. MYC insertions in diffuse-type gastric adenocarcinoma. Anticancer Res. 2009;29:2479-2483. [PubMed] |
| 6. | Panani AD. Cytogenetic and molecular aspects of gastric cancer: clinical implications. Cancer Lett. 2008;266:99-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 7. | Koo SH, Kwon KC, Shin SY, Jeon YM, Park JW, Kim SH, Noh SM. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence In situ hybridization studies. Cancer Genet Cytogenet. 2000;117:97-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 8. | Wu MS, Chang MC, Huang SP, Tseng CC, Sheu JC, Lin YW, Shun CT, Lin MT, Lin JT. Correlation of histologic subtypes and replication error phenotype with comparative genomic hybridization in gastric cancer. Genes Chromosomes Cancer. 2001;30:80-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 9. | Kimura Y, Noguchi T, Kawahara K, Kashima K, Daa T, Yokoyama S. Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: gain of 20q and loss of 18q are associated with tumor progression. Mod Pathol. 2004;17:1328-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Takeno SS, Leal MF, Lisboa LC, Lipay MV, Khayat AS, Assumpção PP, Burbano RR, Smith Mde A. Genomic alterations in diffuse-type gastric cancer as shown by high-resolution comparative genomic hybridization. Cancer Genet Cytogenet. 2009;190:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Seabra AD, Araújo TM, Mello Junior FA, Di Felipe Ávila Alcântara D, De Barros AP, De Assumpção PP, Montenegro RC, Guimarães AC, Demachki S, Burbano RM. High-density array comparative genomic hybridization detects novel copy number alterations in gastric adenocarcinoma. Anticancer Res. 2014;34:6405-6415. [PubMed] |
| 12. | Calcagno DQ, Leal MF, Seabra AD, Khayat AS, Chen ES, Demachki S, Assumpção PP, Faria MH, Rabenhorst SH, Ferreira MV. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World J Gastroenterol. 2006;12:6207-6211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 58] [Cited by in RCA: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Calcagno DQ, Freitas VM, Leal MF, de Souza CR, Demachki S, Montenegro R, Assumpção PP, Khayat AS, Smith Mde A, dos Santos AK. MYC, FBXW7 and TP53 copy number variation and expression in gastric cancer. BMC Gastroenterol. 2013;13:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 14. | de Souza CR, Leal MF, Calcagno DQ, Costa Sozinho EK, Borges Bdo N, Montenegro RC, Dos Santos AK, Dos Santos SE, Ribeiro HF, Assumpção PP. MYC deregulation in gastric cancer and its clinicopathological implications. PLoS One. 2013;8:e64420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 15. | Costa Raiol LC, Figueira Silva EC, Mendes da Fonseca D, Leal MF, Guimarães AC, Calcagno DQ, Khayat AS, Assumpção PP, de Arruda Cardoso Smith M, Burbano RR. Interrelationship between MYC gene numerical aberrations and protein expression in individuals from northern Brazil with early gastric adenocarcinoma. Cancer Genet Cytogenet. 2008;181:31-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Liang L, Fang JY, Xu J. Gastric cancer and gene copy number variation: emerging cancer drivers for targeted therapy. Oncogene. 2016;35:1475-1482. [PubMed] |
| 17. | Leal MF, Martins do Nascimento JL, da Silva CE, Vita Lamarão MF, Calcagno DQ, Khayat AS, Assumpção PP, Cabral IR, de Arruda Cardoso Smith M, Burbano RR. Establishment and conventional cytogenetic characterization of three gastric cancer cell lines. Cancer Genet Cytogenet. 2009;195:85-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Leal MF, Calcagno DQ, Borges da Costa Jde F, Silva TC, Khayat AS, Chen ES, Assumpção PP, de Arruda Cardoso Smith M, Burbano RR. MYC, TP53, and chromosome 17 copy-number alterations in multiple gastric cancer cell lines and in their parental primary tumors. J Biomed Biotechnol. 2011;2011:631268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Ribeiro HF, Alcântara DF, Matos LA, Sousa JM, Leal MF, Smith MA, Burbano RR, Bahia MO. Cytogenetic characterization and evaluation of c-MYC gene amplification in PG100, a new Brazilian gastric cancer cell line. Braz J Med Biol Res. 2010;43:717-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Lima EM, Rissino JD, Harada ML, Assumpção PP, Demachki S, Guimarães AC, Casartelli C, Smith MA, Burbano RR. Conventional cytogenetic characterization of a new cell line, ACP01, established from a primary human gastric tumor. Braz J Med Biol Res. 2004;37:1831-1838. [PubMed] |
| 21. | Batista dos Santos SE, Rodrigues JD, Ribeiro-dos-Santos AK, Zago MA. Differential contribution of indigenous men and women to the formation of an urban population in the Amazon region as revealed by mtDNA and Y-DNA. Am J Phys Anthropol. 1999;109:175-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 22. | Kulikowski LD, Bellucco FT, Nogueira SI, Christofolini DM, Smith Mde A, de Mello CB, Brunoni D, Melaragno MI. Pure duplication 1q41-qter: further delineation of trisomy 1q syndromes. Am J Med Genet A. 2008;146A:2663-2667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Sireteanu A, Covic M, Gorduza EV. [Array CGH: technical considerations and applications]. Rev Med Chir Soc Med Nat Iasi. 2012;116:545-551. [PubMed] |
| 24. | Kamradt J, Jung V, Wahrheit K, Tolosi L, Rahnenfuehrer J, Schilling M, Walker R, Davis S, Stoeckle M, Meltzer P. Detection of novel amplicons in prostate cancer by comprehensive genomic profiling of prostate cancer cell lines using oligonucleotide-based arrayCGH. PLoS One. 2007;2:e769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Knuutila S, Aalto Y, Autio K, Björkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155:683-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 289] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 26. | Nessling M, Solinas-Toldo S, Wilgenbus KK, Borchard F, Lichter P. Mapping of chromosomal imbalances in gastric adenocarcinoma revealed amplified protooncogenes MYCN, MET, WNT2, and ERBB2. Genes Chromosomes Cancer. 1998;23:307-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 27. | Kang JU, Kang JJ, Kwon KC, Park JW, Jeong TE, Noh SM, Koo SH. Genetic alterations in primary gastric carcinomas correlated with clinicopathological variables by array comparative genomic hybridization. J Korean Med Sci. 2006;21:656-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Zhu YQ, Zhu ZG, Liu BY, Chen XH, Yin HR, Wang XH. [Chromosomal alterations analyzed by comparative genomic hybridization in primary gastric carcinoma]. Zhonghua Wei Chang Wai Ke Za Zhi. 2007;10:160-164. [PubMed] |
| 29. | Fan B, Dachrut S, Coral H, Yuen ST, Chu KM, Law S, Zhang L, Ji J, Leung SY, Chen X. Integration of DNA copy number alterations and transcriptional expression analysis in human gastric cancer. PLoS One. 2012;7:e29824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, Sugano S, Nakamura Y, Inazawa J. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000;60:4735-4739. [PubMed] |
| 31. | Italiano A, Attias R, Aurias A, Pérot G, Burel-Vandenbos F, Otto J, Venissac N, Pedeutour F. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23-p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Wu J, Liu S, Liu G, Dombkowski A, Abrams J, Martin-Trevino R, Wicha MS, Ethier SP, Yang ZQ. Identification and functional analysis of 9p24 amplified genes in human breast cancer. Oncogene. 2012;31:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | Towle R, Tsui IF, Zhu Y, MacLellan S, Poh CF, Garnis C. Recurring DNA copy number gain at chromosome 9p13 plays a role in the activation of multiple candidate oncogenes in progressing oral premalignant lesions. Cancer Med. 2014;3:1170-1184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | Sarhadi VK, Lahti L, Scheinin I, Ellonen P, Kettunen E, Serra M, Scotlandi K, Picci P, Knuutila S. Copy number alterations and neoplasia-specific mutations in MELK, PDCD1LG2, TLN1, and PAX5 at 9p in different neoplasias. Genes Chromosomes Cancer. 2014;53:579-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Campbell CL, Jiang Z, Savarese DM, Savarese TM. Increased expression of the interleukin-11 receptor and evidence of STAT3 activation in prostate carcinoma. Am J Pathol. 2001;158:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 117] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 36. | Kiessling S, Muller-Newen G, Leeb SN, Hausmann M, Rath HC, Strater J, Spottl T, Schlottmann K, Grossmann J, Montero-Julian FA. Functional expression of the interleukin-11 receptor alpha-chain and evidence of antiapoptotic effects in human colonic epithelial cells. J Biol Chem. 2004;279:10304-10315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 37. | Zurita AJ, Troncoso P, Cardó-Vila M, Logothetis CJ, Pasqualini R, Arap W. Combinatorial screenings in patients: the interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer Res. 2004;64:435-439. [PubMed] |
| 38. | Hanavadi S, Martin TA, Watkins G, Mansel RE, Jiang WG. Expression of interleukin 11 and its receptor and their prognostic value in human breast cancer. Ann Surg Oncol. 2006;13:802-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Nakayama T, Yoshizaki A, Izumida S, Suehiro T, Miura S, Uemura T, Yakata Y, Shichijo K, Yamashita S, Sekin I. Expression of interleukin-11 (IL-11) and IL-11 receptor alpha in human gastric carcinoma and IL-11 upregulates the invasive activity of human gastric carcinoma cells. Int J Oncol. 2007;30:825-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 40. | Lewis VO, Ozawa MG, Deavers MT, Wang G, Shintani T, Arap W, Pasqualini R. The interleukin-11 receptor alpha as a candidate ligand-directed target in osteosarcoma: consistent data from cell lines, orthotopic models, and human tumor samples. Cancer Res. 2009;69:1995-1999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 41. | Liu T, Ma Q, Zhang Y, Ke S, Yan K, Chen X, Wen Y, Fan Q, Qiu X. Interleukin-11 receptor α is overexpressed in human osteosarcoma, and near-infrared-labeled IL-11Rα imaging agent could detect osteosarcoma in mouse tumor xenografts. Tumour Biol. 2015;36:2369-2375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 42. | Schwertschlag US, Trepicchio WL, Dykstra KH, Keith JC, Turner KJ, Dorner AJ. Hematopoietic, immunomodulatory and epithelial effects of interleukin-11. Leukemia. 1999;13:1307-1315. [PubMed] |
| 43. | Teramura M, Kobayashi S, Yoshinaga K, Iwabe K, Mizoguchi H. Effect of interleukin 11 on normal and pathological thrombopoiesis. Cancer Chemother Pharmacol. 1996;38 Suppl:S99-S102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 44. | Marie SK, Okamoto OK, Uno M, Hasegawa AP, Oba-Shinjo SM, Cohen T, Camargo AA, Kosoy A, Carlotti CG, Toledo S. Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int J Cancer. 2008;122:807-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 116] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 45. | Ganguly R, Hong CS, Smith LG, Kornblum HI, Nakano I. Maternal embryonic leucine zipper kinase: key kinase for stem cell phenotype in glioma and other cancers. Mol Cancer Ther. 2014;13:1393-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 46. | Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One. 2007;2:e594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 231] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 47. | Pickard MR, Green AR, Ellis IO, Caldas C, Hedge VL, Mourtada-Maarabouni M, Williams GT. Dysregulated expression of Fau and MELK is associated with poor prognosis in breast cancer. Breast Cancer Res. 2009;11:R60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 48. | Kuner R, Fälth M, Pressinotti NC, Brase JC, Puig SB, Metzger J, Gade S, Schäfer G, Bartsch G, Steiner E. The maternal embryonic leucine zipper kinase (MELK) is upregulated in high-grade prostate cancer. J Mol Med (Berl). 2013;91:237-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 49. | Du T, Qu Y, Li J, Li H, Su L, Zhou Q, Yan M, Li C, Zhu Z, Liu B. Maternal embryonic leucine zipper kinase enhances gastric cancer progression via the FAK/Paxillin pathway. Mol Cancer. 2014;13:100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 50. | Gray D, Jubb AM, Hogue D, Dowd P, Kljavin N, Yi S, Bai W, Frantz G, Zhang Z, Koeppen H. Maternal embryonic leucine zipper kinase/murine protein serine-threonine kinase 38 is a promising therapeutic target for multiple cancers. Cancer Res. 2005;65:9751-9761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 148] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 51. | Chung S, Nakamura Y. MELK inhibitor, novel molecular targeted therapeutics for human cancer stem cells. Cell Cycle. 2013;12:1655-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 52. | Minata M, Gu C, Joshi K, Nakano-Okuno M, Hong C, Nguyen CH, Kornblum HI, Molla A, Nakano I. Multi-kinase inhibitor C1 triggers mitotic catastrophe of glioma stem cells mainly through MELK kinase inhibition. PLoS One. 2014;9:e92546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 53. | Ganguly R, Mohyeldin A, Thiel J, Kornblum HI, Beullens M, Nakano I. MELK-a conserved kinase: functions, signaling, cancer, and controversy. Clin Transl Med. 2015;4:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 54. | Li S, Li Z, Guo T, Xing XF, Cheng X, Du H, Wen XZ, Ji JF. Maternal embryonic leucine zipper kinase serves as a poor prognosis marker and therapeutic target in gastric cancer. Oncotarget. 2016;7:6266-6280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |