Published online Nov 7, 2016. doi: 10.3748/wjg.v22.i41.9214
Peer-review started: August 11, 2016
First decision: September 12, 2016
Revised: September 27, 2016
Accepted: October 19, 2016
Article in press: October 19, 2016
Published online: November 7, 2016
Processing time: 88 Days and 17 Hours
To compare (1) demographics in urea breath test (UBT) vs endoscopy patients; and (2) the molecular detection of antibiotic resistance in stool vs biopsy samples.
Six hundred and sixteen adult patients undergoing endoscopy or a UBT were prospectively recruited to the study. The GenoType HelicoDR assay was used to detect Helicobacter pylori (H. pylori) and antibiotic resistance using biopsy and/or stool samples from CLO-positive endoscopy patients and stool samples from UBT-positive patients.
Infection rates were significantly higher in patients referred for a UBT than endoscopy (overall rates: 33% vs 19%; treatment-naïve patients: 33% vs 14.7%, respectively). H. pylori-infected UBT patients were younger than H. pylori-infected endoscopy patients (41.4 vs 48.4 years, respectively, P < 0.005), with a higher percentage of H. pylori-infected males in the endoscopy-compared to the UBT-cohort (52.6% vs 33.3%, P = 0.03). The GenoType HelicoDR assay was more accurate at detecting H. pylori infection using biopsy samples than stool samples [98.2% (n = 54/55) vs 80.3% (n =53/66), P < 0.005]. Subset analysis using stool and biopsy samples from CLO-positive endoscopy patients revealed a higher detection rate of resistance-associated mutations using stool samples compared to biopsies. The concordance rates between stool and biopsy samples for the detection of H. pylori DNA, clarithromycin and fluoroquinolone resistance were just 85%, 53% and 35%, respectively.
Differences between endoscopy and UBT patients provide a rationale for non-invasive detection of H. pylori antibiotic resistance. However, the GenoType HelicoDR assay is an unsuitable approach.
Core tip: The successful detection of clarithromycin and/or fluoroquinolone resistant Helicobacter pylori (H. pylori) infections by non-invasive methods would enable a widespread assessment of resistance rates. Here we evaluate the GenoType HelicoDR assay for the detection of clarithromycin and fluoroquinolone resistance using DNA isolated from stool samples compared to biopsy samples. Although results using this assay on biopsy tissue have previously been shown to correspond well with culture and antimicrobial susceptibility testing, there was weak correlation between results obtained using biopsy vs stool samples in our study. Further studies are required to optimise the non-invasive detection of clarithromycin and fluoroquinolone resistant H. pylori infection.
- Citation: Brennan DE, Omorogbe J, Hussey M, Tighe D, Holleran G, O’Morain C, Smith SM, McNamara D. Molecular detection of Helicobacter pylori antibiotic resistance in stool vs biopsy samples. World J Gastroenterol 2016; 22(41): 9214-9221
- URL: https://www.wjgnet.com/1007-9327/full/v22/i41/9214.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i41.9214
Helicobacter pylori (H. pylori) is a gram-negative bacterium that specifically colonizes the epithelium of the human stomach, in particular the gastric antrum. It infects approximately 50% of the world’s population. The prevalence of H. pylori varies globally, increasing with older age and lower socio-economic status. Most infected individuals will not develop any clinically significant complications; however the most common symptoms of infection are gastritis and gastric or duodenal ulcers. The diagnosis and treatment of H. pylori infection are critical factors in the prevention and management of these conditions[1-3]. H. pylori infection can be detected by invasive and non-invasive means, using a variety of diagnostic tests. The Maastricht IV/Florence Consensus Report recommends the “Test and Treat” strategy for patients presenting with uncomplicated dyspepsia with no alarm symptoms associated with an increased risk of gastric cancer[2]. In the Irish healthcare setting, the urea breath test (UBT) is the current gold standard non-invasive test for H. pylori infection in patients managed by the “Test and Treat” strategy. The UBT is highly accurate with a sensitivity of 88%-95% and specificity of 95%-100%[4]. For patients presenting with new onset dyspepsia (above 45 years; European guidelines) or dyspepsia along with accompanying alarm symptoms such as weight loss, gastrointestinal bleeding, abdominal mass or iron deficient anaemia, endoscopy is recommended[2]. When an endoscopy is performed, H. pylori infection can be diagnosed using gastric biopsy specimens. The most common test employed is the rapid urease-test for Campylobacter-like organisms (CLO), which has a sensitivity of > 90% and specificity of > 95%[5].
Treatment for H. pylori infection is recommended in all symptomatic individuals. However, eradication rates have fallen in many countries in recent years[6-8] mainly due to poor patient compliance and the emergence of antibiotic resistant strains of H. pylori, particularly to clarithromycin and levofloxacin[9-11]. The European Helicobacter and Microbiota Study Group (EHMSG) and the most recent Maastricht IV/Florence Consensus recommend local surveillance of existing and emerging antibiotic resistance and that the combination of antibiotics for H. pylori eradication should be chosen according to the local resistance patterns[2,10]. Clarithromycin-based first-line triple therapy is no longer recommended in regions where antibiotic resistance surveillance indicates that clarithromycin resistance is above 15%-20%[2]. Since H. pylori is a fastidious bacterium, culture and antimicrobial sensitivity testing is time-consuming. The sensitivity of culture of H. pylori from gastric biopsy samples has been reported to be as low as 55%[11]. Molecular testing represents an attractive alternative to culture-based methods and has been recommended by the Maastricht Consensus guidelines to detect H. pylori and both clarithromycin and fluoroquinolone resistance when standard culture and sensitivity testing are unavailable[2]. Single point mutations (most commonly A2146C, A2146G and A2147G) within the H. pylori rrl gene that encodes the 23S ribosomal subunit confer clarithromycin resistance[11,12]. The most significant mutations conferring fluoroquinolone resistance are located at positions 87 (N87K) and 91 (D91N, D91G, D91Y) of the H. pylori gyrA gene, which encodes the A subunit of the DNA gyrase enzyme[11,12]. The GenoType HelicoDR assay allows for the molecular genetic identification of H. pylori and its resistance to clarithromycin and fluoroquinolones, such as levofloxacin. The assay has been reported to be efficient at detecting mutations predictive of antibiotic resistance when applied to H. pylori cultures or gastric biopsy specimens[13-16], with a sensitivity and specificity of 94%-100% and 86%-99% for detecting clarithromycin resistance and 83%-87% and 95%-98.5% for detecting fluoroquinolone resistance, respectively[16,17].
Currently, H. pylori antibiotic resistance surveillance is based primarily on patients undergoing invasive testing by means of endoscopy. However, most patients are diagnosed by non-invasive methods such as the UBT. As such, antibiotic resistance data obtained solely from endoscopy patients may not reflect the true prevalence of H. pylori infection and the rates of antibiotic resistance in symptomatic patients. The aims of this study were to (1) compare demographics and prevalence of H. pylori infection in patients referred for endoscopy with those of patients referred for a UBT; and (2) evaluate the potential use of the GenoType HelicoDR assay for the non-invasive detection of H. pylori and antibiotic resistant infection using stool samples.
A prospective study was carried out in a tertiary referral teaching hospital (Adelaide and Meath Hospital, Dublin, Ireland) affiliated with Trinity College Dublin. Patients who had been referred to the endoscopy clinic were included from August 2014 until March 2016. The study received ethical approval from the Adelaide and Meath Hospital Research Ethics Committee. Informed consent was obtained from all patients before enrolment.
Inclusion criteria were (1) ability and willingness to participate in the study and to provide informed consent; and (2) confirmed H. pylori infection by UBT or a positive rapid urease test (TRI-MED Distributors, PTY LTD, Washington, United States) at 60 min performed and/or histology.
Exclusion criteria were (1) age less than 18 years; (2) pregnancy or lactation; (3) severe inter-current illness; (4) current PPI use or recent antibiotic use (within 4 wk); and (5) bleeding problems or use of blood thinning drugs (for endoscopy patients).
A single corpus and antrum biopsy from each patient was placed into DENT transport medium (brain heart infusion broth containing 2.5% (w/v) yeast extract, 5% sterile horse serum and Dent Helicobacter Selective Supplement; Oxoid, Basingstoke, United Kingdom) for transport to the research laboratory. Biopsies were placed into fresh collection tubes and stored at -20 °C until processed for genomic DNA isolation using the QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany) according to manufacturer’s instructions. Patients attending for endoscopy or the UBT were invited to provide a stool sample collected within 24 h of their appointment. Stool samples were stored at 4 °C until processed for genomic DNA isolation using the PSP Spin Stool DNA Plus Kit (STRATEC Molecular GmbH, Berlin, Germany) according to the manufacturers’ instructions. All isolated DNA was stored -20 °C until genotyping for clarithromycin and fluoroquinolone-mediating mutations was performed using the Genotype HelicoDR assay (Hain Lifescience GmbH, Nehren, Germany). Multiplex amplification of DNA regions of interest was performed using the biotinylated primers supplied in the GenoType HelicoDR kit and the Hotstart Taq DNA polymerase kit (Qiagen). PCR products were reverse hybridised to DNA strips containing probes for gene regions of interest, developed and interpreted according to the manufacturers’ instructions. Briefly, all strips were analysed for the presence of a conjugate control band (to indicate successful conjugate binding and substrate reaction), an amplification control band (to indicate a successful amplification reaction), a H. pylori control band (to document the presence of a H. pylori strain) and gene locus control bands for gyrA and 23S (to indicate successful detection of the gene regions of interest). In addition, the strips were analysed for the presence of wild type and/or mutation bands. An infection was considered clarithromycin sensitive when the 23S wild-type probe stained positive and clarithromycin resistant if one of the 23S mutation probes stained positive. As per manufacturers’ instructions, results of both positions of the gyrA gene were combined to draw conclusions about fluoroquinolone resistance. Thus, an infection was only considered fluoroquinolone sensitive when one of the wild-type probes for codon 87 of the gyrA gene stained positive together with a positive wild-type probe for codon 91. Fluoroquinolone resistance was indicated if either the wild-type probes for codon 87 or the wild-type probe for codon 91 stained negative, or if one of the mutant codon 87 or 91 probes stained positive. For all mutations probes, only bands whose intensities were equal to or stronger than the amplification control were considered positive.
Statistical analysis was carried out using GraphPad Prism (GraphPad Software Inc., CA, United States). Continuous variables are presented as arithmetic mean and SD. P values for continuous variables were calculated and compared using the two-tailed independent t-test. Categorical variables are presented as percentages and 95% confidence intervals (95%CI). P values for categorical variables were calculated using the Fisher’s exact test/Pearson χ2 test. In all cases, a P value less than 0.05 was considered significant.
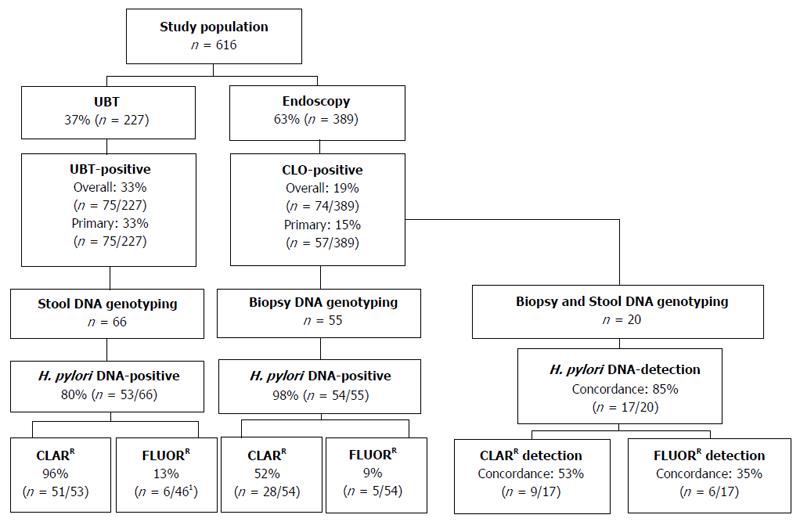
A schematic of patient inclusion and analysis is presented in Figure 1. In all, 616 patients were included in the study between August 2014 and March 2016; 389 patients (mean age 52.3 years, 42.2% male) underwent endoscopy and 227 patients (mean age 39.6 years, 30.4% male) a UBT (Table 1). The overall prevalence of H. pylori infection was significantly higher in the UBT cohort than the endoscopy cohort [33.0% (n = 75) vs 19% (n = 74), P < 0.001; 95%CI: 6.58-21.54] (Figure 1). Of the H. pylori-positive endoscopy patients (CLO-positive), 17 had been previously treated for H. pylori infection, therefore the prevalence of primary H. pylori infection was 14.7% (n = 57). All of the H. pylori-positive UBT patients were treatment naïve, thus the prevalence of primary H. pylori infection was also significantly higher in patients referred for UBT than for endoscopy (33.0% vs 14.7%, P < 0.001, 95%CI: 11.07-25.65; Figure 1 and Table 1). In keeping with the guidelines recommending endoscopy for symptomatic patients over 45 years, H. pylori-positive patients in the endoscopy cohort were significantly older than those in the UBT cohort (48.4 years vs 41.4 years; P < 0.005, 95%CI: 2.19-11.81). There were a greater number of H. pylori-positive men in the endoscopy cohort than the UBT cohort (52.6% vs 33.3%, P = 0.03, 95%CI: 1.23-36.29; Table 1). Taken together, these findings indicate significant differences in demographics and the prevalence of both overall and primary H. pylori infection rates in patients referred for endoscopy and those referred for the UBT.
| Endoscopy | UBT | Pvalue | 95%CI | |
| Total | n = 389 | n = 227 | ||
| Mean age (SD) | 52.3 (16.4) | 39.6 (12.6) | < 0.001 | 10.22-15.18 |
| Male | 42.2% (n = 164) | 30.4% (n = 69) | < 0.005 | 3.68-19.60 |
| Primary H. pylori infection | 14.7% (n = 57) | 33.0% (n = 75) | < 0.001 | 11.07-25.65 |
| Mean age (SD) | 48.4 (14.9) | 41.4 (13.0) | < 0.005 | 2.19-11.81 |
| Male | 52.6% (n = 30) | 33.3% (n = 25) | 0.03 | 1.23-36.29 |
The GenoType HelicoDR assay is based on DNA strip technology that enables the molecular genetic identification of H. pylori and resistance to fluoroquinolones and/or clarithromycin by detecting the most frequent mutations in the gyrA gene (codons 87 and 91) and the 23S gene (positions 2146 and 2147), respectively. Previous studies have demonstrated strong correlations between results obtained using the GenoType HelicoDR assay on biopsy specimens compared to culture and antimicrobial testing[14,16,17]. In order to evaluate the GenoType HelicoDR assay for the non-invasive detection of H. pylori using stool samples, we first set out to compare the detection rate of H. pylori infection using stool samples from H. pylori-positive UBT patients with that obtained using biopsy samples from CLO-positive endoscopy patients. Initial control experiments showed that the assay did not detect H. pylori DNA in stool samples from 2 uninfected UBT-negative patients (not shown). In H. pylori-infected patients, the GenoType HelicoDR assay was significantly more accurate at detecting H. pylori infection using biopsy samples than stool samples [98.2% (n = 54/55) vs 80.3% (n = 53/66), P < 0.005, 95%CI: 6.10-29.66] (Figure 1 and Table 2).
Next, the prevalence of antibiotic resistance-mediating mutations was compared using stool samples from H. pylori-positive UBT patients and biopsy samples from CLO-positive endoscopy patients. Using the GenoType HelicoDR assay, the 23S gene locus control was positive in all of H. pylori-positive DNA samples isolated from either biopsy tissue (100%, n = 54/54) or stool specimens (100%, n = 53/53). A significantly higher level of clarithromycin resistance-mediating mutations was detected using stool samples from UBT-positive patients than biopsy samples from CLO-positive patients [96.2% (n = 51/53) vs 51.9% (n = 28/54), P < 0.001, 95%CI: 27.70-58.71] (Figure 1 and Table 2).
In terms of gyrA genotyping, the gyrA locus control probe was positive in all DNA samples isolated from biopsy tissue (100%, n = 54/54), but only 86.8% (n = 46/53) of H. pylori-positive DNA samples isolated from stool. Fluoroquinolone resistance-mediating mutations were detected in 9.3% (n = 5/54) of biopsy samples from CLO-positive patients compared to 13% (n = 6/46) of stool samples from UBT-positive patients (P = 0.56, 95%CI: -9.99-18.28; Figure 1 and Table 2). For both endoscopy and UBT patients, all samples that were positive for fluoroquinolone resistance mutations were positive for clarithromycin resistance mutations (Table 2). Taken together, these findings indicate that the GenoType HelicoDR assay is more accurate at detecting H. pylori DNA using biopsies from CLO-positive endoscopy patients than stool DNA isolated from UBT-positive patients. In addition, the assay detected a significantly higher rate of clarithromycin resistance using stool samples from patients diagnosed by the UBT than that obtained when biopsy samples from CLO-positive endoscopy patients were analysed.
Given the high rate of clarithromycin resistance detected using stool specimens from UBT-positive patients (96.2%; Table 2) and the lack of published data on the use of the GenoType HelicoDR assay for stool sample analysis, we next set out to directly compare a stool DNA sample with that of a biopsy DNA sample isolated from a subset of the CLO-positive endoscopy patients. In all, stool and biopsy samples from 20 CLO-positive patients were analysed (mean age 46.8 ± 15.8 years, 50% male). H. pylori DNA was detected in 95% (n = 19/20) of biopsy samples and 90% (n = 18/20) of stool samples from the CLO-positive patients. Concordance between results from biopsy and stool samples of individual patients for the detection of H. pylori DNA was 85% (n = 17/20; Figure 1 and Table 3). In terms of antibiotic resistance, results were compared in the 17 patients with concordant results for the presence of H. pylori DNA in both their stool and biopsy samples. Concordance for the analysis of stool and biopsy samples of individual patients was just 52.9% (n = 9/17) for clarithromycin resistance and 35.3% (n = 6/17) for fluoroquinolone resistance (Figure 1, Table 3). Higher rates of both clarithromycin and fluoroquinolone resistance were detected in stool samples compared to biopsy samples obtained from the same patient (Table 3), suggesting a lack of specificity of the assay for the detection of antibiotic resistance-mediating mutations using DNA isolated from stool samples.
As the recommended first-line therapy for H. pylori infection should be guided by the local prevalence of primary clarithromycin resistance and third-line and subsequent treatment regimens should be guided by antimicrobial susceptibility testing[2], methods for detecting antibiotic resistance are of great interest. Antimicrobial susceptibility testing for H. pylori is mainly performed using biopsy specimens obtained by invasive means at endoscopy. As a result, findings on the prevalence of H. pylori infection and antibiotic resistance based solely on this patient cohort may not represent the true rates of resistance in a given population. In order to determine whether H. pylori-infected endoscopy patients are representative of the wider H. pylori-infected population, we compared the prevalence of infection and patient demographics between endoscopy patients with those referred for non-invasive H. pylori diagnoses by the UBT. Indeed we found significant differences between the two patient cohorts. Both the overall infection rate and the prevalence of primary infection in H. pylori treatment-naïve patients were significantly higher in patients referred for a UBT than endoscopy (overall infection rates of 33% vs 19% respectively, and primary infection rates of 33% vs 14.7%, respectively). H. pylori-infected UBT patients were also significantly younger than H. pylori-infected endoscopy patients (41.4 vs 48.4 years, respectively), with a higher percentage of H. pylori infected males in the endoscopy compared to UBT cohort (52.6% vs 33.3%). Both age and sex have been reported as risk factors for H. pylori antibiotic resistance, for example age > 50 years has been reported as a risk factor for levofloxacin resistance and being female has been associated with metronidazole resistance in the most recent pan-European study on antimicrobial resistance[10]. Thus the statistically significant differences in age and sex between endoscopy and UBT patients in our study suggests that H. pylori-infected endoscopy patients are likely not representative of the wider H. pylori-positive cohort, providing a strong rationale for performing more widespread antimicrobial susceptibility testing.
Successfully extending molecular-based methods to diagnose H. pylori non-invasively would greatly enhance our ability to more accurately assess the prevalence of resistance to a range of antibiotics, and enable clinicians to offer personalised antimicrobial susceptibility-based therapy to a wider number of patients. H. pylori DNA has been detected in a number of clinical specimens including blood, stool samples and oral cavity specimens [18-22]. Analysis of stool samples has shown the most promise for the molecular detection of clarithromycin resistance-mediating mutations to date[18,23-28]. Studies have demonstrated sensitivity and specificity values of 83%-98% and 98%-100%, respectively, for the detection clarithromycin resistance using the H. pylori ClariRes Assay (Ingenetix) to analyse stool samples[23-26]. However, data on the detection of H. pylori fluoroquinolone resistance using stool samples is lacking. Although the GenoType HelicoDR assay has proven useful for detecting clarithromycin and fluoroquinolone resistance in biopsy or culture specimens[13-17], evaluation of the assay for the analysis of stool specimens presented herein proved suboptimal. Firstly, the assay detected H. pylori infection in a significantly lower percentage of H. pylori-infected patients when stool rather than biopsy specimens were analysed (80%-90% vs 95%-98.2%; respectively Tables 2 and 3). As H. pylori specifically colonizes the stomach and is not an intestinal bacterium, it is present only in low numbers in the stool, a factor which may have impacted the sensitivity of H. pylori detection using stool samples in our study. Additionally, H. pylori DNA may be exposed to enzymatic or mechanical degradation during transit from the stomach through the intestines[22]. When results using biopsy samples from individual H. pylori-infected patients were directly compared with those obtained from their stool samples, concordance scores for clarithromycin and fluoroquinolone resistance were just 52.9% and 35.6%, respectively. In addition, a higher rate of clarithromycin and fluoroquinolone resistance was detected in DNA isolated from the stool samples compared to DNA isolated from biopsy samples from the same patient (Table 3). Given that previous studies have demonstrated strong correlations between results obtained using the GenoType HelicoDR assay on biopsy specimens compared to culture and antimicrobial testing[14,16,17], this would suggest that stool sample analysis using the GenoType HeliocDR assay is less sensitive than biopsy sample analysis, providing an explanation for the high rates of antibiotic resistance obtained using stool samples from the UBT patients in Table 2. The presence of large amounts of diverse commensal bacteria in the stool may hamper the specificity of the Genotype HelicoDR assay in detection of H. pylori antibiotic resistance-mediating mutations. Our findings suggest it is currently unsuitable for the accurate detection of clarithromycin and fluoroquinolone resistance-mediating mutations in stool specimens. Further studies are required to extend approaches for the non-invasive detection of H. pylori resistance to include multiple antibiotics. Recent advances in next generation DNA sequencing technologies may provide more robust opportunities for the accurate analysis of specific resistance-associated DNA regions. The successful optimisation of molecular-based antimicrobial susceptibility testing methods will enable resistance data obtained from patients managed by the “Test and Treat” strategy to be utilised in choosing effective antibiotics for the treatment of H. pylori. In this way, eradication rates for H. pylori may be improved.
Currently antimicrobial susceptibility testing for Helicobacter pylori (H. pylori) is mainly performed using cultures isolated from tissue biopsy samples obtained at endoscopy by invasive means. However, many patients are diagnosed with H. pylori infection by non-invasive means, such as the urea breath test. As such, antibiotic resistance data based solely on endoscopy patients may not truly reflect the prevalence of antibiotic resistance in the wider H. pylori infected population.
Molecular methods for the detection of H. pylori antibiotic resistance-mediating mutations offer a more rapid alternative to standard culture-based methods. Studies have shown that data generated using molecular methods on tissue biopsy samples correlates well with culture and antimicrobial susceptibility testing. Data on the use of molecular methods, in particular the GenoType HelicoDR assay, for the analysis of stool samples is limited.
The present findings suggest that the GenoType HelicoDR assay is not suitable for the accurate detection of antibiotic resistance-mediating mutations using stool samples from H. pylori infected patients. Alternative PCR or DNA sequencing-based methods may show more potential.
While the GenoType HelicoDR assay has been shown to be accurate for the analysis of clarithromycin- and fluoroquinolone-mediating mutations using biopsy tissue samples, the present findings indicate that this assay is not suitable for the analysis of stool samples.
The authors described an examination of antibiotic resistance in both gastric biopsy and stool samples obtained from patients who underwent testing for a urea breath test or had a gastroscopy performed. The main conclusion is that the Genotype HelicoDR assay is not appropriate for use on stool samples. This seriously limits its use and thus the paper is of importance and deserves to be published. It would have been useful to include formal sensitivity testing to the bacteria isolated on gastric biopsy.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Ireland
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: El-Zahaby SA, Malnick S S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 1. | McColl KE. Clinical practice. Helicobacter pylori infection. N Engl J Med. 2010;362:1597-1604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 550] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 2. | Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1719] [Cited by in RCA: 1588] [Article Influence: 122.2] [Reference Citation Analysis (5)] |
| 3. | Smith SM, Haider RB, O'Connor H, McNamara D, O'Morain C. Practical treatment of Helicobacter pylori: a balanced view in changing times. Eur J Gastroenterol Hepatol. 2014;26:819-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Howden CW, Hunt RH. Guidelines for the management of Helicobacter pylori infection. Ad Hoc Committee on Practice Parameters of the American College of Gastroenterology. Am J Gastroenterol. 1998;93:2330-2338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 220] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 5. | Gisbert JP, Pajares JM. Review article: 13C-urea breath test in the diagnosis of Helicobacter pylori infection -- a critical review. Aliment Pharmacol Ther. 2004;20:1001-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 257] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 6. | Malfertheiner P, Bazzoli F, Delchier JC, Celiñski K, Giguère M, Rivière M, Mégraud F. Helicobacter pylori eradication with a capsule containing bismuth subcitrate potassium, metronidazole, and tetracycline given with omeprazole versus clarithromycin-based triple therapy: a randomised, open-label, non-inferiority, phase 3 trial. Lancet. 2011;377:905-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 373] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 7. | Haider RB, Brennan DE, Omorogbe J, Holleran G, Hall B, O'Morain C, Breslin N, O'Connor HJ, Smith SM, McNamara D. A randomized-controlled study to compare the efficacy of sequential therapy with standard triple therapy for Helicobacter pylori eradication in an Irish population. Eur J Gastroenterol Hepatol. 2015;27:1265-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Smith SM. An update on the treatment of Helicobacter pylori infection. EMJ Gastroenterology. 2015;4:101-107. |
| 9. | Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59:1143-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 652] [Cited by in RCA: 735] [Article Influence: 49.0] [Reference Citation Analysis (1)] |
| 10. | Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62:34-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 632] [Article Influence: 52.7] [Reference Citation Analysis (3)] |
| 11. | Smith SM, O'Morain C, McNamara D. Antimicrobial susceptibility testing for Helicobacter pylori in times of increasing antibiotic resistance. World J Gastroenterol. 2014;20:9912-9921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 114] [Cited by in RCA: 112] [Article Influence: 10.2] [Reference Citation Analysis (1)] |
| 12. | Mégraud F, Bénéjat L, Ontsira Ngoyi EN, Lehours P. Molecular Approaches to Identify Helicobacter pylori Antimicrobial Resistance. Gastroenterol Clin North Am. 2015;44:577-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 13. | Vannarath S, Vilaichone RK, Rasachak B, Mairiang P, Yamaoka Y, Mahachai V. Antibiotic Resistant Pattern of Helicobacter Pylori Infection Based on Molecular Tests in Laos. Asian Pac J Cancer Prev. 2016;17:285-287. [PubMed] |
| 14. | Lee JW, Kim N, Nam RH, Park JH, Choi YJ, Kim JM, Kim JS, Jung HC. GenoType HelicoDR test in the determination of antimicrobial resistance of Helicobacter pylori in Korea. Scand J Gastroenterol. 2014;49:1058-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Tanih NF, Ndip RN. Molecular Detection of Antibiotic Resistance in South African Isolates of Helicobacter pylori. Gastroenterol Res Pract. 2013;2013:259457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Miendje Deyi VY, Burette A, Bentatou Z, Maaroufi Y, Bontems P, Lepage P, Reynders M. Practical use of GenoType® HelicoDR, a molecular test for Helicobacter pylori detection and susceptibility testing. Diagn Microbiol Infect Dis. 2011;70:557-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Cambau E, Allerheiligen V, Coulon C, Corbel C, Lascols C, Deforges L, Soussy CJ, Delchier JC, Megraud F. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J Clin Microbiol. 2009;47:3600-3607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 128] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 18. | Rimbara E, Sasatsu M, Graham DY. PCR detection of Helicobacter pylori in clinical samples. Methods Mol Biol. 2013;943:279-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Mentis A, Lehours P, Mégraud F. Epidemiology and Diagnosis of Helicobacter pylori infection. Helicobacter. 2015;20 Suppl 1:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 120] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 20. | Ismail H, Morgan C, Griffiths P, Williams J, Jenkins G. A Newly Developed Nested PCR Assay for the Detection of Helicobacter pylori in the Oral Cavity. J Clin Gastroenterol. 2016;50:17-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 21. | Ogaya Y, Nomura R, Watanabe Y, Nakano K. Detection of Helicobacter pylori DNA in inflamed dental pulp specimens from Japanese children and adolescents. J Med Microbiol. 2015;64:117-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Puz S, Innerhofer A, Ramharter M, Haefner M, Hirschl AM, Kovách Z, Rotter M, Makristathis A. A novel noninvasive genotyping method of Helicobacter pylori using stool specimens. Gastroenterology. 2008;135:1543-1551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Vécsei A, Innerhofer A, Binder C, Gizci H, Hammer K, Bruckdorfer A, Riedl S, Gadner H, Hirschl AM, Makristathis A. Stool polymerase chain reaction for Helicobacter pylori detection and clarithromycin susceptibility testing in children. Clin Gastroenterol Hepatol. 2010;8:309-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 24. | Scaletsky IC, Aranda KR, Garcia GT, Gonçalves ME, Cardoso SR, Iriya K, Silva NP. Application of real-time PCR stool assay for Helicobacter pylori detection and clarithromycin susceptibility testing in Brazilian children. Helicobacter. 2011;16:311-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 25. | Schabereiter-Gurtner C, Hirschl AM, Dragosics B, Hufnagl P, Puz S, Kovách Z, Rotter M, Makristathis A. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J Clin Microbiol. 2004;42:4512-4518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 147] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 26. | Lottspeich C, Schwarzer A, Panthel K, Koletzko S, Rüssmann H. Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. J Clin Microbiol. 2007;45:1718-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Xiong LJ, Tong Y, Wang Z, Mao M. Detection of clarithromycin-resistant Helicobacter pylori by stool PCR in children: a comprehensive review of literature. Helicobacter. 2013;18:89-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 28. | Noguchi N, Rimbara E, Kato A, Tanaka A, Tokunaga K, Kawai T, Takahashi S, Sasatsu M. Detection of mixed clarithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J Med Microbiol. 2007;56:1174-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |