Published online Jan 28, 2016. doi: 10.3748/wjg.v22.i4.1497
Peer-review started: June 1, 2015
First decision: July 14, 2015
Revised: August 6, 2015
Accepted: October 12, 2015
Article in press: October 13, 2015
Published online: January 28, 2016
Processing time: 236 Days and 19.9 Hours
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer-related death worldwide. Chronic infection of hepatitis B virus (HBV) and/or hepatitis C virus (HCV) is a major risk factor in the development of the HCC, independently from excessive alcohol abuse and metabolic disease. Since the biology of HBV and HCV is different, their oncogenic effect may go through different mechanisms, direct and/or indirect. Viral hepatitis infection is associated with cellular inflammation, oxidative stress, and DNA damage, that may lead to subsequent hepatic injuries such as chronic hepatitis, fibrosis, cirrhosis, and finally HCC. Direct oncogenic properties of these viruses are related with their genotypic characteristics and the ability of viral proteins to interact with host proteins, thus altering the molecular pathways balance of the cells. In addition, the integration of HBV DNA, especially the gene S and X, in a particular site of the host genome can disrupt chromosomal stability and may activate various oncogenic mechanisms, including those in hematopoietic cells. Recently, several studies also had demonstrated that viral hepatitis could trigger the population of hepatic cancer stem cells. This review summarize available pre-clinical and clinical data in literature regarding oncogenic properties of HBV and HCV in the early initiation of HCC.
Core tip: According to the most recent data released by the International Agency for Research on Cancer-World Health Organization, liver cancer is the second most common cause of cancer mortality worldwide. Hepatocellular carcinoma (HCC) accounts for around 90% of liver cancer cases and it is variably distributed according to its main risk factors. The hepatotropic viral [hepatitis B virus (HBV) and hepatitis C virus (HCV)] infection can cause a disarrangement in cellular pathways through an indirect and/or direct mechanism in liver injury. This review summarize available data in literature regarding the oncogenic properties of HBV and HCV in the initiation of HCC, including their role in the activation of hepatic stem cells.
- Citation: Sukowati CH, El-Khobar KE, Ie SI, Anfuso B, Muljono DH, Tiribelli C. Significance of hepatitis virus infection in the oncogenic initiation of hepatocellular carcinoma. World J Gastroenterol 2016; 22(4): 1497-1512
- URL: https://www.wjgnet.com/1007-9327/full/v22/i4/1497.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i4.1497
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancers, it accounts for around 90% of all cases[1]. According to the Globocan 2012 data of the International Agency for Research on Cancer-World Health Organization, it is the fifth most common cancer in men and the ninth in women, and the second most common cause of cancer-related death, estimated to be responsible for around 9% of all cases in 2012[2-4].
The global distribution of HCC is associated with the prevalence of its dominant risk factors. Infection of endemic hepatitis B virus (HBV) is the major cause of HCC in eastern Asia and sub-Saharan Africa for around 70%. In Europe and North America countries, hepatitis C virus (HCV) infection ranges from 50%-70% while excessive alcohol consumption leading to alcohol steatohepatitis (ASH) contributes for around 20% of all cases[1,5,6]. In its development, HCC usually emerges from a long-term chronic disease course with underlying liver cirrhosis (around 80%)[7]. However, it should be noted that HCC can occur in non-cirrhotic liver, accounts for around 20% of all cases[8].
Besides the infection of hepatotropic viruses and alcohol, obesity and diabetes that commonly associated with non-alcoholic steatohepatitis (NASH) increase the risk of HCC. Synergism between hepatitis virus infection and metabolic liver disease seems to worsen the course of the disease. Certain toxins and chemical agents such as aflatoxin B1 and vinyl chloride monomer also contribute in the progression of HCC. The cumulative risk for HCC is higher in male gender compared to female[3,9,10]. It has been thought to occur because of life style, male is more prone to viral infection and alcoholic cirrhosis. However, it is important to note that hormones (testosterone, progesterone, and estrogens) also take part in viral infection and subsequent liver damages[10].
HCC is a heterogeneous disease with various features and prognostic types. HCC is commonly developed in an extended period and different treatment options may vary between individuals. Based on the consensus of the Barcelona Clinic Liver Cancer (BCLC) staging system, liver resection is the best treatment option for very early stage HCC (0), liver transplantation and radiofrequency or percutaneous ethanol injection (PEI) for early stage (A), trans-arterial chemoembolization (TACE) for intermediate stage (B), molecular treatment with Sorafenib for advanced stage (C), and supportive palliative care for terminal stage (D)[11]. Based on continental and geographical policy, Fong and Tanabe compared the guidelines in Asia, Europe, and America, and they showed that most guidelines are similar with some variances in disease surveillance and treatment allocation recommendations[12].
It has been widely known that hepatocarcinogenesis is accompanied with complex aberrations in developmental and oncogenic molecular signaling pathways (excellently reviewed by[13,14]). Until now, a number of molecule-targeted drugs has been developed or under development to treat patients that cannot receive curative intervention. Various novel and promising drugs have been developed and tested in various phases of clinical trials, as a single agent or combined regimen[15]. As mentioned above, Sorafenib is the approved drug for the treatment of advanced HCC. Sorafenib is a multikinase inhibitor agent that targets the molecular pathway alteration commonly observed in HCC, including that of Wnt-β catenin pathway[16,17].
HBV, a member of Hepadnaviridae family, is a partially double-stranded DNA virus with 3.2 kb genome size. HBV genome contains 4 major open reading frames (ORF) that encode for polymerase (pol) for reverse transcriptase activity and replication, surface protein (HBsAg), core that form nucleocapsid and secreted HBeAg, and X that is important in viral replication[18,19]. Hepatitis B is one of the major public health problems that affect approximately 2 billion people globally, with more than 240 million chronic carriers and more than 780000 deaths annually[20]. By 2010, half of HCC cases are HBV-related, with or without history of liver fibrosis[4,21,22].
Based on several studies, HBV-related HCC development is mainly associated with risk factors such as male gender, persistently high HBV DNA levels, hepatitis B e antigen (HBeAg) positivity, presence of liver cirrhosis, older age, persistently high ALT levels, family history of HCC or chronic infection from perinatal transmission, and co-infection with HIV and/or HCV[23-28]. For example, male gender has hazard ratio (HR) 2-8 times more for HCC development compared to female[23-26]. Similarly, higher HBV DNA levels is associated with higher incidence of HCC compared to HBV DNA levels lower than 10000 copies/mL[24]. HBeAg positivity and ALT levels ≥ 45 U/L have HR 4.3 and 4.1, respectively, while liver cirrhosis is associated with 10.8-33.3 increased risk of HCC development compared to chronic hepatitis B patients without cirrhosis[24,25,29]. Based on these analyses, algorithms to screen and monitor high-risk populations have been proposed in many guidelines, which may reduce the incidence of HCC-related mortality because of the poor prognosis of advanced HCC development[30-33].
Prevalence of HBV has been shown to be reduced with the introduction of hepatitis B immunization program in newborns, complemented with administration of hepatitis B immunoglobulin for those born to mothers with chronic HBV infection[34-36]. Due to the commitment to eradicate the vertical transmission of HBV through national mass vaccination policy, the prevalence of hepatitis B can be decreased that leads to reduction of HCC cases, as demonstrated in the successful national program in Taiwan[37,38]. Prevention of HCC development by HBV vaccination is in line with the new Sustained Development Goals proposed by WHO, in which HBV-related HCC is one of the three preventable cancers that make up the bulk of cancer-related mortality globally[22].
HBV naturally-occurring genetic variations such as genotypes and subgenotypes, as well as mutations in some of the HBV genomic regions have been associated with different clinical manifestations such as development of cirrhosis and/or HCC[27,39]. Currently, HBV is classified into 9 genotypes (A to I) and one putative genotype (J) based on genome-wide divergence of more than 7.5%[40,41]. The distribution of the different HBV genotypes is geographically-related, most likely in association with the distribution of the different ethnic populations worldwide[42-45].
Since hepatitis B is endemic mainly in the Asia Pacific regions with HBV genotype B and/or C domination, most reports on the relation between genotype and HCC development concerns these two genotypes. Most reports propose HBV genotype B to be more lenient than genotype C, with some exceptions[39,46,47]. In general, HBV genotype C is commonly associated with later HBe seroconversion, more severe liver diseases, as well as faster progression of liver fibrosis and HCC development, although the life-long risk remain similar between genotype B and C[46-50]. In addition, HBV genotype B is associated with better response to treatment, enhancing the prognosis and reducing the risk of advanced disease progression[45,51]. Even compared to other genotypes, genotype C appeared to have worse prognosis in term of severe advanced liver disease development, with HR 2.05-2.34 times more than HBV genotype B or A and D, the four major HBV genotypes associated with HCC development[52]. This might be because HBV genotype C has higher tendency to induce DNA double-strand breaks and accumulate reactive oxygen species (ROS) that causes endoplasmic reticulum (ER) stress, in addition to more efficient cellular homologous-recombination events that increase the risk of chromosomal rearrangements and DNA damage, stimulating the formation and development of HCC[53]. Thus, genotyping of HBV is an important diagnostic tool in predicting the prognosis and response of therapy in hepatitis B patients.
Based on many reports, a double mutation in the basal core promoter (BCP) region of HBV genome (A1762T/G1764A) is associated with 1.7-10.6 fold increased risk of HCC incidence, particularly for those infected with HBV genotype C compared to genotype B[46,54-57]. In addition, in combination with C1653T and T1753V point mutations, these BCP mutations are associated with increased risk of HCC in HBeAg-positive than HBeAg-negative subjects, which can predict up to 80% of HCC development[24,46]. On the other hand, precore G1896A mutation is associated with HBeAg-negativity, but not with increased risk of liver cirrhosis or HCC development[46,54,55]. A report on novel mutation in genotype D observed the significance of T1858C mutation in the precore region that is associated with HCC progression in HBeAg-negative sample[53]. This report also identified several amino acid changes in HBV core antigen (HBcAg) that is associated with HCC development, namely I116L, P130Q, and T147C; two of which are parts of B- and killer T-cell epitopes[58].
Mutations in other regions of HBV genome have also been linked with disease progression, especially the PreS region. Mutations or deletions in the PreS region accumulated with the progression of chronic hepatitis B, and were associated with significant increase of HCC risk, even in adolescent[46,59]. An amino acid change S98T in the PreS1 region showed significant association with progression of liver fibrosis to cirrhosis and HCC, particularly in HBeAg-negative patients[58].
The accumulation of HBsAg particle in the ER of hepatocytes leads to the histological appearance of ground glass hepatocytes (GGH)[60]. It was repeatedly shown that hepatocytes expressing various forms of PreS2 mutants often developed into type II GGH, which exhibited ER and oxidative stress due to protein retention and DNA damage, while simultaneously created genomic instability and up-regulation of cell cycle progression and proliferation[61-63]. A mutation in codon 38 of X gene was preferentially found in patients with HCC and can be used as an independent risk factor for the development of HCC[64]. These newly defined oncoproteins may be utilized to develop novel biomarkers to predict HCC development[62].
Hepatitis D virus (HDV) is an imperfect RNA virus which needs HBV to be able to replicate[65]. Therefore, HDV always presents as co-infection or super-infection in approximately 5% of HBV-infected individuals, and causes more severe outcome such as fulminant hepatitis, liver cirrhosis, and HCC[65-67]. Super-infection often manifests in a rapidly progressive disease leading to cirrhosis within 2 years in 10%-15% of patients[68]. HBV/HDV co-infected patients usually have higher ALT and bilirubin levels as well as a higher prevalence of liver cirrhosis and HCC[69]. HDV co-infection is considered as a risk factor for HCC (HR 1.4-6.0 fold compared to HBV mono-infection), with lower survival rate[56,70-73]. In addition, HDV co-infection significantly increases the incidence rate of other gastrointestinal-related diseases in enhanced magnitude, and mortality rate of severe hepatitis manifestations[74]. Recently, it has been reported that high serum level of HDV RNA can be used as a predictor of cirrhosis and liver cancer in patients with chronic HDV infection[75]. Interestingly, although HBV/HDV co-infection leads to faster cirrhosis or HCC development in immuno-competent individuals, it may actually take part in lengthening the survival of liver-transplant or graft patients that are immuno-compromised, even though the mechanism is still unclear[76].
Recent advances in the development of highly effective HBV vaccine can be used as a preventive measure not only in the reduction of HBV infection, but also in the decrease of HDV, its associated pathogen[77]. The development of animal model with chronic hepatitis delta infection[78] can be utilized as a tool to study its pathogenesis per se and to discover its significance in the development of HCC. Detailed aspect on HDV viral biology, epidemiology, pathogenesis, and treatment is reviewed in[79,80].
HCV, a member of Flaviviridae family, is a single stranded RNA virus with 9.6 kb genome size. HCV genome is processed into structural proteins core, E1, and E2, and non-structural proteins p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B[18]. Chronic HCV infection affects approximately 170 million people worldwide, and may lead to development of liver fibrosis, cirrhosis, and HCC[81,82]. HCV infection may result in extra hepatic manifestations and metabolic disorder, including insulin resistance, type 2 diabetes and cardiovascular disease[83,84]. The rapidly evolving HCV treatment in the last decade has caused a decline in the incidence rate of viral infection[85]. However, the disease burden of HCV-related liver diseases is predicted to continue increasing in the approaching years[86]. HCV lifecycle occurs mostly in the cytoplasm, with the viral replication complex enclosed within a membranous web structure that is closely associated with the ER membrane, mitochondrial outer membrane, and lipid droplets[87,88].
HCV is highly heterogeneous and can be classified into seven recognized genotypes (genotype 1 to 7) and multiple subtypes based on the differences of the whole viral genome. Genotypes and subtypes can be divided into quasispecies based on genetic diversity[89]. HCV genotypes have different susceptibility to interferon (IFN), thus HCV genotyping is used to determine the type and duration of antiviral therapy[90]. Treatment with IFN-based regiment resulted in high sustained virological response (SVR) (about 80%) in genotype 2 and 3-infected patients, while genotypes 1 and 4 have lower SVR (about 50%) and genotypes 5 and 6 have intermediate response rates[91,92].
HCV genotypes have been associated with distinct pathological features, such as liver steatosis, insulin resistance, inflammation, and hepatitis reactivation[93-98]. In regards on the association between HCV genotype and risk of developing HCC, the available evidences are quite inconsistent[10]. Early study showed that genotype 1b patients have a significantly higher risk of developing HCC[99]. This early observation was supported by the result of a seventeen-year prospective cohort study, which showed 44 out of 104 genotype 1b followed-up patients developed HCC[100]. A meta-analysis study that calculated age-adjusted risk estimated genotype 1b patients had almost double the risk of developing HCC in comparison with patients infected with other genotypes[101]. On the other hand, recent studies suggest the association between genotype 3 and accelerated liver disease progression[102]. A retrospective cohort study involving 353 patients showed that genotype 3 patients develop more HCC compared to non-3 patients (44%-26%)[103]. These data were confirmed by successive studies with large cohort of infected patients that showed, even after adjustment of age, clinical, and antiviral treatment factors, genotype 3 had a higher risk of developing cirrhosis and HCC than genotype 1 patients[104,105].
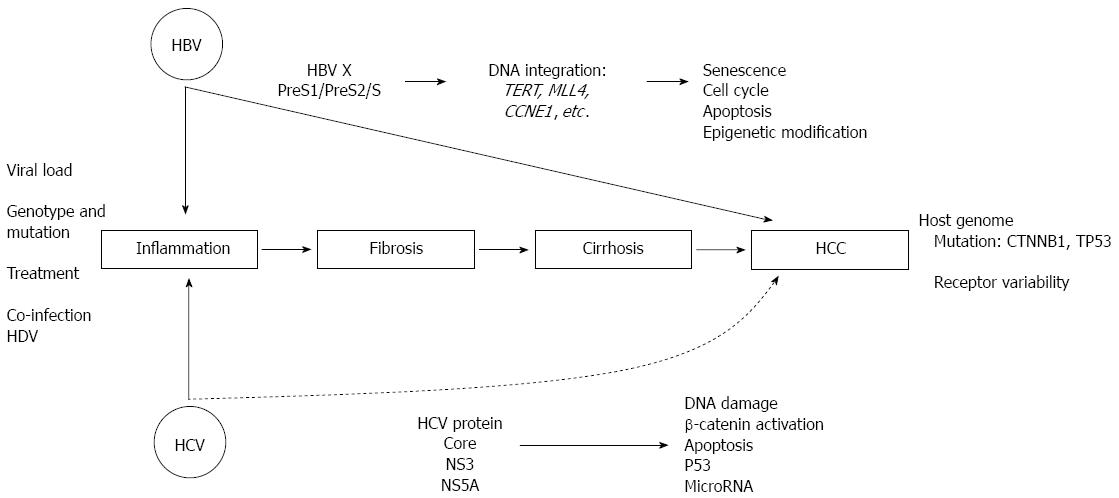
Chronic infection of HBV and/or HCV as a risk factor in the development of HCC is clearly acknowledged, as shown in Figure 1. Advances in in vitro technique and transgenic animal model with the insertion and modification of viral transgene[106] have opened many possibilities to understand the involvement of viral proteins in cellular damage. However, their pathogenesis in the oncogenic initiation of hepatotocarcinogenesis is still unclear. Limitation in the cellular biology, for example a low efficiency in viral replicon in vitro hampers the study of the pathogenicity of these agents.
HBV infection causes immunological response that may lead to oxidative stress and successive DNA damages of the cells (reviewed in[107]). Direct oncogenic property of HBV sequence by integration of its DNA into human genome can explain the incidence of non-cirrhotic HCC[108]. This insertion might involve deletions, cis/trans-activation, translocation, production of fusion transcripts and generalized genomic instability[109,110]. Consequently, it may lead to disruption of the host cellular pathway. Nevertheless, non-cirrhotic HCC with low grade fibrosis can be found also in HCV and NASH-related HCC[111,112], probably through a different oncogenic cascade. HBV DNA integration is present in majority of HBV-related HCC, even though it is also found in non-tumor tissue and chronic hepatitis B without HCC[113,114]. HBV DNA integration is considered as a strong oncogenic effect in hepatocarcinogenesis even though a recent study has proposed a contrasting evidence[115].
Direct oncogenic property of HBV, in particular for HBx and surface protein HBs has been intensely investigated. HBx is composed of 154 amino acids with a molecular mass of approximately 17.5 kDa. It has pleitropic functions as an important regulator in viral life cycle, a transcriptional activator, and a stimulator in the cytoplasmic signal transduction pathways[116]. Expression of HBx protein was found to be present in around 50%-60% of HBV-infected liver cirrhotic and HCC tissues, noticed in the cytoplasm of parenchymal and neoplastic cells[117].
The presence of HBx has been associated with tumor suppressor p53 rescue of apoptosis[118]. A European study showed p53 is a frequent altered pathway in HBV-related HCC. Mutations of host TP53 was associated with shorter survival and interestingly it was also related with genotype B of the patients from Asia and sub-Saharan Africa[119]. The interaction between phosphorylated Ser41-Pro motif of HBx and peptidyl-prolyl isomerase Pin1, a regulator of p53, followed by cis-trans isomerization and stabilization, significantly augmented the expression of HBx downstream target genes, leading to oncogenesis[120]. Based on this study, it was revealed that Ser41-Pro motif was conserved in HBV genotype A and B, but altered in most of the genotype C. Further analysis of eight HBV genotypes A-H showed a correlation between this motif and HBV genotypes, associated with HCC distribution and age[121].
Integration of HBV X sequence into host genome is a common event in HCC[113]. It was reported that HBV X integration occurred more often in HCC than in cirrhosis and it was positively related with the level of cell-cycle and apoptotic protein, including Cyclin A, retinoblastoma protein (Rb), Fas-associated death domain protein (FADD), tumor necrosis factor receptor-associated death domain protein (TRADD), and nuclear factor kappa B (NF-κβ)[122]. Whole genome sequencing of HCC samples have demonstrated HBV X DNA integration within or upstream the sequence of telomerase reverse transcriptase (TERT), epigenetic regulator MLL4, and cell cycle gene CCNE1 as “hot site” breakpoints[123-125]. DNA integration was frequently observed in the tumors (around 85%) compared to adjacent liver tissues (around 30%)[125]. Large numbers of HBV integration (defined as ≥ 3) was positively associated with the serum level of HBsAg and AFP, and importantly, with low survival time compared to those with no or low numbers of integration[125]. HBx is associated with epigenetic modifications through interaction with histone deacetylase 1 and DNA methyltransferase during hepatocarcinogenesis[126-128].
A recent article reported development of HCC in the absence of severe liver damage in a HFE-haemochromatosis patient that was seronegative for hepatitis B and C infections. HBx gene sequence was detected in tumor but not in non-tumor. HBV integration involved a 5’-deleted X gene with an intact enhancer-II/basal-core promoter region and integrated upstream of the partitioning-defective-6-homolog-gamma gene (PARD6G)[129]. The role of HBV X gene in hepatocarcinogenesis is reviewed in[130,131].
ORF S gene region with three translational start sites PreS1, PreS2, and S, encodes for large (L), middle (S) and small (S) surface protein (HBs), respectively. S protein is composed of 226 amino acids, M protein is S with additional 55 amino acids, and L protein is M with additional 108 or 119 amino acids, based on virus genotype[132,133]. S region is conserved while both PreS1 and PreS2 are variable and prone to genetic mutations. Besides genotyping based on whole genome sequence, the variation in PreS2 region has been used to determine HBV subgenotypes[134]. As mentioned previously, different GGH appearances showed different mutated S proteins. Several studies from the group of Su et al[61-63] had shown that type I GGH harbored mutants with deletions within the pre-S1 region while type II GGH contained PreS2 mutants[61-63]. In hepatocarcinogenesis, PreS2 mutant was demonstrated to produce an aberrant Cyclin A expression and centrosome over-duplication through ER stress that led to chromosomal instability[135].
A transgenic animal model with the insertion of PreS/S gene regions expressed high level of HBsAg, showed inflammation and appearance of GGH, preneoplastic lesion, and finally it led to HCC in major number of animals[136], indicating a direct oncogenic input of this gene. Gene expression profile of 3-month old mice showed differentially expressed genes involved in various regulations such as apoptosis, cell cycle, NF-κB signal transduction pathway, and inflammatory response[137].
As that of HBV X DNA, the insertion of S regions into host genome has been widely reported, first noticed in the WHV-infected animals. In their study, Sung et al[125] observed recurrent integration of PreS1, PreS2, and S sequences in human genes TERT and MLL4, even though they did not notice it in CCNE1 gene as HBV X sequence.
A recent study reported that in HCC patients with occult hepatitis B, HBV DNA integration was found in around 75% of cases, in which the inserted viral genes were mainly X and PreS/S, followed by C and Polymerase sequences[138]. Furthermore, in a prospective 12-years study in chronic hepatitis C patients with occult hepatitis B, X integration can be associated with HCC development in the absence of cirrhosis[139]. The HBV DNA integration is not only observed in liver cells but also in blood cells. The transcript of HBsAg coding gene and the integration of HBV DNA in bone marrow haematopoietic stem cells from chronic HBV infection patients was observed[140].
Since HCV RNA cannot integrate into human genome, at the beginning, the mechanism in HCV-related HCC pathogenesis is thought majorly to be indirect pathways via the effects of chronic inflammation and oxidative stress. Subsequently, it leads to fibrosis and eventually cirrhosis as observed in the other HCC etiologies such as ASH, NASH, and obesity-related disorder. However, current literatures also showed a direct oncogenic effect of the viral proteins[141].
Oxidative stress has been implicated as one of the mechanisms of HCV-induced hepatocarcinogenesis[142]. Oxidative stress occurs when there is imbalance in the production and clearance of ROS. ROS is a normal by-product of numerous cell processes including proliferation, apoptosis, and cell senescence[143]. In the liver, ROS is mainly produced by mitochondria in hepatocytes, and from nicotinamide adenine dinucleotide phosphate oxidase and xanthine oxidase reactions in Kupffer cells and inflammatory cells[144]. Long-term oxidative stress may induce DNA damage, and since ROS can also functions as second messenger in cellular signaling, increased ROS level may trigger the activation of oncogenic signaling pathways[107,142].
Increased oxidative stress in chronic hepatitis C patients has been shown through elevated levels of several oxidative stress biomarkers, including 8-hydroxydeoxyguanosine (8-OHdG), malondialdehyde, and thioredoxin in both sera and liver biopsy samples[145-147]. Chronic hepatitis C patients have also been shown to have higher expression of 8-OHdG, also an indicator for DNA damage, in comparison with chronic hepatitis B patients; suggesting that hepatic oxidative DNA damage is more common in chronic hepatitis C[148]. Further, some clinical studies have shown that addition of antioxidant agent could improve oxidative stress-caused liver injury and maybe important for treatment management of HCV patients[149,150].
HCV-effect on increased oxidative stress has been mainly attributed to the expression of viral core protein, although other viral proteins have also been shown to induce oxidative stress[151]. The expression of HCV core protein, either in vitro or in vivo, have been shown to induce alteration of mitochondrial function, increased ROS levels, and increased intrahepatic lipid peroxidation[152-154]. HCV NS5A protein has been shown to alter intracellular calcium level and induces oxidative stress in an in vitro model[155]. Increased ROS is linked to dysregulation of various cells signaling pathways, particularly those that regulate cell survival, apoptosis, and inflammation. Indeed, increased oxidative stress resulted in activation of p38 MAPK, JNK, NF-κβ and STAT3[155-157], which promotes cell survival. Increased ROS have been linked to dysregulation of various cells signaling pathways, particularly those that regulate cell survival, apoptosis, and inflammation. Indeed, increased oxidative stress resulted in activation of p38 MAPK, JNK, NF-κβ and STAT3[155-157], which promote cell survival. These activations induce subsequent activation of TGFβ1[158], a major profibrogenic factor in the liver, causing activation of hepatic stellate cells[159].
HCV core protein has also been shown to induce upregulation of TGFβ1 transcription, showing a more direct role of HCV protein in inducing fibrogenesis[160]. Activated stellate cells could also produce TGFβ1 and other pro-inflammatory cytokines, which facilitate further inflammatory response[161] and demonstrate close association between oxidative stress and inflammation in chronic hepatitis C. In addition, increased ROS production along with persistent viral expressions might also induced cell death, either through TNFα- or mitochondria-mediated apoptosis[162,163]. The resulting apoptotic bodies will release alarming molecules that serve as pro-inflammatory mediators, further aggravating the oxidative stress-associated inflammation. Oxidative stress can also damage telomeres, causing them to shorten[164] that has been reported occurring in the presence of increased oxidative stress marker 8-oHdG expression[165], signifying the effect of HCV-induced oxidative stress on telomere shortening and senescence.
One of the consequences of chronic oxidative stress is oxidative DNA damage. ROS could interact directly with DNA to induce DNA damage[107], and mitochondrial DNA (mtDNA) that has no protective histone protein is more susceptible to the damage[144]. HCV infection has been reported to induce a mutator phenotype by causing dsDNA breaks[166]. In line with this, decreased mtDNA in peripheral blood leukocytes of chronic hepatitis C patients have been reported, and the degree of DNA damage was found to be correlated with increased liver inflammation[167] that may lead to progressively liver damage including HCC.
The close association of the HCV viral replication complex to the ER membrane might cause ER dysfunction. Indeed, HCV has been shown to cause ER stress[168,169]. ER stress occurs when ER function as the site of production and posttranslational modifications of cell proteins is perturbed, which then triggered the unfolded protein response (UPR) pathway to restore protein homeostasis[170,171]. This effect is achieved by initiating transmembrane protein ATF6 proteolytic cleavage[169], subsequently activating the transcription of ER chaperone genes, GRP78[172]. Furthermore, up-regulation of both GRP78 and transcription factor CHOP/GADD153 have been correlated with down-regulation of anti-apoptotic Bcl-2 gene expression, increased NF-κβ, and cleavage of caspase-3 and PARP[173-175]. Increased GADD153 expression has also been linked to cell susceptibility to oxidant injury[173], suggesting that ER stress and oxidative stress is closely related in pathogenesis of HCV-infection.
The expression of singular HCV proteins, particularly core, NS5A, and NS2 proteins and/or HCV subgenomic replicons in in vitro induced the UPR pathways[169,174,176]. These findings were later confirmed using HCV transgenic mice model[175,177]. The HCV-induced ER stress is reduced following treatment with interferon-α 2a treatment[177] or NS3 protease inhibitor[176], resolving UPR response and restoring protein homeostasis. ER is also major site for intracellular calcium storage, and these calcium ions are trafficked to and from the ER to regulate various cellular signal transduction[171]. HCV core, and also NS5A, alters ER calcium homeostasis by inducing ER stress and depleting ER calcium content[174,178], resulting in mitochondrial membrane depolarization and triggering mitochondria-mediated apoptosis. This HCV-core effect is fully diminished by restoring the ER calcium storage[174]. The changes in calcium homeostasis in HCV-infected cells have been suggested as the result of viral-induced increase ROS production and oxidative stress[178], again indicating a strong correlation between oxidative stress, ER stress, and mitochondrial dysfunction.
Recent literatures demonstrated that HCV protein core, NS3, NS4B, and NS5A, can induce cell transformation in vitro and in vivo mice transgenic model[179]. The fact that HCV transgenic mice with the expression of a HCV viral protein can develop HCC, suggests a direct oncogenic effect rather than an inflammatory mechanism[180]. Viral protein in cytoplasm has the ability to interact with host protein and to alter the stability of the cellular mechanism leading to carcinogenesis. Further, direct interactions between these viral proteins with numerous host cell factors have been shown to lead to dysregulation of wide range of cellular signaling, particularly those involved in cell proliferation, apoptosis, cell metabolism, immune responses and also oxidative stress[179,181].
Core protein has been reported to directly deregulate the tumor suppressor p53 pathway based on viral protein over-expression cell culture system. The level of deregulation is unclear, since available reports have shown for both HCV-induced activation and repression of p53-dependent gene expression[182]. Host genetic variation and somatic mutation in TP53, as well as CTNNB1 encoding β-catenin, was found to be significantly associated with young age and moderate and poor differentiated HCV-related HCC[183]. CTNNB1 activating mutations were also found to be more frequent in HCV-related compared to HBV-related HCC[119]. In an in vitro study, HCV core protein activated canonical Wnt signaling via regulations of several important molecules upstream of β-catenin and presumably resulted in promotion of cell proliferation[184]
Recent report showed that expression of HCV protein also increased proto-oncogene c-Myc expression in vivo and in infected human livers. This change is mediated through Akt-dependent activation of β-catenin and might further contribute to HCV-related oxidative stress and genetic damage[185]. Furthermore, it had been demonstrated that the expression of NS5A stabilized and accumulated β-catenin through the phosphorylation and inactivation of GSK3β[186,187].
HCV NS5B RNA-dependent RNA polymerase forms a cytoplasmic complex with Rb, downregulating the Rb expression and its DNA damage responses[188,189]. HCV core has also been reported to inhibit Rb expression[190]. HCV NS3/4A protein directly interacts with ataxia telangiectasia mutated kinase (ATM); a tumor suppressor protein that detects dsDNA breaks, resulting in impaired DNA repair[191]. In addition, HCV core also binds to NBS1 protein to inhibit Mre11/NBs1/Rad50 defective DNA-sensing complex, resulting in impaired ATM activation and inhibition of repair enzymes DNA binding[192]. HCV impairs the expression of NEIL1 DNA-excision glycosylases, as shown in vitro and in liver biopsy specimens of advanced liver disease patients[193]. These observations suggest that HCV induces accumulation of DNA damage by inhibiting multiple DNA repair processes and promoting chromosome instability with consequent malignant transformation.
The association between HCV viral proteins with several tumor suppressor genes as listed above might affect the regulation of cellular senescence. Senescence pathway responds to cellular stress and acts to limit the proliferation of damaged cells[194]. Inflammation, oxidative, and oncogenic stress can induce premature senescence, and this change is characterized by cell-cycle arrest, resistance to apoptosis, and oncogenic epigenetic changes[142,194].
Recent studies had implied the role of several microRNAs in HCV-related HCC. Several studies had shown that miR-122, a liver-specific microRNA, was down-regulated in the majority of HCC samples analyzed, apart from HCV-related HCC (reviewed by Borel et al[195]). Previously it had been shown that the miR-122 regulated cell cycle protein Cyclin G1 that affected the stability of p53 and also altered chemotherapy sensitivity[196]. The involvement of miR-122 in HCV-induced hepatocarcinogenesis is reviewed in[182].
Accumulating evidences highlight the importance of cancer stem cells (CSC) in HCC biology. Different hepatic CSCs have been reported in various subtypes of HCC and they are considered as the master regulators of HCC initiation, progression, and metastasis[197,198]. By using immunostaining and RNA-FISH for stem cells markers OV6, CK19, and CD133, the frequency of positive stem cell markers in liver cirrhosis and HCC roughly correlated with the relatively frequency of HCC that develops in the clinical setting[199].
Despite the increasing importance of this heterogeneous population in driving carcinogenesis, little is known about the effect of viral hepatitis in the biology of CSC. Histological analysis of human tissue found a positive correlation between HBV infection and CD90[200] and an inverse correlation with CD133[201]. However, since co-staining of the CSC markers and the HBV proteins was not performed, is not clear if and how HBV alters the physiology of CD90+ and CD133+ CSC. Furthermore, it will be also important to put virus genotype in consideration to assess the role of a specific virus type with the phenotype of the cells.
Several reports have described the involvement of HBV in the generation of CSC. In particular, a correlation between HBx expression and EpCAM+ CSC appearance was clearly demonstrated since Arzumanyan et al[202] showed that the pluripotent stem cell transcription factors Oct-4, Nanog, and Klf-4, as well as EpCAM and β-catenin, were up-regulated in HBx expressing cells. Phenotypically, HBx stimulated cell migration, growth in soft agar, and spheroid formation. These data were confirmed in HBx transgenic mice fed with 3,5-diethoxycarbonyl-1,4-dihydrocollidine where an elevated number of EpCAM+ cells with characteristics of human progenitor cells was observed[203]. Transformation of rat oval cells with HBx and the subsequent injection in nude mice treated with aflatoxin B1 in vivo, gave rise to tumor that expressed markers of adult hepatocytes as albumin and CK18, undifferentiated marker AFP, and oncoprotein c-Myc[204].
Moreover, clinical evidence showed that high HBx expression in human HBV-related HCC was statistically associated with expansion of EpCAM+ or OV6+ tumor cells, aggressive clinicopathological features[203,205], activated β-catenin signalling, and up-regulation of miR-181[202]. In 2015, Fan et al[206] investigated the molecular mechanism by which HBx induces EpCAM expression, suggesting DNA demethylation as major mechanism driving the re-expression of EpCAM into hepatocytes.
Infection of HCV has also been associated in the induction of CSC, perhaps in a direct oncogenesis manner. The expression of an HCV subgenomic replicon in cultured cells resulted in the acquisition of CSC traits including an enhanced expression of doublecortin and CaM kinase-like-1, Lgr5, CD133, AFP, cytokeratin-19, Lin28, and c-Myc. Conversely, curing of the replicon from these cells results in diminished expression of these factors. The analysis of liver tissues from HCV-positive patients and liver tissue microarrays reiterated these observations[207].
The effect of the HCV nonstructural NS5A protein was studied in a transgenic mouse model. Viral protein, in synergy with alcohol-induced endotoxemia, induced the up-regulation of the Toll-like receptor 4 (TLR4) with the consequent expression of the pluripotency gene Nanog, a downstream gene up-regulated by TLR4, and CD133[208]. On the other side, the CD133+/CD49f+ cells isolated from HCC developed in HCV core transgenic mice were tumorigenic both in vitro and in vivo and the TLR4-Nanog pathway was necessary for the maintenance of tumorigenic properties[209].
Collectively, literature review had demonstrated the significances of hepatotropic HBV and HCV during hepatocarcinogenesis. In the oncogenic initiation, they can induce immunological responses lead to successive damages of the liver cells that may direct the development of HCC. Since the biology of HBV and HCV is different, their oncogenic effect may go through a different mechanism, direct and/or indirect, as had been demonstrated in many pre-clinical and clinical studies. Even though studies in in vitro and transgenic animal model had expanded the knowledge of viral-specific proteins, the mechanism of the viral particle in inducing hepatocarcinogenesis is still unclear and open for discussion. This is partially due to several methodological limitations such as the difficulty on viral culture, and transgenic animal model cannot reflect the entire virus particle and its interaction with host cells receptor (e.g., hepatocyte, immune cells, etc.). Furthermore, genetic characteristic of the virus (genotypes, subgenotypes, and quasispecies) can be related to different disease outcomes, treatment options, and viral susceptibilities.
In order to prevent HCC development in chronic hepatitis patients, antiviral therapy is a treatment choice to suppress viral replication and improve general status of the patients. However, as reviewed by Papatheodoridis et al[210] current nucleos(t)ide analogs against HBV can reduce but not eliminate the risk of HCC. It is of importance to increase awareness among health-care personnel and the public in the urgency to protect new generations, particularly in endemic areas, as well as to raise the population-wide immunity by neonatal immunization program and booster and/or catch-up vaccination.
P- Reviewer: Tashiro F, Tomizawa M S- Editor: Qi Y L- Editor: A E- Editor: Zhang DN
| 1. | El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 3088] [Article Influence: 220.6] [Reference Citation Analysis (0)] |
| 2. | Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1163] [Cited by in RCA: 1226] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 3. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20504] [Article Influence: 2050.4] [Reference Citation Analysis (20)] |
| 4. | Stewart BW, Wild CP, editors . World Cancer Report 2014. Lyon, France: International Agency for Research on Cancer 2014; . |
| 5. | Bosetti C, Levi F, Boffetta P, Lucchini F, Negri E, La Vecchia C. Trends in mortality from hepatocellular carcinoma in Europe, 1980-2004. Hepatology. 2008;48:137-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 181] [Article Influence: 10.6] [Reference Citation Analysis (1)] |
| 6. | Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3249] [Cited by in RCA: 3594] [Article Influence: 276.5] [Reference Citation Analysis (4)] |
| 7. | Caldwell S, Park SH. The epidemiology of hepatocellular cancer: from the perspectives of public health problem to tumor biology. J Gastroenterol. 2009;44 Suppl 19:96-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 155] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 8. | Alkofer B, Lepennec V, Chiche L. Hepatocellular cancer in the non-cirrhotic liver. J Visc Surg. 2011;148:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 9. | Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 400] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 10. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2506] [Article Influence: 192.8] [Reference Citation Analysis (2)] |
| 11. | European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4059] [Cited by in RCA: 4517] [Article Influence: 347.5] [Reference Citation Analysis (2)] |
| 12. | Fong ZV, Tanabe KK. The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: a comprehensive and evidence-based comparison and review. Cancer. 2014;120:2824-2838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 205] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 13. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4264] [Article Influence: 236.9] [Reference Citation Analysis (2)] |
| 14. | Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312-1327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 823] [Cited by in RCA: 831] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 15. | Huynh H. Molecularly targeted therapy in hepatocellular carcinoma. Biochem Pharmacol. 2010;80:550-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 102] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 16. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9016] [Cited by in RCA: 10258] [Article Influence: 603.4] [Reference Citation Analysis (2)] |
| 17. | Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y, Villanueva A, Minguez B, Newell P, Tsai HW. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res. 2012;18:4997-5007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 244] [Article Influence: 18.8] [Reference Citation Analysis (1)] |
| 18. | Shlomai A, de Jong YP, Rice CM. Virus associated malignancies: the role of viral hepatitis in hepatocellular carcinoma. Semin Cancer Biol. 2014;26:78-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 138] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 19. | Dandri M, Locarnini S. New insight in the pathobiology of hepatitis B virus infection. Gut. 2012;61 Suppl 1:i6-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 174] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 20. | World Health Organization. Hepatitis B Fact Sheet no.204, updated July 2015. Available from: http://www.who.int/mediacentre/factsheets/fs204/en/. |
| 21. | Sherman M. Hepatocellular carcinoma: New and emerging risks. Dig Liver Dis. 2010;42 Suppl 3:S215-S222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham A, Adair T, Aggarwal R, Ahn SY. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-2128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9500] [Cited by in RCA: 9574] [Article Influence: 736.5] [Reference Citation Analysis (0)] |
| 23. | Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5-S16. [PubMed] |
| 24. | Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2309] [Cited by in RCA: 2363] [Article Influence: 124.4] [Reference Citation Analysis (0)] |
| 25. | Kim GA, Lee HC, Kim MJ, Ha Y, Park EJ, An J, Lee D, Shim JH, Kim KM, Lim YS. Incidence of hepatocellular carcinoma after HBsAg seroclearance in chronic hepatitis B patients: a need for surveillance. J Hepatol. 2015;62:1092-1099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 26. | Li Y, Zhang Z, Shi J, Jin L, Wang L, Xu D, Wang FS. Risk factors for naturally-occurring early-onset hepatocellular carcinoma in patients with HBV-associated liver cirrhosis in China. Int J Clin Exp Med. 2015;8:1205-1212. [PubMed] |
| 27. | Pollack HJ, Kwon SC, Wang SH, Wyatt LC, Trinh-Shevrin C. Chronic hepatitis B and liver cancer risks among Asian immigrants in New York City: Results from a large, community-based screening, evaluation, and treatment program. Cancer Epidemiol Biomarkers Prev. 2014;23:2229-2239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 28. | Shimakawa Y, Lemoine M, Bottomley C, Njai HF, Ndow G, Jatta A, Tamba S, Bojang L, Taal M, Nyan O. Birth order and risk of hepatocellular carcinoma in chronic carriers of hepatitis B virus: a case-control study in The Gambia. Liver Int. 2015;35:2318-2326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | van Bömmel F, Berg T. Treatment of HBV related cirrhosis. Liver Int. 2013;33 Suppl 1:176-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 30. | European Association For The Study Of The Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167-185. [PubMed] |
| 31. | Liaw YF, Kao JH, Piratvisuth T, Chan HL, Chien RN, Liu CJ, Gane E, Locarnini S, Lim SG, Han KH. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2012 update. Hepatol Int. 2012;6:531-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 742] [Cited by in RCA: 792] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 32. | Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2125] [Cited by in RCA: 2171] [Article Influence: 135.7] [Reference Citation Analysis (0)] |
| 33. | World Health Organization. Guidelines for the prevention, care and treatment of persons with chronic hepatitis B infection. Geneva, Switzerland: WHO Press 2015; . |
| 34. | Ang LW, Tey SH, Cutter J, James L, Goh KT. Seroprevalence of hepatitis B virus infection among children and adolescents in Singapore, 2008-2010. J Med Virol. 2013;85:583-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 35. | Chiang CJ, Yang YW, You SL, Lai MS, Chen CJ. Thirty-year outcomes of the national hepatitis B immunization program in Taiwan. JAMA. 2013;310:974-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 179] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 36. | Hsu HM, Lee SC, Wang MC, Lin SF, Chen DS. Efficacy of a mass hepatitis B immunization program after switching to recombinant hepatitis B vaccine: a population-based study in Taiwan. Vaccine. 2001;19:2825-2829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Chang MH, You SL, Chen CJ, Liu CJ, Lee CM, Lin SM, Chu HC, Wu TC, Yang SS, Kuo HS. Decreased incidence of hepatocellular carcinoma in hepatitis B vaccinees: a 20-year follow-up study. J Natl Cancer Inst. 2009;101:1348-1355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 427] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 38. | Chang MH, Chen DS. Prevention of hepatitis B. Cold Spring Harb Perspect Med. 2015;5:a021493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 39. | Chan HL, Wong ML, Hui AY, Hung LC, Chan FK, Sung JJ. Hepatitis B virus genotype C takes a more aggressive disease course than hepatitis B virus genotype B in hepatitis B e antigen-positive patients. J Clin Microbiol. 2003;41:1277-1279. [PubMed] |
| 40. | Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 323] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 41. | Pourkarim MR, Amini-Bavil-Olyaee S, Kurbanov F, Van Ranst M, Tacke F. Molecular identification of hepatitis B virus genotypes/subgenotypes: revised classification hurdles and updated resolutions. World J Gastroenterol. 2014;20:7152-7168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 147] [Cited by in RCA: 147] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 43. | Kurbanov F, Tanaka Y, Mizokami M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res. 2010;40:14-30. [PubMed] |
| 44. | Thedja MD, Muljono DH, Nurainy N, Sukowati CH, Verhoef J, Marzuki S. Ethnogeographical structure of hepatitis B virus genotype distribution in Indonesia and discovery of a new subgenotype, B9. Arch Virol. 2011;156:855-868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 45. | Westland C, Delaney W, Yang H, Chen SS, Marcellin P, Hadziyannis S, Gish R, Fry J, Brosgart C, Gibbs C. Hepatitis B virus genotypes and virologic response in 694 patients in phase III studies of adefovir dipivoxil1. Gastroenterology. 2003;125:107-116. [PubMed] |
| 46. | Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 328] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 47. | Orito E, Mizokami M. Differences of HBV genotypes and hepatocellular carcinoma in Asian countries. Hepatol Res. 2007;37:S33-S35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 48. | Kao JH. Hepatitis B virus genotypes and hepatocellular carcinoma in Taiwan. Intervirology. 2003;46:400-407. [PubMed] |
| 49. | Orito E, Mizokami M. Hepatitis B virus genotypes and hepatocellular carcinoma in Japan. Intervirology. 2003;46:408-412. [PubMed] |
| 50. | Sumi H, Yokosuka O, Seki N, Arai M, Imazeki F, Kurihara T, Kanda T, Fukai K, Kato M, Saisho H. Influence of hepatitis B virus genotypes on the progression of chronic type B liver disease. Hepatology. 2003;37:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 314] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 51. | Zhao H, Kurbanov F, Wan MB, Yin YK, Niu JQ, Hou JL, Wei L, Wang GQ, Tanaka Y, Mizokami M. Genotype B and younger patient age associated with better response to low-dose therapy: a trial with pegylated/nonpegylated interferon-alpha-2b for hepatitis B e antigen-positive patients with chronic hepatitis B in China. Clin Infect Dis. 2007;44:541-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 52. | Wong GL, Chan HL, Yiu KK, Lai JW, Chan VK, Cheung KK, Wong EW, Wong VW. Meta-analysis: The association of hepatitis B virus genotypes and hepatocellular carcinoma. Aliment Pharmacol Ther. 2013;37:517-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 53. | Datta S, Roychoudhury S, Ghosh A, Dasgupta D, Ghosh A, Chakraborty BC, Ray S, Gupta S, Santra AK, Datta S. Distinct distribution pattern of hepatitis B virus genotype C and D in liver tissue and serum of dual genotype infected liver cirrhosis and hepatocellular carcinoma patients. PLoS One. 2014;9:e102573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 54. | Constantinescu I, Dinu AA, Boscaiu V, Niculescu M. Hepatitis B virus core promoter mutations in patients with chronic hepatitis B and hepatocellular carcinoma in bucharest, romania. Hepat Mon. 2014;14:e22072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 55. | Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 428] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 56. | Khan M, Dong JJ, Acharya SK, Dhagwahdorj Y, Abbas Z, Jafri SMW, Mulyono DH, Tozun N, Sarin SK. Hepatology issues in Asia: Perspectives from regional leaders. J Gastroenterol Hepatol. 2004;19:S419-S430. |
| 57. | Yotsuyanagi H, Hino K, Tomita E, Toyoda J, Yasuda K, Iino S. Precore and core promoter mutations, hepatitis B virus DNA levels and progressive liver injury in chronic hepatitis B. J Hepatol. 2002;37:355-363. [PubMed] |
| 58. | Datta S, Ghosh A, Dasgupta D, Ghosh A, Roychoudhury S, Roy G, Das S, Das K, Gupta S, Basu K. Novel point and combo-mutations in the genome of hepatitis B virus-genotype D: characterization and impact on liver disease progression to hepatocellular carcinoma. PLoS One. 2014;9:e110012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 59. | Oba U, Koga Y, Hoshina T, Suminoe A, Abe K, Hayashida M, Taguchi T, Hara T. An adolescent female having hepatocellular carcinoma associated with hepatitis B virus genotype H with a deletion mutation in the pre-S2 region. J Infect Chemother. 2015;21:302-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 60. | Hadziyannis S, Gerber MA, Vissoulis C, Popper H. Cytoplasmic hepatitis B antigen in “ground-glass” hepatocytes of carriers. Arch Pathol. 1973;96:327-330. [PubMed] |
| 61. | Hsieh YH, Chang YY, Su IJ, Yen CJ, Liu YR, Liu RJ, Hsieh WC, Tsai HW, Wang LH, Huang W. Hepatitis B virus pre-S2 mutant large surface protein inhibits DNA double-strand break repair and leads to genome instability in hepatocarcinogenesis. J Pathol. 2015;236:337-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 62. | Su IJ, Wang LH, Hsieh WC, Wu HC, Teng CF, Tsai HW, Huang W. The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis. J Biomed Sci. 2014;21:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 63. | Wang HC, Huang W, Lai MD, Su IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 64. | Muroyama R, Kato N, Yoshida H, Otsuka M, Moriyama M, Wang Y, Shao RX, Dharel N, Tanaka Y, Ohta M. Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C is associated with an increased risk of hepatocellular carcinoma. J Hepatol. 2006;45:805-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 65. | Smedile A, Farci P, Verme G, Caredda F, Cargnel A, Caporaso N, Dentico P, Trepo C, Opolon P, Gimson A. Influence of delta infection on severity of hepatitis B. Lancet. 1982;2:945-947. [PubMed] |
| 66. | Keshvari M, Alavian SM, Aghaee B, Behnava B, Mahdavi M, Fesharaki MG, Sharafi H. Seroepidemiology and clinical features of hepatitis delta among HBsAg carriers: a study from Hepatitis Clinic of Iranian Blood Transfusion Organization. Transfus Med. 2014;24:411-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 67. | Shirvani-Dastgerdi E, Amini-Bavil-Olyaee S, Alavian SM, Trautwein C, Tacke F. Comprehensive analysis of mutations in the hepatitis delta virus genome based on full-length sequencing in a nationwide cohort study and evolutionary pattern during disease progression. Clin Microbiol Infect. 2015;21:510.e11-510.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 68. | Rizzetto M. Hepatitis D: clinical features and therapy. Dig Dis. 2010;28:139-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 69. | Kiesslich D, Crispim MA, Santos C, Ferreira Fde L, Fraiji NA, Komninakis SV, Diaz RS. Influence of hepatitis B virus (HBV) genotype on the clinical course of disease in patients coinfected with HBV and hepatitis delta virus. J Infect Dis. 2009;199:1608-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 70. | Fattovich G, Giustina G, Christensen E, Pantalena M, Zagni I, Realdi G, Schalm SW. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. The European Concerted Action on Viral Hepatitis (Eurohep). Gut. 2000;46:420-426. [PubMed] |
| 71. | Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35-S50. [PubMed] |
| 72. | Lacombe K, Boyd A, Desvarieux M, Serfaty L, Bonnord P, Gozlan J, Molina JM, Miailhes P, Lascoux-Combe C, Gault E. Impact of chronic hepatitis C and/or D on liver fibrosis severity in patients co-infected with HIV and hepatitis B virus. AIDS. 2007;21:2546-2549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 73. | Liao B, Zhang F, Lin S, He H, Liu Y, Zhang J, Xu Y, Yi J, Chen Y, Liu H. Epidemiological, clinical and histological characteristics of HBV/HDV co-infection: a retrospective cross-sectional study in Guangdong, China. PLoS One. 2014;9:e115888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 74. | Gu XH, Chen Z, Dai RY, Zhang ML, Tang HM, Chen LB, Dong B. Analysis on the clinical features of 507 HDV-infected patients. Cell Biochem Biophys. 2014;70:1829-1832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 75. | Romeo R, Foglieni B, Casazza G, Spreafico M, Colombo M, Prati D. High serum levels of HDV RNA are predictors of cirrhosis and liver cancer in patients with chronic hepatitis delta. PLoS One. 2014;9:e92062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (1)] |
| 76. | Rifai K, Wedemeyer H, Rosenau J, Klempnauer J, Strassburg CP, Manns MP, Tillmann HL. Longer survival of liver transplant recipients with hepatitis virus coinfections. Clin Transplant. 2007;21:258-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 77. | Thomas E, Yoneda M, Schiff ER. Viral hepatitis: past and future of HBV and HDV. Cold Spring Harb Perspect Med. 2015;5:a021345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 78. | Aldabe R, Suárez-Amarán L, Usai C, González-Aseguinolaza G. Animal models of chronic hepatitis delta virus infection host-virus immunologic interactions. Pathogens. 2015;4:46-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 79. | Hughes SA, Wedemeyer H, Harrison PM. Hepatitis delta virus. Lancet. 2011;378:73-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 396] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 80. | Pascarella S, Negro F. Hepatitis D virus: an update. Liver Int. 2011;31:7-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 79] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 81. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1770] [Cited by in RCA: 1846] [Article Influence: 153.8] [Reference Citation Analysis (3)] |
| 82. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 1145] [Article Influence: 114.5] [Reference Citation Analysis (0)] |
| 83. | Negro F. HCV infection and metabolic syndrome: which is the chicken and which is the egg? Gastroenterology. 2012;142:1288-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 84. | Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol. 2014;61:S58-S68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 614] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 85. | Pawlotsky JM. New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology. 2014;146:1176-1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 412] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 86. | Razavi H, Waked I, Sarrazin C, Myers RP, Idilman R, Calinas F, Vogel W, Mendes Correa MC, Hézode C, Lázaro P. The present and future disease burden of hepatitis C virus (HCV) infection with today's treatment paradigm. J Viral Hepat. 2014;21 Suppl 1:34-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 294] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 87. | Bartenschlager R, Cosset FL, Lohmann V. Hepatitis C virus replication cycle. J Hepatol. 2010;53:583-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 88. | Joyce MA, Tyrrell DL. The cell biology of hepatitis C virus. Microbes Infect. 2010;12:263-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 89. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 980] [Article Influence: 89.1] [Reference Citation Analysis (1)] |
| 90. | Chayama K, Hayes CN. Hepatitis C virus: How genetic variability affects pathobiology of disease. J Gastroenterol Hepatol. 2011;26 Suppl 1:83-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 91. | Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55:1350-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 491] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 92. | Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 423] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 93. | Roingeard P. Hepatitis C virus diversity and hepatic steatosis. J Viral Hepat. 2013;20:77-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 94. | Goossens N, Negro F. Is genotype 3 of the hepatitis C virus the new villain? Hepatology. 2014;59:2403-2412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 95. | Leandro G, Mangia A, Hui J, Fabris P, Rubbia-Brandt L, Colloredo G, Adinolfi LE, Asselah T, Jonsson JR, Smedile A. Relationship between steatosis, inflammation, and fibrosis in chronic hepatitis C: a meta-analysis of individual patient data. Gastroenterology. 2006;130:1636-1642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 408] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 96. | Hui JM, Sud A, Farrell GC, Bandara P, Byth K, Kench JG, McCaughan GW, George J. Insulin resistance is associated with chronic hepatitis C virus infection and fibrosis progression [corrected]. Gastroenterology. 2003;125:1695-1704. [PubMed] |
| 97. | Fartoux L, Poujol-Robert A, Guéchot J, Wendum D, Poupon R, Serfaty L. Insulin resistance is a cause of steatosis and fibrosis progression in chronic hepatitis C. Gut. 2005;54:1003-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 278] [Article Influence: 13.9] [Reference Citation Analysis (1)] |
| 98. | Rumi MG, De Filippi F, La Vecchia C, Donato MF, Gallus S, Del Ninno E, Colombo M. Hepatitis C reactivation in patients with chronic infection with genotypes 1b and 2c: a retrospective cohort study of 206 untreated patients. Gut. 2005;54:402-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 99. | Bruno S, Silini E, Crosignani A, Borzio F, Leandro G, Bono F, Asti M, Rossi S, Larghi A, Cerino A. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a prospective study. Hepatology. 1997;25:754-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 244] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 100. | Bruno S, Crosignani A, Maisonneuve P, Rossi S, Silini E, Mondelli MU. Hepatitis C virus genotype 1b as a major risk factor associated with hepatocellular carcinoma in patients with cirrhosis: a seventeen-year prospective cohort study. Hepatology. 2007;46:1350-1356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 150] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 101. | Raimondi S, Bruno S, Mondelli MU, Maisonneuve P. Hepatitis C virus genotype 1b as a risk factor for hepatocellular carcinoma development: a meta-analysis. J Hepatol. 2009;50:1142-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 168] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 102. | Probst A, Dang T, Bochud M, Egger M, Negro F, Bochud PY. Role of hepatitis C virus genotype 3 in liver fibrosis progression--a systematic review and meta-analysis. J Viral Hepat. 2011;18:745-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 103. | Nkontchou G, Ziol M, Aout M, Lhabadie M, Baazia Y, Mahmoudi A, Roulot D, Ganne-Carrie N, Grando-Lemaire V, Trinchet JC. HCV genotype 3 is associated with a higher hepatocellular carcinoma incidence in patients with ongoing viral C cirrhosis. J Viral Hepat. 2011;18:e516-e522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 104. | van der Meer AJ, Veldt BJ, Feld JJ, Wedemeyer H, Dufour JF, Lammert F, Duarte-Rojo A, Heathcote EJ, Manns MP, Kuske L. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA. 2012;308:2584-2593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1165] [Cited by in RCA: 1167] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 105. | Kanwal F, Kramer JR, Ilyas J, Duan Z, El-Serag HB. HCV genotype 3 is associated with an increased risk of cirrhosis and hepatocellular cancer in a national sample of U.S. Veterans with HCV. Hepatology. 2014;60:98-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 240] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 106. | Heindryckx F, Colle I, Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. Int J Exp Pathol. 2009;90:367-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 293] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 107. | Higgs MR, Chouteau P, Lerat H. ‘Liver let die’: oxidative DNA damage and hepatotropic viruses. J Gen Virol. 2014;95:991-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 108. | Nault JC. Pathogenesis of hepatocellular carcinoma according to aetiology. Best Pract Res Clin Gastroenterol. 2014;28:937-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 109. | Bonilla Guerrero R, Roberts LR. The role of hepatitis B virus integrations in the pathogenesis of human hepatocellular carcinoma. J Hepatol. 2005;42:760-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 146] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 110. | Ringelhan M, O’Connor T, Protzer U, Heikenwalder M. The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. J Pathol. 2015;235:355-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 111. | Albeldawi M, Soliman M, Lopez R, Zein NN. Hepatitis C virus-associated primary hepatocellular carcinoma in non-cirrhotic patients. Dig Dis Sci. 2012;57:3265-3270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 112. | Kawada N, Imanaka K, Kawaguchi T, Tamai C, Ishihara R, Matsunaga T, Gotoh K, Yamada T, Tomita Y. Hepatocellular carcinoma arising from non-cirrhotic nonalcoholic steatohepatitis. J Gastroenterol. 2009;44:1190-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 166] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 113. | Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 2010;29:2309-2324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 199] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 114. | Bréchot C, Gozuacik D, Murakami Y, Paterlini-Bréchot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol. 2000;10:211-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 222] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 115. | Jiang S, Yang Z, Li W, Li X, Wang Y, Zhang J, Xu C, Chen PJ, Hou J, McCrae MA. Re-evaluation of the carcinogenic significance of hepatitis B virus integration in hepatocarcinogenesis. PLoS One. 2012;7:e40363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 116. | Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725-12734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 386] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 117. | Su Q, Schröder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 176] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 118. | Knoll S, Fürst K, Thomas S, Villanueva Baselga S, Stoll A, Schaefer S, Pützer BM. Dissection of cell context-dependent interactions between HBx and p53 family members in regulation of apoptosis: a role for HBV-induced HCC. Cell Cycle. 2011;10:3554-3565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 119. | Amaddeo G, Cao Q, Ladeiro Y, Imbeaud S, Nault JC, Jaoui D, Gaston Mathe Y, Laurent C, Laurent A, Bioulac-Sage P. Integration of tumour and viral genomic characterizations in HBV-related hepatocellular carcinomas. Gut. 2015;64:820-829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 120. | Pang R, Lee TK, Poon RT, Fan ST, Wong KB, Kwong YL, Tse E. Pin1 interacts with a specific serine-proline motif of hepatitis B virus X-protein to enhance hepatocarcinogenesis. Gastroenterology. 2007;132:1088-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 105] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 121. | Datta S, Banerjee A, Chandra PK, Chakravarty R. Pin1-HBx interaction: a step toward understanding the significance of hepatitis B virus genotypes in hepatocarcinogenesis. Gastroenterology. 2007;133:727-728; author reply 728-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 122. | Peng Z, Zhang Y, Gu W, Wang Z, Li D, Zhang F, Qiu G, Xie K. Integration of the hepatitis B virus X fragment in hepatocellular carcinoma and its effects on the expression of multiple molecules: a key to the cell cycle and apoptosis. Int J Oncol. 2005;26:467-473. [PubMed] |
| 123. | Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 713] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 124. | Jiang Z, Jhunjhunwala S, Liu J, Haverty PM, Kennemer MI, Guan Y, Lee W, Carnevali P, Stinson J, Johnson S. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 234] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 125. | Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 722] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 126. | Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH, Surzycki SJ, Lee YI. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 246] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 127. | Zheng DL, Zhang L, Cheng N, Xu X, Deng Q, Teng XM, Wang KS, Zhang X, Huang J, Han ZG. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol. 2009;50:377-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 128. | Shon JK, Shon BH, Park IY, Lee SU, Fa L, Chang KY, Shin JH, Lee YI. Hepatitis B virus-X protein recruits histone deacetylase 1 to repress insulin-like growth factor binding protein 3 transcription. Virus Res. 2009;139:14-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 129. | Pollicino T, Vegetti A, Saitta C, Ferrara F, Corradini E, Raffa G, Pietrangelo A, Raimondo G. Hepatitis B virus DNA integration in tumour tissue of a non-cirrhotic HFE-haemochromatosis patient with hepatocellular carcinoma. J Hepatol. 2013;58:190-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 130. | Ng SA, Lee C. Hepatitis B virus X gene and hepatocarcinogenesis. J Gastroenterol. 2011;46:974-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 131. | Motavaf M, Safari S, Saffari Jourshari M, Alavian SM. Hepatitis B virus-induced hepatocellular carcinoma: the role of the virus x protein. Acta Virol. 2013;57:389-396. [PubMed] |
| 132. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. [PubMed] |
| 133. | Liang T, Chen EQ, Tang H. Hepatitis B virus gene mutations and hepatocarcinogenesis. Asian Pac J Cancer Prev. 2013;14:4509-4513. [PubMed] |
| 134. | Nurainy N, Muljono DH, Sudoyo H, Marzuki S. Genetic study of hepatitis B virus in Indonesia reveals a new subgenotype of genotype B in east Nusa Tenggara. Arch Virol. 2008;153:1057-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 135. | Wang LH, Huang W, Lai MD, Su IJ. Aberrant cyclin A expression and centrosome overduplication induced by hepatitis B virus pre-S2 mutants and its implication in hepatocarcinogenesis. Carcinogenesis. 2012;33:466-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 136. | Chisari FV, Klopchin K, Moriyama T, Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL, Palmiter RD. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145-1156. [PubMed] |
| 137. | Barone M, Spano D, D’Apolito M, Centra M, Lasalandra C, Capasso M, Di Leo A, Volinia S, Arcelli D, Rosso N. Gene expression analysis in HBV transgenic mouse liver: a model to study early events related to hepatocarcinogenesis. Mol Med. 2006;12:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 138. | Saitta C, Tripodi G, Barbera A, Bertuccio A, Smedile A, Ciancio A, Raffa G, Sangiovanni A, Navarra G, Raimondo G. Hepatitis B virus (HBV) DNA integration in patients with occult HBV infection and hepatocellular carcinoma. Liver Int. 2015;35:2311-2317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 139. | Toyoda H, Kumada T, Kaneoka Y, Murakami Y. Impact of hepatitis B virus (HBV) X gene integration in liver tissue on hepatocellular carcinoma development in serologically HBV-negative chronic hepatitis C patients. J Hepatol. 2008;48:43-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 140. | Shi Y, Lan Y, Cao F, Teng Y, Li L, Wang F, Li J, Zhou J, Li Y. Infected hematopoietic stem cells and with integrated HBV DNA generate defective T cells in chronic HBV infection patients. J Viral Hepat. 2014;21:e39-e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 141. | Castello G, Scala S, Palmieri G, Curley SA, Izzo F. HCV-related hepatocellular carcinoma: From chronic inflammation to cancer. Clin Immunol. 2010;134:237-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 112] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 142. | Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 633] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 143. | Marra M, Sordelli IM, Lombardi A, Lamberti M, Tarantino L, Giudice A, Stiuso P, Abbruzzese A, Sperlongano R, Accardo M. Molecular targets and oxidative stress biomarkers in hepatocellular carcinoma: an overview. J Transl Med. 2011;9:171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 180] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 144. | Koike K, Miyoshi H. Oxidative stress and hepatitis C viral infection. Hepatol Res. 2006;34:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 145. | Farinati F, Cardin R, Degan P, De Maria N, Floyd RA, Van Thiel DH, Naccarato R. Oxidative DNA damage in circulating leukocytes occurs as an early event in chronic HCV infection. Free Radic Biol Med. 1999;27:1284-1291. [PubMed] |
| 146. | Sumida Y, Nakashima T, Yoh T, Nakajima Y, Ishikawa H, Mitsuyoshi H, Sakamoto Y, Okanoue T, Kashima K, Nakamura H. Serum thioredoxin levels as an indicator of oxidative stress in patients with hepatitis C virus infection. J Hepatol. 2000;33:616-622. [PubMed] |
| 147. | Mahmood S, Kawanaka M, Kamei A, Izumi A, Nakata K, Niiyama G, Ikeda H, Hanano S, Suehiro M, Togawa K. Immunohistochemical evaluation of oxidative stress markers in chronic hepatitis C. Antioxid Redox Signal. 2004;6:19-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 77] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 148. | Fujita N, Sugimoto R, Ma N, Tanaka H, Iwasa M, Kobayashi Y, Kawanishi S, Watanabe S, Kaito M, Takei Y. Comparison of hepatic oxidative DNA damage in patients with chronic hepatitis B and C. J Viral Hepat. 2008;15:498-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 99] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 149. | Gabbay E, Zigmond E, Pappo O, Hemed N, Rowe M, Zabrecky G, Cohen R, Ilan Y. Antioxidant therapy for chronic hepatitis C after failure of interferon: results of phase II randomized, double-blind placebo controlled clinical trial. World J Gastroenterol. 2007;13:5317-5323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 31] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 150. | Gane EJ, Weilert F, Orr DW, Keogh GF, Gibson M, Lockhart MM, Frampton CM, Taylor KM, Smith RA, Murphy MP. The mitochondria-targeted anti-oxidant mitoquinone decreases liver damage in a phase II study of hepatitis C patients. Liver Int. 2010;30:1019-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 295] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 151. | Ivanov AV, Bartosch B, Smirnova OA, Isaguliants MG, Kochetkov SN. HCV and oxidative stress in the liver. Viruses. 2013;5:439-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 152. | Moriya K, Nakagawa K, Santa T, Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Miyazawa T, Ishibashi K, Horie T. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 2001;61:4365-4370. [PubMed] |
| 153. | Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366-375. [PubMed] |
| 154. | Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, Weinman SA. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481-37488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 317] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 155. | Gong G, Waris G, Tanveer R, Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc Natl Acad Sci USA. 2001;98:9599-9604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 498] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 156. | Qadri I, Iwahashi M, Capasso JM, Hopken MW, Flores S, Schaack J, Simon FR. Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication: role of JNK, p38 MAPK and AP-1. Biochem J. 2004;378:919-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 119] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 157. | Waris G, Turkson J, Hassanein T, Siddiqui A. Hepatitis C virus (HCV) constitutively activates STAT-3 via oxidative stress: role of STAT-3 in HCV replication. J Virol. 2005;79:1569-1580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 158. | Lin W, Tsai WL, Shao RX, Wu G, Peng LF, Barlow LL, Chung WJ, Zhang L, Zhao H, Jang JY. Hepatitis C virus regulates transforming growth factor beta1 production through the generation of reactive oxygen species in a nuclear factor kappaB-dependent manner. Gastroenterology. 2010;138:2509-218, 2518.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 159. | Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655-1669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2139] [Cited by in RCA: 2161] [Article Influence: 127.1] [Reference Citation Analysis (0)] |
| 160. | Taniguchi H, Kato N, Otsuka M, Goto T, Yoshida H, Shiratori Y, Omata M. Hepatitis C virus core protein upregulates transforming growth factor-beta 1 transcription. J Med Virol. 2004;72:52-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 105] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 161. | Kisseleva T, Brenner DA. Mechanisms of fibrogenesis. Exp Biol Med (Maywood). 2008;233:109-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 392] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 162. | Nomura-Takigawa Y, Nagano-Fujii M, Deng L, Kitazawa S, Ishido S, Sada K, Hotta H. Non-structural protein 4A of Hepatitis C virus accumulates on mitochondria and renders the cells prone to undergoing mitochondria-mediated apoptosis. J Gen Virol. 2006;87:1935-1945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 163. | Kang SM, Kim SJ, Kim JH, Lee W, Kim GW, Lee KH, Choi KY, Oh JW. Interaction of hepatitis C virus core protein with Hsp60 triggers the production of reactive oxygen species and enhances TNF-alpha-mediated apoptosis. Cancer Lett. 2009;279:230-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 164. | Kawanishi S, Oikawa S. Mechanism of telomere shortening by oxidative stress. Ann N Y Acad Sci. 2004;1019:278-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 379] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 165. | Sekoguchi S, Nakajima T, Moriguchi M, Jo M, Nishikawa T, Katagishi T, Kimura H, Minami M, Itoh Y, Kagawa K. Role of cell-cycle turnover and oxidative stress in telomere shortening and cellular senescence in patients with chronic hepatitis C. J Gastroenterol Hepatol. 2007;22:182-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 166. | Machida K, Cheng KT, Sung VM, Shimodaira S, Lindsay KL, Levine AM, Lai MY, Lai MM. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc Natl Acad Sci USA. 2004;101:4262-4267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 284] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 167. | Yen HH, Shih KL, Lin TT, Su WW, Soon MS, Liu CS. Decreased mitochondrial deoxyribonucleic acid and increased oxidative damage in chronic hepatitis C. World J Gastroenterol. 2012;18:5084-5089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 30] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 168. | Tardif KD, Mori K, Siddiqui A. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J Virol. 2002;76:7453-7459. [PubMed] |
| 169. | Waris G, Tardif KD, Siddiqui A. Endoplasmic reticulum (ER) stress: hepatitis C virus induces an ER-nucleus signal transduction pathway and activates NF-kappaB and STAT-3. Biochem Pharmacol. 2002;64:1425-1430. [PubMed] |
| 170. | Ji C. Dissection of endoplasmic reticulum stress signaling in alcoholic and non-alcoholic liver injury. J Gastroenterol Hepatol. 2008;23 Suppl 1:S16-S24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 171. | Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460-3470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1214] [Cited by in RCA: 1537] [Article Influence: 128.1] [Reference Citation Analysis (1)] |
| 172. | Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741-33749. [PubMed] |
| 173. | Ciccaglione AR, Marcantonio C, Tritarelli E, Equestre M, Vendittelli F, Costantino A, Geraci A, Rapicetta M. Activation of the ER stress gene gadd153 by hepatitis C virus sensitizes cells to oxidant injury. Virus Res. 2007;126:128-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 174. | Benali-Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, Buscail L, Bartenschlager R, Ichas F, Rizzuto R. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene. 2005;24:4921-4933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 214] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 175. | Joyce MA, Walters KA, Lamb SE, Yeh MM, Zhu LF, Kneteman N, Doyle JS, Katze MG, Tyrrell DL. HCV induces oxidative and ER stress, and sensitizes infected cells to apoptosis in SCID/Alb-uPA mice. PLoS Pathog. 2009;5:e1000291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 126] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 176. | von dem Bussche A, Machida R, Li K, Loevinsohn G, Khander A, Wang J, Wakita T, Wands JR, Li J. Hepatitis C virus NS2 protein triggers endoplasmic reticulum stress and suppresses its own viral replication. J Hepatol. 2010;53:797-804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 177. | Merquiol E, Uzi D, Mueller T, Goldenberg D, Nahmias Y, Xavier RJ, Tirosh B, Shibolet O. HCV causes chronic endoplasmic reticulum stress leading to adaptation and interference with the unfolded protein response. PLoS One. 2011;6:e24660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 178. | Dionisio N, Garcia-Mediavilla MV, Sanchez-Campos S, Majano PL, Benedicto I, Rosado JA, Salido GM, Gonzalez-Gallego J. Hepatitis C virus NS5A and core proteins induce oxidative stress-mediated calcium signalling alterations in hepatocytes. J Hepatol. 2009;50:872-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 179. | Bartosch B, Thimme R, Blum HE, Zoulim F. Hepatitis C virus-induced hepatocarcinogenesis. J Hepatol. 2009;51:810-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 180. | Lemon SM, McGivern DR. Is hepatitis C virus carcinogenic? Gastroenterology. 2012;142:1274-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 181. | Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene. 2006;25:3834-3847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 298] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 182. | McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene. 2011;30:1969-1983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 171] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 183. | Tornesello ML, Buonaguro L, Tatangelo F, Botti G, Izzo F, Buonaguro FM. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102:74-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 131] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 184. | Liu J, Wang Z, Tang J, Tang R, Shan X, Zhang W, Chen Q, Zhou F, Chen K, Huang A. Hepatitis C virus core protein activates Wnt/β-catenin signaling through multiple regulation of upstream molecules in the SMMC-7721 cell line. Arch Virol. 2011;156:1013-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 185. | Higgs MR, Lerat H, Pawlotsky JM. Hepatitis C virus-induced activation of β-catenin promotes c-Myc expression and a cascade of pro-carcinogenetic events. Oncogene. 2013;32:4683-4693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 186. | Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, Jung G, Choi KY, Hwang SB. Nonstructural 5A protein activates beta-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J Hepatol. 2009;51:853-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 72] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 187. | Street A, Macdonald A, McCormick C, Harris M. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. J Virol. 2005;79:5006-5016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 115] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 188. | Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2005;102:18159-18164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 189. | Hernando E, Nahlé Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 424] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 190. | Hassan M, Ghozlan H, Abdel-Kader O. Activation of RB/E2F signaling pathway is required for the modulation of hepatitis C virus core protein-induced cell growth in liver and non-liver cells. Cell Signal. 2004;16:1375-1385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 191. | Lai CK, Jeng KS, Machida K, Cheng YS, Lai MM. Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology. 2008;370:295-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 192. | Machida K, McNamara G, Cheng KT, Huang J, Wang CH, Comai L, Ou JH, Lai MM. Hepatitis C virus inhibits DNA damage repair through reactive oxygen and nitrogen species and by interfering with the ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and hepatocytes. J Immunol. 2010;185:6985-6998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 193. | Pal S, Polyak SJ, Bano N, Qiu WC, Carithers RL, Shuhart M, Gretch DR, Das A. Hepatitis C virus induces oxidative stress, DNA damage and modulates the DNA repair enzyme NEIL1. J Gastroenterol Hepatol. 2010;25:627-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 194. | Hoare M, Das T, Alexander G. Ageing, telomeres, senescence, and liver injury. J Hepatol. 2010;53:950-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 195. | Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 196. | Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM, Tavolari S. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761-5767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 321] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 197. | Oishi N, Yamashita T, Kaneko S. Molecular biology of liver cancer stem cells. Liver Cancer. 2014;3:71-84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 198. | Yamashita T, Kaneko S. Orchestration of hepatocellular carcinoma development by diverse liver cancer stem cells. J Gastroenterol. 2014;49:1105-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 199. | Oliva J, French BA, Qing X, French SW. The identification of stem cells in human liver diseases and hepatocellular carcinoma. Exp Mol Pathol. 2010;88:331-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 200. | Lu JW, Chang JG, Yeh KT, Chen RM, Tsai JJ, Hu RM. Overexpression of Thy1/CD90 in human hepatocellular carcinoma is associated with HBV infection and poor prognosis. Acta Histochem. 2011;113:833-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 201. | Yeh CT, Kuo CJ, Lai MW, Chen TC, Lin CY, Yeh TS, Lee WC. CD133-positive hepatocellular carcinoma in an area endemic for hepatitis B virus infection. BMC Cancer. 2009;9:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 202. | Arzumanyan A, Friedman T, Ng IO, Clayton MM, Lian Z, Feitelson MA. Does the hepatitis B antigen HBx promote the appearance of liver cancer stem cells? Cancer Res. 2011;71:3701-3708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 203. | Wang C, Yang W, Yan HX, Luo T, Zhang J, Tang L, Wu FQ, Zhang HL, Yu LX, Zheng LY. Hepatitis B virus X (HBx) induces tumorigenicity of hepatic progenitor cells in 3,5-diethoxycarbonyl-1,4-dihydrocollidine-treated HBx transgenic mice. Hepatology. 2012;55:108-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 204. | Li CH, Wang YJ, Dong W, Xiang S, Liang HF, Wang HY, Dong HH, Chen L, Chen XP. Hepatic oval cell lines generate hepatocellular carcinoma following transfection with HBx gene and treatment with aflatoxin B1 in vivo. Cancer Lett. 2011;311:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 205. | Kimura O, Kondo Y, Kogure T, Kakazu E, Ninomiya M, Iwata T, Morosawa T, Shimosegawa T. Expression of EpCAM increases in the hepatitis B related and the treatment-resistant hepatocellular carcinoma. Biomed Res Int. 2014;2014:172913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 206. | Fan H, Zhang H, Pascuzzi PE, Andrisani O. Hepatitis B virus X protein induces EpCAM expression via active DNA demethylation directed by RelA in complex with EZH2 and TET2. Oncogene. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 207. | Ali N, Allam H, May R, Sureban SM, Bronze MS, Bader T, Umar S, Anant S, Houchen CW. Hepatitis C virus-induced cancer stem cell-like signatures in cell culture and murine tumor xenografts. J Virol. 2011;85:12292-12303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 208. | Machida K, Tsukamoto H, Mkrtchyan H, Duan L, Dynnyk A, Liu HM, Asahina K, Govindarajan S, Ray R, Ou JH. Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci USA. 2009;106:1548-1553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 184] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 209. | Machida K, Chen CL, Liu JC, Kashiwabara C, Feldman D, French SW, Sher L, Hyeongnam JJ, Tsukamoto H. Cancer stem cells generated by alcohol, diabetes, and hepatitis C virus. J Gastroenterol Hepatol. 2012;27 Suppl 2:19-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 210. | Papatheodoridis GV, Chan HL, Hansen BE, Janssen HL, Lampertico P. Risk of hepatocellular carcinoma in chronic hepatitis B: assessment and modification with current antiviral therapy. J Hepatol. 2015;62:956-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 407] [Article Influence: 40.7] [Reference Citation Analysis (0)] |