Published online Jan 28, 2016. doi: 10.3748/wjg.v22.i4.1487
Peer-review started: July 27, 2015
First decision: September 11, 2015
Revised: September 25, 2015
Accepted: October 12, 2015
Article in press: October 13, 2015
Published online: January 28, 2016
Processing time: 184 Days and 1.8 Hours
MicroRNAs (miRNAs) are small noncoding RNAs. More than 2500 mature miRNAs are detected in plants, animals and several types of viruses. Hepatitis C virus (HCV), which is a positive-sense, single-stranded RNA virus, does not encode viral miRNA. However, HCV infection alters the expression of host miRNAs, either in cell culture or in patients with liver disease progression, such as liver fibrosis, cirrhosis, and hepatocellular carcinoma. In turn, host miRNAs regulate HCV life cycle through directly binding to HCV RNAs or indirectly targeting cellular mRNAs. Increasing evidence demonstrates that miRNAs are one of the centered factors in the interaction network between virus and host. The competitive viral and host RNA hypothesis proposes a latent cross-regulation pattern between host mRNAs and HCV RNAs. High loads of HCV RNA sequester and de-repress host miRNAs from their normal host targets and thus disturb host gene expression, indicating a means of adaptation for HCV to establish a persistent infection. Some special miRNAs are closely correlated with liver-specific disease progression and the changed levels of miRNAs are even higher sensitivity and specificity than those of traditional proteins. Therefore, some of them can serve as novel diagnostic/prognostic biomarkers in HCV-infected patients with liver diseases. They are also attractive therapeutic targets for development of new anti-HCV agents.
Core tip: Hepatitis C virus (HCV) infection changes the expression of host miRNAs in vitro and in vivo, while host MicroRNAs (miRNAs) in turn regulate HCV life cycle through directly binding to HCV RNA and/or indirectly targeting cellular mRNAs. The miRNA-centered competitive viral and host RNA network displays a cross-regulation pattern between host mRNAs and HCV genome. Evidence based on the miRNA-mediated host/viral interactions suggests that specific miRNAs can serve as novel diagnostic/prognostic biomarkers in HCV-infected patients with liver diseases and therapeutic targets for development of anti-HCV agents in the future.
- Citation: Li H, Jiang JD, Peng ZG. MicroRNA-mediated interactions between host and hepatitis C virus. World J Gastroenterol 2016; 22(4): 1487-1496
- URL: https://www.wjgnet.com/1007-9327/full/v22/i4/1487.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i4.1487
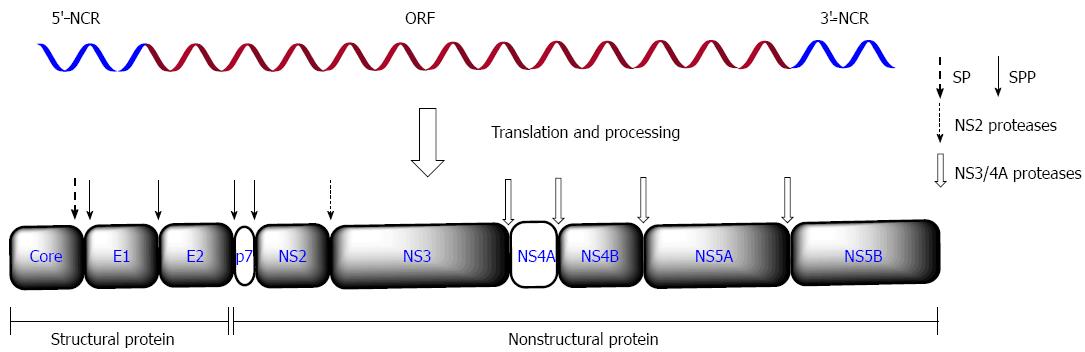
Hepatitis C virus (HCV) is a hepatotropic virus classified in the hepacivirus genus of the Flaviviridae family, and infection can cause acute or chronic hepatitis in humans[1]. To date, about 2.8% of the human population worldwide (> 185 million people) has been infected with HCV, and about 20% of cases eventually lead to liver fibrosis, cirrhosis, or even to hepatocellular carcinoma (HCC)[2]. The HCV genome is a positive-sense, single-stranded RNA about 9.6 kb with a conserved 5’ noncoding region (NCR), one open reading frame (ORF) and following a conserved 3’-NCR. The only ORF encodes a polyprotein of about 3000 amino acids, and its N terminus is cleaved by endoplasmic reticulum (ER) signal peptidase and/or signal peptide peptidase (SPP) into three structural proteins (core, E1 and E2). The other sequence of the polyprotein is processed by the viral NS2 and/or NS3/4A protease into seven nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B), which play important roles in the HCV life cycle[3] (Figure 1). HCV isolates are grouped into seven genotypes and several subtypes with distinct geographic distribution and different response to anti-HCV agents[2,4,5].
MicroRNAs (miRNAs) are small noncoding RNAs (about 22 nt) firstly discovered in Caenorhabditis elegans in 1993[6], which regulate the expression of complementary mRNA. To date, > 2500 mature miRNAs have been identified in humans[7]. A cellular gene encodes primary miRNA transcript (pri-miRNA), which is recognized and cleaved into a 70-100-nt hairpin form precursor miRNA (pre-miRNA) by RNase III enzyme called Drosha[8]. The pre-miRNA is then further processed into a mature miRNA duplex of about 22 bp by Dicer in the cytoplasm. After strand separation, only the single-strand mature miRNA is incorporated into the RNA-induced silencing complex, which then cleaves the mRNA or induces translational repression by binding to the target sequences of complementary mRNA[9-11]. Over the last decade, two regulation modes of miRNA have been clarified. One is that bases of miRNA pair with target sequences of mRNA by precise or nearly precise complementarity in plants, and then trigger the degradation of the target mRNA. The other is that bases of miRNA pair imperfectly with target sequences and generally inhibit the translation of mRNA[12]. The nucleotide sequence from site 2 to 7 at the 5’ end of miRNA, which is called the seed region, determines the specificity of miRNA in miRNA-mediated RNAi[13]. However, any given miRNA could theoretically bind to a broad spectrum of mRNAs, resulting in potentially disturbing enormous regulatory networks.
Increasing evidence shows that HCV infection regulates the expression of many cellular miRNAs, which in turn directly or indirectly up- or down-regulate HCV replication[14]. From the viewpoint of evolution, it is not difficult to predict that HCV utilizes cellular miRNAs for its replication, because HCV is an obligate intracellular microorganism. However, host cells also change the expression of some miRNAs to defend against infection. In this review, we describe the reciprocal interaction mediated by host miRNAs between host cells and HCV, and then summarize the predictive applications of miRNAs.
In 2005, a full-length infectious HCV culture system was established in hepatoma cells[15], which facilitated exploring miRNAs and their roles in HCV infection. After analyzing the HCV-infected human hepatoma line Huh7.5.1 with a comprehensive microarray, Liu et al[16] found that expression of 108 human miRNAs was changed more than twofold. Among them, miR-122 was downregulated 2.8-fold, and conversely, miR-351 and miR-296 were upregulated 15.2- and 4.9-fold at day 4 post-infection, respectively. Similarly, Ishida et al[17] demonstrated that HCV infection upregulated expression of miR-192, miR-194 and miR-215 and downregulated expression of miR-320 and miR-491 in vitro. Further evidence showed that some of these changes might, in turn, enhance HCV replication. Moreover, seven miRNAs (miR-30b, 30c, 130a, 192, 301, 324-5p and 565) were downregulated in Huh7.5 cells infected with HCV genotype 2a, however, those changes were reversed after subsequent treatment with interferon (IFN)-α[18]. Although the detailed mechanism is unclear, expression of many cellular miRNAs is directly changed after HCV infection and these changes might in turn facilitate or inhibit HCV replication.
HCV infection causes liver-specific injury, including progressive liver fibrosis, cirrhosis and HCC. These diseases result from both direct dysregulation of cellular metabolism by viral proteins and indirect consequence of the host response to HCV infection. Cellular miRNAs are important factors in HCV-induced liver disease progression.
Accumulating evidence shows that changes of miRNAs induced by HCV infection subsequently influence liver fibrosis progression. After comparing chronic HCV patients with normal controls, Ramachandran et al[19] identified 22 upregulated and 35 downregulated miRNAs (> 2-fold). Among them, miR-200c, which is highly expressed in chronic HCV patients, down-regulates fas-associated protein (FAP)-1 and subsequently activates Src kinases-mediated profibrotic signaling pathway, suggesting that it is a promoting factor of hepatic fibrosis. In 2012, Ogawa et al[20] reported that miR-222 and its homolog miR-221 were significantly increased during liver fibrosis progression in HCV-infected patients and mice. Further results showed that miR-221/222 inhibited expression of cyclin-dependent kinase inhibitor 1B (CDKN1B) via binding to the 3’UTR of CDKN1B and positively correlated with the expression of fibrosis-related α-1 type I collagen (Col1A1) and α-smooth muscle actin (SMA). Therefore, miR-221/222 might potentially serve as new fibrosis biomarkers in HCV-infected patients. By contrast, in patients infected with HCV, down-regulation of miR-29 in activated hepatic stellate cells (HSCs) might potentiate synthesis of collagen by dramatically derepressing the expression of COL1A1 and COL3A1, which eventually promotes progression of fibrosis[21,22]. In hepatic fibrosis patients infected with HCV, miR-449a is decreased and thus derepressed expression of NOTCH1, which results in stabilization of p65 in nuclear by the NOTCH signaling pathway and eventually activates the expression of inflammatory biomarker YKL40[23]. Recent studies also revealed that, in liver needle biopsies from patients infected with HCV, the level of miR-21 is positively correlated with fibrotic stage and could finally lead to increased fibrogenesis by targeting SMAD7 in the transforming growth factor (TGF)-β signaling pathway, while the level of miR-122 is negatively correlated with fibrotic stage[24].
miRNAs are directly involved in hepatocellular carcinogenesis in HCV-infected patients[25]. In 2006, after analyzing the expression profiles of miRNAs in 25 pairs of HCC and adjacent non-tumorous tissues and nine additional chronic hepatitis specimens with a human miRNA microarray, Murakami et al[26] found that eight miRNAs had significantly different expression patterns. Among them, miR-92, 20, 18 and precursor miR-18 had a significantly negative correlation with differentiation of HCC, while expression of miR-99a was consistent with the degree of tumor differentiation. Likewise, after investigating the expression of 2226 miRNAs in HCC patients infected with HCV, Diaz et al[27] identified 18 HCC-exclusive miRNAs, including three new HCC-related miRNAs (miR-497, 1269 and miR-424-3p). Seventeen of these miRNAs were connected into a regulatory network centered on p53, phosphatase and tensin homolog (PTEN) and all-trans retinoic acid, which are three important modulators in the development of HCC[27,28]. miR-199a-3p, which is derived from its precursor mir-199a-1 or mir-199a-2, contains the same base sequences as miR-199b-3p. Reduction of miR-199a/b-3p in HCC tissues is dramatically correlated with poor survival of HCC patients and thus could be a potential biomarker for predicting prognosis of HCC[29]. miR-199a/b-3p inhibits HCC growth through targeting tumor-promoting PAK4 to inhibit the PAK4/Raf/MEK/ERK pathway[29] or through targeting c-Met and mTOR to inhibit HCC cell cycle progression[30,31].
However, only some abnormal expression of miRNAs was induced directly by HCV, even though altered expression of miRNAs in liver diseases has been reported frequently. HCV-induced changes in expression of several critical miRNAs highlight their clinical implications in liver disease progression (Table 1). Analyzing the changes of host miRNAs and mRNAs in response to HCV infection might provide us with a deeper understanding of HCV-induced liver diseases.
| miRNA | Expression after HCV infection | Target | Biological process | Clinical relevance | Therapeutic strategy | Ref. |
| miR-200c | Up | FAP-1 | Down-regulate FAP1 and subsequently activate Src kinases-mediated profibrotic signaling pathway | Promoting factor of hepatic fibrosis | Antagonist | [19] |
| miR-221/222 | Up | CDKN1B | Inhibit CDKN1B expression and correlate with expression of fibrosis-related Col1A1 and α-SMA | New marker for stellate cell activation and liver fibrosis progression | Antagonist | [20] |
| miR-29 | Down | COL1A1,COL3A1 | Decrease collagen synthesis and thus suppress fibrosis progression | Therapeutic and diagnosis biomarker of liver fibrosis | Mimic | [21,22] |
| miR-449a | Down | NOTCH1 | Repress YKL40 expression by targeting components of NOTCH signaling pathway and then inhibit liver fibrosis | Negative correlation with HCV-induced liver inflammation and fibrosis | Mimic | [23] |
| miR-21 | Up | SMAD7 | Enhance TGF-β-mediated fibrosis | Positive correlation with fibrotic stage | Antagonist | [24] |
| miR-122 | Down | HCV RNA, Bach1/HO-1 | Directly promote HCV replication and translation; Enhance HCV replication indirectly through Bach1/HO-1 | Inverse correlation with fibrosis stages | Antagonist | [24] |
| miR-199a/b-3p | Down | PAK4, c-Met, mTOR | Suppress HCC growth through inhibiting PAK4/Raf/MEK/ERK pathway or HCC cell cycle progression | Potential diagnostic/prognostic marker and therapeutic target in HCC | Mimic | [29-31] |
| miR-196 | Down | Bach1, HCV RNA | Inhibit HCV replication directly by targeting HCV NS5A and indirectly through Bach1 mRNA suppression | Potential predictive/prognostic factor in HCV infection | Mimic | [36,68,73] |
| miR-20a | Up | - | - | Predictive biomarker in HCV-mediated early to late stage fibrosis | - | [69] |
| miR-618 | Up | LPR12 | Suppress LPR12 and lead to progression of HCC | Promising biomarker for early detection of HCC among high-risk HCV-positive population | - | [70] |
| miR-650 | Down | TRAF4 | Down-regulate tumorigenesis-associated TRAF4 | Promising biomarker for the early detection of HCC among high-risk HCV-positive population | - | [70] |
| miR-155 | Up | APC | Promote hepatocarcinogenesis by activating Wnt signaling pathway | Negative predictive/prognostic marker and therapeutic target in HCV infection | Antagonist | [71-74] |
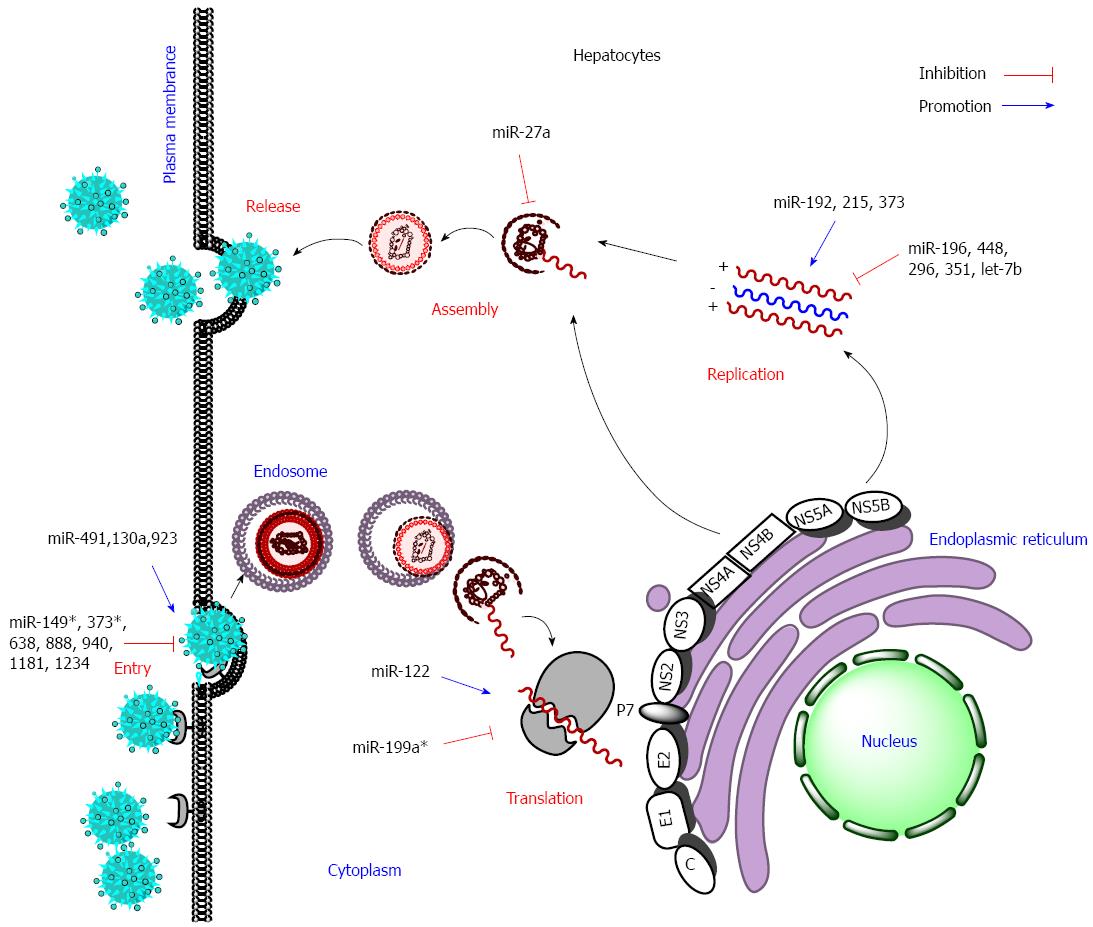
Virus-encoded and host-gene-derived miRNAs are both involved in the regulation of viral life cycle. However, it is now commonly believed that HCV, which is a single-stranded RNA virus that replicates in the cytoplasm, does not encode viral miRNAs[32,33]. Host miRNAs inhibit or stimulate viral replication in almost all stages of the viral life cycle by targeting viral RNAs and/or host mRNAs (Figure 2).
The single-stranded HCV RNA genome serves as a template for its replication and translation and as a binding site for cellular miRNAs. So, host miRNAs could theoretically influence HCV replication through directly targeting the HCV genome.
Most host miRNAs prevent HCV replication through directly targeting HCV RNAs. The highly conserved internal ribosome entry site (IRES) is in the 5’-untranslated region (UTR) of the HCV genome, and it recruits ribosomes to initiate translation of the HCV genome into a single polyprotein. Several miRNAs target the 5’-UTR region and thus inhibit translation of HCV polyprotein. Representatively, miR-199a* targets domain II of the HCV 5’-UTR IRES region, overexpression of miR-199a* decreases HCV replication in HCV 1b or 2a replicon cells while silencing expression of miR-199a* by specific antisense oligonucleotides facilitates viral replication[34]. Similarly, let-7b suppresses HCV replication and downregulates HCV accumulation by targeting the 5’-UTR and NS5B region in the HCV genome[35]. Pedersen et al[36] identified about 30 miRNAs differentially expressed in interferon-stimulated lymphocytes using miRNA array analysis. Among them, eight miRNAs (miR-1, 30, 128, 196, 296, 351, 431 and miR-448) displayed nearly perfect complementarities in their seed sequences with the HCV genome. The following study verified that miR-448 and miR-196 directly targeted the core and NS5A coding region in the HCV genome, respectively, and thus showed antiviral activity[36].
In contrast, a few miRNAs can facilitate HCV replication by directly targeting HCV RNA. For instance, liver-specific miRNA-122 positively modulates HCV replication, mainly through cooperatively binding to two targets in the 5’-UTR[37]. Although it has not been confirmed, miR-192/miR-215 might bind to the HCV genome and thus enhance HCV replication[17]. In addition, overexpression of miR-923 or inhibition of miR-149*, 373*, 638, 888, 940, 1181 and 1234 enhances the entry into cells of HCV pseudotype particles, though the detailed mechanism remains unclear[16].
Generally, drug treatment or HCV infection induces host physiological changes and alters miRNA expression. The changes in miRNAs partly act on regulatory genes associated with host signaling pathways to regulate HCV life cycle indirectly.
Abnormal expression of miRNAs is closely related to innate immunity, in which type I IFN signaling pathway and inflammatory cytokines play essential roles[38,39]. IFN-induced transmembrane (IFITM) 1 protein disrupts the process of viral entry by interacting with HCV co-receptors, such as CD81 and occluding[40]. miR-130a was the first reported miRNA to be directly involved in HCV-mediated innate immune response. Upregulation of miR-130a induced by HCV infection blocks expression of IFITM1 protein and thus subsequently facilitates HCV entry into host cells through an IFN signaling pathway[27,40,41]. The JAK/STAT signaling pathway plays critical roles in transcription of IFN-stimulating gene and initiation of host antiviral response[42]. The expression level of Janus kinase (JAK) 1 and the phosphorylation status of signal transducer and activator of transcription (STAT) 1 were significantly decreased because upregulation of miR-373 resulted from HCV infection. On the contrary, knockdown of miR-373 in hepatocytes enhances type-I-IFN-mediated antiviral response through directly targeting JAK1 and IRF9 and thus reduces HCV RNA replication[42]. miR-30 targets SOCS1 and SOCS3 and influences the helper T cell differentiation and the cytokine-activated JAK/STAT signaling pathway[18,43]. Therefore, down-regulation of miR-30 in HCV-infected Huh7.5 cells might finally create a permissive environment for viral replication[18].
However, miRNAs also affect HCV replication through other signaling pathways. For instance, miR-491 promotes HCV replication possibly through regulating the viral-entry-associated PI3K/Akt pathway, although the target gene has not been verified[17,44]. Putative target genes of miR-320c and miR-483-5p are involved in the PI3K/Akt, NF-κB and MAPK signaling pathways, which contribute to HCV pathogenesis or participate in host defense against infection[45]. HCV infection or lipid overload up-regulates expression of miR-27a, and this change in turn inhibits the expression of many genes related to lipid metabolism signaling pathways, such as ApoA1, ApoB100 and ApoE3, and consequently represses lipid storage in cells and production of infectious HCV particles[46-49]. This negative feedback mechanism has the benefit of maintaining a low viral load to escape host immune surveillance and thus establish persistent HCV infection.
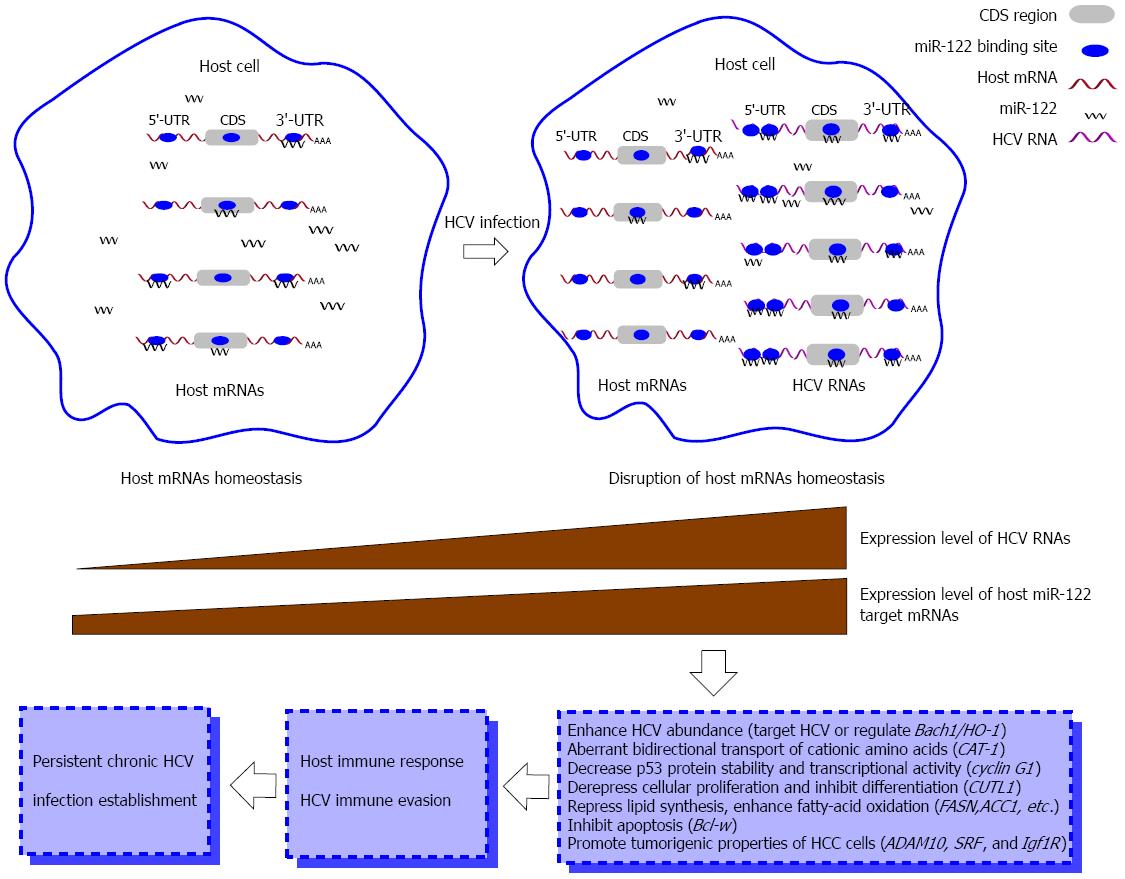
Although the biological functions of miRNAs in regulating HCV replication have been intensely studied, the potential roles of miRNAs in the genome-wide gene regulatory networks are not exactly defined. Many miRNAs, such as miR-122, can both positively and negatively modulate tens or hundreds of mRNAs, and several miRNAs can also regulate the same mRNA[50-52]. During HCV infection, many cellular miRNAs are involved in regulatory networks. After reviewing experimental evidence of reciprocal interactions between viral and host mRNAs during viral infections, Li et al[53] proposed a cvhRNA hypothesis that exogenous viral RNAs harbor the same miRNA-binding sites as cellular RNAs and act as competitors with host RNAs for the common miRNA pools in infected cells. The crosstalk mechanisms of cvhRNAs are dependent on the ability to sequester or degrade the common miRNAs by binding to viral RNAs and the extent of de-repression for host mRNAs by miRNA down-regulation[53]. Many factors might contribute to cvhRNA interactions, such as recognition/pairing ability, relative abundance, and intracellular localization of common miRNAs and the number of miRNA recognition elements in virus/host RNAs[53,54]. In fact, cvhRNA networks are present in many viral infections, such as hepatitis B virus, herpesvirus saimiri, lytic murine cytomegalovirus and human cytomegalovirus infections[55-58].
miRNA-mediated cvhRNA networks exist in HCV-infected hepatocytes. HCV RNA can specially sequester and de-repress miR-122 from its normal host targets[59]. miR-122 positively modulates HCV replication by cooperatively binding to two sites on the 5’-UTR sequence, while the sequence binding with it in the 3’-UTR does not appear to have any functional relevance[37]. However, the well-conserved fourth miR-122 recognition element in the NS5B coding region of the HCV genome potently inhibits HCV replication at the level of both translation and replication. This negative feedback regulation of miR-122 might contribute to immune evasion of HCV infection and progression to chronic HCV infection[60]. As the key element of the miR-122-centered cvhRNA network, miR-122 also regulates replication of HCV and host gene expression by targeting host mRNAs. Shan et al[61] showed that miR-122 indirectly promoted HCV replication partly through Bach1-mediated upregulation of heme oxygenase-1. miR-122 can also lead to low expression of endogenous cationic amino acid transporter (CAT)-1 by targeting the 3’- of CAT-1 mRNA[62,63]. Through targeting cyclin G1, miR-122 indirectly influences the stability and transcriptional activity of p53 protein and thus reduces the invasiveness of HCC-derived cells[64]. miR-122 participates in the whole regulatory network through targeting other non-liver-specific genes, such as the prominent proliferation- and differentiation-associated transcriptional repressor gene CUTL1, lipid-metabolism-associated genes FASN, ACC1, ACC2, SCD1 and ACLY, and apoptosis- and cell-cycle-associated genes Bcl-w, ADAM10, SRF and Igf1R[51,65-67]. All the targets serve as endogenous competitors for miR-122 and thus jointly form the miR-122-centered cvhRNA network, which can alter host homeostasis and thus contribute to the establishment of persistent viral infection (Figure 3).
Analogous to miR-122, miR-196 inhibits HCV replication by directly targeting the HCV genome or indirectly binding to two sites of the Bach1 3’-UTR, and might also form the cvhRNA network[68]. In principle, numerous host miRNAs, which could bind to host and HCV RNAs, potentially form cvhRNA networks. The phenomenon that virus-specific host miRNAs regulate the HCV life cycle through the cvhRNA networks might also reflect a means of immune evasion and adaptation for HCV to establish a persistent infection.
Expression profiles of miRNAs are significantly changed after HCV infection and can be partly reversed by drug treatment[36]. Therefore, it is possible to define specific miRNAs as diagnostic/prognostic biomarkers, and assessment indicators of drug therapeutic effects as well (Table 1).
Clinically, alterations of circulating miRNAs are associated with HCV infection and disease progression. For instance, upregulation of miR-134, 198, 320c and 483-5p in serum might be predictive biomarkers reflecting the pathophysiology after HCV infection[45]. Likewise, miR-20a might serve as a good predictive biomarker in HCV-mediated early to late fibrosis, because its expression is significantly elevated in the fibrotic stage of HCV infection[69]. The putative targets of miR-650 and miR-618 are tumorigenesis-associated gene TRAF4 and tumor-suppressor gene LRP12, respectively. Down-regulated miR-650 and up-regulated miR-618 in urine are strongly associated with HCC in high-risk HCV patients and thus might serve as early diagnostic biomarkers, whose changes show even higher sensitivity and specificity than those of traditional α-fetoprotein[70]. Likewise, HCV-induced miR-155 expression promotes hepatocarcinogenesis by inhibiting expression of adenomatous polyposis coli (APC) and activating Wnt/β-catenin signaling pathway[71,72]. Increased expression of miR-155 in serum, peripheral mononuclear cells, and liver tissues of patients with chronic HCV genotypes 1, 2 and 3 infection could be a negative prognostic marker, which suggests incomplete virus elimination, and increased risk of liver damage and HCC[71-74]. Also, miRNAs might be biomarkers for evaluation of drug therapeutic effects. Kałuzna reviewed a large number of studies and suggested that overexpression of miR-196, in response to IFN-β treatment, might serve as a positive prognostic marker indicating decreased inflammation, inhibition of HCV replication, and an efficacious response to IFN-based therapy[36,73].
miRNAs have gradually emerged as promising drug targets. Miravirsen (SPC3649), a locked nucleic acid form of anti-miR-122 oligonucleotides, decreases HCV viral load in a genotype-independent manner through inhibiting the biogenesis of miR-122, and it has been approved for clinical trial[75]. This provides a molecular rationale for combination therapy with silencing of miR-122 function during HCV infection. In contrast to the high level of miR-122 in the liver, miR-199a* is expressed at a low level in liver tissues, but in high quantities in extrahepatic tissues. miR-199a* also precludes efficient HCV replication through multiple mechanisms and thus might potentially be another anti-HCV therapeutic target[34,76,77]. In recent years, investigators have proposed a miRNA-based decoy mechanism in cvhRNA networks as therapeutic approaches for human diseases[78-80].
The replication of HCV in liver cells differentially triggers extensive expression of host miRNAs, which in turn positively or negatively regulate HCV life cycle through directly targeting the HCV genome and/or genes associated with cellular signaling pathways. This interaction makes miRNAs regulatory intermediates and finally develops a genome-scale regulatory network. miRNAs play important roles in disease progression and drug treatment, and they can be predictive and prognostic biomarkers and even drug targets. However, research on miRNAs associated with HCV is still in the early stage, and their functions are still to be clarified, especially in cvhRNA networks. The complicated cvhRNA networks mediated by miRNAs possibly indicate an adaption of HCV infection and host resistance. miRNAs will undoubtedly be useful tools for selecting agents to combat HCV infection in the future.
P- Reviewer: Khattab MA, Larrubia JR S- Editor: Qi Y L- Editor: A E- Editor: Zhang DN
| 1. | Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105-1113. [PubMed] |
| 2. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1770] [Cited by in RCA: 1847] [Article Influence: 153.9] [Reference Citation Analysis (3)] |
| 3. | Suzuki T, Ishii K, Aizaki H, Wakita T. Hepatitis C viral life cycle. Adv Drug Deliv Rev. 2007;59:1200-1212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 1146] [Article Influence: 114.6] [Reference Citation Analysis (0)] |
| 5. | Yang Z, Yu Y, Zhang H, Shang G, Gao J, Jiang JD, Peng Z. Replication priority of hepatitis C virus genotype 2a in a Chinese cohort. Acta Pharm Sin B. 2014;4:266-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] |
| 7. | Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68-D73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3819] [Cited by in RCA: 3860] [Article Influence: 321.7] [Reference Citation Analysis (0)] |
| 8. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3632] [Article Influence: 165.1] [Reference Citation Analysis (0)] |
| 9. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16089] [Article Influence: 1005.6] [Reference Citation Analysis (2)] |
| 10. | Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1638] [Cited by in RCA: 1781] [Article Influence: 127.2] [Reference Citation Analysis (0)] |
| 11. | Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 783] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 12. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7919] [Cited by in RCA: 8601] [Article Influence: 409.6] [Reference Citation Analysis (0)] |
| 13. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9285] [Article Influence: 464.3] [Reference Citation Analysis (0)] |
| 14. | Lee CH, Kim JH, Lee SW. The role of microRNAs in hepatitis C virus replication and related liver diseases. J Microbiol. 2014;52:445-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 1850] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 16. | Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 108] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 17. | Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, Shimizu S, Hikita H, Hiramatsu N, Kanto T, Hayashi N. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 2011;412:92-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Zhang X, Daucher M, Armistead D, Russell R, Kottilil S. MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-α. PLoS One. 2013;8:e55733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 19. | Ramachandran S, Ilias Basha H, Sarma NJ, Lin Y, Crippin JS, Chapman WC, Mohanakumar T. Hepatitis C virus induced miR200c down modulates FAP-1, a negative regulator of Src signaling and promotes hepatic fibrosis. PLoS One. 2013;8:e70744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 20. | Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600-1609. [PubMed] |
| 21. | Bandyopadhyay S, Friedman RC, Marquez RT, Keck K, Kong B, Icardi MS, Brown KE, Burge CB, Schmidt WN, Wang Y. Hepatitis C virus infection and hepatic stellate cell activation downregulate miR-29: miR-29 overexpression reduces hepatitis C viral abundance in culture. J Infect Dis. 2011;203:1753-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 22. | Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 656] [Cited by in RCA: 661] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 23. | Sarma NJ, Tiriveedhi V, Subramanian V, Shenoy S, Crippin JS, Chapman WC, Mohanakumar T. Hepatitis C virus mediated changes in miRNA-449a modulates inflammatory biomarker YKL40 through components of the NOTCH signaling pathway. PLoS One. 2012;7:e50826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 546] [Reference Citation Analysis (0)] |
| 24. | Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, Christensen RN, Schmidt WN, McCaffrey AP. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727-1736. [PubMed] |
| 25. | Negrini M, Gramantieri L, Sabbioni S, Croce CM. microRNA involvement in hepatocellular carcinoma. Anticancer Agents Med Chem. 2011;11:500-521. [PubMed] |
| 26. | Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537-2545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 876] [Cited by in RCA: 896] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 27. | Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, Farci P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133:816-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 28. | Chen JS, Wang Q, Fu XH, Huang XH, Chen XL, Cao LQ, Chen LZ, Tan HX, Li W, Bi J. Involvement of PI3K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: Association with MMP-9. Hepatol Res. 2009;39:177-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 277] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 29. | Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 588] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 30. | Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J Biol Chem. 2008;283:18158-18166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 172] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 31. | Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L, Gramantieri L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184-5193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 336] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 32. | Fukuhara T, Matsuura Y. Role of miR-122 and lipid metabolism in HCV infection. J Gastroenterol. 2013;48:169-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 33. | Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 34. | Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 35. | Cheng JC, Yeh YJ, Tseng CP, Hsu SD, Chang YL, Sakamoto N, Huang HD. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci. 2012;69:2621-2633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 36. | Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919-922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 722] [Cited by in RCA: 703] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 37. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 1987] [Article Influence: 99.4] [Reference Citation Analysis (0)] |
| 38. | Huang MH, Jiang JD, Peng ZG. Recent advances in the anti-HCV mechanisms of interferon. Acta Pharmaceutica Sinica B. 2014;4:241-247. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 445] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 40. | Wilkins C, Woodward J, Lau DT, Barnes A, Joyce M, McFarlane N, McKeating JA, Tyrrell DL, Gale M. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology. 2013;57:461-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 41. | Bhanja Chowdhury J, Shrivastava S, Steele R, Di Bisceglie AM, Ray R, Ray RB. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J Virol. 2012;86:10221-10225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 42. | Mukherjee A, Di Bisceglie AM, Ray RB. Hepatitis C virus-mediated enhancement of microRNA miR-373 impairs the JAK/STAT signaling pathway. J Virol. 2015;89:3356-3365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 43. | Yoshimura A, Suzuki M, Sakaguchi R, Hanada T, Yasukawa H. SOCS, Inflammation, and Autoimmunity. Front Immunol. 2012;3:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 196] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 44. | Liu Z, Tian Y, Machida K, Lai MM, Luo G, Foung SK, Ou JH. Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J Biol Chem. 2012;287:41922-41930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 45. | Shwetha S, Gouthamchandra K, Chandra M, Ravishankar B, Khaja MN, Das S. Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep. 2013;3:1555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 46. | Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci USA. 2007;104:5848-5853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 429] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 47. | Hishiki T, Shimizu Y, Tobita R, Sugiyama K, Ogawa K, Funami K, Ohsaki Y, Fujimoto T, Takaku H, Wakita T. Infectivity of hepatitis C virus is influenced by association with apolipoprotein E isoforms. J Virol. 2010;84:12048-12057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 109] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 48. | Mancone C, Steindler C, Santangelo L, Simonte G, Vlassi C, Longo MA, D’Offizi G, Di Giacomo C, Pucillo LP, Amicone L. Hepatitis C virus production requires apolipoprotein A-I and affects its association with nascent low-density lipoproteins. Gut. 2011;60:378-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 49. | Shirasaki T, Honda M, Shimakami T, Horii R, Yamashita T, Sakai Y, Sakai A, Okada H, Watanabe R, Murakami S. MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol. 2013;87:5270-5286. [PubMed] |
| 50. | Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3005] [Cited by in RCA: 3070] [Article Influence: 153.5] [Reference Citation Analysis (0)] |
| 51. | Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87-98. [PubMed] [DOI] [Full Text] |
| 52. | Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 278] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 53. | Li C, Hu J, Hao J, Zhao B, Wu B, Sun L, Peng S, Gao GF, Meng S. Competitive virus and host RNAs: the interplay of a hidden virus and host interaction. Protein Cell. 2014;5:348-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 54. | Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Léopold V, Tay Y, Provero P, Zecchina R, Pandolfi PP. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci USA. 2013;110:7154-7159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 273] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 55. | Li C, Wang Y, Wang S, Wu B, Hao J, Fan H, Ju Y, Ding Y, Chen L, Chu X. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol. 2013;87:2193-2205. [PubMed] |
| 56. | Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, Ahn K. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe. 2013;13:678-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 57. | Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012;8:e1002510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 58. | Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563-1566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 421] [Cited by in RCA: 391] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 59. | Luna JM, Scheel TK, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 276] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 60. | Nasheri N, Singaravelu R, Goodmurphy M, Lyn RK, Pezacki JP. Competing roles of microRNA-122 recognition elements in hepatitis C virus RNA. Virology. 2011;410:336-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology. 2007;133:1166-1174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 141] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 62. | Hatzoglou M, Fernandez J, Yaman I, Closs E. Regulation of cationic amino acid transport: the story of the CAT-1 transporter. Annu Rev Nutr. 2004;24:377-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 151] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 63. | Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol. 2006;71:369-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 64. | Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM, Tavolari S. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761-5767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 321] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 65. | Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 227] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 66. | Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 206] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 67. | Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015-32027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 413] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 68. | Hou W, Tian Q, Zheng J, Bonkovsky HL. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology. 2010;51:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 69. | Shrivastava S, Petrone J, Steele R, Lauer GM, Di Bisceglie AM, Ray RB. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013;58:863-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 70. | Abdalla MA, Haj-Ahmad Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:19-31. [PubMed] |
| 71. | Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. [PubMed] |
| 72. | Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, Sun S. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 257] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 73. | Kałużna EM. MicroRNA-155 and microRNA-196b: promising biomarkers in hepatitis C virus infection? Rev Med Virol. 2014;24:169-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 74. | Riad SE, El-Ekiaby N, Mekky RY, Ahmed R, El Din MA, El-Sayed M, Abouelkhair MM, Salah A, Zekri AR, Esmat G. Expression signature of microRNA-155 in hepatitis C virus genotype 4 infection. Biomed Rep. 2015;3:93-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 75. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1665] [Cited by in RCA: 1702] [Article Influence: 141.8] [Reference Citation Analysis (0)] |
| 76. | Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 761] [Cited by in RCA: 841] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 77. | Pietschmann T. Regulation of hepatitis C virus replication by microRNAs. J Hepatol. 2009;50:441-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 78. | Almeida MI, Reis RM, Calin GA. Decoy activity through microRNAs: the therapeutic implications. Expert Opin Biol Ther. 2012;12:1153-1159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 79. | Seo H, Kim W, Lee J, Youn B. Network-based approaches for anticancer therapy (Review). Int J Oncol. 2013;43:1737-1744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 80. | Giza DE, Vasilescu C, Calin GA. MicroRNAs and ceRNAs: therapeutic implications of RNA networks. Expert Opin Biol Ther. 2014;14:1285-1293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |