Published online Sep 14, 2016. doi: 10.3748/wjg.v22.i34.7767
Peer-review started: May 17, 2016
First decision: June 20, 2016
Revised: July 5, 2016
Accepted: July 31, 2016
Article in press: August 1, 2016
Published online: September 14, 2016
Processing time: 115 Days and 14.7 Hours
To investigate the association of tumor necrosis factor alpha (TNFα) -G308A polymorphism with different liver pathological changes in treatment-naïve Egyptian patients infected with hepatitis C virus (HCV) genotype 4.
This study included 180 subjects, composed of 120 treatment-naïve chronic HCV patients with different fibrosis grades (F0-F4) and 60 healthy controls. The TNFα -G308A region was amplified by PCR and the different genotypes were detected by restriction fragment length polymorphism analysis. The TNFα protein was detected by enzyme-linked immunosorbent assay. The influence of different TNFα -G308A genotypes on TNFα expression and liver disease progression were statistically analyzed. The OR and 95%CI were calculated to assess the relative risk confidence.
Current data showed that the TNFα -G308A SNP frequency was significantly different between controls and HCV infected patients (P = 0.001). Both the AA genotype and A allele were significantly higher in late fibrosis patients (F2-F4, n = 60) than in early fibrosis patients (F0-F1, n = 60) (P = 0.05, 0.04 respectively). Moreover, the GA or AA genotypes increased the TNFα serum level greater than the GG genotype (P = 0.002). The results showed a clear association between severe liver pathological conditions (inflammation, steatosis and fibrosis) and (GA + AA) genotypes (P = 0.035, 0.03, 0.04 respectively). The stepwise logistic regression analysis showed that the TNFα genotypes (GA + AA) were significantly associated with liver inflammation (OR = 3.776, 95%CI: 1.399-10.194, P = 0.009), severe steatosis (OR = 4.49, 95%CI: 1.441-14.0, P = 0.010) and fibrosis progression (OR = 2.84, 95%CI: 1.080-7.472, P = 0.034). Also, the A allele was an independent risk factor for liver inflammation (P = 0.003), steatosis (P = 0.003) and fibrosis (P = 0.014).
TNFα SNP at nucleotide -308 represents an important genetic marker that can be used for the prognosis of different liver pathological changes in HCV infected patients
Core tip: Tumor necrosis factor alpha (TNFα) is a proinflammatory and antiviral cytokine that plays a major role in liver injury and initiation of the fibrosis process. We investigated the association of TNFα -G308A polymorphism with liver pathological changes in treatment-naïve Egyptian patients infected with hepatitis C virus (HCV) genotype 4. The results showed that the TNFα A allele produced high circulating TNFα in the body, which induces inflammatory response. The TNFα A allele was an independent risk factor for liver inflammation, steatosis and fibrosis. TNFα -G308A represents an important genetic marker that can be used for the prognosis of different liver pathological changes in HCV infected patients.
- Citation: Bader El Din NG, Farouk S, El-Shenawy R, Ibrahim MK, Dawood RM, Elhady MM, Salem AM, Zayed N, Khairy A, El Awady MK. Tumor necrosis factor-α -G308A polymorphism is associated with liver pathological changes in hepatitis C virus patients. World J Gastroenterol 2016; 22(34): 7767-7777
- URL: https://www.wjgnet.com/1007-9327/full/v22/i34/7767.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i34.7767
Hepatic fibrosis is the liver wound healing process for many injuries, such as excess vitamin A, inherited metabolic disorders, alcohol, drugs, cholestatic disorders, viral hepatitis and autoimmune disorders[1]. In Egypt, hepatitis C virus (HCV) is a major public health problem, with a 15% prevalence rate and the predominant genotype being genotype 4[2]. Chronic HCV patients are usually at risk of developing cirrhosis and hepatocellular carcinoma. Complications of HCV infection in Egypt are responsible for 67% of the liver disease death rate[3].
Exacerbated immune response against HCV, along with the consequent excess production of chronic inflammatory mediators, is the underlying cause of liver injury and development of fibrosis. Chronically infected patients may have a slow, intermediate or rapid fibrosis progression rate. Multiple factors have been reported to affect the liver disease progression rate[4]. Single nucleotide polymorphisms (SNPs) in the non-coding or coding regions of cytokine genes have been documented to affect their expression and secretion, which can cause chronic inflammation and disease progression[5,6].
Altered expression of cytokines, such as tumor necrosis factor alpha (TNFα), transforming growth factor beta, interferon gamma (IFNγ), platelet-derived growth factor, interleukin (IL)-1, -10 and -28B, and the IL-1 receptor antagonist, were found to be associated with liver fibrosis[1,7,8]. The expression of these cytokines plays important roles in the regulation of liver cell functions and the fibrotic process in vivo. TNFα is a proinflammatory and antiviral cytokine secreted by macrophage and cytotoxic T lymphocytes in the liver, where it regulates the immune response, cell growth and apoptosis. TNFα is considered the immune-mediated hepatic cytokine that plays a major role in liver injury and the initiation of the fibrosis process. It stimulates hepatic stellate cells’ proliferation, chemoattraction, extracellular matrix components’ secretion and connective tissue growth factors’ expression. Therefore, TNFα expression is considered a key molecular link between liver inflammation, steatosis and fibrosis[9,10].
The TNFα gene is located on chromosome 6p21.3[11]. Six polymorphisms at nucleotides -1031, -863, -857, -376, -308 and -238 in the TNF promoter were thought to influence the TNFα expression[12,13]. Several studies showed that the TNFα -G308A polymorphism affects the TNFα transcription level. In the normal population, the wild genotype is GG and AA is the mutant genotype. The A allele has a significant functional effect on TNF transcription and is associated with higher inducible TNFα transcription levels than the G allele[14].
Many inflammatory disorders and infectious diseases are associated with TNFα -G308A polymorphism, such as systemic lupus erythematosus, inflammatory bowel disease, insulin-dependent diabetes mellitus, malaria and leishmaniasis[13,15-17]. In chronic HCV patients, elevated levels of TNFα were detected in liver and serum[8]. TNFα increases fat deposition in the liver by affecting the hepatic lipogenesis[18]. Furthermore, Hooper et al[19] showed that increased levels of TNFα and reduced anti-inflammatory cytokines correlate with the severity of liver injury in non-alcoholic steatohepatitis (NASH) patients.
The role of TNFα -G308A polymorphism in HCV pathogenesis has been examined in different studies, with divergent results[6,20-22]. These variations may be due to racial background of the studied populations, who have different cytokine gene polymorphisms[23]. Also, these studies focused on the role of TNFα -G308A in HCV susceptibility and treatment response rate but none of these studies examined the role of TNFα -G308A polymorphism in HCV liver steatosis and fibrosis progression rate. The current study was designed to investigate the association of TNFα -G308A polymorphism with liver inflammation, steatosis and different grades of fibrosis in treatment-naïve Egyptian patients infected with HCV genotype 4.
The study was approved by the Ethics Committee of the National Research Centre, Giza, Egypt according to the Helsinki Declaration of 1975 revised in 2008. A total of 180 subjects, including 120 treatment-naïve HCV infected patients with different fibrosis grades grade who did not receive any treatment and 60 healthy controls, were enrolled in this study. Informed consents were obtained from all study subjects before enrollment in the study and collection of blood samples. The healthy controls (mean age, 42.6 ± 9.4 years) had normal liver function tests, no history of liver injury and no viral hepatitis, particularly HCV (negative HCV Ab and negative viremia). The control subjects also did not suffer from any other metabolic dysfunctions or bacterial, viral or malignant diseases. The HCV patients (mean age, 43.1 ± 10.1 years) were admitted to the Endemic Medicine Department of Kasr El Ainy Hospitals at Cairo University. Patients were excluded if they had decompensated cirrhosis, metabolic liver disease, hepatitis B virus, schistosomiases, alcohol, drug-induced hepatitis or any significant coexisting medical conditions. Laboratory tests, including measurements of alanine aminotransferase (ALT), aspartate transaminase (AST), bilirubin, alkaline phosphatase (ALP), creatinine, complete blood count, body mass index (BMI) and HCV viral load were performed, as well as liver biopsy, for all patients. The extents of liver inflammation, steatosis and different grades of fibrosis were determined according to the METAVIR scoring system[24].
Total RNA was extracted from serum using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) to detect the presence of HCV RNA. HCV RNA was detected quantitatively using the Artus HCV RT-PCR Kit (Qiagen) according to the manufacturer’s instructions. Genotype of HCV RNA was detected using the INNOLiPA HCV genotyping Kit (Innogenetics, Ghent, Belgium).
Total DNA was extracted from whole blood of all subjects using the QIAamp Blood Kit (Qiagen) according to the manufacturer’s instructions. The TNFα -G308A genotyping was performed using PCR-restriction fragment length polymorphism (RFLP) analysis. The TNFα -308 G/A promoter polymorphism was amplified by PCR using 5’-AGGCAATAGGTTTTGAGGGCCAT-3’ as forward primer and 5’-CCTCCCTGCTCCGATTCCG-3’ as reverse primer, as previously described by Shmarina et al[10] and Ho et al[25]. These primers cover the TNFα -308 polymorphism region and produced a 107 bp PCR fragment. The PCR amplification was carried out in 25 μL, containing 5-10 μg DNA, 4 mmol/L MgCl2 (Promega, Madison, WI, United States), 1 μmol/L of each primer, 200 μmol/L dNTPs (Promega), 1 × PCR buffer (Promega), and 1 U Go Taq DNA polymerase (Promega). The PCR thermal cycling was carried out in an MJ Research cycler instrument. The thermal cycling conditions consisted of initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 1 min, with a final extension at 72 °C for 10 min. The PCR fragments were detected by electrophoresis in a 3% agarose gel stained with ethidium bromide.
The G/A allelic polymorphism at TNFα -308 was detected by RFLP analysis. The PCR (107 bp) fragments from all subjects were digested with NcoI restriction enzyme. The digestion reaction was carried out in a total volume of 20 μL, containing 1 × restriction buffer, 8 μL PCR product, and 5 U NcoI (Promega) according to the manufacturer’s recommendations. The NcoI restriction digestion was performed at 37 °C for 4 h. Afterward, 10 μL of the digested products were run on a 4% agarose gel stained with ethidium bromide.
To confirm the results of TNFα -308 PCR-RFLP analysis, some TNFα -308 PCR products were sequenced using the Sanger dideoxynucleotide chain termination method. The TNFα -308 PCR products were purified using the QIAquick PCR Purification Kit (Qiagen). Then, the PCR products were sequenced with the TNFα reverse primer using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems Inc, Carlsbad, CA, United States). After the cycle sequencing reaction, the products were purified using the BigDye XTerminator Purification Kit (Applied Biosystems Inc) and analyzed on the ABI 3500 Genetic Analyzer.
Serum samples were collected and stored at -80 °C. TNFα level in serum was determined in 180 subjects (120 HCV patients and 60 healthy individuals) by TNF enzyme-linked immunosorbent assay (ELISA) kit (Biosource Europe S.A, Nivelles, Belgium) according to the manufacturer’s instructions. The concentration of TNFα in serum was determined using a double antibody sandwich ELISA. All samples were analyzed and recombinant standards were included on every plate.
The statistical methods of this study were reviewed by a specialized IT development and biomedical statistician. Data were collected, prepared and analyzed using the SPSS software, version 15. Data were expressed as number and percentage for qualitative parameters and as mean ± SD for quantitative parameters. The frequency of genotypes and alleles in patients and controls were analyzed by χ2 test. Comparisons of the clinical parameters of different groups and genotypes were performed by t test. Then, stepwise logistic regression analysis was used to identify predictors associated with degree of fibrosis in chronic HCV patients. The OR and 95%CI were calculated to assess the relative risk confidence. P value ≤ 0.05 was considered significant, while was considered P < 0.01 highly significant.
The biochemical, virological and histopathological parameters of 120 HCV-infected patients with different fibrosis grade (F0: n = 30, F1: n = 30, F2-F3: n = 30, F4: n = 30) are summarized in Table 1. There were no significant differences within the different fibrosis groups for age, sex, BMI and HCV RNA viral load. Patients with liver fibrosis grades F2-3 or F4 had statistically significant higher levels of AST, ALT, ALP and total bilirubin and significantly lower platelet count and albumin level, as compared to patients with liver fibrosis grade F0 or F1.
| HCV patients with different fibrosis grade (n = 120) | ||||
| F0 (n = 30) | F1 (n = 30) | F2-3 (n = 30) | F4 (n = 30) | |
| Age (yr) | 38.4 ± 7.1 | 41.5 ± 8.7 | 43.5 ± 8.4 | 48.3 ± 9.3 |
| Sex (female/male) | 13/17 | 12/18 | 10/20 | 9/21 |
| BMI (kg/m2) | 25.5 ± 2.3 | 27.6 ± 3.4 | 28.7 ± 2.7 | 29.5 ± 2.7 |
| ALT (U/L) | 23.4 ±7.1 | 39.8 ± 23.7 | 51.1 ± 20.7 | 60.1 ± 20.3 |
| AST (U/L) | 22.4 ± 6.5 | 34.9 ± 20.9 | 48.9 ± 25.9 | 58.7 ± 18.6 |
| ALP (U/L) | 113.6 ± 32.8 | 122.5 ± 37.1 | 158.7 ± 47.7 | 153.8 ± 53.5 |
| Total bilirubin (mg/dL) | 0.53 ± 0.32 | 0.61 ± 0.23 | 0.96 ± 0.37 | 1.60 ± 0.65 |
| Albumin (g/dL) | 4.10 ± 0.33 | 4.00 ± 0.38 | 3.80 ± 0.45 | 3.50 ± 0.39 |
| Platelets (× 109/L) | 300.5 ± 56.2 | 250.2 ± 48.4 | 239.6 ± 55.6 | 159.4 ± 69.5 |
| HCV RNA (× 103 IU/mL) | 684.3 ± 124.8 | 860.1 ± 1042 | 1241.5 ± 1286.7 | 1501.4 ± 1661.0 |
| Fibroscan | 4.70 ± 0.75 | 6.30 ± 0.36 | 9.90 ± 1.78 | 23.27 ± 9.30 |
| Activity | ||||
| A0-A1 | 20 (66.7) | 13 (43.3) | 10 (33.3) | 1 (3.3) |
| A2-A3 | 10 (33.3) | 17 (56.7) | 20 (66.7) | 29 (96.7) |
| Steatosis | ||||
| ≤ 33% | 28 (93.3) | 26 (86.7) | 16 (53.3) | 4 (13.3) |
| > 33% | 2 (6.7) | 4 (13.3) | 14 (46.7) | 26 (86.7) |
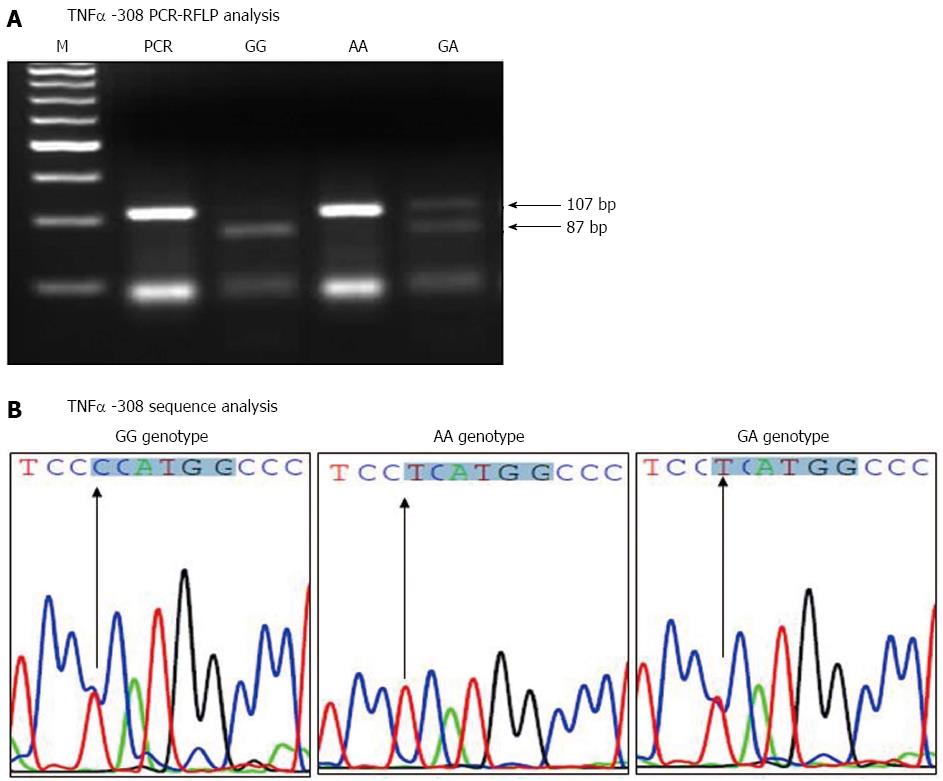
The amplified TNFα -308 PCR products were digested with NcoI restriction enzyme, and run on 4% agarose gel, as shown in Figure 1A. The homozygote AA genotype at TNFα -308 showed the original 107 bp fragment intact (lacking the NcoI site), while the homozygote GG genotype showed two fragments of 87 and 20 bp. The heterozygote GA genotype showed three fragments of 87, 20 and 107 bp. Moreover, the sequence results confirmed the integrity of the NcoI restriction site’s surrounding area and verified the results of the TNFα -308 PCR-RFLP analysis. The sequence results for the TNFα -308 different genotypes are shown in Figure 1B.
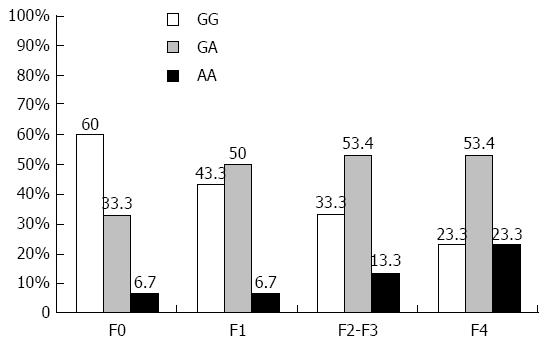
Distribution of TNFα -G308A genotypes in HCV infected patients and controls are shown in Table 2. The results showed that the TNFα -G308A SNP frequency was significantly different between controls and HCV infected patients (P = 0.001). The TNFα -G308A genotypes in the controls were 66.6% GG, 30% GA and 3.3% AA. The GG genotype in controls (66.6%) is higher than in chronic HCV patients (40%), while the chronic HCV patients had higher GA and AA genotypes (47.5% and 12.5%) compared to controls (30% and 3.3%). Moreover, the G allele is more frequent than the A allele in both controls (81.7% vs 18.3%) and HCV-infected patients (63.8% vs 36.3%), with statistically significant difference (P = 0.002). The distribution of TNFα genotypes in HCV patients with different fibrosis grade is shown in Figure 2. The TNFα GG genotype was 60% in F0, 43.3% in F1, 33.3% in F2-F3, and 23.3% in F4 patients, while the (GA + AA) genotypes were 40% in F0, 56.7% in F1, 66.6% in F2-F3, and 76.6% in F4 patients.
| Polymorphism of TNFα -308 | Controls (n = 60) | HCV Patients (n = 120) | F0 patients (n = 30) | F1 patients (n = 30) | F2-3 patients (n = 30) | F4 patients (n = 30) |
| GG genotype | 40 (66.6) | 48 (40.0) | 18 (60.0) | 13 (43.3) | 10 (33.3) | 7 (23.3) |
| GA genotype | 18 (30.0) | 57 (47.5) | 10 (33.3) | 15 (50.0) | 16 (53.4) | 16 (53.4) |
| AA genotype | 2 (3.3) | 15 (12.5) | 2 (6.7) | 2 (6.7) | 4 (13.3) | 7 (23.3) |
| GA + AA | 20 (33.3) | 72 (60.0) | 12 (40.0) | 17 (56.7) | 20 (66.6) | 23 (76.6) |
| G allele | 98 (81.7) | 153 (63.8) | 46 (76.7) | 41 (68.3) | 36 (60.0) | 30 (50.0) |
| A allele | 22 (18.3) | 87 (36.3) | 23 (23.3) | 19 (31.7) | 24 (40.0) | 30 (50.0) |
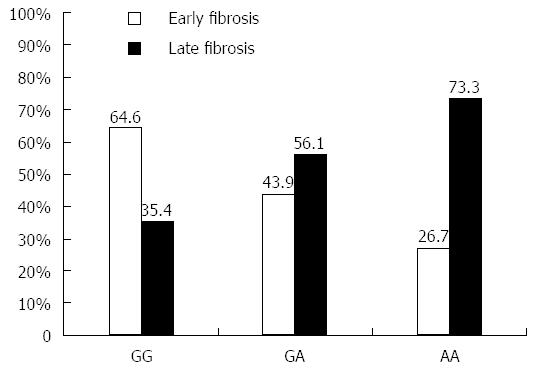
The 120 chronic HCV patients with different fibrosis grade were classified into 60 patients with early fibrosis grade (F0-F1) and 60 patients with late fibrosis grade (F2-F4). Analysis of the frequency of each TNFα genotype showed that early fibrosis patients have 26.7% of AA, 43.9% of AG and 64.6% of GG genotype, which indicates an increasing trend of having the G allele. The AA genotype was detected in 73.3% of late fibrosis patients vs 26.7% of early fibrosis patients (P = 0.05), as shown in Figure 3. In general, the late fibrosis patients had a statistically significant higher rate of A allele than the early fibrosis patients (62.1% vs 37.9% respectively, P = 0.04).
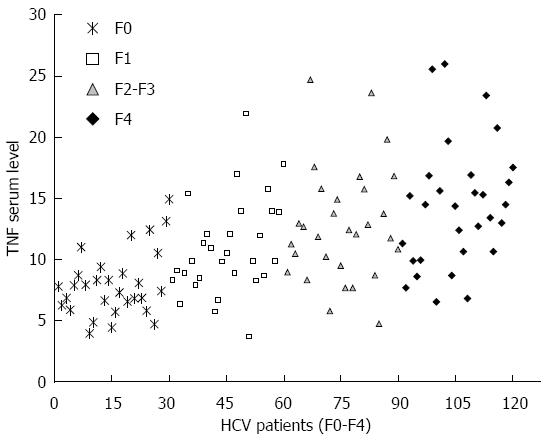
The TNFα serum levels were determined in healthy controls and HCV patients with different fibrosis grade. Compared with that in healthy controls, serum TNFα level was gradually elevated in HCV infected patients with different fibrosis grade, as shown in Figure 4. The results showed that the TNFα serum level was 4.01 ± 1.1 pg/mL in controls, 7.9 ± 2.6 pg/mL in F0 patients, 11.3 ± 4.8 pg/mL in F1 patients, 12.9 ± 4.7 pg/mL in F2-F3 patients, and 14.1 ± 4.9 pg/mL in F4 patients. Patients with late fibrosis (F2-F4) had higher TNF levels (13.4 ± 4.8 pg/mL) than patients with early fibrosis (F0-F1) (8.7 ± 2.9 pg/mL). The results showed that the TNFα serum level was positively correlated to HCV liver fibrosis progression (P < 0.001). Moreover, the TNFα serum concentration was significantly higher in patients with A containing genotypes (GA or AA) than in those with GG genotype (10.9 ± 4.9 pg/mL vs 8.3 ± 2.6 pg/mL, P = 0.01) in early fibrosis (F0-F1) patients and (14.4 ± 4.9 pg/mL vs 10.4 ± 2.1 pg/mL, P = 0.002) in late fibrosis (F2-F4) HCV patients, as shown in Table 3.
| TNFαserum levels | P value | ||||
| GG | GA | AA | GA + AA | GG vs AA | |
| All fibrosis patients (F0-F4, n = 120) | 9.1 ± 2.6 | 12.1 ± 4.9 | 16.1 ± 4.9 | 13.1 ± 5.1 | 0.0001 |
| early fibrosis patients (F0-F1, n = 60) | 8.3 ± 2.6 | 10.1 ± 4.5 | 16.2 ± 4.02 | 10.9 ± 4.9 | 0.010 |
| late fibrosis patients (F2-F4, n = 60) | 10.4 ± 2.1 | 13.8 ± 4.5 | 16.1 ± 5.6 | 14.4 ± 4.9 | 0.002 |
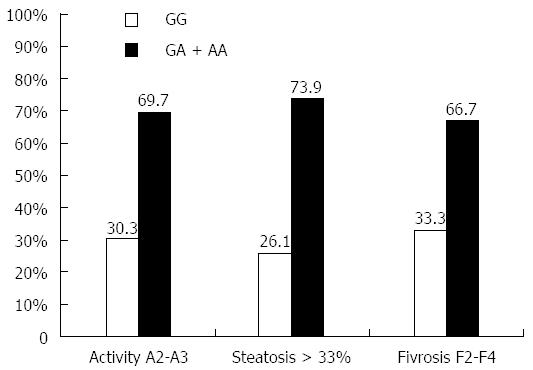
The influence of different TNFα genotypes on hepatic parameters and liver pathology (liver inflammation, steatosis, and fibrosis) is shown in Table 4. The results demonstrated that TNFα AA genotype patients have significantly higher levels of AST, ALT, ALP and total bilirubin than GG patients (P = 0.0003, 0.0001, 0.002 and 0.0002 respectively). Patients with AA genotype have significantly lower level of albumin and platelets count (P = 0.01 and 0.036 respectively). Moreover, the effect of serum TNFα level expressed by genotypes (GA and AA) on liver inflammation, steatosis and fibrosis are shown in Figure 5. The results showed that genotypes GA or AA which express higher TNF serum level were found in 69.7% of high liver inflammation (A2-A3) patients, in 73.9% of severe steatotic patients, and in 66.7% of late fibrotic (F2-F4) patients.
| TNF genotypes (n = 120) | P value | ||||
| GG (n = 48) | AG (n = 57) | AA (n = 15) | GG vs AA | GG vs GA + AA | |
| AST (U/L) | 31.9 ± 20.6 | 44.7 ± 21.6 | 57.7 ± 28.7 | 0.0003 | 0.0003 |
| ALT (U/L) | 34.5 ± 20.9 | 46.5 ± 20.5 | 62.0 ± 28.2 | 0.0001 | 0.0004 |
| ALP (U/L) | 113.7 ± 36.8 | 138.3 ± 41.5 | 190.1 ± 46.5 | 0.0020 | 0.3500 |
| Albumin (g/dL) | 3.9 ± 0.46 | 3.9 ± 0.39 | 3.6 ± 0.59 | 0.0100 | 0.2000 |
| Total bilirubin (mg/dL) | 0.73 ± 0.41 | 0.89 ± 0.51 | 1.3 ± 0.68 | 0.0002 | 0.0100 |
| Platelets (× 109/L) | 254.9 ± 66.2 | 224.5 ± 72.3 | 205.8 ± 96.6 | 0.0360 | 0.0220 |
| HCV RNA (× 103 IU/mL) | 862.6 ± 1295.6 | 1083.9 ± 1187.2 | 1675.5 ± 1477.6 | 0.0450 | 0.2200 |
| Activity | |||||
| A0-A1 (n = 44) | 25 (56.8) | 18 (40.9) | 1 (2.3) | 0.0290 | 0.0350 |
| A2-A3 (n = 76) | 23 (30.3) | 39 (51.3) | 14 (18.4) | ||
| Steatosis | |||||
| ≤ 33% (n = 74) | 36 (48.7) | 34 (45.9) | 4 (5.4) | 0.0280 | 0.0300 |
| > 33% (n = 46) | 12 (26.1) | 23 (50.0) | 11 (23.9) | ||
| Fibrosis | |||||
| F0-F1 (n = 60) | 28 (46.7) | 30 (50.0) | 2 (3.3) | 0.0310 | 0.0400 |
| F2-F4 (n = 60) | 20 (33.3) | 27 (45.0) | 13 (21.6) | ||
Different TNF genotypes were investigated to determine their significance in liver disease progression using stepwise logistic regression analysis. The outcome of liver progression was significantly influenced by different TNF genotypes and alleles, as shown in Table 5. Patients with TNFα GA and AA genotypes have increased risk of liver inflammation (A2-A3) (OR = 3.776, 95%CI: 1.399-10.194, P = 0.009), severe steatosis (> 33%) (OR = 4.49, 95%CI: 1.441-14.0, P = 0.010) and fibrosis progression (F2-F4) (OR = 2.84, 95%CI: 1.080-7.472, P = 0.034) than those with GG genotype. In general, patients with A allele have more risk of liver inflammation, steatosis and fibrosis than those carrying the G allele (P = 0.003, 0.003 and 0.014 respectively).
| TNF genotype | Regression coefficient | SE | OR (95%CI) | P value |
| Activity | ||||
| GG vs GA + AA | 1.329 | 0.507 | 3.776 (1.399-10.194) | 0.009 |
| G vs A allele | 1.171 | 0.400 | 3.226 (1.474-7.060) | 0.003 |
| Steatosis | ||||
| GG vs GA + AA | 1.502 | 0.580 | 4.491 (1.441-14.000) | 0.010 |
| G vs A allele | 1.099 | 0.373 | 3.000 (1.445-6.228) | 0.003 |
| Fibrosis | ||||
| GG vs GA + AA | 1.044 | 0.493 | 2.841 (1.080-7.472) | 0.034 |
| G vs A allele | 0.895 | 0.366 | 2.446 (1.195-5.008) | 0.014 |
Cytokines represent a large family that includes the IFNs, ILs, chemokines and TNF proteins. Cytokines play important roles in the activation and regulation of human immune responses and their production are variable due to SNPs[26]. Several studies have reported the impact of different cytokines’ polymorphisms on the pathogenesis and outcomes of many diseases. Competent cytokine responses to HCV infection are very important to control virus replication, disease progression and treatment response.
TNFα is a diverse multifunctional, proinflammatory and immuno-mediator cytokine. Several studies have shown that the A allele at the TNFα promoter region -308 polymorphism affects the transcriptional level of TNFα gene[27,28]. There are few data regarding the role of TNFα polymorphism in disease progression and response to antiviral therapy in patients infected with HCV genotype 4. In this study, we examined the association between TNFα -G308A polymorphism and pathological parameters of liver such as inflammation, steatosis and fibrosis in 120 treatment-naïve HCV genotype 4 Egyptian patients.
Current findings showed that TNFα GG genotype is significantly higher in controls than in HCV infected patients (66.6% vs 40%, P = 0.001), while the A allele tends to be more frequent in HCV-infected patients than in controls (36.3% vs 18.3%, P = 0.002). The (GA + AA) genotypes exhibited gradual increase with progressive fibrosis, with 40% in (F0), 56.7% in (F1), 66.6% in (F2-F3), and 76.6% in (F4). In general, 62.1% of the A allele were found in late fibrosis patients (F2-F4) compared to 37.9% in early fibrosis patients (F0-F1) (P = 0.015). Similar results were found by Dogra et al[29], who reported that the frequency of TNFα -308 genotypes was significantly different between healthy controls and HCV patients. The association between TNFα -308A allele and HCV persistence and failure of response to IFN treatment was reported by Radwan et al[30] and Dai et al[31]. Nevertheless, other studies failed to find association between the TNFα -G308A polymorphism and chronicity of HCV infection[6,20,32].
On the other hand, there were significantly high serum TNFα mean values in late fibrosis (F2-F4) patients, with higher levels in individuals with GA or AA genotypes than those with GG genotype. Similar findings were reported in patients with Crohn’s disease, where the TNFα -308A allele was associated with increased TNFα production and inflammatory activity[33]. Also, Roth-Isigkeit et al[34] reported elevated TNFα serum levels in GA heterozygote individuals undergoing cardiac surgery. Further in vitro support of these findings was reported by Abraham and Kroeger[14], who found that A allele induced mRNA expression and TNFα production up to 5-fold more than did the G allele.
Several studies have shown that TNFα -308A polymorphism alters TNFα expression levels and changes the immune response. It was reported that TNFα -308A polymorphism can worsen the clinical outcome of many inflammatory and infectious diseases and can increase the risk of some autoimmune diseases. Abraham and Kroeger[14] defined TNFα as a genetic susceptibility factor for some autoimmune, inflammatory and infectious diseases. High TNFα expression level was also reported to inhibit insulin signaling and decrease adiponectin levels, leading to development of insulin resistance and liver steatosis. Furthermore, it was demonstrated that high circulating TNFα is a potent risk factor for steatosis[35].
In the present study, we examined the link between high TNFα serum levels and liver inflammation, steatosis and fibrosis. Our results showed that TNFα -308AA genotype patients have significantly higher levels of AST, ALT, ALP and total bilirubin than GG patients. Also, HCV patients with severe liver pathological conditions (inflammation, steatosis and fibrosis) have high frequencies of TNFα (GA or AA) genotypes. These results showed a clear association between severe liver pathological conditions (inflammation, steatosis and fibrosis) and (GA + AA) genotypes (P = 0.035, 0.03 and 0.04 respectively). These results suggest that A containing genotypes (GA + AA) express higher TNFα levels than GG genotype and consequently induce inflammatory response that leads to liver inflammation, injury and severe fibrosis[36].
Our results confirmed the previous studies in which a significant elevation in TNFα serum level was detected in cirrhotic liver patients and was correlated with progression to hepatocellular carcinoma[22,37,38]. It was reported that TNFα -308AA genotype is associated with the HCV pathogenesis and viral persistence[21,31]. Aller et al[35] showed that patients with the TNFα mutant genotype (GA or AA) have moderate to severe portal inflammation and fibrosis, contrasting those patients with wild genotype (GG). Also, high TNFα plays a role in fatty liver disease pathogenesis[35] and leads to severe liver injury in NASH patients[19]. However, other studies were unable to find any association between TNFα genetic polymorphisms and severity of liver disease or response to antiviral therapy[6,26,39].
The stepwise logistic regression analysis showed that the TNFα genotypes (GA + AA) were significantly associated with liver inflammation (OR = 3.776, 95%CI: 1.399-10.194, P = 0.009), severe steatosis (OR = 4.49, 95%CI: 1.441-14.0, P = 0.010) and fibrosis progression (OR = 2.84, 95%CI: 1.080-7.472, P = 0.034). The A allele was an independent risk factor for liver inflammation (P = 0.003), steatosis (P = 0.003) and fibrosis (P = 0.014). Therefore, the current results confirm that HCV infected patients carrying one or two A alleles at TNFα -308 have a risk factor for development of severe liver pathological grades.
Several experimental studies showed that the inhibition of TNFα signaling by anti-TNFα antibodies or compounds could reduce inflammation, liver injury and improve fibrosis[40,41]. However, complete neutralization of TNFα in alcoholic hepatitis patients was associated with serious infectious complications[42]. Therefore, it was recommended to use pentoxifylline (PTX), which partially attenuates TNFα level and has lower infectious complication rates. It was demonstrated that PTX therapy effectively reduces the liver biochemical parameters and improves the histological injury in NASH patients[43].
Although liver biopsy is the conventional way to determine the grade of liver fibrosis, it is an invasive method and there is too much sampling. Therefore, there is an increasing need for biomarkers that are specific, sensitive and respond quickly to changes in fibrosis and activity grades. Based on the results of the current study and our previous work on IL28B[44,45], OAS1 and MxA[7,46,47], we have presented some potential genetic markers that can be useful in the determination of liver disease progression. In conclusion, the TNFα SNP at nucleotide -308 represents an important genetic marker for developing strategies of prognosis for liver inflammation, steatosis and fibrosis in HCV infected patients.
Hepatitis C virus (HCV) infection is highly endemic in Egypt and the mortality rate of HCV-associated liver diseases reaches 67%. Both viral and host factors play important roles in controlling HCV liver disease progression and response to treatment. Host genetic factors and immunological response to HCV infection can affect the pathogenesis of liver diseases. The single nucleotide polymorphisms (SNPs) are responsible for inter-individual differences in cytokines’ and cellular antiviral proteins’ production and secretion. Tumor necrosis factor alpha (TNFα) is a proinflammatory and antiviral cytokine that plays a major role in liver injury and initiation of the fibrosis process. The TNFα expression is considered a key molecular link between liver inflammation, steatosis, and fibrosis. TNFα transcription level is affected by the TNFα -G308A polymorphism. In Egypt, more than 90% of patients are infected with HCV genotype 4. Therefore, we designed the current study to investigate the association of the TNFα -G308A polymorphism with liver pathological changes in treatment-naïve chronic HCV genotype 4 patients with different fibrosis grades (F0-F4).
Chronic hepatitis C infection can cause severe liver diseases, such as fibrosis, cirrhosis, liver failure and hepatocellular carcinoma. Liver fibrosis and cirrhosis could not be reversed but the liver scarring can be improved with HCV treatment. However, timing and rules for HCV genotype 4 treatment without major complications and relapse are under investigation by research studies and clinical trails. The research hotspot is currently how to determine and understand which HCV patients are likely to have chronic infection, progressive liver disease or to not respond significantly to HCV treatment. Therefore, there is a high need for a prognostic model that contains many genetic factors for staging and prediction of HCV liver disease progression, representing an approach that guides therapeutic interventions and prevents further spread of liver fibrosis and hepatic failure.
The TNFα SNP at nucleotide -308 represents an important genetic marker to predict the liver disease progression rate in treatment-naïve chronic HCV genotype 4 infected patients. There is a demanding need for biological and genetic markers’ application for making a decision as to whether the HCV patient will have progressive liver disease or not. These data clearly improve the predication by detecting the TNFα -308 polymorphism and assessing the TNFα serum level. The data presented in this study demonstrated that the TNFα expression level provide a predictive value for liver disease progression rate.
The data of this study will help hepatologists and gastroenterologists to better predict liver disease progression in chronic HCV infected patients, to make better decisions on liver disease treatment, and to design novel therapeutic interventions that will control HCV-associated liver disease complications.
SNPs in HCV infected patients lead to different immune responses, which significantly influence the progression of chronic HCV infection and response to therapy. TNFα is a diverse multifunctional, proinflammatory and immuno-mediator cytokine. The A allele at the TNFα promoter region -308 polymorphism affects the transcriptional level of the TNFα gene. High TNFα level in HCV infected patients may accelerate liver disease progression and complications.
This study added new information regarding a subtyping (4) of HCV and its relation with the TNFα cytokine.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Egypt
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: De Re V S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Ma S
| 1. | Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 188] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 2. | Mohamoud YA, Mumtaz GR, Riome S, Miller D, Abu-Raddad LJ. The epidemiology of hepatitis C virus in Egypt: a systematic review and data synthesis. BMC Infect Dis. 2013;13:288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 251] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 3. | de Torres M, Poynard T. Risk factors for liver fibrosis progression in patients with chronic hepatitis C. Ann Hepatol. 2003;2:5-11. [PubMed] |
| 4. | WHO. 2014. Available from: http://www.worldlifeexpectancy.com/egypt-liver-disease. |
| 5. | Hold GL, Untiveros P, Saunders KA, El-Omar EM. Role of host genetics in fibrosis. Fibrogenesis Tissue Repair. 2009;2:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Amini M, Poustchi H. Hepatitis C virus spontaneous clearance: immunology and genetic variance. Viral Immunol. 2012;25:241-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | El Awady MK, Anany MA, Esmat G, Zayed N, Tabll AA, Helmy A, El Zayady AR, Abdalla MS, Sharada HM, El Raziky M. Single nucleotide polymorphism at exon 7 splice acceptor site of OAS1 gene determines response of hepatitis C virus patients to interferon therapy. J Gastroenterol Hepatol. 2011;26:843-850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Pasha HF, Radwan MI, Hagrass HA, Tantawy EA, Emara MH. Cytokines genes polymorphisms in chronic hepatitis C: impact on susceptibility to infection and response to therapy. Cytokine. 2013;61:478-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 9. | McCaughan GW, George J. Fibrosis progression in chronic hepatitis C virus infection. Gut. 2004;53:318-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Shmarina G, Pukhalsky A, Petrova N, Zakharova E, Avakian L, Kapranov N, Alioshkin V. TNF gene polymorphisms in cystic fibrosis patients: contribution to the disease progression. J Transl Med. 2013;11:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Hajeer AH, Hutchinson IV. TNF-alpha gene polymorphism: clinical and biological implications. Microsc Res Tech. 2000;50:216-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 12. | Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195-3199. [PubMed] |
| 13. | Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF, Oksenberg J, McNicholl J, Pociot F, Hardt C. Cytokine gene polymorphism in human disease: on-line databases, supplement 1. Genes Immun. 2001;2:61-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 197] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 14. | Abraham LJ, Kroeger KM. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66:562-566. [PubMed] |
| 15. | McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994;371:508-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 809] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 16. | Cabrera M, Shaw MA, Sharples C, Williams H, Castes M, Convit J, Blackwell JM. Polymorphism in tumor necrosis factor genes associated with mucocutaneous leishmaniasis. J Exp Med. 1995;182:1259-1264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 346] [Cited by in RCA: 348] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 17. | Bouma G, Xia B, Crusius JB, Bioque G, Koutroubakis I, Von Blomberg BM, Meuwissen SG, Peña AS. Distribution of four polymorphisms in the tumour necrosis factor (TNF) genes in patients with inflammatory bowel disease (IBD). Clin Exp Immunol. 1996;103:391-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 102] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Endo M, Masaki T, Seike M, Yoshimatsu H. TNF-alpha induces hepatic steatosis in mice by enhancing gene expression of sterol regulatory element binding protein-1c (SREBP-1c). Exp Biol Med (Maywood). 2007;232:614-621. [PubMed] |
| 19. | Hooper AJ, Adams LA, Burnett JR. Genetic determinants of hepatic steatosis in man. J Lipid Res. 2011;52:593-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 20. | Vidigal PG, Germer JJ, Zein NN. Polymorphisms in the interleukin-10, tumor necrosis factor-alpha, and transforming growth factor-beta1 genes in chronic hepatitis C patients treated with interferon and ribavirin. J Hepatol. 2002;36:271-277. [PubMed] |
| 21. | Thio CL. Host genetic factors and antiviral immune responses to hepatitis C virus. Clin Liver Dis. 2008;12:713-726, xi. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Talaat RM. Soluble angiogenesis factors in sera of Egyptian patients with hepatitis C virus infection: correlation with disease severity. Viral Immunol. 2010;23:151-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Conjeevaram HS, Fried MW, Jeffers LJ, Terrault NA, Wiley-Lucas TE, Afdhal N, Brown RS, Belle SH, Hoofnagle JH, Kleiner DE. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 366] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 24. | Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2860] [Cited by in RCA: 3081] [Article Influence: 106.2] [Reference Citation Analysis (0)] |
| 25. | Ho SY, Wang YJ, Chen HL, Chen CH, Chang CJ, Wang PJ, Chen HH, Guo HR. Increased risk of developing hepatocellular carcinoma associated with carriage of the TNF2 allele of the -308 tumor necrosis factor-alpha promoter gene. Cancer Causes Control. 2004;15:657-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Ollier WE. Cytokine genes and disease susceptibility. Cytokine. 2004;28:174-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 27. | Hajeer AH, Hutchinson IV. Influence of TNFalpha gene polymorphisms on TNFalpha production and disease. Hum Immunol. 2001;62:1191-1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 28. | Cheong JY, Cho SW, Hwang IL, Yoon SK, Lee JH, Park CS, Lee JE, Hahm KB, Kim JH. Association between chronic hepatitis B virus infection and interleukin-10, tumor necrosis factor-alpha gene promoter polymorphisms. J Gastroenterol Hepatol. 2006;21:1163-1169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 104] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 29. | Dogra G, Chakravarti A, Kar P, Chawla YK. Polymorphism of tumor necrosis factor-α and interleukin-10 gene promoter region in chronic hepatitis C virus patients and their effect on pegylated interferon-α therapy response. Hum Immunol. 2011;72:935-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Radwan MI, Pasha HF, Mohamed RH, Hussien HI, El-Khshab MN. Influence of transforming growth factor-β1 and tumor necrosis factor-α genes polymorphisms on the development of cirrhosis and hepatocellular carcinoma in chronic hepatitis C patients. Cytokine. 2012;60:271-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Dai CY, Chuang WL, Chang WY, Chen SC, Lee LP, Hsieh MY, Hou NJ, Lin ZY, Huang JF, Hsieh MY. Tumor necrosis factor- alpha promoter polymorphism at position -308 predicts response to combination therapy in hepatitis C virus infection. J Infect Dis. 2006;193:98-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Höhler T, Kruger A, Gerken G, Schneider PM, Meyer zum Büschenfelde KH, Rittner C. Tumor necrosis factor alpha promoter polymorphism at position -238 is associated with chronic active hepatitis C infection. J Med Virol. 1998;54:173-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 33. | González S, Rodrigo L, Martínez-Borra J, López-Vázquez A, Fuentes D, Niño P, Cadahía V, Saro C, Dieguez MA, López-Larrea C. TNF-alpha -308A promoter polymorphism is associated with enhanced TNF-alpha production and inflammatory activity in Crohn’s patients with fistulizing disease. Am J Gastroenterol. 2003;98:1101-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 34. | Roth-Isigkeit A, Hasselbach L, Ocklitz E, Brückner S, Ros A, Gehring H, Schmucker P, Rink L, Seyfarth M. Inter-individual differences in cytokine release in patients undergoing cardiac surgery with cardiopulmonary bypass. Clin Exp Immunol. 2001;125:80-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 35. | Aller R, de Luis DA, Izaola O, González Sagrado M, Conde R, Alvarez Gago T, Pacheco D, González JM, Velasco MC. G308A polymorphism of TNF-alpha gene is associated with insulin resistance and histological changes in non alcoholic fatty liver disease patients. Ann Hepatol. 2010;9:439-444. [PubMed] |
| 36. | Chuang E, Del Vecchio A, Smolinski S, Song XY, Sarisky RT. Biomedicines to reduce inflammation but not viral load in chronic HCV--what’s the sense? Trends Biotechnol. 2004;22:517-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Kishta SA, Abd-Alhade AA, Hamam O, Raoof EA, Abeya S. Prognostic value of TNF a mRNA and VEGF mRNA expression in patients with chronic hepatitis C genotype-4, with and without cirrhosis and hepatocellular carcinoma to predict disease outcome. J Egypt Soc Parasitol. 2010;40:515-530. [PubMed] |
| 38. | Talaat RM, Esmail AA, Elwakil R, Gurgis AA, Nasr MI. Tumor necrosis factor-alpha -308G/A polymorphism and risk of hepatocellular carcinoma in hepatitis C virus-infected patients. Chin J Cancer. 2012;31:29-35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 39. | Kusumoto K, Uto H, Hayashi K, Takahama Y, Nakao H, Suruki R, Stuver SO, Ido A, Tsubouchi H. Interleukin-10 or tumor necrosis factor-alpha polymorphisms and the natural course of hepatitis C virus infection in a hyperendemic area of Japan. Cytokine. 2006;34:24-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 40. | Bahcecioglu IH, Koca SS, Poyrazoglu OK, Yalniz M, Ozercan IH, Ustundag B, Sahin K, Dagli AF, Isik A. Hepatoprotective effect of infliximab, an anti-TNF-alpha agent, on carbon tetrachloride-induced hepatic fibrosis. Inflammation. 2008;31:215-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 41. | Koca SS, Bahcecioglu IH, Poyrazoglu OK, Ozercan IH, Sahin K, Ustundag B. The treatment with antibody of TNF-alpha reduces the inflammation, necrosis and fibrosis in the non-alcoholic steatohepatitis induced by methionine- and choline-deficient diet. Inflammation. 2008;31:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 119] [Article Influence: 6.6] [Reference Citation Analysis (1)] |
| 42. | Frazier TH, Stocker AM, Kershner NA, Marsano LS, McClain CJ. Treatment of alcoholic liver disease. Therap Adv Gastroenterol. 2011;4:63-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 43. | Satapathy SK, Sakhuja P, Malhotra V, Sharma BC, Sarin SK. Beneficial effects of pentoxifylline on hepatic steatosis, fibrosis and necroinflammation in patients with non-alcoholic steatohepatitis. J Gastroenterol Hepatol. 2007;22:634-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 44. | El-Awady MK, Mostafa L, Tabll AA, Abdelhafez TH, Bader El Din NG, Zayed N, Shenawy RE, El Abd Y, Hasan RM, Zaghlol H. Association of IL28B SNP With Progression of Egyptian HCV Genotype 4 Patients to End Stage Liver Disease. Hepat Mon. 2012;12:271-277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 45. | El Awady MK, Bader El Din NG, Tabll A, El Hosary Y, Abdel Aziz AO, El Khayat H, Salama M, Abdelhafez TH. IL28B polymorphism and cytomegalovirus predict response to treatment in Egyptian HCV type 4 patients. World J Gastroenterol. 2013;19:290-298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 46. | Bader El Din NG, Anany MA, Dawood RM, Ibrahim MK, El-Shenawy R, El Abd YS, El Awady MK. Impact of OAS1 Exon 7 rs10774671 Genetic Variation on Liver Fibrosis Progression in Egyptian HCV Genotype 4 Patients. Viral Immunol. 2015;28:509-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 47. | Bader El Din NG, Salum GM, Anany MA, Ibrahim MK, Dawood RM, Zayed N, El Abd YS, El-Shenawy R, El Awady MK. Association of Myxovirus Resistance Gene Promoter Polymorphism with Response to Combined Interferon Treatment and Progression of Liver Disease in Chronic HCV Egyptian Patients. J Interferon Cytokine Res. 2015;35:641-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |